Abstract
Background: Ribonucleoside-diphosphate reductase subunit M2 (RRM2) is the catalytic subunit of ribonucleotide reductase and modulates the enzymatic activity, which is essential for DNA replication and repair. However, the role of RRM2 in lung adenocarcinoma (LUAD) remains unclear.
Methods: In this study, we explored the expression pattern and prognostic value of RRM2 in LUAD across TCGA, GEO, Oncomine, UALCAN, PrognoScan, and Kaplan-Meier Plotter, and confirmed its independent prognostic value via Cox analyses. LinkedOmics and GEPIA2 were applied to investigate co-expression and functional networks associated with RRM2. Besides, we used TIMER to assess the correlation between RRM2 and the main six types of tumor-infiltrating immune cells. Lastly, the correlations between immune signatures of immunomodulators, chemokines, and 28 tumor-infiltrating lymphocytes (TILs) and RRM2 were examined by tumor purity-corrected partial Spearman's rank correlation coefficient through TIMER portal.
Results: RRM2 was found upregulated in tumor tissues in TCGA-LUAD, and validated in multiple independent cohorts. Moreover, whether in TCGA or other cohorts, high RRM2 expression was found to be associated with poor survival. Cox analyses showed that high RRM2 expression was an independent risk factor for overall survival, disease-specific survival, and progression-free survival of LUAD. Functional network analysis suggested that RRM2 regulates RNA transport, oocyte meiosis, spliceosome, ribosome biogenesis in eukaryotes, and cellular senescence signaling through pathways involving multiple cancer-related kinases and E2F family. Also, RRM2 expression correlated with infiltrating levels of B cells, CD4+ T cells, and neutrophils. Subsequent analysis found that B cells and dendritic cells could predict the outcome of LUAD. B cells were identified as an independent risk factor among six types of immune cells through Cox analyses. At last, the correlation analysis showed RRM2 correlated with 67.68% (624/922) of the immune signatures we performed.
Conclusion: Our research showed that RRM2 could independently predict the prognosis of LUAD and was associated with immune infiltration. In particular, the tight relationship between RRM2 and B cell marker genes are the potential epicenter of the immune response and one of the critical factors affecting the prognosis. Our findings laid the foundation for further research on the immunomodulatory role of RRM2 in LUAD.
Keywords: RRM2, lung cancer, lung adenocarcinoma, prognosis, immune infiltrates, TIL
Introduction
Lung cancer is the leading cause of cancer-associated deaths worldwide 1-3. The survival rate of lung cancer depends mainly on the stage of diagnosis. In general, the current 5-year survival rate is about 18%; however, the prognosis can be improved when confirmed early 4. Unfortunately, only about 15% of cases were at the early stage when diagnosed, while the vast majority (57%) were already at the advanced stage 4. Lung adenocarcinoma (LUAD) is a subclass of non-small cell lung cancer (NSCLC), which develops along the outer edge of the lungs within glandular cells in the small airways. LUAD accounted for approximately 40% of all lung cancer cases being the most common type of histology 5.
Whereas, due to the combination of adverse factors that span a range of different biological and clinical behaviors and the increased resistance to anti lung cancer drugs, existing targeted drugs have shown unsatisfactory efficacy 6. In NSCLC, little is known about the genomic and host factors that drive the progression of pre-invasive lesions. Investigating these factors can enhance our understanding of lung cancer biology, help to develop better screening strategies, and improve patient prognosis 7. Furthermore, the lack of specific markers for disease stages or tumor types represents a fundamental gap in the current understanding and treatment of LUAD.
Ribonucleoside-diphosphate reductase subunit M2 (RRM2) is the catalytic subunit of ribonucleotide reductase and modulates the enzymatic activity, which is essential for DNA replication and repair 8. According to recent reports, RRM2 is involved in the progression of various cancers, including glioma, colorectal cancer, and bladder cancer 9, 10. Compared with normal tissues, RRM2 is overexpressed in breast cancer patients and is associated with poor survival 11. Recent studies have shown that RRM2 upregulation occurs in multiple myeloma tumors, and RRM2 inhibition can inhibit multiple myeloma cell proliferation 12. Rahman et al. demonstrated that alteration of RRM2 induces apoptosis by modulating Bcl-2 expression in lung cancer 13. Low expression of RRM2 has been reported can be used to value the treatment response to platinum-based chemotherapy of lung cancer 14. Immunohistochemical evaluation of RRM2 indicates that it has strong prognostic significance in some subsets of NSCLC patients (primarily women, non-smokers, and former smokers quitting longer than ten years) 15. Previous researches on the relationship between RRM2 and lung cancer were too specific but short of a comprehensive view 16-18. Moreover, whether RRM2 is a robust biomarker for LUAD, existing studies do not present a clear answer. Furthermore, the biological function of RRM2 in LUAD remains to be established.
In this study, we examined the expression and prognostic value of RRM2 in LUAD patients in the Cancer Genome Atlas (TCGA) and validated them in multiple independent cohorts. Moreover, using multidimensional analysis, we assessed the co-expression and functional network associated with RRM2 in LUAD and studied its part in tumor immunity. The present study may potentially reveal new direction, biological targets, and strategies for the diagnosis, treatment, and prognosis assessment of LUAD.
Materials and Methods
Data mining from TCGA
LUAD patients' gene expression profiles, along with their clinical data such as age, gender, tumor stage, TNM classification, and survival status, were downloaded from the TCGA portal (v22.0, https://portal.gdc.cancer.gov/) with project ID: TCGA-LUAD.
RRM2 differential expression
In TCGA-LUAD cohort, the analysis of differential mRNA expression of RRM2 in tumor and healthy tissues were examined by the Wilcoxon test, including unpaired or paired test. Oncomine (version 4.5, https://www.oncomine.org/) is a cancer microarray database and web-based data-mining platform. The mRNA expression level or copy number of RRM2 in LUAD and normal tissue were examined in Oncomine. In order to select the dataset to be included in this study from Oncomine, the screening parameters were set as follows: 1) Set “Analysis Type” as “Cancer vs. Normal Analysis” and “Cancer Type” as “Lung Adenocarcinoma”; 2) Set “THRESHOLD” as “P-value<0.05”, “FOLD CHANGE” as “ALL”, and “GENE RANK” as “ALL”. UALCAN (http://ualcan.path.uab.edu/) is an online tool for analyzing cancer transcriptome data, which is based on public cancer transcriptome data (TCGA and MET500 transcriptome sequencing) 19. The "CPTAC analysis" module of UALCAN provides protein expression analysis option using data from Clinical Proteomic Tumor Analysis Consortium (CPTAC) Confirmatory/Discovery dataset 20. The comparison of RRM2 protein expression between LUAD and normal lung was examined in UALCAN. Analyses in TCGA-LUAD and Oncomine were visualized through R package “beeswarm”. A difference was defined as significant at p-value < 0.05.
RRM2 expression in clinical characteristics sub-groups
The associations of RRM2 expression with clinical were examined by a non-parametric test (i.e., if the data were divided into two groups, the Wilcoxon test was performed; if the data were divided into three groups or more, the Kruskal-Wallis test was performed). R software was used for the visualization.
Survival analyses of RRM2
Survival analyses in the TCGA-LUAD cohort were conducted between high and low RRM2 expression groups through Kaplan-Meier analysis with the log-rank test. PrognoScan (http://dna00.bio.kyutech.ac.jp/PrognoScan/) is a database for meta-analysis of the prognostic value of genes 21. In order to select the dataset to be included in this study from PrognoScan, the screening parameters were set as follows: 1) Set “Cancer Type” as “Lung cancer” and “Subtype” as “Adenocarcinoma”; 2) Set “THRESHOLD” as “P-value<0.05”. Kaplan-Meier Plotter (https://kmplot.com/) is a web application developed for meta-analysis-based biomarker assessment that can be used for breast, ovarian, lung, gastric, and liver cancer 22. The correlations between RRM2 expression and survival in LUAD were additionally analyzed in PrognoScan and Kaplan-Meier Plotter. Analyses from TCGA-LUAD and PrognoScan were visualized through “survminer” and “survival” packages in R.
The independent prognostic value of RRM2
To identify the independent prognostic value of RRM2 in LUAD and assess the correlation between important clinical characteristics and prognosis, we performed Cox analyses. First, we conducted univariate Cox analysis on every variable, in turn, to check their correlation with prognosis. Then, all variables were gathered for a multivariate Cox analysis to evaluate whether each of them has an independent prognostic value.
LinkedOmics and GEPIA2 databases analysis
LinkedOmics (http://www.linkedomics.org) is a publicly available portal that includes multi-omics data from all 32 TCGA Cancer types23. In the “LinkFinder” module of LinkedOmics, we used the Pearson test to perform statistical analysis on RRM2 co-expression, and the results were displayed in the form of volcano, heat, or scatter plots. “LinkInterpreter” module of LinkedOmics was applied to conducted analyses of Gene Ontology (Biological Process), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, kinase-target enrichment, miRNA-target enrichment and transcription factor-target enrichment through Gene Set Enrichment Analysis (GSEA). The rank criterion was false discovery rate (FDR) < 0.05, and simulations was 1000. The Gene Expression Profiling Interactive Analysis (GEPIA2) database (http://gepia2.cancer-pku.cn/) is a web server for analyzing the RNA sequencing expression data of 9,736 tumors and 8,587 normal samples from the TCGA and the GTEx projects, using a standard processing pipeline24. GEPIA2 was applied to plot survival heatmaps of top co-expression genes and survival curves of top kinase regulators.
The correlation between RRM2 and six types of infiltrating immune cells
The Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) is a comprehensive resource for systematical analysis of immune infiltrates across diverse cancer types 25, 26. “Gene” module of TIMER was applied to explore the correlation between RRM2 expression and abundance of six types of immune cells infiltrates, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells, in LUAD, by tumor purity-corrected partial Spearman's correlation. Kaplan-Meier analysis was conducted to assess the prognostic value of each immune infiltrate. Multivariate Cox analysis was used to evaluate how RRM2 and these six types of immune cells together affect outcomes.
Correlation between RRM2 and immune signatures
TISIDB is a central portal for tumor and immune system interactions, which integrates multiple heterogeneous data types 27. This online tool contained various immune gene signatures categorized by type of immune or their function. Gene signatures of chemokine, receptor, major histocompatibility complex (MHC), immunoinhibitor, immunostimulator, and 28 tumor-infiltrating lymphocytes (TILs) 28 were downloaded. The correlations between RRM2 and these gene signatures were calculated via the “Correlation” module of TIMER with tumor purity-corrected partial Spearman's correlation.
Results
Patient characteristics
The clinical characteristics of 522 cases were obtained from the TCGA-LUAD cohort, including age, gender, tumor stage and TNM classification. As shown in Table 1, 241 (46.17%) cases were younger than or equal to 65 years old, 262 (50.19%) were older than 60 years old, while 19 (3.64%) were unknown. 280 (53.64%) were females, and 242 (46.36%) were males. Stage I was found in 279 patients (53.45%), stage II in 124 (23.75%), stage III in 85 (16.28%), stage IV in 26 (4.98%), and unknown in 8 (1.53%). The patients with T1 (32.95%) and T2 (53.83%) made up the majority of the total patients, and the remaining are T3 (9.00%), T4 (3.64%), and unknown (0.57%). 353 (67.62%) were at M0, 25 (4.79%) were at M1, while the rest 144 cases (27.59%) were unknown. In the distribution of N classification, N0, N1, N2, N3, and unknown accounted for 64.18%, 18.77%, 14.37%, 0.38%, and 2.30%, respectively.
Table 1.
The characteristics of patients in the TCGA-LUAD cohort
| Characteristic | Total (522) | Percentage (%) |
|---|---|---|
| Age | ||
| ≤65 | 241 | 46.17 |
| >60 | 262 | 50.19 |
| unknown | 19 | 3.64 |
| Gender | ||
| Female | 280 | 53.64 |
| Male | 242 | 46.36 |
| Tumor stage | ||
| Stage I | 279 | 53.45 |
| Stage II | 124 | 23.75 |
| Stage III | 85 | 16.28 |
| Stage IV | 26 | 4.98 |
| unknown | 8 | 1.53 |
| T classification | ||
| T1 | 172 | 32.95 |
| T2 | 281 | 53.83 |
| T3 | 47 | 9.00 |
| T4 | 19 | 3.64 |
| unknown | 3 | 0.57 |
| N classification | ||
| N0 | 335 | 64.18 |
| N1 | 98 | 18.77 |
| N2 | 75 | 14.37 |
| N3 | 2 | 0.38 |
| unknown | 12 | 2.30 |
| M classification | ||
| M0 | 353 | 67.62 |
| M1 | 25 | 4.79 |
| unknown | 144 | 27.59 |
TCGA: The Cancer Genome Atlas; LUAD: lung adenocarcinoma.
High RRM2 expression in LUAD
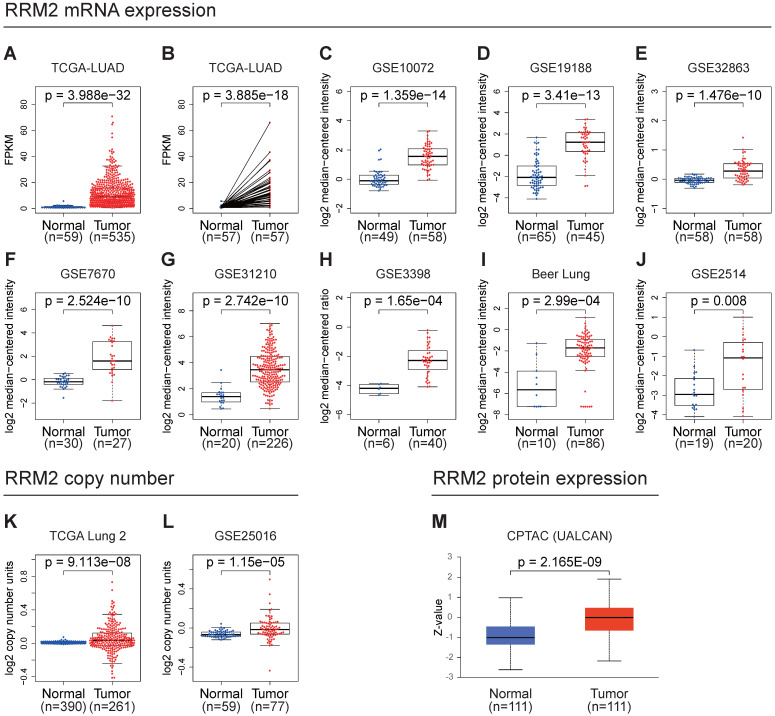
In the TCGA-LUAD cohort, we compared the mRNA expression of RRM2 in tumor and normal or their adjacent tissues. The unpaired and paired tests both indicated that the mRNA expression of RRM2 in LUAD was elevated (Figure 1A, 1B). Moreover, we checked the Oncomine online database finding that many datasets suggested that the mRNA expression of RRM2 increased in tumor tissues (Figure 1C-J). Besides, we found that the copy number of RRM2 also increased in tumor tissue (Figure 1K, 1L). We examined RRM2 protein expression in UALCAN database, discovering it was highly expressed in tumor tissues (Figure 1M).
Figure 1.
RRM2 is highly expressed in LUAD. (A) RRM2 mRNA expression comparison between normal and tumor tissues in the TCGA-LUAD cohort (unpaired Wilcoxon test). (B) RRM2 mRNA expression comparison between normal and adjacent tissues in the TCGA-LUAD cohort (paired Wilcoxon test). (C - J) RRM2 mRNA expression comparisons between normal and tumor tissues obtained from the Oncomine web tool (Wilcoxon test). (K - L) RRM2 copy number comparisons between normal and tumor tissues obtained from the Oncomine web tool (Wilcoxon test). (M) RRM2 protein expression comparison between normal and tumor tissues obtained from the UALCAN web tool (Wilcoxon test). TCGA: The Cancer Genome Atlas; LUAD: lung adenocarcinoma; CPTAC: Clinical Proteomic Tumor Analysis Consortium; The title of each graphic refers to the project name in TCGA, GEO, Oncomine, or UALCAN; P-value < 0.05 was used to assess differences.
Distribution of RRM2 expression in clinical characteristics sub-groups
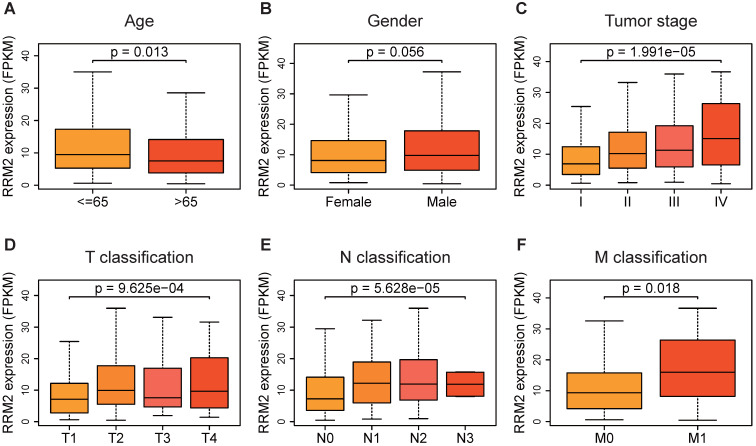
Based on the gene profile and clinical data extracted from TCGA-LUAD, the expression of RRM2 in patients with younger age (≤ 65 years old) was found significantly higher compared to patients who were older than 65 years old (p-value = 0.013, Figure 2A). The distribution of RRM2 showed a significant difference among the tumor stages. RRM2 was highly expressed as tumor stage increased (p-value = 1.991e-05, Figure 2C). Similarly, RRM2 increased with the N classification (p-value = 9.625e-04, Figure 2E). Moreover, RRM2 became increased expression in patients with tumor metastasis (p-value = 0.018, Figure 2F). As shown in Figure 2D, RRM2 was significantly differently distributed in the sub-groups of T classification, with highly expressed in T2 and T4 classification (p-value = 9.625e-04). Furthermore, different genders were not associated with the expression of RRM2 (Figure 2B).
Figure 2.
RRM2 expression in sub-groups of clinical characteristics. (A, B, F) RRM2 expression distribution analyses stratified based on age, gender, and M classification (Wilcoxon test). (C - E) RRM2 expression distribution analyses stratified based on tumor stage and TN classification (Kruskal-Wallis test). P-value < 0.05 was used to assess differences.
High RRM2 expression indicates significant worse survival in LUAD
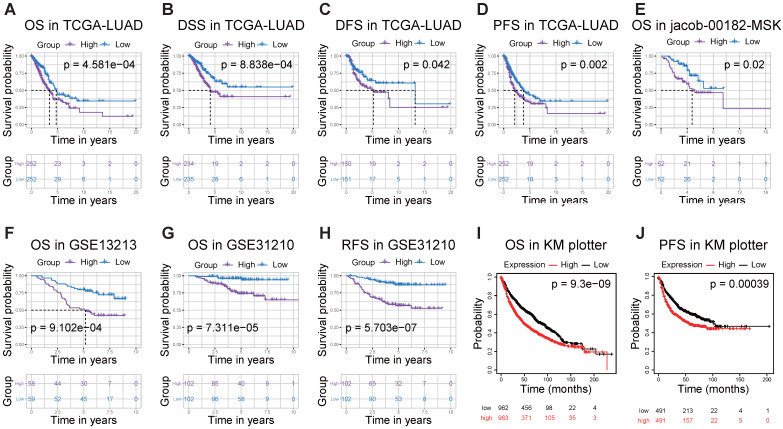
Then, to understand the correlation between RRM2 expression and patients' outcomes, we used the Kaplan-Meier survival curves to evaluate and compare the survival differences between patients with high and low (grouped according to median) expression of RRM2 (Figure 3). In the TCGA-LUAD cohort, the high RRM2 expression group had significantly shorter overall survival, and the median overall survival of group of high expression vs. low expression was 3.47 years vs. 4.73 years (log-rank test, p-value = 4.581e-04, Figure 3A). The high expression group also had a significant unfavorable disease-specific survival (log-rank test, p-value = 8.838e-04, Figure 3B) and disease-free survival (log-rank test, p-value = 0.042, Figure 3C) than that in low expression group. Furthermore, in the comparison of progression-free survival between high and low expression patients, the high expression group had a worse median survival than that in low expression group (2.13 years vs. 3.78 years, log-rank test, p-value = 0.002, Figure 3D). Besides, we checked the PrognoScan and Kaplan-Meier Plotter finding that high RRM2 expression associated with poor overall survival, relapse-free survival, and progression-free survival (Figure 3E-J).
Figure 3.
RRM2 is associated with survival outcome. (A - D) Survival analyses of RRM2 in the TCGA-LUAD cohort by Kaplan-Meier estimator with a log-rank test. (E - H) Survival analyses of RRM2 by Kaplan-Meier estimator with log-rank test obtained from PrognoScan web tool. (I, J) Survival analyses of RRM2 by Kaplan-Meier estimator with log-rank test obtained from the Kaplan Meier plotter web tool. Survival differences are compared between patients with high and low (grouped according to median) expression of RRM2; The numbers below the figures denote the number of patients at risk in each group; The title of each graphic refers to the project name in TCGA, GEO, PrognoScan, or Kaplan Meier plotter web tool; TCGA: The Cancer Genome Atlas; LUAD: lung adenocarcinoma; OS: overall survival; DSS: disease-specific survival; DFS: disease-free survival; PFS: progression-free survival; RFS: Relapse free survival; P-value < 0.05 was used to assess differences.
High expression of RRM2 is a potential independent risk factor
As shown above, a higher expression of RRM2 was related to a higher tumor stage (Figure 2). Kaplan-Meier analysis indicated that high expression of RRM2 was associated with poor survival (Figure 3). To find more evidence, the Cox proportional-hazards model was constructed. Univariate Cox analyses in overall survival showed that tumor stage, TNM classification, and RRM2 were acting potential risk roles in LUAD. Additionally, the multivariate Cox analyses confirmed the critical value of T classification and RRM2, proving that they can predict tumor prognosis independently of other factors in overall survival (Table 2). Also, the Cox analysis based on disease-specific survival only showed that RRM2 was the riskiest factor. Consistently, in the progression-free survival Cox analysis, RRM2 was the only marker that can predict survival independently. Taking together, RRM2 is a potential independent risk factor in LUAD.
Table 2.
Univariate analysis and multivariate analysis of the correlation of RRM2 expression and important clinical characteristics with survival among lung adenocarcinoma patients
| Parameter | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Overall survival | ||||||
| Age | 1.000 | 0.983-1.018 | 9.685E-01 | 1.012 | 0.994-1.030 | 2.059E-01 |
| Gender | 1.074 | 0.768-1.503 | 6.757E-01 | 0.939 | 0.668-1.321 | 7.191E-01 |
| Tumor stage | 1.584 | 1.355-1.852 | 7.826E-09 | 1.469 | 0.975-2.215 | 6.616E-02 |
| T classification | 1.607 | 1.322-1.954 | 1.953E-06 | 1.293 | 1.030-1.624 | 2.667E-02 |
| N classification | 1.724 | 1.422-2.091 | 3.055E-08 | 1.083 | 0.755-1.552 | 6.661E-01 |
| M classification | 1.825 | 1.028-3.240 | 3.982E-02 | 0.737 | 0.261-2.082 | 5.642E-01 |
| RRM2 | 1.028 | 1.015-1.041 | 1.542E-05 | 1.286 | 1.118-1.480 | 4.324E-04 |
| Disease-specific survival | ||||||
| Age | 0.981 | 0.960-1.002 | 7.232E-02 | 0.995 | 0.973-1.017 | 6.300E-01 |
| Gender | 0.900 | 0.576-1.406 | 6.424E-01 | 0.781 | 0.497-1.226 | 2.832E-01 |
| Tumor stage | 1.597 | 1.301-1.960 | 7.515E-06 | 1.473 | 0.840-2.580 | 1.763E-01 |
| T classification | 1.514 | 1.157-1.982 | 2.534E-03 | 1.242 | 0.918-1.680 | 1.602E-01 |
| N classification | 1.628 | 1.267-2.094 | 1.428E-04 | 1.014 | 0.623-1.650 | 9.562E-01 |
| M classification | 2.258 | 1.126-4.530 | 2.184E-02 | 0.818 | 0.183-3.653 | 7.922E-01 |
| RRM2 | 1.035 | 1.020-1.051 | 5.702E-06 | 1.330 | 1.106-1.601 | 2.483E-03 |
| Progression-free survival | ||||||
| Age | 0.993 | 0.977-1.010 | 4.155E-01 | 0.998 | 0.981-1.015 | 8.134E-01 |
| Gender | 0.987 | 0.708-1.375 | 9.384E-01 | 0.920 | 0.657-1.287 | 6.255E-01 |
| Tumor stage | 1.320 | 1.123-1.551 | 7.468E-04 | 1.475 | 0.975-2.231 | 6.606E-02 |
| T classification | 1.356 | 1.112-1.653 | 2.653E-03 | 1.185 | 0.932-1.505 | 1.660E-01 |
| N classification | 1.299 | 1.060-1.591 | 1.152E-02 | 0.824 | 0.570-1.191 | 3.026E-01 |
| M classification | 1.432 | 0.771-2.658 | 2.551E-01 | 0.508 | 0.168-1.536 | 2.304E-01 |
| RRM2 | 1.022 | 1.009-1.035 | 6.605E-04 | 1.215 | 1.064-1.388 | 4.101E-03 |
HR: hazard ratio; CI: confidence interval. Bold values indicate p-value < 0.05.
RRM2 co-expression networks in LUAD
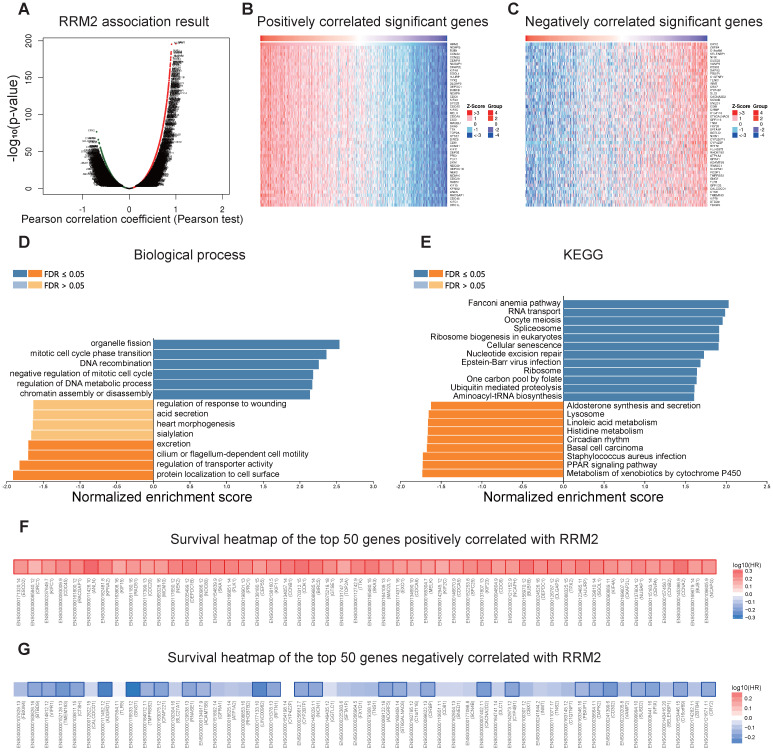
In order to better understand the biological meaning of RRM2 in LUAD, the “LinkFinder” module in LinkedOmics was applied to check the co-expression pattern of RRM2. As plotted in Figure 4A, it shows that 6152 genes (red dots) positively correlated with RRM2, and 7399 genes (green dots) negatively correlated (p-value < 0.05). Figures 4B and 4C show the heatmaps of the top 50 genes positively and negatively associated with RRM2, respectively. Moreover, Table S1 detailed lists the co-expressed genes.
Figure 4.
RRM2 co-expression genes in LUAD (LinkedOmics). (A) The global RRM2 highly correlated genes identified by the Pearson test in LUAD. Red and green dots represent positively and negatively significantly correlated genes with RRM2, respectively. (B and C) Heatmaps showing the top 50 genes positively and negatively correlated with RRM2 in LUAD. (D and E) Significantly enriched GO: Biological process annotations and KEGG pathways of RRM2 in LUAD. (F and G) Survival heatmaps of the top 50 genes positively and negatively correlated with RRM2 in LUAD. The survival heatmaps show the hazard ratios in the logarithmic scale (log10) for different genes. The red and blue blocks denote higher and lower risks, respectively. The rectangles with frames mean the significant unfavorable and favorable results in prognostic analyses (p-value < 0.05). FDR: false discovery rate; KEGG: Kyoto Encyclopedia of Genes and Genomes; GO: Gene Ontology; LUAD: Lung adenocarcinoma.
Significant Gene Ontology term annotation by GSEA showed that RRM2 co-expressed genes involved mainly in the organelle fission, mitotic cell cycle phase transition, DNA recombination, negative regulation of mitotic cell cycle, regulation of DNA metabolic process, and chromatin assembly or disassembly. In contrast, the protein localization to cell surface, regulation of transporter activity, cilium or flagellum-dependent cell motility, and excretion were inhibited (Figure 4D and Table S2). KEGG analysis showed genes were primarily enriched in the Fanconi anemia pathway, RNA transport, oocyte meiosis, spliceosome, ribosome biogenesis in eukaryotes, and cellular senescence pathways, etc. (Figure 4E and Table S3).
RRM2 expression displayed a strong positive association with the expression of NCAPG (positive rank #1, r = 0.908, p-value = 5.28E-196), BUB1 (r = 0.899, p-value = 8.98E-186), and CCNA2 (r = 0.897, p-value = 3.79E-184), etc. Remarkably, the top 50 positively genes owned highly probability of becoming high-risk markers in LUAD, interesting, of which 50/50 genes had high hazard ratio (HR, p-value < 0.05) (Figure 4F). In comparison, 26 of the top 50 negatively correlated genes had low HR (p-value <0.05) (Figure 4G).
Regulators of RRM2 networks in LUAD
To understand the regulatory factors of RRM2 in LUAD, we further analyzed the enrichment of kinases, miRNAs, and transcription factors of RRM2 co-expressed genes. The top 5 kinases related mainly to CDK1, PLK1, CDK2, AURKB, and ATM (Table 3 and Table S4). In fact, 3 of the top 5 kinase genes include CDK1, PLK1, and AURKB, were significantly highly expressed in tumor tissues and significantly related to the overall survival of LUAD (Figure S1). Interestingly, the co-expressed genes of RRM2 were not enriched on any miRNA targets significantly (Table 3 and Table S5). Transcription factor enrichment results showed that the co-expressed genes of RRM2 were mainly enriched in the E2F transcription factor family (Table 3 andTable S6), including V$E2F_Q6, V$E2F_Q4, V$E2F1_Q6, V$E2F4DP1_01, and V$E2F1DP1_01.
Table 3.
The kinases, miRNAs, and transcription factors-target networks of RRM2 in LUAD
| Enriched category | Gene set | Leading edge number | NES | FDR |
|---|---|---|---|---|
| Kinase target | Kinase_CDK1 | 67 | 2.3829 | 0 |
| Kinase_PLK1 | 31 | 2.2374 | 0 | |
| Kinase_CDK2 | 84 | 2.2281 | 0 | |
| Kinase_AURKB | 31 | 2.1503 | 0 | |
| Kinase_ATM | 38 | 2.1011 | 0 | |
| miRNA Target | AGCGCTT,MIR-518F,MIR-518E,MIR-518A | 5 | -1.4072 | 0.48209 |
| CCCAGAG,MIR-326 | 30 | -1.4088 | 0.55154 | |
| AGGGCAG,MIR-18A | 34 | -1.4499 | 0.55987 | |
| GAGCTGG,MIR-337 | 34 | -1.2417 | 0.61109 | |
| ACACTGG,MIR-199A,MIR-199B | 36 | -1.3139 | 0.61239 | |
| Transcription Factor | V$E2F_Q6 | 87 | 2.1966 | 0 |
| V$E2F_Q4 | 87 | 2.1905 | 0 | |
| V$E2F1_Q6 | 91 | 2.1891 | 0 | |
| V$E2F4DP1_01 | 91 | 2.1752 | 0 | |
| V$E2F1DP1_01 | 90 | 2.1731 | 0 |
LUAD: Lung adenocarcinoma; NES: normalized enrichment score; FDR: false discovery rate.
Correlation analysis between RRM2 expression and six main infiltrating immune cells
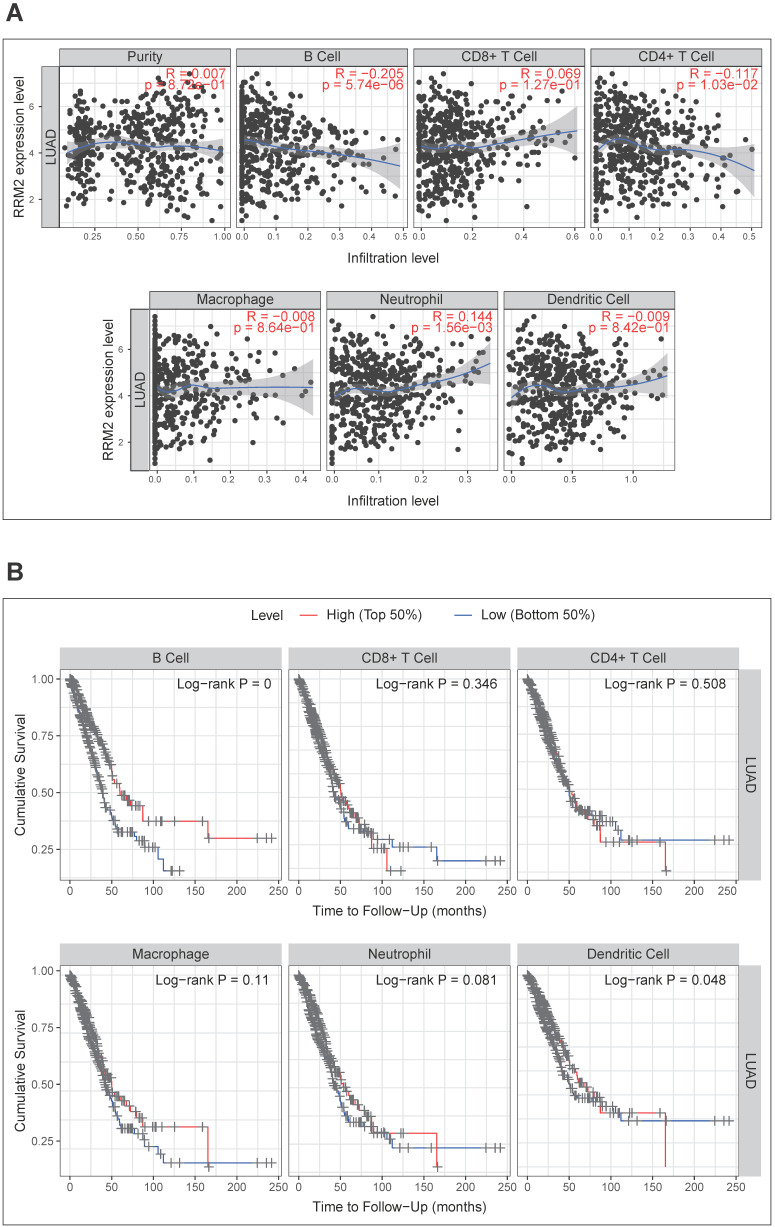
Then, we investigated whether RRM2 expression was correlated with six main infiltrating immune cells (B cells, CD4 T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) in LUAD using TIMER database. The analysis showed that RRM2 expression levels correlated with B cells (r = -0.205, p-value = 5.74e-06), CD4+ T cells (r = -0.117, p-value = 1.03e-02), and neutrophils (r = 0.144, p-value = 1.56e-03) (Figure 5A). Moreover, we evaluated the prognostic value of each of the six types of immune cells via Kaplan-Meier analysis, finding B cells (p-value = 0 in log-rank test) and dendritic cells (p-value = 0.048 in log-rank test) can predict the outcome of LUAD (Figure 5B). At last, Cox proportional hazard models were applied to assess the impacts of RRM2 expression and the six types of immune cells on the overall survival of LUAD. RRM2 showed significant risk in univariate analyses (HR = 1.291, 95% CI = 1.150-1.450, p-value = 0), and multivariate analyses (HR = 1.255, 95% CI = 1.112-1.415, p-value = 0), indicating it can predict tumor outcomes independently of the other six immune cells. Interestingly, B cells also displayed the similar performance in univariate (HR = 0.024, 95% CI = 0.004-0.142, p-value = 0) and multivariate analyses (HR = 0.008, 95% CI = 0.001-0.106, p-value = 0) (Table 4). Taking together, the significantly infiltrating with B cells seemed like one of the critical factors that RRM2 holds to influence the outcome of LUAD pronounced.
Figure 5.
Correlation analysis between RRM2 expression and six types of infiltrating immune cells in LUAD. (A) Correlation of RRM2 expression with six types of immune infiltration cells obtained from TIMER (purity-corrected Spearman test). (B) Overall survival curve of each of the six types of immune cells produced by Kaplan-Meier estimator from TIMER. Survival differences are compared between patients with high and low (grouped according to median) infiltrating of each kind of immune cells; LUAD: Lung adenocarcinoma; TIMER: The Tumor Immune Estimation Resource.
Table 4.
Cox analysis of the correlation between RRM2 expression and six types of immune cells and prognosis in patients with lung adenocarcinoma
| Variables | Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| coef | HR | 95% CI | P-value | coef | HR | 95% CI | P-value | |
| B cell | -3.715 | 0.024 | 0.004-0.142 | 0 | -4.822 | 0.008 | 0.001-0.106 | 0 |
| CD8+ T cell | -1.209 | 0.299 | 0.083-1.074 | 0.064 | 0.382 | 1.466 | 0.244-8.813 | 0.676 |
| CD4+ T cell | -1.061 | 0.346 | 0.078-1.541 | 0.164 | 3.101 | 22.212 | 1.494-330.241 | 0.024 |
| Macrophage | -0.55 | 0.577 | 0.094-3.539 | 0.552 | 0.385 | 1.469 | 0.112-19.184 | 0.769 |
| Neutrophil | -1.006 | 0.366 | 0.055-2.452 | 0.3 | -2.146 | 0.117 | 0.002-5.856 | 0.283 |
| Dendritic | -0.592 | 0.553 | 0.308-0.993 | 0.047 | -0.026 | 0.974 | 0.258-3.683 | 0.97 |
| RRM2 | 0.255 | 1.291 | 1.150-1.450 | 0 | 0.227 | 1.255 | 1.112-1.415 | 0 |
coef: regression coefficient; HR: hazard ratio; CI: confidence interval; Bold values indicate p-value < 0.05.
Correlation between RRM2 expression and immune signatures
Lastly, to expand the understanding of the crosstalk between RRM2 and multiple immune marker genes of 28 TILs, immune inhibitory or stimulatory, cytokine-related, cancer-testis antigen, and MHC, we did correlation analysis between them. The analysis showed that the expression level of RRM2 in LUAD was significantly correlated with 67.68% (624/922) immune marker genes (Table S7). In the significant correlated immune markers, 352/624 (56.41%) were positively, while, 272/624 (43.59%) were negatively related. On the whole, the top 5 positively correlated marker genes with RRM2 were CCNA2 (r = 0.901976588, p-value = 3.2894E-181), CCNB1 (r = 0.874181398, p-value = 3.775E-156), EXO1 (r = 0.868718344, p-value = 6.3113E-152), PRC1 (r = 0.862049011, p-value = 5.0769E-147), and KIF11 (r = 0.858239682, p-value = 2.474E-144). Besides, the top 5 negatively correlated markers with RRM2 were CD302 (r = -0.563236799, p-value = 1.30947E-42), DAPK2 (r = -0.550326103, p-value = 2.23251E-40), GNG7 (r = -0.542454019, p-value = 4.59789E-39), DLC1 (r = -0.525310139, p-value = 2.55007E-36), and GPRC5C (r = -0.524798161, p-value = 3.06273E-36).
As for immunoinhibitory genes, results showed CD274 LAG3, PDCD1LG2, PDCD1, IDO1, KIR2DL3, PVRL2, TIGIT, IL10RB, HAVCR2, CTLA4, IL10, TGFBR1 have positively correlations with RRM2 expression, while, ADORA2A and BTLA have negatively correlations with RRM2 expression. Moreover, the top 5 immunostimulatory genes positively correlated RRM2 expression were PVR, CD276, MICB, TNFSF4, and TNFRSF9, besides, the top 5 negative markers were TNFSF13, TMEM173, IL6R, TNFRSF13B, and CD40LG (Table S7).
In the previous section, we found that B cell infiltration may be one of the key reasons that caused RRM2 to become a prognostic factor. Thus, we were very interested in the correlation between RRM2 and B cell marker genes. Table 5, which was extracted from Table S7, shows the purity-corrected partial Spearman's correlation between RRM2 and B cell markers. In B cells, RRM2 is highly correlated with CCNA2 (#1, r = 0.901976588, p-value = 3.2894E-181), CDKN3 (#2, r = 0.828306212, p-value = 1.2672E-125), GNG7 (#3, r = -0.542454019, p-value = 4.59789E-39), FCER1A (#4, r = -0.498295422, p-value = 2.66219E-32), and MICAL3 (#5, r = 0.344177052, p-value = 3.71747E-15). In total, 35/57 of the B cell marker genes associated significantly to RRM2 expression, of which the number of positive correlations was 9/35 (25.71%), and the negative was 26/35 (74.29%). We plotted the survival heatmaps of the significant B cell markers correlated significantly with RRM2 expression in Figure S2. Notably, almost all of the positively related markers showed a high probability of becoming high-risk factors in LUAD, of which 3/9 markers had elevated HR (p-value < 0.05) (Figure S2A). In comparison, there were 20/26 genes with low HR (p-value < 0.05) in negatively related markers (Figure S2B).
Table 5.
Correlation analysis between RRM2 and of B cell markers in LUAD
| Variables | None adjusted | Tumor purity adjusted | ||
|---|---|---|---|---|
| Cor | P-value | Cor | P-value | |
| Activated B cell | ||||
| GNG7 | -0.526 | 0.000E+00 | -0.542 | 4.598E-39 |
| MICAL3 | 0.340 | 2.102E-15 | 0.344 | 3.717E-15 |
| CLEC9A | -0.292 | 1.334E-11 | -0.300 | 1.108E-11 |
| BLK | -0.233 | 9.074E-08 | -0.255 | 9.844E-09 |
| HLA-DOB | -0.226 | 2.137E-07 | -0.240 | 6.752E-08 |
| MS4A1 | -0.189 | 1.720E-05 | -0.207 | 3.462E-06 |
| SPIB | -0.167 | 1.407E-04 | -0.181 | 5.204E-05 |
| BACH2 | 0.157 | 3.387E-04 | 0.178 | 6.919E-05 |
| CD79B | -0.159 | 3.009E-04 | -0.173 | 1.157E-04 |
| CR2 | -0.174 | 7.130E-05 | -0.170 | 1.548E-04 |
| CLECL1 | -0.163 | 1.945E-04 | -0.166 | 2.140E-04 |
| CLEC17A | -0.128 | 3.637E-03 | -0.143 | 1.490E-03 |
| AKNA | -0.135 | 2.078E-03 | -0.142 | 1.535E-03 |
| CD27 | -0.132 | 2.703E-03 | -0.139 | 1.934E-03 |
| TNFRSF17 | -0.140 | 1.499E-03 | -0.139 | 1.990E-03 |
| ARHGAP25 | -0.122 | 5.639E-03 | -0.126 | 5.098E-03 |
| CD19 | -0.107 | 1.507E-02 | -0.119 | 8.208E-03 |
| FCRL2 | -0.097 | 2.726E-02 | -0.095 | 3.513E-02 |
| PNOC | -0.072 | 1.046E-01 | -0.069 | 1.235E-01 |
| TCL1A | -0.060 | 1.770E-01 | -0.058 | 1.986E-01 |
| CCL21 | 0.023 | 6.025E-01 | 0.047 | 2.948E-01 |
| ADAM28 | 0.026 | 5.587E-01 | 0.029 | 5.238E-01 |
| CD38 | -0.001 | 9.755E-01 | 0.014 | 7.587E-01 |
| CD180 | -0.022 | 6.135E-01 | -0.003 | 9.491E-01 |
| Immature B cell | ||||
| TXNIP | -0.338 | 4.403E-15 | -0.340 | 8.067E-15 |
| CD22 | -0.257 | 3.589E-09 | -0.285 | 1.217E-10 |
| KIAA0226 | 0.278 | 1.540E-10 | 0.278 | 3.555E-10 |
| FCRL1 | -0.232 | 9.644E-08 | -0.257 | 7.175E-09 |
| STAP1 | -0.199 | 5.553E-06 | -0.223 | 5.747E-07 |
| FAM129C | -0.190 | 1.495E-05 | -0.203 | 5.791E-06 |
| SP100 | 0.142 | 1.190E-03 | 0.155 | 5.319E-04 |
| FCRLA | -0.134 | 2.245E-03 | -0.143 | 1.401E-03 |
| HLA-DQA1 | -0.131 | 2.895E-03 | -0.137 | 2.230E-03 |
| HDAC9 | 0.096 | 2.981E-02 | 0.087 | 5.282E-02 |
| FCRL3 | -0.084 | 5.697E-02 | -0.082 | 6.954E-02 |
| CYBB | 0.027 | 5.347E-01 | 0.054 | 2.292E-01 |
| TAGAP | -0.047 | 2.859E-01 | -0.044 | 3.262E-01 |
| ZCCHC2 | 0.038 | 3.895E-01 | 0.042 | 3.524E-01 |
| HVCN1 | -0.052 | 2.396E-01 | -0.038 | 3.953E-01 |
| NCF1 | 0.020 | 6.561E-01 | 0.037 | 4.110E-01 |
| FCRL5 | 0.017 | 7.015E-01 | 0.027 | 5.563E-01 |
| NCF1B | -0.028 | 5.302E-01 | -0.019 | 6.759E-01 |
| P2RY10 | 0.000 | 9.974E-01 | 0.018 | 6.836E-01 |
| Memory B cell | ||||
| CCNA2 | 0.901 | 0.000E+00 | 0.902 | 3.289E-181 |
| CDKN3 | 0.824 | 0.000E+00 | 0.828 | 1.267E-125 |
| FCER1A | -0.490 | 2.167E-32 | -0.498 | 2.662E-32 |
| ENPP1 | 0.326 | 4.798E-14 | 0.334 | 2.405E-14 |
| MYC | 0.264 | 1.275E-09 | 0.263 | 2.929E-09 |
| SOX5 | -0.092 | 3.720E-02 | -0.103 | 2.249E-02 |
| SORL1 | -0.096 | 2.959E-02 | -0.099 | 2.866E-02 |
| RUNX2 | 0.083 | 5.829E-02 | 0.092 | 4.067E-02 |
| AICDA | 0.045 | 3.095E-01 | 0.069 | 1.252E-01 |
| FCRL4 | -0.042 | 3.400E-01 | -0.050 | 2.667E-01 |
| CLCN5 | -0.038 | 3.852E-01 | -0.037 | 4.185E-01 |
| STAT5B | -0.032 | 4.666E-01 | -0.036 | 4.298E-01 |
| TLR9 | 0.013 | 7.642E-01 | 0.030 | 5.049E-01 |
| STAT5A | -0.033 | 4.490E-01 | -0.021 | 6.418E-01 |
Cor: Correlation coefficient; LUAD: Lung adenocarcinoma; Bold values indicate p-value < 0.05.
Discussion
The present study found that RRM2 was highly expressed in LUAD tumor tissue and significantly predicts a poor prognosis; also, the higher tumor stage got a higher expression. Univariate and multivariate Cox analyses indicated the RRM2 might be a potential independent biomarker for LUAD prognosis. Then we examined the co-expression and regulators networks of RRM2. At last, we conducted a correlation analysis between RRM2 and immune infiltration or immune signatures, finding that RRM2 was related to most of the immune marker genes, and its infiltration in B cells may be one of the factors for its prognostic ability. Such work we have done aimed to guide future research in LUAD.
The dysregulated cell cycle has been identified in many types of cancer 29. Ribonucleotide reductase is an enzyme involved in the cell cycle. It consists of two subunits, namely the regulatory subunit RRM1 and the catalytic subunit RRM2, which is essential for DNA replication and repair 30, 31. RRM2 is a rate-limiting enzyme used for DNA synthesis and repair, plays a vital role in many critical cellular processes, such as cell proliferation, invasiveness, migration, angiogenesis, and aging 8. The present bioinformatics analysis showed that RRM2 was overexpressed in breast cancer patients to normal tissues and was associated with worse survival 32. Overexpression of RRM2 was shown to be associated with an unfavorable prognosis in HER-2 positive breast cancer patients 33. A recent study indicated that RRM2 upregulation occurred in multiple myeloma tumors and that RRM2 knockdown inhibited multiple myeloma cell proliferation 12. Li et al. illustrated that RRM2 was overexpressed in human glioblastoma cells, and promoted proliferation, migration, and invasion of human glioblastoma cells 34. Suppression of RRM2 inhibits cell proliferation, causes cell cycle arrest, and promotes the apoptosis of human neuroblastoma cells 35. In our story, the investigation of differential expression in LUAD found that RRM2 was highly expressed in tumor tissues, which was subsequently examined in multiple independent cohorts.
Then, we found that distinct histologic staging was associated with RRM2 expression. High expression of RRM2 happened in the more upper stage, which indicated RRM2 is mainly involved in the advanced period in LUAD, indicating a possible relationship existed between RRM2 expression and disease outcomes in LUAD. Thus, we carried out survival analysis in TCGA-LUAD, revealing that high RRM2 expression was associated with poor outcomes, which was also checked in the other independent cohorts. Besides, the Cox analyses further proved that RRM2 was an independent risk factor in LUAD. Therefore, our results indicate that RRM2 upregulation occurs in LUAD, and as a potential diagnostic and prognostic marker, it is worthy of further clinical verification.
We explored the regulators responsible for RRM2 dysregulation and found that RRM2 was related to kinase networks, such as CDK1, PLK1, CDK2, AURKB, and ATM. These kinases mainly regulated mitosis, genome stability, and cell cycle, and showed survival prognosis value and differential expression in LUAD. CDK1 is a prototype kinase, a central regulator that drives cells through G2 phase and mitosis 36. CDK1 orchestrates the transition from the G2 phase into mitosis, and as cancer cells often display enhanced CDK1 activity, it has been proposed as a tumor-specific anti-cancer target 37. Data mining from different databases demonstrated CDK1 upregulation in LUAD. Furthermore, CDK1 upregulation is associated with poor prognosis 38. However, the molecular mechanism and potential application of CDK1 in lung cancer have not been determined 39. PLK1 is indispensable for finely regulating cell division and maintenance of genomic stability in mitosis, spindle assembly, and DNA damage response 40. Studies have shown that PLK1 is highly expressed in most human carcinoma, and its overexpression is associated with an unfavorable prognosis 41-43. In human tumors, the overexpression of AURKB is associated with poor prognosis. AURKB inhibitors are in clinical trials for stage I-II leukemia 44. AURKB is also involved in resistance to specific anti-tumor agents, such as paclitaxel in NSCLC 45. Bertran-Alamillo et al. revealed that AURKB is related to acquired resistance to EGFR TKIs, and AURKB can become a potential biological target for anti-EGFR therapy of NSCLC without carrying resistance mutations 46.
In this study, we found that the E2F family was the main transcription factor constituting RRM2 dysregulation. E2F is a group of genes that encodes a family of transcription factors in advanced eukaryotes. They participate in regulating the cell cycle and DNA synthesis in mammalian cells 47. Our analysis did not find miRNAs that are significantly associated with RRM2, which may be since RRM2 is involved in the role of mRNA spliceosomes and is far away from miRNA cellular. Our results indicate that E2F1 is a vital regulator of RRM2, and RRM2 may play a role in regulating the cell cycle and proliferation ability of LUAD through this factor.
Our study found that RRM2 expression levels had significant correlations with B cells, CD4+ T cells, and neutrophils infiltrating (Figure 5A). Moreover, the subsequent Kaplan-Meier analysis found that B cells and dendritic cells could predict the outcome of LUAD (Figure 5B). Cox analyses showed that B cells and RRM2 were significant independent risk factors among all variables (Table 4). These findings indicated that B cell infiltration might be one of the critical factors of RRM2 with prognostic value.
Next, we conducted correlation analyses between RRM2 and several immune signatures. We detailed analyzed the correlation between RRM2 and B cell signatures finding 35/57 (61.40%) of the B cell marker genes associated significantly to RRM2 expression, including CCNA2, CDKN3, and GNG7. CCNA2, also known as cyclin A2, belongs to the highly conserved cyclin family and plays a key role in cell cycle control 48. A recent study demonstrated that CCNA2 is a crucial regulator of NSCLC cells metastasis promoting invasion and migration of NSCLC cells through integrin αVβ3 signaling pathway 49. CDKN3 gene encodes a dual-specificity protein phosphatase, which was previously thought to suppress tumors by controlling mitosis via CDK1/CDK2 50. It is well known that CDKN3 is overexpressed in multiple human tumor tissues and cell lines 51, 52. The high expression of CDKN3 in human cancer tissue may reflect the increased proportion of mitotic cells in the tumor 53. Elevated CDKN3 expression is associated with the adverse outcome of LUAD. Overexpression of CDKN3 in LUAD is not because of alternative splicing or mutation, but increased mitotic activity, which is related to CDKN3 as a tumor suppressor 53. GNG7 is a subunit of heterotrimeric G protein, which is commonly expressed in various tissues, but low in cancer 54. It has been speculated that GNG7 may be involved in cell contact-induced growth arrest and thus blocks uncontrolled cell proliferation in multicellular organisms 55. Correlate analysis provides an exhaustive characterization of the association between RRM2 and immune signatures in LUAD patients, indicating that RRM2 is a crucial player in immune escape in the tumor microenvironment. Also, the correlation between RRM2 and B cell markers is particularly vital to the prognosis of LUAD patients. It is worth noting that RRM2 may be a key factor mediating B cell therapy, which is needed to be clarified in further research.
At present, how RRM2 affects the prognosis of LUAD and the biological function of RRM2 in LUAD is still in its infancy. Souglakos' study revealed that the efficacy of docetaxel/gemcitabine in lung adenocarcinoma patients was associated with RRM2 mRNA expression from 42 patients 18. Huang and colleagues studied 44 patient samples and found that the overexpression of RRM2 promoted proliferation, inhibited apoptosis, and increased the chemotherapy resistance of NSCLC cells through upregulating EGFR expression and AKT phosphorylation 17. Recently, Yang's team worked on 30 patients and found that RRM2 was upregulated in NSCLC tumors and cell lines, leading to poor prognosis 16. Nevertheless, these are not enough, and there are still many unknowns to be elaborated. Insufficient sample inclusion in previous researches will lead to statistical bias. So, in our study, we attempted a comprehensive and multi-angle analysis of RRM2 by combining multiple databases to explain the role of RRM2 in LUAD. The previous studies did not highlight the independent prognostic capabilities of RRM2, which has been confirmed in our study. Immunity plays a crucial role in the development of tumors. Existing research is failed to declare how RRM2 affects LUAD immunity. To this end, we not only elaborated the relationship between various immune cells and RRM2, but also discussed in detail the immune signature genes related to RRM2 that affect the prognosis. Such a research design for RRM2 and LUAD has never reported before, which are aiming at providing one more possibility and direction for future LUAD research.
Our research also has some limitations, described as follows. 1) The results came from retrospective data, and more prospective data were needed for proving the clinical utility of it. 2) There is lacking wet experimental data in this study explaining the relationship between RRM2 and their mechanism in LUAD samples. More effort is needed to clarify the potential relationship between RRM2 and LUAD.
Conclusions
In summary, this study provided all-round evidence for the value of RRM2 in the progress of lung cancer and its potential as a bio-target and prognostic predictor of LUAD. Our results showed that the up-regulation of RRM2 in LUAD indicates an adverse outcome, which may be caused by multiple steps that weaken genomic stability or disturb the cell cycle. Furthermore, we find that RRM2 has a significant correlation with mostly immune signatures. Moreover, the connection between RRM2 and B cell markers needs to be noted, which may be the new direction of future LUAD research.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
Chao Ma and Huan Luo thank Zhengzhou University Overseas Virtual Research Institute for financing support studying abroad. Chao Ma thanks the China Scholarship Council (No. 201708410121) for its financial support.
Author Contributions
Chao Ma and Huan Luo organized and wrote the manuscript. Chao Ma designed and produced the figures. Huan Luo contributed to the literature search for the manuscript. Jing Cao grammar edited the manuscript. Chengshan Gao, Xianen Fa, and Guangsuo Wang revised the manuscript. All authors reviewed the manuscript and approved the manuscript for publication.
Funding
This work is supported by Shenzhen Key Medical Discipline Construction Fund (No. SZXK018) and Shenzhen Overseas High-level Talents Innovation and Entrepreneurship Plan (No. KQTD2016113015442590).
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: TCGA: https://portal.gdc.cancer.gov/; GEO: https://www.ncbi.nlm.nih.gov/geo/; CPTAC: https://cptac-data-portal.georgetown.edu/.
Abbreviations
- RRM2
ribonucleoside-diphosphate reductase subunit M2
- LUAD
lung adenocarcinoma
- NSCLC
non-small cell lung cancer
- TCGA
The Cancer Genome Atlas
- GEO
Gene Expression Omnibus
- CPTAC
Clinical Proteomic Tumor Analysis Consortium
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GO
Gene Ontology
- BP
Biological Process
- GSEA
Gene Set Enrichment Analysis
- GEPIA
The Gene Expression Profiling Interactive Analysis
- TIMER
The Tumor Immune Estimation Resource
- TILs
tumor-infiltrating lymphocytes
- MHC
major histocompatibility complex
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.The L. Lung cancer: some progress, but still a lot more to do. Lancet. 2019;394:1880. doi: 10.1016/S0140-6736(19)32795-3. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 5.Senosain MF, Massion PP. Intratumor Heterogeneity in Early Lung Adenocarcinoma. Front Oncol. 2020;10:349. doi: 10.3389/fonc.2020.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan M, Huang LL, Chen JH, Wu J, Xu Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct Target Ther. 2019;4:61. doi: 10.1038/s41392-019-0099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devarakonda S, Govindan R. Untangling the evolutionary roots of lung cancer. Nat Commun. 2019;10:2979. doi: 10.1038/s41467-019-10879-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida Y, Tsunoda T, Doi K, Tanaka Y, Fujimoto T, Machida T. et al. KRAS-mediated up-regulation of RRM2 expression is essential for the proliferation of colorectal cancer cell lines. Anticancer Res. 2011;31:2535–9. [PubMed] [Google Scholar]
- 10.Morikawa T, Maeda D, Kume H, Homma Y, Fukayama M. Ribonucleotide reductase M2 subunit is a novel diagnostic marker and a potential therapeutic target in bladder cancer. Histopathology. 2010;57:885–92. doi: 10.1111/j.1365-2559.2010.03725.x. [DOI] [PubMed] [Google Scholar]
- 11.Chen W-x, Yang L-g, Xu L-y, Cheng L, Qian Q, Sun L. et al. Bioinformatics analysis revealing prognostic significance of RRM2 gene in breast cancer. Bioscience Reports. 2019;39:BSR20182062. doi: 10.1042/BSR20182062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Peng J, Zhou Y, Xie B, Wang J. Silencing RRM2 inhibits multiple myeloma by targeting the Wnt/betacatenin signaling pathway. Mol Med Rep. 2019;20:2159–66. doi: 10.3892/mmr.2019.10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahman MA, Amin AR, Wang D, Koenig L, Nannapaneni S, Chen Z. et al. RRM2 regulates Bcl-2 in head and neck and lung cancers: a potential target for cancer therapy. Clin Cancer Res. 2013;19:3416–28. doi: 10.1158/1078-0432.CCR-13-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Meng L, Wang XW, Ma GY, Chen JH. Expression of RRM1 and RRM2 as a novel prognostic marker in advanced non-small cell lung cancer receiving chemotherapy. Tumour Biol. 2014;35:1899–906. doi: 10.1007/s13277-013-1255-4. [DOI] [PubMed] [Google Scholar]
- 15.Mah V, Alavi M, Marquez-Garban DC, Maresh EL, Kim SR, Horvath S. et al. Ribonucleotide reductase subunit M2 predicts survival in subgroups of patients with non-small cell lung carcinoma: effects of gender and smoking status. PLoS One. 2015;10:e0127600. doi: 10.1371/journal.pone.0127600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Li S, Cao J, Li Y, Hu H, Wu Z. RRM2 Regulated By LINC00667/miR-143-3p Signal Is Responsible For Non-Small Cell Lung Cancer Cell Progression. Onco Targets Ther. 2019;12:9927–39. doi: 10.2147/OTT.S221339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang N, Guo W, Ren K, Li W, Jiang Y, Sun J. et al. LncRNA AFAP1-AS1 Supresses miR-139-5p and Promotes Cell Proliferation and Chemotherapy Resistance of Non-small Cell Lung Cancer by Competitively Upregulating RRM2. Front Oncol. 2019;9:1103. doi: 10.3389/fonc.2019.01103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Souglakos J, Boukovinas I, Taron M, Mendez P, Mavroudis D, Tripaki M. et al. Ribonucleotide reductase subunits M1 and M2 mRNA expression levels and clinical outcome of lung adenocarcinoma patients treated with docetaxel/gemcitabine. Br J Cancer. 2008;98:1710–5. doi: 10.1038/sj.bjc.6604344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B. et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–58. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen F, Chandrashekar DS, Varambally S, Creighton CJ. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat Commun. 2019;10:5679. doi: 10.1038/s41467-019-13528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D63. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W60. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS. et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108–e10. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J. et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC. et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35:4200–2. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 28.Chen X, Xu C, Hong S, Xia X, Cao Y, McDermott J. et al. Immune Cell Types and Secreted Factors Contributing to Inflammation-to-Cancer Transition and Immune Therapy Response. Cell Rep. 2019;26:1965–77. doi: 10.1016/j.celrep.2019.01.080. e4. [DOI] [PubMed] [Google Scholar]
- 29.Sun H, Yang B, Zhang H, Song J, Zhang Y, Xing J. et al. RRM2 is a potential prognostic biomarker with functional significance in glioma. Int J Biol Sci. 2019;15:533–43. doi: 10.7150/ijbs.30114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrents E, Aloy P, Gibert I, Rodriguez-Trelles F. Ribonucleotide reductases: divergent evolution of an ancient enzyme. J Mol Evol. 2002;55:138–52. doi: 10.1007/s00239-002-2311-7. [DOI] [PubMed] [Google Scholar]
- 31.Chabes A, Thelander L. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J Biol Chem. 2000;275:17747–53. doi: 10.1074/jbc.M000799200. [DOI] [PubMed] [Google Scholar]
- 32.Chen WX, Yang LG, Xu LY, Cheng L, Qian Q, Sun L, Bioinformatics analysis revealing prognostic significance of RRM2 gene in breast cancer. Biosci Rep. 2019. 39. [DOI] [PMC free article] [PubMed]
- 33.Lin Y, Fu F, Lv J, Wang M, Li Y, Zhang J. et al. Identification of potential key genes for HER-2 positive breast cancer based on bioinformatics analysis. Medicine (Baltimore) 2020;99:e18445. doi: 10.1097/MD.0000000000018445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, Zheng J, Chen S, Huang B, Li G, Feng Z. et al. RRM2 promotes the progression of human glioblastoma. J Cell Physiol. 2018;233:6759–67. doi: 10.1002/jcp.26529. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Pang J, Liu Y, Zhang J, Zhang C, Shen G. et al. Suppression of RRM2 inhibits cell proliferation, causes cell cycle arrest and promotes the apoptosis of human neuroblastoma cells and in human neuroblastoma RRM2 is suppressed following chemotherapy. Oncol Rep. 2018;40:355–60. doi: 10.3892/or.2018.6420. [DOI] [PubMed] [Google Scholar]
- 36.Diril MK, Ratnacaram CK, Padmakumar VC, Du T, Wasser M, Coppola V. et al. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc Natl Acad Sci U S A. 2012;109:3826–31. doi: 10.1073/pnas.1115201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prevo R, Pirovano G, Puliyadi R, Herbert KJ, Rodriguez-Berriguete G, O'Docherty A. et al. CDK1 inhibition sensitizes normal cells to DNA damage in a cell cycle dependent manner. Cell Cycle. 2018;17:1513–23. doi: 10.1080/15384101.2018.1491236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi YX, Zhu T, Zou T, Zhuo W, Chen YX, Huang MS. et al. Prognostic and predictive values of CDK1 and MAD2L1 in lung adenocarcinoma. Oncotarget. 2016;7:85235–43. doi: 10.18632/oncotarget.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuang Y, Guo W, Ling J, Xu D, Liao Y, Zhao H. et al. Iron-dependent CDK1 activity promotes lung carcinogenesis via activation of the GP130/STAT3 signaling pathway. Cell Death Dis. 2019;10:297. doi: 10.1038/s41419-019-1528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–41. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 41.Ramani P, Nash R, Sowa-Avugrah E, Rogers C. High levels of polo-like kinase 1 and phosphorylated translationally controlled tumor protein indicate poor prognosis in neuroblastomas. J Neurooncol. 2015;125:103–11. doi: 10.1007/s11060-015-1900-4. [DOI] [PubMed] [Google Scholar]
- 42.Tut TG, Lim SH, Dissanayake IU, Descallar J, Chua W, Ng W. et al. Upregulated Polo-Like Kinase 1 Expression Correlates with Inferior Survival Outcomes in Rectal Cancer. PLoS One. 2015;10:e0129313. doi: 10.1371/journal.pone.0129313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang R, Shi H, Ren F, Liu H, Zhang M, Deng Y. et al. Misregulation of polo-like protein kinase 1, P53 and P21WAF1 in epithelial ovarian cancer suggests poor prognosis. Oncol Rep. 2015;33:1235–42. doi: 10.3892/or.2015.3723. [DOI] [PubMed] [Google Scholar]
- 44.Tang A, Gao K, Chu L, Zhang R, Yang J, Zheng J. Aurora kinases: novel therapy targets in cancers. Oncotarget. 2017;8:23937–54. doi: 10.18632/oncotarget.14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Khafaji AS, Davies MP, Risk JM, Marcus MW, Koffa M, Gosney JR. et al. Aurora B expression modulates paclitaxel response in non-small cell lung cancer. Br J Cancer. 2017;116:592–9. doi: 10.1038/bjc.2016.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bertran-Alamillo J, Cattan V, Schoumacher M, Codony-Servat J, Gimenez-Capitan A, Cantero F. et al. AURKB as a target in non-small cell lung cancer with acquired resistance to anti-EGFR therapy. Nat Commun. 2019;10:1812. doi: 10.1038/s41467-019-09734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer. 2019;19:326–38. doi: 10.1038/s41568-019-0143-7. [DOI] [PubMed] [Google Scholar]
- 48.Gao T, Han Y, Yu L, Ao S, Li Z, Ji J. CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen resistance. PLoS One. 2014;9:e91771. doi: 10.1371/journal.pone.0091771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruan JS, Zhou H, Yang L, Wang L, Jiang ZS, Wang SM. CCNA2 facilitates epithelial-to-mesenchymal transition via the integrin alphavbeta3 signaling in NSCLC. Int J Clin Exp Pathol. 2017;10:8324–33. [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Che W, Wang W, Su G, Zhen T, Jiang Z. CDKN3 promotes tumor progression and confers cisplatin resistance via RAD51 in esophageal cancer. Cancer Manag Res. 2019;11:3253–64. doi: 10.2147/CMAR.S193793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Espinosa AM, Alfaro A, Roman-Basaure E, Guardado-Estrada M, Palma I, Serralde C. et al. Mitosis is a source of potential markers for screening and survival and therapeutic targets in cervical cancer. PLoS One. 2013;8:e55975. doi: 10.1371/journal.pone.0055975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu Y, Jiang X, Schoch BS, Carroll RS, Black PM, Johnson MD. Aberrant splicing of cyclin-dependent kinase-associated protein phosphatase KAP increases proliferation and migration in glioblastoma. Cancer Res. 2007;67:130–8. doi: 10.1158/0008-5472.CAN-06-2478. [DOI] [PubMed] [Google Scholar]
- 53.Fan C, Chen L, Huang Q, Shen T, Welsh EA, Teer JK. et al. Overexpression of major CDKN3 transcripts is associated with poor survival in lung adenocarcinoma. Br J Cancer. 2015;113:1735–43. doi: 10.1038/bjc.2015.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, Ji X, Li Z, Yang X, Wang W, Zhang X. G protein gamma subunit 7 induces autophagy and inhibits cell division. Oncotarget. 2016;7:24832–47. doi: 10.18632/oncotarget.8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shibata K, Tanaka S, Shiraishi T, Kitano S, Mori M. G-protein gamma 7 is down-regulated in cancers and associated with p 27kip1-induced growth arrest. Cancer Res. 1999;59:1096–101. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: TCGA: https://portal.gdc.cancer.gov/; GEO: https://www.ncbi.nlm.nih.gov/geo/; CPTAC: https://cptac-data-portal.georgetown.edu/.