Abstract
Non-small cell lung cancer is the most common type of lung cancer, accounting for more than 80% of this tumor. Ubiquitin-specific protease (USP) 14 is one of the 100 deubiquitinating enzymes that is overexpressed in lung cancer and has been validated as a therapeutic target. The aim of this study is to determine whether the accumulation of ubiquitinated proteins results in endoplasmic reticulum (ER) stress-mediated autophagy. To inhibit USP-14, A549 lung cancer cells were treated with USP-14 siRNA and IU1-47 (20 μM). The protein level, mRNA expression, and cell cycle analysis were evaluated using Western blot, real-time PCR, and flow cytometry, respectively. We found that treating A549 cells with USP14 inhibitors significantly reduced the proliferation rate and induced cell cycle arrest at G2/M phase. We also found that USP14 inhibitors did not induce apoptosis but actually induced autophagy through accumulation of ubiquitinated proteins/ER stress/unfolded protein response (UPR) axis. Moreover, we have for the first time demonstrated that the USP14 inhibition induces ER stress–mediated autophagy in A549 cells by activation of c-Jun N-terminal kinase 1 (JNK1). In conclusion, the current investigation represents a new mechanism by which inhibition of USP14 triggers autophagy via ER stress–mediated UPR in A549 cells.
Keywords: Ubiquitin-proteasome, Endoplasmic reticulum stress, Autophagy, JNK, NSCLC
Introduction
Lung cancer is the most frequent cause of cancer-associated death across the world and non-small cell lung cancer (NSCLC) accounts for 85 percent of lung cancers (Goldstraw et al. 2011). The ubiquitin-specific protease (UPS) is a selective proteolytic system in which misfolded or unfolded proteins are tagged with ubiquitin and targeted to the proteasome for degradation (Ciechanover 2015). Autophagy is a cellular degradation process whose primary function is to maintain cellular homeostasis by removing long-lived proteins and damaged organelles. When the level of misfolded or unfolded proteins overwhelms the folding capacity of the endoplasmic reticulum (ER), the unfolded protein response (UPR) is then activated as an adaptive or protective process (Zhao et al. 2002). The proteasome most exclusively used in human is the cytosolic 26S proteasome which comprises a proteolytic 20S core particle capped by two 19S regulatory particles (Wang and Maldonado 2006).
Malignant cancer cells often demonstrate increasing protein synthesis rates; thus, folding capacity of the endoplasmic reticulum is exceeded and accumulated unfolded or misfolded proteins tend to initiate ER stress response followed by cell death (Mofers et al. 2017). Neoplastic cells, however, resist against cell death by activating the ubiquitin-proteasome system (UPS) that degrades the unfolded proteins (Shen et al. 2013). In this context, targeting UPS has emerged as an innovative strategy for developing new anti-neoplastic agents, as malignant cells are presumed to be highly dependent on functional UPS (D’Arcy and Linder 2012). Bortezomib, a potent and selective inhibitor of the 26S proteasome, apart from exhibiting protective effects against human NSCLC cell lines via inducing G2/M cell cycle arrest and apoptosis, has successfully retarded the NSCLC progression in clinical settings (Piperdi et al. 2012). Moreover, bortezomib promotes the cytotoxic effects of inducible caspase-9 suicide gene system in NSCLC cell lines (Ando et al. 2014).
In mammalian, there are two major degradation systems: the UPS and the autophagy-lysosome system, which are responsible for approximately 80–90% (soluble short-lived ubiquitinated proteins) and 10–20% (insoluble long-lived ubiquitinated proteins) of cellular proteolysis, respectively (Kwon and Ciechanover 2017). The crosstalk between these systems revealed that the autophagy-lysosome system can compensatively clear soluble short-lived ubiquitinated proteins when UPS system is suppressed; the UPS can also act in a compensatory fashion (Mooneyham and Bazzaro 2017). Autophagy is a process by which cancer cells can be adapted either to survive through upregulation of cell cycle and the escape from apoptosis or to elimination through induction of apoptosis and cell cycle arrest (Liang et al. 1999). In addition, autophagy can be activated in response to various physiological and pathological conditions such as starving, reactive oxygen species, cellular energy depletion, ER stress, and accumulation of ubiquitinated proteins. Inhibition of deubiquitinating enzyme USP14, a key enzyme of proteasomal pathway, accumulates ubiquitinated proteins and results in ER stress (Ding et al. 2018). Therefore, in the present investigation, we aimed to study whether USP14 inhibition is capable of inducing autophagy via ER stress–induced unfolded protein response in A549 cells.
Materials and methods
Materials
Potent USP14 inhibitor IU1-47 was obtained from AOBIOUS INC (Gloucester, MA, USA). Potent selective JNK inhibitor SP600125 was purchased from Cell Signaling Technology (Beverly, MA). Dulbecco’s Modified Eagle’s Medium (DMEM), fetal bovine serum (FBS), and penicillin-streptomycin were obtained from Gibco Invitrogen.
Cell culture
The A549 human lung carcinoma epithelial-like cell line was purchased from Pasteur Institute (Tehran, Iran). Cells were cultured in DMEM supplemented with penicillin (100 μg/ml), streptomycin (100 U/ml), and 10% FBS. Cells were grown and maintained in a humidified atmosphere at 37 °C and 5% CO2.
USP14 gene silencing
A549 cells were transfected with specific USP14 siRNA duplex oligonucleotides (sense: 5′-UCAGCAUCGUAACACCAGAAGAUAU-3′, antisense: 5′-AUCUUCUGGUGUUACGAUGCUGACA-3′) or scrambled negative control oligonucleotides (sense: 5′- UUCUCCGAACGUGUCACGUTT-3′, antisense: 5′- ACGUGACACGUUCGGAGAATT-3′) using Lipofectamine 2000 regent (Invitrogen) according to manufacturer’s instructions. Briefly, cells were randomly grown in 60-mm dishes to about 50% confluence, and were washed twice in Opti-MEM medium. The mixture was prepared by mixing 60 nM USP14 siRNA duplex oligonucleotides or 60 nM scrambled negative control oligonucleotides and 10 μM Lipofectamine 2000 in 1-ml Opti-MEM medium. After 8-h incubation with the above-mentioned mixture, 3 ml of DMEM medium containing 10% FBS was added and incubations were continued at 37 °C for another 40 h.
Cell viability assay
Cell survival was evaluated by MTT assay. Cells (5 × 103 cells/well) were seeded in 96-well plates for 24 h and then treated with different doses of USP14 inhibitor IU1-47 (5, 10, 20, 30, and 40 μM) or JNK inhibitor SP600125 (5, 10, 20, and 40 μM) for 48 h. Afterwards, the medium was removed and 20 μl of MTT solution (2 mg/ml, Sigma Aldrich, Germany) was added to each well. Cells were covered by aluminum foil and incubated at 37 °C for 4 h and then 200 μl of dimethylsulfoxide (DMSO, Merck) was added to dissolve the formazan crystals. The plates were shaken for 15 min, and the optical density (OD) values were measured at 570 nm using a microplate reader (BDSL Immunoskan, Finland). The percentage of viable cells was calculated using the following formula: % viability = (ODtest − ODblank/ODcontrol − ODblank) × 100%.
Proliferation assay with bromodeoxyuridine incorporation test
The cell proliferation of A549 cells was measured by a bromodeoxyuridine (BrdU) Cell Proliferation ELISA Kit (Abcam, UK). At the first step, cells were seeded at a density of 2 × 104 cells/well in 96-well plates, and treated with IU1-47 and USP14 siRNA for 48 h. Next, 20 μl/well of BrdU solution was added for additional 24 h prior to fixing the cells with fixing solution (200 μl/well). After the washing step, anti-BrdU monoclonal antibody and HRP-conjugated secondary antibody were incubated for 60 and 30 min at room temperature, respectively. 100 μl/well of substrate solution was pipetted, and incubated for 30 min in dark condition. Ultimately, stop solution was added and OD values were determined at 450 nm.
Real-time qPCR
Total RNA was isolated using Trizol (Sigma-Aldrich, MO, USA). Complementary DNA was synthesized using Prime Script RT reagent kit (TaKaRa, Japan). RT-qPCR was performed using SYBR Green (Amplicon, Brighton, UK) detection method with Mic thermocycler (BioMolecular Systems, Upper Coomera, Australia). The primer sequences for Beclin-1 and GAPDH genes have been presented in Table 1.
Table 1.
List of RT-qPCR primer sequences
| Gene name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| Beclin-1 | CAGGAACTCACAGCTCCATTAC | GAATCTGCGAGAGACACCATC |
| GAPDH | CACCCACTCCTCCACCTTTG | CCACCACCCTGTTGCTGTAG |
Western blotting
Whole cell extracts were prepared using radioimmunoprecipitation assay buffer. The extracted protein samples were separated by electrophoresis on SDS-PAGE. The protein bands were transferred onto a PVDF membrane and probed with specific antibodies of anti-USP14 (sc-100630, Santa Cruz), anti-Ubiquitin (sc-8017, Santa Cruz), anti-ATF-6 (PA5-20216, Thermo Fisher), anti-phospho-IRE1 alpha (Ser724) (PA1-16927, Thermo Fisher), anti-phospho-JNK (sc-6254, Santa Cruz), anti phospho-PERK (Thr980) (MA5-15033, Thermo Fisher), anti phospho-eIF2α (sc-133132, Santa Cruz), anti-ATF4 (ab23760, Abcam), anti-CHOP (sc-7351, Santa Cruz), anti-LC3-I/II (ab51520, Abcam), anti-cyclin B1 (sc-245, Santa Cruz), anti-cdc-2 (sc-8395, Santa Cruz), anti-caspase-3 (sc-56053, Santa Cruz), anti-caspase-8 (sc-73526, Santa Cruz), anti-caspase-9 (sc-73548, Santa Cruz), anti β-tubulin (ab15568, Abcam), and anti β-actin (sc-47778, Santa Cruz), and with appropriate horseradish peroxide–conjugated secondary antibodies (Santa Cruz, USA). The intensity of each band was normalized to that of β-actin. Values are reported as the fold change relative to the untreated control. Calculations were carried out by the ImageJ picture analysis software (version 1.41).
Flow cytometry–based apoptosis detection
Fluorescein Annexin V-FITC/PI double labeling was performed using Annexin V-FITC Apoptosis Detection Kit (eBioscience, San Diego, CA) to detect the possible apoptotic effects of USP-14 inhibition on the cells. The cells were stained according to the manufacturer’s instructions and the apoptotic cells were determined with a flow cytometer (Miltenyi Biotec, CA) and analyzed with FlowJo 7.6.1 Software (FlowJo Systems, Tree Star, OR).
Cell cycle analysis
Cell cycle assay was carried out by using cell cycle phase determination kit (Cayman Chemical Company, Ann Arbor, MI, USA). Initially, cells were seeded in 6-well plates at a concentration of 2 × 105 cells/well. Next, cells were collected after treatment with USP-14 siRNA and IU1-47 (20 μM), and rinsed with assay buffer. Following the fixation and permeabilization of cells, PI solution in the presence of RNase A was added to stain cells. Finally, flow cytometer (Miltenyi Biotec, CA, USA) was applied to detect PI fluorescence. Flow cytometry data were analyzed by Flowing software 2.5.1 (Turku Centre for Biotechnology, University of Turku, Turku, Finland).
Statistical analysis
All values were expressed as means ± SD. Data were analyzed using Student’s t test, one-way ANOVA or two-way ANOVA followed by Tukey’s post hoc test, where appropriate. Each experiment has been done in triplicate. The p values < 0.05 were considered significant.
Results
Inhibition of USP14 suppresses proliferation without apoptosis induction
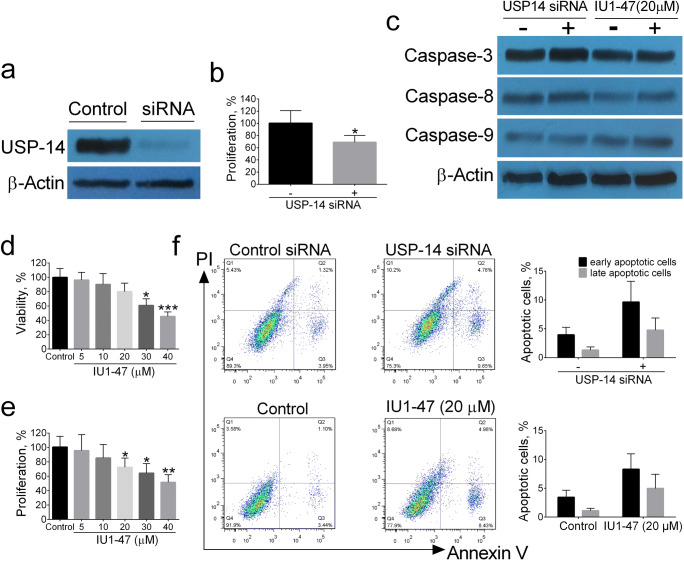
At the first, A549 cells were transfected with USP14 siRNA for 40 h and assayed for USP14 by Western blotting. As shown in Fig. 1a, USP14 siRNA transfection led to an almost complete knockdown of USP14 compared with control siRNA. We also used the pharmacological USP14 inhibitor IU1-47 at different doses (5, 10, 20, 30, 40 μM). Next, we investigated the effect of USP14 inhibition on cell viability and proliferation rate of A549 cells. Compared with the control siRNA, knocking down of USP14 significantly reduced proliferation rate of A549 cells (Fig. 1b). Similarly, compared with DMSO-treated cells, the IU1-47-treated cells markedly reduced both cell viability and proliferation rate of A549 cells in a dose-dependent manner (Fig. 1d, e). These data suggest that the proliferation of A549 cells is associated with USP14 inhibition.
Fig. 1.
Assessment of USP-14 inhibition on cell viability, proliferation, and apoptosis of lung cancer cell line A549. The protein levels of USP14 were assessed by Western blotting (a). The effect of USP-14 siRNA on the percentage of proliferating cells (b). Assessment of pro-apoptotic markers by Western blotting (c). MTT assay in different concentrations of IU1-47 (5, 10, 20, 30, 40 μM) for 48 h (d). The proliferating cell percentage by BrdU assay (e). The Annexin-V/PI flow cytometry analysis for apoptosis (f). Data are shown as mean ± SD of three independent replicates. *p value < 0.05, **p value < 0.01 versus control
In order to investigate whether the anti-proliferative effect of USP14 inhibition was correlated with apoptosis induction of A549 cells, the apoptosis was evaluated by Annexin V/PI flow cytometric analysis; as shown in Fig. 1f, flow cytometry results revealed no significant differences in apoptotic cells between USP14 inhibitors and their controls. Furthermore, the protein levels of pro-apoptotic caspase-3, -9, and -8 were quantified by Western blotting. As shown in Fig. 1c, siRNA knockdown and pharmacological USP14 inhibitor IU1-47 (20 μM) did not change the protein levels of caspase-3, -9, and -8 in A549 cell line. These data suggest that the intrinsic and extrinsic apoptosis pathways are not responsible for anti-proliferative effects of USP14 inhibition in A549 cells.
Inhibition of USP14 arrests cell cycle at G2/M phase
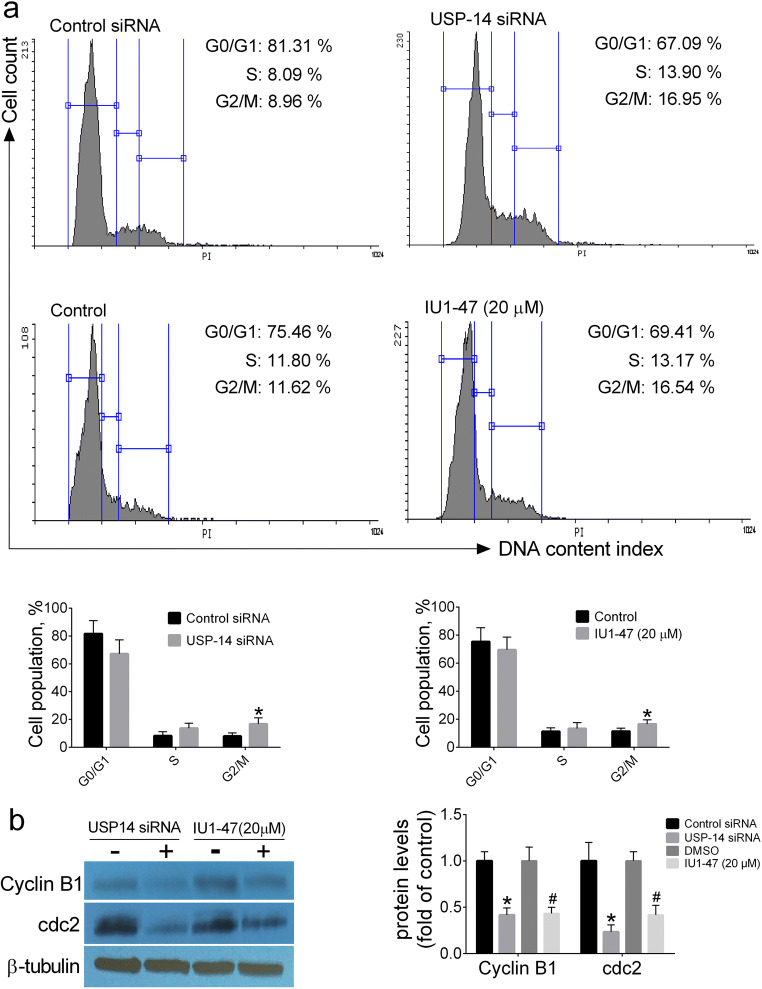
In order to clarify whether the growth-inhibitory effects of USP14 inhibition may be related to its ability in inducing cell cycle arrest, the cell cycle analysis and expression of G2/M proteins including cyclin B1 and cdc2 were assessed by flow cytometry and Western blotting, respectively. Our results revealed that knockdown of USP14 arrested A549 cells at G2/M phase as compared with control siRNA; flow cytometry analysis revealed that siRNA knockdown and pharmacological USP14 inhibitor IU1-47 (20 μM) resulted in a significant increase in the distribution of A549 cells at G2/M phase, a decrease in the distribution at G0/G1 phase, and no significant changes in the cell distribution at S phase (Fig. 2a). Consistent with flow cytometry results, Western blot analysis of G2/M phase regulatory proteins revealed that siRNA knockdown and pharmacological USP14 inhibitor IU1-47 (20 μM) resulted in a significant decrease in the protein levels of cyclin B1 and cdc2 in A549 cells compared with controls (Fig. 2b). These findings propose that inhibition of USP14 arrests A549 cells at G2/M phase and this perturbation can be responsible for growth-inhibitory effects of USP14 inhibition in A549 cells.
Fig. 2.
The effect of USP-14 inhibition on cell cycle progression. Cell cycle analysis (a). Western blotting analysis of G2/M phase-related proteins cyclin B1 and cdc2 (b). Data are shown as mean ± SD of three independent replicates. *p value < 0.05 versus control siRNA and #p value < 0.05 versus DMSO-treated group
Inhibition of USP14 induces accumulation of ubiquitinated proteins and ER stress
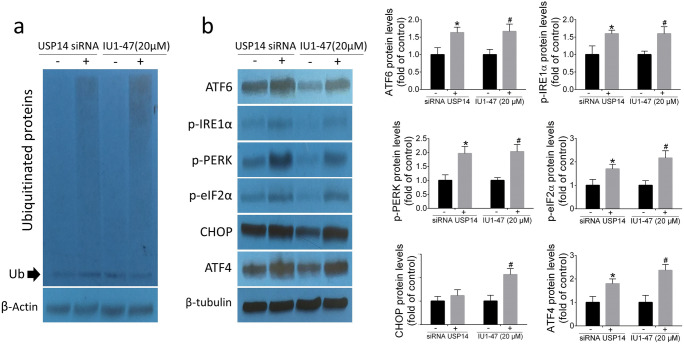
To investigate the mechanism underlying the effect of USP14 inhibition on ER stress, A549 cells were exposed to siRNA knockdown and pharmacological USP14 inhibitor IU1-47 (20 μM) for 48 h. The effect of USP14 inhibition on ER stress–related proteins and accumulation of ubiquitinated proteins were monitored by Western blotting. As shown in Fig. 3a, USP14 inhibition induced accumulation of total ubiquitinated proteins in A549 cells. As reported previously, USP14 inhibition triggers ER stress response (Cai et al. 2017). Consistently, here we found that USP14 knockdown and USP14 inhibitor IU1-47 (20 μM) markedly increased ER stress–related proteins including ATF6, p-IRE1α, p-PERK, p-eIF2α, CHOP, and ATF4 in A549 cell (Fig. 3b). These findings clearly indicate that the accumulation of ubiquitinated proteins induced by USP14 inhibition results in ER stress response in A549 cell line.
Fig. 3.
Analysis of ubiquitinated and ER stress–related proteins. Western blotting of total ubiquitinated proteins (a). Western blotting analysis of ER stress–related proteins ATF6, p-IRE1α, p-PERK, p-eIF2α, CHOP, and ATF4 (b). Data are shown as mean ± SD of three independent replicates. *p value < 0.05 versus control siRNA and #p value < 0.05 versus DMSO-treated group
Inhibition of USP14 induces autophagy
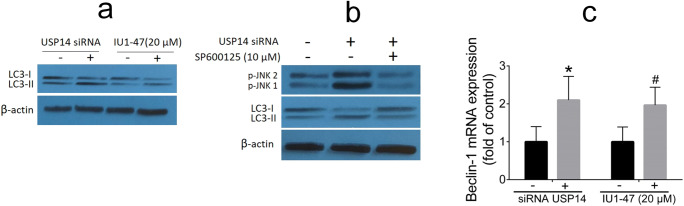
Previous findings revealed that activation of JNK pathway is critically involved in autophagy and cell death (Byun et al. 2009; Xie et al. 2011). It was well documented that autophagy (i.e., induced by increased ER stress) can lead to cell survival or cell death depending on the duration and severity of the process (Klionsky and Emr 2000). To clarify the mechanism underlying the effect of USP14 inhibition on autophagy, A549 cells were exposed to siRNA knockdown and pharmacological USP14 inhibitor IU1-47 (20 μM) for 48 h; the conversion of LC3-I to LC3-II was used as an autophagy marker. Herein, we found that USP14 inhibition results in an increase in the LC3-II levels (Fig. 4a). We then asked whether USP14 inhibition–induced autophagy is mediated by activation of JNK pathway. We found that pretreatment with JNK inhibitor SP600125 could reverse the upregulation of LC3-II levels induced by USP14 siRNA knockdown in A549 cells (Fig. 4b). The mRNA expression of the Beclin-1, as an autophagic gene, was quantified using qRT-PCR; our results revealed that USP14 inhibition can result in an increase in expression of Beclin-1 compared with their controls (Fig. 4c). Taken together, these results show that inhibition of USP14 leads to increased JNK-mediated autophagy in A549 cell line.
Fig. 4.
Analysis of autophagy-related markers. Western blotting analysis of LC3-I to LC3-II conversion (a). A549 cells were treated with JNK inhibitor SP600125 (10 μM) at the presence or absence of USP14 siRNA for 48 h, followed by Western blotting analysis of LC3-I to LC3-II conversion and p-JNK1,2 (b). The RT-PCR analysis of mRNA expression of Beclin-1 (c). Data are shown as mean ± SD of three independent replicates. *p value < 0.05 versus control siRNA and #p value < 0.05 versus DMSO-treated group
Discussion
Ubiquitin-specific protease 14 (USP14), a 19S proteasome–associated deubiquitinating enzyme, is rapidly emerging as a novel target for cancer therapy. USP14 is highly expressed in solid tumors and hematologic malignancies (Liu et al. 2019a). Indeed, a recent study shows the high expression of USP14 in lung cancer tissue (Han et al. 2019). This study was aimed to clarify whether the USP14 inhibition induces autophagy via accumulation of ubiquitinated proteins/ER stress/UPR axis in human lung cancer cell line A549.
Firstly, our results have shown that USP14 inhibition accumulates ubiquitinated proteins in the A549 cell line and causes ER stress–mediated UPR. Under physiological conditions, deubiquitinating enzyme USP14 regulates the protein degradation process by removing ubiquitin tags from mistakenly tagged substrates, thereby playing critical role in protein quality control and homeostasis (Yuan et al. 2018). Thus, it is not surprising that specific USP14 inhibitors can potentially result in accumulation of ubiquitinated proteins. In line with our result, pharmacologic or genetic inhibition of USP14 results in accumulation of ubiquitinated proteins and provides a promising anticancer therapeutic strategy (Yuan et al. 2018). A variety of stress responses such as oxidative stress, mitochondrial dysfunction, and ER stress is presented when ubiquitinated proteins are accumulated in the cells (Didier et al. 2018). In this study, we observed that the expression of ER stress markers ATF6, p-IRE1α, p-PERK, p-eIF2α, CHOP, and ATF4 increases as the A549 cells are exposed to USP14 inhibitors. Similarly, supplementation with b-AP15, a potent and selective USP14 inhibitor, induces UPR through activating ER stress in vivo and in vitro (Ding et al. 2018).
Secondly, we demonstrated that USP14 inhibition arrests A549 cells at G2/M phase. Interestingly, USP14 inhibition also caused a marked reduction in the expression of G2/M phase–related proteins including cyclin B1 and cdc2. Cell cycle progression is essential for rapid proliferation of cancer cells, and in fact is regulated by the combinations of cyclins and cyclin-dependent kinases that are periodically expressed during cell cycle phases. USP14 clearly functions in cell cycle progression. USP14 silencing resulted in cell cycle arrest and β-catenin degradation in NSCLC cell line A549, while over-expression of USP14 promoted tumor cell proliferation (Wu et al. 2013). Similarly, USP14 inhibition delayed G0/G1 to S transition during cell cycle progression in AR-responsive breast cancer cells (Liao et al. 2018). In line with our results, USP14 has a role in ubiquitination of cyclin B1 (Lee et al. 2016); and silencing of USP14 arrested breast cancer cells at G2/M phase through the ubiquitination and downregulation of cyclin B1 (Liu et al. 2019b).
Thirdly, we have for the first time found that the USP14 inhibition induces autophagy in A549 cells by phosphorylation/activation of c-Jun N-terminal kinase 1 (JNK1); and the upregulation of Beclin-1 expression as well as the conversion of LC3I to LC3II verified the existence of autophagy. Furthermore, we also indicated that USP14 inhibition–induced autophagy has not been accompanied with significant induction of intrinsic or extrinsic apoptosis in A549 cells. As mentioned above, the intracellular accumulation of ubiquitinated proteins triggers ER stress and UPR activation. Once UPR is initiated, it activates three major ER stress sensors termed IRE1, PERK, and ATF6 to recover ER homeostasis (Yan et al. 2015). IRE1-XBP1 pathway is indirectly able to induce autophagy through Beclin-1 transcriptional activation (Margariti et al. 2013); conversely, XBP1s deficiency does not induce autophagy in neurons by affecting FOXO1 transcriptional activity (Vidal et al. 2012). Furthermore, activation of JNK1 promotes dissociation of Beclin-1/Bcl-2 complex and initiates autophagy (Wei et al. 2008). Our results showing the pivotal role of PERK/eIF2α/ATF4/CHOP axis in autophagy are consistent with previous studies. On the other hand, radiation-induced ER stress mediates autophagy in caspase-3/7-deficient cells via PERK/eIF2α pathway (Kim et al. 2010). Similarly, severe hypoxia results in upregulation of ATF4 in a PERK-dependent manner and ATF4 directly binds to cAMP response element-binding protein to promote the expression of light chain 3 β (LC3β), a vital constituent of autophagosomal membrane (Rzymski et al. 2010). Paradoxically, inhibition of PERK can block autophagy in MYCN-amplified neuroblastoma cells, as manifested by decreased LC3 II/ LC3 I ratio (Wang et al. 2018). Autophagy is a process in which cell survival or apoptosis can occur and it depends on the cell type and context (Klionsky 2008). Inconsistent with our observation, previous study has shown that USP14 inhibition–mediated autophagy is not accompanied with increase in caspase-3 activity in A549 cell line (Han et al. 2019). Conversely, USP14 inhibition induced apoptosis in epithelial ovarian cancer cells (Wang et al. 2015). However, although targeting ubiquitin proteasome system provides new anticancer therapeutic strategy, UPR-induced autophagy can result in cell death or cell survival in a context-dependent manner. For example, accumulation of ubiquitinated proteins can lead to significant ER stress in malignant glioma cells followed by apoptosis or autophagy; however, autophagy was cytoprotective in this context (Ciechomska et al. 2013). In conclusion, this investigation proposes new mechanism in which USP14 inhibition induces autophagy via ER stress–dependent upregulation of PERK, IRE1α, and JNK1 pathways, arrests A549 cells at G2/M phase, and does not induce significant apoptosis.
Funding information
This study was funded by the grant obtained from the Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran (grant no.: 60424).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ali-Asghar Moghadami and Elmira Aboutalebi Vand Beilankouhi contributed equally to this work.
References
- Ando M, et al. Bortezomib sensitizes non-small cell lung cancer to mesenchymal stromal cell-delivered inducible caspase-9-mediated cytotoxicity. Cancer Gene Ther. 2014;21:472. doi: 10.1038/cgt.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun J-Y, Yoon C-H, An S, Park I-C, Kang C-M, Kim M-J, Lee S-J. The Rac1/MKK7/JNK pathway signals upregulation of Atg5 and subsequent autophagic cell death in response to oncogenic Ras. Carcinogenesis. 2009;30:1880–1888. doi: 10.1093/carcin/bgp235. [DOI] [PubMed] [Google Scholar]
- Cai J, et al. A novel deubiquitinase inhibitor b-AP15 triggers apoptosis in both androgen receptor-dependent and-independent prostate cancers. Oncotarget. 2017;8:63232. doi: 10.18632/oncotarget.18774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The unravelling of the ubiquitin system. Nat Rev Mol Cell Biol. 2015;16:322–324. doi: 10.1038/nrm3982. [DOI] [PubMed] [Google Scholar]
- Ciechomska I, Gabrusiewicz K, Szczepankiewicz A, Kaminska B. Endoplasmic reticulum stress triggers autophagy in malignant glioma cells undergoing cyclosporine a-induced cell death. Oncogene. 2013;32:1518–1529. doi: 10.1038/onc.2012.174. [DOI] [PubMed] [Google Scholar]
- D’Arcy P, Linder S. Proteasome deubiquitinases as novel targets for cancer therapy. Int J Biochem Cell Biol. 2012;44:1729–1738. doi: 10.1016/j.biocel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- Didier R, et al. Targeting the proteasome-associated deubiquitinating enzyme USP14 impairs melanoma cell survival and overcomes resistance to MAPK-targeting therapies. Mol Cancer Ther. 2018;17:1416–1429. doi: 10.1158/1535-7163.MCT-17-0919. [DOI] [PubMed] [Google Scholar]
- Ding Y, Chen X, Wang B, Yu B, Ge J. Deubiquitinase inhibitor b-AP15 activates endoplasmic reticulum (ER) stress and inhibits Wnt/Notch1 signaling pathway leading to the reduction of cell survival in hepatocellular carcinoma cells. Eur J Pharmacol. 2018;825:10–18. doi: 10.1016/j.ejphar.2018.02.020. [DOI] [PubMed] [Google Scholar]
- Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, Shepherd FA. Non-small-cell lung cancer. Lancet. 2011;378:1727–1740. doi: 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- Han KH, et al. USP14 inhibition regulates tumorigenesis by inducing autophagy in lung cancer in vitro. Int J Mol Sci. 2019;20:5300. doi: 10.3390/ijms20215300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Moretti L, Mitchell LR, Jung DK, Lu B (2010) Endoplasmic reticulum stress mediates radiation-induced autophagy by perk-eIF2α in caspase-3/7-deficient cells. Oncogene 29(22):3241–51 [DOI] [PMC free article] [PubMed]
- Klionsky DJ. Autophagy revisited: a conversation with Christian de Duve. Autophagy. 2008;4:740–743. doi: 10.4161/auto.6398. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT, Ciechanover A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem Sci. 2017;42:873–886. doi: 10.1016/j.tibs.2017.09.002. [DOI] [PubMed] [Google Scholar]
- Lee B-H, Lu Y, Prado MA, Shi Y, Tian G, Sun S, Elsasser S, Gygi SP, King RW, Finley D (2016) USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature 532(7599):398–401 [DOI] [PMC free article] [PubMed]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Liao Y, et al. Growth arrest and apoptosis induction in androgen receptor-positive human breast cancer cells by inhibition of USP14-mediated androgen receptor deubiquitination. Oncogene. 2018;37:1896–1910. doi: 10.1038/s41388-017-0069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Chen J, Zhang S (2019a) Emerging role of ubiquitin-specific protease 14 in oncogenesis and development of tumor: therapeutic implication Life sciences:116875 [DOI] [PubMed]
- Liu B, Liu Y, Wang Y, Xie C, Gan M, Han T, Cao J, Wang J (2019) CyclinB1 deubiquitination by USP14 regulates cell cycle progression in breast cancer. Pathology-Research and Practice 215(10):152592 [DOI] [PubMed]
- Margariti A, Li H, Chen T, Martin D, Vizcay-Barrena G, Alam S, Karamariti E, Xiao Q, Zampetaki A, Zhang Z, Wang W, Jiang Z, Gao C, Ma B, Chen YG, Cockerill G, Hu Y, Xu Q, Zeng L. XBP1 mRNA splicing triggers an autophagic response in endothelial cells through BECLIN-1 transcriptional activation. J Biol Chem. 2013;288:859–872. doi: 10.1074/jbc.M112.412783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mofers A, Pellegrini P, Linder S, D’Arcy P. Proteasome-associated deubiquitinases and cancer. Cancer Metastasis Rev. 2017;36:635–653. doi: 10.1007/s10555-017-9697-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooneyham A, Bazzaro M (2017) Targeting deubiquitinating enzymes and autophagy in cancer. In: Cancer Gene Networks. Springer, pp 49-59 [DOI] [PubMed]
- Piperdi B, et al. Phase-I/II study of bortezomib in combination with carboplatin and bevacizumab as first-line therapy in patients with advanced non–small-cell lung cancer. J Thorac Oncol. 2012;7:1032–1040. doi: 10.1097/JTO.0b013e31824de2fa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzymski T, Milani M, Pike L, Buffa F, Mellor H, Winchester L, Pires I, Hammond E, Ragoussis I, Harris AL (2010) Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 29(31):4424–35 [DOI] [PubMed]
- Shen M, Schmitt S, Buac D, Dou QP. Targeting the ubiquitin–proteasome system for cancer therapy. Expert Opin Ther Targets. 2013;17:1091–1108. doi: 10.1517/14728222.2013.815728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal RL, Figueroa A, Court FA, Thielen P, Molina C, Wirth C, Caballero B, Kiffin R, Segura-Aguilar J, Cuervo AM, Glimcher LH, Hetz C. Targeting the UPR transcription factor XBP1 protects against Huntington’s disease through the regulation of FoxO1 and autophagy. Hum Mol Genet. 2012;21:2245–2262. doi: 10.1093/hmg/dds040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol. 2006;3:255–261. [PubMed] [Google Scholar]
- Wang Y, Wang J, Zhong J, Deng Y, Xi Q, He S, Yang S, Jiang L, Huang M, Tang C, Liu R. Ubiquitin-specific protease 14 (USP14) regulates cellular proliferation and apoptosis in epithelial ovarian cancer. Med Oncol. 2015;32:379. doi: 10.1007/s12032-014-0379-8. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. The protective autophagy activated by GANT-61 in MYCN amplified neuroblastoma cells is mediated by PERK. Oncotarget. 2018;9:14413. doi: 10.18632/oncotarget.24214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Liu C, Bai C, Han Y-P, Cho W, Li Q. Over-expression of deubiquitinating enzyme USP14 in lung adenocarcinoma promotes proliferation through the accumulation of β-catenin. Int J Mol Sci. 2013;14:10749–10760. doi: 10.3390/ijms140610749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C-M, Chan WY, Yu S, Zhao J, Cheng CH. Bufalin induces autophagy-mediated cell death in human colon cancer cells through reactive oxygen species generation and JNK activation. Free Radic Biol Med. 2011;51:1365–1375. doi: 10.1016/j.freeradbiomed.2011.06.016. [DOI] [PubMed] [Google Scholar]
- Yan MM, Ni JD, Song D, Ding M, Huang J. Interplay between unfolded protein response and autophagy promotes tumor drug resistance. Oncol Lett. 2015;10:1959–1969. doi: 10.3892/ol.2015.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T, Yan F, Ying M, Cao J, He Q, Zhu H, Yang B. Inhibition of ubiquitin-specific proteases as a novel anticancer therapeutic strategy. Front Pharmacol. 2018;9:1080. doi: 10.3389/fphar.2018.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]