Abstract
High-throughput proteomic technologies are widely used for understanding the disease mechanism, drug-resistant mechanism, and to identify drug targets and markers for diagnostics. Studies with proteomics applications, relating to Leishmaniasis, are being constantly reported in the literature. However, from such studies, a readily accessible knowledge of differentially modulated proteins associated with Leishmaniasis is lacking. Hence, we performed a systematic review concerning differentially modulated proteins (DMP) in Leishmania as well as host infected with Leishmania from the published articles between the years 2000 and 2019. This review is classified into five different sections, namely, DMP in the host after Leishmania infection, DMP between different strains of Leishmania, DMP in drug-resistant Leishmania, DMP in Leishmania under stress, and DMP in different life stages of Leishmania. A lot of consensuses could be observed among the DMP in drug-resistant and stressed Leishmania. In addition to the review, a database was constructed with the data collected in this study (protein accession ID, protein name, gene name, host organism, experimental conditions, fold change, and regulatory data). A total of 2635 records are available in the database. We believe this review and the database will help the researcher in understanding the disease better and provide information for the targeted proteomics study related to Leishmaniasis. Database availability: http://ldepdb.biomedinformri.com/.
Electronic supplementary material
The online version of this article (10.1007/s11033-020-05936-z) contains supplementary material, which is available to authorized users.
Keywords: Proteomics, Differentially modulated proteins, Leishmania, Database, Leishmaniasis
Introduction
Leishmaniasis, a neglected tropical disease, is caused by the protozoa parasite of the genus Leishmania. More than 20 species of Leishmania are responsible for the disease in humans and it is mainly transmitted to humans by the sand flies [1]. Leishmania is a unicellular eukaryote with a well-defined nucleus, kinetoplast, and flagella [2]. It adapts two different structural variants in their life cycle, namely promastigote and amastigote. An infected sandfly injects promastigotes into a human during their blood meal, where they are mostly phagocytized by macrophages and transform themselves into amastigotes. Further, the amastigotes divide and infect other phagocytic cells [3]. Hence, amastigotes are the cause of disease in the mammalian host [4]. Leishmaniasis is mainly associated with the weak immune system, malnutrition, and poor housing [5], thus majorly affecting the poor population of Africa, Asia, and Latin America. Since more than 90 countries are endemic for leishmaniasis [6], it is considered one of the major neglected tropical diseases of serious concern worldwide. There are three main forms of leishmaniasis, cutaneous leishmaniasis (CL), visceral leishmaniasis (VL), and mucocutaneous leishmaniasis (MCL). CL, the most common form of the disease, is majorly reported in the new world countries and is caused by L. mexicana, L. amazonensis, L. braziliensis, L. infantum chagasi, and L. major [7]. VL, the most serious form, is mainly caused by Leishmania donovani and Leishmania infantum [8]. While the rare form, MCL, is mostly caused by L. braziliensis complex, L. guyanensis, and L. panamenensis [9]. Countries like India, Bangladesh, Sudan, Ethiopia, and Brazil have reported more VL cases (> 90% in total) than the rest [10]. According to the 2017 World Health Organization report, 75 countries are endemic for visceral leishmaniasis [11]. Also, Post-Kala-Azar Dermal Leishmaniasis (PKDL) [12] is of more serious concern in India and Sudan [13]. Chemotherapy is the preferred choice for treating VL [14]. Drugs available for the treatment of VL include Amphotericin B, Miltefosine, Paromomycin, Pentamidine, and Sodium stibogluconate [15]. Amphotericin B is considered an effective drug for VL in the Indian subcontinent [16–18]. However, increased cases of resistance to Amphotericin B in VL patients [19] and the rate of relapse in PKDL patients (after treatment) have increased substantially [20, 21]. At the same time, combination therapy proved to be more effective in treating VL than monotherapy [22]. However, there are also reports of resistance to combination therapy [23–25]. Efforts were made in the past to identify a lot of small-molecule inhibitors against leishmaniasis [26]. However, the number of well-established drug targets for VL is limited [27]. Hence, identifying new drug target(s) becomes a constant requirement in the process of combating VL. Although several VL vaccine candidates are under different phases of the trial [28, 29], none are yet available. These facts indicate the necessity of alternate drugs, drug targets and vaccine candidates to treat VL.
The advancement in proteomic technologies [30] acts as a valuable tool for potential drug target identification, understanding the disease progression, diagnostic biomarker identification [31], and characterization of antigens in vaccine development [32]. Specifically, oncological studies effectively employ the advanced proteomic approaches for the identification of drug targets, the discovery of downstream effectors, protein-protein interaction study, and biomarker discovery [31]. Also, many studies involving high-throughput proteomic techniques (related to leishmaniasis) report the comparison of high-resolution protein data under different experimental conditions. Hence, we conducted a systematic review of such studies involving high-throughput proteomic techniques (mainly focusing on comparable experimental conditions) and provided data that is easy to compare and access. This review is majorly divided into the following sections: (a) data collection, (b) differentially modulated proteins (DMP) in the host after Leishmania infection, (c) DMP between different strains of Leishmania sp., (d) DMP in drug-resistant Leishmania sp., (e) DMP in stressed Leishmania sp., (f) DMP in different life stages of Leishmania sp., and (g) database development. In addition to the information presented in this review, a freely-accessible and searchable database (http://ldepdb.biomedinformri.com/) is also made available. PubMed ID, protein ID, protein name, gene name, fold change, the experimental organism, experimental conditions, and the experimental methods of corresponding studies (collected for this review) are accessible via the database. Hence, we believe this study includes a comprehensive review of differential protein expression studies related to Leishmaniasis and presented the data to the research community in an easy to access way.
Data collection
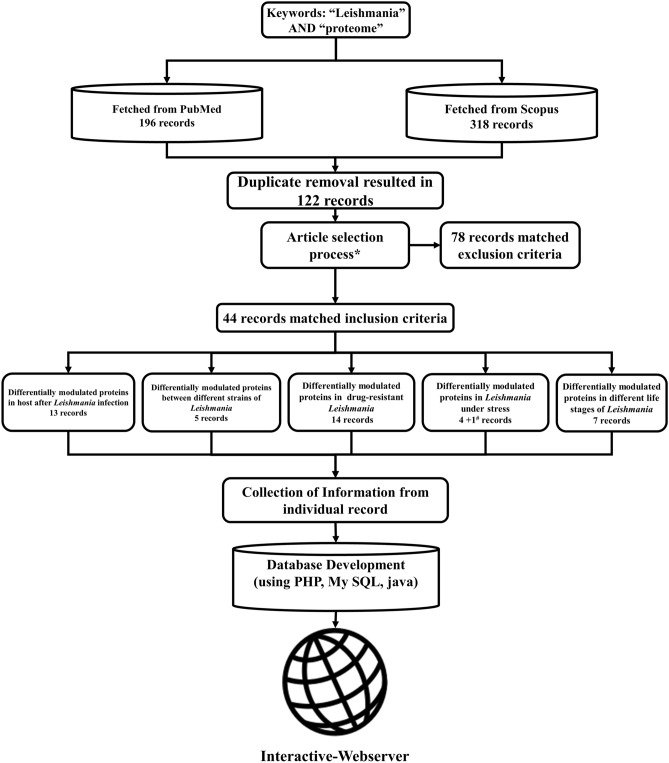
PubMed and Scopus literature databases are used for searching the articles. The keywords “Leishmania” and “proteome”, present in “title” or “abstract”, were searched in the literature databases. The search was performed on 24th October 2019 and the search year timeline was between 2000 and 2019. The search resulted in a total of 196 records in PubMed and 318 records in Scopus. The duplicate entries were removed, resulting in a total of 122 unique records. These 122 records were included for the further manual screening process. An overview of the data collection process is depicted in Fig. 1. The inclusion and exclusion criteria for this study are briefed below.
Fig. 1.
Schematic of systematic procedure adopted for the review. *Refer data collection section; #not directly related to stress
Inclusion and exclusion criteria
Only the published research articles were considered. The full-text of all the 122 unique records were obtained and read manually to identify the relevancy. Two authors independently verified the articles for relevancy. Articles that report DMP and protein identification studies (in addition to 2-Dimentional electrophoresis) relating to Leishmania or leishmaniasis were only considered. Review articles were not considered. Studies that focused only on one protein were not considered. Studies only with genomics or transcriptomics experiments were not considered. Studies involving only the in-silico methods were not considered. Studies that do not involve any proteomic techniques were not considered. Full articles that cannot be obtained through any means (including personal correspondence to the authors of the article) were not considered. Based on the inclusion and exclusion criteria, a total of 44 articles are found relevant and used for this review.
Differentially modulated proteins in the host after Leishmania sp. infection
Comparing the host protein expression levels before and after infection will provide information about the disease mechanism, host-response, and the potential markers for diagnosis [33, 34]. Concerning Leishmaniasis, out of articles matching the selection criteria of this review, a total of 13 articles reported differential proteome analysis between infected and healthy states (Tables 1 and 2). Based on the type of host, the 13 articles are further grouped into three categories (Homo sapiens: 3, Mus musculus: 7, and Canis lupus: 3) and discussed below.
Table 1.
Details of studies included in this review
| Organism | Proteomics methodology | Experiment | No. of differentially regulated proteins | Year | References | Study No. | |
|---|---|---|---|---|---|---|---|
| Up | Down | ||||||
| Proteins of Homo sapiens | |||||||
| Infection based studies | |||||||
| L. donovani | 2DE; 2D-DIGE; MALDI-TOF/TOF MS/MS; Western blot; ELISA |
VL vs endemic control VL vs non-endemic control |
38 | 10 | 2014 | [35] | 1 |
| L. braziliensis | 2DE-MALDI-TOF/TOF MS | CL vs normal skin | 9 | 4 | 2015 | [38] | 2 |
| L. panamensis | SDS-PAGE; LC ESI–MS/MS | Start of treatment day vs 7th day of treatment | 7 | 5 | 2019 | [37] | 3 |
| Proteins of Mus Musculus | |||||||
|---|---|---|---|---|---|---|---|
| Infection based studies | |||||||
|
L. amazonensis and L. major |
Nano LC–MS/MS; Western blot | Infected vs control | 4 | 11 | 2013 | [39] | 4 |
| L. mexicana | SDS-PAGE; LC MS/MS; Western blot; EMSA | Macrophage infected with L. mexicana vs un-infected macrophage as control | 27 | 7 | 2013 | [45] | 5 |
|
L. amazonensis, L. major, and L. infantum |
Western blot; Nano LC–MS/MS; LTQ-OrbitrapVelos MS | Infected vs healthy | 107 | 42 | 2019 | [40] | 6 |
|
L. amazonensis and L. major |
Western blot; Nano LC–MS/MS; LTQ-OrbitrapVelos MS | Infected vs healthy | 69 | 51 | 2019 | [41] | 7 |
| L. infantum | Flow Cytometry; ITRAQ; Nano LC–MS/MS | Protein Malnourished infected vs well nutritioned un-infected | 38 | 71 | 2019 | [43] | 8 |
| L. infantum | ITRAQ; Nano LC–MS/MS |
Malnourished infected vs well nutritioned un-infected and Well nutritioned infected vs well nutritioned un-infected |
35 | 60 | 2019 | [44] | 9 |
|
L. amazonensis and L. major |
Tandem Mass Tag; LC–MS/MS |
LPS stimulated macrophage vs L. major infected macrophage and LPS stimulated macrophage vs L. amazonensis infected macrophage |
51 | – | 2019 | [42] | 10 |
| Proteins of Canis lupus | |||||||
|---|---|---|---|---|---|---|---|
| Infection based studies | |||||||
| Leishmania | IFAT nano LC; nano LC–MS/MS | High titre immunofluorescent antibody test vs negative IFAT | 17 | 5 | 2010 | [46] | 11 |
| L. infantum | ELISA; LC–MS/MS | Post infection vs pre infection | 19 | 12 | 2019 | [48] | 12 |
| L. infantum | ELISA; RP‐ LC‐MS/MS | After infection 168 days vs before infection, 211 days after infection vs before infection and 211 days after infection vs 168 days after infection | 15 | 15 | 2020 | [47] | 13 |
| Proteins of Leishmania | |||||||
|---|---|---|---|---|---|---|---|
| Between infective strain | |||||||
| L. infantum | 2DE; MALDI-TOF/TOF MS | Abundance volume ratio in BH 400 strain of L. infantum vs BH 46 strain | 32 | 31 | 2014 | [50] | 14 |
| L. amazonensis | IEF; 2DE; ESI-TOF MS/MS; Western blot | Day 0 vs day 150 | 19 | 37 | 2014 | [51] | 15 |
| L. donovani | SDS-PAGE; Nano HPLC–MS/MS; Western blot | VL vs CL | 29 | 109 | 2015 | [53] | 16 |
| L. tropica | 2DE-MALDI-TOF/TOF MS | Percent volume of spot in gel of VL vs CL | 10 | 35 | 2015 | [54] | 17 |
| L. amazonensis | LC–MS/MS; Western blot | LV79 strain vs PH8 strain | 25 | 12 | 2017 | [52] | 18 |
| Drug Resistant | |||||||
| L. donovani | 2DE; MS/MS | Antimony resistant Sb(v)9518 strain vs Antimony sensitive Sb(v)9551 strain | 4 | 3 | 2007 | [56] | 19 |
| L. infantum | 2DE; Western blot; MS/MS | Antimony resistant strain Sb2000.1 vs WT | 1 | 6 | 2009 | [57] | 20 |
| L. donovani | 2DE; MALDI TOF/TOF MS | Sodium antimony gluconate (SAG) resistant vs Sensitive | 20 | – | 2010 | [58] | 21 |
| L. donovani | ITRAQ; LC–MS/MS | Antimony resistant strain GE1-R vs Antimony sensitive strain AG83-S and Antimony resistant strain NR3A-R vs Antimony sensitive strain AG83-S | 77 | 16 | 2011 | [59] | 22 |
| L. panamensis | 2DE; LC–ESI–MS/MS; Western blot |
Antimony resistant 12.3R strain vs WT (np-40 vs urea-soluble proteome), Antimony resistant 12.3R strain vs WT (triton x-114 soluble proteome), Antimony resistant 1000.1R strain vs WT (triton x-114 soluble proteome), and Antimony resistant 1000.1R strain vs WT (np-40 vs urea-soluble proteome) |
12 | – | 2012 | [60] | 23 |
|
L. infantum chagasi and L. braziliensis |
2DE; LC–MS/MS; Western blot |
Antimony resistant L. braziliensis vs WT strain of L. braziliensis, Antimony resistant L. infantum chagasi vs WT strain of L. infantum chagasi |
55 | – | 2013 | [61] | 24 |
| L. braziliensis | 2D-DIGE; MS/MS; Western blot |
Lbwts 0.025 vs Lbwts 0, Lbwts 0.025 vs Lbsbr 2, Lbwts 0.025 vs Lbsbr 0.025, Lbwts 0 vs Lbwts 0.025, Lbwts 0 vs Lbsbr 0.025, Lbsbr 2 vs Lbwts 0.025, Lbsbr 0.025 vs Lbwts 0.025, and Lbsbr 0.025 vs Lbwts 0 |
103 | – | 2015 | [62] | 25 |
| L. Infantum | SDS-PAGE; MS/MS | Sb200 strain vs WT | 42 | 18 | 2013 | [63] | 26 |
| L. infantum | SDS PAGE; 2DE; Western blot |
Miltefosine resistant vs Wild type, Antimony resistant vs Wild type |
20 | 36 | 2015 | [64] | 27 |
| L. major | 2DE; LC–MS/MS |
LV39 mutant vs Wild type, LV39_60.2 mutant vs Wild type |
26 | – | 2004 | [65] | 28 |
| L. donovani | ITRAQ; SCX fractionation; Nano LC–MS/MS | DMFO resistant vs Sensitive wild type | 82 | 19 | 2014 | [66] | 29 |
| L. infantum chagasi | 2D-GE; SDS-PAGE; Western blot; MALDI-TOF/TOF MS | Miltefosine sensitive vs Miltefosine resistant and Miltefosine resistant vs Miltefosine sensitive | 32 | – | 2014 | [67] | 30 |
| L. donovani | 2D-DIGE; MALDI TOF/TOF MS | Miltefosine sensitive vs Resistant | 4 | 8 | 2019 | [68] | 31 |
| L. donovani | SDS-PAGE; Nano LC–MS/MS | Amphotericin B resistant vs Sensitive | 32 | 22 | 2019 | [70] | 32 |
| Stress | |||||||
| L. donovani | SDS-PAGE; LC–MS/MS; Western blot |
Exoproteome at 37 °C vs 26 °C and Exoproteome at pH 7 vs pH 5.5 |
309 | 99 | 2010 | [75] | 33 |
| L. donovani | 2DE; MS; Western blot | Average ratio of abundance in splenic vs Axenic amastigotes | 8 | – | 2011 | [77] | 34 |
| L. donovani | SDS-PAGE; ITRAQ; FPLC-MS/MS; Western blot |
Oxidative stress vs Un-treated control, Oxidative and nitrosative stress vs Un-treated control, and Nitrosative stress vs Un-treated control |
36 | 12 | 2010 | [78] | 35 |
| L. amazonensis | 2DE; MALDI TOF MS; Western blot | Balb/c nude vs Balb/c | 16 | 5 | 2015 | [79] | 36 |
| Life stages | |||||||
| L. panamensis | 2DE; LC-ES-MS/MS | Expression ratio amastigotes vs Promastigotes | 3 | – | 2006 | [81] | 37 |
| L. infantum | SCX Chromatography; ICAT; MALDI TOF/TOF MS | ICAT labelled amastigote proteins: Promastigote labelled proteins | 8 | 35 | 2007 | [82] | 38 |
| L. infantum | FFE; 2DE; MS/MS; Western blot | Promastigote vs Amastigote | 86 | – | 2010 | [83] | 39 |
| L. infantum | 2DE; MALDI-TOF/TOF MS/MS | Stationary vs logarithmic phase L. infantum promastigotes | 16 | 12 | 2011 | [86] | 40 |
| L. donovani | ITRAQ; Nano LC–MS/MS; Western blot |
NR3a-R (antimony resistant) amastigotes vs Promastigotes, GEL-R (antimony resistant) amastigotes vs Promastigotes and Ag83-S (antimony sensitive) amastigotes vs Promastigotes |
83 | 98 | 2012 | [84] | 41 |
|
L. infantum and L.mexicana |
SCX Chromatography; LC-MALDI TOF/TOF MS/MS; Western blot | Membrane fraction of promastigotes: Membrane fraction of amastigotes | 67 | – | 2013 | [85] | 42 |
| L. amazonensis | 2DE MALDI-TOF/TOF MS | Stationary phase vs Early logarithmic phase | 12 | 14 | 2016 | [87] | 43 |
| Misc | |||||||
| L. donovani | SDS-PAGE; LC–MS/MS; Western blot | Conditioned medium vs Cell associated proteins | 10 | – | 2008 | [76] | 44 |
Table 2.
Additional experimental and data accession details of each study
| Study No | Peptide/Protein identification parameters | No. of total identified protein/ Reported protein | No. of differentially regulated protein reporteda | No. of differentially regulated protein | Repository and Accession No. or contact detailsb | |
|---|---|---|---|---|---|---|
| Up | Down | |||||
| 1 | 5-kDa Molecular weight cut-off | 39 proteins from two experimental conditions | Cond.1: 24 | 19 | 5 | chitra_mandal@yahoo.com |
| Cond.2: 24 | 19 | 5 | ||||
| 2 | Trypsin; missed cleavage 1; modification: alkylation, iodoacetamide, and partial oxidation of methionine; mass tolerance: < 0.1 Da | 59 | 13 | 9 | 4 | cler_ss@hotmail.com |
| 3 | Trypsin; missed cleavages 2; Modification: carbamidomethyl (C) as a fixed modification and acetyl (Protein N-term) and Oxidation (M); peptide mass tolerance ± 25 ppm for precursors and 0.02 Da for the fragment masses. Score distribution models were used to compute peptide-spectrum match p-values, and spectra recovered by a false discovery rate (FDR) < = 0.01 | 267 | 12 | 7 | 5 | sara.robledo@udea.edu.co |
| 4 | Trypsin; missed cleavage 2; minimum peptides 1; modification: + 57.02146 for Cysteine alkylation, and + 15.99492 for methionine oxidation. Mass deviation: 3 amu; 1% FDR | 1352 proteins in uninfected and infected cases | Cond.1: 13 | 2 | 11 | pveras@bahia.fiocruz.br |
| Cond.2: 2 | 2 | – | ||||
| 5 | Trypsin; missed cleavages: 2; modification: carbamidomethyl and oxidation of methionine; mass tolerances: 10 ppm and 0.6 Da for precursor and fragment ions respectively; proteins with minimum 3 peptides and peptide score higher than 20 were considered as true hits | 248 | 34 | 27 | 7 | martin.olivier@mcgill.ca |
| 6 | Trypsin; mass tolerance: 50 ppm for precursor and 600 for fragment ion; | 149 proteins after infection from three different species | Cond.1: 53 | 43 | 10 | negraosf@gmail.com |
| Cond.2: 47 | 38 | 9 | ||||
| Cond.3: 49 | 26 | 23 | ||||
| 7 | Trypsin; mass tolerance: 50 ppm for precursor and 600 ppm for fragment ion; modification: carbamidomethylation of cysteine | 342 proteins after infection with L.amazonensis or L.major | Cond.1: 60 | 43 | 17 | negraosf@gmail.com |
| Cond.2: 60 | 26 | 34 | ||||
| 8 | Trypsin; minimum peptides 2; missed cleavage 3; mass tolerance: 40 ppm for precursor; modification: carbamidomethylation of cysteine | 280 | 109 | 38 | 71 | https://www.ebi.ac.uk/pride/archive/projects/PXD010414 |
| 9 | Trypsin; minimum peptides 2; missed cleavage 3; mass tolerance: 40 ppm for precursor; modification: carbamidomethylation of cysteine | 344 proteins after infection with either L.amazonensis or L.major | Cond.1: 74 | 27 | 47 | patricia.cuervo@fiocruz.br |
| Cond.2: 21 | 8 | 13 | ||||
| 10 | Trypsin; minimum peptides 1; mass tolerance: 50 ppm for precursor and 600 ppm for fragment ion; modification: carbamidomethylation of cysteine | 482 | Cond.1: 26 | 26 | – | negraosf@gmail.com |
| Cond.2: 25 | 25 | – | ||||
| 11 | Trypsin; minimum peptides 1; iTRAQ ratios below 0.7 or above 1.5 were considered significant | 70 | 22 | 17 | 5 | britti@unicz.it |
| 12 | Trypsin; values of P < 0.05 were considered to be significant | 47 | 31 | 19 | 12 | silviams@um.es |
| 13 | Trypsin; minimum peptides 3; 15 ppm ion library tolerance, 5 min XIC extraction window, 0.01 Da XIC width | 169 | Cond.1: 20 | 12 | 8 | https://www.peptideatlas.org/PASS/PASS01352 |
| Cond.2: 9 | 3 | 6 | ||||
| Cond.3: 1 | - | 1 | ||||
| 14 | Trypsin; missed cleavage 1; modification: methionine oxidation, cysteine carbamidomethylation, pyroglutamate formation at N-terminal glutamine; mass tolerance: 0.6 Da for MS spectra and 0.4 Da for MS/MS spectra | 63 | 63 | 32 | 31 | https://www.peptideatlas.org/PASS/PASS00393 |
| 15 | Trypsin; minimum peptides 1; missed cleavage 1; modification: methionine oxidation, cysteine carbamidomethylation; peptide tolerance: ± 0.1 Da; MS/MS tolerance: ± 0.1 Da | 56 | 56 | 19 | 37 | eduardoferrazcoelho@yahoo.com |
| 16 | Trypsin; minimum peptides 2; missed cleavage 1; mass tolerance: 0.100 Da for the fragment ion and 10.0 ppm for parent ion; modification: carbamidomethylation of cysteine and oxidation of methionine | 2428 from two strains | 138 | 29 | 109 | greg.matlashewski@mcgill.ca |
| 17 | Trypsin; MASCOT score > 1 and P-value: > 0.05 | 56 | 45 | 10 | 35 | hajaranh@tums.ac.ir |
| 18 | Trypsin; minimum peptides 1; missed cleavage 2 at both ends; mass tolerance: 4.5 ppm and 0.5 Da for MS and MS/MS; modification: carbamidomethylation of cysteine, oxidation of methionine | 301 | 37 | 25 | 12 | bstolf@usp.br |
| 19 | Missed cleavage 2; minimum peptides 2; modification: Carbamidomethylation of cysteine, partial oxidation of methionine; error tolerance of 2.0 Da for peptides and 0.5 Da for fragments | 7 | 7 | 4 | 3 | marc.ouellette@crchul.ulaval.ca |
| 20 | Trypsin; minimum peptides 2; missed cleavage 2; error tolerance; 2.0 Da for peptides and 0.5 Da for fragments | 7 | 7 | 1 | 6 | marc.ouellette@crchul.ulaval.ca |
| 21 | Trypsin; minimum peptides 2; maximum peptide precursor tolerance: 40 ppm and MS/MS fragment tolerance: 0.2 kDa; modification: carbamidomethylation of cysteine, oxidation of methionine | 20 proteins form two different fractions | 20 | 20 | – | anuradha_dube@hotmail.com |
| 22 | Trypsin; minimum peptides 2; the ‘Search Effort’ parameter ‘Thorough ID’ which provides a broad search of various protein modifications and multiple mass cleavages | 269 | Cond.1: 38 | 30 | 8 | madhubala@mail.jnu.ac.in |
| Cond.2: 55 | 47 | 8 | ||||
| 23 | Trypsin; minimum peptides 2; modification: Partial carboxamidomethylation of cysteine and oxidation of methionine | 12 | Cond.1: 6 | 6 | – | mgomez@cideim.org.co |
| Cond.2: 6 | 6 | – | ||||
| 24 | Trypsin; minimum peptides 1; missed cleavage 2; mass tolerance; 0.3 Da and 0.1 Da; modification: Partial carboxamidomethylation of cysteine and oxidation of methionine | 57 | Cond.1: 28 | 28 | – | silvane@cpqrr.fiocruz.br |
| Cond.2: 27 | 27 | – | ||||
| 25 | Trypsin; fold change above 1.5 and p-values < 0.05 were considered significant | 48 | Cond.1: 12 | 12 | – | silvane@cpqrr.fiocruz.br |
| Cond.2:18 | 18 | – | ||||
| Cond.3:16 | 16 | – | ||||
| Cond.4:10 | 10 | – | ||||
| Cond.5:3 | 3 | – | ||||
| Cond.6: 18 | 18 | – | ||||
| Cond.7: 22 | 22 | – | ||||
| Cond.8: 4 | 4 | – | ||||
| 26 | Trypsin; carbamidomethyl (C) as a fixed modification and standard SILAC (Lys + 8, Arg + 10) settings for QSTAR or Triple TOF 5600 instruments | 60 | 60 | 42 | 18 | https://www.ebi.ac.uk/ena/browser/view/PRJEB3283 |
| 27 | Trypsin; minimum peptides 2; missed cleavage 2; mass tolerance: 2.0 Da for peptides and 0.5 Da for fragments; modification: Carbamidomethylation of cysteine and partial oxidation of methionine | 28 | Cond.1: 28 | 10 | 18 | Marc.Ouellette@crchul.ulaval.ca |
| Cond.2: 28 | 10 | 18 | ||||
| 28 | Minimum peptides 2; modification: complete carboxamidomethylation of cysteine, partial methionine oxidation; mass deviation < 60 ppm; score > 51 was considered significant (p < 0.05) | 13 Proteins from three experimental conditions | Cond.1: 13 | 13 | – | marcouellette@crchul.ulaval.ca |
| Cond.2: 12 | 12 | – | ||||
| Cond.3: 1 | 1 | – | ||||
| 29 | Trypsin; minimum peptides 1; missed cleavage 2; fixed modification: methyl methane thiosulfonate- labeled cysteine parameter | 686 | 101 | 82 | 19 | rentala@outlook.com |
| 30 | Trypsin; missed cleavage 2; modification: carbamidomethylation of cysteine, oxidation of methionine, deamination of asparagine and glutamine, pyroglumate formation at N-terminal glutamine of peptide | 22 | Cond.1: 11 | 11 | – | lemosem@ndi.ufes.br |
| Cond.2: 21 | 21 | – | ||||
| 31 | Trypsin; missed cleavage 1; modification: carbamidomethylation of cysteine (fixed), oxidation of methionine (variable); peptide charge: + 2, and ‘other eukaryota’ was selected as the taxonomy. P < 0.05; scores above 20 were considered as significant | 12 | 12 | 4 | 8 | rmusl@uohyd.ernet.in |
| 32 | Trypsin; missed cleavage 1; modification: carbamidomethylation of cysteine (fixed), oxidation of methionine (variable); tolerance: 50 ppm for peptide and 100 ppm for fragment | 406 | 54 | 32 | 22 | vahab_ali@yahoo.com |
| 33 | Trypsin; minimum peptides 1; missed cleavage 1; modification: carbamidomethylation of cysteine (fixed), heavy and light dimethylated lysine side chains and peptide amino termini (variable); mass tolerance: 3 ppm for precursor ion and 0.8 Da for the fragment ion | 329 | Cond.1: 233 | 218 | 15 | ethan@interchange.ubc.ca |
| Cond.2: 175 | 91 | 84 | ||||
| 34 | Trypsin; MASCOT Protein score ≥ 50 and a GPS Explorer Protein confidence index ≥ 95% | 8 | 8 | 8 | – | gerald.spaeth@pasteur.fr |
| 35 | Trypsin; protein identified at ≥ 95% confidence | 1653 | Cond. 1: 16 | 12 | 4 | drpradeep.das@gmail.com |
| Cond. 2: 16 | 12 | 4 | ||||
| Cond. 3: 16 | 12 | 4 | ||||
| 36 | Trypsin; missed cleavage1; mass tolerance: 0.05 Da for monoisotopic and 0.25 Da for the fragment; modification: methionine oxidation | 21 | 21 | 16 | 5 | bstolf@usp.br Abstract |
| 37 | Trypsin; minimum peptides 2; modification: partial carboxamidomethylation of cysteine and oxidation of methionine; MASCOT score of greater than 51 | 7 | 3 | 3 | – | johnwalker@cideim.org.co |
| 38 | Trypsin; minimum peptides 1; mass tolerance: 50 ppm for MS and 0.20 Da for MS/MS | 91 | 43 | 8 | 35 | robm@interchange.ubc.ca |
| 39 | Trypsin; missed cleavage 2; mass tolerance: 2.0 Da for peptides and 0.5 Da for fragments; modification: Carbamidomethylation of cysteine and partial oxidation of methionine | 86 proteins from two experimental conditions | Cond. 1: 50 | 50 | – | marc.ouellette@crchul.ulaval.ca |
| Cond. 2: 36 | 36 | – | ||||
| 40 | Trypsin digestion | 28 | 28 | 16 | 12 | vlarraga@cib.csic.es |
| 41 | Trypsin; minimum peptides 1; mass range: 140–1600 amu; | 215 | 181 | 83 | 98 | madhubala@mail.jnu.ac.in |
| 42 | Trypsin; modification: cysteine alkylation; mass range: 800–3500 Da | 296 proteins from two species | Cond. 1: 49 | 49 | – | rmcmaster@exchange.ubc.ca |
| Cond. 2: 18 | 18 | – | ||||
| 43 | Trypsin; missed cleavage 1; modification: carbamidomethyl cysteine (fixed) and oxidation of methionine(variable); mass tolerance: ± 50 ppm for precursor and ± 0.3 Da for MS/MS fragment ions | 339 | 26 | 12 | 14 | https://www.ebi.ac.uk/pride/archive/projects/PXD002939 |
| 44 | Trypsin; minimum peptides 2; mascot score > 25 | 358 proteins from two samples | 10 | 10 | – | ethan@interchange.ubc.ca |
Cond. Different experimental conditions of the corresponding study
aDifference in experimental conditions or difference in organism with same experimental setup
bData accession link for the repository submitted data (in absence of it, corresponding author contact email was provided)
Bag et al. [35] conducted the differential proteomic study between VL patients (L. donovani) and healthy human subjects. In this study, with endemic and non-endemic healthy controls, a comparison of proteins and glycoproteins from the plasma of VL patients (belonging to various age groups) were performed. The study identified 39 differentially expressed spots and reported eight DMP in VL infection. The five up-regulated proteins include amyloid A1 precursor, alpha-1-antitrypsin precursor, fibrinogen gamma-β chain precursor, haptoglobin precursor, and alpha 1-β glycoprotein. The three down-regulated proteins include vitamin-D binding protein, apo A-I protein, and transthyretin. Since glycoproteins are the preferred markers for diagnosing diseases [36], differentially expressed glycoproteins from the Bag et al. [35] study can be used as prognostic and diagnostic markers for VL. Two differential protein expression studies [37, 38], involving human subjects with CL, were reported. Da Silva Santos et al. [38] compared the proteome of L. braziliensis infected skin lesion against normal skin biopsy and reported 13 DMP. The nine up-regulated proteins include tetratricopeptide repeat domain, fas (TNFRSF6) binding factor, chromosome 1 open reading frame, Caspase-9, MIF4G domain-containing protein isoform, T-cell receptor-β, transcription factor IIIB, cTAGE family, and Na/K-ATPase. The four down-regulated proteins include basal cell adhesion molecule, ankyrin repeat and LEM domain-containing protein, phosphodiesterase, and solute carrier family protein. In another recent study, Montoya et al. [37] conducted a similar study comparing the proteome of L. panamensis infected human skin lesion against normal human skin and reported 12 DMP. The seven up-regulated proteins include keratin type I and II, lysosome-associated membrane glycoprotein, protein S100-A8 and A9, leukocyte elastase inhibitor, and copper-transporting ATPase. The five down-regulated proteins include fatty acid-binding protein, protein S100-A2, peroxiredoxin, hemoglobin subunit-α, and hemoglobin subunit-δ. However, both studies have reported no common DMP. This may be because of differences in the location of the skin biopsies (border and middle of ulcer) and the infectious parasite of corresponding study.
In addition to studies on humans, proteomics studies have been reported on Mus musculus, infected with different Leishmania species. Among such studies (matching the selection criteria), four studies involved L. major and L. amazonensis infection, two studies involved L. infantum infection, and one study involved L. mexicana infection. Menezes et al. [39] reported 15 DMP in infected mice (L. major or L. amazonensis) compared to the control. The four up-regulated proteins of infected mice include phospholipase D1, RAS-related protein, glucuronidase β, and phosphotransferase regulation domain. Negrão et al. [40] performed a proteomic study among mice infected with L. amazonensis, L. major, L. infantum, mice inoculated with lipopolysaccharide (as positive control), and healthy mice (as negative control). The study reported 31 proteins to be differentially regulated only in L. amazonensis, 25 proteins to be differentially regulated only in L. major, and 27 proteins to be differentially regulated only in L. infantum. The study also reported 22 DMP in all three infections compared to healthy. The RAS-related protein was up-regulated in this study as well as in the Menezes et al. [39] study. The same group [41] performed a similar experiment with skin biopsies and reported 17 DMP in infected mice. Further, they [42] adopted the labeled (tandem mass tag) proteomic approach and confirmed their earlier findings. Also, the study reported U6 snRNA-associated Sm-like protein and heat repeat-containing protein 3 to be precisely up-regulated in infection with L. major, while protein tweety homolog 3, phosphatidate cytidylyltransferase, and macrophage metalloelastase to be precisely up-regulated in infection with L. amazonensis. Overall, the studies by Negrão et al. [40, 42, 41] demonstrated the species-specific protein modulation in Leishmania infection as well as differentiated the host response through LPS inoculation and Leishmania infection. To observe the immune response effects of malnutrition in Leishmania infection, Losada-Barragán et al. [43, 44] conducted the differential proteomic studies involving malnourished mice. In both their studies [43, 44], they compared the proteomes of protein-malnourished mice (infected with L. infantum) and uninfected well-nourished mice. In the study involving thymic tissue samples [43], 38 and 71 proteins are up-regulated and down-regulated in infected malnourished subjects, respectively. In the study involving splenic interstitial fluid [44], 35 and 60 proteins are up-regulated and down-regulated in infected malnourished subjects, respectively. These studies concluded that the protein malnutrition reduces the pro-inflammatory response towards a Leishmania infection. Hassani and Olivier [45] studied the immunomodulatory effects of L. mexicana infection in macrophages using comparative proteomics and identified 248 proteins. In the infected macrophages, 27 and 7 proteins were up-regulating and down-regulating, respectively. The majority of the up-regulated proteins have involvement in immunological pathways. Moreover, this study claims that it was the first to report surface protease GP63 to be present in exosome released by L. mexicana infected J774 macrophage. The study concluded that the Leishmania parasite has the potential to modulate the host exosome machinery during infection.
Three comparative proteomic studies, involving Canis lupus with and without canine leishmaniasis, were reported [46–48]. Britti et al. [46] compared the proteins in the serum of naturally infected and uninfected dogs and reported 17 and 5 up-regulated and down-regulated proteins, respectively. Lorena et al. [48] compared salivary proteins in experimentally infected and uninfected dogs and reported 19 up-regulated proteins and 12 down-regulated proteins. More recently, Franco-Martínez et al. [47] compared the serum proteins of the subclinical and clinical stage of infected dogs and reported 15 up-regulated and down-regulated proteins, each. Contrsatingly, the up-regulated apolipoprotein in serum samples [47] was found down-regulating in salivary samples [48]. Similarly, the protein haptoglobin up-regulated in natural infection was found down-regulating in the experimental infection [47].
Among the studies with the different hosts, one could observe consensus as well as contrasting regulation of the same protein. For instance, the transferrin protein down-regulated in dogs [47] was up-regulating in mice [43, 40]. On the other hand, the amyloid protein is up-regulating in both dogs and humans [35, 46].
Differentially modulated proteins between different strains of Leishmania sp.
The severity and virulence of infection may vary widely between the different strains [49]. In this aspect, out of articles matching the selection criteria of this review, a total of five articles were reported involving differential proteome analysis between different infective strains of Leishmania (Tables 1 and 2).
Da Fonseca Pires et al. [50] compared two strains of L. infantum BH400 (Canis lupus) and BH46 (human) proteome to observe the virulence factor among the two strains. The study reported 32 and 31 proteins to be up-regulated and down-regulated, respectively, in the BH400 strain. Most of the virulence-related proteins like heat-shock protein, elongation factor 2, kinetoplastid membrane protein-11, and enolase were up-regulating. However, the confirmatory western blot analysis revealed that the kinetoplastid membrane protein-11 to be up-regulated and elongation factor 1β to be down-regulated in BH400. Despite the presence of a similar abundance level of other virulence-related proteins in both BH400 and BH46, the study reported higher signal intensity in the proteins of BH400. On the other hand, Magalhães et al. [51] compared the protein modulations between severe (day 0 of culture) and less virulent (day 150 of culture) L. amazonensis and reported 56 differentially regulated proteins. Out of them, 19 and 37 proteins were up-regulated and down-regulated, respectively. However, validation by western blot analysis was performed only for four of the proteins. In western blot, the down-regulated α-tubulin and paraflagellar rod proteins were identifiable, while the up-regulated heat-shock protein 83 and glucose-regulated protein 78 were not. Also, the up-regulated proteins (enolase and elongation factor 2) in L. infantum BH400 strain [50] are down-regulated in virulent L. amazonensis. In another study [52] involving the proteome comparison of virulent strains of L. amazonensis, 12 proteins are up-regulating in the highly virulent strain, which includes the virulent protein GP63. However, in contrast, the GTP-binding protein Rab1 reported to be up-regulated in this study as well as in Da Fonseca Pires et al. [50] study was down-regulated in virulent L. amazonensis [51]. McCall et al. [53] and Hajjaran et al. [54] performed proteomic profiling of L. donovani and L. tropica, respectively. McCall et al. [53] McCall compared the proteomes of L. donovani from CL and VL patients, while Hajjaran et al. [54] compared L. tropica from cutaneous and visceral tissue of the same infected host. McCall et al. [53] reported 38 and 148 proteins to be up-regulated in VL and CL strain, respectively. Proteins involved in stress responses, like mitochondrial peroxidoxin and superoxide dismutase, were up-regulated in VL strain. While proteins involved in transport and trafficking processes, like ABC transporters, kinesin, and dynein, were up-regulated in CL strain. However, the majority of up-regulated proteins in CL strain were uncharacterized protein. Overall, the study hypothesized that the visceral environment could be more stressful for the parasite. Similarly, Hajjaran et al. [54] also reported ubiquitin, co-chaperonin, and tryparedoxin (that aids the parasite survival in stress condition) proteins to be up-regulated in visceral form, indicating the fact that visceral environment could be more stressful.
Differentially modulated proteins in drug-resistant Leishmania sp.
Leishmaniasis, like other infectious diseases, is also prone to drug resistance [55]. Quantitative and comparative proteomic studies are of great tool to understand the drug resistance mechanism of the organism in comparison to drug-sensitive organisms [30]. Fourteen such articles, addressing the comparative proteomic study between drug-sensitive and drug resistance in Leishmaniasis, are discussed below.
Nine studies compared the proteomic profiles of antimony resistant Leishmania and antimony sensitive Leishmania (Tables 1 and 2). Vergnes et al. [56] reported only seven DMP between antimony resistant and sensitive L. donovani. Among the resistant strains, 14-3-3 and heat shock 83-1 protein were significantly up-regulated, while the small kinetosome calpain-related protein was significantly down-regulated. The study concluded, up-regulated heat shock 83-1 protein increased the antimony resistance and reduced the drug-mediated programmed cell death, on the other hand, the down-regulated small kinetoplastid calpain-related protein promoted the antimonial induced programmed cell death. However, a similar study on antimony resistant L. infantum [57] reported the up-regulation of only one protein (argininosuccinate synthase) in the resistant strain. Since the kinetoplastid membrane protein-11 (KMP-11) was down-regulated in both the resistant strains studied, the study concluded that the KMP-11 could be responsible for the drug resistance mechanism. However, the study did not report the implication of KMP-11 in the drug resistance mechanism. Moreover, Kumar et al. [58] reported several proteins to be overexpressed in sodium antimony gluconate (SAG) resistant L. donovani promastigotes. The study reported the up-regulation of more cytosolic proteins than proteins localized in the membrane. The heat shock protein 83 is highly expressed in SAG-sensitive promastigotes (based on fold change). This is in consonance with the study by Vergnes et al. [56]. In a study similar to Kumar et al. [58] (where antimony resistant and antimony sensitive L. donovani promastigotes are studied), Biyani et al. [59] also reported 77 proteins to be up-regulated in antimony resistant promastigotes. The aldose 1-epimerase-like protein was up-regulated highly (based on fold change), while the proteins like beta-tubulin, heat shock protein, and fructose-1,6-bisphosphate aldolase were up-regulated moderately. These moderately up-regulated proteins are also up-regulating in the Kumar et al. [58] study. In contrast, 60S ribosomal protein L23a was down-regulated in the Biyani et al. [59] study, while it was up-regulated in the Kumar et al. [58] study. In trivalent antimony resistant L. panamensis promastigotes, 12 proteins were reported to be up-regulated [60]. Out of twelve proteins, three proteins had fold change of > 2, which includes ATPase-beta subunit, prostaglandin F2-a synthase, and GTP-binding proteins. Another similar study in promastigotes of L. infantum chagasi and L. braziliensis [61] reported 28 and 27 proteins to be up-regulated in trivalent antimony resistant parasite, respectively. In addition to several up-regulated proteins (like heat shock proteins, enolase, tubulin proteins, etc.), the study reported enzymes of trypanothione synthesis pathway (like treparodoxin and trypanothione reductase) to be up-regulating. The same group [62] performed the phosphoproteomic analysis in wild-type and antimony-resistant L. braziliensis promastigotes. The study reported 48 proteins to be differentially abundant in wild-type and resistant (treated with different concentrations of antimony). Nine of the proteins including proliferating cell nuclear antigen protein, heat shock protein 70, heat shock protein 83-1, α and β-tubulin, tryparedoxin peroxidase, pteridine reductase, iron-sulfur protein, and paraflagellar rod protein are differentially expressed in both total protein and phosphoprotein analysis, indicating the modulation in both protein pools. A combined genomic and proteomic study [63], for the first time, reported the antimony resistant marker (ABC transporter MRPA) to be up-regulated in trivalent antimonial resistant L. infantum promastigotes. They also reported 42 proteins of several important pathways upregulated in antimonial resistant promastigotes, including trypanosome thiol synthesis and sterol biosynthesis. For the first time, Vincent et al. [64] compared the mitochondrial proteomic profile of antimony resistant L. infantum promastigote with the wild type. Out of 28 DMP reported in the drug-resistant parasite, only ten proteins were up-regulating. Most of the up-regulated proteins are hypothetical proteins, while others are flavoprotein subunit like protein, i/6 autoantigen like protein, succinyl co-A, and axoneme central apparatus protein.
Beside, antimonial drug resistance in Leishmania, proteomic studies were also reported on several other drug resistance, including methotrexate, difluoromethylornithine (DMFO), miltefosine, and Amphotericin B (AmB). Drummelsmith et al. [65] conducted the differential proteomic study between methotrexate resistant and sensitive L. major and reported 13 DMP. Enolase, beta-tubulin, and heat shock proteins are overexpressed, in addition to several other proteins. However, the study concluded that methionine adenosyltransferase (AdoMets) overexpression contributed to the methotrexate resistance. The same study confirmed the role of AdoMets in drug resistance through gene transfection and metabolite analysis. Singh et al. [66] studied the differential protein expression between DMFO resistant and sensitive L. donovani promastigotes. Out of 101 DMP, 82 and 19 proteins were up-regulated and down-regulated, respectively. Despite the up-regulation of several proteins from different pathways, the study claimed S-adenosylmethionine synthetase and Cystathionine beta-lyase-like protein up-regulation playing a major role in DMFO resistance. Two differential proteomic studies compared miltefosine resistant and sensitive parasite. One involves promastigotes of L. infantum chagasi [67] and other L. donovani [68]. A total of 21 and four proteins were up-regulated in drug-resistant L. infantum chagasi and L. donovani, respectively. Carnielli et al. [67] concluded that the up-regulation of proteins NADP-dependent alcohol dehydrogenase and translation elongation factor 1-beta play a key role in redox homeostasis and thereby could aid in miltefosine resistance. Vacchina et al. [69] reported that the mitochondrial HSP70 is involved in miltefosine resistance as well as in the stress response of the parasite. In addition, Veronica et al. [68] reported that the protein iron superoxide dismutase contributes to the miltefosine resistance. The same study reported proteins enolase and tryparedoxin (up-regulated in antimony resistant) to be down-regulating in miltefosine resistant. Garg et al. [70] have recently reported a proteomic study between AmB resistant and sensitive promastigotes of L. donovani secretome. The study reported 54 DMP, among them 32 and 22 proteins were up-regulated and down-regulated, respectively, in AmB resistant promastigotes. The study suggested a majority of up-regulated proteins have a role in parasite adaptiveness to stress condition. The same study also reported proteins tryparedoxin, heat shock protein, and tubulin protein (up-regulated in other drug resistance) to be up-regulated in AmB resistant promastigotes.
Differentially modulated proteins in Leishmania sp. under stress
Stress can be defined as the difference in the biological responses due to intrinsic or extrinsic stimulus. Different types of stress may affect the gene expression, protein expression and metabolism of organisms to varying degrees [71]. The different types of stress experienced by Leishmania includes oxidative, nitrosative, temperature, pH, and drug-induced stress [72–74]. The knowledge of protein expression levels during such a condition plays a vital role in understanding the parasite survival under stress. Four such articles (Tables 1 and 2) are discussed below.
Silverman et al. [75] conducted a quantitative proteomic comparison of Leishmania donovani promastigotes (exosomes) under varying temperature (26 ºC and 37 ºC) and pH (5.5 and 7.5). They identified 329 proteins, among which 79, 154, and 96 proteins are differentially expressed in a condition having a change in temperature and pH, the temperature only, and pH only, respectively. The study reported, at 37 ºC and acidic pH, an increase in exosome release. Since the temperature and pH mimic the infection, the study concluded that Leishmania up-regulates and modify exosome production and thereby modulate the regulation and function of macrophages. The 60 s acidic-ribosomal protein and β-fructofuranosidase up-regulated under pH stress are also up-regulating in parasite secretome [76]. Pescher et al. [77] compared the protein expression levels between splenic (nutrition poor condition) and axenic (nutrient-rich) amastigotes and reported the up-regulation of eight proteins in nutrition deprived conditions. Among them, the protein methylthioadenosine phosphorylase was strongly up-regulating. Since methylthioadenosine phosphorylase involves the metabolism of polyamines and purines, they suspected that this enzyme could help in the survival of parasite under nutrition-deprived conditions. The same study confirmed this by culturing the splenic and axenic amastigotes in nutrient-poor medium and found the splenic amastigotes survived while the axenic amastigotes died between 5 and 7 days. Later, Sardar et al. [78] exposed L. donovani promastigotes to three different stress conditions viz., oxidative stress, nitrosative stress, and both oxidative and nitrosative stress, and performed the comparative proteomic study. The study revealed 13 and 3 DMP to be involved in redox homeostasis and reactive oxygen species, respectively. Among those 16 DMP, only four proteins down-regulated in all three stress conditions. The rest of the 12 proteins up-regulated at least by one-fold. The study concluded that the differentially expressed proteins in different stress conditions were mainly involved in trypanothione metabolism, superoxide metabolism, mitochondrial respiration process, and oxidative phosphorylation. In another study, Teixeria et al. [79] compared the protein expression between L. amazonensis amastigotes from BALB/c nude (lack of thymus mediated immunity) and BALB/c mice. The study reported 21 DMP, among which 16 and five proteins were up-regulated and down-regulated, respectively, in amastigotes from BALB/c nude. Proteins like trypanothione reductase, peroxidoxin, tryparedoxin peroxidase, tryparedoxin, and heat shock proteins (associated with oxidative/nitrosative stress condition) were up-regulated at lease by one-fold. From the results it is apparent that the proteins involved in the synthesis and metabolism of trypanothione are differentially modulating during stress. Hence, these proteins can be a potential target for drug development.
Differentially modulated proteins in different life stages of Leishmania sp.
Leishmania parasite has two different forms, namely promastigotes and amastigotes. In general, the amastigotes are infectious in mammals [4], while the promastigotes are mostly present in sand flies [80]. Hence, several proteomic studies have been conducted in the past to study the protein expression levels between the promastigotes and amastigotes. Also, a protein profiling study has reported on promastigotes during different growth phase expression. Seven such articles (Tables 1 and 2) are discussed below.
In L. panamenensis amastigotes, Walker et al. [81] analyzed 51 overexpressed protein spots obtained from 2-Dimentional electrophoresis (2DE) for the differential expression. They identified four proteins, namely beta-tubulin, cysteine synthase, pyruvate dehydrogenase, and uncharacterized protein, to be present only in amastigotes. In addition, heat-shock protein 83, glucose-6-phosphate dehydrogenase, and ran-binding protein were up-regulated by more than onefold in amastigotes, with respect to promastigotes. Later, Leifso et al. [82] conducted differential mRNA and protein expression studies between the L. infantum promastigotes and amastigotes. They identified 91 proteins (protein expression study), out of which 8 and 35 proteins were up-regulated and down-regulated, respectively, in the amastigotes while the rest of the proteins were neutrally regulated. The up-regulated proteins in amastigotes include hypothetical proteins, chaperonin HSP60, precursor hexokinase, ribosomal protein 13, histone h3, and carboxypeptidase. However, they observed only 1.4% of differentially expressed mRNAs in amastigote. Also, Leifso et al. [82] concluded that the majority of mRNAs are constitutively expressed in both amastigotes as well as promastigotes and thereby emphasizing that the differentiation, virulence, and pathogenesis of Leishmania may not be depending on the regulation of genes or proteins. Brotherton et al. [83] used free-flow electrophoresis to analyze the specific expression of the basic protein in Promastigote and amastigote of Leishmania infantum. A total of 36 and 50 proteins were up-regulated in amastigotes and promastigotes, respectively. Moreover, 37 and 42 proteins are identified only in amastigotes and promastigotes, respectively. Elongation initiation factor-2 alpha subunit, cysteinyl-tRNA synthetase, and glutathione peroxidase-like protein were up-regulated by four-fold in amastigotes than promastigotes. Interestingly, both Leifso et al. and Brotherton et al. [82, 83] reported the up-regulation of chaperonin HSP60 protein in L. infantum amastigotes. In contrast, Leifso et al. [82] reported the up-regulation of carboxypeptidase in L. infantum amastigotes (with respect to promastigotes) while Brotherton et al. [83] reported the same in L. infantum promastigotes (with respect to amastigotes). Biyani and Madhubala [84] profiled the proteins of three different strains of L. donovani promastigotes and amastigotes for comparison. Among the three strains, two strains (GEI-R and NR3A-R) are resistant to antimony, and the one is sensitive (AG83-S). The study identified 28, 29, and 26 proteins to be up-regulated and 34, 33, and 31 proteins to be down-regulated in amastigotes of AG83-S, GEI-R, and NR3A-R, respectively. Six proteins, namely histone h2b, glucose-regulated protein 78, ADP-ATP carrier protein, GTP-binding protein, and 2 hypothetical proteins, were significantly up-regulated in amastigotes of resistant strains (with respect to sensitive strains). Leifso et al. [82] reported histone h2b to be down-regulating while Biyani et al. [84] reported the same to be up-regulating in L. infantum amastigotes. However, Lynn et al. [85] reported histone h2 protein to be up-regulating in L. infantum amastigotes. In addition, Lynn et al. [85] reported the up-regulation of GTP-binding protein in L. infantum promastigotes, which agrees with the Biyani et al. [84] study. Also, Lynn et al. [85] identified arginase and alpha-adaptin like protein to be up-regulated by two-fold in L. infantum amastigotes (with respect to promastigotes). Alcolea et al. [86] studied the regulation pattern of mRNA and protein in L. infantum promastigotes logarithmic phase and stationary phase. Through differential proteomic techniques, the study identified 16 up-regulated and 12 down-regulated proteins in the stationary phase. Among the 28 differentially regulated proteins, ETF and eEF1-alpha were differentially regulated even at the mRNA transcript level. Alcolea et al. [87] also studied the protein expression level of oxidative and nitrosative resistant L. amazonensis promastigotes in the stationary phase and early logarithmic phase. They reported 12 and 14 proteins to be up-regulated and down-regulated, respectively, in the stationary phase. The study concluded that the up-regulation of arginase, tryparedoxin peroxidase, heat shock protein, and protein kinase A (involved in survival of parasite under stress conditions) in the stationary phase favours the pre-adaptative hypothesis.
Database development and usage
All the differential protein data collected in this study was made available as a database. Supplementary Table 1 describes the structure of the database. The database includes a total of 2635 records from 44 studies. The database includes a dynamic search feature with multiple field filters and includes active links to Uniprot (https://www.uniprot.org/) and TriTrypDB [88]. The link in the “TriTrypDB Search” column of the database will perform a “Protein Keyword search” in the TriTrypDB for the corresponding Leishmania proteins and will return the genomic information as a “.csv” file (since TriTrypDB is specific to Leishmania, the proteins of host organisms will have “NA” in the corresponding column). The database has two different panels, one for setting the filters and other for the display of records (Supplementary Fig. 1). The filter panel is self-descriptive. The display panel contains top-section to export data and report the filtered number of records, a table section to display the data and sort data, and bottom-section to adjust the visibility of the records in table and navigation of records. Also, the database includes provisions for exporting the filtered records in multiple formats. PHP, MySQL, CSS, W3CSS, and JavaScript are used to develop the database and the access interface. The database is freely accessible at http://ldepdb.biomedinformri.com/. The following section describes the database usage.
Consider a researcher who is interested in studying a protein (for example, trypanothione reductase in L. donovani) wants to know whether the protein is differentially regulating during stress conditions or not. To query this in the database, a user can select the “STRESS” option in the “Test” filter and “L. donovani” in the “Organism” filter. This will display four records in the display panel containing the information that the protein trypanothione reductase was up-regulating during oxidative stress, nitrosative stress, and temperature stress from two different studies. In addition, for a particular test condition, the database is used to retrieve information like proteins commonly and uniquely up or down-regulated.
Discussion
Advanced proteomics techniques are widely been utilized in Leishmania related studies to understand the disease mechanism, to identify the regulation of proteins or pathways during stress, to study the survival strategy of the drug-resistant parasite, and to study the host response due to the different forms of infection. In this study, to our knowledge, a comprehensive review covering all the above aspects was reported. Understanding drug resistance (14 studies) was the primary focus, while the secondary focus is on understanding the host response during infection (13 studies). Despite the different experimental conditions and setup, heat-shock and Tryparedoxin related proteins were differentially regulated in 17 studies, indicating that those proteins are often modulated in the parasite. Moreover, several proteins were consistently identifiable and also to be similarly modulating in studies with similar experimental conditions [42, 40, 41]. Only three studies explored the host response (human subjects) during Leishmania infection. However, no consensus results exist among those studies. More studies with similar experimental conditions are required to holistically understand the host (human) response upon Leishmania infection. Studies involving drug-resistant mechanisms and stress reported a lot of consensuses results. However, focus on new studies should be towards targeted proteomics, where the protein of interest is detectable with quantitative accuracy and high sensitivity [89]. This practically assists the researcher in identifying new drug targets and diagnostics. With this in mind, besides the systematic review, this study also collected differentially modulated proteins of different experimental conditions and made them as a database with an easy-to-access interface. In addition, the developed database was also actively linked to Uniprot (https://www.uniprot.org/) as well as TriTrypDB [88]. We strongly believe that this review and the database will help the research community in understanding the Leishmania infection, drug resistance, and host-response, and will help provide the information for the future study of targeted-proteomics related to Leishmania.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Indian Council for Medical Research (ICMR) New Delhi and Rajendra Memorial Research Institute of Medical Sciences (RMRIMS) for providing necessary computational facility for carrying out this work.
Data availability
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M, Team WHOLC et al. Leishmaniasis worldwide and global estimates of its incidence. PloS one. 2012;7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pulvertaft RJV, Hoyle GF. Stages in the life-cycle of Leishmania donovani. Trans R Soc Trop Med Hyg. 1960;54:191–196. doi: 10.1016/0035-9203(60)90057-2. [DOI] [PubMed] [Google Scholar]
- 3.Esch KJ, Petersen CA. Transmission and epidemiology of zoonotic protozoal diseases of companion animals. Clin Microbiol Rev. 2013;26:58–85. doi: 10.1128/CMR.00067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gossage SM, Rogers ME, Bates PA. Two separate growth phases during the development of Leishmania in sand flies: implications for understanding the life cycle. Int J Parasitol. 2003;33:1027–1034. doi: 10.1016/s0020-7519(03)00142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Losada-Barragán M, Umaña-Pérez A, Rodriguez-Vega A, Cuervo-Escobar S, Azevedo R, Morgado FN, de Frias Carvalho V, Aquino P, Carvalho PC, Porrozzi R, Sánchez-Gómez M, Padron G, Cuervo P. Proteomic profiling of splenic interstitial fluid of malnourished mice infected with Leishmania infantum reveals defects on cell proliferation and pro-inflammatory response. J Proteom. 2019;208:103492. doi: 10.1016/j.jprot.2019.103492. [DOI] [PubMed] [Google Scholar]
- 6.Kevric I, Cappel MA, Keeling JH. New world and old world Leishmania infections: a practical review. Dermatol Clin. 2015;33:579–593. doi: 10.1016/j.det.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Pearson RD, De Queiroz Sousa A. Clinical spectrum of leishmaniasis. Clin Infect Dis. 1996;22:1–13. doi: 10.1093/clinids/22.1.1. [DOI] [PubMed] [Google Scholar]
- 8.van Griensven J, Diro E. Visceral leishmaniasis. Infect Dis Clin. 2012;26:309–322. doi: 10.1016/j.idc.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Handler MZ, Patel PA, Kapila R, Al-Qubati Y, Schwartz RA. Cutaneous and mucocutaneous leishmaniasis: clinical perspectives. J Am Acad Dermatol. 2015;73:897–908. doi: 10.1016/j.jaad.2014.08.051. [DOI] [PubMed] [Google Scholar]
- 10.Ls Anversa, Tiburcio MGS, VnB Richini-Pereira, Ramirez LE. Human leishmaniasis in Brazil: a general review. Rev da Assoc Med Bras. 2018 doi: 10.1590/1806-9282.64.03.281. [DOI] [PubMed] [Google Scholar]
- 11.WHO Weekly epidemiological record. In 2017
- 12.Zijlstra EE, Musa AM, Khalil EAG, El Hassan IM, El-Hassan AM. Post-kala-azar dermal leishmaniasis. Lancet Infect Dis. 2003;3:87–98. doi: 10.1016/s1473-3099(03)00517-6. [DOI] [PubMed] [Google Scholar]
- 13.Noiri E, Jha TK (2017) Kala Azar in South Asia
- 14.Ghorbani M, Farhoudi R. Leishmaniasis in humans: drug or vaccine therapy? Drug Design Dev Ther. 2018;12:25. doi: 10.2147/DDDT.S146521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uliana SRB, Trinconi CT, Coelho AC. Chemotherapy of leishmaniasis: present challenges. Parasitology. 2018;145:464–480. doi: 10.1017/S0031182016002523. [DOI] [PubMed] [Google Scholar]
- 16.Rodrigo C, Weeratunga P, Fernando SD, Rajapakse S. Amphotericin B for treatment of visceral leishmaniasis: systematic review and meta-analysis of prospective comparative clinical studies including dose-ranging studies. Clin Microbiol Infect. 2018;24:591–598. doi: 10.1016/j.cmi.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Sinha PK, Bhattacharya S. Single-dose liposomal amphotericin B: an effective treatment for visceral leishmaniasis. Lancet Global Health. 2014;2:e7–e8. doi: 10.1016/S2214-109X(13)70151-7. [DOI] [PubMed] [Google Scholar]
- 18.Sundar S. Drug resistance in Indian visceral leishmaniasis. Trop Med Int Health. 2001;6:849–854. doi: 10.1046/j.1365-3156.2001.00778.x. [DOI] [PubMed] [Google Scholar]
- 19.Sundar S, Chakravarty J, Meena LP. Leishmaniasis: treatment, drug resistance and emerging therapies. Expert Opin Orphan Drugs. 2019;7:1–10. [Google Scholar]
- 20.Purkait B, Kumar A, Nandi N, Sardar AH, Das S, Kumar S, Pandey K, Ravidas V, Kumar M, De T, et al. Mechanism of amphotericin B resistance in clinical isolates of Leishmania donovani. Antimicrobial Agents Chemother. 2012;56:1031–1041. doi: 10.1128/AAC.00030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramesh V, Singh R, Avishek K, Verma A, Deep DK, Verma S, Salotra P. Decline in clinical efficacy of oral miltefosine in treatment of post kala-azar dermal leishmaniasis (PKDL) in India. PLoS Negl Trop Dis. 2015;9:1–12. doi: 10.1371/journal.pntd.0004093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sundar S, Sinha PK, Rai M, Verma DK, Nawin K, Alam S, Chakravarty J, Vaillant M, Verma N, Pandey K, et al. Comparison of short-course multidrug treatment with standard therapy for visceral leishmaniasis in India: an open-label, non-inferiority, randomised controlled trial. Lancet. 2011;377:477–486. doi: 10.1016/S0140-6736(10)62050-8. [DOI] [PubMed] [Google Scholar]
- 23.García-Hernández R, Manzano JI, Castanys S, Gamarro F. Leishmania donovani develops resistance to drug combinations. PLoS Negl Trop Dis. 2012;6:e1974. doi: 10.1371/journal.pntd.0001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrickx S, Beyers J, Mondelaers A, Eberhardt E, Lachaud L, Delputte P, Cos P, Maes L. Evidence of a drug-specific impact of experimentally selected paromomycin and miltefosine resistance on parasite fitness in Leishmania infantum. J Antimicrob Chemother. 2016;71:1914–1921. doi: 10.1093/jac/dkw096. [DOI] [PubMed] [Google Scholar]
- 25.Ponte-Sucre A, Gamarro F, Dujardin J-C, Barrett MP, López-Vélez R, Garc\‘\ia-Hernández R, Pountain AW, Mwenechanya R, Papadopoulou B. Drug resistance and treatment failure in leishmaniasis: a 21st century challenge. PLoS Negl Trop Dis. 2017;11:e0006052. doi: 10.1371/journal.pntd.0006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijayakumar S, Kant V, Das P. LeishInDB: a web-accessible resource for small molecule inhibitors against Leishmania sp. Acta Trop. 2019;190:375–379. doi: 10.1016/j.actatropica.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 27.Vijayakumar S, Das P. Recent progress in drug targets and inhibitors towards combating leishmaniasis. Acta Trop. 2018;181:95–104. doi: 10.1016/j.actatropica.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Chakravarty J, Kumar S, Trivedi S, Rai VK, Singh A, Ashman JA, Laughlin EM, Coler RN, Kahn SJ, Beckmann AM, et al. A clinical trial to evaluate the safety and immunogenicity of the LEISH-F1 + MPL-SE vaccine for use in the prevention of visceral leishmaniasis. Vaccine. 2011;29:3531–3537. doi: 10.1016/j.vaccine.2011.02.096. [DOI] [PubMed] [Google Scholar]
- 29.Osman M, Mistry A, Keding A, Gabe R, Cook E, Forrester S, Wiggins R, Di Marco S, Colloca S, Siani L, et al. A third generation vaccine for human visceral leishmaniasis and post kala azar dermal leishmaniasis: first-in-human trial of ChAd63-KH. PLoS Negl Trop Dis. 2017;11:e0005527. doi: 10.1371/journal.pntd.0005527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundar S, Singh B. Understanding Leishmania parasites through proteomics and implications for the clinic. Expert Rev Proteom. 2018 doi: 10.1080/14789450.2018.1468754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belczacka I, Latosinska A, Metzger J, Marx D, Vlahou A, Mischak H, Frantzi M. Proteomics biomarkers for solid tumors: current status and future prospects. Mass Spectr Rev. 2019;38:49–78. doi: 10.1002/mas.21572. [DOI] [PubMed] [Google Scholar]
- 32.Donnarumma D, Faleri A, Costantino P, Rappuoli R, Norais N. The role of structural proteomics in vaccine development: recent advances and future prospects. Expert Rev Proteom. 2016;13:55–68. doi: 10.1586/14789450.2016.1121113. [DOI] [PubMed] [Google Scholar]
- 33.Banks RE, Dunn MJ, Hochstrasser DF, Sanchez JC, Blackstock W, Pappin DJ, Selby PJ. Proteomics: new perspectives, new biomedical opportunities. Lancet. 2000;356:1749–1756. doi: 10.1016/S0140-6736(00)03214-1. [DOI] [PubMed] [Google Scholar]
- 34.Capelli-Peixoto J, Mule SN, Tano FT, Palmisano G, Stolf BS. Proteomics and leishmaniasis: potential clinical applications. Proteom Clin Appl. 2019;13:1–16. doi: 10.1002/prca.201800136. [DOI] [PubMed] [Google Scholar]
- 35.Bag AK, Saha S, Sundar S, Saha B, Chakrabarti A, Mandal C. Comparative proteomics and glycoproteomics of plasma proteins in Indian visceral leishmaniasis. Proteome Sci. 2014;12:1–14. doi: 10.1186/s12953-014-0048-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang S, Wang PG. Method development of glycoprotein biomarkers for cancers. Bioanalysis. 2017;9:903–906. doi: 10.4155/bio-2017-0077. [DOI] [PubMed] [Google Scholar]
- 37.Montoya A, López MC, Vélez ID, Robledo SM. Label-free quantitative proteomic analysis reveals potential biomarkers for early healing in cutaneous leishmaniasis. PeerJ. 2019;6:e6228. doi: 10.7717/peerj.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Da Silva Santos C, Attarha S, Saini RK, Boaventura V, Costa J, Khouri R, Barral-Netto M, Brodskyn CI, Souchelnytskyi S. Proteome profiling of human cutaneous leishmaniasis lesion. J Investig Dermatol. 2015;135:400–410. doi: 10.1038/jid.2014.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menezes JPBD, Almeida TFD, Petersen ALDOA, Guedes CES, Mota MSV, Lima JGB, Palma LC, Buck GA, Krieger MA, Probst CM, et al. Proteomic analysis reveals differentially expressed proteins in macrophages infected with Leishmania amazonensis or Leishmaniamajor. Microbes Infect. 2013;15:579–591. doi: 10.1016/j.micinf.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Negrão F, Fernandez-Costa C, Zorgi N, Giorgio S, Nogueira Eberlin M, Yates JR., III Label-free proteomic analysis reveals parasite-specific protein alterations in macrophages following Leishmania amazonensis, Leishmania major, or Leishmania infantum Infection. ACS Infect Dis. 2019;5:851–862. doi: 10.1021/acsinfecdis.8b00338. [DOI] [PubMed] [Google Scholar]
- 41.Negrão F, Giorgio S, Eberlin MN, Yates JR. Comparative proteomic analysis of murine cutaneous lesions induced by Leishmania amazonensis or Leishmania major. ACS Infect Dis. 2019;5:1295–1305. doi: 10.1021/acsinfecdis.8b00370. [DOI] [PubMed] [Google Scholar]
- 42.Negrão F, Diedrich JK, Giorgio S, Eberlin MN, Yates JR., 3rd Tandem mass tag proteomic analysis of in vitro and in vivo models of cutaneous leishmaniasis reveals parasite-specific and nonspecific modulation of proteins in the host. ACS Infect Dis. 2019;5:2136–2147. doi: 10.1021/acsinfecdis.9b00275. [DOI] [PubMed] [Google Scholar]
- 43.Losada-Barragán M, Umaña-Pérez A, Durães J, Cuervo-Escobar S, Rodríguez-Vega A, Ribeiro-Gomes FL, Berbert LR, Morgado F, Porrozzi R, Mendes-da-Cruz DA, Aquino P, Carvalho PC, Savino W, Sánchez-Gómez M, Padrón G, Cuervo P. Thymic microenvironment is modified by malnutrition and Leishmania infantum Infection. Front Cell Infect Microbiol. 2019;9:1–19. doi: 10.3389/fcimb.2019.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Losada-Barragán M, Umaña-Pérez A, Rodriguez-Vega A, Cuervo-Escobar S, Azevedo R, Morgado FN, de Frias Carvalho V, Aquino P, Carvalho PC, Porrozzi R, Sánchez-Gómez M, Padron G, Cuervo P. Proteomic profiling of splenic interstitial fluid of malnourished mice infected with Leishmania infantum reveals defects on cell proliferation and pro-inflammatory response. J Proteom. 2019 doi: 10.1016/j.jprot.2019.103492. [DOI] [PubMed] [Google Scholar]
- 45.Hassani K, Olivier M. Immunomodulatory impact of leishmania-induced macrophage exosomes: a comparative proteomic and functional analysis. PLoS Negl Trop Dis. 2013;7:e2185. doi: 10.1371/journal.pntd.0002185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Britti D, Gaspari M, Massimini G, Casalinuovo F, Morittu VM, Cuda G. Proteomic analysis in canine leishmaniasis. Vet Res Commun. 2010;34:91–96. doi: 10.1007/s11259-010-9410-5. [DOI] [PubMed] [Google Scholar]
- 47.Franco-Martínez L, Villar M, Tvarijonaviciute A, Escribano D, Bernal LJ, Cerón JJ, Thomas MDC, Mateos-Hernández L, Tecles F, de la Fuente J, López MC, Martínez-Subiela S. Serum proteome of dogs at subclinical and clinical onset of canine leishmaniosis. Transbound Emerg Dis. 2020;67:318–327. doi: 10.1111/tbed.13354. [DOI] [PubMed] [Google Scholar]
- 48.Lorena F-M, Tvarijonaviciute A, Horvatić A, Guillemin N, Bernal LJ, Barić Rafaj R, Cerón JJ, Thomas MDC, López MC, Tecles F, Martínez-Subiela S, Mrljak V. Changes in saliva of dogs with canine leishmaniosis: a proteomic approach. Vet Parasitol. 2019;272:44–52. doi: 10.1016/j.vetpar.2019.06.014. [DOI] [PubMed] [Google Scholar]
- 49.Manabe YC, Dannenberg AM, Tyagi SK, Hatem CL, Yoder M, Woolwine SC, Zook BC, Pitt MLM, Bishai WR. Different strains of Mycobacterium tuberculosis cause various spectrums of disease in the rabbit model of tuberculosis. Infect Immun. 2003;71:6004–6011. doi: 10.1128/IAI.71.10.6004-6011.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Da Fonseca Pires S, Fialho LC, Silva SO, Melo MN, De Souza CC, Tafuri WL, Bruna Romero O, De Andrade HM. Identification of virulence factors in leishmania infantum strains by a proteomic approach. J Proteome Res. 2014;13:1860–1872. doi: 10.1021/pr400923g. [DOI] [PubMed] [Google Scholar]
- 51.Magalhães RDM, Duarte MC, Mattos EC, Martins VT, Lage PS, Chávez-Fumagalli MA, Lage DP, Menezes-Souza D, Régis WCB, Manso Alves MJ, Soto M, Tavares CAP, Nagen RAP, Coelho EAF. Identification of differentially expressed proteins from Leishmania amazonensis associated with the loss of virulence of the parasites. PLoS Negl Trop Dis. 2014 doi: 10.1371/journal.pntd.0002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Rezende E, Kawahara R, Peña MS, Palmisano G, Stolf BS. Quantitative proteomic analysis of amastigotes from Leishmania (L.) amazonensis LV79 and PH8 strains reveals molecular traits associated with the virulence phenotype. PLoS Negl Trop Dis. 2017;11:e0006090. doi: 10.1371/journal.pntd.0006090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCall L-I, Zhang W-W, Dejgaard K, Atayde VD, Mazur A, Ranasinghe S, Liu J, Olivier M, Nilsson T, Matlashewski G. Adaptation of Leishmania donovani to cutaneous and visceral environments: in vivo selection and proteomic analysis. J Proteome Res. 2015;14:1033–1059. doi: 10.1021/pr5010604. [DOI] [PubMed] [Google Scholar]
- 54.Hajjaran H, Mousavi P, Burchmore R, Mohebali M, Mohammadi Bazargani M, Hosseini Salekdeh G, Kazemi-Rad E, Khoramizadeh MR. Comparative proteomic profiling of Leishmania tropica: investigation of a case infected with simultaneous cutaneous and viscerotropic leishmaniasis by 2-dimentional electrophoresis and mass spectrometry. Iran J Parasitol. 2015;10:366–380. [PMC free article] [PubMed] [Google Scholar]
- 55.Maltezou HC (2009) Drug resistance in visceral leishmaniasis. BioMed Research International 2010
- 56.Vergnes B, Gourbal B, Girard I, Sundar S, Drummelsmith J, Ouellette M. A proteomics screen implicates HSP83 and a small kinetoplastid calpain-related protein in drug resistance in Leishmania donovani clinical field isolates by modulating drug-induced programmed cell death. Mol Cell Proteom. 2007;6:88–101. doi: 10.1074/mcp.M600319-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.El Fadili K, Drummelsmith J, Roy G, Jardim A, Ouellette M. Down regulation of KMP-11 in Leishmania infantum axenic antimony resistant amastigotes as revealed by a proteomic screen. Exp Parasitol. 2009;123:51–57. doi: 10.1016/j.exppara.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 58.Kumar A, Sisodia B, Misra P, Sundar S, Shasany AK, Dube A. Proteome mapping of overexpressed membrane-enriched and cytosolic proteins in sodium antimony gluconate (SAG) resistant clinical isolate of Leishmania donovani. Br J Clin Pharmacol. 2010;70:609–617. doi: 10.1111/j.1365-2125.2010.03716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biyani N, Singh AK, Mandal S, Chawla B, Madhubala R. Differential expression of proteins in antimony-susceptible and -resistant isolates of Leishmania donovani. Mol Biochem Parasitol. 2011;179:91–99. doi: 10.1016/j.molbiopara.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 60.Walker J, Gongora R, Vasquez JJ, Drummelsmith J, Burchmore R, Roy G, Ouellette M, Gomez MA, Saravia NG. Discovery of factors linked to antimony resistance in Leishmania panamensis through differential proteome analysis. Mol Biochem Parasitol. 2012;183:166–176. doi: 10.1016/j.molbiopara.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 61.Matrangolo FSV, Liarte DB, Andrade LC, De Melo MF, Andrade JM, Ferreira RF, Santiago AS, Pirovani CP, Silva-Pereira RA, Murta SMF. Comparative proteomic analysis of antimony-resistant and-susceptible Leishmania braziliensis and Leishmania infantum chagasi lines. Mol Biochem Parasitol. 2013;190:63–75. doi: 10.1016/j.molbiopara.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 62.de Souza Moreira D, Pescher P, Laurent C, Lenormand P, Späth GF, Murta SMF. Phosphoproteomic analysis of wild-type and antimony-resistant Leishmania braziliensis lines by 2D-DIGE technology. Proteomics. 2015;15:2999–3019. doi: 10.1002/pmic.201400611. [DOI] [PubMed] [Google Scholar]
- 63.Brotherton M-C, Bourassa S, Leprohon P, Légaré D, Poirier GG, Droit A, Ouellette M. Proteomic and genomic analyses of antimony resistant Leishmania infantum mutant. PloS one. 2013;8:e81899. doi: 10.1371/journal.pone.0081899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vincent IM, Racine G, Légaré D, Ouellette M. Mitochondrial proteomics of antimony and miltefosine resistant Leishmania infantum. Proteomes. 2015;3:328–346. doi: 10.3390/proteomes3040328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Drummelsmith J, Girard I, Trudel N, Ouellette M. Differential protein expression analysis of Leishmania major reveals novel roles for methionine adenosyltransferase and S-adenosylmethionine in methotrexate resistance. J Biol Chem. 2004;279:33273–33280. doi: 10.1074/jbc.M405183200. [DOI] [PubMed] [Google Scholar]
- 66.Singh AK, Roberts S, Ullman B, Madhubala R. A quantitative proteomic screen to identify potential drug resistance mechanism in $α$-difluoromethylornithine (DFMO) resistant Leishmania donovani. J Proteom. 2014;102:44–59. doi: 10.1016/j.jprot.2014.02.030. [DOI] [PubMed] [Google Scholar]
- 67.Carnielli JBT, de Andrade HM, Pires SF, Chapeaurouge AD, Perales J, Monti-Rocha R, SiFG Carvalho, Ribeiro LP, Dietze R, Figueiredo SG, et al. Proteomic analysis of the soluble proteomes of miltefosine-sensitive and-resistant Leishmania infantum chagasi isolates obtained from Brazilian patients with different treatment outcomes. J Proteom. 2014;108:198–208. doi: 10.1016/j.jprot.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 68.Veronica J, Chandrasekaran S, Dayakar A, Devender M, Prajapati VK, Sundar S, Maurya R. Iron superoxide dismutase contributes to miltefosine resistance in Leishmania donovani. FEBS J. 2019;286:3488–3503. doi: 10.1111/febs.14923. [DOI] [PubMed] [Google Scholar]
- 69.Vacchina P, Norris-Mullins B, Carlson ES, Morales MA. A mitochondrial HSP70 (HSPA9B) is linked to miltefosine resistance and stress response in Leishmania donovani. Parasites Vectors. 2016;9:621. doi: 10.1186/s13071-016-1904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garg G, Ali V, Singh K, Gupta P, Ganguly A, Sahasrabuddhe AA, Das P. Quantitative secretome analysis unravels new secreted proteins in Amphotericin B resistant Leishmania donovani. J Proteom. 2019;207:103464. doi: 10.1016/j.jprot.2019.103464. [DOI] [PubMed] [Google Scholar]
- 71.Yaribeygi H, Panahi Y, Sahraei H, Johnston TP, Sahebkar A. The impact of stress on body function: a review. EXCLI J. 2017;16:1057. doi: 10.17179/excli2017-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dolai S, Adak S. Endoplasmic reticulum stress responses in Leishmania. Mol Biochem Parasitol. 2014;197:1–8. doi: 10.1016/j.molbiopara.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 73.Mathur R, Das RP, Ranjan A, Shaha C. Elevated ergosterol protects Leishmania parasites against antimony-generated stress. FASEB J. 2015;29:4201–4213. doi: 10.1096/fj.15-272757. [DOI] [PubMed] [Google Scholar]
- 74.Ravanan P, Srikumar IF, Talwar P. Autophagy: the spotlight for cellular stress responses. Life Sci. 2017;188:53–67. doi: 10.1016/j.lfs.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 75.Silverman JM, Clos J, de’Oliveira CC, Shirvani O, Fang Y, Wang C, Foster LJ, Reiner NE. An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J Cell Sci. 2010;123:842–852. doi: 10.1242/jcs.056465. [DOI] [PubMed] [Google Scholar]
- 76.Maxwell MJ, Chan SK, Robinson DP, Dwyer DM, Nandan D, Foster LJ, Reiner NE. Proteomic analysis of the secretome of Leishmania donovani. Genome Biol. 2008 doi: 10.1186/gb-2008-9-2-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pescher P, Blisnick T, Bastin P, Späth GF. Quantitative proteome profiling informs on phenotypic traits that adapt Leishmania donovani for axenic and intracellular proliferation. Cell Microbiol. 2011;13:978–991. doi: 10.1111/j.1462-5822.2011.01593.x. [DOI] [PubMed] [Google Scholar]
- 78.Sardar AH, Kumar S, Kumar A, Purkait B, Das S, Sen A, Kumar M, Sinha KK, Singh D, Equbal A, et al. Proteome changes associated with Leishmania donovani promastigote adaptation to oxidative and nitrosative stresses. J Proteom. 2013;81:185–199. doi: 10.1016/j.jprot.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 79.Teixeira PC, Velasquez LG, Lepique AP, de Rezende E, Bonatto JMC, Barcinski MA, Cunha-Neto E, Stolf BS. Regulation of Leishmania (L.) amazonensis protein expression by host T cell dependent responses: differential expression of oligopeptidase B, tryparedoxin peroxidase and HSP70 isoforms in amastigotes isolated from BALB/c and BALB/c nude mice. PLoS Negl Trop Dis. 2015;9:e0003411. doi: 10.1371/journal.pntd.0003411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dostálová A, Volf P. Leishmania development in sand flies: parasite-vector interactions overview. Parasites Vectors. 2012;5:276. doi: 10.1186/1756-3305-5-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walker J, Vasquez JJ, Gomez MA, Drummelsmith J, Burchmore R, Girard I, Ouellette M. Identification of developmentally-regulated proteins in Leishmania panamensis by proteome profiling of promastigotes and axenic amastigotes. Mol Biochem Parasitol. 2006;147:64–73. doi: 10.1016/j.molbiopara.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 82.Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol Biochem Parasitol. 2007;152:35–46. doi: 10.1016/j.molbiopara.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 83.Brotherton MC, Racine G, Foucher AL, Drummelsmith J, Papadopoulou B, Ouellette M. Analysis of stage-specific expression of basic proteins in Leishmania infantum. J Proteome Res. 2010;9:3842–3853. doi: 10.1021/pr100048m. [DOI] [PubMed] [Google Scholar]
- 84.Biyani N, Madhubala R. Quantitative proteomic profiling of the promastigotes and the intracellular amastigotes of Leishmania donovani isolates identifies novel proteins having a role in Leishmania differentiation and intracellular survival. Biochim et Biophys Acta Proteins Proteom. 2012;1824:1342–1350. doi: 10.1016/j.bbapap.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 85.Lynn MA, Marr AK, McMaster WR. Differential quantitative proteomic profiling of Leishmania infantum and Leishmania mexicana density gradient separated membranous fractions. J Proteom. 2013;82:179–192. doi: 10.1016/j.jprot.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 86.Alcolea PJ, Alonso A, Larraga V. Proteome profiling of Leishmania infantum promastigotes. J Eukaryot Microbiol. 2011;58:352–358. doi: 10.1111/j.1550-7408.2011.00549.x. [DOI] [PubMed] [Google Scholar]
- 87.Alcolea PJ, Alonso A, García-Tabares F, Mena MC, Ciordia S, Larraga V. Increased abundance of proteins involved in resistance to oxidative and nitrosative stress at the last stages of growth and development of Leishmania amazonensis promastigotes revealed by proteome analysis. PLoS ONE. 2016;11:1–20. doi: 10.1371/journal.pone.0164344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38(suppl_1):D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Borràs E, Sabidó E. What is targeted proteomics? A concise revision of targeted acquisition and targeted data analysis in mass spectrometry. Proteomics. 2017;17:17–18. doi: 10.1002/pmic.201700180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.