Viruses commonly antagonize the antiviral type I interferon response by targeting signal transducer and activator of transcription 1 (STAT1) and STAT2, key mediators of interferon signaling. Other STAT family members mediate signaling by diverse cytokines important to infection, but their relationship with viruses is more complex. Importantly, virus-STAT interaction can be antagonistic or stimulatory depending on diverse viral and cellular factors. While STAT antagonism can suppress immune pathways, many viruses promote activation of specific STATs to support viral gene expression and/or produce cellular conditions conducive to infection.
KEYWORDS: STAT signaling, STAT transcription factors, host-pathogen interactions, immune evasion, virus-host interactions
ABSTRACT
Viruses commonly antagonize the antiviral type I interferon response by targeting signal transducer and activator of transcription 1 (STAT1) and STAT2, key mediators of interferon signaling. Other STAT family members mediate signaling by diverse cytokines important to infection, but their relationship with viruses is more complex. Importantly, virus-STAT interaction can be antagonistic or stimulatory depending on diverse viral and cellular factors. While STAT antagonism can suppress immune pathways, many viruses promote activation of specific STATs to support viral gene expression and/or produce cellular conditions conducive to infection. It is also becoming increasingly clear that viruses can hijack noncanonical STAT functions to benefit infection. For a number of viruses, STAT function is dynamically modulated through infection as requirements for replication change. Given the critical role of STATs in infection by diverse viruses, the virus-STAT interface is an attractive target for the development of antivirals and live-attenuated viral vaccines. Here, we review current understanding of the complex and dynamic virus-STAT interface and discuss how this relationship might be harnessed for medical applications.
THE JAK-STAT PATHWAY
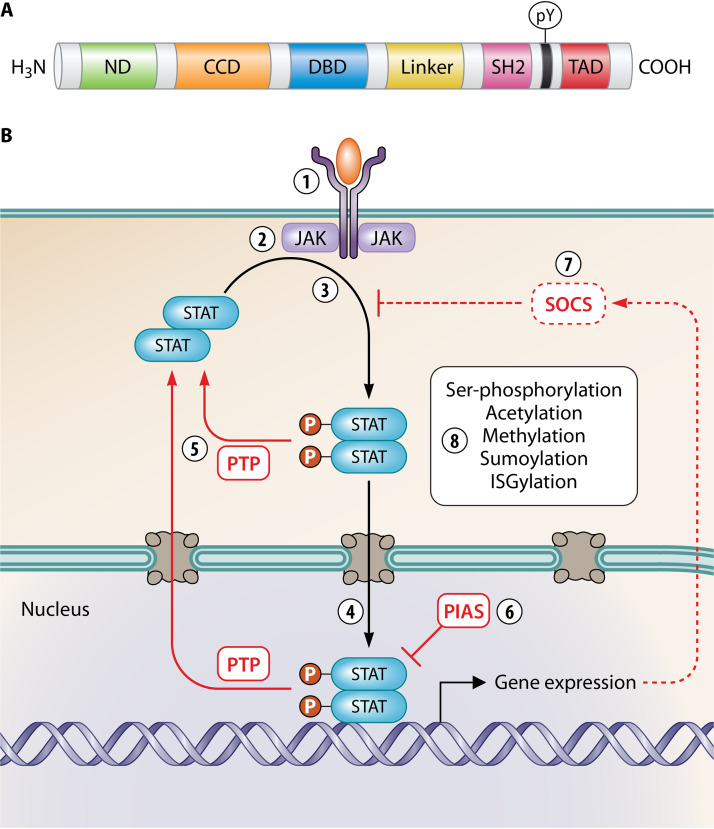
The antiviral immune response requires an array of cytokines and chemokines, many of which signal through STATs (Table 1). The STAT family has seven members in mammals—STAT1 to -4, STAT5a, STAT5b, and STAT6—which share common structural domains that mediate intracellular signaling (Fig. 1A) (1). In the canonical Janus kinase (JAK)-STAT pathway, STATs are activated by receptor-associated kinases of the JAK family (comprising JAK1 to -3 and Tyk2) that undergo autophosphorylation following binding of cytokines, growth factors, etc., to cognate receptors (Fig. 1B) (1, 2). Latent STATs are recruited to receptors and phosphorylated at a conserved tyrosine (pY) by JAKs, resulting in formation of parallel pY-STAT dimers through reciprocal interactions of the pY and Src homology 2 (SH2) domains. The dimers accumulate in the nucleus and bind specific promoter elements to regulate the transcription of large numbers of target genes. Signaling is then deactivated by protein tyrosine phosphatases (PTPs) and members of the protein inhibitor of activated STAT (PIAS) and suppressor of cytokine signaling (SOCS) families (Fig. 1B) (1–3). STATs are further regulated by posttranslational modifications (PTMs) that enhance or inhibit signaling, including serine phosphorylation, acetylation, methylation, sumoylation, and ISGylation (2, 3).
TABLE 1.
Examples of STAT activators and heterodimer partners
| STAT | Principal activator(s)a | Heterodimer partners/cytokines shown to promote heterodimer formation | References |
|---|---|---|---|
| STAT1 | All IFNs |
STAT2 (type I and III IFNs) STAT3 (IFNs, IL-6, IL-27, EGF) STAT4 (IL-35) |
1, 4, 25, 216 |
| STAT2 | Type I and III IFNs |
STAT1 (type I and III IFNs) STAT6 (type I IFNs) |
1, 4 |
| STAT3 |
IL-6 family (e.g., IL-6, LIF, OSM) IL-10, G-CSF, EGF |
STAT1 (IFNs, IL-6, IL-27, EGF) STAT5 (IL-2, IL-7 M-CSF, PDGF) STAT4 (IL-23) |
1, 4, 25, 151, 216 |
| STAT4 | IL-12, IL-23, IL-35 |
STAT1 (IL-35) STAT3 (IL-23) |
1, 4 |
| STAT5a/b | Gamma chain cytokine family (e.g. IL-2, IL-7, IL-15) IL-3, GM-CSF, prolactin, EPO | STAT3 (IL-2, IL-7 M-CSF, PDGF) | 1, 4, 97, 151 |
| STAT6 | IL-4, IL-13, IL-15, PDGF | STAT2 (type I IFNs) | 1, 4, 126 |
EGF, epidermal growth factor; PDGF, platelet-derived growth factor.
FIG 1.
Structure and signaling pathways of STATs. (A) Schematic representation of the conserved domain structure of STATs: N-terminal domain (ND), coiled-coil domain (CCD), DNA-binding domain (DBD), linker domain, Src homology 2 (SH2) domain, and C-terminal transactivation domain (TAD). The conserved C-terminal tyrosine phosphorylated by JAKs (pY) is indicated. (B) Diverse cytokines signal via a classical JAK-STAT paradigm. (1 and 2) Cytokine, growth factor, etc., bind to the cognate receptor (1), activating specific receptor-associated JAKs (2). (3) JAKs phosphorylate latent antiparallel STAT dimers (P in circles), inducing the formation of parallel dimers through reciprocal pY-SH2 interactions. (4) pY-STAT dimers translocate into the nucleus and bind specific DNA elements, activating gene expression. (5) pY-STATs are dephosphorylated by nuclear and cytoplasmic PTPs to turn off signaling. (6 and 7) STAT signaling is also negatively regulated (blunt arrows) by PIAS proteins (which bind STATs and prevent DNA binding, recruit corepressors, or mediate sumoylation) (6) and SOCS proteins (which are induced by pY-STATs and inhibit JAK kinase activity, bind competitively to STAT-binding sites on receptors, or target cytokine receptors and JAKs for proteasomal degradation; dotted line) (7). Red line, negative regulation; black line, positive regulation. (8) STATs are also positively or negatively regulated by PTMs.
Cytokine-specific transcriptional responses derive from the activation of specific subsets of STATs by different cytokines due to interactions of receptors with specific JAKs and STATs (1); this induces an array of STAT homo- and heterodimers (Table 1) (4). Viral targeting of specific STATs and/or STAT complexes can thus have major impacts on cell biology and immunity.
STAT1/2 ARE COMMON VIRAL TARGETS
The interferon (IFN) response is the principal line of defense of mammalian cells against viral infection (3, 5). Type I (e.g., IFN-α and IFN-β), type II (IFN-γ), and type III (e.g., IFN-λ1) IFNs are produced by immune and nonimmune cells following detection of virus and/or infection and signal in autocrine and paracrine fashion by binding to broadly expressed receptors to activate STAT1 or STAT1 and -2. Type I and III IFNs primarily activate STAT1-STAT2 heterodimers, which, with IFN regulatory factor 9 (IRF-9), form the IFN-stimulated gene factor 3 (ISGF3) complex and bind genomic IFN-stimulated response elements (ISREs). Type II IFNs primarily activate STAT1 homodimers that bind gamma-activated sequence (GAS) elements. ISRE/GAS sequences regulate the transcription of hundreds of IFN-stimulated genes (ISGs), many of which encode proteins with antiviral, antiproliferative, proapoptotic, or immune-modulatory functions to suppress viral replication and promote adaptive responses (5).
To prosper within cells, viruses must counteract IFN responses. This is commonly mediated by viral IFN antagonist proteins, hundreds of which have been described. Among them, antagonists of STAT1 and/or -2 are common (reviewed in references 3, 5, and 6), with effector mechanisms that include cytoplasmic sequestration, dephosphorylation, or degradation of STATs; inhibition of upstream signaling components; and induction of negative regulators (Table 2). Notably, a number of viruses show selective targeting of STAT1 or STAT2, resulting in differential antagonism of IFN cytokines. Among paramyxoviruses, simian virus 5 V protein targets STAT1 and has broad impact on IFNs (7), while human parainfluenza virus 2 V protein preferentially antagonizes STAT2, thereby inhibiting type I and III but not type II IFNs (8–10). Similarly, Zika virus (ZIKV) NS5 mediates the degradation of STAT2, suppressing signaling by type I/III but not type II IFNs (11, 12). Notably, loss of STAT2 expression also promotes the formation of STAT1 homodimers, enhancing IFN-γ-induced ISG expression (13). Since IFN-γ positively affects ZIKV replication (13), STAT2 degradation may represent a mechanism to simultaneously suppress antiviral signaling while promoting proviral signaling. Thus, STAT1/2 targeting is a common strategy for antagonism and, potentially, modulation of innate immune responses.
TABLE 2.
Examples of STAT1/2-antagonistic strategies of viruses
| Mechanism | Virus/IFN antagonista (reference) |
|---|---|
| Interaction of IFN antagonists with STAT1/2 | |
| Cytoplasmic sequestration | RABV P1 (187) |
| NIV V (95) | |
| Huaiyangshan banyangvirus NSs (217) | |
| Inhibition of DNA binding | HCMV IE1 (218) |
| RABV P (153, 214) | |
| Porcine bocavirus NP1 (219) | |
| Dephosphorylation of STATs | Vaccinia virus VH1 (220) |
| Inhibition of ISGF3 complex formation | MEV V (221) |
| Targeting of STATs for proteasomal degradation | Simian virus 5 V (222) |
| Dengue virus NS5 (223) | |
| ZIKV NS5 (11, 12) | |
| PEDV (224) | |
| Indirect antagonism | |
| Inhibition of transcription from STAT gene | HPV E6/E7 (225) |
| Inhibition of STAT nuclear import machinery | Ebola virus VP24 (226) |
| HBV Pol (227) | |
| PRRSV Nsp1β (228) | |
| Reduced IFN receptor expression | Respiratory syncytial virus NS1 (229) |
| EBV LMP2A/B (230) | |
| Inhibition of JAK expression/activity | Marburg virus VP40 (231) |
| EBV LMP1 (36) | |
| Murine polyomavirus T antigen (232) | |
| Adenovirus (233) | |
| Upregulation of negative regulators of STAT | IAV NS1 (154) |
| HBV X (234) | |
| HCV core (235) | |
PRRSV, porcine reproductive and respiratory syndrome virus.
STAT3: A PRO- AND ANTIVIRAL MOLECULE
STAT3 is a pleiotropic signaling molecule involved in processes including growth and development, consistent with embryonic lethality of STAT3-deficient mice (14, 15). STAT3 is generally considered a prosurvival or proliferation factor, consistent with association with many cancers (16), but also has proapoptotic roles (17, 18). STAT3 mediates signaling by diverse cytokines (Table 1), including as the major mediator of Gp130 receptor signaling by interleukin-6 (IL-6) family cytokines (1, 15). IL-6 cytokines are important to immunity, including the acute-phase inflammatory response, antibody production, and T-cell differentiation (19). Interestingly, STAT3 also mediates anti-inflammatory IL-10 signaling (1, 15).
STAT3 is also implicated in IFN responses, although its precise role is complicated by reports indicating distinct or opposing effects of STAT3 depletion. Specifically, some data indicate that STAT3 has antiviral activity as an important component of IFN signaling (20–24), while others indicate a negative regulatory role toward IFN (25–28). It has been suggested that this disparity might be due in part to the use of different cell types in the studies (21). Indeed, the diverse functions attributed to STAT3 likely derive to a great extent from distinct roles in different cell types, as genomic binding patterns of STAT3 are largely cell-type specific, relating to cell-specific interactions with coactivators and repressors (29, 30). Nevertheless, STAT3 binds and regulates a “universal” gene subset, independent of cell type (29, 30). Notably, STAT3 has also been reported to have opposing roles within the same cell type. For example, the STAT3-activating cytokines IL-6, IL-10, and IL-21 produce proinflammatory, anti-inflammatory, and proapoptotic outcomes, respectively, in dendritic cells (29, 31, 32). Thus, STAT3 function appears to be determined by several factors, potentially involving PTMs and heterodimerization with other STATs (4, 29).
VIRAL ACTIVATION OF STAT3
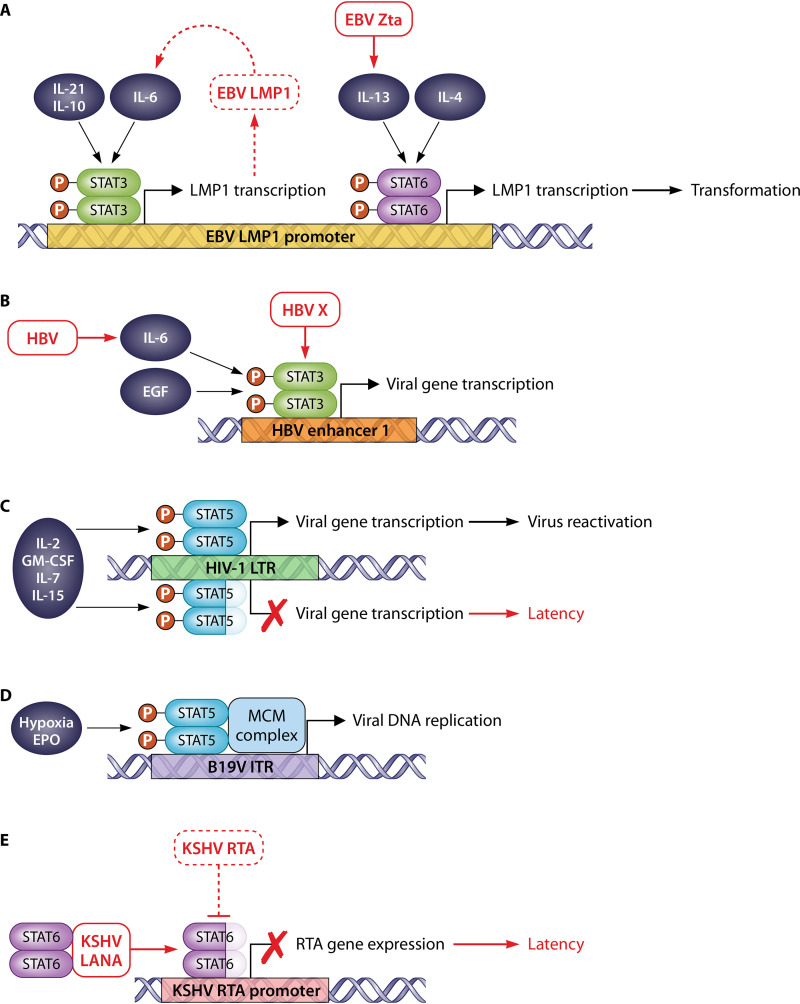
The relationship of STAT3 with viruses is complex, relating to the diversity of STAT3 functions and the diverse nature of viruses (33–35). Many oncoviruses promote STAT3 activation to upregulate prosurvival and proproliferation genes through direct interaction of viral proteins with STAT3 and/or activation of upstream signaling (Table 3). The Epstein-Barr virus (EBV) oncoprotein LMP1, which blocks STAT2 activation by IFN-α (36), promotes STAT3 activation and STAT3-dependent gene expression via upregulation of IL-6 (37, 38), highlighting the distinct and opposing roles that STATs can play in infection. Interestingly, the LMP1 promoter contains a STAT3-binding site and so is induced in response to STAT3-activating cytokines (Fig. 2A) (38–41). This appears to form a positive-feedback loop that promotes cellular transformation during infection (38, 39). The feedback loop likely also promotes latency, since STAT3 is implicated in maintaining latency of EBV and other herpesviruses (Kaposi’s sarcoma-associated herpesvirus [KSHV] and herpes simplex virus 1 [HSV-1]) by inducing the expression of host-encoded transcriptional repressors of lytic genes (42–45). Notably, the HSV-1 genome also contains STAT3-binding elements. During HSV-1 infection, STAT3 induces NEAT1, a noncoding cellular RNA involved in forming intranuclear paraspeckles. STAT3 is also recruited to the paraspeckles and interacts with the viral genome to induce expression of the HSV-1 ICP0 and TK genes (46). Thus, STAT3 appears to be an important proviral factor for multiple herpesviruses.
TABLE 3.
Examples of viral regulators of STAT3a
| Virus | Effect on STAT3 | Reference(s) |
|---|---|---|
| EBV | EBV proteins LMP1, LMP2A, and EBNA2 upregulate STAT3 signaling to promote cell growth/transformation and viral gene expression/latency and inhibit autophagy. STAT3 binds and activates transcription from promoters of LMP1 and EBNA1 in the viral genome. | 37–41, 73, 74, 236–239 |
| HBV | HBV X protein promotes STAT3 activation through interaction with JAK1, deregulation of cellular miRNA expression, and induction of reactive oxygen species. HBV infection activates STAT3 through IL-6-dependent pathways. STAT3 activation impacts viral gene expression/replication, cell survival/proliferation, and apoptosis. STAT3 binds and activates transcription from enhancer 1 in the viral genome. | 47–53 |
| HPV | HPV E6 upregulates IL-6/STAT3 signaling to promote viral gene expression and tumorigenesis. STAT3 binds the upstream regulatory region in the viral genome. | 240–243 |
| Varicella-zoster virus | Varicella-zoster virus upregulates miR-21 to enhance STAT3 signaling, promoting cell survival and virus replication. | 192, 244 |
| Rift Valley fever virus | Rift Valley fever virus NSs induces STAT3 activation to promote cell survival and inhibit virus-induced cell death. | 245 |
| PEDV | PEDV upregulates EGFR/STAT3 signaling to inhibit type I IFN signaling and promote viral replication. | 55 |
| KSHV | KSHV upregulates STAT3 signaling, including through binding of KSHV LANA and RTA to STAT3 and through JAK-dependent pathways. KSHV kaposin B induces monophosphorylation of STAT3 at S727. STAT3 signaling promotes cell transformation, latency, and inflammatory responses and inhibits autophagy. | 43, 75, 141–146, 181 |
| KSHV cyclin K binds STAT3 and inhibits growth-suppressive effects of OSM. KSHV encodes miRNAs that inhibit IL-6/IFN-α-induced STAT3 activation/target gene expression. STAT3 inhibition may induce lytic reactivation. | 106, 246, 247 | |
| HCV | HCV upregulates STAT3 signaling, including through binding of HCV core to STAT3, upregulation of IL-6, and induction of reactive oxygen species. STAT3 activation deregulates inflammatory responses and promotes cell proliferation/transformation and viral replication. | 63–68 |
| HCV blocks STAT3-DNA binding and induces proteasomal degradation of STAT3, inhibiting IFN-α/LIF signaling. | 69, 70 | |
| HCMV | HCMV US28 upregulates IL-6/STAT3 signaling to promote cell proliferation/transformation. STAT3 activity is required for efficient viral gene expression/replication. | 71, 193, 248 |
| HCMV IE1 binds and sequesters unphosphorylated STAT3 in the nucleus, inhibiting IL-6/STAT3-dependent gene expression and rewiring IL-6 signaling into an IFN-γ-like response. HCMV inhibits STAT3 activation in response to IFN-α/γ. | 71, 72, 249 | |
| HIV-1 | HIV-1 Nef and gp120 upregulate soluble factors that activate STAT3, potentially promoting cell survival and deregulating inflammatory responses. HIV-1 induces monophosphorylation of STAT3 at S727. HIV-1 upregulates IL-10/STAT3 signaling in bystander cells to inhibit autophagy. | 76, 156, 180, 250 |
| HIV-1 Vif induces proteasomal degradation of STAT3, inhibiting IFN-α signaling. | 251 | |
| EV71 | EV71 induces STAT3 activation to inhibit type I IFN signaling and promote viral replication. | 54 |
| EV71 induces miRNA-124, which suppresses IL-6/STAT3 signaling to inhibit anti-EV71 activity. EV71 induces proteasomal degradation of STAT3. | 54, 252 | |
| RABV | RABV induces miRNA-455, which suppresses SOCS3 and enhances STAT3 activation to promote viral replication. | 253 |
| RABV P protein selectively binds IFN-α/OSM-activated STAT3-STAT1 heterodimers to inhibit nuclear accumulation and target gene expression. | 155, 189 | |
| IAV | IAV induces STAT3 activation to delay apoptosis. | 254 |
| IAV inhibits STAT3 activation in response to IFN-β/IL-6 by upregulating SOCS3 and downregulating IFN receptor. Inhibition of IL-6/STAT3 signaling is associated with disease severity in vivo. | 154, 255 | |
| Severe acute respiratory syndrome coronavirus 1 | Severe acute respiratory syndrome coronavirus 1 PLpro promotes STAT3 signaling to upregulate the Egr-1/TGF-β1b fibrotic pathway. | 256 |
| Severe acute respiratory syndrome coronavirus 1 reduces STAT3 phosphorylation, potentially promoting apoptosis. | 257 | |
| MUV | MUV V protein induces proteasomal degradation of STAT3, inhibiting IL-6/v-Src/STAT3-dependent gene expression. STAT3 antagonism is associated with disease severity in vivo. | 57, 59 |
| MEV | MEV V protein binds STAT3, inhibiting IL-6/v-Src/STAT3-dependent gene expression. | 58 |
| Tioman virus | Tioman virus V protein binds STAT3, inhibiting IL-6/STAT3-dependent gene expression. | 62 |
| PRRSV | PRRSV NSP5 induces proteasomal degradation of STAT3, inhibiting OSM/STAT3-dependent gene expression. | 258 |
| Hepatitis E virus | Hepatitis E virus ORF3 disrupts EGF receptor trafficking, inhibiting EGF/STAT3-dependent gene expression. | 259 |
| Marburg virus | Marburg virus VP40 inhibits JAK1 phosphorylation, suppressing IL-6-induced STAT3 activation. | 231 |
| Human metapneumovirus | Human metapneumovirus inhibits JAK2 phosphorylation, suppressing IL-6/STAT3-dependent gene expression. | 260 |
| Adenovirus | Adenovirus inhibits JAK1 phosphorylation, suppressing IL-6/OSM-induced STAT3 activation. | 233 |
Lightface and boldface indicate stimulatory and inhibitory effects, respectively, on STAT3.
TGF-β1, transforming growth factor β1.
FIG 2.
Examples of viruses with genomic STAT-binding sites. (A) The EBV LMP1 promoter contains STAT3 and STAT6 sites. Cytokines activating STAT3 (IL-6, IL-21, and IL-10) or STAT6 (IL-13 and IL-4) induce LMP1, promoting cell transformation. LMP1 upregulates IL-6, resulting in a positive-feedback loop. EBV Zta protein induces IL-13, and IL-13 induces LMP1 via STAT6. (B) HBV enhancer 1 contains a STAT3 site. Cytokines activating STAT3 (IL-6 and epidermal growth factor [EGF]) induce viral gene expression. HBV (including HBV X protein) induces STAT3 activation. (C) The HIV-1 LTR contains a STAT5 site that is activated by STAT5-activating cytokines (e.g., IL-2, GM-CSF, IL-7, and IL-15) to induce gene expression and viral reactivation; this is inhibited in cells expressing C-terminally truncated STAT5, which binds to the LTR site. (D) The B19V replication origin in inverted terminal repeats (ITRs) contains a STAT5 site, where EPO/hypoxia-activated pY-STAT3 recruits the minichromosome maintenance (MCM) complex to initiate replication. (E) The KSHV RTA promoter contains a STAT6 site; LANA mediates cleavage of the STAT6 C terminus, and cleaved STAT6 binds to the RTA promoter, repressing RTA expression to promote latency. During lytic infection, expressed RTA induces STAT6 degradation.
Hepatitis B virus (HBV) also hijacks STAT3 to activate viral gene expression (Fig. 2B) (47) and, consistent with this, promotes STAT3 activation through several mechanisms (Table 3) (48–50). HBV-induced STAT3 activation also upregulates cellular genes, which appears to promote a prosurvival (49, 51) or proapoptotic (52, 53) state, depending on the cell type. Certain nononcogenic viruses also exploit STAT3 to promote cell survival (Table 3), while the reported capability of STAT3 to downregulate the antiviral IFN response (see above) appears to be hijacked by enterovirus 71 (EV71) (54) and porcine epidemic diarrhea virus (PEDV) (55). Clearly, STAT3 is an important proviral factor for diverse viruses, despite differences in the specific cellular outcomes of STAT3 activation.
VIRAL ANTAGONISM OF STAT3
Despite evident proviral activities, inhibition of STAT3 signaling also forms part of the immune evasion strategy of many viruses. The paramyxoviruses mumps virus (MUV) and measles virus (MEV) antagonize IFN/IL-6-dependent STAT signaling via the V protein; MUV V protein targets STAT3 and STAT1 for proteasomal degradation (56, 57), while MEV V protein binds STAT1, -2, and -3 and inhibits nuclear translocation (58). Disruption of STAT3 antagonism by mutation of MUV V protein reduced neurovirulence in vivo (59, 60), indicative of an important role in disease. Interestingly, V proteins of the paramyxoviruses Hendra virus, Nipah virus (NIV), and simian virus 5 target STAT1 and/or -2 but not STAT3 (57, 61), while Tioman bat paramyxovirus V protein does not target STAT1 or -2 but can bind STAT3 and impair IL-6 signaling (62). Thus, STAT3/IL-6 antagonism appears to have highly specific roles in the biology of certain paramyxoviruses. Precisely how the differential targeting of STATs impacts the transcriptome in response to specific cytokines and the consequences for host and viral biology are not resolved.
The list of viruses that antagonize STAT3 is growing in number and diversity (Table 3). Interestingly, while activation of STAT3 by hepatitis C virus (HCV) is well established (63–67), consistent with a proviral role (67, 68), STAT3 also has anti-HCV activity (24) and is inhibited or degraded during infection (69, 70). The reason for these apparently contrasting findings is unclear, but a number of viruses that can induce STAT3 signaling with apparently proviral effects also encode STAT3-antagonistic mechanisms (Table 3). In an interesting variation on this theme, human cytomegalovirus (HCMV) IE1 protein binds and sequesters unphosphorylated STAT3 within the nucleus, preventing activation and target gene expression in response to IL-6 (71). However, depletion or pharmacological inhibition of STAT3 impairs HCMV gene expression and replication, correlating with impaired nuclear relocalization. Thus, it seems that STAT3 nuclear sequestration is important for both immune evasion and HCMV replication (71). Notably, STAT3 antagonism by IE1 also results in enhanced activation of STAT1 in response to IL-6 cytokines, so that IE1 “rewires” IL-6 signaling into an IFN-γ-like response (72), similar to the modulation of STAT1/2 responses by ZIKV (see above).
The balance between activation and antagonism of STAT3 by different viruses, and even by the same virus, likely depends on a range of host and viral factors, including differing roles for STAT3 in different cell types and cytokine pathways, virus-specific requirements for infection (e.g., cell lysis to release progeny or promotion of cell survival to facilitate replication and reduce inflammation), and distinct requirements through stages of infection (e.g., early versus late and lytic versus latent). Indeed, for many viruses, different infection stages are characterized by expression of distinct viral protein subsets, which might have specific relationships with STAT3. Virus infection can also activate or regulate other cellular pathways (e.g., stress responses) that modulate STAT3-regulatory proteins or PTMs, potentially defining in dynamic fashion whether STAT3 is pro- or antiviral. For example, EBV, KSHV, and human immunodeficiency virus type 1 (HIV-1) induce STAT3 phosphorylation at serine 727 (S727) (73–76), which is required for maximal transactivation function (2). Despite these insights, it is clear that our understanding of the potentially nuanced and dynamic relationship of viruses and STAT3 is still in its infancy.
STAT4
STAT4 is the key mediator of signaling by IL-12, which is produced by a number of innate immune cells in response to infection (77). IL-12 primarily acts on natural killer (NK) and T cells to induce IFN-γ and other genes through STAT4, so that IL-12-induced activation of NK cells and differentiation of T helper 1 cells is impaired in STAT4-deficient mice (78, 79). STAT4 is also activated by other cytokines (Table 1), including type I IFNs that induce expression of IFN-γ (80–82) and specific ISGs (83, 84) via STAT4.
It is not surprising that STAT4 is implicated in antiviral immunity, including roles in IFN-γ induction and CD4+ and CD8+ T cell responses during lymphocytic choriomeningitis virus infection (80, 84–86), and in efficient NK cell responses to murine cytomegalovirus (87). Meta-analysis identified a link between a STAT4 polymorphism and chronic HBV infection (88). STAT4 was also implicated in effective IFN-γ responses to HSV-2 (89). However, for HSV-1 ocular infection, the importance of STAT4 is unclear, with one study indicating that STAT4-deficient mice are more susceptible to encephalitis and facial lesions (90) while another indicated no impact on disease severity, although higher viral titers were detected in the eyes of STAT4-deficient mice at early time points (91). STAT4-deficient mice could also mount T-cell and antibody responses against influenza A virus (IAV) as effectively as wild-type mice, resulting in viral clearance (92). Thus, the extent to which STAT4 contributes to immunity might be virus specific. Interestingly, a noncanonical role for unphosphorylated STAT4 in type I IFN induction in response to RNA viruses has been reported in macrophages. STAT4 interacts with the E3 ligase CHIP and inhibits CHIP-mediated proteasomal degradation of retinoic-acid-inducible gene I (RIG-I), promoting RIG-I-dependent IFN induction (93).
Despite apparent antiviral activities, evidence of direct viral antagonism of STAT4 is scarce, possibly reflecting restricted tissue distribution. However, STAT4 was recently identified as a binding partner of the NIV IFN antagonist protein isoforms V and W (94). Since V and W antagonize STAT1 by direct binding (95, 96) and mutation of V/W residue G121 disrupts binding to STAT1 and STAT4 (94), it seems likely that V/W-STAT4 interaction is antagonistic, affecting signaling by STAT4-activating cytokines. Confirmation of the role of this interaction awaits further investigation.
STAT5a/b
STAT5a and -b are highly homologous proteins that arose from gene duplication and function in overlapping and distinct processes as STAT5a/b homo- or heterodimers. STAT5a/b mediate signaling by multiple cytokines and growth factors (Table 1) and are implicated in growth; erythropoiesis; lymphoid development and differentiation; and proliferation and activity of myeloid, NK, and T cells (97). By mediating signaling by gamma chain cytokines, STAT5a/b have key roles in expansion and differentiation of T cells and enhancing the cytolytic activity of NK cells (98). STAT5a/b are also activated by and contribute to the antiviral IFN response (99–102).
In common with STAT3, STAT5 is associated with many cancers due to prosurvival and proliferation functions (97), including cancers caused by human T-lymphotropic virus type I (HTLV-1), EBV, and human papillomavirus (HPV) (Table 4). Moreover, the E7 protein of high-risk HPV promotes STAT5 activation to induce genes involved in DNA damage responses, which are required for viral genome replication in differentiating epithelial cells (103, 104). Similar to its relationship with STAT3, KSHV can promote and inhibit STAT5 pathways. Upregulation of the IL-3/STAT5 pathway to induce PROX1 expression is critical for KSHV reprogramming of blood vascular endothelial cell fate (105), while KSHV-encoded microRNAs (miRNAs) target erythropoietin (EPO) receptor mRNA in endothelial cells, downregulating EPO-dependent STAT5 signaling (106). These data are consistent with STAT5 being pro- or antiviral for KSHV, depending on the cellular and signaling context.
TABLE 4.
Examples of viral regulators of STAT5a
| Virus | Effect on STAT5 | Reference(s) |
|---|---|---|
| EBV | EBV LMP1 upregulates STAT5 signaling to promote cell growth/transformation. | 38, 39, 261 |
| HTLV-1 | HTLV-1 p12(I) and Tax upregulate STAT5 signaling to promote cell proliferation/transformation. | 262–265 |
| HPV | HPV upregulates STAT5 signaling to promote cell survival/proliferation and induce ATM/ATR DNA damage responses, promoting viral gene expression/replication. STAT5 binds the regulatory region in the viral genome. | 103, 104, 266–269 |
| B19V | STAT5 binds inverted terminal repeats in the viral genome, promoting B19V genome replication. | 122, 123 |
| KSHV | KSHV upregulates IL-3/STAT5 signaling to reprogram cell fate. | 105 |
| KSHV encodes miRNAs that target EPO receptor mRNA, inhibiting EPO/STAT5 signaling. | 106 | |
| HIV-1 | HIV-1 increases basal STAT5 activation. Cytokine (e.g. IL-2, IL-15, and IL-7)-activated full-length or C-terminally truncated STAT5 binds LTRs in the viral genome, upregulating or inhibiting, respectively, virus production and gene expression. | 110–113, 115–120 |
| HIV-1 inhibits STAT5 signaling pathways (e.g., IL-2, IL-7, and GM-CSF activated). HIV-1 Nef inhibits STAT5 expression in hematopoietic progenitors to inhibit multipotency. | 107–111, 114, 121 | |
| WNV | WNV inhibits STAT5 activation in response to IFN-β/IL-4. | 124 |
| ZIKV | ZIKV inhibits JAK1/TYK2 and STAT5 activation in response to IFN-β/IL-4. | 124 |
Lightface and boldface indicate stimulatory and inhibitory effects, respectively, on STAT5.
The relationship between HIV-1 and STAT5 is complex, with reports of impaired and enhanced STAT5 responses (Table 4) (107–114). These discrepancies may arise in part from differences in viral strains and experimental approaches but are also likely to be related to the roles of STAT5 in controlling latency. In response to cytokines (e.g., IL-2 and IL-15), pY-STAT5 activates transcription from a STAT5-binding site within HIV-1 long terminal repeats (LTRs), inducing reactivation (Fig. 2C) (113, 115–119). Interestingly, a naturally occurring C-terminally truncated STAT5 isoform is present in HIV-1-infected peripheral blood mononuclear cells and binds the LTR but represses gene expression and virus reactivation, likely due to truncation of the transactivation domain (Fig. 1A) (112, 120). Further complicating the STAT5 story, secreted HIV-1 Nef protein deregulates the multipotency of uninfected early hematopoietic progenitors by downregulating STAT5 expression via the PPARγ pathway, contributing to hematopoietic deficiency and immunodeficiency in AIDS patients (121). Thus, STAT5 activity appears to be dynamically modulated by HIV-1 in infected and bystander cells to control cellular fate and support or reverse latency. The genome of human parvovirus B19 (B19V) also contains a pY-STAT5-binding element, which functions in replication initiation by recruiting the minichromosome maintenance complex (Fig. 2D) (122, 123).
Reports of STAT5 antagonism are limited (Table 4), but the flaviviruses West Nile virus (WNV) and ZIKV were recently shown to inhibit STAT5 phosphorylation in dendritic cells in response to type I IFNs and IL-4, but not granulocyte-macrophage colony-stimulating factor (GM-CSF) (124). Interestingly, these viruses appear to use distinct antagonistic mechanisms, while other flaviviruses, dengue virus and yellow fever virus, had no effect on STAT5 phosphorylation, indicating virus-specific roles (124).
STAT6
STAT6 is primarily implicated in signaling by the functionally related cytokines IL-4 and IL-13, which have cooperative and nonredundant roles in T helper 2 (Th2) differentiation, immunoglobulin class switching, macrophage activation, and major histocompatibility complex class II (MHC-II) expression and are key players in defense against extracellular parasites (125, 126). STAT6 is also activated by other cytokines (Table 1), including type I IFNs (126). A noncanonical role for STAT6 in innate immune responses has also been reported, involving stimulator of interferon genes (STING) (127), a sensor of cytosolic DNA and adaptor protein for signaling by certain sensors of viral RNA. Typically, STING recruits TANK-binding kinase 1 (TBK1) and IRF-3, leading to induction of type I IFNs, but STING was shown to also interact with and promote phosphorylation of STAT6 independently of cytokine and JAK signaling, via TBK1 and an unknown kinase (127). This causes upregulation of a specific set of STAT6 target genes different from those of the canonical JAK-STAT6 pathway, including certain chemokines. Consistent with an antiviral role, STAT6-deficient mice were more susceptible to infection by DNA and RNA viruses (127). Interestingly, earlier studies found that STAT6-deficient mice were less susceptible to disease caused by HSV-1 (128) or ectromelia virus (129). The differences may be related to the distinct disease models used and the complex outcomes of Th1/Th2 dysregulation (due to the lack of STAT6) in an in vivo setting, highlighting the multifaceted nature of STAT6 function during infection.
Persistent STAT6 activation is associated with several cancers (130–132), and similar to STAT3 and STAT5, STAT6 can be activated by viral proteins directly or via upstream pathways (e.g., upregulation of IL-13 by EBV Zta protein [133]), promoting transformation (Table 5). Since the EBV LMP1 promoter contains a STAT6-binding site, which is activated by IL-13 and IL-4 (Fig. 2A) (134), this indicates a proviral role for IL-13. Perhaps the best-characterized STAT6-virus interaction is with KSHV, where infection induces constitutive, albeit modest, phosphorylation via upregulation of IL-13 and downregulation of the PTP SHP1, promoting cell survival and proliferation (135, 136). However, recent studies have indicated that modulation of STAT6 through the KSHV infectious cycle is dynamic and complex, with differential regulation of STAT6 expression and activity important in the lytic and latent phases characteristic of herpesviruses.
TABLE 5.
Examples of viral regulators of STAT6a
| Virus | Effect on STAT6 | Reference(s) |
|---|---|---|
| Herpesvirus saimiri | Herpesvirus saimiri Tip interacts with STAT6 and promotes STAT6 activation/gene expression, promoting cell transformation. | 270, 271 |
| HTLV-1 | HTLV-1 Tax upregulates transcription from the IL-13 promoter, promoting IL-13/STAT6 signaling and cell survival/transformation. | 272 |
| EBV | EBV Zta upregulates transcription from the IL-13 promoter, promoting IL-13-induced cell proliferation. IL-13/IL-4-activated STAT6 binds and activates transcription from the LMP1 promoter in viral genome. | 133, 134 |
| STAT6 expression is reduced during lytic EBV infection. | 138 | |
| KSHV | KSHV induces basal STAT6 phosphorylation through upregulation of IL-13 and downregulation of SHP1, promoting cell proliferation. | 135, 136 |
| KSHV LANA binds and induces truncation of STAT6, inhibiting IL-4/STAT6 signaling. Truncated STAT6 binds the RTA promoter in the viral genome, inhibiting RTA expression/virus reactivation. KSHV RTA binds STAT6 and induces STAT6 proteasomal/lysosomal degradation to promote virion production. | 135, 137, 138 |
Lightface and boldface indicate stimulatory and inhibitory effects, respectively, on STAT6.
It was originally reported that during latent infection the KSHV-encoded LANA protein inhibits STAT6 phosphorylation and transactivation activity in response to IL-4, thereby inhibiting IL-4-induced B cell activation and proliferation (135). However, a subsequent study indicated that LANA binds to STAT6 independently of tyrosine phosphorylation and induces accumulation in the nucleus, where STAT6 is cleaved to generate a “dominant-negative” protein with a truncated transactivation domain (137). The truncated STAT6 binds to a canonical STAT6-binding site in the promoter of KSHV replication and transcription activator (RTA), the key mediator of viral reactivation, preventing RTA transcription and maintaining latency (Fig. 2E) (137). Consistent with prolatency functions, STAT6 depletion induced RTA expression and virion production (135, 137, 138). Notably, IL-4/STAT6 signaling is also reported to induce RTA expression and reactivation of KSHV (139, 140), suggesting that full-length STAT6 may be able to activate transcription from the RTA promoter. Delineation of the precise biological relationship of these observations awaits further investigation.
During lytic replication of KSHV, RTA interacts with and ubiquitylates STAT6, inducing degradation (138), presumably to release the repressive activity of truncated STAT6 on RTA expression and reactivation. The effect of STAT6 degradation on cytokine signaling has not been examined, but it would be expected to be inhibitory and so contribute to both the latent-lytic switch and immune evasion, the latter being important during productive infection. Notably, the STAT6 degradation mechanism appears to be conserved among the human herpesviruses EBV, HCMV, and HSV-1 (138). For EBV, degradation of STAT6 during lytic infection is consistent with roles of STAT6 in inducing the prolatency factor LMP1 (see above).
KSHV encodes multiple STAT6-binding proteins, indicating the importance of STAT6 subversion to infection. Interestingly, STAT3 is also implicated in the KSHV latent-lytic switch, as it is activated during latency (141), with inhibition and depletion resulting in reactivation (43, 106). A number of KSHV proteins and mechanisms activate STAT3 (75, 141–145), while STAT3 repression for reactivation might involve KSHV-encoded miRNAs (106). RTA and LANA also interact with STAT3, although these interactions contrast with those for STAT6 (see above), as they promote STAT3 activity (138, 145, 146), highlighting how individual viral proteins can target different STATs with distinct outcomes. In addition to regulating STAT3 and STAT6, KSHV promotes and inhibits different STAT5 cytokine signaling pathways (see above) and blocks STAT1/2 signaling to antagonize type I IFN responses (147, 148). KSHV thus provides a clear example of how viruses can dynamically modulate STAT biology and nonredundant STAT pathways to induce conditions favoring infection and spread.
STAT HETERODIMERS
Many cytokines activate multiple STATs to induce an array of homo- and heterodimers (Table 1), presenting an additional level of complexity in viral targeting. For example, while STAT3 signals as a homodimer, several cytokines also induce STAT3 heterodimers with STAT1, STAT4, or STAT5 (Table 1) (4). The factors defining the composition of the cellular pool of activated STAT homodimers and heterodimers are poorly understood but likely depend on the stimulating cytokine(s) and receptor(s), the concentration and stoichiometry of different STATs, and the cell type. The role of heterodimers also remains controversial (4), including the function of STAT3 in IFN signaling (see above); in one study, STAT3 was proposed to effect inhibitory sequestration of STAT1 into STAT3-STAT1 heterodimers (25), while others reported positive roles in ISG expression (20–23).
Different STATs have differing affinities for promoter sites (149, 150), suggesting that heterodimers activate distinct gene subsets compared to the homodimeric counterparts. This is supported by reports that monocyte colony-stimulating factor (M-CSF)-activated STAT5-STAT3 heterodimers bind to a DNA element distinct from that bound by STAT5 homodimers (151) and that IL-35-activated STAT1-STAT4 heterodimers bind to promoters of Ebi3 and Il12a but not Irf1 (a classical STAT1 target) or Il18ra (a classical STAT4 target) (152). STAT heterodimers may also have distinct interactions with other transcription factors, coactivators, and corepressors. This is well understood for STAT1, homodimers of which bind to GAS sites, while STAT1-STAT2 heterodimers bind IRF9 and target ISREs (5). Thus, the various homo- and heterodimers formed by specific STATs are likely to have distinct biological significances, so that dimer-selective targeting by viruses would be expected to significantly impact transcriptional responses and might contribute to the apparent capacity of some STATs to be pro- and antiviral.
Despite this, analysis of viral targeting of specific homo- and heterodimers has remained somewhat limited. Electrophoretic mobility shift assays have indicated that rabies virus (RABV) P protein, which interacts with IFN-activated STAT1 and -2, blocks DNA binding of STAT1 homodimers and STAT1-STAT2 heterodimers (153). IAV NS1 protein impaired DNA binding of STAT1 and STAT3 homodimers and STAT3-STAT1 heterodimers following IFN-β activation, as expected, since IAV inhibits STAT activation (154).
Selective STAT targeting can also be inferred through analysis using STAT1-deficient cells. Such assays indicated that MUV V protein targets STAT1 and STAT3 independently to evade signaling by IFN and IL-6, so all STAT1- and STAT3-containing complexes are likely to be affected (57, 60). In contrast, recent data have indicated that antagonism of IL-6-activated STAT3 by RABV P protein is dependent on STAT1, so that P protein targets STAT3 only in the context of STAT3-STAT1 heterodimers (155). This appears to represent a mechanism to differentially regulate target genes of STAT3 homo- and heterodimers (155). A similar selectivity in viral stimulation of STATs was reported for HIV-1 Nef protein, which induced DNA binding of STAT1 homodimers and STAT3-STAT1 heterodimers, but not STAT3 homodimers, in monocyte-derived macrophages (156). A greater focus on resolving viral specificity for STAT complexes and consequences for the transcriptome should give valuable insights into viral modulation of the cell and the specific roles of distinct STAT dimers in cellular physiology, a poorly understood area in biology.
NONCANONICAL STAT FUNCTIONS
It is becoming increasingly clear that STATs have functions distinct from canonical JAK/STAT signaling (Fig. 3), so virus-STAT interactions are likely to have impacts beyond cytokine responses. It is generally accepted that non-tyrosine-phosphorylated “U-STATs” can dimerize, shuttle between the nucleus and cytoplasm, and bind DNA (157, 158) to activate specific subsets of the genes regulated by their pY-STAT counterparts (159–161) or distinct gene sets (162–165) or to effect gene repression (166–168). Importantly, STAT1 and STAT3 gene expression is induced by pY-STAT1 and pY-STAT3, so many cytokine responses are characterized by an initial transient pY-STAT-mediated gene expression profile followed by a “second wave” of genes activated by upregulated U-STATs (169).
FIG 3.
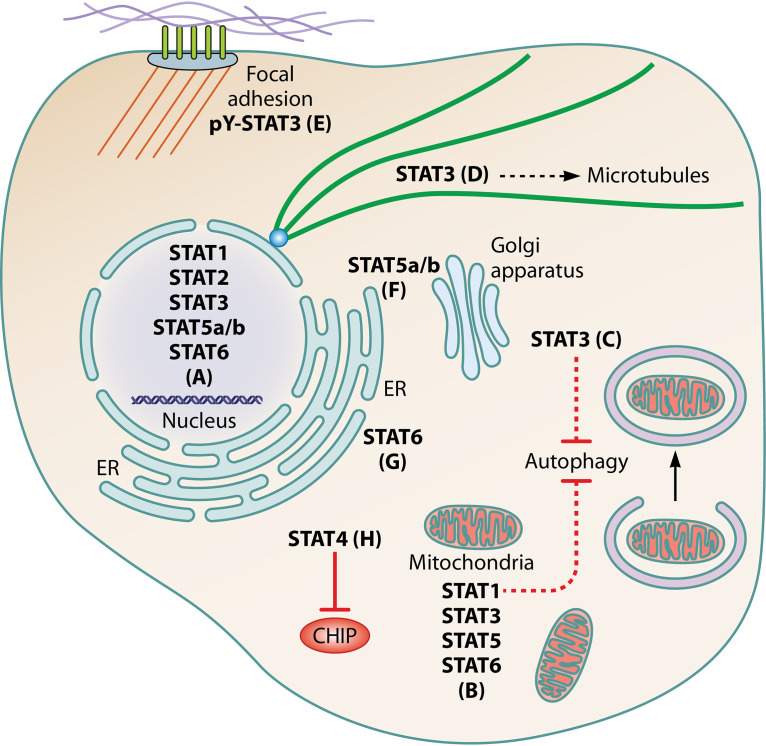
Noncanonical STAT functions. (A) U-STAT1 to -3, -5a/b, and -6 are reported to have intranuclear gene-regulatory function. (B) STAT1, -3, -5, and -6 localize to mitochondria and regulate metabolism, cancer cell growth and proliferation, the mitochondrial permeability transition pore, mitochondrial gene expression, and autophagy. (C and D) Cytoplasmic STAT3 inhibits PKR to repress autophagy induction (C) and stathmin to stabilize microtubules (D). (E) pY-STAT3 accumulates at focal adhesions in ovarian cancer cells, potentially contributing to invasiveness. (F) U-STAT5a/b localize to the ER and Golgi apparatus and promote structural integrity. (G) STAT6 is recruited to the ER by STING following viral nucleic acid detection and is phosphorylated by non-JAK kinases before translocation to the nucleus to activate gene expression. (H) Cytoplasmic STAT4 promotes type I IFN induction in response to RNA viruses by binding the E3 ligase CHIP to protect RIG-I from proteasomal degradation.
Outside the nucleus, STAT1, -3, -5, and -6 can localize to mitochondria to mediate STAT-specific functions (170–172), while STAT2 is implicated in mitochondrial fission (173). The best-characterized mitochondrial association is that of STAT3, which regulates the electron transport chain and the mitochondrial permeability transition pore (172). Cytoplasmic STAT3 can also inhibit autophagy and stabilize microtubules via interaction with protein kinase R (PKR) (174) and stathmin (175, 176), respectively. pY-STAT3 was additionally reported to localize to focal adhesions in cancer cells (177). Interestingly, U-STAT5 is found in the Golgi apparatus and endoplasmic reticulum (ER), with a role in structural integrity (178, 179). As discussed above, STAT6 (127) and STAT4 (93) have roles distinct from JAK/STAT signaling in innate antiviral responses.
This raises important questions as to how viral targeting of STATs affects noncanonical functions and whether these functions are pro- or antiviral. Since pY-STATs activate expression of their corresponding U-STATs, viruses that induce and enhance pY-STAT signaling are likely to upregulate U-STAT functions. The proviral role of STAT3 toward HCV has been partially attributed to microtubule stabilization (68), providing a possible reason for the promotion of STAT3 activity by HCV (Table 3). Moreover, viral activation of STAT3 is implicated in inhibition of autophagy by HIV-1 (180), EBV (74), and KSHV (181). Autophagy can be anti- or proviral, depending on the virus (182), so distinct requirements might contribute to the differing nature of STAT3 targeting by different viruses. Mitochondrial function is also commonly impacted by viruses (183). The extent to which this involves virus-STAT interactions is unclear, although phosphorylation of STAT3 S727, induced by several viruses (73, 75, 76), is associated with mitochondrial roles. Notably, a number of viruses induce S727 phosphorylation of STAT3 and/or STAT1 in the absence of tyrosine phosphorylation (75, 76, 184, 185). This noncanonical “monophosphorylation” is thought to promote aberrant inflammatory responses, contributing to pathogenesis (186). For example, S727-monophosphorylated STAT3 induced by KSHV kaposin B protein drives the expression of a specific subset of STAT3 target genes, including those encoding the inflammatory mediators IL-6 and CCL5 (75).
This diversity of STAT functions might underpin the array of strategies by which different viruses, or different proteins of the same virus, antagonize STATs. Many paramyxoviruses use V protein to target STAT1, -2, and/or -3 for degradation (56, 57), which would be expected to impact canonical and noncanonical functions with broad effects on cell biology. While many antagonists bind and inhibit STATs irrespective of cytokine stimulation, RABV and adenovirus effect mislocalization of STATs only in cytokine-stimulated cells (187–190). Notably, RABV P protein efficiently binds STAT1, -2, and -3 only following cytokine activation (187–189), presumably ensuring that P protein (which has key functions in replication) is diverted to STAT antagonism only as required. This might also permit important cellular functions of U-STATs, but not those of antiviral cytokines and pY-STATs. This diversity likely evolved due to distinct properties of viruses, including cell tropism and infectious-cycle duration. Clearly, the effects of viruses on broader noncanonical STAT functions is likely an important, but largely overlooked, aspect of infection.
THERAPEUTIC POTENTIAL OF VIRUS-STAT INTERFACES
A detailed understanding of the virus-STAT interface should provide opportunities for therapeutic or prophylactic approaches. Where STATs are proviral, pharmacological inhibition is a promising avenue, particularly given extensive research on inhibitors of STATs and STAT pathways (primarily STAT3 and STAT5) for diverse malignancies (191) that could be repurposed. Importantly, in vitro, ex vivo, and animal studies have indicated the potential of such compounds: inhibition of specific STATs or upstream components (e.g., JAKs) can inhibit replication (e.g., B19V [122, 123], HCV [67, 68], HPV [103], HCMV [71], and varicella-zoster virus [192]) and virus-induced cell proliferation and transformation (e.g., HCMV [193], HBV [51], and HTLV-1 [194]). The FDA-approved JAK inhibitors tofacitinib and ruxolitinib block HIV-1 replication and reactivation by preventing STAT5 phosphorylation and LTR binding (113, 195). While this disrupts virion production and dissemination, it cannot eradicate latent reservoirs. However, benzotriazoles were identified as promising inducers of reactivation, which could potentially be used in a “shock and kill” strategy (117). Notably, benzotriazoles block STAT5 sumoylation, a PTM that negatively regulates STAT5 activity, highlighting PTMs other than tyrosine phosphorylation as potential targets. Among these, serine phosphorylation of STATs may be significant based on reports of viral induction (73, 75, 76, 184, 185) and potential roles in inflammation and pathogenesis (186).
Inhibition of STAT3 induces reactivation of latent herpesviruses (42–45), an important finding given the challenges latent reservoirs present for therapy. The utility of this approach was recently indicated by the finding that the combination of the STAT3 inhibitor Stattic with an antiviral that blocks KSHV replication potently inhibited virion production (106). Since the latent and lytic switches of EBV and HSV-1 can also be induced by STAT3 inhibitors (42, 44), this represents an attractive potential treatment strategy for herpesviruses more generally (106). Modulating STAT function is thus a promising avenue for management of diverse viruses, although its effectiveness in the clinic remains to be determined.
Given the critical roles of IFN antagonists in shutting down antiviral responses and the consequent significance to pathogenesis, they present attractive targets for vaccine and antiviral drug development. Screening approaches have identified IFN antagonist inhibitors (196–199), and the development of viral reverse genetics systems enables site-directed mutagenesis for the design of potential “live” attenuated vaccines (3, 6). A number of important viruses are attenuated by deletion of STAT1/2 antagonists (e.g., respiratory syncytial virus NS2 [200] and MEV V and C [201] proteins). Influenza A and B viruses are attenuated by deletion or truncation of the gene encoding NS1, which blocks STAT1 to -3 signaling (154), and potential application for vaccines has been explored in animal models and clinical trials (202–206). Notably, these antagonists are multifunctional, so STAT signaling would not be the only process impacted by deletion.
Many IFN antagonists, however, have additional critical functions in replication, so deletion or truncation can render a virus nonviable. Attenuation thus requires mutations that can selectively disrupt antagonistic function. “Forward genetic” approaches comparing virus strains and species of differing virulences have enabled the identification of mutations and substitutions impacting STAT1/2 antagonism that correlate with pathogenesis (e.g., Sindbis virus [207], WNV [208], RABV [209], and Ebola virus [210, 211]). Reverse genetics analysis has directly supported STAT antagonism as a specific pathogenicity factor. Mutations in the V or P gene of MEV or MUV that inhibit STAT1 or STAT3 antagonism attenuated virus in rhesus monkeys (212) and reduced neurovirulence in rats (59), respectively. Different mutations impacting STAT1 binding by the RABV P protein rendered a fixed pathogenic strain nonlethal and significantly attenuated a pathogenic street strain, although the latter remained lethal, consistent with important roles of other immune evasion mechanisms (213, 214). Similarly, a single mutation in the NIV V or P gene that disrupted STAT1 binding did not significantly affect lethality in ferrets, although it altered the disease course (215). Thus, targeting of virus-STAT interactions has potential to provide methods for attenuation, but generation of live vaccines clearly requires a combination of several attenuating mutations, potentially including mutations impacting other immune evasion mechanisms, to achieve a sufficient attenuation and safety profile.
CONCLUDING REMARKS
The interface of viruses with STATs is clearly highly diverse and dynamic, reflecting the diversity and evolutionary complexity of viruses and the array of functions associated with STATs. Furthermore, it appears that idiosyncrasies of the virus and host cell, including the duration of the infectious cycle, whether the virus can become latent, cellular expression of STAT family members, and exposure to specific cytokines, are critical determinants of the significance of particular STATs, including their propensities for anti- or proviral roles. Importantly, for many viruses, these requirements appear to change throughout infection, necessitating the evolution of strategies to both induce and suppress the functions of a specific STAT.
As our understanding of the scope of STAT functions expands, so do the implications for the viral interface. In particular, increasing evidence of targeting of STATs other than STAT1/2 suggests that much of the virus-STAT interface has been overlooked in terms of targeting of both individual STATs and specific complexes, which might enable exquisite regulation of the transcriptome. Viral modulation of noncanonical STAT functions also remains poorly defined. Delineation of these elements is significant for the potential development of new antiviral and vaccine approaches.
Finally, as in many areas of virological research, the virus-STAT interface has implications beyond infection. Through the evolution of specific mechanisms to modulate cell biology, viruses have provided invaluable tools to dissect many complex cellular processes. Elucidation of the molecular mechanisms underlying viral modulation of specific STAT pathways might have far-reaching implications for our understanding of cellular signaling, transcriptional regulation, and immunity, with significance for other pathologies, including cancer.
ACKNOWLEDGMENTS
This work was supported by National Health and Medical Research Council Australia project grant 1125704 (G.W.M.) and an Australian Government Research Training Program Scholarship (A.R.H.).
We thank Patrick Lane (ScEYEnce Studios) for graphical enhancement of the figures.
Biographies
Angela R. Harrison obtained her Bachelor of Science degree from Monash University (Australia) as a part of the competitive Science Scholar Program, focusing on Microbiology and Biochemistry. She then obtained First Class Honors from the University of Melbourne (Australia) in the laboratory of Dr. Gregory Moseley, where she was awarded the AB-SCIEX Honours Prize, before moving with Dr. Moseley to Monash University to undertake her Ph.D. Ms. Harrison’s research focuses on the immune evasion strategies of highly pathogenic lyssaviruses and Ebola virus, including targeting of STAT3 complexes to suppress responses to interleukin-6 family cytokines.
Gregory W. Moseley did his undergraduate studies in Biochemistry at the University of York (United Kingdom) before undertaking research toward a Ph.D. at The University of Sheffield (United Kingdom) and Walter and Eliza Hall Institute (Australia) on the roles of tetraspanin proteins in immunity. Under the auspices of a Royal Society postdoctoral fellowship, he pursued research in immunology at the Austin Research Institute (Australia). He subsequently moved to Monash University (Australia) to pursue research on protein trafficking, and established a laboratory investigating the molecular mechanisms of viral immune evasion and pathogenesis. He also undertook visiting positions in Gifu University (Japan) and CNRS (France). In 2013, he was awarded the Grimwade Fellowship at the University of Melbourne and in 2017 took a tenured position in the Department of Microbiology, Monash University, where his laboratory continues to probe the mechanisms underlying viral immune evasion, including targeting of STAT proteins, the topic of this review.
REFERENCES
- 1.Morris R, Kershaw NJ, Babon JJ. 2018. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci 27:1984–2009. doi: 10.1002/pro.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark GR, Darnell JE. 2012. The JAK-STAT pathway at twenty. Immunity 36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nan Y, Wu C, Zhang YJ. 2017. Interplay between Janus kinase/signal transducer and activator of transcription signaling activated by type i interferons and viral antagonism. Front Immunol 8:1758. doi: 10.3389/fimmu.2017.01758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delgoffe GM, Vignali DA. 2013. STAT heterodimers in immunity: a mixed message or a unique signal? JAKSTAT 2:e23060. doi: 10.4161/jkst.23060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 6.Fleming SB. 2016. Viral inhibition of the IFN-induced JAK/STAT signalling pathway: development of live attenuated vaccines by mutation of viral-encoded IFN-antagonists. Vaccines 4:23. doi: 10.3390/vaccines4030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didcock L, Young DF, Goodbourn S, Randall RE. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J Virol 73:9928–9933. doi: 10.1128/JVI.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parisien JP, Lau JF, Rodriguez JJ, Sullivan BM, Moscona A, Parks GD, Lamb RA, Horvath CM. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230–239. doi: 10.1006/viro.2001.0856. [DOI] [PubMed] [Google Scholar]
- 9.Young DF, Didcock L, Goodbourn S, Randall RE. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383–390. doi: 10.1006/viro.2000.0240. [DOI] [PubMed] [Google Scholar]
- 10.Precious B, Young DF, Andrejeva L, Goodbourn S, Randall RE. 2005. In vitro and in vivo specificity of ubiquitination and degradation of STAT1 and STAT2 by the V proteins of the paramyxoviruses simian virus 5 and human parainfluenza virus type 2. J Gen Virol 86:151–158. doi: 10.1099/vir.0.80263-0. [DOI] [PubMed] [Google Scholar]
- 11.Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez-Seco MP, Evans MJ, Best SM, Garcia-Sastre A. 2016. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar A, Hou S, Airo AM, Limonta D, Mancinelli V, Branton W, Power C, Hobman TC. 2016. Zika virus inhibits type-I interferon production and downstream signaling. EMBO Rep 17:1766–1775. doi: 10.15252/embr.201642627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhary V, Yuen KS, Chan JFW, Chan CP, Wang PH, Cai JP, Zhang S, Liang M, Kok KH, Chan CP, Yuen KY, Jin DY. 2017. Selective activation of type II interferon signaling by Zika virus NS5 protein. J Virol 91:e00163-17. doi: 10.1128/JVI.00163-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. 1997. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A 94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy DE, Lee CK. 2002. What does Stat3 do? J Clin Invest 109:1143–1148. doi: 10.1172/JCI0215650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson DE, O'Keefe RA, Grandis JR. 2018. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol 15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamero AM, Potla R, Wegrzyn J, Szelag M, Edling AE, Shimoda K, Link DC, Dulak J, Baker DP, Tanabe Y, Grayson JM, Larner AC. 2006. Activation of Tyk2 and Stat3 is required for the apoptotic actions of interferon-beta in primary pro-B cells. J Biol Chem 281:16238–16244. doi: 10.1074/jbc.M509516200. [DOI] [PubMed] [Google Scholar]
- 18.Abell K, Bilancio A, Clarkson RW, Tiffen PG, Altaparmakov AI, Burdon TG, Asano T, Vanhaesebroeck B, Watson CJ. 2005. Stat3-induced apoptosis requires a molecular switch in PI(3)K subunit composition. Nat Cell Biol 7:392–398. doi: 10.1038/ncb1242. [DOI] [PubMed] [Google Scholar]
- 19.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. 2003. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J 374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang CH, Murti A, Pfeffer LM. 1998. STAT3 complements defects in an interferon-resistant cell line: evidence for an essential role for STAT3 in interferon signaling and biological activities. Proc Natl Acad Sci U S A 95:5568–5572. doi: 10.1073/pnas.95.10.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahony R, Gargan S, Roberts KL, Bourke N, Keating SE, Bowie AG, O'Farrelly C, Stevenson NJ. 2017. A novel anti-viral role for STAT3 in IFN-alpha signalling responses. Cell Mol Life Sci 74:1755–1764. doi: 10.1007/s00018-016-2435-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeffer SR, Fan M, Du Z, Yang CH, Pfeffer LM. 2017. Unphosphorylated STAT3 regulates the antiproliferative, antiviral, and gene-inducing actions of type I interferons. Biochem Biophys Res Commun 490:739–745. doi: 10.1016/j.bbrc.2017.06.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsia HC, Stopford CM, Zhang Z, Damania B, Baldwin AS. 2017. Signal transducer and activator of transcription 3 (Stat3) regulates host defense and protects mice against herpes simplex virus-1 (HSV-1) infection. J Leukoc Biol 101:1053–1064. doi: 10.1189/jlb.4A1016-199RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu H, Shang X, Terada N, Liu C. 2004. STAT3 induces anti-hepatitis C viral activity in liver cells. Biochem Biophys Res Commun 324:518–528. doi: 10.1016/j.bbrc.2004.09.081. [DOI] [PubMed] [Google Scholar]
- 25.Ho HH, Ivashkiv LB. 2006. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J Biol Chem 281:14111–14118. doi: 10.1074/jbc.M511797200. [DOI] [PubMed] [Google Scholar]
- 26.Wang WB, Levy DE, Lee CK. 2011. STAT3 negatively regulates type I IFN-mediated antiviral response. J Immunol 187:2578–2585. doi: 10.4049/jimmunol.1004128. [DOI] [PubMed] [Google Scholar]
- 27.Lu L, Zhu F, Zhang M, Li Y, Drennan AC, Kimpara S, Rumball I, Selzer C, Cameron H, Kellicut A, Kelm A, Wang F, Waldmann TA, Rui L. 2018. Gene regulation and suppression of type I interferon signaling by STAT3 in diffuse large B cell lymphoma. Proc Natl Acad Sci U S A 115:E498–E505. doi: 10.1073/pnas.1715118115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai MH, Lee CK. 2018. STAT3 cooperates with phospholipid scramblase 2 to suppress type i interferon response. Front Immunol 9:1886. doi: 10.3389/fimmu.2018.01886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchins AP, Diez D, Miranda-Saavedra D. 2013. Genomic and computational approaches to dissect the mechanisms of STAT3's universal and cell type-specific functions. JAKSTAT 2:e25097. doi: 10.4161/jkst.25097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutchins AP, Diez D, Takahashi Y, Ahmad S, Jauch R, Tremblay ML, Miranda-Saavedra D. 2013. Distinct transcriptional regulatory modules underlie STAT3's cell type-independent and cell type-specific functions. Nucleic Acids Res 41:2155–2170. doi: 10.1093/nar/gks1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun DA, Fribourg M, Sealfon SC. 2013. Cytokine response is determined by duration of receptor and signal transducers and activators of transcription 3 (STAT3) activation. J Biol Chem 288:2986–2993. doi: 10.1074/jbc.M112.386573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan CK, Oh J, Li P, West EE, Wong EA, Andraski AB, Spolski R, Yu ZX, He J, Kelsall BL, Leonard WJ. 2013. The cytokines IL-21 and GM-CSF have opposing regulatory roles in the apoptosis of conventional dendritic cells. Immunity 38:514–527. doi: 10.1016/j.immuni.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuchipudi SV. 2015. The complex role of STAT3 in viral infections. J Immunol Res 2015:272359. doi: 10.1155/2015/272359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roca Suarez AA, Van Renne N, Baumert TF, Lupberger J. 2018. Viral manipulation of STAT3: evade, exploit, and injure. PLoS Pathog 14:e1006839. doi: 10.1371/journal.ppat.1006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang Z, Wang Y, Zhou X, Long JE. 2018. STAT3 roles in viral infection: antiviral or proviral? Future Virol 13:557–574. doi: 10.2217/fvl-2018-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geiger TR, Martin JM. 2006. The Epstein-Barr virus-encoded LMP-1 oncoprotein negatively affects Tyk2 phosphorylation and interferon signaling in human B cells. J Virol 80:11638–11650. doi: 10.1128/JVI.01570-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu YP, Tan YN, Wang ZL, Zeng L, Lu ZX, Li LL, Luo W, Tang M, Cao Y. 2008. Phosphorylation and nuclear translocation of STAT3 regulated by the Epstein-Barr virus latent membrane protein 1 in nasopharyngeal carcinoma. Int J Mol Med 21:153–162. [PubMed] [Google Scholar]
- 38.Chen H, Hutt-Fletcher L, Cao L, Hayward SD. 2003. A positive autoregulatory loop of LMP1 expression and STAT activation in epithelial cells latently infected with Epstein-Barr virus. J Virol 77:4139–4148. doi: 10.1128/jvi.77.7.4139-4148.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H, Lee JM, Zong Y, Borowitz M, Ng MH, Ambinder RF, Hayward SD. 2001. Linkage between STAT regulation and Epstein-Barr virus gene expression in tumors. J Virol 75:2929–2937. doi: 10.1128/JVI.75.6.2929-2937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konforte D, Simard N, Paige CJ. 2008. Interleukin-21 regulates expression of key Epstein-Barr virus oncoproteins, EBNA2 and LMP1, in infected human B cells. Virology 374:100–113. doi: 10.1016/j.virol.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 41.Kis L, Takahara M, Nagy N, Klein G, Klein E. 2006. IL-10 can induce the expression of EBV-encoded latent membrane protein-1 (LMP-1) in the absence of EBNA-2 in B lymphocytes and in Burkitt lymphoma- and NK lymphoma-derived cell lines. Blood 107:2928–2935. doi: 10.1182/blood-2005-06-2569. [DOI] [PubMed] [Google Scholar]
- 42.Hill ER, Koganti S, Zhi J, Megyola C, Freeman AF, Palendira U, Tangye SG, Farrell PJ, Bhaduri-McIntosh S. 2013. Signal transducer and activator of transcription 3 limits Epstein-Barr virus lytic activation in B lymphocytes. J Virol 87:11438–11446. doi: 10.1128/JVI.01762-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.King CA, Li X, Barbachano-Guerrero A, Bhaduri-McIntosh S. 2015. STAT3 regulates lytic activation of Kaposi's sarcoma-associated herpesvirus. J Virol 89:11347–11355. doi: 10.1128/JVI.02008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du T, Zhou G, Roizman B. 2013. Modulation of reactivation of latent herpes simplex virus 1 in ganglionic organ cultures by p300/CBP and STAT3. Proc Natl Acad Sci U S A 110:E2621–E2628. doi: 10.1073/pnas.1309906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koganti S, Clark C, Zhi J, Li X, Chen EI, Chakrabortty S, Hill ER, Bhaduri-McIntosh S, Longnecker RM. 2015. Cellular STAT3 functions via PCBP2 to restrain Epstein-Barr virus lytic activation in B lymphocytes. J Virol 89:5002–5011. doi: 10.1128/JVI.00121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Fan P, Zhao Y, Zhang S, Lu J, Xie W, Jiang Y, Lei F, Xu N, Zhang Y. 2017. NEAT1 modulates herpes simplex virus-1 replication by regulating viral gene transcription. Cell Mol Life Sci 74:1117–1131. doi: 10.1007/s00018-016-2398-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waris G, Siddiqui A. 2002. Interaction between STAT-3 and HNF-3 leads to the activation of liver-specific hepatitis B virus enhancer 1 function. J Virol 76:2721–2729. doi: 10.1128/jvi.76.6.2721-2729.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee YH, Yun Y. 1998. HBx protein of hepatitis B virus activates Jak1-STAT signaling. J Biol Chem 273:25510–25515. doi: 10.1074/jbc.273.39.25510. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Lu Y, Toh S, Sung WK, Tan P, Chow P, Chung A, Jooi L, Lee C. 2010. Lethal-7 is down-regulated by the hepatitis B virus X protein and targets signal transducer and activator of transcription 3. J Hepatol 53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 50.Waris G, Huh KW, Siddiqui A. 2001. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-kappa B via oxidative stress. Mol Cell Biol 21:7721–7730. doi: 10.1128/MCB.21.22.7721-7730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hosel M, Quasdorff M, Ringelhan M, Kashkar H, Debey-Pascher S, Sprinzl MF, Bockmann JH, Arzberger S, Webb D, von Olshausen G, Weber A, Schultze JL, Buning H, Heikenwalder M, Protzer U. 2017. Hepatitis B virus activates signal transducer and activator of transcription 3 supporting hepatocyte survival and virus replication. Cell Mol Gastroenterol Hepatol 4:339–363. doi: 10.1016/j.jcmgh.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He P, Zhang D, Li H, Yang X, Li D, Zhai Y, Ma L, Feng G. 2013. Hepatitis B virus X protein modulates apoptosis in human renal proximal tubular epithelial cells by activating the JAK2/STAT3 signaling pathway. Int J Mol Med 31:1017–1029. doi: 10.3892/ijmm.2013.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lei XY, Chen XX, Sun YH, Gao MD, Hu XX, Suo YH. 2019. Hepatitis B virus X protein decreases nephrin expression and induces podocyte apoptosis via activating STAT3. Exp Ther Med doi: 10.3892/etm.2019.7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H, Yuan M, Wang S, Zhang L, Zhang R, Zou X, Wang X, Chen D, Wu Z. 2019. STAT3 regulates the type I IFN-mediated antiviral response by interfering with the nuclear entry of STAT1. Int J Mol Sci 20:4870. doi: 10.3390/ijms20194870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang L, Xu J, Guo L, Guo T, Zhang L, Feng L, Chen H, Wang Y. 2018. Porcine epidemic diarrhea virus-induced epidermal growth factor receptor activation impairs the antiviral activity of type I interferon. J Virol 92:e02095-17. doi: 10.1128/JVI.02095-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kubota T, Yokosawa N, Yokota S, Fujii N. 2001. C terminal CYS-RICH region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem Biophys Res Commun 283:255–259. doi: 10.1006/bbrc.2001.4764. [DOI] [PubMed] [Google Scholar]
- 57.Ulane CM, Rodriguez JJ, Parisien JP, Horvath CM. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J Virol 77:6385–6393. doi: 10.1128/jvi.77.11.6385-6393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palosaari H, Parisien JP, Rodriguez JJ, Ulane CM, Horvath CM. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J Virol 77:7635–7644. doi: 10.1128/jvi.77.13.7635-7644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malik T, Ngo L, Bosma T, Rubin S. 2019. A single point mutation in the mumps V protein alters targeting of the cellular STAT pathways resulting in virus attenuation. Viruses 11:1016. doi: 10.3390/v11111016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Puri M, Lemon K, Duprex WP, Rima BK, Horvath CM. 2009. A point mutation, E95D, in the mumps virus V protein disengages STAT3 targeting from STAT1 targeting. J Virol 83:6347–6356. doi: 10.1128/JVI.00596-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez JJ, Wang LF, Horvath CM. 2003. Hendra virus V protein inhibits interferon signaling by preventing STAT1 and STAT2 nuclear accumulation. J Virol 77:11842–11845. doi: 10.1128/jvi.77.21.11842-11845.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caignard G, Lucas-Hourani M, Dhondt KP, Labernardiere JL, Petit T, Jacob Y, Horvat B, Tangy F, Vidalain PO. 2013. The V protein of Tioman virus is incapable of blocking type I interferon signaling in human cells. PLoS One 8:e53881. doi: 10.1371/journal.pone.0053881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida T, Hanada T, Tokuhisa T, Kosai K, Sata M, Kohara M, Yoshimura A. 2002. Activation of STAT3 by the hepatitis C virus core protein leads to cellular transformation. J Exp Med 196:641–653. doi: 10.1084/jem.20012127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou JJ, Chen RF, Deng XG, Zhou Y, Ye X, Yu M, Tang J, He XY, Cheng D, Zeng B, Zhou QB, Li ZH. 2014. Hepatitis C virus core protein regulates NANOG expression via the Stat3 pathway. FEBS Lett 588:566–573. doi: 10.1016/j.febslet.2013.11.041. [DOI] [PubMed] [Google Scholar]
- 65.Tacke RS, Tosello-Trampont A, Nguyen V, Mullins DW, Hahn YS. 2011. Extracellular hepatitis C virus core protein activates STAT3 in human monocytes/macrophages/dendritic cells via an IL-6 autocrine pathway. J Biol Chem 286:10847–10855. doi: 10.1074/jbc.M110.217653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gong G, Waris G, Tanveer R, Siddiqui A. 2001. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-κB. Proc Natl Acad Sci U S A 98:9599–9604. doi: 10.1073/pnas.171311298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waris G, Turkson J, Hassanein T, Siddiqui A. 2005. Hepatitis C virus (HCV) constitutively activates STAT-3 via oxidative stress: role of STAT-3 in HCV replication. J Virol 79:1569–1580. doi: 10.1128/JVI.79.3.1569-1580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.McCartney EM, Helbig KJ, Narayana SK, Eyre NS, Aloia AL, Beard MR. 2013. Signal transducer and activator of transcription 3 is a proviral host factor for hepatitis C virus. Hepatology 58:1558–1568. doi: 10.1002/hep.26496. [DOI] [PubMed] [Google Scholar]
- 69.Stevenson NJ, Bourke NM, Ryan EJ, Binder M, Fanning L, Johnston JA, Hegarty JE, Long A, O'Farrelly C. 2013. Hepatitis C virus targets the interferon-alpha JAK/STAT pathway by promoting proteasomal degradation in immune cells and hepatocytes. FEBS Lett 587:1571–1578. doi: 10.1016/j.febslet.2013.03.041. [DOI] [PubMed] [Google Scholar]
- 70.Heim MH, Moradpour D, Blum HE. 1999. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol 73:8469–8475. doi: 10.1128/JVI.73.10.8469-8475.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reitsma JM, Sato H, Nevels M, Terhune SS, Paulus C. 2013. Human cytomegalovirus IE1 protein disrupts interleukin-6 signaling by sequestering STAT3 in the nucleus. J Virol 87:10763–10776. doi: 10.1128/JVI.01197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harwardt T, Lukas S, Zenger M, Reitberger T, Danzer D, Ubner T, Munday DC, Nevels M, Paulus C. 2016. Human cytomegalovirus immediate-early 1 protein rewires upstream STAT3 to downstream STAT1 signaling switching an IL6-type to an IFNgamma-like response. PLoS Pathog 12:e1005748. doi: 10.1371/journal.ppat.1005748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kung CP, Raab-Traub N. 2008. Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor through effects on Bcl-3 and STAT3. J Virol 82:5486–5493. doi: 10.1128/JVI.00125-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilardini Montani MS, Santarelli R, Granato M, Gonnella R, Torrisi MR, Faggioni A, Cirone M. 2019. EBV reduces autophagy, intracellular ROS and mitochondria to impair monocyte survival and differentiation. Autophagy 15:652–667. doi: 10.1080/15548627.2018.1536530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.King CA. 2013. Kaposi's sarcoma-associated herpesvirus kaposin B induces unique monophosphorylation of STAT3 at serine 727 and MK2-mediated inactivation of the STAT3 transcriptional repressor TRIM28. J Virol 87:8779–8791. doi: 10.1128/JVI.02976-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chaudhuri A, Yang B, Gendelman HE, Persidsky Y, Kanmogne GD. 2008. STAT1 signaling modulates HIV-1-induced inflammatory responses and leukocyte transmigration across the blood-brain barrier. Blood 111:2062–2072. doi: 10.1182/blood-2007-05-091207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zundler S, Neurath MF. 2015. Interleukin-12: functional activities and implications for disease. Cytokine Growth Factor Rev 26:559–568. doi: 10.1016/j.cytogfr.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 78.Kaplan M, Sun YL, Hoey T, Grusby M. 1996. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature 382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 79.Thierfelder W, van Deursen J, Yamamoto K, Tripp R, Sarawar S, Carson R, Sangster M, Vignali DA, Doherty P, Grosveld G, Ihle JN. 1996. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature 382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 80.Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O'Shea JJ, Biron CA. 2002. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science 297:2063–2066. doi: 10.1126/science.1074900. [DOI] [PubMed] [Google Scholar]
- 81.Berenson LS, Gavrieli M, Farrar JD, Murphy TL, Murphy KM. 2006. Distinct characteristics of murine STAT4 activation in response to IL-12 and IFN-alpha. J Immunol 177:5195–5203. doi: 10.4049/jimmunol.177.8.5195. [DOI] [PubMed] [Google Scholar]
- 82.Cho S, Bacon C, Sudarshan C, Rees R, Finbloom D, Pine R, O’Shea J. 1996. Activation of STAT4 by IL-12 and IFN-α. J Immunol 157:4781–4789. [PubMed] [Google Scholar]
- 83.Iida K, Suzuki K, Yokota M, Nakagomi D, Wakashin H, Iwata A, Kawashima H, Takatori H, Nakajima H. 2011. STAT4 is required for IFN-beta-induced MCP-1 mRNA expression in murine mast cells. Int Arch Allergy Immunol 155(Suppl 1):71–76. doi: 10.1159/000327300. [DOI] [PubMed] [Google Scholar]
- 84.Gil MP, Ploquin MJY, Watford WT, Lee S-H, Kim K, Wang X, Kanno Y, O'Shea JJ, Biron CA. 2012. Regulating type 1 IFN effects in CD8 T cells during viral infections—changing STAT4 and STAT1 expression for function. Blood 120:3718–3728. doi: 10.1182/blood-2012-05-428672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suarez-Ramirez JE, Tarrio ML, Kim K, Demers DA, Biron CA. 2014. CD8 T cells in innate immune responses: using STAT4-dependent but antigen-independent pathways to gamma interferon during viral infection. mBio 5:e01978-14. doi: 10.1128/mBio.01978-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weinstein JS, Laidlaw BJ, Lu Y, Wang JK, Schulz VP, Li N, Herman EI, Kaech SM, Gallagher PG, Craft J. 2018. STAT4 and T-bet control follicular helper T cell development in viral infections. J Exp Med 215:337–355. doi: 10.1084/jem.20170457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Adams NM, Lau CM, Fan X, Rapp M, Geary CD, Weizman OE, Diaz-Salazar C, Sun JC. 2018. Transcription factor IRF8 orchestrates the adaptive natural killer cell response. Immunity 48:1172–1182. doi: 10.1016/j.immuni.2018.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shi H, He H, Ojha SC, Sun C, Fu J, Yan M, Deng C, Sheng Y. 2019. Association of STAT3 and STAT4 polymorphisms with susceptibility to chronic hepatitis B virus infection and risk of hepatocellular carcinoma: a meta-analysis. Biosci Rep 39:BSR20190783. doi: 10.1042/BSR20190783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Svensson A, Tunback P, Nordstrom I, Shestakov A, Padyukov L, Eriksson K. 2012. STAT4 regulates antiviral gamma interferon responses and recurrent disease during herpes simplex virus 2 infection. J Virol 86:9409–9415. doi: 10.1128/JVI.00947-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Banerjee K, Biswas PS, Rouse BT. 2007. Role of Stat4-mediated signal transduction events in the generation of aggressor CD4+ T cells in herpetic stromal keratitis pathogenesis. J Interferon Cytokine Res 27:65–75. doi: 10.1089/jir.2007.0077. [DOI] [PubMed] [Google Scholar]
- 91.Allen S, Mott K, Ghiasi H. 2010. Involvement of STAT4 in IgG subtype switching and ocular HSV-1 replication in mice. Mol Vis 16:98–104. [PMC free article] [PubMed] [Google Scholar]
- 92.Bot A, Rodrigo E, Wolfe T, Bot S, Von Herrath MG. 2003. Infection-triggered regulatory mechanisms override the role of STAT 4 in control of the immune response to influenza virus antigens. J Virol 77:5794–5800. doi: 10.1128/jvi.77.10.5794-5800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao K, Zhang Q, Li X, Zhao D, Liu Y, Shen Q, Yang M, Wang C, Li N, Cao X. 2016. Cytoplasmic STAT4 promotes antiviral type I IFN production by blocking CHIP-mediated degradation of RIG-I. J Immunol 196:1209–1217. doi: 10.4049/jimmunol.1501224. [DOI] [PubMed] [Google Scholar]
- 94.Martinez-Gil L, Vera-Velasco NM, Mingarro I. 2017. Exploring the human-Nipah virus protein-protein interactome. J Virol 91:e01461-17. doi: 10.1128/JVI.01461-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rodriguez JJ, Parisien JP, Horvath CM. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J Virol 76:11476–11483. doi: 10.1128/jvi.76.22.11476-11483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shaw ML, Garcia-Sastre A, Palese P, Basler CF. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J Virol 78:5633–5641. doi: 10.1128/JVI.78.11.5633-5641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rani A, Murphy JJ. 2016. STAT5 in cancer and immunity. J Interferon Cytokine Res 36:226–237. doi: 10.1089/jir.2015.0054. [DOI] [PubMed] [Google Scholar]
- 98.Rochman Y, Spolski R, Leonard WJ. 2009. New insights into the regulation of T cells by γc family cytokines. Nat Rev Immunol 9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Uddin S, Lekmine F, Sassano A, Rui H, Fish EN, Platanias LC. 2003. Role of Stat5 in type I interferon-signaling and transcriptional regulation. Biochem Biophys Res Commun 308:325–330. doi: 10.1016/s0006-291x(03)01382-2. [DOI] [PubMed] [Google Scholar]
- 100.Tanabe Y, Nishibori T, Su L, Arduini RM, Baker DP, David M. 2005. Cutting edge: role of STAT1, STAT3, and STAT5 in IFN-alpha beta responses in T lymphocytes. J Immunol 174:609–613. doi: 10.4049/jimmunol.174.2.609. [DOI] [PubMed] [Google Scholar]
- 101.Fish EN, Uddin S, Korkmaz M, Majchrzak B, Druker B, Platanias LC. 1999. Activation of a CrkL-Stat5 signaling complex by type I interferons. J Biol Chem 274:571–573. doi: 10.1074/jbc.274.2.571. [DOI] [PubMed] [Google Scholar]
- 102.Meinke A, Barahmand-Pour F, Wohrl S, Stoiber D, Decker T. 1996. Activation of different Stat5 isoforms contributes to cell-type restricted signaling in response to interferons. Mol Cell Biol 16:6937–6944. doi: 10.1128/mcb.16.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hong S, Laimins LA. 2013. The JAK-STAT transcriptional regulator, STAT-5, activates the ATM DNA damage pathway to induce HPV 31 genome amplification upon epithelial differentiation. PLoS Pathog 9:e1003295. doi: 10.1371/journal.ppat.1003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hong S, Cheng S, Iovane A, Laimins LA, Goff SP. 2015. STAT-5 regulates transcription of the topoisomerase IIβ-binding protein 1 (TopBP1) gene to activate the ATR pathway and promote human papillomavirus replication. mBio 6:e02006-15. doi: 10.1128/mBio.02006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Damania B, Yoo J, Lee HN, Choi I, Choi D, Chung HK, Kim KE, Lee S, Aguilar B, Kang J, Park E, Lee YS, Maeng Y-S, Kim NY, Koh CJ, Hong Y-K. 2012. Opposing regulation of PROX1 by interleukin-3 receptor and NOTCH directs differential host cell fate reprogramming by Kaposi sarcoma herpes virus. PLoS Pathog 8:e1002770. doi: 10.1371/journal.ppat.1002770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ramalingam D, Ziegelbauer JM. 2017. Viral microRNAs target a gene network, inhibit STAT activation, and suppress interferon responses. Sci Rep 7:40813. doi: 10.1038/srep40813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pericle F, Pinto L, Kirken R, Sconocchia G, Rusnak J, Dolan M, Shearer G, Segal DM. 1998. Cutting edge- HIV-1 infection induces a selective reduction in STAT5 protein expression. J Immunol 160:28–31. [PubMed] [Google Scholar]
- 108.Zheng CF, Jones GJ, Shi M, Wiseman JCD, Marr KJ, Berenger BM, Huston SM, Gill MJ, Krensky AM, Kubes P, Mody CH. 2008. Late expression of granulysin by microbicidal CD4+ T cells requires PI3K- and STAT5-dependent expression of IL-2Rβ that is defective in HIV-infected patients. J Immunol 180:7221–7229. doi: 10.4049/jimmunol.180.11.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Selliah N, Finkel TH. 2001. HIV-1 NL4-3, but not IIIB, inhibits JAK3/STAT5 activation in CD4(+) T cells. Virology 286:412–421. doi: 10.1006/viro.2001.0994. [DOI] [PubMed] [Google Scholar]
- 110.Juffroy O, Bugault F, Lambotte O, Landires I, Viard JP, Niel L, Fontanet A, Delfraissy JF, Theze J, Chakrabarti LA. 2010. Dual mechanism of impairment of interleukin-7 (IL-7) responses in human immunodeficiency virus infection: decreased IL-7 binding and abnormal activation of the JAK/STAT5 pathway. J Virol 84:96–108. doi: 10.1128/JVI.01475-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Landires I, Bugault F, Lambotte O, de Truchis P, Slama L, Danckaert A, Delfraissy JF, Theze J, Chakrabarti LA. 2011. HIV infection perturbs interleukin-7 signaling at the step of STAT5 nuclear relocalization. AIDS 25:1843–1853. doi: 10.1097/QAD.0b013e32834a3678. [DOI] [PubMed] [Google Scholar]
- 112.Bovolenta C, Camorali L, Lorini A, Ghezzi S, Vicenzi E, Lazzarin A, Poli G. 1999. Constitutive activation of STATs upon in vivo human immunodeficiency virus infection. Blood 94:4202–4209. [PubMed] [Google Scholar]
- 113.Gavegnano C, Brehm JH, Dupuy FP, Talla A, Ribeiro SP, Kulpa DA, Cameron C, Santos S, Hurwitz SJ, Marconi VC, Routy JP, Sabbagh L, Schinazi RF, Sekaly RP. 2017. Novel mechanisms to inhibit HIV reservoir seeding using Jak inhibitors. PLoS Pathog 13:e1006740. doi: 10.1371/journal.ppat.1006740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Warby TJ, Crowe SM, Jaworowski A. 2003. Human immunodeficiency virus type 1 infection inhibits granulocyte-macrophage colony-stimulating factor-induced activation of STAT5A in human monocyte-derived macrophages. J Virol 77:12630–12638. doi: 10.1128/jvi.77.23.12630-12638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Crotti A, Chiara G, Ghezzi S, Lupo R, Jeeninga R, Liboi E, Lievens P, Vicenzi E, Bovolenta C, Berkhout B, Poli G. 2007. Heterogeneity of signal transducer and activator of transcription binding sites in the long-terminal repeats of distinct HIV-1 subtypes. Open Virol J 1:26–32. doi: 10.2174/1874357900701010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Selliah N, Zhang M, DeSimone D, Kim H, Brunner M, Ittenbach RF, Rui H, Cron RQ, Finkel TH. 2006. The gammac-cytokine regulated transcription factor, STAT5, increases HIV-1 production in primary CD4 T cells. Virology 344:283–291. doi: 10.1016/j.virol.2005.09.063. [DOI] [PubMed] [Google Scholar]
- 117.Bosque A, Nilson KA, Macedo AB, Spivak AM, Archin NM, Van Wagoner RM, Martins LJ, Novis CL, Szaniawski MA, Ireland CM, Margolis DM, Price DH, Planelles V. 2017. Benzotriazoles reactivate latent HIV-1 through inactivation of STAT5 SUMOylation. Cell Rep 18:1324–1334. doi: 10.1016/j.celrep.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Scripture-Adams DD, Brooks DG, Korin YD, Zack JA. 2002. Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype. J Virol 76:13077–13082. doi: 10.1128/jvi.76.24.13077-13082.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jones RB, Mueller S, O'Connor R, Rimpel K, Sloan DD, Karel D, Wong HC, Jeng EK, Thomas AS, Whitney JB, Lim SY, Kovacs C, Benko E, Karandish S, Huang SH, Buzon MJ, Lichterfeld M, Irrinki A, Murry JP, Tsai A, Yu H, Geleziunas R, Trocha A, Ostrowski MA, Irvine DJ, Walker BD. 2016. A subset of latency-reversing agents expose HIV-infected resting CD4+ T-cells to recognition by cytotoxic T-lymphocytes. PLoS Pathog 12:e1005545. doi: 10.1371/journal.ppat.1005545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Crotti A, Lusic M, Lupo R, Lievens PM, Liboi E, Della Chiara G, Tinelli M, Lazzarin A, Patterson BK, Giacca M, Bovolenta C, Poli G. 2007. Naturally occurring C-terminally truncated STAT5 is a negative regulator of HIV-1 expression. Blood 109:5380–5389. doi: 10.1182/blood-2006-08-042556. [DOI] [PubMed] [Google Scholar]
- 121.Prost S, Le Dantec M, Auge S, Le Grand R, Derdouch S, Auregan G, Deglon N, Relouzat F, Aubertin AM, Maillere B, Dusanter-Fourt I, Kirszenbaum M. 2008. Human and simian immunodeficiency viruses deregulate early hematopoiesis through a Nef/PPARgamma/STAT5 signaling pathway in macaques. J Clin Invest 118:1765–1775. doi: 10.1172/JCI33037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen AY, Kleiboeker S, Qiu J. 2011. Productive parvovirus B19 infection of primary human erythroid progenitor cells at hypoxia is regulated by STAT5A and MEK signaling but not HIFalpha. PLoS Pathog 7:e1002088. doi: 10.1371/journal.ppat.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ganaie SS, Zou W, Xu P, Deng X, Kleiboeker S, Qiu J. 2017. Phosphorylated STAT5 directly facilitates parvovirus B19 DNA replication in human erythroid progenitors through interaction with the MCM complex. PLoS Pathog 13:e1006370. doi: 10.1371/journal.ppat.1006370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zimmerman MG, Bowen JR, McDonald CE, Young E, Baric RS, Pulendran B, Suthar MS. 2019. STAT5: a target of antagonism by neurotropic flaviviruses. J Virol 93:e00665-19. doi: 10.1128/JVI.00665-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Goenka S, Kaplan MH. 2011. Transcriptional regulation by STAT6. Immunol Res 50:87–96. doi: 10.1007/s12026-011-8205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hebenstreit D, Wirnsberger G, Horejs-Hoeck J, Duschl A. 2006. Signaling mechanisms, interaction partners, and target genes of STAT6. Cytokine Growth Factor Rev 17:173–188. doi: 10.1016/j.cytogfr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 127.Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, Xia W, Gu J, Ishikawa H, Gutman D, Barber G, Qin Z, Jiang Z. 2011. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 128.Ghiasi H, Osorio Y, Nesburn AB, Wechsler SL. 2002. Enhanced clearance of herpes simplex virus type 1 and reduced herpetic eye disease in STAT6 knockout mice is associated with increased IL-2. Virology 302:286–293. doi: 10.1006/viro.2002.1611. [DOI] [PubMed] [Google Scholar]
- 129.Mahalingam S, Karupiah G, Takeda K, Akira S, Matthaei KI, Foster PS. 2001. Enhanced resistance in STAT6-deficient mice to infection with ectromelia virus. Proc Natl Acad Sci U S A 98:6812–6817. doi: 10.1073/pnas.111151098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Skinnider B, Elia A, Gascoyne R, Patterson B, Trumper L, Kapp U, Mak T. 2002. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 99:618–626. doi: 10.1182/blood.v99.2.618. [DOI] [PubMed] [Google Scholar]
- 131.Guiter C, Dusanter-Fourt I, Copie-Bergman C, Boulland ML, Le Gouvello S, Gaulard P, Leroy K, Castellano F. 2004. Constitutive STAT6 activation in primary mediastinal large B-cell lymphoma. Blood 104:543–549. doi: 10.1182/blood-2003-10-3545. [DOI] [PubMed] [Google Scholar]
- 132.Ni Z, Lou W, Lee S, Dhir R, DeMiguel F, Grandis J, Gao A. 2002. Selective activation of members of the signal transducers and activators of transcription family in prostate carcinoma. J Urol 167:1859–1862. [PubMed] [Google Scholar]
- 133.Tsai SC, Lin SJ, Chen PW, Luo WY, Yeh TH, Wang HW, Chen CJ, Tsai CH. 2009. EBV Zta protein induces the expression of interleukin-13, promoting the proliferation of EBV-infected B cells and lymphoblastoid cell lines. Blood 114:109–118. doi: 10.1182/blood-2008-12-193375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kis L, Gerasimcik N, Salamon D, Persson E, Nagy N, Klein G, Severinson E, Klein E. 2011. STAT6 signaling pathway activated by the cytokines IL-4 and IL-13 induces expression of the Epstein-Barr virus-encoded protein LMP-1 in absence of EBNA-2: implications for the type II EBV latent gene expression in Hodgkin lymphoma. Blood 117:165–174. doi: 10.1182/blood-2010-01-265272. [DOI] [PubMed] [Google Scholar]
- 135.Cai Q, Verma SC, Choi JY, Ma M, Robertson ES. 2010. Kaposi's sarcoma-associated herpesvirus inhibits interleukin-4-mediated STAT6 phosphorylation to regulate apoptosis and maintain latency. J Virol 84:11134–11144. doi: 10.1128/JVI.01293-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang C, Zhu C, Wei F, Zhang L, Mo X, Feng Y, Xu J, Yuan Z, Robertson E, Cai Q, Longnecker RM. 2015. Constitutive activation of interleukin-13/STAT6 contributes to Kaposi's sarcoma-associated herpesvirus-related primary effusion lymphoma cell proliferation and survival. J Virol 89:10416–10426. doi: 10.1128/JVI.01525-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang C, Zhu C, Wei F, Gao S, Zhang L, Li Y, Feng Y, Tong Y, Xu J, Wang B, Yuan Z, Robertson ES, Cai Q. 2017. Nuclear localization and cleavage of STAT6 is induced by Kaposi’s sarcoma-associated herpesvirus for viral latency. PLoS Pathog 13:e1006124. doi: 10.1371/journal.ppat.1006124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gu F, Wang C, Wei F, Wang Y, Zhu Q, Ding L, Xu W, Zhu C, Cai C, Qian Z, Yuan Z, Robertson E, Cai Q. 2018. STAT6 degradation and ubiquitylated TRIML2 are essential for activation of human oncogenic herpesvirus. PLoS Pathog 14:e1007416. doi: 10.1371/journal.ppat.1007416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Reese TA, Wakeman BS, Choi HS, Hufford MM, Huang SC, Zhang X, Buck MD, Jezewski A, Kambal A, Liu CY, Goel G, Murray PJ, Xavier RJ, Kaplan MH, Renne R, Speck SH, Artyomov MN, Pearce EJ, Virgin HW. 2014. Helminth infection reactivates latent gamma-herpesvirus via cytokine competition at a viral promoter. Science 345:573–577. doi: 10.1126/science.1254517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zeng Y, Zhang X, Huang Z, Cheng L, Yao S, Qin D, Chen X, Tang Q, Lv Z, Zhang L, Lu C. 2007. Intracellular Tat of human immunodeficiency virus type 1 activates lytic cycle replication of Kaposi's sarcoma-associated herpesvirus: role of JAK/STAT signaling. J Virol 81:2401–2417. doi: 10.1128/JVI.02024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Punjabi AS, Carroll PA, Chen L, Lagunoff M. 2007. Persistent activation of STAT3 by latent Kaposi's sarcoma-associated herpesvirus infection of endothelial cells. J Virol 81:2449–2458. doi: 10.1128/JVI.01769-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Burger M, Hartmann T, Burger JA, Schraufstatter I. 2005. KSHV-GPCR and CXCR2 transforming capacity and angiogenic responses are mediated through a JAK2-STAT3-dependent pathway. Oncogene 24:2067–2075. doi: 10.1038/sj.onc.1208442. [DOI] [PubMed] [Google Scholar]
- 143.Chen M, Sun F, Han L, Qu Z. 2016. Kaposi’s sarcoma herpesvirus (KSHV) microRNA K12-1 functions as an oncogene by activating NF-κB:IL-6:STAT3 signaling. Oncotarget 7:33363–33373. doi: 10.18632/oncotarget.9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Molden J, Chang Y, You Y, Moore PS, Goldsmith MA. 1997. A Kaposi’s sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J Biol Chem 272:19625–19631. doi: 10.1074/jbc.272.31.19625. [DOI] [PubMed] [Google Scholar]
- 145.Muromoto R, Okabe K, Fujimuro M, Sugiyama K, Yokosawa H, Seya T, Matsuda T. 2006. Physical and functional interactions between STAT3 and Kaposi's sarcoma-associated herpesvirus-encoded LANA. FEBS Lett 580:93–98. doi: 10.1016/j.febslet.2005.11.057. [DOI] [PubMed] [Google Scholar]
- 146.Gwack Y, Hwang S, Lim C, Won YS, Lee CH, Choe J. 2002. Kaposi's sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J Biol Chem 277:6438–6442. doi: 10.1074/jbc.M108289200. [DOI] [PubMed] [Google Scholar]
- 147.Bisson SA, Page AL, Ganem D. 2009. A Kaposi's sarcoma-associated herpesvirus protein that forms inhibitory complexes with type I interferon receptor subunits, Jak and STAT proteins, and blocks interferon-mediated signal transduction. J Virol 83:5056–5066. doi: 10.1128/JVI.02516-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mutocheluh M, Hindle L, Areste C, Chanas SA, Butler LM, Lowry K, Shah K, Evans DJ, Blackbourn DJ. 2011. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor-2 inhibits type 1 interferon signalling by targeting interferon-stimulated gene factor-3. J Gen Virol 92:2394–2398. doi: 10.1099/vir.0.034322-0. [DOI] [PubMed] [Google Scholar]
- 149.Horvath CM, Wen Z, Darnell JE Jr. 1995. A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev 9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 150.Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, Nabholz M, Bucher P. 2001. DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J Biol Chem 276:6675–6688. doi: 10.1074/jbc.M001748200. [DOI] [PubMed] [Google Scholar]
- 151.Novak U, Mui A, Miyajima A, Paradiso L. 1996. Formation of STAT5-containing DNA binding complexes in response to colony-stimulating factor-1 and platelet-derived growth factor. J Biol Chem 271:18350–18354. doi: 10.1074/jbc.271.31.18350. [DOI] [PubMed] [Google Scholar]
- 152.Collison LW, Delgoffe GM, Guy CS, Vignali KM, Chaturvedi V, Fairweather D, Satoskar AR, Garcia KC, Hunter CA, Drake CG, Murray PJ, Vignali DA. 2012. The composition and signaling of the IL-35 receptor are unconventional. Nat Immunol 13:290–299. doi: 10.1038/ni.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vidy A, El Bougrini J, Chelbi-Alix MK, Blondel D. 2007. The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. J Virol 81:4255–4263. doi: 10.1128/JVI.01930-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jia D, Rahbar R, Chan RW, Lee SM, Chan MC, Wang BX, Baker DP, Sun B, Peiris JS, Nicholls JM, Fish EN. 2010. Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling. PLoS One 5:e13927. doi: 10.1371/journal.pone.0013927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Harrison AR, Lieu KG, Larrous F, Ito N, Bourhy H, Moseley GW. 2020. Lyssavirus P-protein selectively targets STAT3-STAT1 heterodimers to modulate cytokine signalling. PLoS Pathog 16(9):e1008767. doi: 10.1371/journal.ppat.1008767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Percario Z, Olivetta E, Fiorucci G, Mangino G, Peretti S, Romeo G, Affabris E, Federico M. 2003. Human immunodeficiency virus type 1 (HIV-1) Nef activates STAT3 in primary human monocyte/macrophages through the release of soluble factors: involvement of Nef domains interacting with the cell endocytotic machinery. J Leukoc Biol 74:821–832. doi: 10.1189/jlb.0403161. [DOI] [PubMed] [Google Scholar]
- 157.Sehgal PB. 2008. Paradigm shifts in the cell biology of STAT signaling. Semin Cell Dev Biol 19:329–340. doi: 10.1016/j.semcdb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Reich NC. 2013. STATs get their move on. JAKSTAT 2:e27080. doi: 10.4161/jkst.27080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chatterjee-Kishore M, Wright K, Ting J, Stark GR. 2000. How STAT1 mediates constitutive gene expression: a complex of unphosphorylated STAT1 and IRF1 supports transcription of the LMP2 gene. EMBO J 19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cheon H, Stark GR. 2009. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci U S A 106:9373–9378. doi: 10.1073/pnas.0903487106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cheon H, Holvey-Bates EG, Schoggins JW, Forster S, Hertzog P, Imanaka N, Rice CM, Jackson MW, Junk DJ, Stark GR. 2013. IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J 32:2751–2763. doi: 10.1038/emboj.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. 2007. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev 21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cui X, Zhang L, Luo J, Rajasekaran A, Hazra S, Cacalano N, Dubinett SM. 2007. Unphosphorylated STAT6 contributes to constitutive cyclooxygenase-2 expression in human non-small cell lung cancer. Oncogene 26:4253–4260. doi: 10.1038/sj.onc.1210222. [DOI] [PubMed] [Google Scholar]
- 164.Kumar A, Commane MA, Flickinger TW, Horvath CM, Stark GR. 1997. Defective TNF-alpha-lnduced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science 278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- 165.Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, Stark GR. 2005. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res 65:939–947. [PubMed] [Google Scholar]
- 166.Park HJ, Li J, Hannah R, Biddie S, Leal-Cervantes AI, Kirschner K, Flores Santa Cruz D, Sexl V, Gottgens B, Green AR. 2016. Cytokine-induced megakaryocytic differentiation is regulated by genome-wide loss of a uSTAT transcriptional program. EMBO J 35:580–594. doi: 10.15252/embj.201592383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hu X, Dutta P, Tsurumi A, Li J, Wang J, Land H, Li WX. 2013. Unphosphorylated STAT5A stabilizes heterochromatin and suppresses tumor growth. Proc Natl Acad Sci U S A 110:10213–10218. doi: 10.1073/pnas.1221243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan SJ, Li WX. 2008. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol 10:489–496. doi: 10.1038/ncb1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cheon H, Yang J, Stark GR. 2011. The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. J Interferon Cytokine Res 31:33–40. doi: 10.1089/jir.2010.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bourke LT, Knight RA, Latchman DS, Stephanou A, McCormick J. 2013. Signal transducer and activator of transcription-1 localizes to the mitochondria and modulates mitophagy. JAKSTAT 2:e25666. doi: 10.4161/jkst.25666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Macias E, Rao D, Carbajal S, Kiguchi K, DiGiovanni J. 2014. Stat3 binds to mtDNA and regulates mitochondrial gene expression in keratinocytes. J Invest Dermatol 134:1971–1980. doi: 10.1038/jid.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Meier JA, Larner AC. 2014. Toward a new STATe: the role of STATs in mitochondrial function. Semin Immunol 26:20–28. doi: 10.1016/j.smim.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shahni R, Cale CM, Anderson G, Osellame LD, Hambleton S, Jacques TS, Wedatilake Y, Taanman J-W, Chan E, Qasim W, Plagnol V, Chalasani A, Duchen MR, Gilmour KC, Rahman S. 2015. Signal transducer and activator of transcription 2 deficiency is a novel disorder of mitochondrial fission. Brain 138:2834–2846. doi: 10.1093/brain/awv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shen S, Niso-Santano M, Adjemian S, Takehara T, Malik SA, Minoux H, Souquere S, Marino G, Lachkar S, Senovilla L, Galluzzi L, Kepp O, Pierron G, Maiuri MC, Hikita H, Kroemer R, Kroemer G. 2012. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell 48:667–680. doi: 10.1016/j.molcel.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 175.Ng DC, Lin BH, Lim CP, Huang G, Zhang T, Poli V, Cao X. 2006. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J Cell Biol 172:245–257. doi: 10.1083/jcb.200503021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Verma NK, Dourlat J, Davies AM, Long A, Liu WQ, Garbay C, Kelleher D, Volkov Y. 2009. STAT3-stathmin interactions control microtubule dynamics in migrating T-cells. J Biol Chem 284:12349–12362. doi: 10.1074/jbc.M807761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Silver D, Naora H, Liu J, Cheng W, Montell D. 2004. Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res 64:3550–3558. doi: 10.1158/0008-5472.CAN-03-3959. [DOI] [PubMed] [Google Scholar]
- 178.Lee JE, Yang YM, Liang FX, Gough DJ, Levy DE, Sehgal PB. 2012. Nongenomic STAT5-dependent effects on Golgi apparatus and endoplasmic reticulum structure and function. Am J Physiol Cell Physiol 302:C804–C820. doi: 10.1152/ajpcell.00379.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lee JE, Yang YM, Yuan H, Sehgal PB. 2013. Definitive evidence using enucleated cytoplasts for a nongenomic basis for the cystic change in endoplasmic reticulum structure caused by STAT5a/b siRNAs. Am J Physiol Cell Physiol 304:C312–C323. doi: 10.1152/ajpcell.00311.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Van Grol J, Subauste C, Andrade RM, Fujinaga K, Nelson J, Subauste CS. 2010. HIV-1 Inhibits autophagy in bystander macrophage/monocytic cells through Src-Akt and STAT3. PLoS One 5:e11733. doi: 10.1371/journal.pone.0011733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Santarelli R, Gonnella R, Di Giovenale G, Cuomo L, Capobianchi A, Granato M, Gentile G, Faggioni A, Cirone M. 2015. STAT3 activation by KSHV correlates with IL-10, IL-6 and IL-23 release and an autophagic block in dendritic cells. Sci Rep 4:4241. doi: 10.1038/srep04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chiramel AI, Brady NR, Bartenschlager R. 2013. Divergent roles of autophagy in virus infection. Cells 2:83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Anand SK, Tikoo SK. 2013. Viruses as modulators of mitochondrial functions. Adv Virol 2013:738794. doi: 10.1155/2013/738794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yu Y, Wang R, Nan Y, Zhang L, Zhang Y. 2013. Induction of STAT1 phosphorylation at serine 727 and expression of proinflammatory cytokines by porcine reproductive and respiratory syndrome virus. PLoS One 8:e61967. doi: 10.1371/journal.pone.0061967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.McLaren J, Rowe M, Brennan P. 2007. Epstein-Barr virus induces a distinct form of DNA-bound STAT1 compared with that found in interferon-stimulated B lymphocytes. J Gen Virol 88:1876–1886. doi: 10.1099/vir.0.82741-0. [DOI] [PubMed] [Google Scholar]
- 186.Nan Y, Wu C, Zhang YJ. 2018. Interferon independent non-canonical STAT activation and virus induced inflammation. Viruses 10:196. doi: 10.3390/v10040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Vidy A, Chelbi-Alix M, Blondel D. 2005. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J Virol 79:14411–14420. doi: 10.1128/JVI.79.22.14411-14420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Brzozka K, Finke S, Conzelmann KK. 2006. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J Virol 80:2675–2683. doi: 10.1128/JVI.80.6.2675-2683.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lieu KG, Brice A, Wiltzer L, Hirst B, Jans DA, Blondel D, Moseley GW. 2013. The rabies virus interferon antagonist P protein interacts with activated STAT3 and inhibits Gp130 receptor signaling. J Virol 87:8261–8265. doi: 10.1128/JVI.00989-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sohn SY, Hearing P. 2011. Adenovirus sequesters phosphorylated STAT1 at viral replication centers and inhibits STAT dephosphorylation. J Virol 85:7555–7562. doi: 10.1128/JVI.00513-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Orlova A, Wagner C, de Araujo ED, Bajusz D, Neubauer HA, Herling M, Gunning PT, Keserű GM, Moriggl R. 2019. Direct targeting options for STAT3 and STAT5 in cancer. Cancers 11:1930. doi: 10.3390/cancers11121930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sen N, Che X, Rajamani J, Zerboni L, Sung P, Ptacek J, Arvin AM. 2012. Signal transducer and activator of transcription 3 (STAT3) and survivin induction by varicella-zoster virus promote replication and skin pathogenesis. Proc Natl Acad Sci U S A 109:600–605. doi: 10.1073/pnas.1114232109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Slinger E, Maussang D, Schreiber A, Siderius M, Rahbar A, Fraile-Ramos A, Lira S, Soderberg-Naucler C, Smit M. 2010. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6–STAT3 axis. Sci Signal 3:ra58. doi: 10.1126/scisignal.2001180. [DOI] [PubMed] [Google Scholar]
- 194.Tomita M, Kawakami H, Uchihara JN, Okudaira T, Masuda M, Matsuda T, Tanaka Y, Ohshiro K, Mori N. 2006. Inhibition of constitutively active Jak-Stat pathway suppresses cell growth of human T-cell leukemia virus type 1-infected T-cell lines and primary adult T-cell leukemia cells. Retrovirology 3:22. doi: 10.1186/1742-4690-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 195.Gavegnano C, Detorio M, Montero C, Bosque A, Planelles V, Schinazi RF. 2014. Ruxolitinib and tofacitinib are potent and selective inhibitors of HIV-1 replication and virus reactivation in vitro. Antimicrob Agents Chemother 58:1977–1986. doi: 10.1128/AAC.02496-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Vasou A, Paulus C, Narloch J, Gage ZO, Rameix-Welti MA, Eleouet JF, Nevels M, Randall RE, Adamson CS. 2018. Modular cell-based platform for high throughput identification of compounds that inhibit a viral interferon antagonist of choice. Antiviral Res 150:79–92. doi: 10.1016/j.antiviral.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Basu D, Walkiewicz MP, Frieman M, Baric RS, Auble DT, Engel DA. 2009. Novel influenza virus NS1 antagonists block replication and restore innate immune function. J Virol 83:1881–1891. doi: 10.1128/JVI.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lemm JA, O'Boyle D, Liu M, Nower PT, Colonno R, Deshpande MS, Snyder LB, Martin SW, St Laurent DR, Serrano-Wu MH, Romine JL, Meanwell NA, Gao M. 2010. Identification of hepatitis C virus NS5A inhibitors. J Virol 84:482–491. doi: 10.1128/JVI.01360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Emert-Sedlak L, Kodama T, Lerner EC, Dai W, Foster C, Day BW, Lazo JS, Smithgall TE. 2009. Chemical library screens targeting an HIV-1 accessory factor/host cell kinase complex identify novel antiretroviral compounds. ACS Chem Biol 4:939–947. doi: 10.1021/cb900195c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wright PF, Karron RA, Madhi SA, Treanor JJ, King JC, O'Shea A, Ikizler MR, Zhu Y, Collins PL, Cutland C, Randolph VB, Deatly AM, Hackell JG, Gruber WC, Murphy BR. 2006. The interferon antagonist NS2 protein of respiratory syncytial virus is an important virulence determinant for humans. J Infect Dis 193:573–581. doi: 10.1086/499600. [DOI] [PubMed] [Google Scholar]
- 201.Devaux P, Hodge G, McChesney MB, Cattaneo R. 2008. Attenuation of V- or C-defective measles viruses: infection control by the inflammatory and interferon responses of rhesus monkeys. J Virol 82:5359–5367. doi: 10.1128/JVI.00169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wacheck V, Egorov A, Groiss F, Pfeiffer A, Fuereder T, Hoeflmayer D, Kundi M, Popow-Kraupp T, Redlberger-Fritz M, Mueller C, Cinatl J, Michaelis M, Geiler J, Bergmann M, Romanova J, Roethl E, Morokutti A, Wolschek M, Ferko B, Seipelt J, Dick-Gudenus R, Muster T. 2010. A novel type of influenza vaccine: safety and immunogenicity of replication-deficient influenza virus created by deletion of the interferon antagonist NS1. J Infect Dis 201:354–362. doi: 10.1086/649428. [DOI] [PubMed] [Google Scholar]
- 203.Wang P, Zheng M, Lau SY, Chen P, Mok BW, Liu S, Liu H, Huang X, Cremin CJ, Song W, Chen Y, Wong YC, Huang H, To KK, Chen Z, Xia N, Yuen KY, Chen H. 2019. Generation of DelNS1 influenza viruses: a strategy for optimizing live attenuated influenza vaccines. mBio 10:e02180-19. doi: 10.1128/mBio.02180-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mossler C, Groiss F, Wolzt M, Wolschek M, Seipelt J, Muster T. 2013. Phase I/II trial of a replication-deficient trivalent influenza virus vaccine lacking NS1. Vaccine 31:6194–6200. doi: 10.1016/j.vaccine.2013.10.061. [DOI] [PubMed] [Google Scholar]
- 205.Blanco-Lobo P, Nogales A, Rodriguez L, Martinez-Sobrido L. 2019. Novel approaches for the development of live attenuated influenza vaccines. Viruses 11:190. doi: 10.3390/v11020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Talon J, Salvatore M, O'Neill RE, Nakaya Y, Zheng H, Muster T, Garcia-Sastre A, Palese P. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc Natl Acad Sci U S A 97:4309–4314. doi: 10.1073/pnas.070525997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Simmons JD, Wollish AC, Heise MT. 2010. A determinant of Sindbis virus neurovirulence enables efficient disruption of Jak/STAT signaling. J Virol 84:11429–11439. doi: 10.1128/JVI.00577-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Laurent-Rolle M, Boer EF, Lubick KJ, Wolfinbarger JB, Carmody AB, Rockx B, Liu W, Ashour J, Shupert WL, Holbrook MR, Barrett AD, Mason PW, Bloom ME, Garcia-Sastre A, Khromykh AA, Best SM. 2010. The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling. J Virol 84:3503–3515. doi: 10.1128/JVI.01161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ito N, Moseley GW, Blondel D, Shimizu K, Rowe CL, Ito Y, Masatani T, Nakagawa K, Jans DA, Sugiyama M. 2010. Role of interferon antagonist activity of rabies virus phosphoprotein in viral pathogenicity. J Virol 84:6699–6710. doi: 10.1128/JVI.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ebihara H, Takada A, Kobasa D, Jones S, Neumann G, Theriault S, Bray M, Feldmann H, Kawaoka Y. 2006. Molecular determinants of Ebola virus virulence in mice. PLoS Pathog 2:e73. doi: 10.1371/journal.ppat.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pappalardo M, Collu F, Macpherson J, Michaelis M, Fraternali F, Wass MN. 2017. Investigating Ebola virus pathogenicity using molecular dynamics. BMC Genomics 18:566. doi: 10.1186/s12864-017-3912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Devaux P, Hudacek AW, Hodge G, Reyes-Del Valle J, McChesney MB, Cattaneo R. 2011. A recombinant measles virus unable to antagonize STAT1 function cannot control inflammation and is attenuated in rhesus monkeys. J Virol 85:348–356. doi: 10.1128/JVI.00802-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wiltzer L, Okada K, Yamaoka S, Larrous F, Kuusisto HV, Sugiyama M, Blondel D, Bourhy H, Jans DA, Ito N, Moseley GW. 2014. Interaction of rabies virus P-protein with STAT proteins is critical to lethal rabies disease. J Infect Dis 209:1744–1753. doi: 10.1093/infdis/jit829. [DOI] [PubMed] [Google Scholar]
- 214.Hossain MA, Larrous F, Rawlinson SM, Zhan J, Sethi A, Ibrahim Y, Aloi M, Lieu KG, Mok YF, Griffin MDW, Ito N, Ose T, Bourhy H, Moseley GW, Gooley PR. 2019. Structural elucidation of viral antagonism of innate immunity at the STAT1 interface. Cell Rep 29:1934–1945. doi: 10.1016/j.celrep.2019.10.020. [DOI] [PubMed] [Google Scholar]
- 215.Satterfield BA, Borisevich V, Foster SL, Rodriguez SE, Cross RW, Fenton KA, Agans KN, Basler CF, Geisbert TW, Mire CE. 2019. Antagonism of STAT1 by Nipah virus P gene products modulates disease course but not lethal outcome in the ferret model. Sci Rep 9:16710. doi: 10.1038/s41598-019-53037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhong Z, Wen Z, Darnell JE Jr. 1994. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 217.Ning YJ, Feng K, Min YQ, Cao WC, Wang M, Deng F, Hu Z, Wang H. 2015. Disruption of type I interferon signaling by the nonstructural protein of severe fever with thrombocytopenia syndrome virus via the hijacking of STAT2 and STAT1 into inclusion bodies. J Virol 89:4227–4236. doi: 10.1128/JVI.00154-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Paulus C, Krauss S, Nevels M. 2006. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc Natl Acad Sci U S A 103:3840–3845. doi: 10.1073/pnas.0600007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zhang R, Fang L, Wang D, Cai K, Zhang H, Xie L, Li Y, Chen H, Xiao S. 2015. Porcine bocavirus NP1 negatively regulates interferon signaling pathway by targeting the DNA-binding domain of IRF9. Virology 485:414–421. doi: 10.1016/j.virol.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Najarro P, Traktman P, Lewis JA. 2001. Vaccinia virus blocks gamma interferon signal transduction: viral VH1 phosphatase reverses Stat1 activation. J Virol 75:3185–3196. doi: 10.1128/JVI.75.7.3185-3196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nagano Y, Sugiyama A, Kimoto M, Wakahara T, Noguchi Y, Jiang X, Saijo S, Shimizu N, Yabuno N, Yao M, Gooley PR, Moseley GW, Tadokoro T, Maenaka K, Ose T. 2020. The measles virus V protein binding site to STAT2 overlaps with that of IRF9. J Virol 94:e01169-20. doi: 10.1128/JVI.01169-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ulane CM, Horvath CM. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160–166. doi: 10.1006/viro.2002.1773. [DOI] [PubMed] [Google Scholar]
- 223.Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol 83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Guo L, Luo X, Li R, Xu Y, Zhang J, Ge J, Bu Z, Feng L, Wang Y. 2016. Porcine epidemic diarrhea virus infection inhibits interferon signaling by targeted degradation of STAT1. J Virol 90:8281–8292. doi: 10.1128/JVI.01091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hong S, Mehta KP, Laimins LA. 2011. Suppression of STAT-1 expression by human papillomaviruses is necessary for differentiation-dependent genome amplification and plasmid maintenance. J Virol 85:9486–9494. doi: 10.1128/JVI.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF. 2007. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J Virol 81:13469–13477. doi: 10.1128/JVI.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Chen J, Wu M, Zhang X, Zhang W, Zhang Z, Chen L, He J, Zheng Y, Chen C, Wang F, Hu Y, Zhou X, Wang C, Xu Y, Lu M, Yuan Z. 2013. Hepatitis B virus polymerase impairs interferon-alpha-induced STAT activation through inhibition of importin-alpha5 and protein kinase C-delta. Hepatology 57:470–482. doi: 10.1002/hep.26064. [DOI] [PubMed] [Google Scholar]
- 228.Wang R, Nan Y, Yu Y, Zhang YJ. 2013. Porcine reproductive and respiratory syndrome virus Nsp1 inhibits interferon-activated JAK/STAT signal transduction by inducing karyopherin-1 degradation. J Virol 87:5219–5228. doi: 10.1128/JVI.02643-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zhang Y, Yang L, Wang H, Zhang G, Sun X. 2016. Respiratory syncytial virus non-structural protein 1 facilitates virus replication through miR-29a-mediated inhibition of interferon-alpha receptor. Biochem Biophys Res Commun 478:1436–1441. doi: 10.1016/j.bbrc.2016.08.142. [DOI] [PubMed] [Google Scholar]
- 230.Shah KM, Stewart SE, Wei W, Woodman CB, O'Neil JD, Dawson CW, Young LS. 2009. The EBV-encoded latent membrane proteins, LMP2A and LMP2B, limit the actions of interferon by targeting interferon receptors for degradation. Oncogene 28:3903–3914. doi: 10.1038/onc.2009.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Valmas C, Grosch MN, Schumann M, Olejnik J, Martinez O, Best SM, Krahling V, Basler CF, Muhlberger E. 2010. Marburg virus evades interferon responses by a mechanism distinct from ebola virus. PLoS Pathog 6:e1000721. doi: 10.1371/journal.ppat.1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Weihua X, Ramanujam S, Lindner DJ, Kudaravalli RD, Freund R, Kalvakolanu DV. 1998. The polyoma virus T antigen interferes with interferon-inducible gene expression. Proc Natl Acad Sci U S A 95:1085–1090. doi: 10.1073/pnas.95.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Shi L, Ramaswamy M, Manzel LJ, Look DC. 2007. Inhibition of Jak1-dependent signal transduction in airway epithelial cells infected with adenovirus. Am J Respir Cell Mol Biol 37:720–728. doi: 10.1165/rcmb.2007-0158OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Tsunematsu S, Suda G, Yamasaki K, Kimura M, Izumi T, Umemura M, Ito J, Sato F, Nakai M, Sho T, Morikawa K, Ogawa K, Tanaka Y, Watashi K, Wakita T, Sakamoto N. 2017. Hepatitis B virus X protein impairs alpha-interferon signaling via up-regulation of suppressor of cytokine signaling 3 and protein phosphatase 2A. J Med Virol 89:267–275. doi: 10.1002/jmv.24643. [DOI] [PubMed] [Google Scholar]
- 235.Bode JG, Ludwig S, Ehrhardt C, Albrecht U, Erhardt A, Schaper F, Heinrich PC, Haussinger D. 2003. IFN-α antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J 17:488–490. doi: 10.1096/fj.02-0664fje. [DOI] [PubMed] [Google Scholar]
- 236.Chen H, Lee JM, Wang Y, Huang DP, Ambinder RF, Hayward SD. 1999. The Epstein-Barr virus latency BamHI-Q promoter is positively regulated by STATs and Zta interference with JAK/STAT activation leads to loss of BamHI-Q promoter activity. Proc Natl Acad Sci U S A 96:9339–9344. doi: 10.1073/pnas.96.16.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Muromoto R, Ikeda O, Okabe K, Togi S, Kamitani S, Fujimuro M, Harada S, Oritani K, Matsuda T. 2009. Epstein-Barr virus-derived EBNA2 regulates STAT3 activation. Biochem Biophys Res Commun 378:439–443. doi: 10.1016/j.bbrc.2008.11.053. [DOI] [PubMed] [Google Scholar]
- 238.Xu Y, Shi Y, Yuan Q, Liu X, Yan B, Chen L, Tao Y, Cao Y. 2013. Epstein-Barr virus encoded LMP1 regulates cyclin D1 promoter activity by nuclear EGFR and STAT3 in CNE1 cells. J Exp Clin Cancer Res 32:90. doi: 10.1186/1756-9966-32-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Incrocci R, Barse L, Stone A, Vagvala S, Montesano M, Subramaniam V, Swanson-Mungerson M. 2017. Epstein-Barr virus latent membrane protein 2A (LMP2A) enhances IL-10 production through the activation of Bruton's tyrosine kinase and STAT3. Virology 500:96–102. doi: 10.1016/j.virol.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Arany I, Grattendick KG, Tyring SK. 2002. Interleukin-10 induces transcription of the early promoter of human papillomavirus type 16 (HPV16) through the 5′-segment of the upstream regulatory region (URR). Antiviral Res 55:331–339. doi: 10.1016/s0166-3542(02)00070-0. [DOI] [PubMed] [Google Scholar]
- 241.Ren C, Cheng X, Lu B, Yang G. 2013. Activation of interleukin-6/signal transducer and activator of transcription 3 by human papillomavirus early proteins 6 induces fibroblast senescence to promote cervical tumourigenesis through autocrine and paracrine pathways in tumour microenvironment. Eur J Cancer 49:3889–3899. doi: 10.1016/j.ejca.2013.07.140. [DOI] [PubMed] [Google Scholar]
- 242.Johannsen E, Lambert PF. 2013. Epigenetics of human papillomaviruses. Virology 445:205–212. doi: 10.1016/j.virol.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Morgan EL, Wasson CW, Hanson L, Kealy D, Pentland I, McGuire V, Scarpini C, Coleman N, Arthur JSC, Parish JL, Roberts S, Macdonald A. 2018. STAT3 activation by E6 is essential for the differentiation-dependent HPV18 life cycle. PLoS Pathog 14:e1006975. doi: 10.1371/journal.ppat.1006975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Li Y, Wu R, Liu Z, Fan J, Yang H. 2014. Enforced expression of microRNA-21 influences the replication of varicella-zoster virus by triggering signal transducer and activator of transcription 3. Exp Ther Med 7:1291–1296. doi: 10.3892/etm.2014.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pinkham C, An S, Lundberg L, Bansal N, Benedict A, Narayanan A, Kehn-Hall K. 2016. The role of signal transducer and activator of transcription 3 in Rift Valley fever virus infection. Virology 496:175–185. doi: 10.1016/j.virol.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lundquist A, Barre B, Bienvenu F, Hermann J, Avril S, Coqueret O. 2003. Kaposi sarcoma-associated viral cyclin K overrides cell growth inhibition mediated by oncostatin M through STAT3 inhibition. Blood 101:4070–4077. doi: 10.1182/blood-2002-07-1994. [DOI] [PubMed] [Google Scholar]
- 247.Gallaher AM, Das S, Xiao Z, Andresson T, Kieffer-Kwon P, Happel C, Ziegelbauer J. 2013. Proteomic screening of human targets of viral microRNAs reveals functions associated with immune evasion and angiogenesis. PLoS Pathog 9:e1003584. doi: 10.1371/journal.ppat.1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lepiller Q, Abbas W, Kumar A, Tripathy MK, Herbein G. 2013. HCMV activates the IL-6-JAK-STAT3 axis in HepG2 cells and primary human hepatocytes. PLoS One 8:e59591. doi: 10.1371/journal.pone.0059591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Le VT, Trilling M, Wilborn M, Hengel H, Zimmermann A. 2008. Human cytomegalovirus interferes with signal transducer and activator of transcription (STAT) 2 protein stability and tyrosine phosphorylation. J Gen Virol 89:2416–2426. doi: 10.1099/vir.0.2008/001669-0. [DOI] [PubMed] [Google Scholar]
- 250.Del Corno M, Donninelli G, Varano B, Da Sacco L, Masotti A, Gessani S. 2014. HIV-1 gp120 activates the STAT3/interleukin-6 axis in primary human monocyte-derived dendritic cells. J Virol 88:11045–11055. doi: 10.1128/JVI.00307-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Gargan S, Ahmed S, Mahony R, Bannan C, Napoletano S, O'Farrelly C, Borrow P, Bergin C, Stevenson NJ. 2018. HIV-1 promotes the degradation of components of the type 1 IFN JAK/STAT pathway and blocks anti-viral ISG induction. EBioMedicine 30:203–216. doi: 10.1016/j.ebiom.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Chang Z, Wang Y, Bian L, Liu Q, Long JE. 2017. Enterovirus 71 antagonizes the antiviral activity of host STAT3 and IL-6R with partial dependence on virus-induced miR-124. J Gen Virol 98:3008–3025. doi: 10.1099/jgv.0.000967. [DOI] [PubMed] [Google Scholar]
- 253.Tu Z, Xu M, Zhang J, Feng Y, Hao Z, Tu C, Liu Y. 2019. Pentagalloylglucose inhibits the replication of rabies virus via mediation of the miR-455:SOCS3:STAT3:IL-6 pathway. J Virol 93:e00539-19. doi: 10.1128/JVI.00539-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Hui KP, Li HS, Cheung MC, Chan RW, Yuen KM, Mok CK, Nicholls JM, Peiris JS, Chan MC. 2016. Highly pathogenic avian influenza H5N1 virus delays apoptotic responses via activation of STAT3. Sci Rep 6:28593. doi: 10.1038/srep28593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Liu S, Yan R, Chen B, Pan Q, Chen Y, Hong J, Zhang L, Liu W, Wang S, Chen JL. 2019. Influenza virus-induced robust expression of SOCS3 contributes to excessive production of IL-6. Front Immunol 10:1843. doi: 10.3389/fimmu.2019.01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Li S-W, Wang C-Y, Jou Y-J, Yang T-C, Huang S-H, Wan L, Lin Y-J, Lin C-W. 2016. SARS coronavirus papain-like protease induces Egr-1-dependent up-regulation of TGF-β1 via ROS/p38 MAPK/STAT3 pathway. Sci Rep 6:25754. doi: 10.1038/srep25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Mizutani T, Fukushi S, Murakami M, Hirano T, Saijo M, Kurane I, Morikawa S. 2004. Tyrosine dephosphorylation of STAT3 in SARS coronavirus-infected Vero E6 cells. FEBS Lett 577:187–192. doi: 10.1016/j.febslet.2004.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Yang L, Wang R, Ma Z, Xiao Y, Nan Y, Wang Y, Lin S, Zhang YJ. 2017. Porcine reproductive and respiratory syndrome virus antagonizes JAK/STAT3 signaling via nsp5, which induces STAT3 degradation. J Virol 91:e02087-16. doi: 10.1128/JVI.02087-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Chandra V, Kar-Roy A, Kumari S, Mayor S, Jameel S. 2008. The hepatitis E virus ORF3 protein modulates epidermal growth factor receptor trafficking, STAT3 translocation, and the acute-phase response. J Virol 82:7100–7110. doi: 10.1128/JVI.00403-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Mitzel DN, Jaramillo RJ, Stout-Delgado H, Senft AP, Harrod KS. 2014. Human metapneumovirus inhibits the IL-6-induced JAK/STAT3 signalling cascade in airway epithelium. J Gen Virol 95:26–37. doi: 10.1099/vir.0.055632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ando S, Kawada JI, Watanabe T, Suzuki M, Sato Y, Torii Y, Asai M, Goshima F, Murata T, Shimizu N, Ito Y, Kimura H. 2016. Tofacitinib induces G1 cell-cycle arrest and inhibits tumor growth in Epstein-Barr virus-associated T and natural killer cell lymphoma cells. Oncotarget 7:76793–76805. doi: 10.18632/oncotarget.12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Migone TS, Lin JX, Cereseto A, Mulloy JC, O'Shea JJ, Franchini G, Leonard WJ. 1995. Constitutively activated Jak-STAT pathway in T Cells transformed with HTLV-I. Science 269:79–81. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- 263.Nakamura N, Fuji M, Tsukahara T, Arai M, Ohashi T, Wakao H, Kannagi M, Yamamoto N. 1999. Human T-cell leukemia virus type 1 Tax protein induces the expression of STAT1 and STAT5 genes in T-cells. Oncogene 18:2667–2675. doi: 10.1038/sj.onc.1202608. [DOI] [PubMed] [Google Scholar]
- 264.Nicot C, Mulloy J, Ferrari M, Johnson J, Fu K, Fukumoto R, Trovato R, Fullen J, Leonard WJ, Franchini G. 2001. HTLV-1 p12I protein enhances STAT5 activation and decreases the interleukin-2 requirement for proliferation of primary human peripheral blood mononuclear cells. Blood 98:823–829. doi: 10.1182/blood.v98.3.823. [DOI] [PubMed] [Google Scholar]
- 265.Fung MM, Chu YL, Fink JL, Wallace A, McGuire KL. 2005. IL-2- and STAT5-regulated cytokine gene expression in cells expressing the Tax protein of HTLV-1. Oncogene 24:4624–4633. doi: 10.1038/sj.onc.1208507. [DOI] [PubMed] [Google Scholar]
- 266.Carson A, Khan SA. 2006. Characterization of transcription factor binding to human papillomavirus type 16 DNA during cellular differentiation. J Virol 80:4356–4362. doi: 10.1128/JVI.80.9.4356-4362.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Sobti RC, Singh N, Hussain S, Suri V, Bharadwaj M, Das BC. 2010. Deregulation of STAT-5 isoforms in the development of HPV-mediated cervical carcinogenesis. J Recept Signal Transduct Res 30:178–188. doi: 10.3109/10799891003786218. [DOI] [PubMed] [Google Scholar]
- 268.Valle-Mendiola A, Weiss-Steider B, Rocha-Zavaleta L, Soto-Cruz I. 2014. IL-2 enhances cervical cancer cells proliferation and JAK3/STAT5 phosphorylation at low doses, while at high doses IL-2 has opposite effects. Cancer Invest 32:115–125. doi: 10.3109/07357907.2014.883526. [DOI] [PubMed] [Google Scholar]
- 269.Morgan EL, MacDonald A. 2019. JAK2 inhibition impairs proliferation and sensitises cervical cancer cells to cisplatin-induced cell death. Cancers 11:1934. doi: 10.3390/cancers11121934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Kim Y, Kwon EK, Jeon JH, So I, Kim IG, Choi MS, Kim IS, Choi JK, Jung JU, Cho NH. 2012. Activation of the STAT6 transcription factor in Jurkat T-cells by the herpesvirus saimiri Tip protein. J Gen Virol 93:330–340. doi: 10.1099/vir.0.036087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Mazumder ED, Jardin C, Vogel B, Heck E, Scholz B, Lengenfelder D, Sticht H, Ensser A. 2012. A molecular model for the differential activation of STAT3 and STAT6 by the herpesviral oncoprotein tip. PLoS One 7:e34306. doi: 10.1371/journal.pone.0034306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Waldele K, Schneider G, Ruckes T, Grassmann R. 2004. Interleukin-13 overexpression by tax transactivation: a potential autocrine stimulus in human T-cell leukemia virus-infected lymphocytes. J Virol 78:6081–6090. doi: 10.1128/JVI.78.12.6081-6090.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]