Abstract
The African clawed frog (Xenopus laevis) naturally tolerates severe dehydration using biochemical adaptation, one of which is the elevation of antioxidant defenses during whole-body dehydration. The present study investigated the role and regulation of a pathway known to regulate oxidative stress response, the Akt-FoxO signaling pathway, in clawed frog skeletal muscle, responding to medium (15%) and high (30%) dehydration. Protein levels of total and phosphorylated Akt, FoxO1, and FoxO3 were assessed via immunoblotting, in addition to the levels of the E3 ubiquitin ligase known to be associated with muscle atrophy, MAFbx. Akt activity/phosphorylation in addition to its total protein levels were decreased in the skeletal muscle during dehydration, and this corresponded with decreases in the relative phosphorylation of FoxO1 and FoxO3 as well on several residues. Akt is an inhibitor of FoxO1 and FoxO3 activity via phosphorylation, suggesting that FoxO activities were increased during dehydration stress. Furthermore, MAFbx showed decreased protein expression during high dehydration as well, suggesting that the clawed frog may exhibit some natural resistance to skeletal muscle atrophy during severe dehydration conditions. In addition to identifying that the suppression of Akt could lead to an activation of FoxO transcription factors in X. laevis during dehydration, these investigations suggest that X. laevis dehydration may implicate FoxO1 and FoxO3 in controlling skeletal muscle atrophy in X. laevis exposed to dehydration. This study implicates the Akt signaling pathway, its regulation of FoxO transcription factors, and FoxO-controlled targets, in stress adaptation against dehydration.
Keywords: Xenopus laevis, Akt, FoxO, Dehydration, Immunoblot
Introduction
Many animals live in environments that can change to create conditions that can be stressful to the animal. Therefore, through evolution animals have developed various strategies to increase their chances of survival during periods of extreme environmental stress. The African clawed frog, Xenopus laevis, is a primarily aquatic frog that was described in 1803 by Daudin. Wagler and Daudin both named the frog Xenopus for its peculiar clawed foot and laevis for its smooth body (Deuchar 1975). Despite preferring aquatic environments, the African clawed frog lives in the sub-Saharan areas where the swamps, lakes, and rivers are subject to seasonal drought (Tinsley and Kobel 1996). Being an aquatic animal, dehydration is a major stressor that impacts their chances of survival during the dry season. To minimize water loss, they burrow down into the wet mud of drying ponds. In addition to increasing soil to skin contact, these frogs also elevate the concentrations of osmolytes in their tissues to reduce their water potential (Feder and Burggren 1985). The main osmolyte used by amphibians to reduce water loss during aestivation and dehydration is urea (Grundy and Storey 1994). For example, X. laevis naturally dehydrated for 2–3 months showed plasma amino acid levels that tripled and plasma urea concentrations that increased by 15–20-fold (Balinsky et al. 1967). In addition, findings from our lab have indicated that there are various other molecular and biochemical pathways activated during dehydration stress, in particular microRNAs and heat shock proteins (Luu and Storey 2015; Luu et al. 2018; Zhang et al. 2018).
One of the molecular pathways activated during dehydration stress in X. laevis is antioxidant defenses (Malik and Storey 2009b, 2011). This antioxidant defense mechanism is activated during dehydration in X. laevis because the stress of dehydration leads to an accumulation of ROS products, probably due to the reduced turnover of oxidative-damaged macromolecules (Storey and Storey 2012). Previous work from our lab showed that two antioxidant enzymes, catalase and Mn2+-dependent superoxide dismutase (MnSOD), showed increased mRNA and protein expression during dehydration in X. laevis (Malik and Storey 2011). Both proteins are known to be downstream targets regulated by the Forkhead box class O (FoxO) family of transcription factors during oxidative stress (Malik and Storey 2011; Judge et al. 2014). Four FoxO proteins have been identified (FoxO1, FoxO3, FoxO4, and FoxO6), and the first three are major substrates of Akt/protein kinase B (PKB). Akt-dependent phosphorylation is responsive to growth factor and insulin stimulation, and it reduces the DNA-binding ability of FoxO transcription factors via phosphorylation. FoxO phosphorylation by Akt at conserved residues leads to cytoplasmic localization, export from the nucleus, and enhanced FoxO degradation (Lam et al. 2006; Salih and Brunet 2008).
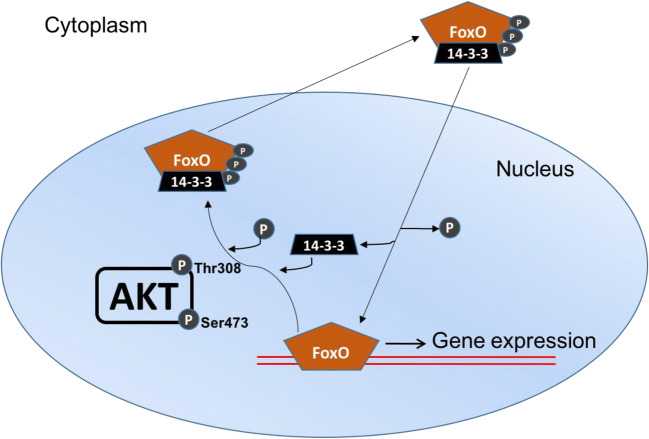
As shown in Fig. 1, the activation of Akt depends on phosphorylation on two residues: serine (Ser)-473 and threonine (Thr)-308 (Alessi et al. 1997). The Thr-308 residue is found within the kinase domain, so the protein kinase activity of Akt is stimulated by phosphorylation at this site. On the other hand, Ser-473 phosphorylation serves a regulatory role, such that when it is phosphorylated by mammalian target of rapamycin, complex 2 (mTOR2), this enhances Akt kinase activity as well (Yang et al. 2002; Hresko and Mueckler 2005; Sarbassov et al. 2005). When Akt becomes active, it moves away from the plasma membrane to phosphorylate proteins at multiple locations throughout the cell, including the nucleus (Andjelkovic et al. 1997). As shown in Fig. 1, nuclear Akt phosphorylates FoxO1 and FoxO3 on several sites (Thr-24 and Ser-319/Ser-256 for FoxO1, Thr-32, Ser-318/Ser-321 for FoxO3) to promote FoxO binding to the 14-3-3 protein, resulting in nuclear expulsion and degradation (Zhao et al. 2004; Furukawa-Hibi et al. 2005; Huang et al. 2005; Aune et al. 2014; Das et al. 2016). In X. laevis, FoxO1 transcriptional activity was shown to be enhanced in the liver of highly dehydrated frogs, where FoxO1 nuclear protein levels increased by 1.8-fold relative to control, and inactive, phosphorylated FoxO1 Ser-256 in total and nuclear extracts decreased as well (Malik and Storey 2011). Therefore, we are interested in furthering our understanding of the role that Akt-mediated FoxO regulation plays in X. laevis undergoing dehydration stress by studying this pathway in skeletal muscle.
Fig. 1.
Schematic of FoxO transcription factor nuclear/cytoplasmic shuttling. Active/phosphorylated Akt at Thr-308 and Ser-473 result in Akt nuclear localization, where it can phosphorylate and regulate the nuclear/cytoplasmic shuttling of FoxO transcription factors. Dephosphorylated FoxOs are able to translocate into the nucleus, bind to the promoters of target genes, and regulate their gene expression. However, active Akt phosphorylate FoxOs at various consensus sites to promote FoxO binding to the 14-3-3 protein, causing nuclear expulsion into the cytoplasm, where it can be degraded
In skeletal muscle, FoxO transcription factors play roles other than regulating oxidative stress response proteins. For instance, it has been well-studied in mammalian models that the FoxOs regulate the expression of the E3 ubiquitin ligases – muscle atrophy F-box (MAFbx/atrogin-1) and muscle ring finger (MuRF1) (Sandri et al. 2004; Stitt et al. 2004; Waddell et al. 2008). E3 ubiquitin ligases are part of a protein degradation mechanism called the ubiquitin proteasome system (UPS), where E3 ligases recognize specific proteins and ligate them to ubiquitin groups (Herrmann et al. 2007). The ligases MAFbx and MuRF1 have been shown to be essential for skeletal muscle atrophy in various mammalian models used to study skeletal muscle atrophy, including the thirteen-lined ground squirrel (Ictidomys tridecemlineatus) (Bodine et al. 2001; Foletta et al. 2011; Bodine and Baehr 2014; Zhang et al. 2016). However, in X. laevis, although the UPS has been identified in its egg and embryos (McDowell and Philpott 2016), the role of the UPS, specifically of MAFbx and MuRF1, in adult frogs has not been studied. Studies in other amphibians such as the green-striped burrowing frogs (Cyclorana alboguttata) that tolerate stresses like aestivation for months at a time have shown that there is specific resistance to muscle atrophy in certain muscle groups, namely, slow-twitch, oxidative muscles that are not used for jumping (Hudson and Franklin 2002; Mantle et al. 2009). Therefore, we were interested in identifying whether the cellular signaling pathways associated with muscle atrophy are activated during dehydration in X. laevis.
The present study analyzes protein expression patterns and phosphorylation states of the FoxO transcription factors as well as their upstream regulator, Akt, and their downstream effectors, the E3 ubiquitin ligases MAFbx and MuRF1, in clawed frog skeletal muscle in response to dehydration. We hypothesize that FoxO1 and FoxO3 are tightly controlled by Akt in X. laevis during dehydration stress, as FoxO1 and FoxO3 are important regulators of oxidative stress response genes and possibly of MAFbx and MuRF1 as well during dehydration. Our results indicated that there were decreases in active Akt and nuclear Akt protein levels during dehydration, resulting in increased activation of FoxO1 and FoxO3. However, FoxO1 and FoxO3 total protein levels decreased throughout dehydration as well. Furthermore, MAFbx showed moderately decreased protein levels at high dehydration. Therefore, this study demonstrates that the FoxO transcription factors are dynamically regulated during exposure to dehydration and potentially control downstream proteins involved in both oxidative stress and muscle atrophy in X. laevis skeletal muscle.
Materials and methods
Animals
Animal treatment procedures were approved by the Carleton University Animal Care Committee (animal protocol no. 13683) in accordance with the guidelines set by the Canadian Council on Animal Care. Adult African clawed frogs, X. laevis, were purchased from the Department of Zoology at the University of Toronto. Animals were held in tanks of dechlorinated water at 22 °C with aeration for 1–2 weeks. The frogs were fed CU Adult Frog diet (PMI Nutrition International) three times per day, and the tank water was changed the day following each feeding. Animals were divided into two groups and remained unfed: controls maintained in the conditions described above and dehydration groups. For the dehydration group, frogs were placed in closed containers held at 22 °C that had a layer of silica gel desiccant on the bottom, with a perforated divider to prevent frogs from touching the desiccant. Water loss was determined by body mass measurements at approximately 12-h time intervals and at shorter intervals as water loss reached higher rates until the desired amount of water loss was reached. Percentage of total body water lost was calculated using the following equation:
Mi is the initial mass of the animal, Md is the mass of the dehydrated frog at each weighing, and BWCi is the initial body water content of the frog, which is estimated to be 0.741 ± 0.019 g of water per gram of body mass (Malik and Storey 2009a, 2009b). Medium dehydration was achieved when approximately 15% water loss was obtained, at which time this group of frogs was stored in a glass vacuum desiccator. High dehydration was achieved when % water loss reached approximately 30%. For sampling, all frogs were euthanized by pithing, and then skeletal muscle tissues were quickly dissected out and flash frozen in liquid nitrogen prior to transfer to -80 °C for long-term storage. Four biological replicates of skeletal muscle from the sartorius, triceps, and rectus femoris were used for control and dehydration experiments.
Preparation of total protein extracts
The preparation of total protein extracts was performed by taking 60–90 mg of frozen skeletal muscle samples, weighing them, and storing them in liquid nitrogen until they were homogenized in 1:5 w/v ice-cold homogenization buffer (50 mM Tris, pH 8.0 with 10 mM NaF, 2.5 mM EDTA, 2.5 mM EGTA 10% v/v glycerol, 5 mM 2-mercaptoethanol), phosphatase (1 mM Na3VO4 and 10 mM ß-glycerophosphate), and protease inhibitors (BioShop) with 1 mM phenylmethylsulfonyl fluoride using a Polytron PT10 homogenizer. Protein homogenates were centrifuged at 13,000×g for 30 min at 4 °C, and supernatants were removed and stored in ice. Soluble protein concentrations were determined using the Bio-Rad reagent (Bio-Rad Laboratories) at 595 nm on a MR5000 microplate reader. Protein homogenates were adjusted to 10 μg/μL by adding homogenization buffer, then 2x SDS loading buffer (100 mM Tris-base, pH 6.8, 4% w/v SDS, 20% v/v glycerol, 0.2% w/v bromophenol blue, 10% v/v 2-mercaptoethanol) was added 1:1 v/v, and the samples were boiled to attain a final protein concentration of 5 μg/μL. Samples were then stored at − 20 °C until they were used for immunoblotting.
Nuclear extracts
To prepare nuclear extracts, samples of frozen skeletal muscle (1.0 g) were broken up and homogenized by hand using a Dounce homogenizer. Homogenization was performed in a 1:2 w/v ratio of homogenization buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 10 mM EDTA, 1 mM DTT, 20 mM 2-glycerophosphate) with the addition of 10 μL protease inhibitor cocktail (Bio-Rad Laboratories) just prior to homogenization. Samples were centrifuged at 10,000×g for 10 min at 4 °C. The supernatant (cytoplasmic extract) for each sample was removed to a separate tube. The pellet was then resuspended in 150 μL of extraction buffer (20 mM HEPES, pH 7.9; 400 mM NaCl; 1 mM EDTA, 10% v/v glycerol, 10 mM 2-glycerophosphate, 1 mM DTT) with 1.5 μL of protease inhibitor cocktail added just prior to the addition of the buffer to the pellet. Samples were incubated on ice horizontally on a rocking platform for 1 h. After the incubation, samples were centrifuged at 10,000×g for 10 min at 4 °C. The supernatant was collected (nuclear extract) and placed into a separate tube. As described for total soluble protein extraction, the protein concentrations of nuclear extracts were determined with the Bio-Rad protein assay, and concentrations were standardized with extraction buffer. For immunoblotting analysis of nuclear proteins, 2 × SDS sample buffer was added to aliquots of the samples as described in total soluble protein extraction. The integrity of the nuclei was confirmed by immunoblotting of nuclear fractions and probing with a highly conserved eukaryotic protein found exclusively in the nucleus (anti-histone H3 antibody; cell signaling, CS 9175; diluted 1:1000 in TBST).
Immunoblotting
SDS-PAGE was carried out with a 4% stacking gel and a 6–12% resolving gels (depending on target molecular weight) in a Bio-Rad Mini-Protean III apparatus filled with running buffer (25 mM Tris, 190 mM glycine, 0.1% w/v SDS) at 180 V until the protein of interest neared the base of the gel. Pre-stained molecular weight standards (FroggaBio) and 20 μg of total soluble protein extracts or nuclear protein extracts (n = 4 biological replicates for each experimental condition) were loaded onto gels. Proteins were then transferred from the polyacrylamide gels onto polyvinylidene fluoride (PVDF; BioTrace, PALL Life Sciences) membranes using a Bio-Rad Mini Trans-Blot cell with transfer buffer (25 mM Tris, 192 mM glycine, and 10% v/v methanol). The transfer process was run for 60–120 min (depending on target molecular weight) at 160 mA and at room temperature. After protein transfer, all membranes were equilibrated in TBST (10 mM Tris, pH 7.5, 150 mM NaCl, 0.05% v/v Tween-20) for 10 min. Membranes were then blocked with milk by equilibrating in a 2.5–7.5% (depending on target) w/v nonfat milk in TBST for 30 min. After incubation with blocking solution, membranes were washed with fresh TBST for 3 × 5 min.
Membranes were incubated with primary antibody (1:1000 v/v in TBST) overnight at 4 °C with gentle rocking. Primary antibodies were purchased from Cell Signaling Technology for p-Akt Ser-473 (CS 9271), p-Akt Thr-308 (CS 4056), p-FoxO1 Ser-319 (CS 9471), p-FoxO3 Ser-318/321 (CS 9465), and p-FoxO1/3 Thr-24/32 (CS 9464P); Genscript for total Akt (Cat#: A00275) and p-FoxO1 Ser-256 (Cat#: A00395); Genetex for total FoxO1 (GTX110724), total FoxO3 (GTX100277), and MuRF1 (GTX110475); and Santa Cruz Biotechnology for MAFbx (SC27845). After washing with fresh TBST for another 3 × 5 min, the membranes were incubated with anti-mouse or anti-rabbit secondary antibody (1:6000–8000 v/v depending on the target) for 30 min at room temperature with gentle rocking. Fresh TBST was used again to wash the membrane for 4 × 5 min at room temperature with gentle rocking. Enhanced chemiluminescent reagents were used to expose each immunoblot, and band densities were visualized and quantified. The PVDF membrane was then stained with Coomassie Blue dye (0.25% w/v Coomassie brilliant blue, 7.5% v/v acetic acid, 50% v/v methanol). Immunoblot bands corresponded to the molecular weights indicated on the respective antibody specification sheets and their respective X. laevis proteins on the NCBI database as confirmed by the molecular weight standards.
Quantification and statistics
Imaging of immunoblots was done using a Chemi-Genius Bio-Imager (Syngene), and band intensities were quantified using the accompanying GeneTools software. Immunoblot band densities in each lane were standardized against the summed intensity of a group of Coomassie-stained protein bands in the same lane; this group of bands was chosen because it was not located near the band of interest and was prominent and constant across all samples. This method of immunoblot standardization against a total protein loading control has been shown to be more accurate than standardizing against housekeeping proteins such as tubulin (Eaton et al. 2013). Both the quantified intensity of the bands and the density of the protein loaded were corrected against the background of the membrane, and immunoblot band densities were normalized at each condition relative to control. Immunoblotting data were expressed as means ± SEM, n = 4 independent samples from different animals. Statistical testing used the one-way ANOVA and the Tukey post hoc functions from the Prism software (GraphPad).
Results
Total and phosphorylated Akt protein levels
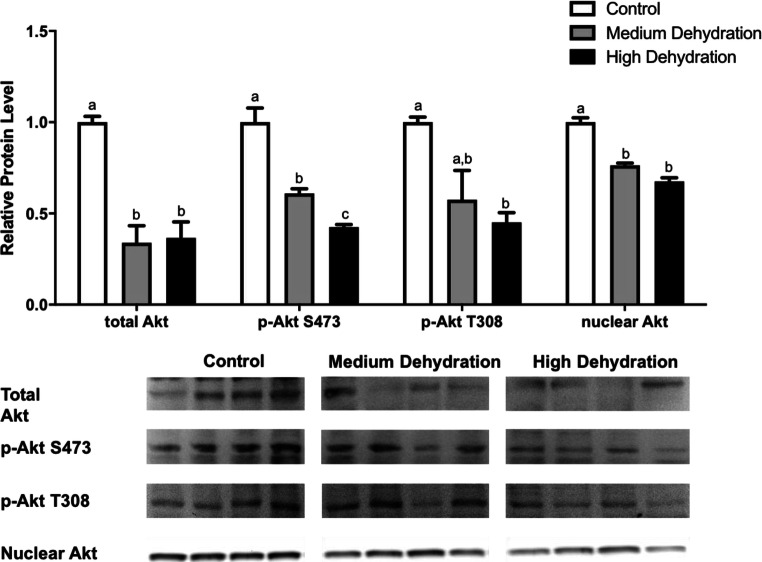
Protein levels of Akt as well as levels of phosphorylated Akt at Ser-473 and Thr-308 were analyzed using immunoblotting in X. laevis skeletal muscle comparing control animals to medium dehydration (15% water loss) and high dehydration (30% water loss) animals. Antibodies detecting total Akt, p-Akt Ser-473, and p-Akt Thr-308 cross-reacted with frog proteins with single bands. Total Akt protein levels decreased by 66% and 63% from control to medium and high dehydration conditions, respectively (Fig. 2). There was no change in total Akt protein levels from medium to high dehydration. In addition to decreased total Akt levels, phosphorylated Akt at Thr-308 protein levels decreased during medium (42%) and high (55%) dehydration relative to control as well (Fig. 2). The 13% decrease in p-Akt Thr-308 levels from medium to high dehydration was not statistically significant. Phosphorylated Akt Ser-473 protein levels decreased significantly by 39% during medium dehydration relative to control (p < 0.05). During high dehydration, p-Akt Ser-473 levels decreased by 58% relative to control, and the 19% decrease from medium to high dehydration was statistically significant as well (Fig. 2) (p < 0.05).
Fig. 2.
Changes in the protein levels of total, phosphorylated, and nuclear Akt over medium and high dehydration in the skeletal muscle of X. laevis. Total Akt, p-Akt Ser-473, p-Akt Thr-308, and nuclear Akt protein levels were visualized at control, medium dehydration (15%), and high dehydration (30%) experimental conditions. Representative immunoblots are shown for all sampling points for all targets. Histograms show mean band densitometries (± SEM, n = 4 independent protein isolations from different animals). Data were analyzed using one-way ANOVA with post hoc Tukey’s test (p < 0.05); for each parameter measured, values that are not statistically different from each other share the same letter notation
Both p-Akt Ser-473 and p-Akt Thr-308 protein levels are reflective of Akt activation. In addition, we traced the downstream effects from Akt activation in the skeletal muscle of clawed frogs, and the nuclear content of total Akt protein within the nucleus was analyzed during control, medium dehydration, and high dehydration experimental conditions. Nuclear Akt protein levels decreased significantly from control to medium dehydration by 24% in comparison with control (p < 0.05). During high dehydration, nuclear Akt levels further declined by 8% relative to medium dehydration and by 32% relative to control, and only the decrease from control to high dehydration was statistically significant (p < 0.05) (Fig. 2).
Total and phosphorylated FoxO1 and FoxO3 protein levels decreased during dehydration stress
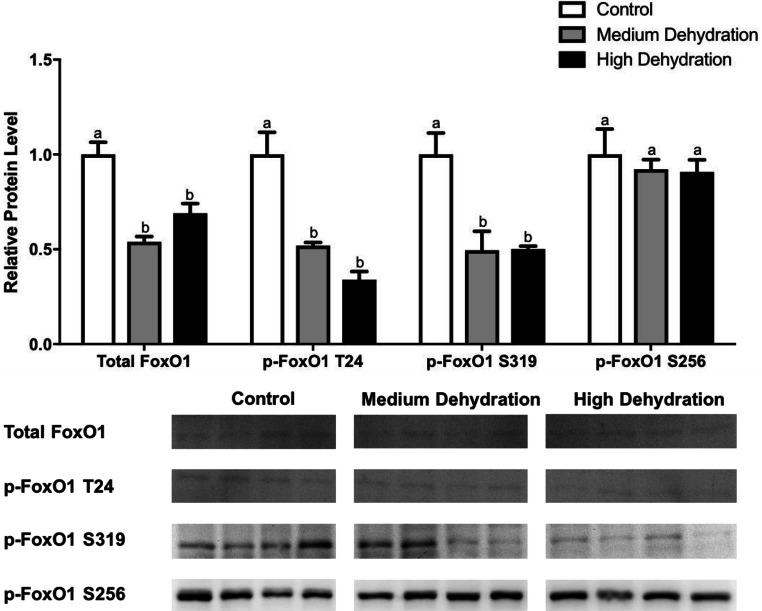
FoxO1 and FoxO3 are target substrates downstream of Akt. FoxO1 is phosphorylated by Akt at Thr-24, Ser-319, and Ser-256, whereas FoxO3 is phosphorylated at Thr-32 and Ser-318/Ser-321 (Zhao et al. 2004; Furukawa-Hibi et al. 2005; Huang et al. 2005; Aune et al. 2014; Das et al. 2016). Dehydration significantly decreased total FoxO1, p-FoxO1 Thr-24, p-FoxO1 Ser-319, FoxO3, p-FoxO3 Thr-32, and p-FoxO3 Ser-318/321 expression. Total FoxO1 protein levels decreased significantly by 46% during medium dehydration relative to control and by 31% during high dehydration compared with control (p < 0.05), although there was no significant difference between medium and high dehydration (Fig. 3). Similarly, p-FoxO1 Thr-24 levels decreased significantly by 48% at medium dehydration and by 66% at high dehydration relative to control (p < 0.05) although the decrease from medium to high dehydration was not significant. Phosphorylated FoxO1 Ser-319 protein levels decreased significantly as well during medium and high dehydration by 51% and 50%, respectively, compared with control (p < 0.05), although there appears to be some variability between medium dehydration samples. On the other hand, p-FoxO1 Ser-256 protein levels did not change during medium and high dehydration relative to each other and relative to control (Fig. 3).
Fig. 3.
Changes in the protein levels of total and phosphorylated FoxO1 over medium and high dehydration in the skeletal muscle of X. laevis. Total FoxO1, p-FoxO1 Thr-24, p-FoxO1 Ser-319, and p-FoxO1 Ser-256 protein levels were visualized at control, medium dehydration (15%), and high dehydration (30%) experimental conditions. Representative immunoblots are shown for all sampling points for all targets. Histograms show mean band densitometries (± SEM, n = 4 independent protein isolations from different animals). Data were analyzed using one-way ANOVAwith post hoc Tukey’s test (p < 0.05); for each parameter measured, values that are not statistically different from each other share the same letter notation
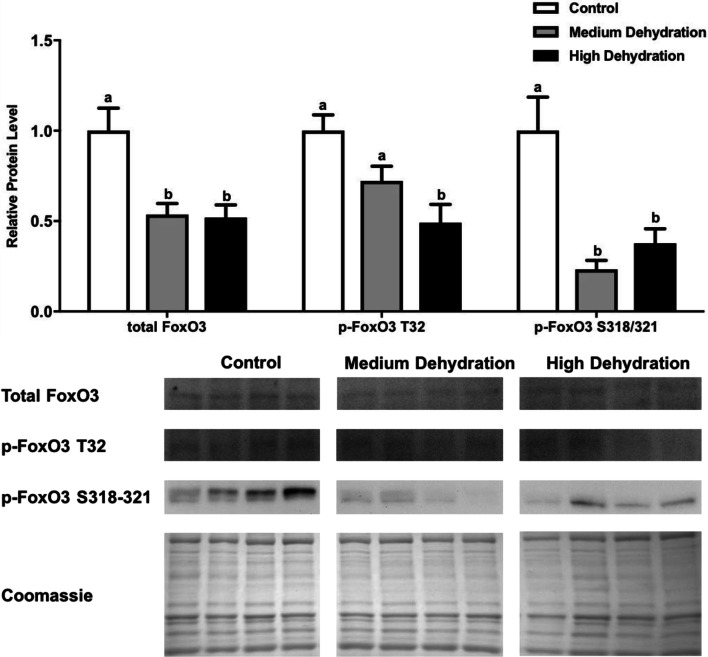
The patterns of protein expression and phosphorylation seen with FoxO3 resemble those seen with FoxO1. Total FoxO3 protein expression decreased significantly from control to medium and high dehydration by 49% and 48%, respectively (p < 0.05). Phosphorylated FoxO3 Thr-32 protein levels decreased at medium dehydration by 20% relative to control, although this decreased was not significant (Fig. 4). At high dehydration, p-FoxO3 Thr-32 levels decreased significantly by 51% relative to control, and this decrease was significant relative to medium dehydration as well (p < 0.05). Furthermore, p-FoxO3 Ser-318/321 levels decreased significantly by 73% from medium dehydration to control and by 62% from high dehydration to control (p < 0.05), although the difference between medium to high dehydration was not significant (Fig. 4).
Fig. 4.
Changes in the protein levels of total and phosphorylated FoxO3 over medium and high dehydration in the skeletal muscle of X. laevis. Total FoxO3, p-FoxO3 Thr-32, and p-FoxO3 Ser-318/321 protein levels were visualized at control, medium dehydration (15%), and high dehydration (30%) experimental conditions. Representative immunoblots and Coomassie total protein loading controls are shown for all sampling points for all targets. Histograms show mean band densitometries (± SEM, n = 4 independent protein isolations from different animals). Data were analyzed using one-way ANOVA with post hoc Tukey’s test (p < 0.05); for each parameter measured, values that are not statistically different from each other share the same letter notation
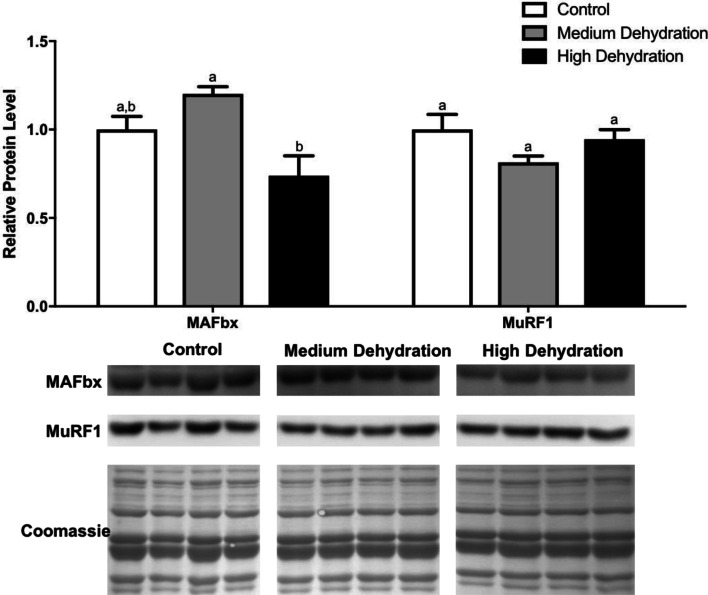
Protein levels of MAFbx and MuRF1
MAFbx and MuRF1 are two E3 ubiquitin ligases that are integral parts of the UPS, which is the primary mechanism of muscle atrophy (Herrmann et al. 2007). The expression of these ligases is controlled by the FoxO family of transcription factors via transcriptional regulation (Sandri et al. 2004; Stitt et al. 2004; Waddell et al. 2008). Therefore, we characterized the protein expression of MAFbx and MuRF1 during dehydration stress in X. laevis skeletal muscle. There was no change in MAFbx protein levels from control to medium dehydration, but its levels did decrease during high dehydration (Fig. 5). This decrease (37% from medium dehydration) was significant relative to medium dehydration (p < 0.05) but not control. Protein levels of MuRF1 did not change with exposure to dehydration (Fig. 5).
Fig. 5.
Changes in the protein levels of the E3 ubiquitin ligases MAFbx and MuRF1 over medium and high dehydration in the skeletal muscle of X. laevis. MAFbx and MuRF1 protein levels were visualized at control, medium dehydration (15%), and high dehydration (30%) experimental conditions. Representative immunoblots and Coomassie total protein loading controls are shown for all sampling points for all targets. Histograms show mean band densitometries (± SEM, n = 4 independent protein isolations from different animals). Data were analyzed using one-way ANOVAwith post hoc Tukey’s test (p < 0.05); for each parameter measured, values that are not statistically different from each other share the same letter notation
Discussion
For African clawed frogs, X. laevis, to survive dehydration stress, various adaptive strategies need to be coordinated. Our lab is interested in the signaling pathways that are activated by dehydration in X. laevis, which include the mitogen-activated protein kinase (MAPK) signaling cascades and various antioxidant stress response pathways (Cowan 2003; Malik and Storey 2009a, 2009b, 2011). We predict that antioxidant defense is a mechanism utilized by X. laevis and other amphibians undergoing environmental stresses as these animals enter a state of hypometabolism (Hillman 1978; Storey and Storey 2012). For instance, after 2 months of aestivation in spadefoot toads (Scaphiopus couchii), reduced amounts of reactive oxygen species (ROS) products such as glutathione, and lipid peroxidation products like conjugated dienes and lipid hydroperoxides, were found in several tissues (Grundy and Storey 1998). It is hypothesized that there was an increase in oxidative stress response because there is a decrease in the turnover of oxidative damaged molecules during metabolic rate suppression (Storey and Storey 2012). A previous study from our lab assessed the role of the antioxidant defense proteins, MnSOD and catalase, and showed increased protein levels during dehydration in X. laevis liver. Its upstream regulator, FoxO1, also displayed increased transcriptional activity in the liver during dehydration. Furthermore, this study also demonstrated that catalase protein levels increased during high dehydration in X. laevis skeletal muscle as well (Malik and Storey 2011), so we predicted that the FoxO transcription factors, namely, FoxO1 and FoxO3, may be involved in its regulation in skeletal muscle. Therefore, we were interested in assessing the role of these two FoxOs, as well as their regulation through the Akt pathway.
We analyzed the Akt pathway by assessing total protein and phosphorylated protein levels. Generally, we found that the Akt pathway was inhibited in response to dehydration, where total and nuclear Akt protein levels in addition to the phosphorylation/activation of Akt at Ser-473 and Thr-308 decreased significantly during medium and high dehydration (Fig. 2). The decrease in total Akt protein suggests a transcriptional suppression of Akt transcript or increased degradation of Akt protein. Therefore, it is expected that the levels of phosphorylated Akt proteins at Thr-308 and Ser-473 would decrease as well. However, total Akt protein levels do not decrease further from medium to high dehydration, whereas the relative levels of Akt phosphorylated at Ser-473 decrease further from medium to high dehydration. This finding indicates that there is a decrease in the activation status of Akt from medium to high dehydration. This finding also suggests that the dehydration response by Akt in muscle may be regulated in part by mTORC2, which is an upstream kinase that phosphorylates Akt at Ser-473 (Hresko and Mueckler 2005). Also, it is known that this occurs before phosphoinositide-dependent kinase 1 (PDK1) phosphorylates Akt at Thr-308 (Dann et al. 2007). Based on the decrease in p-Akt Thr-308 levels, PDK1 may be involved in the suppression of Akt during dehydration as well. In mammalian models of environmental stress response, a similar suppression of Akt was observed in I. tridecemlineatus skeletal muscle during torpor, which facilitated the inhibition of protein translation machinery via the Akt/mTOR pathway (Wu and Storey 2012). Also, in the wood frog (Rana sylvatica) another amphibian that can endure 40% dehydration, there is a strong suppression of Akt total protein, p-Akt Ser-473, and p-Akt Thr-308 levels during dehydration in the liver. When this animal encounters freezing stress, it only decreases the phosphorylation of Akt at Ser-473 in skeletal muscle (Zhang and Storey 2013). This stress adaptation employed by R. sylvatica during freezing is very similar to the strategy employed by X. laevis during dehydration in muscle.
The decrease in Akt activity during dehydration (Fig. 2) could allow for the dephosphorylation and activation of FoxO transcription factors. Indeed, we observed that this was the case since FoxO1 and FoxO3 phosphorylation levels generally decreased during dehydration compared with control. It is well known that FoxO1 nuclear localization is regulated by Akt phosphorylation at Thr-24, Ser-319, and Ser-256, and Akt phosphorylates FoxO3 at Thr-32 and Ser-318/321 (Zhao et al. 2004; Furukawa-Hibi et al. 2005; Huang et al. 2005; Aune et al. 2014; Das et al. 2016) (Fig. 1). We observed that phosphorylation of FoxO1 at the Thr-24 and Ser-319 residues decreased during dehydration (Fig. 3) and p-FoxO3 Thr-32 and Ser-318/321 levels decreased as well (Fig. 4). These results suggest that the activity of both FoxO1 and FoxO3 increased during dehydration stress in the skeletal muscle of X. laevis, which corresponds with the finding that the expression of its downstream target, catalase, was also increased during dehydration to enhance antioxidant response. Therefore, our findings indicate that the dephosphorylation of FoxO1 and FoxO3 at Akt phosphorylation sites plays an important role during dehydration stress in X. laevis. However, our findings also indicated that the total protein levels of FoxO1 and FoxO3 decreased during medium and high dehydration as well (Figs. 3 and 4). The one possible explanation is that since dehydration is an environmental stressor that requires X. laevis to suppress its energy expenditure, the transcription and translation of many proteins are going to be suppressed. It may be that FoxO1 and FoxO3 are two of those proteins, whose expressions are decreased during dehydration despite increases in activity. Activation of proteins via posttranslational modifications is a less energy expensive process than upregulating transcription or translation.
Another objective of this study was to investigate the role of FoxO1 and FoxO3 in regulating the expression of the two E3 ubiquitin ligases, MAFbx and MuRF1, during dehydration in X. laevis skeletal muscle. MAFbx and MuRF1 are integral parts of the UPS that regulates the process of muscle atrophy in mammalian models (Herrmann et al. 2007). We were interested in assessing whether there is a molecular basis behind muscle atrophy during dehydration in X. laevis as findings from other frogs subjected to environmental stress, like C. alboguttata, have shown that certain muscle groups resist disuse-induced muscle atrophy (Hudson and Franklin 2002; Mantle et al. 2009). We found that there was only a decrease at high dehydration in MAFbx protein levels, whereas MuRF1 levels did not change during dehydration (Fig. 5). These results suggest that during more severe and longer periods of dehydration, there may be a resistance to skeletal muscle atrophy as evidenced by the decrease expression of MAFbx. This decrease in MAFbx expression may be explained in part by a decrease in the total protein levels of its upstream regulators, FoxO1 and FoxO3. However, given the increase in FoxO1 and FoxO3 activity, it is more likely that other regulators of MAFbx and MuRF1 are involved in controlling its expression during dehydration. One such candidate is the transcription factor, myogenin (MyoG), which regulates the expression of E3 ubiquitin ligases like MAFbx and MuRF1 to contribute to muscle atrophy in mammalian models (Moresi et al. 2010; Schiaffino et al. 2013). Therefore, further investigation is warranted to build on our understanding of muscle atrophy dynamics during dehydration in X. laevis and the mechanisms involved in regulating atrophy.
In summary, our findings indicate that the dephosphorylation of FoxO1 and FoxO3 at Akt phosphorylation sites plays an important role in increasing antioxidant defenses and other pro-survival mechanisms during dehydration stress in X. laevis. Specifically, phosphorylation of Akt at Ser-473 and Thr-308 in addition to total and nuclear Akt levels were all decreased during dehydration in skeletal muscle, and this likely contributed to a decrease in Akt-mediated phosphorylation of FoxO1 and FoxO3 at its various residues. Throughout medium and high dehydration, we observed an overall dephosphorylation and activation of FoxO1 and FoxO3 despite the decrease in total protein levels of both FoxOs. Furthermore, we also found that a downstream target of FoxO1 and FoxO3, the E3 ubiquitin ligase MAFbx, showed decreased expression during high dehydration in skeletal muscle, suggesting that X. laevis may demonstrate some resistance to atrophy during dehydration. Therefore, in addition to characterizing the regulation and signaling of Akt, FoxO1, and FoxO3 in the skeletal muscle of X. laevis during dehydration, we also identified a novel role for the Akt-FoxO pathway in promoting resistance to muscle atrophy that potentially involves the ubiquitin ligase, MAFbx. Future studies should consider using the dehydrated X. laevis as a model for the analysis of muscle metabolism and function under extreme environmental stress.
Funding information
This work was supported by a Discovery grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (No. 6793) to Kenneth B. Storey. Kenneth B. Storey holds the Canada Research Chair in Molecular Physiology; Yichi Zhang held a postgraduate Queen Elizabeth II Graduate Scholarship in Science and Technology. Bryan E. Luu held a NSERC Canada Graduate Scholarship.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PRJ, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7:261–269. doi: 10.1016/S0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- Andjelkovic M, Alessi DR, Meier R, Fernandez A, Lamb NJC, Frech M, Cron P, Cohen P, Lucocq JM, Hemmings BA. Role of translocation in the activation and function of protein kinase B. J Biol Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- Aune SE, Herr DJ, Mani SK, Menick DR. Selective inhibition of class I but not class IIb histone deacetylases exerts cardiac protection from ischemia reperfusion. J Mol Cell Cardiol. 2014;72:138–145. doi: 10.1016/j.yjmcc.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balinsky JB, Choritz EL, Coe CGL, van der Schans GS. Amino acid metabolism and urea synthesis in naturally aestivating Xenopus laevis. Comp Biochem Physiol. 1967;22:59–68. doi: 10.1016/0010-406X(67)90166-1. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Baehr LM. Skeletal muscle atrophy and the E3 ubiquitin ligases, MuRF1 and MAFbx/atrogin-1. Am J Physiol Endocrinol Metab. 2014;307:E469–E484. doi: 10.1152/ajpendo.00204.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science (80-) 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Cowan KJ. Mitogen-activated protein kinases: new signaling pathways functioning in cellular responses to environmental stress. J Exp Biol. 2003;206:1107–1115. doi: 10.1242/jeb.00220. [DOI] [PubMed] [Google Scholar]
- Dann SG, Selvaraj A, Thomas G. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med. 2007;13:252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Das TP, Suman S, Alatassi H, Ankem MK, Damodaran C. Inhibition of AKT promotes FOXO3a-dependent apoptosis in prostate cancer. Cell Death Dis. 2016;7:e2111. doi: 10.1038/cddis.2015.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuchar EM. Regeneration of the tail of axolotl. J Exp Zool. 1975;192:381–390. doi: 10.1002/jez.1401920311. [DOI] [PubMed] [Google Scholar]
- Eaton SL, Roche SL, Llavero Hurtado M, Oldknow KJ, Farquharson C, Gillingwater TH, Wishart TM. Total protein analysis as a reliable loading control for quantitative fluorescent western blotting. PLoS One. 2013;8:e72457. doi: 10.1371/journal.pone.0072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder ME, Burggren WW. Skin breathing in vertebrates. Sci Am. 1985;253:126–142. doi: 10.1038/scientificamerican1185-126. [DOI] [Google Scholar]
- Foletta VC, White LJ, Larsen AE, Léger B, Russell AP. The role and regulation of MAFbx/atrogin-1 and MuRF1 in skeletal muscle atrophy. Pflugers Arch. 2011;461:325–335. doi: 10.1007/s00424-010-0919-9. [DOI] [PubMed] [Google Scholar]
- Furukawa-Hibi Y, Kobayashi Y, Chen C, Motoyama N. FOXO transcription factors in cell-cycle regulation and the response to oxidative stress. Antioxid Redox Signal. 2005;7:752–760. doi: 10.1089/ars.2005.7.752. [DOI] [PubMed] [Google Scholar]
- Grundy JE, Storey KB. Antioxidant defenses and lipid peroxidation damage in estivating toads, Scaphiopus couchii. J Comp Physiol - B Biochem Syst Environ Physiol. 1998;168:132–142. doi: 10.1007/s003600050129. [DOI] [PubMed] [Google Scholar]
- Grundy JE, Storey KB. Urea and salt effects on enzymes from estivating and non-estivating amphibians. Mol Cell Biochem. 1994;131:9–17. doi: 10.1007/BF01075719. [DOI] [PubMed] [Google Scholar]
- Herrmann J, Lerman LO, Lerman A. Ubiquitin and ubiquitin-like proteins in protein regulation. Circ Res. 2007;100:1276–1291. doi: 10.1161/01.RES.0000264500.11888.f0. [DOI] [PubMed] [Google Scholar]
- Hillman SS. The roles of oxygen delivery and electrolyte levels in the dehydrational death of Xenopus laevis. J Comp Physiol B. 1978;128:169–175. doi: 10.1007/BF00689481. [DOI] [Google Scholar]
- Hresko RC, Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J Biol Chem. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JMA, Tindall DJ. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson NJ, Franklin CE. Effect of aestivation on muscle characteristics and locomotor performance in the green-striped burrowing frog, Cyclorana alboguttata. J Comp Physiol B Biochem Syst Environ Physiol. 2002;172:177–182. doi: 10.1007/s00360-001-0242-z. [DOI] [PubMed] [Google Scholar]
- Judge SM, Wu CL, Beharry AW, Roberts BM, Ferreira LF, Kandarian SC, Judge AR. Genome-wide identification of FoxO-dependent gene networks in skeletal muscle during C26 cancer cachexia. BMC Cancer. 2014;14:1–17. doi: 10.1186/1471-2407-14-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam EW, Francis RE, Petkovic M. FOXO transcription factors : key regulators of cell fate. Biochem Soc Trans. 2006;34:722–726. doi: 10.1042/BST0340722. [DOI] [PubMed] [Google Scholar]
- Luu BE, Storey KB. Dehydration triggers differential microRNA expression in Xenopus laevis brain. Gene. 2015;573:64–69. doi: 10.1016/j.gene.2015.07.027. [DOI] [PubMed] [Google Scholar]
- Luu BE, Wijenayake S, Malik AI, Storey KB. The regulation of heat shock proteins in response to dehydration in Xenopus laevis. Cell Stress Chaperones. 2018;23:45–53. doi: 10.1007/s12192-017-0822-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AI, Storey KB. Activation of antioxidant defense during dehydration stress in the African clawed frog. Gene. 2009;442:99–107. doi: 10.1016/j.gene.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Malik AI, Storey KB. Transcriptional regulation of antioxidant enzymes by FoxO1 under dehydration stress. Gene. 2011;485:114–119. doi: 10.1016/j.gene.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Malik AI, Storey KB. Activation of extracellular signal-regulated kinases during dehydration in the African clawed frog, Xenopus laevis. J Exp Biol. 2009;212:2595–2603. doi: 10.1242/jeb.030627. [DOI] [PubMed] [Google Scholar]
- Mantle BL, Hudson NJ, Harper GS, Cramp RL, Franklin CE. Skeletal muscle atrophy occurs slowly and selectively during prolonged aestivation in Cyclorana alboguttata (Gunther 1867) J Exp Biol. 2009;212:3664–3672. doi: 10.1242/jeb.033688. [DOI] [PubMed] [Google Scholar]
- McDowell GS, Philpott A. Ubiquitin-mediated proteolysis in Xenopus extract. Int J Dev Biol. 2016;60:263–270. doi: 10.1387/ijdb.160186gm. [DOI] [PubMed] [Google Scholar]
- Moresi V, Williams AH, Meadows E, Flynn JM, Potthoff MJ, McAnally J, Shelton JM, Backs J, Klein WH, Richardson JA, Bassel-Duby R, Olson EN. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/S0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013;280:4294–4314. doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/S1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM. Aestivation: signaling and hypometabolism. J Exp Biol. 2012;215:1425–1433. doi: 10.1242/jeb.054403. [DOI] [PubMed] [Google Scholar]
- Tinsley RC, Kobel HR (1996) The biology of Xenopus. Zool Soc London:317–328
- Waddell DS, Baehr LM, van den Brandt J, Johnsen SA, Reichardt HM, Furlow JD, Bodine SC. The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am J Physiol Endocrinol Metab. 2008;295:E785–E797. doi: 10.1152/ajpendo.00646.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-W, Storey KB. Regulation of the mTOR signaling network in hibernating thirteen-lined ground squirrels. J Exp Biol. 2012;215:1720–1727. doi: 10.1242/jeb.066225. [DOI] [PubMed] [Google Scholar]
- Yang J, Cron P, Thompson V, Good VM, Hess D, Hemmings BA, Barford D. Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation. Mol Cell. 2002;9:1227–1240. doi: 10.1016/S1097-2765(02)00550-6. [DOI] [PubMed] [Google Scholar]
- Zhang J, Storey KB. Akt signaling and freezing survival in the wood frog, Rana sylvatica. Biochim Biophys Acta, Gen Subj. 2013;1830:4828–4837. doi: 10.1016/j.bbagen.2013.06.020. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Luu BE, Storey KB. FoxO4 activity is regulated by phosphorylation and the cellular environment during dehydration in the African clawed frog, Xenopus laevis. Biochim Biophys Acta, Gen Subj. 2018;1862:1721–1728. doi: 10.1016/j.bbagen.2018.05.002. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tessier SN, Storey KB. Inhibition of skeletal muscle atrophy during torpor in ground squirrels occurs through downregulation of MyoG and inactivation of Foxo4. Cryobiology. 2016;73:112–119. doi: 10.1016/j.cryobiol.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Zhao X, Gan L, Pan H, et al. Multiple elements regulate nuclear/cytoplasmic shuttling of FOXO1: characterization of phosphorylation- and 14-3-3-dependent and -independent mechanisms. Biochem J. 2004;378:839–849. doi: 10.1042/BJ20031450. [DOI] [PMC free article] [PubMed] [Google Scholar]