Abstract
Mesenchymal cells in the crypt play indispensable roles in the maintenance of intestinal epithelial homeostasis through their contribution to the preservation of stem cells. However, the acquisition properties of the production of stem cell niche factors by the mesenchymal cells have not been well elucidated, due to technical limitations regarding the isolation and subsequent molecular and cellular analyses of cryptal mesenchymal cells. To evaluate the function of mesenchymal cells located at the large intestinal crypt, we established a novel method through which cells are harvested according to the histologic layers of mouse colon, and we compared cellular properties between microenvironmental niches, the luminal mucosa and crypts. The gene expression pattern in the cryptal mesenchymal cells showed that receptors of the hormone/cytokine leptin were highly expressed, and we found a decrease in Wnt2b expression under conditions of leptin receptor deficiency, which also induced a delay in cryptal epithelial proliferation. Our novel stratified layer isolation strategies thus revealed new microenvironmental characteristics of colonic mesenchymal cells, including the intrinsic involvement of leptin in the control of mucosal homeostasis.
Subject terms: Gastrointestinal diseases, Gastrointestinal hormones, Gastrointestinal system, Cell biology, Immunology, Gastroenterology, Anatomy, Cells, Gastrointestinal system
Introduction
The intestinal mucosa plays critical physical roles in the ingestion and absorption of necessary nutrients; at the same time, it is exposed to diverse potentially adverse agents, including infectious pathogens and other harmful substances such as allergens and toxins1,2. Therefore, intestinal mucosa is equipped with physical (e,g., tight junctions), chemical (e.g., antimicrobial peptides), and immunological (e.g., secretary IgA) barrier systems, with the mucosal epithelial regeneration process executing well-organized and prompt responses to tissue damage or inflammation1. Through accumulated past studies, the unique features and dynamics of the mucosal immune system have been well characterized, and immune–epithelial crosstalk for the generation of functional epithelial barriers has been well evaluated1. Furthermore, recent evidence has revealed the indispensable roles of mesenchymal cells (e.g., fibroblasts, myofibroblasts, and telocytes) in epithelial and cryptal regeneration3,4. However, the molecular and cellular mechanisms underlying the regulation of intestinal mesenchymal cells are relatively unknown, and an understanding of these mechanisms is essential to revealing the mucosal regeneration process and disclosing fundamental therapeutic strategies against intestinal inflammation (e.g., inflammatory bowel diseases, IBD)1,5,6.
Accumulated evidence had indicated the importance of the mesenchymal network-based stem cell microenvironment for the creation and maintenance of intestinal homeostasis6. Epithelial regeneration is achieved through the proliferation of LGR-5+ stem cells in the epithelial cryptal base7,8, and the maintenance and differentiation of those stem cells are tightly regulated by the mesenchymal cells around those stem cells8–10. Various subpopulations of mesenchymal cells (e.g., intra-sub-epithelial myofibroblasts, GLI family zinc finger 1+ [Gli1+] fibroblasts, and telocytes) provide niche factors involved in the Wnt/β-catenin pathway, including WNTs, Gremlin (GREM), and Bone Morphogenetic Proteins (BMP)s, that are essential for intestinal renewal and stem cell maintenance11,12. However, the mechanisms how the cryptal mesenchymal cells acquire the ability to maintain the stem cell niche are not well elucidated due to the limited methods for isolating cryptal mesenchymal cells for molecular and cellular analyses. To evaluate the precise function of mesenchymal cells located at the large intestinal crypt, reliable isolation methods of cryptal cells based on the histological layers of the colon are required.
The alimentary canal is anatomically and histologically composed of four broad layers: the mucosa, submucosa, muscularis externa (MEx), and serosa13. The mucosal layer is subdivided further into three layers: epithelium, lamina propria, and muscularis mucosae (MM)13. The compositions of these layers are similar throughout the digestive tract, with partial specialization among mucosal tissues13. Importantly, in gut mucosa, mesenchymal cells or stroma cells exist in each layer of the intestinal compartments; therefore, a new technique that facilitates stratified isolation is needed for the study of cryptal mesenchymal cells6.
In this study, we sought to uncover the anatomical and histological characteristics of the mesenchymal cells in colonic cryptal microenvironments by using a newly developed intestinal layer isolation method with experimental verification. The subsequent molecular and cellular analysis newly identified several receptors highly expressed in cryptal mesenchymal cells, including receptors for leptin, which is a homeostatic hormone/cytokine derived from adipose tissues. Here we present our novel gut mucosal layer isolation method for the characterization of the mesenchymal cell population in murine colon. Using this strategy, we discovered important roles of intrinsic leptin and its involvement in intestinal mucosal regeneration.
Results
Stratified isolation of mouse colon for cryptal cell analysis
That unique mesenchymal populations support cryptal niches in the small intestine and colon is widely accepted14. In addition, the roles of mesenchymal cells differ among their histological and microenvironmental locations, such as villus tips and crypt8,15. To evaluate the characteristics and function of crypt mesenchymal cells in colon as a first step in uncovering mesenchymal cell-mediated mucosal homeostatic pathways, we developed a stratified isolation method for murine colon that achieved optimal separation of the lumen and crypt parts of the colonic mucosa. To this end, murine colon tissues were isolated according to the 6 histologic components: epithelium, upper mucosa with lamina propria, crypt, MM, submucosa, and MEx13.
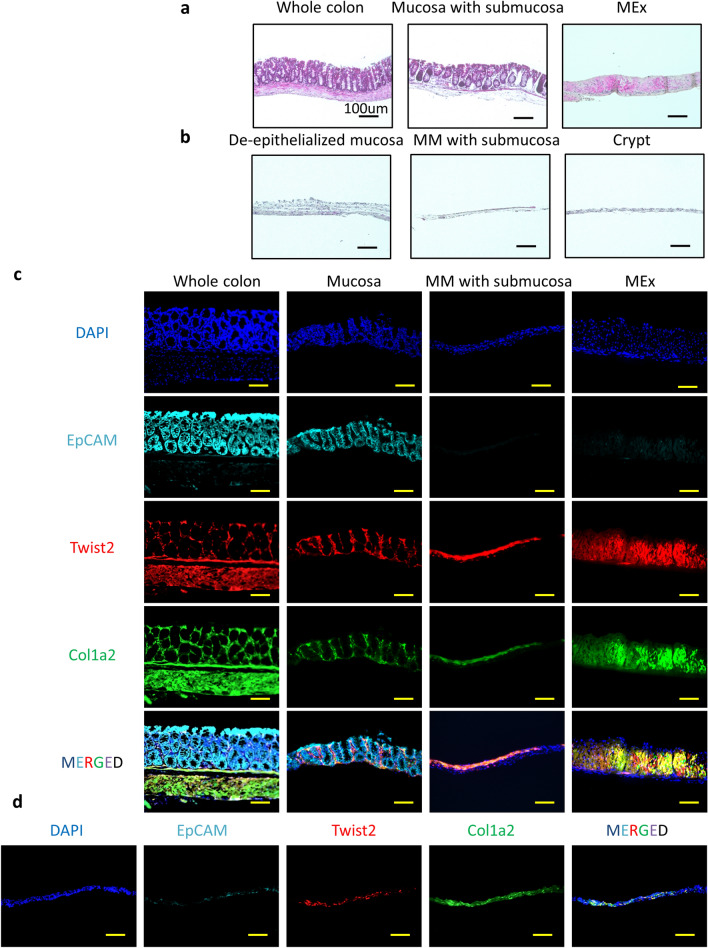
First, we separated the MEx from the mucosa and submucosa by using microtweezers (Fig. 1a, Supplemental Fig. 1, and Supplemental Video); these tissues were partly de-epithelialized through incubation in cell dissociation solution (Fig. 1b). Under a stereo-microscope, we then used a scraper and microtweezers to sub-divide the upper mucosal and cryptal compartments with the MM and submucosal compartments (Fig. 1b, Supplemental Fig. 1, and Supplemental Video). Finally, we isolated the cryptal regions from the MM and submucosal compartment (Fig. 1b, Supplemental Fig. 1, and Supplemental Video). Staining with hematoxylin and eosin confirmed the isolation of the various layers and revealed that, through the separation procedure, the various layers of the colonic mucosa had been anatomically and histologically correctly dissected (Fig. 1a,b).
Figure 1.
Stratified isolation of mouse colon for microenvironmental cellular analysis. (a) Representative images of hematoxylin and eosin (H and E) staining of whole colon, peeled layers containing mucosa and submucosa, and muscularis externa (MEx) are shown. (b) Representative images of H and E staining of de-epithelialized mucosa, muscularis mucosae (MM) with submucosa, and crypt layer are shown. Bar, 100 μm. (c) Representative images of whole colon, mucosa, MM with submucosa, and MEx from Twist2-Cre tdTomato × Col1a2 GFP mice and stained by using EpCAM are shown. (d) Crypt region of Twist2-Cre tdTomato × Col1a2 GFP mice is shown. Bar, 100 μm. Images representative of at least three independent experiments are shown.
To further validate the experimental procedure, we applied the isolation protocols to tissues from Twist2-Cre-td Tomato–type I Collagen (Col1a2)–EGFP mice which targeted two mesenchymal marker genes (Twist2 and Col1a2)5. In these mice, mesenchymal cell populations were stained red in color, and collagen-producing cells in the colonic tissues were green; epithelial cells were stained by the marker EpCAM (Fig. 1c). Whole-colon tissues were separated into 3 parts (mucosa, MM with submucosa, and MEx) (Fig. 1c). Twist2+ Col1a2+ (yellow-colored) cell populations occurred in the mucosal, MM, and MEx compartments of the mouse colon (Fig. 1c and Supplemental Fig. 2). After the upper mucosa was removed, Twist2+ Col1a2+ cell populations remained in the isolated cryptal region (Fig. 1d, Supplemental Fig. 2). These histological results confirmed the validity of the stratified isolation method for cells from the murine colonic mucosa.
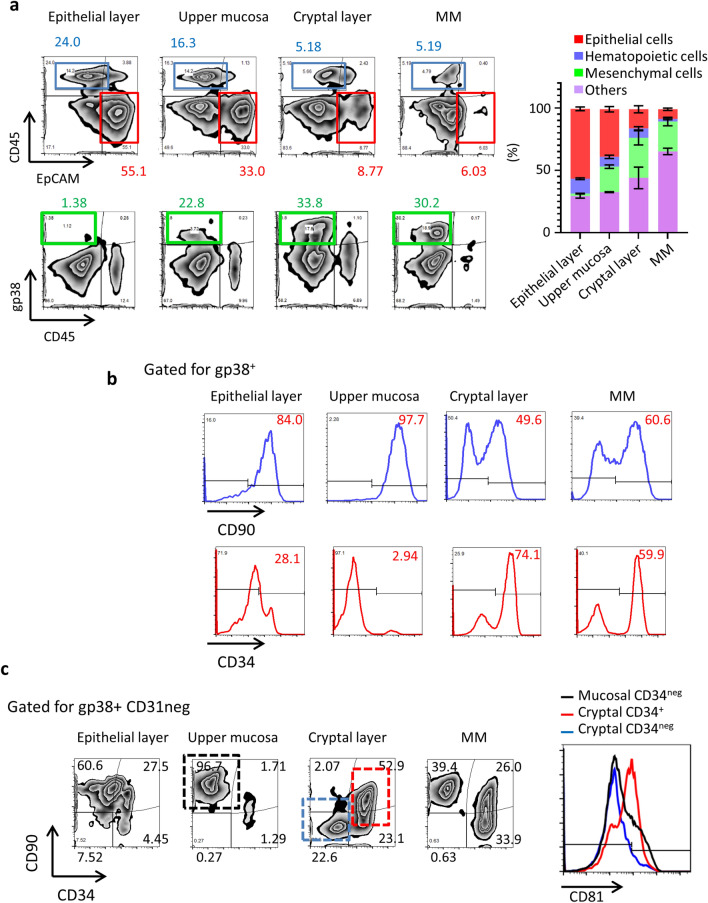
Figure 2.
Cellular profiling of stratified isolated cell populations. (a) Flow cytometry of stratified isolated cells stained with CD45, EpCAM, and gp38. The number indicates percentages of gated populations. Right data indicates the percentages of each cellular component in the colon microenvironment. Data are shown as means ± SEM. (b) Flow cytometry of cells in each layer is shown. The gp38+ cells in the negative-gated areas for EpCAM, TER119, and CD31 are shown. The number indicates the size (percentage) of the gated population. CD90 and CD34 expression of gated gp38+ cells are shown. (c) gp38+ cells were stained with CD90 and CD34. The numbers indicate the size (percentage) of the gated populations. CD81 expressions were shown in the right panel. All data are representative of at least three independent experiments.
Stratified isolation for spatial and phenotypic mesenchymal cellular profiling in the colon microenvironment
To confirm the validity of our novel isolation method by using an alternative means of assessment, we next used FACS to confirm the immunological cellular landscape of the isolated layers (Fig. 2). Epithelial and hematopoietic cells were defined as EpCAM+ and CD45+ cells, respectively. FACS analysis revealed that EpCAM+ cells were rich in the epithelial layer that had been removed through de-epithelialization, and the proportion of epithelial cells gradually decreased in the histological preparations of the crypt and MM compartments (Fig. 2a). CD45+ cells were consistently present throughout the mucosa, but the submucosal microenvironment with the MM contained a relatively lower proportion of CD45+ cells (Fig. 2a). Even though the stratified isolation procedure extended the tissue processing time, cell viability in each layer was not critically decreased compared with that after traditional (unseparated whole-colon) cell-isolation methods (Supplemental Fig. 3a). Next, to clarify the methodological robustness, we assessed the cellular properties in independent experiments (Supplemental Fig. 3b). Importantly, the ratio of TCRγδ cells, recognized by their localization in the intraepithelial compartment, within CD45+ population was high in the epithelial layer, whereas CD4+ cell counts were higher in both the upper mucosa and cryptal layer16 (Supplemental Fig. 4). These results further proved the methodological validity of our stratified isolation strategies and indicate its versatility for further research.
Importantly, gp38+ cells, including most of the mesenchymal cells and lymphatic endothelial cells17, were preferentially distributed in the preparations from the upper mucosa compared with the MM with submucosal compartment (Fig. 2). Together, these results further indicate that this method achieved sufficient stratified isolation for spatial and phenotypic characterization of mesenchymal cells.
To precisely evaluate the colonic mesenchymal cellular populations by flow cytometry, we focused on gp38-expressing cellular populations. Specifically, we gated on gp38+ cells and then examined the expression of CD90 and CD34 (Fig. 2b). According to previous studies, CD90 and CD34 are expressed in the mesenchymal cells located in the crypt8,18; however, those markers were expressed in the mesenchymal cells in other microenvironments, including MM, which mostly consists of smooth muscle cells. Indeed, no previous study has examined the murine colon with exclusion of the submucosa, MM, and MEx; therefore, our results represent the first cellular profiling of the histologically different layers of the colonic mucosa.
Furthermore, CD34– CD90+ mesenchymal cell populations, which contain myofibroblasts8, resided in the upper mucosa and MM but were less abundant in the crypt (Fig. 2b). The sub-epithelial compartment, crypt, and MM contained prominent CD34+ CD90+ and CD90– mesenchymal cell populations but, comparably, those populations in the upper mucosal compartments were relatively small (Fig. 2c). In addition, recently CD81 expression in small intestinal cryptal mesenchymal cells has been reported19. Identically, our cryptal CD34+ cell highly expressed CD81 (Fig. 2c).
Our current evaluation uncovered enrichment of CD34+ mesenchymal cells in not only the crypt but also in the MM (Fig. 2c), indicating the need for cellular profiling of the cryptal region after the use of stratified isolation methods to separate the upper mucosa, crypt, and MM.
Gene profiling of mesenchymal cells in the colonic crypt and luminal mucosa
To characterize crypt mesenchymal cells, we first examined the cell preparation obtained from mouse colon by use of the stratified isolation method. Because the upper mucosal compartment lacked a CD34+ population (Fig. 2c), to compare mesenchymal cells in the upper layers with those in the cryptal layers, we decided to further evaluate mesenchymal cells isolated by FACS sorting as CD45− CD90+ gp38+17 and elucidated their gene expression profiles by using a gene microarray analysis. First, the two layers evaluated (cryptal layer and upper mucosal layer) showed identical expression of collagen genes (Col1a1 and Col1a2) (Supplemental Fig. 5a) and, identical to the FACS data, CD34 expression was dominant in the cryptal mesenchymal cells (Supplemental Fig. 5a). In addition, the genes related to cryptal niches—including Wnt5a, Wnt2b, Gli1, forkhead box l1 (Foxl1), and Sema3a8,11,12,18,20—were highly expressed in the cryptal mesenchymal cells (Supplemental Fig. 5a).
We then compared ex vivo-expanded mesenchymal cells, morphologically fibroblasts or myofibroblasts from the cryptal and luminal regions of the colonic mucosa (Supplemental Fig. 5b), with cells isolated in vivo by using the stratified isolation method and evaluated the genes expressed (Fig. 3a). Among those genes, 546 genes showed higher expression in the cryptal mesenchymal cells compared with the lumen regions of the colonic mucosa (Fig. 3a). The genes reported as highly expressed in cryptal mesenchymal cells, such as Gli1, Foxl1, Bmp4, and Cd34, are listed among the 546 genes (Fig. 3a,b) and top 30 genes in Table 1.
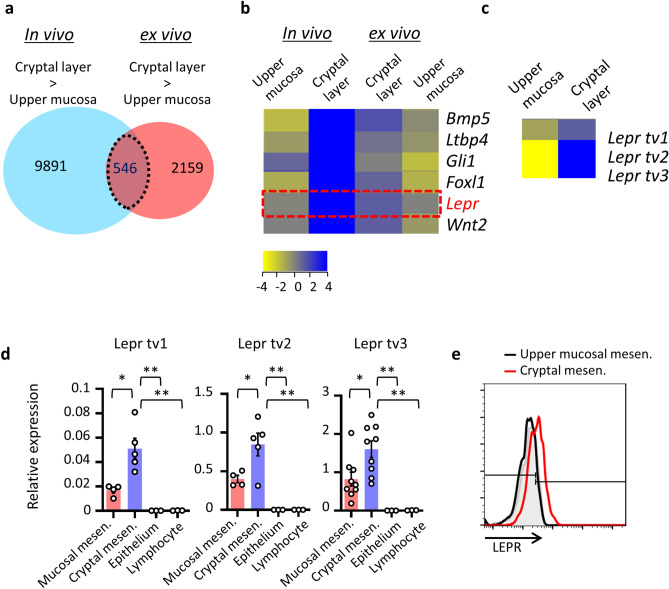
Figure 3.
LepR expression in cryptal mesenchymal cells. (a–c) gp38+ mesenchymal cells isolated from the cryptal and upper mucosal layers underwent gene microarray analysis (n = 1). (d) Gene expression of leptin receptors in mesenchymal cells sorted from upper mucosa or crypt were examined with that of EpCAM+ epithelial cells and CD45+ lymphocytes by quantitative RT-PCR analysis. *P < 0.05. Each result was normalized against the expression of Gapdh. n.s. indicates not significant. (e) Leptin receptor expressions in upper mucosa or crypt were examined by flow cytometry. Gray histogram indicates isotype control. Data from a single representative experiment among three independent experiments are shown.
Table 1.
Top 30 genes expressed in cryptal mesenchymal cells.
| Rank | In vivo upper mucosa | In vivo cryptal layer | Ex vivo cryptal layer | Ex vivo upper mucosa | GeneSymbol | Description |
|---|---|---|---|---|---|---|
| 1 | − 0.1352253 | 10.349305 | 1.0953941 | − 0.032011986 | Lepr | Leptin receptor (Lepr), transcript variant 3, mRNA [NM_001122899] |
| 2 | − 1.7839279 | 9.163433 | 1.4320621 | − 0.32384896 | Bmp5 | Bone morphogenetic protein 5 (Bmp5), mRNA [NM_007555] |
| 0 | 8.42976 | 0.75516224 | − 1.6026034 | Bmp5 | Bone morphogenetic protein 5 (Bmp5), mRNA [NM_007555] | |
| 3 | − 5.845847 | 8.349145 | 0 | − 1.0225489 | Adamdec1 | ADAM-like, decysin 1 (Adamdec1), mRNA [NM_021475] |
| 4 | − 0.036146164 | 7.296133 | 1.1356153 | − 0.10349226 | Pcdh12 | Protocadherin 12 (Pcdh12), mRNA [NM_017378] |
| 5 | 0.8064318 | 7.193366 | 0 | − 1.9055712 | Gli1 | GLI-Kruppel family member GLI1 (Gli1), mRNA [NM_010296] |
| 6 | − 0.31227016 | 7.19128 | 0 | − 1.1451445 | Col15a1 | Collagen, type XV, alpha 1 (Col15a1), mRNA [NM_009928] |
| 7 | − 0.25886297 | 6.7288184 | 1.0249934 | − 0.65438557 | Syne1 | Spectrin repeat containing, nuclear envelope 1 (Syne1), transcript variant 5, mRNA [NM_001347732] |
| 8 | 0.40697193 | 6.3136005 | 0 | − 1.2896981 | Lin7a | Lin-7 homolog A (C. elegans) (Lin7a), transcript variant 1, mRNA [NM_001039354] |
| 9 | 4.5943704 | 6.2623343 | 0 | − 2.2177994 | Stxbp6 | Syntaxin binding protein 6 (amisyn) (Stxbp6), mRNA [NM_144552] |
| 10 | 0 | 6.2549396 | 1.8562856 | − 1.847434 | Grrp1 | Glycine/arginine rich protein 1 (Grrp1), mRNA [NM_001099296] |
| 11 | 1.4523826 | 5.9267335 | 0 | − 1.1692836 | Cadm3 | Cell adhesion molecule 3 (Cadm3), mRNA [NM_053199] |
| 12 | 0.21515155 | 5.9007926 | 0 | − 2.616846 | – | UI-M-EX0-byj-o-02-0-UI.r1 NIH_BMAP_EX0 cDNA clone IMAGE: 5,719,129 5′, mRNA sequence [CB248850] |
| 13 | 0.17255163 | 5.841345 | 0 | − 1.2357674 | Agtr1b | Angiotensin II receptor, type 1b (Agtr1b), mRNA [NM_175086] |
| 14 | 2.6327944 | 5.812486 | 0 | − 1.8589058 | Irf8 | Interferon regulatory factor 8 [Source:MGI Symbol;Acc:MGI:96395] [ENSMUST00000162775] |
| 15 | − 2.760572 | 5.737793 | 1.2296844 | − 0.068935394 | Prdm6 | PR domain containing 6 (Prdm6), mRNA [NM_001033281] |
| 16 | − 1.346303 | 5.5345974 | 0.86455584 | − 1.5482342 | Foxl1 | Forkhead box L1 (Foxl1), mRNA [NM_008024] |
| 17 | − 1.9157376 | 5.5081615 | 1.5659752 | − 0.92323875 | Negr1 | Neuronal growth regulator 1 (Negr1), transcript variant 1, mRNA [NM_001039094] |
| 18 | − 2.9770136 | 5.4516544 | 1.3360395 | − 0.24065113 | Bmp4 | Bone morphogenetic protein 4 (Bmp4), transcript variant 1, mRNA [NM_007554] |
| 19 | − 3.0122108 | 5.441319 | 0.7840228 | − 0.26249528 | Bean1 | Brain expressed, associated with Nedd4, 1 (Bean1), transcript variant 1, mRNA [NM_001141922] |
| 20 | − 0.8304815 | 5.440546 | 0.45523453 | − 0.61619663 | Ltbp4 | Latent transforming growth factor beta binding protein 4 (Ltbp4), transcript variant 1, mRNA [NM_175641] |
| 21 | − 4.281811 | 5.415837 | 0.8549876 | − 0.26267695 | Tcf21 | Transcription factor 21 (Tcf21), mRNA [NM_011545] |
| 22 | 0.30940437 | 5.3867254 | 0 | − 1.0120187 | Cntn4 | Contactin 4 (Cntn4), transcript variant 1, mRNA [NM_001109749] |
| 23 | − 0.0352149 | 5.2685137 | 0.8375006 | − 0.8934727 | Hcar1 | Hydrocarboxylic acid receptor 1 (Hcar1), mRNA [NM_175520] |
| 24 | − 1.1227269 | 5.265641 | 0.48159218 | − 1.4391389 | Negr1 | Neuronal growth regulator 1 (Negr1), transcript variant 2, mRNA [NM_177274] |
| 25 | 0.41755295 | 5.2641068 | 0 | − 1.005084 | Tril | TLR4 interactor with leucine-rich repeats (Tril), mRNA [NM_025817] |
| 26 | − 0.65405416 | 5.2553177 | 0.27284336 | − 1.1687546 | Bend5 | BEN domain containing 5 (Bend5), transcript variant 1, mRNA [NM_026279] |
| 27 | 0.6547599 | 5.2338214 | 0 | − 1.1381359 | Ripply3 | Ripply transcriptional repressor 3 (Ripply3), mRNA [NM_133229] |
| 28 | 0 | 5.189296 | 1.0285192 | − 0.7719319 | Wnt2 | Wingless-type MMTV integration site family, member 2 (Wnt2), mRNA [NM_023653] |
| 29 | 0 | 5.1865263 | 0.31024742 | − 1.5084703 | Efcc1 | EF hand and coiled-coil domain containing 1 [Source:MGI Symbol;Acc:MGI:3611451] [ENSMUST00000160743] |
| 30 | 1.6226387 | 5.186082 | 0 | − 1.0252223 | Plac9a | Placenta specific 9a (Plac9a), mRNA [NM_207229] |
Results from in vivo- and ex vivo-expanded mesenchymal cells from the upper mucosa and crypt are shown. Top 30 genes of in vivo mesenchymal cells isolated from cryptal layers are shown.
We next focused on genes with “receptor/channel” function and speculated that these genes might be developmentally and functionally essential for cryptal mesenchymal cells21. Among the extracted genes, Lepr, Hcar1, Vipr2, Agtra1b, and Adora1, all showed higher expression in cryptal mesenchymal cells based on the gene array (Fig. 3b and data not shown). The ligands of these receptors are leptin, lactate, vasoactive intestinal peptide, angiotensin II, and adenosine, respectively22–25, and some of the receptors (e.g., Lepr, Hcar1, and Adora1) are involved in mucosal homeostasis22,26,27. Among those receptors, leptin is an important adipokine, derived from adipose tissues, and is involved in obesity, immune responses, and inflammation28,29. Several receptor isoforms exist23,30, and indeed trans-variants 1 through 3 were highly expressed in cryptal mesenchymal cells (Fig. 3c). Collectively, gene profiling of the mesenchymal cells in the colonic crypt and luminal mucosa yielded previously unknown cryptal mesenchymal cell-receptors, which—especially in the case of receptors for leptin—might be important for mesenchymal cell function in crypts. To confirm this issue, we used qRT-PCR to further examine the expressions of the receptors (Fig. 3d). We used our stratified cell isolation method to obtain isolated cells that were CD45− EpCAM− and positive for the general mesenchymal cell markers, CD90 and gp3817, from the luminal mucosa and crypts. The expression levels of leptin receptors (LepRs) 1 through 3 were significantly higher in the cryptal CD90+ gp38+ mesenchymal cells than the luminal mesenchymal cells compared with other cellular components (Fig. 3d). In addition, surface leptin receptor expressions were higher in crypt mesenchymal cells (Fig. 3e).
These evidences possibly indicate the critical role of intrinsic leptin for functional regulation of mesenchymal cells, especially in crypts. Intriguingly, when cryptal mesenchymal cells were isolated from germ-free mice and examined for the expression of LepR, the degree of the receptor expression was unaltered regardless of the presence or absence of commensal microbiota (Supplemental Fig. 6). This result indicated the de novo programmed expression of the LepR in mesenchymal cells in the crypt and the expressions were dispensable of signaling from commensal bacteria.
Wnt2b expression is retained in cryptal mesenchymal cells through leptin and LepR
Because the receptors for leptin were dominantly expressed in the colonic cryptal mesenchymal cells (Fig. 3), we further evaluated the biological role of leptin and its receptors in terms of the function of cryptal mesenchymal cells. Cryptal mesenchymal cells are considered to be important in mucosal regeneration and homeostasis18; therefore, we examined whether the leptin system is involved in the production of cryptal niche factors.
Lactate has been reported to have a regulatory function in mucosal regeneration and mobility24,27. After intestinal injury due to either radiation or chemotherapy, lactate stimulation of receptors promotes production of niche factors, including WNTs, and stimulates stem cell proliferation27. We thus first evaluated whether leptin stimulation resulted in the production of crypt niche-related factors, such as Wnt2b, Wnt5a, Rspo3, Grem1, and Bmp1, by comparing the results before and after leptin stimulation. We found that Wnt2b was indeed induced after leptin stimulation, thus indicating the potential involvement of leptin in mucosal protection and repair from injury (Fig. 4a).
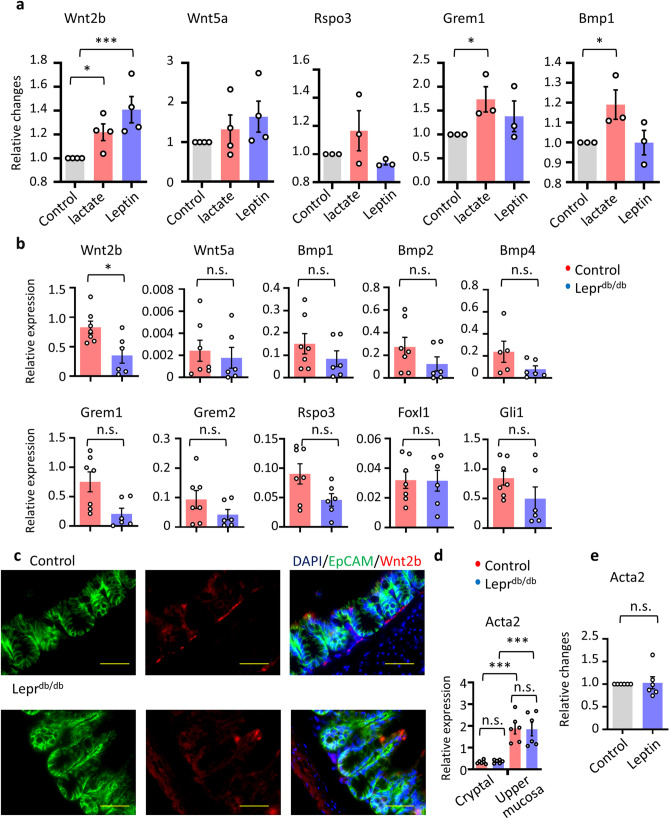
Figure 4.
Wnt2b expression is retained in cryptal mesenchymal cells through the LepR. (a) ex vivo cryptal mesenchymal cells were stimulated by leptin or lactate and the expression of cryptal stem niches genes was examined. Relative changes normalized by normalized against the expression of Gapdh were shown. Each dot represents an individual subject (n = 3 or 4) (b,c) Cryptal mesenchymal cells were sorted from control and Leprdb/db mice. (b) The expression of cryptal stem niches genes was examined by qRT-PCR analysis. Each result was normalized against the expression of Gapdh. Each dot represents an individual subject (n = 6 or 7). (c) Colon tissues of control and Leprdb/db mice were stained by Wnt2b and EpCAM. Representative data from 3 mice are shown. Scale bar, 50 μm. (d) The expression of Acta2a was examined by qRT-PCR analysis. Each result was normalized against the expression of Gapdh. Each dot represents an individual subject (n = 6). (e) ex vivo cryptal mesenchymal cells were stimulated by leptin and the expression of Acta2 was examined (n = 6). Data are shown as means ± SEM; *P < 0.05, ***P < 0.01. n.s. indicates not significant.
We next evaluated LepR-deficient mice (Leprdb/db) regarding their gene expression levels of representative intestinal stem cell niche factors derived from cryptal CD90+ gp38+ mesenchymal cells, including WNTs, BMPs, GREMs, or transcription factors (e.g., Gli1 and Foxl1) (Fig. 4b)23. The expression of BMPs, Grem1, Grem2, Rspo3, transcription factors (Gli1 and Foxl1), and Wnt5a in cryptal mesenchymal cells was not significantly different between Leprdb/db and control strains (Fig. 4b)11,20. In agreement with the stimulation analysis (Fig. 4a), the expression of Wnt2b was significantly reduced in the cryptal mesenchymal cells from LepR-deficient Leprdb/db mice compared with controls, indicating the importance of LepR signaling for Wnt2b expression (Fig. 4b).
Importantly, histological analysis revealed that a strong Wnt2b signal was present in the cryptal region of control mice (Fig. 4c), whereas this signal was much weaker in the Leprdb/db mice. These findings are consistent with the gene expression data in Fig. 4b.
Recently it has been indicated that LepR+ mesenchymal cells found in the bone marrow are α-smooth muscle actin (Acta2)+ myofibroblasts and are considered as a source of osteoblasts and adipocytes31,32. We examined the expression of Acta2 in the cryptal and upper mucosal CD90+ gp38+ mesenchymal cells isolated by our stratified cell isolation methods. Acta2 expression was low in the cryptal mesenchymal cells, indicating these cells are not myofibroblasts but fibroblasts (Fig. 4d). In addition, Acta2 expression levels in WT mice were comparable to those in the cells from LepR-deficient mice (Fig. 4d). In addition, in vitro stimulation with leptin of CD90+ gp38+ mesenchymal cells isolated from LepR-deficient mice caused no change in Acta2 expression (Fig. 4e), indicating the indispensable role of leptin signals in Acta2 expression and myofibroblast development in the steady state (Fig. 4d).
These results collectively indicate the involvement of leptin in the production of Wnt2b, which is essential for the survival and proliferation of intestinal epithelial cells, for gut homeostasis after mucosal inflammation, indicating that leptin signals through the colonic cryptal mesenchymal cells are possibly involved in mucosal protection.
Leptin suppresses intestinal inflammation
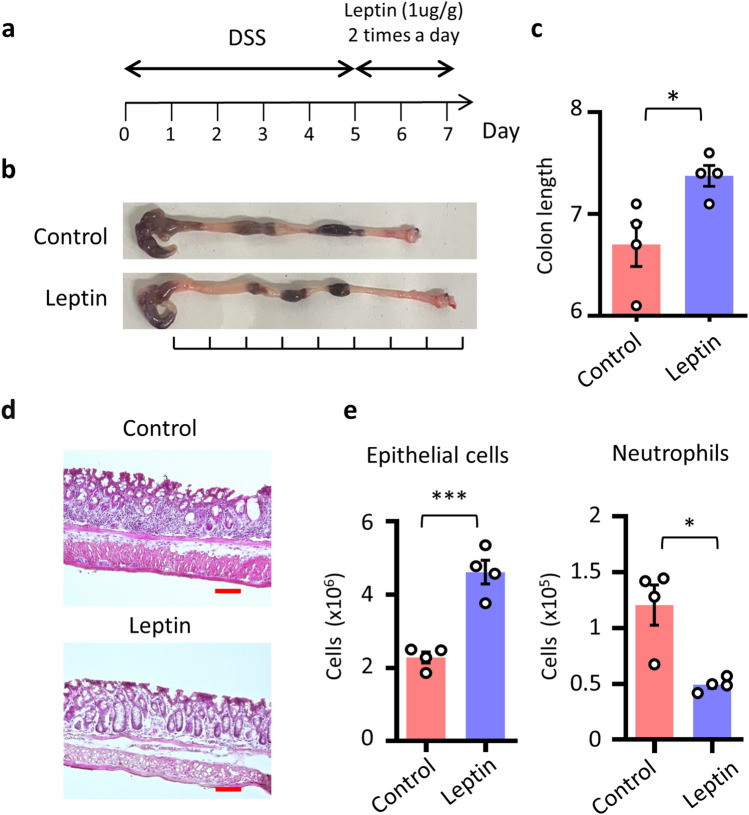
Our present study demonstrated that intrinsic leptin signals in cryptal mesenchymal cells are involved in the expression of Wnt2b, which is considered to be important for mucosal regeneration12. To begin with, we evaluated the protective effect of leptin in mucosal damage. We administered leptin to mice with dextran sodium sulfate (DSS)-induced colitis mice under conditions of reduced endogenous leptin signaling due to food deprivation after DSS administration33 (Fig. 5a): 1 μg/g (body weight) of leptin was administered intraperitoneally during the fasting period28 (Fig. 5a). Mice that received leptin showed reduction of colon shortening, which is one of the markers in the assessment of colonic inflammation (Fig. 5b,c). In addition, crypt morphology was revealed through hematoxylin-and-eosin staining of colonic sections, and FACS analysis was performed to evaluated epithelial cell number and neutrophil accumulation throughout the colon (Fig. 5d,e). These analyses revealed that the epithelial layer and crypt structure were similar between the leptin-treated and control groups but that the accumulation of neutrophils was decreased after leptin administration (Fig. 5d,e). These results suggested the important roles of leptin in the mucosal protection.
Figure 5.
Leptin administration protects intestinal inflammation. (a) Time course of this experiment. Mice underwent DSS treatment, and leptin (1 μg/g) was administered twice daily under fasting conditions to limit intrinsic leptin. (b–d) Representative images and colon length are shown (n = 4 per group). Scale bar, 100 μm. (e) Whole colon cells are isolated and the cell number of EPCAM+ epithelial cells and infiltrated CD11b+ Gr-1+ neutrophils in the colon were shown. *P < 0.05, ***P < 0.01. All data are representative of two independent experiments.
Intrinsic leptin controls mucosal homeostasis
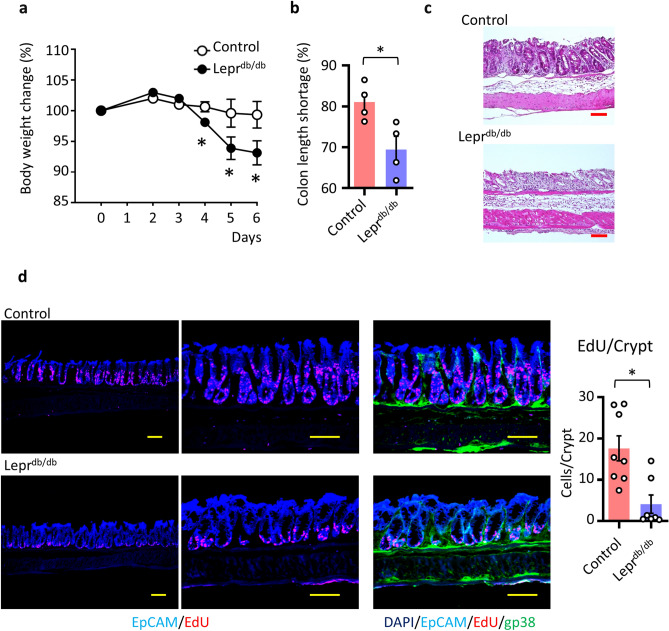
Wnt2b is involved in mucosal epithelial and cryptal regeneration during colitis8. Therefore, we sought to elucidate whether the leptin–cryptal mesenchymal pathway plays a critical role at the colonic mucosal epithelial barrier. To elucidate intrinsic leptin and LepR signaling in mucosal homeostasis, we exposed Leprdb/db and littermate control mice to DSS-induced colonic inflammation and examined them for sensitivity to colitis development (Fig. 6a,b). DSS-treated Leprdb/db mice showed more severe body weight loss and shortened colon length, representing well-known disease symptoms34 (Fig. 6a,b). In addition, histological assessment revealed that the mucosa of DSS-treated Leprdb/db mice showed necrosis and epithelial damage with erosion, loss of epithelial cells, and crypt ectasias (Fig. 6c). Epithelial damage involved deeper colonic crypts in Leprdb/db mice compared with control mice (Fig. 6c).
Figure 6.
Intrinsic leptin controls mucosal healing through LepR+ cryptal mesenchymal cells. (a) Body weight change (n = 4 per group). *P < 0.05. (b) Ratio of control and DSS treated mice colon length are shown. Data are shown as means ± SEM; *P < 0.05. (c) H and E staining of colon on day 6 of DSS treatment is shown. Scale bar, 100 μm. (d) Immunohistochemical analysis of DSS-treated mice on day 6. EdU was administered intraperitoneally 3 h before euthanasia. Tissues were stained with DAPI, EpCAM, and gp38 with detection of proliferated EdU+ cells. Representative data from 4 mice are shown. Scale bar, 100 μm. EdU+ cells counted in per crypt were shown. All data are representative of two independent experiments.
Epithelial morphology and the proportion of Ki67+ proliferated cells (indicative of epithelial turnover) in the steady state did not differ between Leprdb/db and control mice (Supplemental Fig. 7). However, during colitis, the proliferative zone of 5-ethynyl-2′-deoxyuridine (EdU)-stained (red cells) in the crypt was significantly reduced compared with that of control mice (Fig. 6d). These results collectively indicated the importance of Wnt2b from cryptal mesenchymal cell populations through intrinsic hormones, especially leptin, for the maintenance of mucosal homeostasis. Thus, our novel stratified layer isolation strategies uncovered novel and unique characteristics of cryptal mesenchymal cells, sensing intrinsic leptin for controlling mucosal homeostasis.
Discussion
Mesenchymal cells in gut play important roles in the orchestration of immune cells for lymphoid organogenesis, regulation of immune responses, and regeneration of the epithelial barrier5. Epithelial regeneration is an important process for mucosal homeostasis and is consistent with consequent processes such as coverage of denuded compartments, and stem cell proliferation and development of a functional epithelial layer, including columnar epithelial cells, secretory type cells (e,g., goblet cells and Paneth cells), and enteroendocrine cells17. Cytokines involved in epithelial development, such as EGF and WNTs, are produced from Paneth cells and mesenchymal cells, located in the crypt of the small or large intestines5,9. Especially in the colon, which lacks Paneth cells, mesenchymal cells have major roles in crypt and epithelial development and homeostasis11.
Accumulated evidence has revealed the involvement of mesenchymal cells, such as fibroblasts and myofibroblasts, which are located in the cryptal or pericryptal regions, in the regulation of epithelial–immunological homeostasis17. LepR+ mesenchymal cells have been shown to possess specific functions in other organs, such as bone marrow31,32; however, LepR signaling is not involved in the certain function of this specific cell populations31. In colon, recent single-cell RNA-Seq analysis revealed that at least four subsets of fibroblasts are present in the mice colon4,35. We have not evaluated whether LepR+ pericryptal mesenchymal cells are a specific sub-population in the CD34+ population. Future studies will have to define whether LepR+ cells are a specific sub-population of fibroblasts.
In this context, a unique type of interstitial cell, with long projections, or telopodes, has recently been found; these cells are called telocytes and are located surrounding crypts36. Telocytes express CD34 but not c-kit, unlike another type of intestinal crypt cell, Cajal cells, which are the pacemakers of peristaltic movement5. Cryptal mesenchymal cells obtained by the stratified isolation protocol contained crypt cells that expressed CD34 and Foxl1, hallmarks of fibroblasts and telocytes, thus supporting the methodological validity and reliability of the present study (Fig. 3). Conditional deletion of Wnt production from Foxl1+ cells led to the abolishment of stem and transit-amplifying cell proliferation20. However, our finding revealed that other CD34+ cell populations are also located in the other layers, such as the epithelial layers and MM, in the colon (Fig. 2). The past studies seemed to not consider the potential for the contamination of smooth-muscle cells and mesenchymal cells located in MM, submucosa, and MEx, because whole-cell isolation methods with digestive enzymes generally were used8,18. In fact, CD34+ CD90+ mesenchymal cells exist in the MM fraction as well (Fig. 2)8,18. Therefore, here we have shown the methodological significance and compelling utility of our stratified isolation method in designing experiments that analyze various histologically distinct parts of the colonic mucosa (e.g., lumen to crypt).
From our analysis of cryptal mesenchymal cells, we learned the beneficial feature of leptin in promoting epithelial turnover in the inflammatory state. Even though the current histological analysis of the intestine showed no modification in the architecture of the epithelium in Leprdb/db mice (Supplemental Fig. 7)37, a recent study has indicated that physiological intestinal barrier functions are weakened during decreased leptin signaling38. In particular, hyperglycemia in Leprdb/db mice caused disruption of tight junction integrity, leading to enhanced influx of luminal molecules, increased electrical current across the epithelial layer, and heightened susceptibility to enteric infection38. In addition, the induction of hyperleptinemia through subcutaneous administration of exogeneous leptin increased the mucosal barrier and lessened damage severity39, indicating the possible involvement of leptin–mesenchymal pathways in novel regulatory pathways of mucosal protection. However, the regulation of epithelial regeneration via indirect pathways of cryptal mesenchymal cells with alternative leptin pathways might be involved in the phenotype of Leprdb/db mice. For example, leptin is involved in gastric motility as well as in the migration of CCR7+ dendritic cells and differentiation of T cells (e.g., Th1/Th2 cytokine balance and Th17 differentiation)26,40–42. We found that the expression level of LepRs was higher in colonic cryptal mesenchymal cells compared with epithelial cells and CD45+ hematopoietic cells (Fig. 3d). Therefore, we emphasize a critical role of leptin and its receptor interaction on colonic cryptal mesenchymal cells; however, we cannot ignore the possible involvement or crosstalk of leptin to or with other cellular populations.
Leptin has also been shown to be associated with obesity, a form of chronic inflammation, considered in the etiopathogenesis of intestinal inflammation43. Obesity is the hallmark phenotype of Leprdb/db mice, and it may be that obesity causes delayed mucosal regeneration. However, obesity is a proinflammatory state in mice and humans, and adipose tissue produces inflammatory cytokines; therefore, these cytokines may contribute to delayed epithelial growth via another mechanism44,45.
For example, various adipocytokines (e.g., adiponectin, apelin, chemerin, leptin) are secreted from adipocytes, but in obesity, the regulation of the production of these adipocytokines is aberrant due to enlarged adipocytes46,47. Furthermore, inflammation-associated pathological alterations of adipocytes in mesenteric fat have previously been reported46, as have reduced expression levels of adiponectin in colitis mice44,45. Our group and others have reported on the functional transition of mesenchymal fat (e.g., increase of TNF-α and reduction of adipokines) and its effects on immune and epithelial integrity in colitis45,48. Adiponectin has an anti-inflammatory reaction against TNF-α pathways, such as NF-κB activation49. Indeed, we found a reduction of leptin expression in the adipose tissue of colitis (Supplemental Fig. 8). The use of the stratified isolation method opens new areas of investigation into the molecular and cellular interactions between anatomically and histologically different colonic crypt cells and adipose tissues.
Our findings from gene profiling revealed that several receptors in addition to LepR might contribute to the functioning of crypt mesenchymal cells (Fig. 3, Table 1). In particular, receptors for lactate and adenosine have been reported in regard to their regulatory function in mucosal regeneration and mobility22,24,27. In addition, lactate produced from Bifidobacterium and Lactobacillus spp. stimulates the lactate receptor expressed in the Paneth cells and fibroblasts of the small intestine27. After intestinal injury due to radiation and chemotherapy drugs, lactate stimulation of receptors promotes Wnt3 production and stimulates stem cell proliferation27. Further investigation to examine and elucidate the corporative regulatory systems through the multiple receptors that exist in the crypt region is warranted.
Taking together all of these previous and current findings, our new and reliable stratified isolation method revealed the unique characteristics of various types of mesenchymal cells isolated from different histological locations in the colonic mucosa and disclosed a novel pathway for intrinsic control of leptin-mediated mucosal protection (Supplemental Fig. 9). Our initial findings from using this stratified method for isolating mesenchymal cells yield insight into mechanisms of epithelial regeneration and treatment strategies for intestinal inflammation.
Materials and methods
Mice
Male C57Bl/6 mice, BKS.Cg- + Leprdb/ + Leprdb/Jcl (Leprdb/db mice), and BKS.Cg-m+/m+ + Jcl (control litter) mice (age, 7 to 10 weeks) purchased from CLEA (Tokyo, Japan) were used throughout the study. Twist2-Cre and Rosa26-tdTomato mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA), Col1a2-GFP mice were a gift, as previously described50. Colitis was induced by using 2.25% DSS, and body weight was measured as described previously51. For the leptin administration, mice 1ug/g body weight of leptin or PBS as control were administered intraperitoneally during fasting period for two days according to the previous publication33.
All mice were maintained under specific-pathogen-free (SPF) conditions at the experimental animal facility of the Institute of Medical Science, The University of Tokyo and Chiba University. All experiments were approved by the Animal Care and Use Committee of the University of Tokyo (PA17-89 and PA17-96) and Chiba University (A30-112). All procedures were performed in accordance with relevant guidelines and regulations.
Stratified isolation of mouse colon tissues
Colons were collected from mice, washed with PBS, opened vertically, and shaken 40 times with 25 ml of PBS to remove fecal contents and mucus. These processes were repeated twice. Harvested colon tissues were incubated in cell recovery solution (Corning, High Wycomb, UK)45,52 for 1 h on ice (Supplemental Fig. 1). Then, the epithelial and sub-epithelial cells were scraped off gently by using the round part of a J-shaped microtweezer until the honeycomb structures of the mucosal lamina propria were seen under microscopy (model S9D, Leica, Wetzlar, Germany); simultaneously MEx was peeled off (Supplementary Video). After the MEx was peeled from the colon tissue, each colonic layer was isolated as follows. To isolate the upper mucosal cells, upper mucosa was scraped gently away by using the round part of J-shaped microtweezers and scraper (Supplemental Fig. 1 and Supplementary Video). Then, the colonic tissues were turned over, and the MM and submucosal compartments were peeled off. The layer remaining is consistent with the cryptal layer (Supplemental Fig. 1 and Supplementary Video). All procedures are available in the technical video.
Cell collection and FACS analysis
Mononuclear cells were isolated from each colonic layer, according to slight modifications from previously described protocols51. Briefly, epithelium was dissociated by using a cell recovery solution (Becton Dickinson, San Jose, CA, USA) as described previously52, and the isolated de-epithelialized layers of whole colon were digested in 1.25 mg/ml collagenase at 37 °C (WAKO, Takasaki, Gunma, Japan) for 80 min51. The collected mononuclear cells were subsequently filtered through a 100-μm cell strainer (BD Biosciences, San Jose, CA, USA) and examined further.
For flow cytometric analysis, cells were incubated for 5 min with anti-CD16/32 antibody (5 μg/ml; Fc Block, BD Pharmingen, Franklin Lakes, NJ) and stained for 30 min at 4 °C with 7-AAD and fluorescent-labeled antibodies specific for CD11b, CD34, CD45, CD81, CD90.2, EpCAM, gp38, Gr-1, and TER119 (BioLegend, San Diego, CA, USA) and leptin receptor (Abcam, Cambridge, MA, USA). Cells were analyzed by flow cytometry (FACS Canto II, Becton Dickinson) or Attune NxT Acoustic Focusing Flow Cytometer (Thermo Fisher Scientific, MA, USA).
To expand mesenchymal cells ex vivo, CD90+ gp38+ CD45− EpCAM− cells were isolated by using a FACS Aria III (Becton Dickinson) and cultured with 10 ng/ml of EGF (PeproTech, Rocky Hill, NJ, USA). Cells were stimulated by adding 200 ng/ml leptin (Sigma, St. Louis, MO, USA) and 5 mM lactate (Sigma) to the media and incubating overnight; stimulated cells were collected for further analysis27,53.
Quantitative RT-PCR and gene microarray analyses
Total RNA was prepared by using TRIZOL (Thermo Fisher Scientific, Waltham, MA, USA) and reverse-transcribed by using Superscript IV VILO (Thermo Fisher Scientific), as described previously54. Quantitative RT-PCR analysis used the LightCycler 96 (Thermo Fisher Scientific) and the Universal Probe Library (Roche, Basel, Switzerland). The primer set are listed in Table 2.
Table 2.
Primer lists for the experiments.
| Forward 5′–3′ | Reverse 5′–3′ | |
|---|---|---|
| Gapdh | TGTCCGTCGTGGATCTGAC | CCTGCTTCACCACCTTCTTG |
| Wnt2b | CCGGGACCACACTGTCTTT | GCTGACGAGATAGCATAGACGA |
| Wnt5a | ACGCTTCGCTTGAATTCCT | CCCGGGCTTAATATTCCAA |
| Rspo3 | TCAAAGGGAGAGCGAGGA | CAGAGGAGGAGCTTGTTTCC |
| Grem1 | GACCCACGGAAGTGACAGA | CCCTCAGCTGTTGGCAGTAG |
| Grem2 | GAGGAGAGGGACAGGGAGAC | AGCGAGAGCTTCCAGAACAT |
| Bmp1 | GAAGGCCATCCATCAAAGC | CCACTAGTGCCCTGACCAC |
| Bmp2 | AGATCTGTACCGCAGGCACT | GTTCCTCCACGGCTTCTTC |
| Bmp4 | GAGGAGTTTCCATCACGAAGA | GCTCTGCCGAGGAGATCA |
| Gli1 | TGGAGGTCTGCGTGGTAGA | TTGAACATGGCGTCTCAGG |
| Foxl1 | CCATGAAGAAGGGACAAAGC | CCACCGGGGAGTCCTAAG |
| Lepr1 | GTTCCAAACCCCAAGAATTG | TGATTCTGCATGCTTGGTAAA |
| Lepr2 | CATTTCCGCTTCAATATCAGG | CCAGCAGAGATGTAGCTGAGAC |
| Lepr3 | GTTCCAAACCCCAAGAATTG | GACTTCAAAGAGTGTCCGTTCTC |
| Leptin | CAGGATCAATGACATTTCACACA | GCTGGTGAGGACCTGTTGAT |
| Hcar1 | GGCTGAGAAAAGCGGTATGA | TCGTTAACTCTCTCCGAGCTAGA |
| Vipr2 | GGCCTGGTGGTAGCAGTTC | TCGCCATCTTCTTTTCAGTTC |
| Adora1 | GTCAAGATCCCTCTCCGGTA | CAAGGGAGAGAATCCAGCAG |
| Agtr1b | CGCCAGCAGCACTGTAGA | GGGGGTGAATTCAAAATGC |
Sequence of primers for the experiments are listed.
Microarray analysis was performed as in our previous report54. Briefly, mononuclear cells were sorted, and total RNA was extracted from them by using TRIZOL (Thermo Fisher Scientific). cRNA was hybridized with DNA probes on a GeneChip Mouse Genome array (Agilent, Santa Clara, CA, USA). Data were analyzed by using GeneSpring software (Silicon Genetics, Redwood City, CA, USA).
Histological analysis
Hematoxylin and eosin staining (Muto Pure Chemicals, Tokyo, Japan) and immunohistochemical analysis were performed as previously described51. For immunohistochemistry, samples of large intestine were fixed in 4% paraformaldehyde (Wako) and incubated in 20% sucrose. The tissues were embedded in OCT compound (Sakura Fine Technical Company, Tokyo, Japan). Tissue sections were stained with anti-mouse EpCAM (Biolegend, San Diego, CA, USA), anti-mouse gp38 (Biolegend), Ki67 (Biolegend), and Wnt2b (Abcam). Anti-rabbit IgG conjugated with Alexa 488 (Thermo Fisher Scientific) served as a secondary antibody.
EdU analysis
At 3 h before euthanasia, mice were intraperitoneally injected with 100 µg/200 µl of EdU (Thermo Fisher Scientific). EdU staining was performed in accordance with the manufacturer’s instructions.
Statistical analysis
Statistical analysis was performed by using unpaired, two-tailed Student’s t-tests (Prism 8, GraphPad Software, San Diego, CA, USA). A P value < 0.05 was considered to be statistically significant. Data are reported as mean ± SEM (error bars in figures).
Supplementary information
Acknowledgements
We thank Kumiko Fujisawa, Akie Inami, Daiki Yamamoto, Sean Nelson, Kaoru Shimada, Akie Arakawa, and Akiko Naito (University of Tokyo) for technical support. We also thank Dr. Satoru Nagata (Department of Pediatrics, Tokyo Women’s Medical University) for support and Kanako Shimizu, Hiroko Yamamoto (Chiba University), Kayo Satonaga, and Keiko Warren (University of Tokyo) for their assistance. We acknowledge Dr. Sohtaro Mine (NIH) for discussions regarding histology. This work was supported by grants from The Ministry of Education, Culture, Sports, Science, and Technology for the Translational Research Network Program (at the University of Tokyo) Seeds A (Y.K.), B (H.K.), and C (H.K.) and LEADER (Y.K.); the Japan Agency for Medical Research and Development for CREST (H.K.) and PRIME (Y.K.); the Japan Society for the Promotion of Science for a Grant-in Aid for Scientific Research S (18H05280 to H.K.) and Scientific Research B (19H03450 to Y.K.), Young Scientists A (16H06243 to Y.K.), Challenging Exploratory Research (17K19550 and 19K22634 to Y.K.), Funds for the Promotion of Joint International Research (18KK0432 to Y.K.), Science and Technology Research Partnership for Sustainable Development (H.K.); the GSK Japan Research Grant (Y.K.), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (Y.K.), the Takeda Science Foundation (Y.K.), the Uehara Memorial Foundation (Y.K.), the Sumitomo Foundation (Y.K.), the Naito Foundation (Y.K), the Kato Memorial Bio Science Foundation (Y.K.), the Yakult Bio-Science Foundation, and NipponHam Foundation (Y.K.); and the Chiba University–UC San Diego Center for Mucosal Immunology, Allergy, and Vaccines (H.K.) and RO1 (RO1 AI079145 to P.B.E), Center (DK12515 to P.B.E).
Author contributions
S.M. and Y.K. conducted the research and performed experiments; S.Mura., M.M., F.A., Y.S., N.K., and D.K. conducted experiments; Y.I. developed mice; T.K., P.B.E., T.S., and H.K. contributed to the experimental design and data analysis; and Y.K., and H.K. supervised the project and wrote the manuscript. All authors approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Seiichi Matsumura and Yosuke Kurashima.
Supplementary information
is available for this paper at 10.1038/s41598-020-75186-3.
References
- 1.Kurashima Y, Kiyono H. Mucosal ecological network of epithelium and immune cells for gut homeostasis and tissue healing. Annu. Rev. Immunol. 2017;35:119–147. doi: 10.1146/annurev-immunol-051116-052424. [DOI] [PubMed] [Google Scholar]
- 2.Lazar V, et al. Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front. Immunol. 2018;9:1830. doi: 10.3389/fimmu.2018.01830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andoh A, Bamba S, Brittan M, Fujiyama Y, Wright NA. Role of intestinal subepithelial myofibroblasts in inflammation and regenerative response in the gut. Pharmacol. Ther. 2007;114:94–106. doi: 10.1016/j.pharmthera.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Kinchen J, et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell. 2018;175:372–386.e317. doi: 10.1016/j.cell.2018.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurashima Y, Tokuhara D, Kamioka M, Inagaki Y, Kiyono H. Intrinsic control of surface immune and epithelial homeostasis by tissue-resident gut stromal cells. Front. Immunol. 2019;10:1281. doi: 10.3389/fimmu.2019.01281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powell DW, et al. Myofibroblasts. II. Intestinal subepithelial myofibroblasts. Am J Physiol. 1999;277:C183–C201. doi: 10.1152/ajpcell.1999.277.2.C183. [DOI] [PubMed] [Google Scholar]
- 7.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stzepourginski I, et al. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl. Acad. Sci. USA. 2017;114:E506–E513. doi: 10.1073/pnas.1620059114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farin HF, Van Es JH, Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 2012;143:1518–1529.e1517. doi: 10.1053/j.gastro.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 10.Kabiri Z, et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development. 2014;141:2206–2215. doi: 10.1242/dev.104976. [DOI] [PubMed] [Google Scholar]
- 11.Degirmenci B, Valenta T, Dimitrieva S, Hausmann G, Basler K. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature. 2018;558:449–453. doi: 10.1038/s41586-018-0190-3. [DOI] [PubMed] [Google Scholar]
- 12.Valenta T, et al. Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep. 2016;15:911–918. doi: 10.1016/j.celrep.2016.03.088. [DOI] [PubMed] [Google Scholar]
- 13.Pangtey B, Kaul JM, Mishra S. histogenesis of muscularis mucosa and muscularis externa of stomach: A human foetal study. J. Clin. Diagn. Res. 2017;11:AC01–AC03. doi: 10.7860/JCDR/2017/26219.10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gehart H, Clevers H. Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 2019;16:19–34. doi: 10.1038/s41575-018-0081-y. [DOI] [PubMed] [Google Scholar]
- 15.Kosinski C, et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc. Natl. Acad. Sci. USA. 2007;104:15418–15423. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen MM, Witherden DA, Havran WL. gammadelta T cells in homeostasis and host defence of epithelial barrier tissues. Nat. Rev. Immunol. 2017;17:733–745. doi: 10.1038/nri.2017.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurashima Y, et al. Mucosal mesenchymal cells: Secondary barrier and peripheral educator for the gut immune system. Front. Immunol. 2017;8:1787. doi: 10.3389/fimmu.2017.01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karpus ON, et al. Colonic CD90+ crypt fibroblasts secrete semaphorins to support epithelial growth. Cell Rep. 2019;26:3698–3708.e3695. doi: 10.1016/j.celrep.2019.02.101. [DOI] [PubMed] [Google Scholar]
- 19.McCarthy N, et al. Distinct mesenchymal cell populations generate the essential intestinal BMP signaling gradient. Cell Stem Cell. 2020;26:391–402.e395. doi: 10.1016/j.stem.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoshkes-Carmel M, et al. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature. 2018;557:242–246. doi: 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grande MT, Lopez-Novoa JM. Fibroblast activation and myofibroblast generation in obstructive nephropathy. Nat. Rev. Nephrol. 2009;5:319–328. doi: 10.1038/nrneph.2009.74. [DOI] [PubMed] [Google Scholar]
- 22.Antonioli L, et al. Differential recruitment of high affinity A1 and A2A adenosine receptors in the control of colonic neuromuscular function in experimental colitis. Eur. J. Pharmacol. 2011;650:639–649. doi: 10.1016/j.ejphar.2010.10.041. [DOI] [PubMed] [Google Scholar]
- 23.Chen H, et al. Evidence that the diabetes gene encodes the leptin receptor: Identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84:491–495. doi: 10.1016/s0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 24.El-Zaatari M, Kao JY. Role of dietary metabolites in regulating the host immune response in gastrointestinal disease. Front. Immunol. 2017;8:51. doi: 10.3389/fimmu.2017.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paz Ocaranza M, et al. Counter-regulatory renin-angiotensin system in cardiovascular disease. Nat. Rev. Cardiol. 2019 doi: 10.1038/s41569-019-0244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madan R, et al. Role of leptin-mediated colonic inflammation in defense against clostridium difficile colitis. Infect. Immun. 2014;82:341–349. doi: 10.1128/IAI.00972-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YS, et al. Microbiota-derived lactate accelerates intestinal stem-cell-mediated epithelial development. Cell Host Microbe. 2018;24:833–846.e836. doi: 10.1016/j.chom.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Abella V, et al. Leptin in the interplay of inflammation, metabolism and immune system disorders. Nat. Rev. Rheumatol. 2017;13:100–109. doi: 10.1038/nrrheum.2016.209. [DOI] [PubMed] [Google Scholar]
- 29.Pan WW, Myers MG., Jr Leptin and the maintenance of elevated body weight. Nat. Rev. Neurosci. 2018;19:95–105. doi: 10.1038/nrn.2017.168. [DOI] [PubMed] [Google Scholar]
- 30.Lee GH, et al. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379:632–635. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 31.Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Decker M, et al. Leptin-receptor-expressing bone marrow stromal cells are myofibroblasts in primary myelofibrosis. Nat. Cell Biol. 2017;19:677–688. doi: 10.1038/ncb3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faggioni R, Moser A, Feingold KR, Grunfeld C. Reduced leptin levels in starvation increase susceptibility to endotoxic shock. Am. J. Pathol. 2000;156:1781–1787. doi: 10.1016/S0002-9440(10)65049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wirtz S, et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 2017;12:1295–1309. doi: 10.1038/nprot.2017.044. [DOI] [PubMed] [Google Scholar]
- 35.Roulis M, et al. Paracrine orchestration of intestinal tumorigenesis by a mesenchymal niche. Nature. 2020;580:524–529. doi: 10.1038/s41586-020-2166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ibba-Manneschi L, Rosa I, Manetti M. Telocyte implications in human pathology: An overview. Semin. Cell Dev. Biol. 2016;55:62–69. doi: 10.1016/j.semcdb.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 37.Tavernier A, et al. Intestinal deletion of leptin signaling alters activity of nutrient transporters and delayed the onset of obesity in mice. FASEB J. 2014;28:4100–4110. doi: 10.1096/fj.14-255158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thaiss CA, et al. Hyperglycemia drives intestinal barrier dysfunction and risk for enteric infection. Science. 2018;359:1376–1383. doi: 10.1126/science.aar3318. [DOI] [PubMed] [Google Scholar]
- 39.Rivero-Gutierrez B, et al. Exogenous leptin reinforces intestinal barrier function and protects from colitis. Pharmacol. Res. 2019;147:104356. doi: 10.1016/j.phrs.2019.104356. [DOI] [PubMed] [Google Scholar]
- 40.Al-Hassi HO, et al. A mechanistic role for leptin in human dendritic cell migration: Differences between ileum and colon in health and Crohn's disease. Mucosal Immunol. 2013;6:751–761. doi: 10.1038/mi.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lord GM, et al. Leptin modulates the T-cell immune response and reverses starvation-induced immunosuppression. Nature. 1998;394:897–901. doi: 10.1038/29795. [DOI] [PubMed] [Google Scholar]
- 42.Yarandi SS, Hebbar G, Sauer CG, Cole CR, Ziegler TR. Diverse roles of leptin in the gastrointestinal tract: Modulation of motility, absorption, growth, and inflammation. Nutrition. 2011;27:269–275. doi: 10.1016/j.nut.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khalili H, et al. The role of diet in the aetiopathogenesis of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2018;15:525–535. doi: 10.1038/s41575-018-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karagiannides I, et al. Induction of colitis causes inflammatory responses in fat depots: Evidence for substance P pathways in human mesenteric preadipocytes. Proc. Natl. Acad. Sci. USA. 2006;103:5207–5212. doi: 10.1073/pnas.0600821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi Y, et al. Reciprocal inflammatory signaling between intestinal epithelial cells and adipocytes in the absence of immune cells. EBioMedicine. 2017;23:34–45. doi: 10.1016/j.ebiom.2017.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peyrin-Biroulet L, et al. Mesenteric fat in Crohn’s disease: A pathogenetic hallmark or an innocent bystander? Gut. 2007;56:577–583. doi: 10.1136/gut.2005.082925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weidinger C, Ziegler JF, Letizia M, Schmidt F, Siegmund B. Adipokines and their role in intestinal inflammation. Front. Immunol. 2018;9:1974. doi: 10.3389/fimmu.2018.01974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, et al. Interleukin-22 alleviates metabolic disorders and restores mucosal immunity in diabetes. Nature. 2014;514:237–241. doi: 10.1038/nature13564. [DOI] [PubMed] [Google Scholar]
- 49.Ouchi N, et al. Adiponectin, an adipocyte-derived plasma protein, inhibits endothelial NF-kappaB signaling through a cAMP-dependent pathway. Circulation. 2000;102:1296–1301. doi: 10.1161/01.cir.102.11.1296. [DOI] [PubMed] [Google Scholar]
- 50.Higashiyama R, et al. Negligible contribution of bone marrow-derived cells to collagen production during hepatic fibrogenesis in mice. Gastroenterology. 2009;137:1459–1466.e1451. doi: 10.1053/j.gastro.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 51.Kurashima Y, et al. Extracellular ATP mediates mast cell-dependent intestinal inflammation through P2X7 purinoceptors. Nat. Commun. 2012;3:1034. doi: 10.1038/ncomms2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rana B, Mischoulon D, Xie Y, Bucher NL, Farmer SR. Cell-extracellular matrix interactions can regulate the switch between growth and differentiation in rat hepatocytes: Reciprocal expression of C/EBP alpha and immediate-early growth response transcription factors. Mol. Cell Biol. 1994;14:5858–5869. doi: 10.1128/mcb.14.9.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feldman DE, Chen C, Punj V, Tsukamoto H, Machida K. Pluripotency factor-mediated expression of the leptin receptor (OB-R) links obesity to oncogenesis through tumor-initiating stem cells. Proc. Natl. Acad. Sci. USA. 2012;109:829–834. doi: 10.1073/pnas.1114438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurashima Y, et al. The enzyme Cyp26b1 mediates inhibition of mast cell activation by fibroblasts to maintain skin-barrier homeostasis. Immunity. 2014;40:530–541. doi: 10.1016/j.immuni.2014.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.