Abstract
The human sweet taste receptor (T1R2) monomer—a member of the G-protein coupled receptor family that detects a wide variety of chemically and structurally diverse sweet tasting molecules, is known to pose a significant threat to human health. Protein that lack crystal structure is a challenge in structure-based protein design. This study focused on the interaction of the T1R2 monomer with rebaudioside A (Reb-A), a steviol glycoside with potential use as a natural sweetener using in-silico and biosensing methods. Herein, homology modelling, docking studies, and molecular dynamics simulations were applied to elucidate the interaction between Reb-A and the T1R2 monomer. In addition, the electrochemical sensing of the immobilised T1R2-Reb-A complex with zinc oxide nanoparticles (ZnONPs) and graphene oxide (GO) were assessed by testing the performance of multiwalled carbon nanotube (MWCNT) as an adsorbent experimentally. Results indicate a strong interaction between Reb-A and the T1R2 receptor, revealing the stabilizing interaction of the amino acids with the Reb-A by hydrogen bonds with the hydroxyl groups of the glucose moieties, along with a significant amount of hydrophobic interactions. Moreover, the presence of the MWCNT as an anchor confirms the adsorption strength of the T1R2-Reb-A complex onto the GO nanocomposite and supported with electrochemical measurements. Overall, this study could serve as a cornerstone in the development of electrochemical immunosensor for the detection of Reb-A, with applications in the food industry.
Subject terms: Carbon nanotubes and fullerenes, Electrochemistry
Introduction
The two steviol glycosides, rebaudioside A and stevioside, are natural sweeteners extracted from Stevia rebaudiana which belongs to the Asteraceae family1. Stevia sweeteners are obtained from the dried stevia leaves and the extracts are popularly used as a non-caloric substitute for sugar. Stevia contain steviol glycosides that are approximately 150–300 times sweeter than sugar. This plant-based sweetener is also believed to have a positive effect on blood sugar levels, with steviol glycoside as one of the natural sweeteners responsible for the increased blood sugar level in most diabetic patients2. Furthermore, the potential of steviol glycosides to induce sweet and bitter taste sensations have been reported3, however, its bitter aftertaste has long prevented it from becoming widely marketed as food and beverage sweeteners4.
Stevia extracts, besides having therapeutic properties, contain a high level of sweetening compounds known as steviol glycosides, which possess antioxidant, antimicrobial and antifungal activities5. It has also been reported that rebaudioside A (Reb-A) are mostly used as food additives for dairy products, confectionaries and beverages6,7. Most of the artificial sweeteners are low-calorie sweeteners responsible for some of the health-related threats posed by sucrose. The stability of these sweeteners at even higher temperatures have increased their presence in food application. Although there are no reports of stevia glycosides on the health risks of steviol glycosides, its widespread applications in the food industry needs to be regulated.
Reb-A is a diterpene glycoside, which contains a total of four glucose molecules wherein the central glucose is connected to the main steviol backbone, and the remaining glucose at its carboxyl group end, forming an ester bond (Fig. 1A), with 13 rotatable bonds, 14 hydrogen bond donors, and 23 hydrogen bond acceptors. On the other hand, Stevioside (Stv) is a glycoside consisting of the aglycone steviol and three glucose molecules (Fig. 1B)7.
Figure 1.
Chemical structures of (A) Rebaudioside A (Reb A) and (B) Stevioside (Stv).
The human sweet taste receptor (STR) is a heterodimer consisting of the T1R2 and TIR3 subunits which belongs to the class C G-protein coupled receptor (C-GPCR) family. Sucrose, glucose, sucralose and related sugars were reported to interact with the active sites of the amino terminal domain (ATD) of both T1R2 and T1R3 subunits8,9. For example, the dipeptidyl sweeteners such as aspartame and neotame appear to find a binding site in the T1R2 N-terminal domain, which can be considered as orthosteric agonists10,11. Similarly, Neoculin and brazzein are thought to interact with the N-terminal domain12 and the cysteine-rich area13 of human T1R3, respectively.
To date, few studies have reported the modelled structure of the T1R2 and T1R3 subunits of the STR family for comparative activities of the sweeteners10,14. However, the interaction between Reb-A and proteins such as bovine serum albumin, hT2R4 and hT2R14 bitter taste receptors have also been reported15,16.
Although the development of a cytochrome C biosensor for the detection of Reb-A has been previously reported in our laboratory using electrochemical and computational methods17, to the best of our knowledge, the development of a human T1R2 biosensor for the detection of Reb-A is yet to be reported in literature. In this study, the selection of a member of the T1R family as a candidate taste receptor was based on their physiology of taste perception embodied in the characteristics of T1R function. The T1R2/T1R3 receptor has a broad ligand specificity, recognizing monosaccharides to oligosaccharides, artificial sweeteners without saccharide groups, some D-amino acids and even proteins. However, T1R3 undergo mutagenesis and resulting in the lowering of its activity due to cysteine-rich domain of T1R3, but not in the case of T1R218. Therefore, T1R2 receptor has been chosen as a selective bio-recognition element for effective sensing platform. Accordingly, computational tools were employed to assess the feasibility of using T1R2 receptor in the development of an electrochemical immunosensor with unique features including higher sensitivity and selectivity.
Literature studies revealed that diverse analytical methods have been employed in the food industry to quantify steviol glycosides, such as Reb-A. Some of these instruments include liquid chromatography-mass spectrometry19,20, high performance liquid chromatography (HPLC)21,22, high performance thin layer chromatography23, and capillary electrophoretic methods24,25, among others26. In contrast, noteworthy advantages such as low cost, higher sensitivity and selectivity with quick response times, are attributed to electrochemical methods. One such method was reported by Komorsky-Lovrić et al.27. However, the non-availability of the human T1R2 crystal structure or the lack of information about its mechanism at an atomistic level may have influenced the lack of a human T1R2 immunosensor for the detection of Reb-A in literature. Therefore, this is a fertile ground for research and it is deemed timely to develop an immunosensor for the detection of Reb-A that have improved attributes such as better selectivity, sensitivity, robustness and first-rate catalytic activity.
Recently, nanomaterials have been employed in the modification of electrodes due to their recognized sensitive, reliable and selective determination of organic compounds. Accordingly, the incorporation of multiwalled carbon nanotubes (MWCNTs) could prove useful as a result of their widespread capabilities in sensing applications28 when used for the analysis of a wide range of analytes29–32. Generally, the surface sensitivity of MWCNTs further confirms their choice as an ideal material for nanoscale biosensor devices that require high sensitivity33,34. Similarly, graphene oxide (GO) is commonly used due to the attachment of the –OH, –COOH and –C–O–C– functional groups to its plane35. More recently, GO was used for the fabrication of biosensors such as the detection of organic molecules with pharmaceutical, food and environmental importance35–37.
Likewise, ZnONPs, being one of the most important metal oxide nanoparticles, are popularly employed in diverse fields due to their unique physical and chemical properties38,39. ZnONPs have attracted significant attention in the development of electrochemical biosensors39–41. This is due to their unique properties such as a suitable micro-environment for the immobilization/loading of enzymes and proteins while retaining their biological activity, electronic properties supporting different device categories, low cost, ease of preparation, biocompatibility, and catalytic surface activity. Thus, this has prompted their expanded use in the construction of electrochemical biosensors with enhanced analytical performance41–44. Thus far, the immobilization of two types of nanovesicles, which have human T1R2/T1R3 for the umami taste and sweet taste on their membranes, onto graphene surfaces for the simultaneous detection of umami and sweet tastants have been reported45. However, the immobilization of human T1R2 onto GO in the presence of ZnONPs and MWCNT for the detection of Reb-A is still unreported in literature.
Hence, the motivation and the need to use in-silico approaches, which have been shown to rapidly and accurately estimate the activities and properties of nanoparticles46,47, reduce costs and to determine predictive variables that can be used in guiding wet lab experiments48. In-silico methods accurately describe the molecular surface interactions for a better understanding of the reactions on surfaces. Surface studies are essential because they offer useful insights on the surface reactivity with likely changes of the nanoparticles, due to interactions with different elements49.
Herein, we provide information on the interaction between Reb-A and human T1R2 monomer as well as their physical interactions with MWCNT, ZnONPs and GO through the application of an array of methodologies such as homology modelling, molecular docking, molecular dynamics simulations, adsorption studies, and electrochemical biosensing. This study could serve as a cornerstone in the development of electrochemical immunosensor with enhanced selectivity and better performance.
Computational models and methods
Template selection
Due to the absence of a crystal structure of the human taste receptor 1 (T1R2), the 3D structure was modelled using the human taste receptor type 1 member 2 sequence obtained from Uniprot (Uniprot ID: Q8TE23) as template. Homology modelling was performed using the Chimera software50 as an interface to the Modeller Software version 9.151. The model was validated using the Ramachandran plot of the Schrodinger Maestro Suite52 and the active site residues were determined using the Site-Hound web program53.
The sequence of the target protein was uploaded unto PSIPRED V3.354,55 in order to obtain a predicted 3D secondary structure of the human T1R2. The assessment of the bond angles and torsional strain shows the evaluation of the predicted 3D structure. A Ramachandran plot for the analyses of bond angles and torsional strain was generated using Maestro52. MolProbity56 result shows 86.7% (726/837) of all residues were present in favoured (98%) regions, while 94.9% (794/837) of all residues were in the allowed (> 99.8%) regions, with 5% (43/837) as outliers. The list also confirms that none of the active site residues were part of these outliers.
Molecular docking of the human T1R2-Reb-A complex
The 3D structure of Reb-A was retrieved from the PubChem database57 and docked onto the human T1R2 monomer homolog using Autodock Vina58–61. This software performs the prediction of the bound conformation based on free energy, calculated on the basis of the empirical force field and the Lamarckian Genetic Algorithm62. The Grid module was used to create a grid box of dimensions X = 56, Y = 70 and Z = 56 along the center X = 14.542, Y = − 16.714 and Z = 63.662 with a spacing of 0.375 Å. The efficiency of the predictions was amplified by setting the parameters associated with the Lamarckian genetic algorithm to the maximum efficiency values. The docked T1R2-Reb-A conformation with the highest docking score was selected for the molecular dynamics (MD) simulations. The resulting human T1R2-Reb-A complex was used for all subsequent methods performed in this study.
Molecular dynamics (MD) simulations
All atom molecular dynamics simulations have been performed on the docked complex using the AMBER 18 molecular dynamics package63,64. The bonded and non-bonded description of the interactions between the various atoms have been described using the AMBER18 force fields which include the GAFF65 force field parameters. Due to the lack of parameters needed for Reb-A in the Cornell et al. force field66, the missing parameters were generated. Optimization of the Reb-A was performed at the HF/6-31G* level with the Gaussian 09 package67. The human T1R2 homolog was energy minimized and equilibrated via molecular dynamics simulations and discussed as follows. The standard AMBER force field for bioorganic systems (ff03)68 was used to define the topology and parameter files for the T1R2 homolog and Reb-A using “GAFF”65 force field parameters based on the atom types of the force field model developed by Cornell et al.63. The topology and coordinate files were generated using the antechamber module of AMBER 18, and the missing hydrogen atoms were added and saved for the T1R2 homolog using the LEaP module in AMBER 18. Thereafter, the docked T1R2-Reb-A complex was solvated in the octahedron box of TIP3P water model64 with buffering distance of 10 Å between the protein surface and the box boundary as well as the neutralization was performed by adding 9 Na+ counterions to mimic the biological pH. Cubic periodic boundary conditions were imposed, and the long-range electrostatic interactions were treated with the particle-mesh Ewald method implemented in AMBER 18 with a non-bonding cut-off distance of 10 Å.
Initially, a series of energy minimization steps were performed to eliminate any bad contacts in the initially built structures while restraining the solute with 500 kcal/mol/Å2 harmonic force constant. The above system was minimized for 1000 cycles of steepest descent followed by 2000 cycles of conjugate gradient. The entire system was freely minimized for 1000 iterations. Heating was performed for 50 ps from 0 to 300 K with harmonic restraints of 5 kcal/mol Å2 using a Langevin thermostat with a coupling coefficient of 1/ps. The entire system was then equilibrated at 300 K with a 2 fs time step in the NPT ensemble for 500 ps and Berendsen temperature coupling69 was used to maintain a constant pressure at 1 bar. The SHAKE algorithm70 was employed on all atoms so as to constrain the bonds of all hydrogen atoms.
With no restraints imposed, a production run was performed for 100 ns in an isothermal isobaric (NPT) ensemble using a Berendsen barostat with a target pressure of 1 bar and a pressure-coupling constant of 2 ps for analysis of the energy stabilization and RMSD values of the complex. The coordinate file was saved every 1 ps and the trajectory was analyzed every 1 ps using the Ptraj module implemented in AMBER 18.
The thermodynamic property of the system was calculated. The free binding energy of Reb-A to the human T1R2 homolog active site was analysed by the Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) method71–73. A single trajectory approach was used with 100,000 snapshots at 100 ps interval of each simulation. From each snapshot, free binding energy (ΔGbind) was computed from the following equation:
| 1 |
where ΔGbind is the free binding energy; GhT1R2-Reb-A is the energy of the human T1R2-Reb-A complex; GhT1R2 is the energy of the human T1R2; and GReb-A is the energy of the Reb-A. Due to the high computational cost in the entropy calculation, 100 snapshots were extracted from the last equilibrated 100 ns trajectory of the simulation with 100 ps time intervals.
To theoretically evaluate the reliability of the calculated ΔG values, the standard error (SE) of the calculated free binding energy was estimated by using Eq. (2), which is related to the number (N) of snapshots chosen for the calculations60.
| 2 |
Nanomaterials modelling
All studied nanoclusters were built using Materials Studio (MS) BIOVIA74. Geometry optimization of the nanomaterials were performed with the Forcite module75 as implemented in the MS software. Forcite in the MS software is a classical molecular mechanics tool, designed to perform a range of tasks including fast energy calculations and geometry optimizations for single molecules as well as periodic systems. A detailed knowledge of surface interactions play a key role in the design of many materials and processes. An important first step in the preparation of a model of molecules adsorbed onto the surface, is to ensure that the geometries are fully optimized. Among the different steps involved in the modelling approach are the construction of the surface from the pure crystal, the addition of the molecules near the surface, the selection of an appropriate forcefield to study the nanomaterial interaction, followed by initial calculations of the energy and geometry optimization. Modelling and visualization were carried out with VMD76, GaussView77, and Materials studio (MS)74.
Herein, a 3-D model of the GO surface comprising of a pristine graphite structure, conforming to the standard structural database in the MS software was built with oxygen atoms manually added. Likewise, a 3-D model of ZnONPs was built to conform to the standard structural database in MS. A supercell structure (6 × 6 × 1) of ZnO was built and the symmetry was set to have a non-periodic boundary. The unit cell of the bulk ZnO (110) was geometry optimized using the Forcite module, with a 8 × 8 × 8 Monkhorst-Pack78 mesh k-points and a kinetic energy cut-off of 4.5 Å. The unit cell was then relaxed using the conjugate gradient method until the total forces acting on each atom were 8.401e−04 kcal/mol/A. The ZnO (110) surface was geometry optimized by solving the Kohn–Sham equation self-consistently under spin-unrestricted conditions79. The double-layered MWCNT (n = 10, m = 10) was built using the Build option in the MS software. To observe the feasibility of all structures, their primitive structures were optimized by Forcite module with fine-COMPASS force field75. The convergence criteria for the maximum values of energy alteration, force, stress, and displacement were set at 2 × 10−5 kcal/mol, 0.001 kcal/mol/Å, 0.001 GPa, and 10−5 Å respectively. Frequency calculations were carried out to determine if the compounds are at the lowest surface energy and to deduce physical interaction energies i.e. adsorption.
Next, the Adsorption Locator (AL) module as implemented in the MS software was used as a preparatory and screening tool with the forcefield method to obtain a ranking of the energies for each generated configuration, thereby indicating the preferred adsorption sites. Since the adsorbate can be adsorbed at different locations on the GO surface, the AL module was used to deduce the best adsorption site with the lowest energy on the surface. The AL applies Monte Carlo simulations within a lattice dynamics approach to obtain the best configuration. The generated adsorbate–substrate conformations for GO/ZnONPs/T1R2-RebA and GO/MWCNT/ZnONPs/T1R2-RebA were ranked and the lowest adsorption energy conformers were each optimized using the Forcite-Geometry optimization to reach a stable conformation as shown in Table 1.
Table 1.
Optimised geometries of nanomaterials used along with the minimized energies.
| Nanomaterial | Optimized 3-D structures | Total optimized energy kcal/mol |
|---|---|---|
| GO |

|
491.239 |
| MWCNT |

|
221,045.658 |
| ZnONPs |
|
288.891 |
The adsorption energies were calculated using the following equation:
| 3 |
where Eadsorbate is the energy of the adsorbate energy without the surface, Esurface is the surface energy of the ZnO nanoparticles and E adsorbate/surface is the total energy of the surface and the adsorbate. In this case, if Eads < 0; the adsorption energy implies stability and the chemical process is exothermic, with a more negative value implying a stronger adsorption energy.
Experimental method
Procedure for the fabrication of Pt-E with nanocomposite GO/ZnONPs
In this study, ZnONPs were synthesised from zinc gluconate using hydrothermal method. Then the GO solution was prepared according to Bathinapatla et al.17, by quantitatively measuring 1 ml of GO into 10 ml of deionized distilled water. The ZnONPs/GO nanocomposite was achieved by mixing ZnONPs dispersion and GO dispersion in a ratio of 1:1 and ultrasonicated for 1 h to obtain a homogenized mixture. T1R2 which serves as a biological recognition element for Reb A was drop-casted on the surface of GO modified Pt-E and left to stand at room temperature for 1 h before analysis. The finally prepared electrode was utilized to detect Reb A at lower concentration.
GO/MWCNTs/ZnONPs
To evaluate the significance of MWCNTs as an effective adsorbent, DMF was used as a dispersion medium. 0.5 g of MWCNTs was dispersed in a 2.5 ml of DMF and ultrasonicated for 1 h to make a thick solution. Then ZnONPs nanoparticles were added to the MWCNTs dispersion and sonicated at room temperature for 1 h to complete physio-adsorption of ZnONPs onto the surface of MWCNTs. This nanocomposite was coated on the Pt-E modified GO. Finally, T1R2 subunits were immobilized on the Pt-E/GO/MWCNTs to evaluate the efficiency of adsorbent in accommodating the Reb A.
Electrochemical evaluation of interaction of Reb-A with T1R2/ZnONPs/MWCNTs/GO/Pt-E
Electrochemical measurements to predict the interaction of Reb A with T1R2/ZnONPs/MWCNTs/GO/Pt-E was performed with differential pulse voltammetry studies at room temperature using three-electrode system electrochemical cell (VA797 Computrace, Metrohm, South Africa). The three-electrode system consist of platinum as the working electrode, graphite electrode as the counter electrode, and silver/silver chloride as the reference electrode. For measurements, the 20 ml electrochemical cell was filled with 10 ml of 0.1 M pH 11.0 borate buffer, 20 µ/of 100 mg/l of Reb A working standard.
Results and discussion
Modelling of the human sweet T1R2 taste receptor
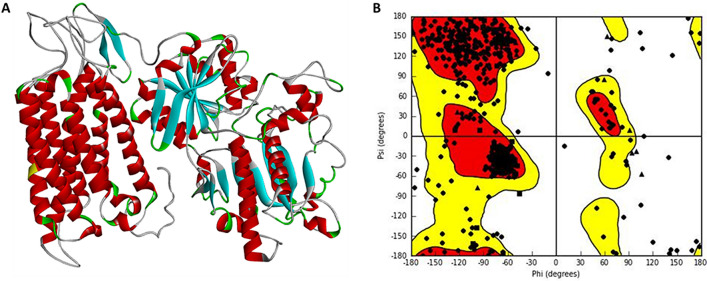
The generated 3-D structure of T1R2 monomer (Fig. 2A) was further subjected to optimization tools for energy minimization and the active site residues were determined and validated using the Site-Hound web program53. Thereafter, the stereo-chemical qualities of the predicted structures were assessed using the Ramachandran plot (Fig. 2B) in Maestro Schrodinger suite52, affirming the validation of the homolog.
Figure 2.
(A) Homology model of the Human Taste Receptor (T1R2) containing 839 amino acid residues (Image generated on Discovery studio v201680 license accessed through CHPC, Cape Town) (B) The Ramachandran Plot showing the validation of the T1R2 monomer (Image generated using the Schrodinger Maestro Suite v201852) .
Closest active site residues were selected and used for further modelling studies. The obtained active site residues for the human T1R2 homolog are Thr183, Pro185, Ile327, Gln328, Ser329, Val330, Arg383, Val384, Tyr386, Ser387, His434, Gln435, Ile436, Phe437, Val444, Ala445, His447, and Arg837. Gln328, Asp433, His434, His447, Tyr469, Gln472, Leu575, Arg837, Arg838, Asp839. The MolProbity56 model validation show 86.7% (726/837) of the residues are in the favoured regions, while 94.9% (794/837 of the residues are in the allowed regions, with 5% (43/837) of the residues as outliers. This list confirms that none of the active site residues reported in this study are part of the outliers (see Supplementary Material). Thus, the above analysis serves as a basis that the predicted 3D structure of the human T1R2 monomer is of a good quality and can be used in this study.
Molecular docking
As docking is the most appropriate tool to provide a better description of the binding theme of inhibitors81,82, molecular docking was performed to elucidate the binding modes of the human T1R2 and Reb-A complex. To this end, the binding affinity energy with 0 Å distance of deviation from the active site was − 8.7 kcal/mol (Fig. 3A,B).
Figure 3.
(A) The docked structure of the T1R2-Reb-A complex. (B) The Reb-A interaction with the human T1R2 monomer showing the distances between the active site residues and Reb-A in green and red colours, corresponding to a binding affinity of − 8.7 kcal/mol. (Images generated on Discovery studio v201680 license accessed through CHPC, Cape Town).
Figure 4 shows the binding modes of Reb-A with the active site residues of the T1R2 receptor depicting major hydrophobic interactions from the active site residues such as Ala43, Tyr62, Val64, Val66, and Met249; hydrogen bond interactions from Asn312, Asn361, Arg378, and Asn460.
Figure 4.
Reb A-T1R2 complex interaction at the predicted binding sites. Major contributions are from the residues that exhibit hydrophobic interactions (in green bubbles), corresponding to a binding affinity of − 8.7 kcal/mol (Image generated using the Schrodinger Maestro Suite v201852).
The hydrophobic interactions are located around the insoluble region of the Reb-A while the hydrogen bonds are formed in the most soluble regions. In addition, π–π stacking interactions and non-covalent bonds between the two aromatic rings have been observed in the respective residues Leu313–Leu316, Leu279–Tyr282, and Thr375–Arg378. The contribution of each of the active site residues in terms of their energy contribution are depicted in Fig. 5.
Figure 5.
Per residue energy contributions of the active site residue to the T1R2-Reb-A complex generated on the Origin data analysis tool v6.0 (https://www.originlab.com/).
Likewise, the major energy contributions (kcal/mol) were observed from hydrophobic residues such as Tyr62 (− 16.265), Pro308 (− 13.969), Val309 (− 13.612), and polar residue such as Asn312 (− 17.100) and Asn360 (− 12.864).
Thereafter, the structural dynamics of the Reb-A complexed with the human T1R2 homolog was analysed by performing molecular dynamics simulations83–87 and free binding energy calculations88,89, which have proved to be very useful and successful in understanding the molecular basis of small molecules interacting with different biological targets including GPCR agonists90,91 and sweet taste receptors86,87. An MD simulation provides useful structural and energetic information about the interaction between the inhibitors (or molecules) and the target receptor. Studies have reported the interaction of steviol glycosides with human taste receptors experimentally92 and from a modelling perspective16,93,94. As previously reported in a study by Zhang et al.95, the Reb-A binds with the hinge region of the T1R2 venus flytrap domain. Foremost, the template chosen by Zhang et al. has low overall sequence identity of T1R2 to mGluR1 (~ 30%), yet, this study showed significant binding activity by residues closer to the hinge side of the T1R2 receptor. In this study, Reb-A binds to the human T1R2 homolog at the hinge region, very close to the V-shaped domain (discussed in Zhang et al.). As there is little information about the precise binding site of the human T1R2 experimentally, it is believed that the reported binding site by Zhang F. et al., with slight variation in the angle of the active site residues, further support the findings observed in the current study.
Molecular dynamics (MD) simulations analysis
In the present study, MM/GBSA was applied as it has been demonstrated to be more efficient for protein-drug systems96–98, carbohydrates99 and nucleic acids100. It utilizes a fully pairwise potential that is useful for decomposing the total binding free energy into atomic or group contributions101. This method explores the type of established interactions, calculating separately the components of the internal energy, the interaction energy and the free energy of Gibbs of solvation102. For the analysis of the binding free energy, 100 snapshots at a time interval of 10 ps were extracted from 100 ns of the MD trajectories. The system showed high interaction as evident from its binding affinity value (− 8.7 kcal/mol) and its free binding energy of − 60.44 kcal/mol.
In addition, the system stability and overall convergence of simulations were monitored in terms of Root Mean-Square Deviation (RMSD) and fluctuations of the human T1R2 backbone atoms. The RMSD and enabled us to verify that equilibration was achieved as illustrated in Fig. 6A. Similarly, the Root Mean Square Fluctuations (RMSF) of the average structure from the trajectory were computed to assess if the simulation results are in accordance with the crystal structure, and the results are shown in Fig. 6B.
Figure 6.
Root Mean Square Deviation (RMSD) and Root mean Square Fluctuation (RMSF) plots of the apo and bound T1R2 complex generated on the Origin data analysis tool v6.0 (https://www.originlab.com/).
The RMSD plots for T1R2 Apo and T1R2-Reb-A-complex shown in Fig. 6A, illustrates slight variations > 2 Å, confirming the stability of the equilibration after 40 ns. At approximately 70 ns, both systems reached a plateau convergence, which suggests that the delay experienced by the complex was due to the presence of the Reb-A. Overall, this plot showed the stability of the human T1R2 backbone.
Similarly, the RMSF plots shown in Fig. 6B indicate the temperature profiles of each amino acid residue. The RMSF plot of both apo and bound show a similar trend in the fluctuations of the amino acid residues. While it may be suggested that some residues were more mobile than the rest, probably due to the absence of a ligand in the apo, the T1R2-Reb-A complex demonstrated a stable structure upon binding.
The radius of gyration (Rg), which studies the kinetics and thermodynamics of protein folding, is an indicator of the compactness of a protein and the stability of biomolecular structures103, was calculated for the T1R2-Reb-A complex. Topology of the Cα backbone has been found to play a role in protein folding104. Therefore, to measure the overall protein folding/unfolding and the compactness of the T1R2 protein, Rg change over time was analysed (see Fig. 7).
Figure 7.
Radius of Gyration (Rg) plot for the apo and bound T1R2 complex generated on the Origin data analysis tool v6.0 (https://www.originlab.com/).
The average Rg of apo and bound T1R2 were compared and the overall Rg difference was observed between 5 and 100 ns for both systems are illustrated in Fig. 7. On average, the apo conformation showed an Rg value of 32.33 Å and 31.96 Å for the bound conformation. This explains the less compactness of the apo conformation of the T1R2. Generally, a decrease in Rg over time, as demonstrated by the T1R2-Reb-A complex, showed the stability of the system and its compactness. This indicates that the T1R2 demonstrates structural/conformational stability when bound to Reb-A. Generally, the RMSD, RMSF and the Rg indicate the conformational stability of the T1R2 homolog and the stability of the T1R2-Reb-A complex.
Following an in-depth study of the interaction of the T1R2-Reb-A complex, the second aspect of this work deals with their biomolecular interactions for sensing applications. The performance of these materials is often dependent on the properties of the interface, i.e. the degree of interaction with biomolecules at a molecular or atomistic level105. Several studies have also reported the role of nano surfaces in protein adsorption106–112.
Materials modelling
All geometry optimized structures of the individual nanomaterials used in this study along with the energy values are tabulated in Table 1.
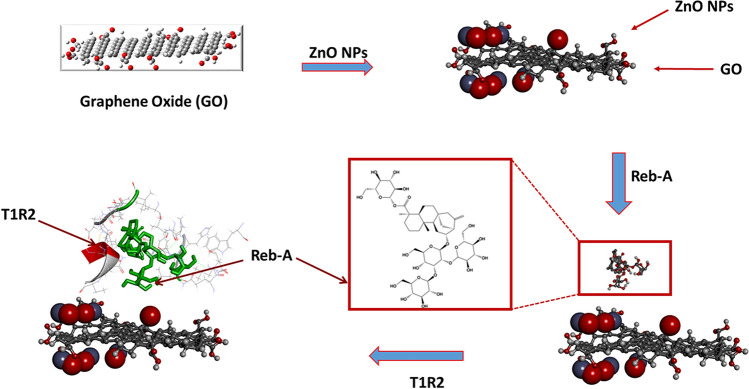
In line with the second key objective of this study, the performance of the interaction of the human T1R2-Reb-A complex on the surface of a (GO/ZnONPs) nanocomposite in the presence of MWCNT was assessed. Accordingly, a layer-by-layer approach was implemented with the interaction of GO/MWCNT/ZnONPs/T1R2-Reb-A described in Fig. 8 and for comparative purposes, the interaction in the absence of MWCNT for GO/ZnONPs/T1R2-Reb-A.
Figure 8.
The steps followed in the modelling of GO/MWCNT/ZnONPs with MWCNT.
Modelling of GO/MWCNT/ZnONPs with MWCNT
Using the optimized structures in Table 1 above, the modelling for Fig. 8 was undertaken in a stepwise fashion, with the ZnONPs adsorbed onto the surface of the GO, resulting in an adsorption energy of the ZnONP/GO complex (see Table 2) stabilized at − 13.852 kcal/mol. The adsorption performance and sensitivity of ZnONPs have been recently reported in literature113,114. As much as this study attempted to mimic experimental conditions, it is evident in Fig. 9A that only some of the ZnONPs take part in the interaction while others were a couple of angstroms away from the GO.
Table 2.
Layer-by-layer system with multiwalled carbon nanotube (MWCNT) along with the calculated adsorption energies for Scheme 1.
| Nano-systems | ∆G (adsorption energy) kcal/mol |
|---|---|
| GO/ZnONPs | − 13.852 |
| GO/MWCNT | − 170.447 |
| GO/MWCNT/ZnONPs | − 76.642 |
| GO/MWCNT/ZnONPs/T1R2-Reb-A | − 44.286 |
Figure 9.
(A) ZnONPs adsorbed onto GO. (B) Zoomed in representation of the interaction between the closest ZnONPs to the GO surface. ∆G = − 13.852 kcal/mol (Images generated on Materials studio74 license accessed through CHPC, Cape Town).
On closer inspection of Fig. 9B, it is clear that the inter-atomic distances between the ZnONPs and GO ranges between 3.072 and 5.340 Å. However, the modification of GO with MWCNT resulted in a considerable stabilization of the GO/MWCNT-complex as shown in Fig. 10.
Figure 10.
The physical adsorption of MWCNT onto GO. ∆G = − 170.447 kcal/mol (Image generated on Materials studio74 license accessed through CHPC, Cape Town).
According to Fig. 11, the atom–atom distances within 3.003 Å and 3.813 Å were measured with respect to the nearest atoms from MWCNT and GO surface. These observed interatomic distances for adsorbent–adsorbate interaction is comparable to recent theoretical115 and experimental investigations on the adsorption performance of GO116 and functionalised MWCNT117, wherein the result of the FTIR spectra established the presence of several functional groups. Farghali et al.117 reported that numerous chemical sorption sites on MWCNTs surface could have been due to these functional groups. This further validated the reason behind the high adsorption energy of GO/MWCNT in the current study (∆G = − 170.447 kcal/mol).
Figure 11.
(A) Top view of the adsorption of MWCNT and ZnONPs on Graphene oxide. (B) A zoomed in representation of the adsorption of MWCNT and ZnONPs on Graphene oxide showing the distances between ZnONPs, MWCNT, and Graphene oxide. ∆G = − 76.642 kcal/mol (Images generated on Materials studio74 license accessed through CHPC, Cape Town).
Figure 11 illustrates a further adsorption with the ZnONPs and the MWCNT with an energy value of − 76.642 kcal/mol.
According to Fig. 11A, the surface area of the GO was adequately utilised by the MWCNT and ZnONPs. As depicted in Fig. 11B, the shortest atom–atom distance between ZnONPs and GO was 2.632 Å. In comparison, the distance between the closest atoms of ZnONPs and MWCNT was 3.052 Å, while it was 3.004 Å for GO and MWCNT. The large difference between the adsorption energy when ZnONPs are included (∆G = − 76.642 kcal/mol) and excluded (∆G = − 170.447 kcal/mol) could be due to a better interaction of the ZnONPs at the surface of the GO. On a closer inspection of Fig. 11B, it is noticeable that the ZnONPs are adsorbed more on the side where MWCNT is absent. To find a suitable explanation for this, another calculation was carried out to provide an insight into the adsorption of ZnONPs and MWCNT only, resulting in an adsorption energy of -18.458 kcal/mol.
Figure 12 illustrates the interaction of the MWCNT, ZnONPs and human T1R2-Reb-A complex onto the graphene oxide surface, with an adsorption energy of − 44.286 kcal/mol. When Reb-A was adsorbed onto the existing layer of GO/MWCNT/ZnONPs, the shortest atom–atom distance between GO and Reb-A was 2.72 Å while for GO and MWCNT, a distance of 5.024 Å was reported between the nearest atoms. The Reb-A seem to propel itself to interact more with the MWCNT by folding alongside its plane.
Figure 12.
(A) Top view of the adsorption of the MWCNT, ZnO nanoparticles, and human T1R2 homolog-Reb-A complex onto graphene oxide surface. (B) Side view of showing the distances of the Reb-A to the MWCNT and ZnONPs respectively. ∆G = − 44.286 kcal/mol (Images generated on Materials studio74 license accessed through CHPC, Cape Town).
Figure 12A shows the top view of the overall interaction study (see Fig. 8) while Fig. 12B shows the side view along with the distances between the Reb-A and GO (14.357 Å); and Reb-A-MWCNT (14.498 Å). It is evident from the adsorption energy (∆G = − 44.286 kcal/mol) that more hydrophobic interaction exists between the biomolecular part (T1R2 homolog and Reb-A) as opposed to the nanomaterials part. This change in ∆G could be because of intermolecular interactions i.e. when highly hydrophobic molecules interact with each other, the enthalpy increases because some of the hydrogen bonds will be broken to form an O–H group with the other molecule. That is why a high binding affinity and free binding energy values were reported for the human T1R2 homolog-Reb-A in this study. Thus, it is safe to say that the reduction in ∆G, observed in Fig. 12, is as a result of the more hydrophobic interaction between the human T1R2 homolog and Reb-A described earlier in Fig. 4. The performance of MWCNT when immobilised onto nanocomposites have been studied in different systems and have been reported to enhance the reactivity and/or the sensitivity of the systems studied118–122. Therefore, a key finding of this study, is the enhanced adsorption on the T1R2-Reb-A complex in the presence of MWCNT.
For comparative purposes, the performance was tested in the absence of MWCNT as proposed in Fig. 13 below.
Figure 13.
The steps followed in the modelling of GO/ZnONPs without MWCNT.
Modelling of GO/ZnONPs without MWCNT
Using the optimized structures in Tables 1 and 3, the modelling for Fig. 13 was undertaken in a similar stepwise fashion as described in Fig. 8, except in the absence of MWCNT. Figure 14 illustrates the adsorption of Reb-A onto ZnONPs.
Table 3.
The layer-by-layer system without multiwalled carbon nanotube (MWCNT) along with calculated adsorption energies (kcal/mol).
| Nano-system | ∆G (adsorption energy) kcal/mol |
|---|---|
| ZnONPs/Reb-A | − 16.725 |
| GO/ZnONPs/T1R2-Reb-A | − 38.328 |
Figure 14.

Adsorption of Reb-A onto ZnONPs. Adsorption energy = − 16.725 kcal/mol (Image generated on Materials studio74 license accessed through CHPC, Cape Town).
There was an increase in adsorption energy when Reb-A interacted with the ZnONPs (Fig. 14), resulting in an adsorption energy of − 16.725 kcal/mol. This could be due to the formation of a hydrogen bond interaction between the H on the Reb-A and the O on the ZnONPs. Figure 15 further illustrates the ZnONPs adsorbed onto GO followed by direct adsorption of the T1R2-Reb-A complex onto the GO surface.
Figure 15.
The adsorption of ZnONPs, T1R2-Reb-A complex onto GO surface ∆G = − 38.328 kcal/mol (Image generated on Materials studio74 license accessed through CHPC, Cape Town).
According to Fig. 15A, a good bio-molecular interaction exists between the human T1R2 and Reb-A, which indicates an enhanced sensitivity of Reb-A in the presence of the human T1R2. Furthermore, Fig. 15B better explains the inter-atomic distances of Reb-A to the GO (− 10.870 Å) and the ZnONPs (23.059 Å) respectively.
Therefore, this study emphasises the role of nanomaterials in the layer-by-layer adsorption and we could say that the different surface arrangements of the nanomaterials play a vital role in its interaction pattern with the T1R2-Reb-A complex.
Comparison of experimental data with computational results
Differential pulse voltammetry (DPV) is an analytical tool comprising of advantages such as short pulse time and sensitivity for both quantitative analysis and understanding chemical reaction kinetics, thermodynamics, and mechanisms. In this study, due to its sensitivity, DPV was used to accurately detect the interaction of Reb A at T1R2/GO/Pt and T1R2/MWCNTs/Pt under optimized electrochemical parameters (pH 11.0, scan rate: 0.09 V s−1, and deposition time: 40 s).
Figure 16A,B displays obtained anodic peak currents with T1R2/GO/Pt and T1R2/MWCNTs/Pt at 0.2 V vs Ag/AgCl with a linear relationship between anodic currents. It was demonstrated that the peak currents obtained with T1R2/MWCNTs/Pt were greater compared to T1R2/GO/Pt, indicating the rapid electron transfer capability and higher surface area of MWCNTs. The modelled MWCNTs demonstrated similar trends in terms of adsorption energies where the ΔG of the systems with MWCNTs was higher compared to systems without MWCNTs.
Figure 16.
DPV of Reb A at pH 11.0 borate buffer using (A) T1R2/GO/Pt and (B) T1R2/MWCNTs/Pt immunosensor.
Conclusions
This study present insights into the interaction between biomolecules at the nanostructure interface by exploring the modification of GO/MWCNT with ZnONPs and its application as an adsorbent for the human T1R2-Reb-A complex from both computational and electrochemical perspectives. Various theoretical calculations and analysis assisted in the understanding of the mechanism of bio-molecular interaction and adsorption at the molecular level through the estimation and comparison of different properties of the two systems (with and without MWCNT). This layer-by-layer adsorption mechanism allows for the structural elucidation for systems modified with and without MWCNT, in which the former adsorbent showed a better performance than the latter. For all the systems studied, the presence of MWCNT showed higher adsorption capacities in systems modified with MWCNT as opposed to unmodified systems. In addition, the bio-molecular interaction between T1R2-Reb-A complex showed higher binding affinity as well as higher free binding energy. Also, hydrophobic interactions played a major role in the interaction between Reb-A and the T1R2 homolog. Different metrics, such as the biomolecular interaction and the adsorption at the materials interface, used in this study provided the best explanation for the overall adsorption mechanism. The high adsorption capacity reported in this study are in agreement with previously reported experimental work117. Comparing the adsorption energies of the two systems, GO/MWCNT/ZnONPs/T1R2-Reb-A (∆G = − 76.642 kcal/mol) shows a better adsorption energy due to the presence of MWCNT in contrast to GO/ZnONPs/T1R2-Reb-A (∆G = − 13.852 kcal/mol), which is a key finding of this work. The obtained experimental results were in good agreement with our reported computational results in terms of adsorption energies. This study demonstrates the usefulness of a layer-by-layer strategy to assess the performance of the T1R2-Reb-A complex by the incorporation of MWCNT with enhanced selectivity, sensitivity and robustness of the proposed immunosensor.
Future work
The obtained modelling results of T1R2 and Reb A on GO as well as MWCNTs surface has given new insights in choosing T1R2/T1R3 subunit as a new biomarker for the development of additional electrochemical immunosensor that could result in more opportunities in terms of enhanced selectivity in quality control laboratories in the food beverage industries.
Supplementary information
Acknowledgements
OA acknowledges the support from the Durban University of Technology (DUT). KB acknowledges the Durban University of Technology and Centre for High Performance Computing (CHPC), Cape Town, South Africa for all the computational resources.
Author contributions
The manuscript was written by O.A.A. through contributions of all authors. O.A.A.I, A.M.A., P.H. and S.K. conceived ideas and K.B. designed and supervised the study. O.A.A. ran all computational calculations and all three authors participated in data interpretation and contributed to the manuscript write-up. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Olayide A. Arodola, Email: Olayide.arodola@gmail.com
Krishna Bisetty, Email: bisettyk@dut.ac.za.
Inamuddin, Email: inamuddin@zhcet.ac.in.
Supplementary information
is available for this paper at 10.1038/s41598-020-75123-4.
References
- 1.Toyomasu T, Sassa T. Chemistry and biology. In: Liu H-W, Mander L, editors. Comprehensive Natural Products II. Amsterdam: Elsevier; 2010. pp. 643–672. [Google Scholar]
- 2.Philippaert K, et al. Steviol glycosides enhance pancreatic beta-cell function and taste sensation by potentiation of TRPM5 channel activity. Nat. Commun. 2017;8:14733. doi: 10.1038/ncomms14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hellfritsch C, Brockhoff A, Stahler F, Meyerhof W, Hofmann T. Human psychometric and taste receptor responses to steviol glycosides. J. Agric. Food Chem. 2012;60:6782–6793. doi: 10.1021/jf301297n. [DOI] [PubMed] [Google Scholar]
- 4.Carakostas MC, Curry L, Boileau A, Brusick D. Overview: The history, technical function and safety of rebaudioside A, a naturally occurring steviol glycoside, for use in food and beverages. Food Chem. Toxicol. 2008;46:S1–S10. doi: 10.1016/j.fct.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Ramya M, et al. Studies on biochemical and medicinal properties of Stevia Rebaudiana grown in vitro. Int. J. Res. Ayurveda Pharm. 2014;5:169–174. doi: 10.7897/2277-4343.05234. [DOI] [Google Scholar]
- 6.González C, Tapia M, Pérez E, Pallet D, Dornier M. Main properties of steviol glycosides and their potential in the food industry: A review. Fruits. 2014;69:127–141. doi: 10.1051/fruits/2014003. [DOI] [Google Scholar]
- 7.Wald JP, Morlock GE. Quantification of steviol glycosides in food products, Stevia leaves and formulations by planar chromatography, including proof of absence for steviol and isosteviol. J. Chromatogr. A. 2017;1506:109–119. doi: 10.1016/j.chroma.2017.05.026. [DOI] [PubMed] [Google Scholar]
- 8.Treesukosol Y, Smith KR, Spector AC. The functional role of the T1R family of receptors in sweet taste and feeding. Physiol. Behav. 2011;105:14–26. doi: 10.1016/j.physbeh.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nie Y, Vigues S, Hobbs JR, Conn GL, Munger SD. Distinct contributions of T1R2 and T1R3 taste receptor subunits to the detection of sweet stimuli. Curr. Biol. 2005;15:1948–1952. doi: 10.1016/j.cub.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 10.Mayank, Vikas J. Interaction model of steviol glycosides from Stevia rebaudiana (Bertoni) with sweet taste receptors: A computational approach. Phytochemistry. 2015;116:12–20. doi: 10.1016/j.phytochem.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Xu H, et al. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14258–14263. doi: 10.1073/pnas.0404384101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koizumi A, et al. Taste-modifying sweet protein, neoculin, is received at human T1R3 amino terminal domain. Biochem. Biophys. Res. Commun. 2007;358:585–589. doi: 10.1016/j.bbrc.2007.04.171. [DOI] [PubMed] [Google Scholar]
- 13.Jiang P, et al. The cysteine-rich region of T1R3 determines responses to intensely sweet proteins. J. Biol. Chem. 2004;279:45068–45075. doi: 10.1074/jbc.M406779200. [DOI] [PubMed] [Google Scholar]
- 14.Acevedo W, Ramírez-Sarmiento CA, Agosin E. Identifying the interactions between natural, non-caloric sweeteners and the human sweet receptor by molecular docking. Food Chem. 2018;264:164–171. doi: 10.1016/j.foodchem.2018.04.113. [DOI] [PubMed] [Google Scholar]
- 15.Mudgal S, Keresztes I, Feigenson GW, Rizvi SS. Controlling the taste receptor accessible structure of rebaudioside A via binding to bovine serum albumin. Food Chem. 2016;197:84–91. doi: 10.1016/j.foodchem.2015.10.064. [DOI] [PubMed] [Google Scholar]
- 16.Acevedo W, González-Nilo F, Agosin E. Docking and molecular dynamics of steviol glycoside-human bitter receptor interactions. J. Agric. Food Chem. 2016;64:7585–7596. doi: 10.1021/acs.jafc.6b02840. [DOI] [PubMed] [Google Scholar]
- 17.Bathinapatla A, Kanchi S, Singh P, Sabela MI, Bisetty K. An ultrasensitive performance enhanced novel cytochrome c biosensor for the detection of rebaudioside A. Biosens. Bioelectron. 2016;77:116–123. doi: 10.1016/j.bios.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Nuemket N, et al. Structural basis for perception of diverse chemical substances by T1r taste receptors. Nat. Commun. 2017;8:15530. doi: 10.1038/ncomms15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah R, De Jager LS, Begley TH. Simultaneous determination of steviol and steviol glycosides by liquid chromatography-mass spectrometry. Food Addit. Contam. A Chem. Anal. Control Expos. Risk Assess. 2012;29:1861–1871. doi: 10.1080/19440049.2012.725946. [DOI] [PubMed] [Google Scholar]
- 20.Kakigi Y, Suzuki T, Icho T, Uyama A, Mochizuki N. Classification of stevia sweeteners in soft drinks using liquid chromatography and time-of-flight mass spectrometry. Food Addit. Contam. A Chem. Anal. Control Expos. Risk Assess. 2013;30:2043–2049. doi: 10.1080/19440049.2013.843101. [DOI] [PubMed] [Google Scholar]
- 21.Woelwer-Rieck U, Lankes C, Wawrzun A, Wüst M. Improved HPLC method for the evaluation of the major steviol glycosides in leaves of Stevia rebaudiana. Eur. Food Res. Technol. 2010;231:581–588. doi: 10.1007/s00217-010-1309-4. [DOI] [Google Scholar]
- 22.Puri M, Sharma D, Barrow CJ, Tiwary AK. Optimisation of novel method for the extraction of steviosides from Stevia rebaudiana leaves. Food Chem. 2012;132:1113–1120. doi: 10.1016/j.foodchem.2011.11.063. [DOI] [PubMed] [Google Scholar]
- 23.Londhe SV, Nanaware SM. HPTLC method for simultaneous analysis of Stevioside and Rebaudioside-A in Stevia rebaudiana. J. AOAC Int. 2013;96:24–26. doi: 10.5740/jaoacint.11-257. [DOI] [PubMed] [Google Scholar]
- 24.Bathinapatla A, Kanchi S, Singh P, Sabela MI, Bisetty K. Fabrication of copper nanoparticles decorated multiwalled carbon nanotubes as a high performance electrochemical sensor for the detection of neotame. Biosens. Bioelectron. 2015;67:200–207. doi: 10.1016/j.bios.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Li SFY. Separation and determination of Stevia sweeteners by capillary electrophoresis and high performance liquid chromatography. J. Liq. Chromatogr. 1995;18:1703–1719. doi: 10.1080/10826079508010001. [DOI] [Google Scholar]
- 26.Vaněk T, Nepovım A, Valı& ´ček P. Determination of Stevioside in plant material and fruit teas. J. Food Compos. Anal. 2001;14:383–388. doi: 10.1006/jfca.2000.0974. [DOI] [Google Scholar]
- 27.Komorsky-Lovrić Š, Novak I, Novak B. Measurement of Stevioside by square-wave polarography. Electroanalysis. 2010;22:2211–2215. doi: 10.1002/elan.201000130. [DOI] [Google Scholar]
- 28.Ensafi AA, Karimi-Maleh H. Determination of 6-mercaptopurine in the presence of uric acid using modified multiwall carbon nanotubes-TiO2 as a voltammetric sensor. Drug Test. Anal. 2012;4:970–977. doi: 10.1002/dta.286. [DOI] [PubMed] [Google Scholar]
- 29.Eguílaz M, Villalonga R, Agüí L, Yáñez-Sedeño P, Pingarrón JM. Gold nanoparticles: Poly(diallyldimethylammonium chloride)–carbon nanotubes composites as platforms for the preparation of electrochemical enzyme biosensors: Application to the determination of cholesterol. J. Electroanal. Chem. 2011;661:171–178. doi: 10.1016/j.jelechem.2011.07.040. [DOI] [Google Scholar]
- 30.Dalkiran B, Kaçar C, Erden PE, Kiliç E. Amperometric xanthine biosensors based on chitosan-Co3O4-multiwall carbon nanotube modified glassy carbon electrode. Sens. Actuators B Chem. 2014;200:83–91. doi: 10.1016/j.snb.2014.04.025. [DOI] [Google Scholar]
- 31.Xue C, et al. Electrochemical serotonin sensing interface based on double-layered membrane of reduced graphene oxide/polyaniline nanocomposites and molecularly imprinted polymers embedded with gold nanoparticles. Sens. Actuators B Chem. 2014;196:57–63. doi: 10.1016/j.snb.2014.01.100. [DOI] [Google Scholar]
- 32.Zappi D, et al. Biologically friendly room temperature ionic liquids and nanomaterials for the development of innovative enzymatic biosensors. Talanta. 2017;175:566–572. doi: 10.1016/j.talanta.2017.07.081. [DOI] [PubMed] [Google Scholar]
- 33.Wang J. Carbon-nanotube based electrochemical biosensors: A review. Electroanalysis. 2005;17:7–14. doi: 10.1002/elan.200403113. [DOI] [Google Scholar]
- 34.Thirumalraj B, Kubendhiran S, Chen SM, Lin KY. Highly sensitive electrochemical detection of palmatine using a biocompatible multiwalled carbon nanotube/poly-l-lysine composite. J. Colloid Interface Sci. 2017;498:144–152. doi: 10.1016/j.jcis.2017.03.045. [DOI] [PubMed] [Google Scholar]
- 35.Gao F, et al. Highly sensitive and selective detection of dopamine in the presence of ascorbic acid at graphene oxide modified electrode. Sens. Actuators B Chem. 2013;186:380–387. doi: 10.1016/j.snb.2013.06.020. [DOI] [Google Scholar]
- 36.Cheemalapati S, Palanisamy S, Mani V, Chen S-M. Simultaneous electrochemical determination of dopamine and paracetamol on multiwalled carbon nanotubes/graphene oxide nanocomposite-modified glassy carbon electrode. Talanta. 2013;117:297–304. doi: 10.1016/j.talanta.2013.08.041. [DOI] [PubMed] [Google Scholar]
- 37.Zor E, Hatay Patir I, Bingol H, Ersoz M. An electrochemical biosensor based on human serum albumin/graphene oxide/3-aminopropyltriethoxysilane modified ITO electrode for the enantioselective discrimination of d- and l-tryptophan. Biosens. Bioelectron. 2013;42:321–325. doi: 10.1016/j.bios.2012.10.068. [DOI] [PubMed] [Google Scholar]
- 38.Ruszkiewicz JA, et al. Neurotoxic effect of active ingredients in sunscreen products, a contemporary review. Toxicol. Rep. 2017;4:245–259. doi: 10.1016/j.toxrep.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatamie A, et al. Zinc oxide nanostructure-modified textile and its application to biosensing, photocatalysis, and as antibacterial material. Langmuir. 2015;31:10913–10921. doi: 10.1021/acs.langmuir.5b02341. [DOI] [PubMed] [Google Scholar]
- 40.Zhu P, et al. Biomedical applications of functionalized ZnO nanomaterials: From biosensors to bioimaging. Adv. Mater. Interfaces. 2016;3:1500494. doi: 10.1002/admi.201500494. [DOI] [Google Scholar]
- 41.Jiang J, Pi J, Cai J. The advancing of zinc oxide nanoparticles for biomedical applications. Bioinorg. Chem. Appl. 2018;2018:18. doi: 10.1155/2018/1062562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y, Wang Z, Peng F, Fu L. Application of biosynthesized ZnO nanoparticles on an electrochemical H 2 O 2 biosensor. Braz. J. Pharm. Sci. 2016;52:781–786. doi: 10.1590/s1984-82502016000400023. [DOI] [Google Scholar]
- 43.Aini BN, Siddiquee S, Ampon K, Rodrigues KF, Suryani S. Development of glucose biosensor based on ZnO nanoparticles film and glucose oxidase-immobilized eggshell membrane. Sens. Bio-Sens. Res. 2015;4:46–56. doi: 10.1016/j.sbsr.2015.03.004. [DOI] [Google Scholar]
- 44.Tripathy N, Kim D-H. Metal oxide modified ZnO nanomaterials for biosensor applications. Nano Converg. 2018;5:27–27. doi: 10.1186/s40580-018-0159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn SR, et al. Duplex bioelectronic tongue for sensing umami and sweet tastes based on human taste receptor nanovesicles. ACS Nano. 2016;10:7287–7296. doi: 10.1021/acsnano.6b02547. [DOI] [PubMed] [Google Scholar]
- 46.Carbó-Dorca R, Besalú E. Construction of coherent nano quantitative structure–properties relationships (nano-QSPR) models and catastrophe theory. SAR QSAR Environ. Res. 2011;22:661–665. doi: 10.1080/1062936X.2011.623319. [DOI] [PubMed] [Google Scholar]
- 47.Riviere JE, Tran CL. Pharmacokinetics of nanomaterials: An overview of carbon nanotubes, fullerenes and quantum dots. Wiley Interdiscipl. Rev. Nanomed. Nanobiotechnol. 2007;1:26–34. doi: 10.1002/wnan.24. [DOI] [PubMed] [Google Scholar]
- 48.Gleeson MP, et al. The challenges involved in modeling toxicity data in silico: A review. Curr. Pharm. Des. 2012;18:1266–1291. doi: 10.2174/138161212799436359. [DOI] [PubMed] [Google Scholar]
- 49.Brivio GP, Trioni MI. The adiabatic molecule–metal surface interaction: Theoretical approaches. Rev. Mod. Phys. 1999;71:231–265. doi: 10.1103/RevModPhys.71.231. [DOI] [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Eswar N, et al. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2001;15:1–30. doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 52.Schrodinger. Vol. 20 Oct. 2018 (20 Oct. 2018).
- 53.Sitehound-web. https://scbx.mssm.edu/sitehound/sitehound-web/Input.html. Accessed 21 Oct 2018.
- 54.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 55.Buchan DWA, et al. Protein annotation and modelling servers at University College London. Nucleic Acids Res. 2010;38:W563–W568. doi: 10.1093/nar/gkq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swiss PDB. www.spdbv.vital-it.ch/disclaim.html. Accessed 21 Oct 2018.
- 57.National Center for Biotechnology Information. PubChem Database (2018). https://pubchem.ncbi.nlm.nih.gov/compound/6918840. Accessed 10 Aug 2018.
- 58.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dym O, Xenarios I, Ke HM, Colicelli J. Molecular docking of competitive phosphodiesterase inhibitors. Mol. Pharmacol. 2002;61:20–25. doi: 10.1124/mol.61.1.20. [DOI] [PubMed] [Google Scholar]
- 60.Morris GM, et al. Automated docking using a lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. doi: 10.1002/(sici)1096-987x(19981115)19:14<1639::aid-jcc10>3.0.co;2-b. [DOI] [Google Scholar]
- 61.Rao MS, Olson AJ. Modelling of factor Xa-inhibitor complexes: A computational flexible docking approach. Proteins. 1999;34:173–183. doi: 10.1002/(sici)1097-0134(19990201)34:2<173::aid-prot3>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 62.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cornell WD, et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules (vol 117, pg 5179, 1995) J. Am. Chem. Soc. 1996;118:2309–2309. doi: 10.1021/ja955032e. [DOI] [Google Scholar]
- 64.Case DA, et al. The amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 66.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 1995;117:5179–5197. doi: 10.1021/ja00124a002. [DOI] [Google Scholar]
- 67.Frisch, M. J. et al.https://gaussian.com/g09citation/. Gaussian Inc, Wallingford, CT (2016). Accessed 21 Oct 2018.
- 68.Duan Y, et al. point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 69.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 70.Elber R, Ruymgaart AP, Hess B. SHAKE parallelization. Eur. Phys. J. Spec. Top. 2011;200:211–223. doi: 10.1140/epjst/e2011-01525-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Massova I, Kollman PA. Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect. Drug Discov. Des. 2000;18:113–135. doi: 10.1023/a:1008763014207. [DOI] [Google Scholar]
- 72.Tsui V, Case DA. Theory and applications of the generalized born solvation model in macromolecular simulations. Biopolymers. 2000;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::aid-bip10024>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 73.Onufriev A, Bashford D, Case DA. Modification of the generalized Born model suitable for macromolecules. J. Phys. Chem. B. 2000;104:3712–3720. doi: 10.1021/jp994072s. [DOI] [Google Scholar]
- 74.Dassault Systèmes BIOVIA. Materials Studio v2018 (2018). https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-materials-studio/. Accessed 10 Aug 2018.
- 75.Sun H. COMPASS: An ab initio force-field optimized for condensed-phase applications overview with details on alkane and benzene compounds. J. Phys. Chem. B. 1998;102:7338–7364. doi: 10.1021/jp980939v. [DOI] [Google Scholar]
- 76.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 77.Dennington R, et al. In GaussView. Shawnee Mission KS: Semichem Inc.; 2009. [Google Scholar]
- 78.Monkhorst HJ, Pack JD. Special points for Brillouin-zone integrations. Phys. Rev. B. 1976;13:5188–5192. doi: 10.1103/PhysRevB.13.5188. [DOI] [Google Scholar]
- 79.Posvyanskii DV, Shulman AY. Self-consistent solution of Kohn-Sham equations for infinitely extended systems with inhomogeneous electron gas. J. Exp. Theor. Phys. 2009;109:145–168. doi: 10.1134/S1063776109070188. [DOI] [Google Scholar]
- 80.Discovery Studio Modeling Environment (San Diego, Release 2017). https://accelrys.com/products/collaborative-science/biovia-discovery-studio/.
- 81.Meng X-Y, Zhang H-X, Mezei M, Cui M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011;7:146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morris GM, Lim-Wilby M. Molecular docking. Methods Mol. Biol. 2008;443:365–382. doi: 10.1007/978-1-59745-177-2_19. [DOI] [PubMed] [Google Scholar]
- 83.Durrant J, McCammon JA. Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9:71. doi: 10.1186/1741-7007-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blake L, Soliman MS. Identification of irreversible protein splicing inhibitors as potential anti-TB drugs: Insight from hybrid non-covalent/covalent docking virtual screening and molecular dynamics simulations. Med. Chem. Res. 2014;23:2312–2323. doi: 10.1007/s00044-013-0822-y. [DOI] [Google Scholar]
- 85.Ahmed SM, et al. Comparison of the molecular dynamics and calculated binding free energies for nine FDA-approved HIV-1 PR drugs against subtype B and C-SA HIV PR. Chem. Biol. Drug Des. 2013;81:208–218. doi: 10.1111/cbdd.12063. [DOI] [PubMed] [Google Scholar]
- 86.Kashani-Amin E, Sakhteman A, Larijani B, Ebrahim-Habibi A. Introducing a new model of sweet taste receptor, a class C G-protein coupled receptor (C GPCR) Cell Biochem. Biophys. 2019;77:227–243. doi: 10.1007/s12013-019-00872-7. [DOI] [PubMed] [Google Scholar]
- 87.Perez-Aguilar JM, Kang SG, Zhang L, Zhou RA-O. Modeling and structural characterization of the sweet taste receptor heterodimer. ACS Chem. Neurosci. 2019;10:4579–4592. doi: 10.1021/acschemneuro.9b00438. [DOI] [PubMed] [Google Scholar]
- 88.Deng Y, Roux B. Computations of standard binding free energies with molecular dynamics simulations. J. Phys. Chem. B. 2009;113:2234–2246. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang J, Deng Y, Roux B. Absolute binding free energy calculations using molecular dynamics simulations with restraining potentials. Biophys. J. 2006;91:2798–2814. doi: 10.1529/biophysj.106.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zou Y, Ewalt J, Ng H-L. Recent Insights from molecular dynamics simulations for G protein-coupled receptor drug discovery. Int. J. Mol. Sci. 2019;20:4237. doi: 10.3390/ijms20174237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Renault P, Louet M, Marie J, Labesse G, Floquet N. Molecular dynamics simulations of the allosteric modulation of the adenosine A2a receptor by a mini-G protein. Sci. Rep. 2019;9:5495. doi: 10.1038/s41598-019-41980-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nicklasson S, et al. Taste responsiveness to two steviol glycosides in three species of nonhuman primates. Curr. Zool. 2017;64:63–68. doi: 10.1093/cz/zox012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hellfritsch C, Brockhoff A, Stähler F, Meyerhof W, Hofmann T. Human psychometric and taste receptor responses to steviol glycosides. J. Agric. Food Chem. 2012;60:6782–6793. doi: 10.1021/jf301297n. [DOI] [PubMed] [Google Scholar]
- 94.Lin M, Wang F, Zhu Y. Modeled structure-based computational redesign of a glycosyltransferase for the synthesis of rebaudioside D from rebaudioside A. Biochem. Eng. J. 2020;159:107626. doi: 10.1016/j.bej.2020.107626. [DOI] [Google Scholar]
- 95.Zhang F, et al. Molecular mechanism of the sweet taste enhancers. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4752–4757. doi: 10.1073/pnas.0911660107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feig M, Brooks CL. Evaluating CASP4 predictions with physical energy functions. Proteins. 2002;49:232–245. doi: 10.1002/prot.10217. [DOI] [PubMed] [Google Scholar]
- 97.Ylilauri M, Pentikäinen OT. MMGBSA as a tool to understand the binding affinities of filamin-peptide interactions. J. Chem. Inf. Model. 2013;53:2626–2633. doi: 10.1021/ci4002475. [DOI] [PubMed] [Google Scholar]
- 98.Soliman MES, Pernia JJR, Greig IR, Williams IH. Mechanism of glycoside hydrolysis: A comparative QM/MM molecular dynamics analysis for wild type and Y69F mutant retaining xylanases. Org. Biomol. Chem. 2009;7:5236–5244. doi: 10.1039/b911644c. [DOI] [PubMed] [Google Scholar]
- 99.Gourmala C, et al. Elucidation of the LewisX-LewisX carbohydrate interaction with molecular dynamics simulations: A glycosynapse model. J. Mol. Struct.-Theochem. 2007;821:22–29. doi: 10.1016/j.theochem.2007.06.027. [DOI] [Google Scholar]
- 100.Shaikh SA, Jayaram B. A swift all-atom energy-based computational protocol to predict DNA-ligand binding affinity and Delta T(m) J. Med. Chem. 2007;50:2240–2244. doi: 10.1021/jm060542c. [DOI] [PubMed] [Google Scholar]
- 101.Gohlke H, Kiel C, Case DA. Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RaIGDS complexes. J. Mol. Biol. 2003;330:891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 102.Perez M. Gibbs-Thomson effects in phase transformations. Scr. Mater. 2005;52:709–712. doi: 10.1016/j.scriptamat.2004.12.026. [DOI] [Google Scholar]
- 103.Lobanov MY, Bogatyreva NS, Galzitskaya OV. Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 2008;42:623–628. doi: 10.1134/S0026893308040195. [DOI] [PubMed] [Google Scholar]
- 104.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J. Mol. Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 105.Perry CC, Patwardhan SV, Deschaume O. From biominerals to biomaterials: The role of biomolecule–mineral interactions. Biochem. Soc. Trans. 2009;37:687. doi: 10.1042/BST0370687. [DOI] [PubMed] [Google Scholar]
- 106.Roach P, Farrar D, Perry CC. Interpretation of protein adsorption: Surface-induced conformational changes. J. Am. Chem. Soc. 2005;127:8168–8173. doi: 10.1021/ja042898o. [DOI] [PubMed] [Google Scholar]
- 107.Roach P, Farrar D, Perry CC. Surface tailoring for controlled protein adsorption: Effect of topography at the nanometer scale and chemistry. J. Am. Chem. Soc. 2006;128:3939–3945. doi: 10.1021/ja056278e. [DOI] [PubMed] [Google Scholar]
- 108.Deschaume O, Fournier A, Shafran KL, Perry CC. Interactions of aluminium hydrolytic species with biomolecules. New J. Chem. 2008;32:1346–1353. doi: 10.1039/B805406C. [DOI] [Google Scholar]
- 109.Deschaume O, Shafran K, Perry C. Interactions of bovine serum albumin with aluminum polyoxocations and aluminum hydroxide. Langmuir ACS J. Surf. Colloids. 2006;22:10078–10088. doi: 10.1021/la061285h. [DOI] [PubMed] [Google Scholar]
- 110.Berkowitz J, Anderson M, Graham R. Laboratory investigation of aluminum solubility and solid-phase properties following alum treatment of lake waters. Water Res. 2005;39:3918–3928. doi: 10.1016/j.watres.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 111.Bergfors E, Trollfors B, Inerot A. Itching nodules and aluminium allergy induced by vaccination with adsorbed vaccines. J. Allergy Clin. Immunol. 2004;113:S294. doi: 10.1016/j.jaci.2004.01.535. [DOI] [Google Scholar]
- 112.Liang L, Chen E-Y, Shen J-W, Wang Q. Molecular modelling of translocation of biomolecules in carbon nanotubes: Method, mechanism and application. Mol. Simul. 2016;42:827–835. doi: 10.1080/08927022.2015.1107184. [DOI] [Google Scholar]
- 113.Hosseini SA, Mashaykhi S, Babaei S. Graphene oxide/zinc oxide nanocomposite: A superior adsorbent for removal of methylene blue—Statistical analysis by response surface methodology (RSM) S. Afr. J. Chem. 2016;69:105–112. doi: 10.17159/0379-4350/2016/v69a13. [DOI] [Google Scholar]
- 114.Chang C-C, et al. High-sensitivity detection of carbohydrate antigen 15–3 using a gold/zinc oxide thin film surface plasmon resonance-based biosensor. Anal. Chem. 2010;82:1207–1212. doi: 10.1021/ac901797j. [DOI] [PubMed] [Google Scholar]
- 115.Tang H, et al. Theoretical insight into the adsorption of aromatic compounds on graphene oxide. Environ. Sci. Nano. 2018;5:2357–2367. doi: 10.1039/C8EN00384J. [DOI] [Google Scholar]
- 116.Rezazadeh T, Dalali N, Sehati N. Investigation of adsorption performance of graphene oxide/polyaniline reinforced hollow fiber membrane for preconcentration of Ivermectin in some environmental samples. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018;204:409–415. doi: 10.1016/j.saa.2018.06.040. [DOI] [PubMed] [Google Scholar]
- 117.Farghali AA, Abdel Tawab HA, Abdel Moaty SA, Khaled R. Functionalization of acidified multi-walled carbon nanotubes for removal of heavy metals in aqueous solutions. J. Nanostruct. Chem. 2017;7:101–111. doi: 10.1007/s40097-017-0227-4. [DOI] [Google Scholar]
- 118.Selvaganesh, S. V., Dhanasekaran, P. & Bhat Santoshkumar, D. in Electrochemical Energy Technology, Vol. 3, 9 (2017).
- 119.Wang X, Zhou Z, Chen F. Surface modification of carbon nanotubes with an enhanced antifungal activity for the control of plant fungal pathogen. Materials (Basel) 2017;10:1375. doi: 10.3390/ma10121375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Q, Zhao X, Sui G, Yang X. Surface sizing treated MWCNTs and its effect on the wettability, interfacial interaction and flexural properties of MWCNT/epoxy nanocomposites. Nanomaterials (Basel) 2018;8:680. doi: 10.3390/nano8090680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rahman G, et al. An overview of the recent progress in the synthesis and applications of carbon nanotubes. J. Carbon Res. 2019 doi: 10.3390/c5010003. [DOI] [Google Scholar]
- 122.Pothupitiya Gamage SJ, et al. MWCNT coated free-standing carbon fiber fabric for enhanced performance in EMI shielding with a higher absolute EMI SE. Materials. 2017;10:1350. doi: 10.3390/ma10121350. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.