Abstract
The harm of pathogenic bacteria to humans has promoted extensive research on physiological processes of pathogens, such as the mechanism of bacterial infection, antibiotic mode of action, and bacterial antimicrobial resistance. Most of these processes can be better investigated by timely tracking of fluorophore-derived antibiotics in living cells. In this paper, we will review the recent development of fluorescent antibiotics featuring the conjugation with various fluorophores, and focus on their applications in fluorescent imaging and real-time detection for various physiological processes of bacteria in vivo.
Keywords: Fluorescent antibiotics, Live-cell tracking, Fluorophore, Pathogenic bacteria, Fluorescent imaging
Graphical abstract
Highlights
-
•
Profiles of Fluorophores-derived Antibiotics in Development.
-
•
Discussing the influence on antibiotic activity after conjugating fluorophore.
-
•
Fluorescent Tracking to better understand physiological processes of Pathogenic bacteria.
-
•
Live-Cell imaging to investigate bacteria in their native environment.
1. Introduction
Bacterial infections are one of the major causes of human diseases and death in the world. At present, bacterial infections have caused 700,000 deaths worldwide each year, and recent projections indicate that this annual deaths number may reach 10 million by 2050 [1,2]. This is mainly attributed to the emergence of multi-drugs resistant (MDR) bacteria stimulated by long-term overuse of antibacterial drugs for patients, especially in second-world countries [3,4]. For instance, the number of hospital infections caused by antimicrobial resistant bacteria has accounted for about 30% of the total number of pathogenic bacteria infections in hospitalized patients [5,6]. More timely and effective treatment is an urgent need for patients.
Antibiotics, one of the greatest discoveries of the 20th century, can effectively inhibit bacterial infections and reduce mortality. But with the increasing prevalence of MDR bacteria, many antibiotics including some potent broad spectrum drugs are no longer susceptible to most therapeutic regimens [7]. Before developing new antibacterial drugs, we should first improve our fundamental understanding of how bacteria infect the human body, how drugs work with both bacteria and the human body, and the mechanism of bacterial resistance to drugs [8,9].
The fluorescent tracking capability of fluorophore-derived recognition element conjugate makes it an increasingly prevalent strategy used to monitor targets timely within organelles, whole cells, and entire animals. Recently, small-molecule fluorophores have been widely used in covalently linking to antibiotics, generally named fluorescent antibiotics. The monitoring of fluorescent antibiotics in living organisms has the potential in diagnosis of bacterial infections, elucidating antimicrobial agents mode of action, assessing drug toxicity, and examining bacterial antimicrobial resistance [[10], [11], [12], [13], [14], [15], [16]]. Fluorescent antibiotics generally divided into two types according to structural differences. One is autofluorescent antibiotics: antibiotics have functional groups with intrinsic fluorescence; the other is fluorophore-conjugated fluorescent antibiotics: a fluorophore was covalently linked to an existing antibiotic to produce a fluorophore–antibiotic compound.
Recently, Blaskovich’s group [17] reviewed the development of fluorescent antibiotics from the perspective of various antibiotics used for intracellular or cell wall targeting, and especially discussed the biological applications in how to fight antibiotic resistance. In this review, we mainly discuss the classification and composition of fluorescent antibiotics dependent on diverse fluorophores, and review the application in fluorescent imaging and real-time detection of pathogenic bacteria in vivo.
2. Autofluorescent antibiotics
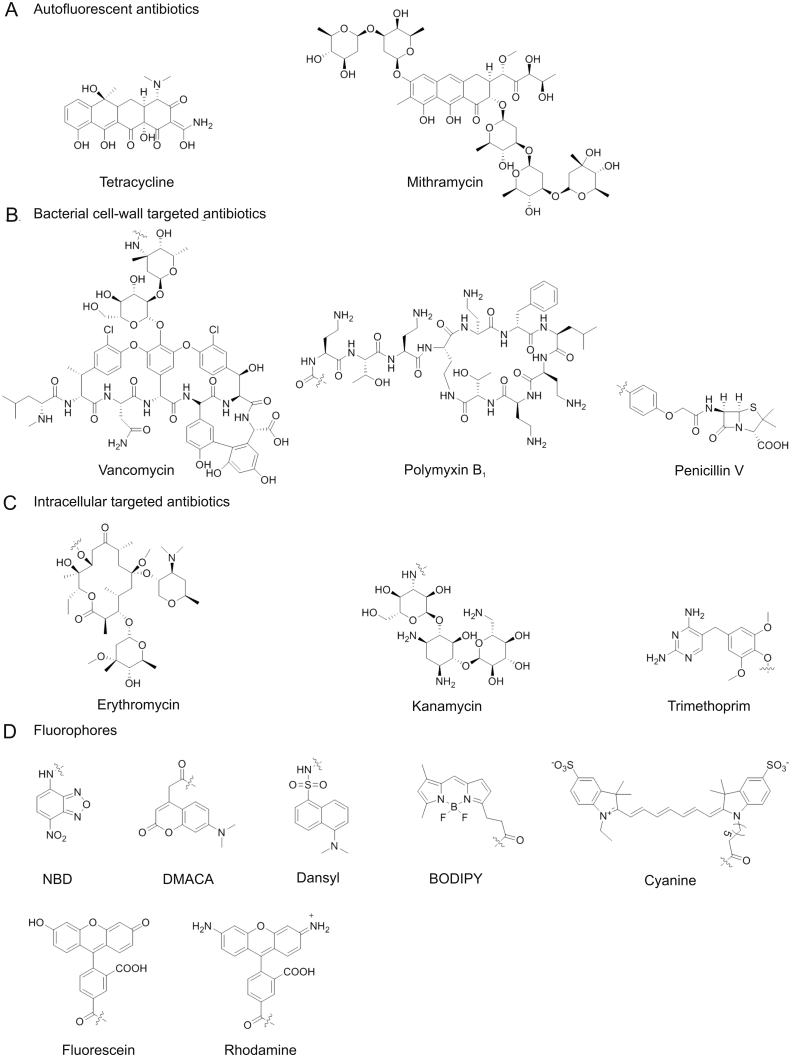
Tetracycline is a classical intrinsic autofluorescent antibiotic (Fig. 1A). Early in the 1960s, tetracycline fluorescence was studied across bacterial membranes [18]. Both Escherichia coli (E. coli) and Bacillus cereus (B. cereus) with tetracycline were observed in dying guinea pig phagocytes under fluorescence microscopy. Furthermore, Glazier’ group [19] reported their initial studies on fermented product monitoring by fluorescence measurements of tetracycline signal in model fermentation media samples containing Streptomyces aureofaciens cell mass. In addition, some studies using autofluorescent antibiotics with fluorescent anthraquinone core, such as olivomycin and mithramycin (Fig. 1A), to stain DNA in cell cultures and for flow cytometry detection [[20], [21], [22], [23], [24]]. However, autofluorescent antibiotics generally showed weak fluorescence and poor stability, which limited their applications, especially in super-resolution fluorescent imaging of bacteria.
Fig. 1.
Structure of various antibiotics and fluorophores. NBD: nitrobenzofurazan, DMACA: 7-(dimethylamino)-coumarin-4-acetic acid, BODIPY: dansyl,boron-dipyrromethene.
3. Fluorophore-antibiotic conjugates
3.1. Antibiotics
Antibiotics with recognition groups of fluorophore-antibiotic conjugates were used to specific recognition of target bacteria. Antibiotics interacted with bacteria by targeting essential survival processes such as inhibiting cell-wall synthesis and interfering with the synthesis of vital proteins or DNA [25]. There are several kinds of bacterial cell-wall related antibiotics, such as β-lactams, glycopeptides, lipopeptides, and polypeptides [[26], [27], [28], [29]]. Among them, β-lactam antibiotics, such as penicillin (Fig. 1B) and ceftaroline, can inhibit the peptidoglycans biosynthesis by covalently binding penicillin-binding proteins (PBPs) [[30], [31], [32], [33], [34]]. Vancomycin and ramoplanin treat Gram-positive infections by targeting lipid II (a precursor for bacterial peptidoglycan biosynthesis) of cell wall [27]. In addition, polymyxins as naturally occurring cyclic antibiotic lipopeptides are typically Gram-negative bacteria antibiotics and bind to lipid A on the bacterial outer membrane through electrostatic interaction (Fig. 1B) [28,35]. Antibiotics that act by targeting DNA or protein intra bacterial cell include macrolides, aminoglycosides, and quinolones etc. For example, erythromycin (macrolides) and kanamycin (aminoglycosides) act with ribosome of bacteria to block protein synthesis in the cells [36]. Trimethoprim (TMP) is the substrate of the intracellular protease dihydrofolate reductase (DHFR) [37]. The structures of them are shown in Fig. 1C. All of the antibiotics above have been linked to a fluorophore for studying in physiological processes of pathogenic bacteria.
3.2. Fluorophores
Commonly used fluorophores for fluorophore-antibiotics conjugates are displayed in Fig. 1D. They include nitrobenzofurazan (NBD), 7-(dimethylamino)-coumarin-4-acetic acid (DMACA), dansyl, boron-dipyrromethene (BODIPY), fluorescein, rhodamine, and cyanine 7(Cy7). Researchers bound these fluorophores or their derivatives to antibiotics for fluorescent monitoring antibiotics in bacteria or tracking target bacteria in live animals. The selection of fluorophore mainly depends on the physicochemical properties of fluorophore, such as size, biocompatibility, brightness, and sensing wavelength. The size and biocompatibility of fluorophores are likely to influence the antibacterial activity of antibiotics, which will be discussed in next section.
A fluorophore with higher brightness is considered to study detail physiological processes of pathogenic bacteria. BODIPY (ε = 9.1 × 104 M−1 cm−1, and ϕ = 0.94), fluorescein (ε = 9.3 × 104 M−1 cm−1, and ϕ = 0.95), or rhodamine (ε = 7.4 × 104 M−1 cm−1, and ϕ = 0.92) derived fluorophores are commercially available fluorophores with excellent extinction coefficient (ε) and quantum yield (ϕ, important parameters of fluorophore brightness, the relative brightness = ε × ϕ), and are widely employed in many different biological imaging applications [38]. Furthermore, near-infrared (sensing wavelength in the region of 650–900 nm) fluorophores conjugated with antibiotic can be used for fluorescent imaging of bacterial infections in vivo (detail description in section 4.2) [39]. In addition, the fluorogenic probe (the probe presenting turn-on or ratiometric signal while identifying target in situ) is created by linking one or two selective fluorophores to an antibiotic for real-time detection of target objects (detail description in section 4.3) [40,41].
3.3. Effect on antibacterial activity of antibiotics
In principle, it is important for fluorophore-derived antibiotics to maintain specificity and affinity to target. Unfortunately, the minimum inhibitory concentration (MIC, a commonly used measure of the efficacy of an antibiotic) of most of fluorophore-derived antibiotics display an inevitable increasing trend (Table 1) [[42], [43], [44], [45], [46], [47]]. This may be due to not only the reduced affinity of the derived antibiotic, but also the fact that the molecules cannot reach the target caused by the change in the physical properties of molecules. While fluorescein is a negatively charged fluorophore, Walker’ group [42] considered that the increased MICs of their fluorescein-derived antibiotics may be caused by the repulsive interactions of the compound to the cell wall (generally with amount of negative charges) and/or poor solubility of the compound. On the contrary, the relatively smaller and neutral BODIPY-derived antibiotics displayed lower MICs (10 μg/mL for BODIPY-Ramoplanin, 2.5 μg/mL for BODIPY-Vancomycin) than fluorescein derivatives (20 μg/mL for fluorescein-Ramoplanin, 20 μg/mL for fluorescein-Vancomycin). However, the MICs of BODIPY-derived antibiotics are still much higher than those of conventional antibiotics.
Table 1.
Minimum inhibitory concentration of fluorescent and conventional antibiotics.
| Fluorescent/Conventional antibiotics | Compound names in the references | MIC (μg/mL) | Species | Ref. |
|---|---|---|---|---|
| Ramoplanin | 5 | Bacillus subtillis | [42] | |
| Fluorescein-Ramoplanin | Ramo-4F1 | 20 | ||
| BODIPY-Ramoplanin | Ramo-4BDP | 10 | ||
| Vancomycin | 0.13 | |||
| Fluorescein-Vancomycin | Van-F1 | 20 | ||
| BODIPY-Vancomycin | Van-BDP | 2.5 | ||
| Polymyxin B | 1 |
Pseudomonas aeruginosa ATCC 27853 |
[43] | |
| Dansyl-polymyxin B | MIPS-9541 | 4 | ||
| Colistin | 1 |
Acinetobacter baumannii ATCC 19606 |
[44] | |
| Dansyl-colistin | FADDI-285 | 4 | ||
| Linezolid | 2 |
Enterococcus faecium Van A ATCC 51559 |
[45] | |
| DMACA-linezolid | Lz-tz-DMACA | 32 | ||
| NBD-linezolid | Lz-tz-NBD | 16 | ||
| Ciprofloxacin | 0.313–0.06 | [46] | ||
| NBD-Ciprofloxacin | Cipro-NBD 3 | 8 | B. subtillis ATCC 6051 | |
| DMACA-Ciprofloxacin | Cipro-DMACA 4 | 4 | ||
| Trimethoprim | 1 |
Staphylococcus aureus ATCC 25923 |
[47] | |
| NBD-Trimethoprim | TMP-3C-TzNBD | 64 | ||
| TMP-4C-TzNBD | 16 | |||
| TMP-5C-TzNBD | 8 | |||
| DMACA-Trimethoprim | TMP-3C-TzDMACA | >64 | ||
| TMP-4C-TzDMACA | >64 | |||
| TMP-5C-TzDMACA | >64 |
DMACA: 7-(dimethylamino)-coumarin-4-acetic acid, NBD: nitrobenzofurazan.
The introduction of fluorophore with smaller size would greatly reduce the influence on the properties of antibiotics. Li’s group [43,44,48] constructed fluorescent polymyxin probes by linking dansyl fluorophore to the N-terminus of polymyxin B. They consider that the small size of the dansyl group helps reduce the likelihood of negative steric effects on the pharmacophore of polymyxins. The MIC of dansyl-polymyxin B was 4 fold higher than that of polymyxin B (Table 1). And polymyxin mechanisms of action were explored by imaging of polymyxin interactions with kidney proximal tubular cells. Furthermore, the smaller fluorophores NBD (M = 164 g/mol) and DMACA (M = 261 g/mol) have been selected to combine with fluoroquinolone antibiotic ciprofloxacin in order to maximize the penetrance of the compound into the cytosol, particularly with Gram-negative bacteria [45,46]. However, not all antibiotics linked to small fluorophores maintain great antibacterial activity; the MIC of NBD-derived Trimethoprim (TMP-3C-TzNBD, MIC = 64 μg/mL) was much higher than that of TMP (MIC = 1 μg/mL) for Staphylococcus aureus (S. aureus) ATCC 25923 [47]. But the MIC value seemed to decrease with the prolonging of the linker (TMP-4C-TzNBD, MIC = 16 μg/mL; TMP-5C-TzNBD, MIC = 8 μg/mL). Nevertheless, DMACA-derived TMP did not display the same phenomenon, and all probes with different linkers showed lower antibacterial activity (MIC > 64 μg/mL). Authors considered the lack of whole cell activity was likely due to compound efflux.
4. Fluorescent tracking of pathogenic bacteria in vivo
Fluorescent tracking of pathogenic bacteria includes monitoring antibiotics in live bacteria or tracking target bacteria in live animals. The fluorescent tracking ability depends on the response fluorescent signals of different fluorophores and the recognition of the antibiotics. Here, we mainly discuss various fluorophores-dependent fluorescent antibiotics utility in fluorescent imaging and real-time detection to better understand physiological processes of pathogenic bacteria in vivo.
4.1. Fluorescent imaging in live cells
Visualization study by fluorescent microscopy is critical to our understanding of bacterial growth and pathogenesis in their native environment. And fluorescent antibiotic is one of the essential tools to identify and track these physiological processes under fluorescent microscopy [[49], [50], [51], [52]]. For example, Fluorescein-derived vancomycins have been used to visualize peptidoglycan biosynthesis in living cells [53,54]. Brightest staining was observed at the division site of Bacillus subtillis (B. subtilis) cells, which was predicted to be the site that peptidoglycan synthesis occurs, along with less apparent sidewall staining in a helical pattern. Fluorescein was further conjugated to ramoplanin (another peptidoglycan-binding antibiotic beside vancomycin) and the probes displayed concentration-dependent staining patterns in B. subtilis [42]. At low concentrations, the probes labeled the nascent division sites, the cell poles, and a helix-like sidewall pattern. The authors also compared the straining effect of the fluorescein and BODIPY-conjugated antibiotics, respectively. BODIPY-conjugated ramoplanin was able to stain both at the poles and the sidewalls of B. subtilis cells, while fluorescein-ramoplanin could only stain the poles. Moreover, many fluorescent antibiotics have been used for live-cell imaging. For example, 9a-NBD-azithromycin, a compound with the most similar cellular pharmacokinetic profile to azithromycin, was used to study azithromycin’s in vivo distribution by confocal microscopy [55]. The fluorescent imaging of BODIPY-daptomycin in bacterial membrane revealed the mechanism of action of daptomycin on cell wall morphology and septation [56]. Lastly, Cy3-polymyxin B and Cy5-vancomycin (Cy3 and Cy5 are analogues of Cy7 fluorophore in Fig. 1D) can selectively labeled Gram-negative and Gram-positive bacteria in various kinds of complex bacterial samples [57].
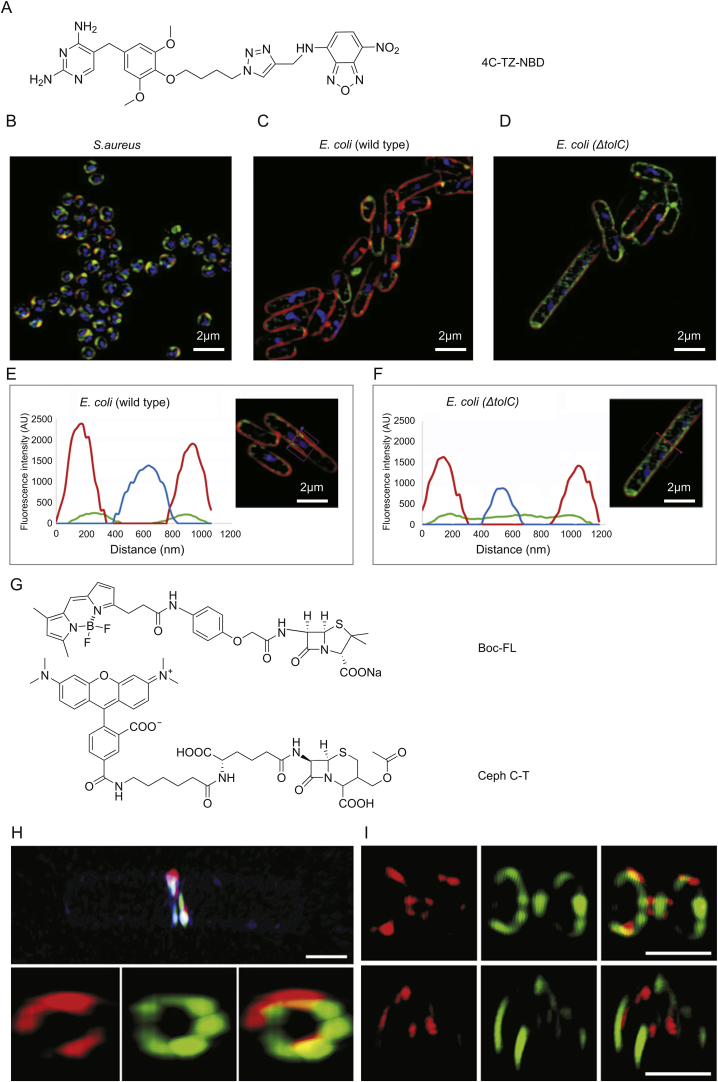
However, the light diffraction of conventional fluorescence microscopes is limited to 200–300 nm and 500–700 nm in the lateral and axial dimensions, respectively. The size of bacteria is so small (1–10 μm) [58], and the spatial resolution of conventional fluorescence microscopes is insufficient for this. Thus, the utility of super-resolution microscopy has become essential to adequately visualizing the subcellular structures of these organisms [59,60]. The super-resolution structured illumination microscopic (SR-SIM) imagings of 4C-Tz-NBD in live S. aureus and E. coli have been reported by Phetsang et al. [47]. 4C-Tz-NBD was created by binding NBD to TMP antibiotic (Fig. 2A), and then S. aureus, wild type E.coli and ΔtolC type E.coli (efflux pump deficient E. coli mutant strain) were strained with FM4-64FX, Hoechst 33342, and 4C-Tz-NBD, respectively. The bacteria were clearly imaged by SR-SIM (Figs. 2B–D), and more details have been observed. Cross sections of representative bacteria demonstrated that the 4C-Tz-NBD was largely localized inside the bacterial membrane when compared to the location of the red FM4-64FX dye (Figs. 2E and F). Moreover, 4C-Tz-NBD displayed higher cellular accumulation in ΔtolC E.coli compared to wild type E.coli, which indicates that in normal bacteria TMP probe accumulation at a sufficiently high concentration to inhibit the DHFR target is prevented due to the removal by the TolC-dependent efflux pump system.
Fig. 2.
Super resolution imaging with different fluorescent antibiotics. (A) Structure of NBD-derived fluorescent antibiotic 4C-Tz-NBD [47]. SR-SIM fluorescence imaging of (B) S. aureus, (C) wild type E. coli, and (D) ΔtolC E. coli. And cross section of fluorescent imaging of (E) wild type E. coli and (F) ΔtolC E. coli: green, 4C-Tz-NBD; red, FM4-64FX; blue, Hoechst 33342. (G) Structures of fluorescent antibiotics Boc-FL and Ceph C-T [76]. 3D-SIM super-resolution microscope imaging of (H) B. subtilis and (I) S. pneumoniae IU 1945 PBPs after dual labeling with Ceph C-T (red) and Boc-FL (green) in live cells. NBD: nitrobenzofurazan, SR-SIM: super-resolution structured illumination microscopy, PBPs: penicillin-binding proteins.
The brightness of NBD (ε = 1.54 × 104 M−1 cm−1, and ϕ = 0.55) fluorophore was relatively lower than that of other probes, which may limit the investigation of detailed information [61]. BODIPY (ε = 9.1 × 104 M−1 cm−1, and ϕ = 0.94), fluorescein (ε = 9.3 × 104 M−1 cm−1, and ϕ = 0.95), or rhodamine (ε = 7.4 × 104 M−1 cm−1, and ϕ = 0.92) are commercially available fluorophores with excellent light brightness, and widely employed in super resolution imaging applications [62]. BODIPY-derived penicillin V (a β-lactam antibiotic), BOCILLIN-FL (Boc-FL), is typical commercial probe. It was first developed in 1999 with the aim of detecting and studying the expression and folding of PBPs [[63], [64], [65]]. PBPs are an important membrane protein involved in cell wall peptidoglycan synthesis and they can be inhibited by β-lactam antibiotics. Thus, Boc-FL is widely used to investigate the structure [66,67] and activity [[68], [69], [70]] of PBPs, and antibiotics mode of action [[71], [72], [73], [74], [75]]. The roles of PBPs in live Gram-positive bacteria were also studied with SR-SIM imagings by combination Boc-FL and rhodamine-derived cephalosporin C (Ceph C-T, structures are shown in Fig. 2G) [76]. SR-SIM imaging of B. subtilis and Streptococcus pneumoniae (S. pneumoniae) divisional septa labeled by Ceph C-T and Boc-FL show greater details compared to the image of wide-field fluorescence microscopy. Comparatively little overlap of the stains was observed as shown in Figs. 2H and I, which indicates that different populations of PBPs are active at discrete locations in the midcell during division of B. subtilis and S. pneumoniae.
4.2. Fluorescent imaging of bacterial infection in live animals
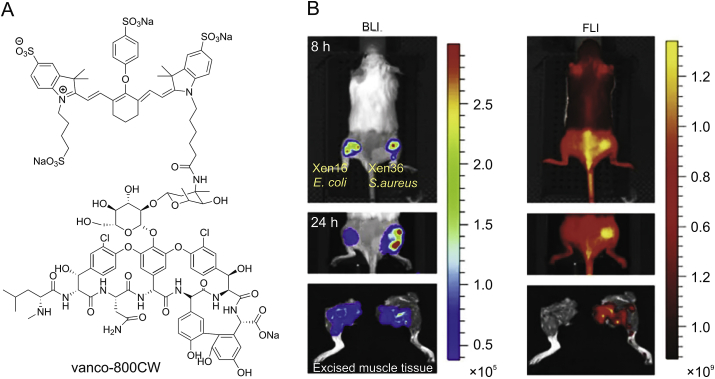
Optical imaging of bacterial infection in living animals is emerging as a powerful tool to study preclinical models of infectious disease [[77], [78], [79]]. Near-infrared (NIR) fluorophores with emission wavelengths in the region of 650–900 nm have been developed to target specific tissues that are otherwise difficult to image [80,81]. Oosten’s group [39] created a NIR probe vanco-800CW by covalently linking IRDye 800CW (a derivative of Cy7 fluorophore) to vancomycin. As shown in Fig. 3, the probe was successfully targeted to Gram-positive bacterial infections in living animals and a human cadaver. And it also can discriminate bacterial infections from sterile inflammation in vivo, indicating the sufficient resolution of the probe. The NIR fluorophores Cy5 and Cy5.5 as fluorescence resonant energy transfer donor-acceptor pair were further attached to cephalosporin constructing ratiometric substrate of β-Lactamase (the mechanism is described in next section), which was used for the real-time imaging of pulmonary infections and rapid quantification of bacteria in live mice [82]. The NIR fluorescent antibiotics were anticipated as a promising clinical optical imaging agent to fight against severe bacterial infections in the future.
Fig. 3.
(A) Near-infrared fluorescent antibiotic vanco-800CW [39] and (B) its real-time imaging of bacterial infections in living mice. Left side: bioluminescence imaging; right side: fluorescence imaging (excitation 745 nm, emission 840 nm).
4.3. Fluorescent real-time detection of bacteria
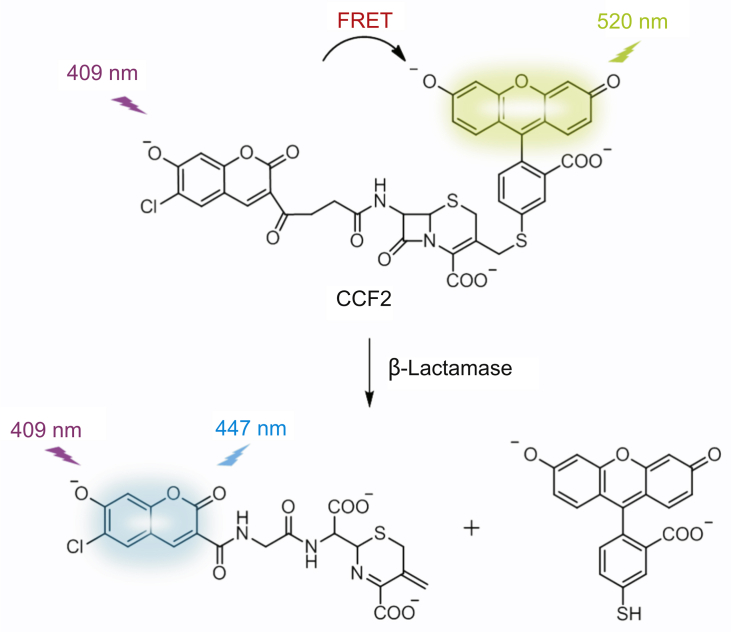
The fluorescent real-time detection requires a turn-on or ratiometric signal of the designed probe while recognizing a target. Turn-on probes exhibit no/weak fluorescence in their native states, and then they display a selective enhanced fluorescence after binding to objects. Rao and colleagues developed turn-on probes with high selectivity for Mycobacterium tuberculosis (Mtb) protein β-Lactamase (BlaC, an enzyme produced by resistant bacteria that degradesβ-lactam antibiotics by destroying the cyclic core) for real-time and accurate detection of very low numbers of Mtb in patient sputum [83,84]. Furthermore, several groups have created ratiometric β-lactam probes by linking fluorescence resonant energy transfer (FRET) donor-accept fluorophores to β-lactam antibiotic. Zlokarnik and coworkers [85] first reported a ratiometric substrate of β-Lactamase in 1998 for real-time monitoring gene expression. 7-hydroxycoumarin and fluorescein as FRET donor-acceptor pair were attached to the 7 and 3′ positions of a cephalosporin, respectively, creating the FRET substrate CCF2 (Fig. 4). It emitted acceptor fluorescein fluorescence at 520 nm when excited at 7-hydroxycoumarin’s wavelength 409 nm. β-Lactamase could cleave CCF2 to separate the donor and acceptor. And then the donor emitted fluorescence at 447 nm. Using this ratiometric probe, β-Lactamase was detected in real-time to measure gene expression in single live mammalian cells with high sensitivity. In addition, various ratiometric fluorescent antibiotics based on cephalosporin were developed by attaching different fluorophores [40,86,87]. Ratiometric fluorescent probe C-2 with fluorescein and rhodamine donor-acceptor pair was used for screening microbes resistant to methoxyimino cephalosporins [87].
Fig. 4.
Schematic representation of ratiometric signal generation process of CCF2 probe while degrated by β-Lactamase.
5. Conclusion
In this review, we have provided a brief introduction of fluorescent antibiotics for physiological studies on pathogenic bacteria in recent years. These studies were done by fluorescent tracking of antibiotics in live cells using autofluorescent antibiotics or fluorophore-antibiotics conjugates. Most probes have potential values for more extensive and in-depth research. Looking forward, the use of bio-specific small molecule probes with high specificity carries both a great potential and significant challenges. To date, the current understanding of microorganisms is still limited, and more excellent fluorophores should try to be incorporated into antibiotics. Meanwhile, super resolution fluorescence imaging is expected to study more details of the physiological processes of pathogenic bacteria. This may help researchers develop new antibiotics to prevent the potential outbreak of resident evil caused by super-resistant bacteria in the near future.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
We are grateful for the financial support from the National Natural Science Foundation of China (21878286, 21908216, 21576043) and Dalian Institute of Chemical Physics (DICPI201938, DICP I202006).
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Contributor Information
Xiaolian Li, Email: zcxu@dicp.ac.cn.
Zhaochao Xu, Email: xlianlid@163.com.
References
- 1.Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature. 2017;543:15. doi: 10.1038/nature.2017.21550. [DOI] [PubMed] [Google Scholar]
- 2.Jones K.E., Patel N.G., Levy M.A. Trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alekshun M.N., Levy S.B. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Piddock L.J.V. Reflecting on the final report of the O’Neill review on antimicrobial resistance. Lancet Infect. Dis. 2016;16:767–768. doi: 10.1016/S1473-3099(16)30127-X. [DOI] [PubMed] [Google Scholar]
- 5.Payne D.J., Gwynn M.N., Holmes D.J. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y.C., Chuang Y.S., Chen Y.Y. Bacteria detection utilizing electrical conductivity. Biosens. Bioelectron. 2008;23:1856–1861. doi: 10.1016/j.bios.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Davies S.C., Fowler T., Watson J. Annual report of the chief medical officer, infection and the rise of antimicrobial resistance. Lancet. 2013;381:1606–1609. doi: 10.1016/S0140-6736(13)60604-2. [DOI] [PubMed] [Google Scholar]
- 8.El Chakhtoura N.G., Saade E., Iovleva A. Therapies for multidrug resistant and extensively drug-resistant non-fermenting gram-negative bacteria causing nosocomial infections: a perilous journey toward ’molecularly targeted’ therapy. Expert Rev. Anti Infect. Ther. 2018;16:89–110. doi: 10.1080/14787210.2018.1425139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alekshun M.N., Levy S.B. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Cama J., Henney A.M., Winterhalter M. Breaching the barrier: quantifying antibiotic permeability across gram-negative bacterial membranes. J. Mol. Biol. 2019;431:3531–3546. doi: 10.1016/j.jmb.2019.03.031. [DOI] [PubMed] [Google Scholar]
- 11.Benito-Pena E., Moreno-Bondi M.C., Orellana G. Development of a novel and automated fluorescent immunoassay for the analysis of beta-lactam antibiotics. J. Agric. Food Chem. 2005;53:6635–6642. doi: 10.1021/jf0511502. [DOI] [PubMed] [Google Scholar]
- 12.Escobedo J.O., Chua Y.H., Wang Q. Live cell imaging of a fluorescent gentamicin conjugate. Nat. Prod. Commun. 2012;7:317–320. [PMC free article] [PubMed] [Google Scholar]
- 13.Vashist J., Tiwari V., Das R. Analysis of penicillin-binding proteins (PBPs) in carbapenem resistant acinetobacter baumannii. Indian J. Med. Res. 2011;133:332–338. [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher S.S., Chaoran J., Darcy S.P. A trimethoprim-based chemical tag for live cell two-photon imaging. Chembiochem. 2010;11:782–784. doi: 10.1002/cbic.200900731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarzembowski T., Wisniewska K., Jozwik A. Heterogeneity of methicillin-resistant staphylococcus aureus strains (MRSA) characterized by flow cytometry. Curr. Microbiol. 2009;59:78–80. doi: 10.1007/s00284-009-9395-x. [DOI] [PubMed] [Google Scholar]
- 16.Jarzembowski T., Wisniewska K., Jozwik A. Flow cytometry as a rapid test for detection of penicillin resistance directly in bacterial cells in enterococcus faecalis and staphylococcus aureus. Curr. Microbiol. 2008;57:167–169. doi: 10.1007/s00284-008-9179-8. [DOI] [PubMed] [Google Scholar]
- 17.Stone M.R.L., Butler M.S., Phetsang W. Fluorescent antibiotics: new research tools to fight antibiotic resistance. Trends Biotechnol. 2018;36:523–536. doi: 10.1016/j.tibtech.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Dubuy H.G., Riley F., Showacre J.L. Tetracycline fluorescence in permeability studies of membranes around intracellular parasites. Science. 1964;145:163–165. doi: 10.1126/science.145.3628.163. [DOI] [PubMed] [Google Scholar]
- 19.Horvath J.J., Glazier S.A. Fluorescence measurement of tetracycline in model fermentation media samples containing streptomyces aureofaciens cell mass. Appl. Spectrosc. 1996;50:327–333. [Google Scholar]
- 20.Crissman H.A., Stevenson A.P., Orlicky D.J. Detailed studies on the application of 3 fluorescent antibiotics for DNA staining in flow cytometry. Stain Technol. 1978;53:321–330. doi: 10.3109/10520297809111954. [DOI] [PubMed] [Google Scholar]
- 21.Crissman H.A., Tobey R.A. Specific staining of DNA with the fluorescent antibiotics, mithramycin, chromomycin, and olivomycin. Trends Cell Biol. 1990;33:97–103. doi: 10.1016/s0091-679x(08)60515-4. [DOI] [PubMed] [Google Scholar]
- 22.Saunders G.C., Martin J.C., Jett J.H. Flow cytometric competitive-binding assay for determination of actinomycin-d concentrations. Cytometry. 1990;11:311–313. doi: 10.1002/cyto.990110213. [DOI] [PubMed] [Google Scholar]
- 23.Mikhailova G.R., Berdnikova Z.E. Use of the DNA-specific fluorochrome olivomycin for cell culture studies. Tsitol. Genet. 1991;25:15–20. [PubMed] [Google Scholar]
- 24.Vekshin N.L., Kovalev V.I. Nucleotide carriers for anti-tumour actinomycin antibiotics. J. Biochem. 2016;159:59–66. doi: 10.1093/jb/mvv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta A., Mumtaz S., Li C.H. Combatting antibiotic-resistant bacteria using nanomaterials. Chem. Soc. Rev. 2019;48:415–427. doi: 10.1039/c7cs00748e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dik D.A., Fisher J.F., Mobashery S. Cell-wall recycling of the gram-negative bacteria and the nexus to antibiotic resistance. Chem. Rev. 2018;118:5952–5984. doi: 10.1021/acs.chemrev.8b00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breukink E., de Kruijff B. Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 2006;5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 28.Velkov T., Roberts K.D., Nation R.L. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol. 2013;8:711–724. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapiro A.B. Investigation of beta-lactam antibacterial drugs, beta-lactamases, and penicillin-binding proteins with fluorescence polarization and anisotropy: a review. Methods Appl. Fluoresc. 2016;4 doi: 10.1088/2050-6120/4/2/024002. [DOI] [PubMed] [Google Scholar]
- 30.Macheboeuf P., Contreras-Martel C., Job V. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 2006;30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 31.Vollmer W., Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Sauvage E., Kerff F., Terrak M. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. Fems. Microbiol. Revi. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 33.Ropy A., Cabot G., Sanchez-Diener I. Role of pseudomonas aeruginosa low-molecular-mass penicillin-binding proteins in AmpC expression, beta-lactam resistance, and peptidoglycan structure. Antimicrob. Agents Chemother. 2015;59:3925–3934. doi: 10.1128/AAC.05150-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moisan H., Pruneau M., Malouin F. Binding of ceftaroline to penicillin-binding proteins of staphylococcus aureus and Streptococcus pneumoniae. J. Antimicrob. Chemother. 2010;65:713–716. doi: 10.1093/jac/dkp503. [DOI] [PubMed] [Google Scholar]
- 35.Clausell A., Rabanal F., Garcia-Subirats M. Membrane association and contact formation by a synthetic analogue of polymyxin B and its fluorescent derivatives. J. Phys. Chem. B. 2006;110:4465–4471. doi: 10.1021/jp0551972. [DOI] [PubMed] [Google Scholar]
- 36.Vince R., Weiss D., Pestka S. Binding of n-substituted erythromycylamines to ribosomes. Antimicrob. Agents Chemother. 1976;9:131–136. doi: 10.1128/aac.9.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calloway N.T., Choob M., Sanz A. Optimized fluorescent trimethoprim derivatives for in vivo protein labeling. Chembiochem. 2007;8:767–774. doi: 10.1002/cbic.200600414. [DOI] [PubMed] [Google Scholar]
- 38.Haugland R.P. 10th Edition. Molecular Probes; Eugene, OR: 2005. The Handbook: A Guide to Fluorescent Probes and Labeling Technologies. [Google Scholar]
- 39.van Oosten M., Schafer T., Gazendam J.A. Real-time in vivo imaging of invasive- and biomaterial-associated bacterial infections using fluorescently labelled vancomycin. Nat. Commun. 2013;4:2584. doi: 10.1038/ncomms3584. [DOI] [PubMed] [Google Scholar]
- 40.Mao W., Qian X., Zhang J. Specific detection of extended-spectrum beta-Lactamase activities with a ratiometric fluorescent Probe. Chembiochem. 2017;18:1990–1994. doi: 10.1002/cbic.201700447. [DOI] [PubMed] [Google Scholar]
- 41.Chen H., Liu C., Chen D. Bacteria-targeting conjugates based on antimicrobial peptide for bacteria diagnosis and therapy. Mol. Pharm. 2015;12:2505–2516. doi: 10.1021/acs.molpharmaceut.5b00053. [DOI] [PubMed] [Google Scholar]
- 42.Tiyanont K., Doan T., Lazarus M.B. Imaging peptidoglycan biosynthesis in bacillus subtilis with fluorescent antibiotics. P. Nati. Acad. Sci. USA. 2006;103:11033–11038. doi: 10.1073/pnas.0600829103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yun B., Azad M.A.K., Nowell C.J. Cellular uptake and localization of polymyxins in renal tubular cells using rationally designed fluorescent probes. Antimicrob. Agents Chemother. 2015;59:7489–7496. doi: 10.1128/AAC.01216-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yun B., Roberts K.D., Thompson P.E. Design and evaluation of novel polymyxin fluorescent probes. Sensors. 2017;17:2598. doi: 10.3390/s17112598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phetsang W., Blaskovich M.A.T., Butler M.S. An azido-oxazolidinone antibiotic for live bacterial cell imaging and generation of antibiotic variants. Bioorg. Med. Chem. 2014;22:4490–4498. doi: 10.1016/j.bmc.2014.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stone M.R.L., Masi M., Phetsang W. Fluoroquinolone-derived fluorescent probes for studies of bacterial penetration and efflux. MedChemComm. 2019;10:901–906. doi: 10.1039/c9md00124g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phetsang W., Pelingon R., Butler M.S. Fluorescent trimethoprim conjugate probes to assess drug accumulation in wild type and mutant escherichia coli. ACS Infect. Dis. 2016;2:688–701. doi: 10.1021/acsinfecdis.6b00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soon R.L., Velkov T., Chiu F. Design, synthesis, and evaluation of a new fluorescent probe for measuring polymyxin-lipopolysaccharide binding interactions. Anal. Biochem. 2011;409:273–283. doi: 10.1016/j.ab.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kocaoglu O., Carlson E.E. Progress and prospects for small-molecule probes of bacterial imaging. Nat. Chem. Biol. 2016;12:472–478. doi: 10.1038/nchembio.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Long S., Qiao Q., Miao L., Xu Z. A self-assembly/disassembly two-photo ratiometric fluorogenic probe for bacteria imaging. Chin. Chem. Lett. 2019;30:573–576. [Google Scholar]
- 51.Chan J., Dodani S.C., Chang C.J. Reaction-based small-molecule fluorescent probes for chemoselective bioimaging. Nat. Chem. 2012;4:973–984. doi: 10.1038/nchem.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li M., Li Y., Wang X. Synthesis and application of near-infrared substituted rhodamines. Chin. Chem. Lett. 2019;30:1682–1688. [Google Scholar]
- 53.Daniel R.A., Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 54.Wang W., Zhu Y., Chen X. Selective imaging of Gram-negative and Gram-positive microbiotas in the mouse Gut. Biochemistry. 2017;56:3889–3893. doi: 10.1021/acs.biochem.7b00539. [DOI] [PubMed] [Google Scholar]
- 55.Matijasic M., Kos V.M., Nujic K. Fluorescently labeled macrolides as a tool for monitoring cellular and tissue distribution of azithromycin. Pharmacol. Res. 2012;66:332–342. doi: 10.1016/j.phrs.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Pogliano J., Pogliano N., Silverman J.A. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J. Bacteriol. 2012;194:4494–4504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang W., Chen X. Antibiotics-based fluorescent probes for selective labeling of Gram-negative and Gram-positive bacteria in living microbiotas. Sci. China Chem. 2018;61:792–796. [Google Scholar]
- 58.Koch A.L. What size should a bacterium be? A question of scale. Annu. Rev. Microbiol. 1996;50:317–348. doi: 10.1146/annurev.micro.50.1.317. [DOI] [PubMed] [Google Scholar]
- 59.Huang B., Bates M., Zhuang X. Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao Liang, Shao Lin, Christopher D., Higgins Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens. Cell. 2012;151:1370–1385. doi: 10.1016/j.cell.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benson S., Fernandez A., Barth N.D. SCOTfluors : Small, conjugatable,orthogonal, and tunable fluorophores for in vivo imaging of cell metabolism. Angew. Chem. Int. Ed. Engl. 2019;58:6911–6915. doi: 10.1002/anie.201900465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng F., Xu Z. Heteroatom-substituted rhodamine dyes: structure and spectroscopic properties, Chin. Chem. Lett. 2019;30:1667–1681. [Google Scholar]
- 63.Zhao G.S., Meier T.I., Kahl S.D. BOCILLIN FL, A sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 1999;43:1124–1128. doi: 10.1128/aac.43.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao G., Meier T.I., Hoskins J. Penicillin-binding protein 2a of streptococcus pneumoniae: expression in escherichia coli and purification and refolding of inclusion bodies into a soluble and enzymatically active enzyme, Protein Expres. Purif. 1999;16:331–339. doi: 10.1006/prep.1999.1080. [DOI] [PubMed] [Google Scholar]
- 65.Gee K.R., Kang H.C., Meier T.I. Fluorescent bocillins: synthesis and application in the detection of penicillin-binding proteins. Electrophoresis. 2001;22:960–965. doi: 10.1002/1522-2683()22:5<960::AID-ELPS960>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 66.Moya B., Beceiro A., Cabot G. Pan-β-lactam resistance development in Pseudomonas aeruginosa clinical strains: molecular mechanisms, penicillin-binding protein profiles, and binding affinities. Antimicrob. Agents Chemother. 2012;56:4771–4778. doi: 10.1128/AAC.00680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maurer P., Todorova K., Sauerbier J. Mutations in streptococcus pneumoniae penicillin-binding protein 2x: importance of the c-terminal penicillin-binding protein and serine/threonine kinase-associateddomains for beta-lactam binding. Microb. Drug Resist. 2012;18:314–321. doi: 10.1089/mdr.2012.0022. [DOI] [PubMed] [Google Scholar]
- 68.Shapiro A.B., Gu R.F., Gao N. Continuous fluorescence anisotropy-based assay of BOCILLIN FL penicillin reaction with penicillin binding protein 3. Anal. Biochem. 2013;43:37–43. doi: 10.1016/j.ab.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 69.Gonzalez-Leiza, de Pedro M.A., Ayala J.A. AmpH, A bifunctional ddendopeptidase and dd-carboxypeptidase of escherichia coli. J. Bacteriol. 2011;193:6887–6894. doi: 10.1128/JB.05764-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rahman M.A., Cheng Z., Matsuo J. Penicillin-binding protein of ehrlichia chaffeensis: cytokine induction through MyD88-dependent pathway. J. Infect. Dis. 2012;206:110–116. doi: 10.1093/infdis/jis313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lemaire S., Glupczynski Y., Duval V. Activities of ceftobiprole and other cephalosporins against extracellular and intracellular (THP-1 macrophages and keratinocytes) forms of methicillin-susceptible and methicillin-resistant staphylococcus aureus. Antimicrob. Agents Chemother. 2009;53:2289–2297. doi: 10.1128/AAC.01135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fedarovich A., Djordjevic K.A., Swanson S.M. High-throughput screening for novel inhibitors of neisseria gonorrhoeae penicillin-binding protein 2. PloS One. 2012;7 doi: 10.1371/journal.pone.0044918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dzhekieva L., Kumar I., Pratt R.F. Inhibition of bacterial DD-peptidases (Penicillin-Binding proteins) in membranes and in vivo by peptidoglycan-mimetic boronic acids. Biochemistry. 2012;51:2804–2811. doi: 10.1021/bi300148v. [DOI] [PubMed] [Google Scholar]
- 74.June C.M., Vaughan R.M., Ulberg L.S. A fluorescent carbapenem for structure function studies of penicillin-binding proteins, beta-lactamases, and beta-lactam sensors. Anal. Biochem. 2014;463:70–74. doi: 10.1016/j.ab.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kocaoglu O., Tsui H.C.T., Winkler M.E. Profiling of beta-lactam selectivity for penicillin-binding proteins in streptococcus pneumoniae D39. Antimicrob. Agents Chemother. 2015;59:3548–3555. doi: 10.1128/AAC.05142-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kocaoglu O., Calvo R.A., Sham L.-T. Selective penicillin-binding protein imaging probes reveal substructure in bacterial cell division. ACS Chem. Biol. 2012;7:1746–1753. doi: 10.1021/cb300329r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bunschoten A., Welling M.M., Termaat M.F. Development and prospects of dedicated tracers for the molecular imaging of bacterial infections. Bioconjugate Chem. 2013;24:1971–1989. doi: 10.1021/bc4003037. [DOI] [PubMed] [Google Scholar]
- 78.Zhou H., Xiao Y., Hong X. New NIR-II dyes without a benzobisthiadiazole core. Chin. Chem. Lett. 2018;29:1425–1428. [Google Scholar]
- 79.Xu J., Shang L. Emerging applications of near-infrared fluorescent metal nanoclusters for biological imaging. Chin. Chem. Lett. 2018;29:1436–1444. [Google Scholar]
- 80.Hong G., Antaris A.L., Dai H. Near-infrared fluorophores for biomedical imaging. Nat. Biomed. Eng. 2017;1 0010. [Google Scholar]
- 81.Chang Z., Liu F., Wang L. Near-infrared dyes, Nanomaterials and proteins. Chin. Chem. Lett. 2019;30 1856–188. [Google Scholar]
- 82.Kong Y., Yao H., Ren H. Imaging tuberculosis with endogenous beta-lactamase reporter enzyme fluorescence in live mice. P. Nati. Acad. Sci. USA. 2010;107:12239–12244. doi: 10.1073/pnas.1000643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xie H., Mire J., Kong Y. Rapid point-of-care detection of the tuberculosis pathogen using a BlaC-specific fluorogenic probe. Nat. Chem. 2012;4:802–809. doi: 10.1038/nchem.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng Y., Xie H., Sule P. Fluorogenic probes with substitutions at the 2 and 7 Positions of cephalosporin are highly BlaC-specific for rapid mycobacterium tuberculosis detection. Angew. Chem. Int. Ed. Engl. 2014;53:9360–9364. doi: 10.1002/anie.201405243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zlokarnik G., Negulescu P.A., Knapp T.E. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science. 1998;279:84–88. doi: 10.1126/science.279.5347.84. [DOI] [PubMed] [Google Scholar]
- 86.Rukavishnikov A., Gee K.R., Johnson I. Fluorogenic cephalosporin substrates for beta-lactamase TEM-1. Anal. Biochem. 2011;419:9–16. doi: 10.1016/j.ab.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 87.Zhang J., Shen Y., May S.L. Ratiometric fluorescence detection of pathogenic bacteria resistant to broad-spectrum beta-lactam antibiotics. Angew. Chem. Int. Ed. Engl. 2012;51:1865–1868. doi: 10.1002/anie.201107810. [DOI] [PubMed] [Google Scholar]