Mononegavirales, known as nonsegmented negative-sense (NNS) RNA viruses, are a class of pathogenic and sometimes deadly viruses that include rabies virus (RABV), human respiratory syncytial virus (HRSV), and Ebola virus (EBOV). Unfortunately, no effective vaccines and antiviral therapeutics against many Mononegavirales are currently available. Viral polymerases have been attractive and major antiviral therapeutic targets. Therefore, Mononegavirales polymerases have been extensively investigated for their structures and functions.
KEYWORDS: cryo-EM structures, Mononegavirales polymerases, RNA-dependent RNA polymerase, human metapneumovirus (HMPV), human respiratory syncytial virus (HRSV), rabies virus (RABV), vesicular stomatitis virus (VSV)
ABSTRACT
Mononegavirales, known as nonsegmented negative-sense (NNS) RNA viruses, are a class of pathogenic and sometimes deadly viruses that include rabies virus (RABV), human respiratory syncytial virus (HRSV), and Ebola virus (EBOV). Unfortunately, no effective vaccines and antiviral therapeutics against many Mononegavirales are currently available. Viral polymerases have been attractive and major antiviral therapeutic targets. Therefore, Mononegavirales polymerases have been extensively investigated for their structures and functions. Mononegavirales mimic RNA synthesis of their eukaryotic counterparts by utilizing multifunctional RNA polymerases to replicate entire viral genomes and transcribe viral mRNAs from individual viral genes as well as synthesize 5′ methylated cap and 3′ poly(A) tail of the transcribed viral mRNAs. The catalytic subunit large protein (L) and cofactor phosphoprotein (P) constitute the Mononegavirales polymerases. In this review, we discuss the shared and unique features of RNA synthesis, the monomeric multifunctional enzyme L, and the oligomeric multimodular adapter P of Mononegavirales. We outline the structural analyses of the Mononegavirales polymerases since the first structure of the vesicular stomatitis virus (VSV) L protein determined in 2015 and highlight multiple high-resolution cryo-electron microscopy (cryo-EM) structures of the polymerases of Mononegavirales, namely, VSV, RABV, HRSV, human metapneumovirus (HMPV), and human parainfluenza virus (HPIV), that have been reported in recent months (2019 to 2020). We compare the structures of those polymerases grouped by virus family, illustrate the similarities and differences among those polymerases, and reveal the potential RNA synthesis mechanisms and models of highly conserved Mononegavirales. We conclude by the discussion of remaining questions, evolutionary perspectives, and future directions.
INTRODUCTION
Mononegavirales, known as nonsegmented negative-sense (NNS) RNA viruses, are a class of viruses infecting numerous plants, animals, and humans, and many of them cause significant diseases and deaths in humans (1–3). There are currently 11 virus families in the order of Mononegavirales, namely, Artoviridae, Bornaviridae, Filoviridae, Lispiviridae, Mymonaviridae, Nyamiviridae, Paramyxoviridae, Pneumoviridae, Rhabdoviridae, Sunviridae, and Xinmoviridae, according to the 2019 taxonomy (4). Recent advances in sequencing technology facilitated the discovery of new families and genera. For example, (i) Pneumoviridae, which used to be the subfamily Pneumovirinae in Paramyxoviridae, became a new virus family (5); and (ii) a new ebolavirus, three new filovirus genera, and a sixth proposed genus were recently added in Filoviridae (6). Within the order, some Mononegavirales circulate within the human population causing respiratory diseases, such as the human respiratory syncytial virus (HRSV) and human metapneumovirus (HMPV) from Pneumoviridae and human parainfluenza virus (HPIV) from Paramyxoviridae (7), and common childhood diseases, such as measles virus (MeV) and mumps virus (MuV) from Paramyxoviridae (8–11). Several emerging and reemerging Mononegavirales often transmit cross-species and cause severe diseases with high mortality rates, such as NIAID category A priority pathogens Ebola virus (EBOV) and Marburg virus (MRAV) from Filoviridae and NIAID category C priority pathogens rabies virus (RABV) from Rhabdoviridae and Nipah virus (NiV) and Hendra virus (HeV) from Paramyxoviridae (1, 12–17). The representative viruses of Mononegavirales are listed in Table 1. Currently, no effective vaccine or antiviral therapy is available to prevent or treat many of those NNS RNA viral pathogens (18–29).
TABLE 1.
Taxonomy of the representative Mononegavirales viruses discussed in this review
| Family | Genus | Species | Virus (abbreviation) |
|---|---|---|---|
| Rhabdoviridae | Vesiculovirus | Indiana vesiculovirus | vesicular stomatitis virus (VSV) |
| Lyssavirus | Rabies lyssavirus | rabies virus (RABV) | |
| Pneumoviridae | Orthopneumovirus | Human orthopneumovirus | human respiratory syncytial virus (HRSV) |
| Metapneumovirus | Human metapneumovirus | human metapneumovirus (HMPV) | |
| Paramyxoviridae | Henipavirus | Hendra henipavirus | Hendra virus (HeV) |
| Nipah henipavirus | Nipah virus (NiV) | ||
| Respirovirus | Human respirovirus | human parainfluenza virus (HPIV) | |
| Murine respirovirus | Sendai virus (SeV) | ||
| Rubulavirus | Mumps rubulavirus | mumps virus (MuV) | |
| Morbillivirus | Measles morbillivirus | measles virus (MeV) | |
| Filoviridae | Ebolavirus | Zaire ebolavirus | Ebola virus (EBOV) |
| Marburgvirus | Marburg marburgvirus | Marburg virus (MARV) |
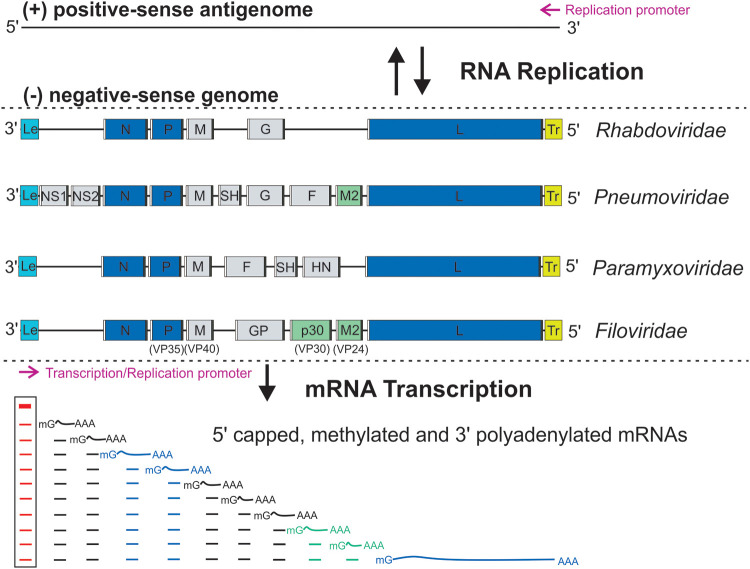
Mononegavirales are enveloped viruses with various morphologies for different families; for example, Rhabdoviridae are bullet-shaped, Paramyxoviridae are pleomorphic or spherical, and Filoviridae are filamentous (30–32). The genome organization and replication of Mononegavirales have been extensively studied for decades (1–3). The NNS RNA viral genomes are linear and single-stranded, and their lengths range from 8.9 to 19.0 kilobases (1–3). Mononegavirales encode 5 to 10 genes, with 4 core genes shared by all members. Those core genes (Fig. 1, blue boxes) encode four shared proteins, nucleoprotein (N or NP), phosphoprotein (P or VP35), matrix protein (M), and large protein (L). Three out of four shared proteins, namely, N, P, and L, constitute the RNA synthesis machine, suggesting the central role of RNA synthesis in the Mononegavirales life cycle (33) (Fig. 1).
FIG 1.
The genome organization and RNA synthesis of Mononegavirales. The negative-sense NNS genome is depicted from the 3′ end to the 5′ end, showing the 3′ leader (Le; cyan box), genes (gray, blue, or green box) flanking with gene start (GS; white box) and gene end (GE; black box), and 5′ trailer (Tr; yellow box). The essential genes (N, P, and L) and necessary cofactors (M2 or p30) for RNA synthesis are colored in blue and green, respectively. The RNA-dependent RNA polymerase (RdRP) sequentially produces a gradient level of Le RNA (red line) and viral mRNAs (black, blue, or green line), with the attenuation of the downstream mRNAs at each gene junction. The Le RNA (red lines inside the box) remains uncapped and nonpolyadenylated, while the viral mRNAs are 5′ capped, methylated, and 3′ polyadenylated. The lines under the Le RNA and representative viral mRNAs indicate the abundancy and gradient levels of the RNA transcripts. The promoters for transcription and replication are shown with magenta arrows.
Mononegavirales initiate viral infection by delivering into the host cell a virus-specific RNA synthesis machine (33–35). The template for RNA synthesis is not RNA alone but rather a complex of the viral genomic RNA completely encapsidated by the N or NP, called nucleocapsid (NC) (36). This NC template is copied by the viral RNA-dependent RNA polymerase (RdRP), which comprises L and cofactor P or VP35 (37–43). Additional viral proteins M2-1 in Pneumoviridae and VP30 and VP24 in Filoviridae are essential for full processivity (44–48). The L protein has all the enzymatic activities necessary for the transcription of the viral mRNAs, including RNA polymerization, 5′ cap addition, cap methylation, and 3′ polyadenylation, as well as the replication of the viral genome (38, 49–57). Thus, L is the catalytic core of a multicomponent and multifunctional RNA synthesis machine.
The RNA polymerase is the sole enzyme of Mononegavirales, and there is a critical need to delineate the molecular and structural basis of the RNA polymerase of Mononegavirales (58). Since the first structure of the L protein alone of vesicular stomatitis virus (VSV) was determined in 2015 (59), multiple structures of RNA polymerases of Mononegavirales, including HRSV, HMPV, RABV, HPIV, and VSV, have been reported in recent months, revealing the architectures of L:P complexes and interactions between L and P (59–64). This review illustrates similarities and differences among the polymerases by comparing the structures of those polymerases and revealing the potential RNA synthesis mechanisms of the highly conserved Mononegavirales polymerases.
RNA SYNTHESIS OF MONONEGAVIRALES
Mononegavirales use the negative-sense genomes as the templates for the following two distinct viral RNA synthesis processes (1–3): (i) transcription to generate 5 to 10 discrete 5′ capped, methylated and 3′ polyadenylated viral mRNAs; and (ii) replication to produce complementary positive-sense antigenomes that act as templates for progeny negative-sense genomes (features highlighted in Fig. 1).
For Mononegavirales transcription, the RdRP initiates de novo RNA synthesis by recognizing a single promoter within the leader (Le) region at the 3′ end of the negative-sense genome and sequentially synthesizes mRNAs of the linear array of genes. The de novo initiation of the RNA synthesis by the RdRP typically involves a priming loop (65, 66). The RdRP first produces a Le RNA that remains uncapped and nonpolyadenylated. After the Le RNA synthesis and before transcription of the first gene, the Le RNA is released by the RdRP. The RdRP then stays on the template, initiates and caps the downstream mRNAs, and terminates and polyadenylates the upstream mRNAs, in response to the cis-acting gene-start (GS) and gene-end (GE) sequences of viral genes, respectively (33, 67–69). Typically, the RdRP produces a gradient level of viral mRNAs with the attenuation of the downstream mRNAs at each gene junction (70, 71). Recent studies showed the nongradient and genotype-dependent transcription in HRSV and EBOV, suggesting alternative gene expression strategies (72, 73) (Fig. 1, bottom part).
For replication in Mononegavirales, the RdRP initiates at the Le region of the genome and ignores all cis-acting regulatory signals to produce a full-length uncapped RNA antigenome. Consequently, the RdRP initiates at the 3′ end of the trailer complementary (TrC) region and replicates the positive-sense antigenome into its negative-sense genome (74). It is known that N protein levels influence the switch from transcription to replication. Unlike transcription, the replication is also dependent on a supply of N protein to encapsidate the nascent antigenome and its progeny genome (75, 76) (Fig. 1, top part).
THE MULTIFUNCTIONAL ENZYME MONOMERIC L
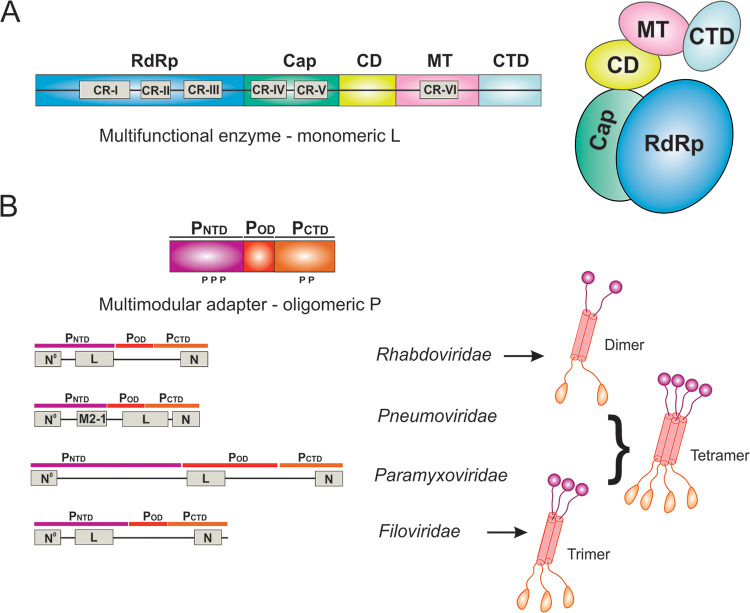
The multifunctional enzyme L protein of Mononegavirales is a single polypeptide of more than 2,000 amino acid residues (except Bornaviridae) long and is larger than 240 kDa in size. The sequence of L is conserved among Mononegavirales, and the sequence alignment reveals six conserved regions (CRs), named CR I to VI (77). The CRs are located within three distinct enzymatic domains of Mononegavirales L, namely, CR I to III in the RNA-dependent RNA polymerization (RdRp) domain, CR IV to V in the cap addition (Cap) domain, and CR VI in the cap methylation (MT) domain (53, 55, 78) (Fig. 2A).
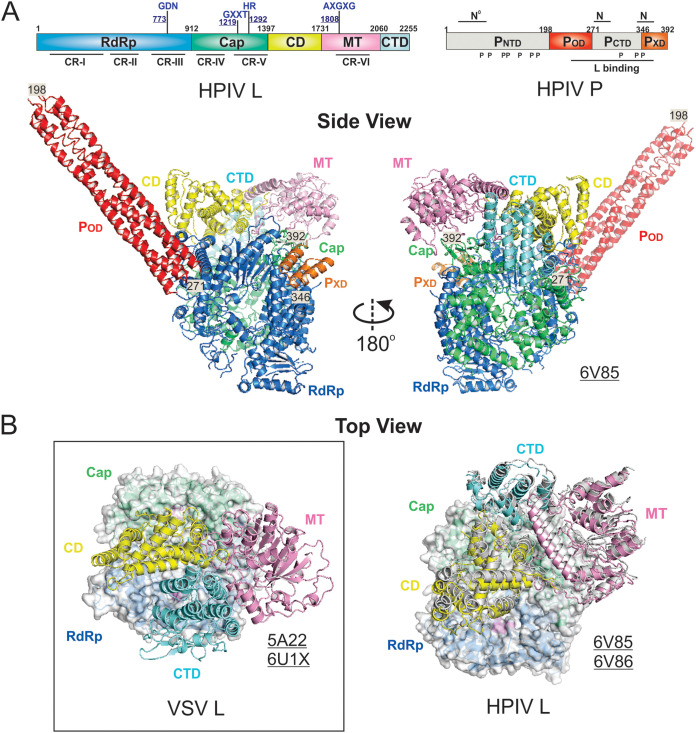
FIG 2.
The domain organization and architecture of L and P. (A) The domain organization and cartoon representation of the multifunctional enzyme monomeric L. The conserved regions (CRs) I to VI are labeled in gray boxes. The RNA-dependent RNA polymerization domain (RdRp), capping domain (Cap), connector domain (CD), methyltransferase domain (MT), and C-terminal domain (CTD) of L are colored in blue, green, yellow, pink, and cyan, respectively. (B) The domain organization and cartoon representation of the multimodular adapter oligomeric P. The intrinsically disordered N-terminal domain (PNTD), oligomerization domain (POD), and C-terminal domain (PCTD) are colored in magenta, red, and orange, respectively. The interaction regions with other viral proteins, including L, N, RNA-free N (N0), and accessory protein (M2-1), are labeled in gray boxes. The representative P oligomers are shown for the representative virus families Rhabdoviridae, Pneumoviridae, Paramyxoviridae, and Filoviridae.
Mononegavirales L contains all the catalytic activities necessary for RNA synthesis. The enzymatic activities of L are coordinated in such a way that the nascent mRNA transcript is synthesized and modified during multiple specific stages. A 5′ cap structure is formed after the mRNA transcript reaches a certain length, and failure to make a 5′ cap for mRNA results in the premature termination of RNA synthesis (53, 79, 80). Cap methylation also influences the RdRP activity, and failure to methylate the 5′ cap of mRNA results in hyperpolyadenylation of mRNA (55, 78, 81, 82). L also synthesizes a poly(A) tail at the 3′ end of mRNA by a “stuttering” mechanism using a short U-rich region within the GE sequence of each gene. Thus, the different enzymatic activities of L are linked. However, how the different activities of the L protein coordinate and influence one another remain mostly unclear.
THE MULTIMODULAR ADAPTER OLIGOMERIC P
The multimodular adapter P protein of Mononegavirales is an oligomeric and nonglobular molecule in solution (83). Although L contains all catalytic functions, P is the essential cofactor required for L to synthesize RNA effectively (38). P not only is the cofactor of L but also acts as an adapter to coordinate and modulate multiple proteins, including RNA-free N protein, NC complex, and additional regulatory proteins (84, 85). Notably, P forms dimers in Rhabdoviridae (83, 86, 87), trimers or tetramers in Filoviridae (88, 89), and tetramers in Paramyxoviridae and Pneumoviridae (90–94). Each P protomer consists of an intrinsically disordered N-terminal domain (PNTD), an oligomerization domain (POD), and a C-terminal domain (PCTD), connecting with a flexible linker (83). Despite a high diversity in length, sequence, and even in the structural folds of individual domains, this modular architecture is conserved among different Mononegavirales (Fig. 2B). The intrinsically disordered PNTD exhibits a substantial conformational heterogeneity and is essential for its dynamic coordination functions. The key features of P can be revealed as the modular architecture with intrinsically disordered domains and structural domains that interact with different proteins that constitute the RNA synthesis machine (95–99). Interestingly, the length difference seems to correlate with additional functions of the adapter P protein. For example, the linker between POD and PCTD of RABV is longer than that of VSV and contains a dynein light chain 8 (LC8) binding site (100); PCTD of EBOV contains an additional region for RNA binding and innate immune escape (101). Furthermore, P is often phosphorylated by the host kinases, and phosphorylation is essential for its regulation of RNA synthesis (102–107).
Together, this information suggests that P plays the following critical roles within the RNA synthesis machine: (i) P is an essential cofactor to regulate the processivity of L. As an adapter, P interacts with NC and bridge in the RNA to thread into the L active sites during transcription and replication (108–113). (ii) P acts as a chaperone to maintain a supply of RNA-free N (N0) and delivers to N0 nascent RNA genome or antigenome during replication (98, 99, 114–118). (iii) P interacts with other essential cofactors, such as M2-1 in Pneumoviridae and VP30 in Filoviridae, to coordinate the RNA synthesis activities of the RdRP (48, 119–122).
OVERVIEW OF THE STRUCTURAL ANALYSES OF THE MONONEGAVIRALES POLYMERASES
The monomeric L and oligomeric P together constitute the RdRP in Mononegavirales. Due to the large size of L and the oligomeric states of P with intrinsically flexible domains, it is challenging to obtain the crystals of the Mononegavirales RdRPs (123). The recent advance of cryo-electron microscopy (cryo-EM) offers an alternative way for a high-resolution structural characterization of such macromolecular complexes (124).
In 2015, the cryo-EM structure of the VSV L was determined at 3.8-Å resolution (PDB: 5A22) (125), and it was the first structure of the Mononegavirales polymerases. Although the VSV L was prepared in the complex of the VSV PNTD, the structure allowed only the de novo model building of the entire L protein but not the model assignment of PNTD, despite extra electron density observed (125). Since 2015, there have been many attempts for the structural characterizations of the Mononegavirales polymerases. For example, crystal structures of NTD and CTD fragments of L have also been reported (126, 127). In recent months, there were multiple successful cases of the structural characterization of the Rhabdoviridae and Pneumoviridae polymerases by cryo-EM, one for RABV (PDB: 6UEB), one for VSV (PNTD visible; PDB: 6U1X), two for HRSV (PDBs: 6PZK and 6UEN), one for HMPV (PDB: 6U5O), and one for HPIV (PDB: 6V85) (59–63). For consistency, the domain organizations and cartoon representations of the individual structures are colored as follows: RdRp (blue), Cap (green), CD (yellow), MT (pink), and CTD (cyan) for L; and PNTD (magenta), POD (red), and PCTD (orange) for P (the same as Fig. 2).
STRUCTURES OF THE RHABDOVIRIDAE POLYMERASES
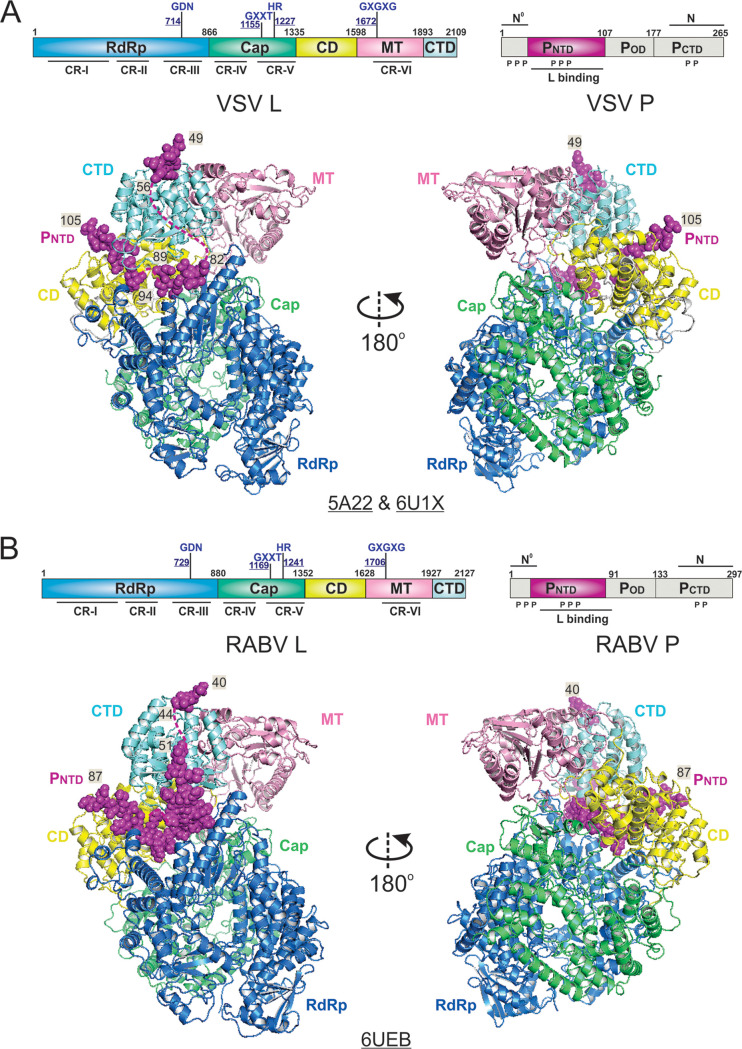
A higher 3.0-Å resolution cryo-EM structure of the VSV polymerase (PDB: 6U1X) was reported that enables the visualization of not only the 2,109-residue VSV L but also the bound PNTD of the 265-residue VSV P (59, 125) (Fig. 3A). The root mean square deviation (RMSD) between 3.8-Å and 3.0-Å structures of VSV L is 1.33 Å (59, 125). All five domains of the VSV L except a few flexible linkers are visible in the structure, including three functional domains, namely, RdRp (35 to 865), Cap (866 to 1334), and MT (1598 to 1892), and two structural domains, namely, the connector domain (CD; 1335 to 1597) and the C-terminal domain (CTD; 1893 to 2109) (59) (Fig. 3A). The RdRp domain resembles the classical RNA polymerase fold. The Cap domain folds next to the RdRp domain, and there was no homology for the Cap domain outside the order of Mononegavirales due to the unique capping mechanism. The CD domain connects the Cap and MT domains, and the CTD domain folds back to be close to the RdRp domain. The three ordered segments 49 to 56, 82 to 89, and 94 to 105 of PNTD (1 to 106) are shown to interact with CTD, RdRp, and CD domains of L, respectively (59, 125) (Fig. 3A).
FIG 3.
The cryo-EM structures of the Rhabdoviridae polymerases. (A) Linear domain representation of the L and P proteins of the vesicular stomatitis virus (VSV) polymerase. The cartoon view of 3.8-Å (PDB: 5A22) and 3.0-Å (PDB: 6U1X) cryo-EM structures of the VSV polymerase are shown. (B) Linear domain representation of the L and P proteins of the rabies virus (RABV) polymerase. The cartoon view of the 3.3-Å (PDB: 6UEB) cryo-EM structure of the RABV polymerase is shown. The RNA-dependent RNA polymerization domain (RdRp), capping domain (Cap), connector domain (CD), methyltransferase domain (MT), C-terminal domain (CTD) of L, and PNTD are colored in blue, green, yellow, pink, cyan, and magenta, respectively. The missing domains are colored in gray. The PNTD is highlighted as spheres, and the terminal residue numbers of the modeled P segments are indicated. The PDB accession codes are underlined.
The 3.3-Å resolution cryo-EM structure of the RABV polymerase closely resembles the VSV polymerase and contains all five domains of the 2,127-residue RABV L and PNTD of the 297-residue RABV P (60) (Fig. 3B). Similar to VSV L, nearly the entire RABV L can be modeled in the map, with a noticeable flexibility of several interdomain linkers. The RMSD between the RABV and VSV L is 2.10 Å. The domain boundaries are as follows: RdRp, 29 to 879; Cap, 880 to 1351; CD, 1352 to 1627; MT, 1628 to 1926; and CTD, 1927 to 2127. The following two segments of PNTD (1–90, 125) have been modeled in the structure of RABV polymerase: a short segment (possibly 40 to 44) interacts with the CTD domain of L; and another long segment (51–86, 125) bridges CTD, RdRp, and CD domains of L (60) (Fig. 3B).
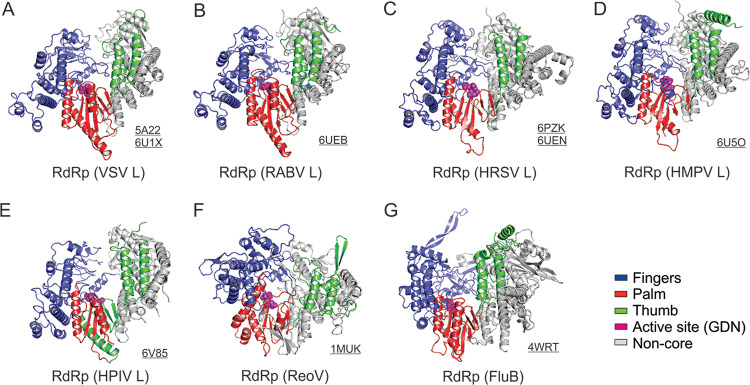
There are 35.05% and 19.22% amino acid identities between VSV and RABV L and P protein, respectively. As expected, VSV and RABV L share high similarity, with a nearly complete conservation of secondary structure elements throughout the protein. Despite having a greater sequence difference, VSV P and RABV P are also structurally similar to each other. Interestingly, there is a flexible loop (1158 to 1172 in VSV and 1171 to 1186 in RABV) in the Cap domain of Rhabdoviridae L that is against the active site of the RdRp domain. This loop is identified as the priming loop responsible for the de novo initiation of RNA synthesis (59, 60, 125). Due to the compact packing of the RdRp and Cap domains, the position of the priming loop appears to block the putative RNA product exit channel. Therefore, it is believed that Rhabdoviridae L adopts an initiation state in the structures, and significant rearrangements of those domains are likely to occur during elongation and other states of RNA synthesis.
STRUCTURES OF THE PNEUMOVIRIDAE POLYMERASES
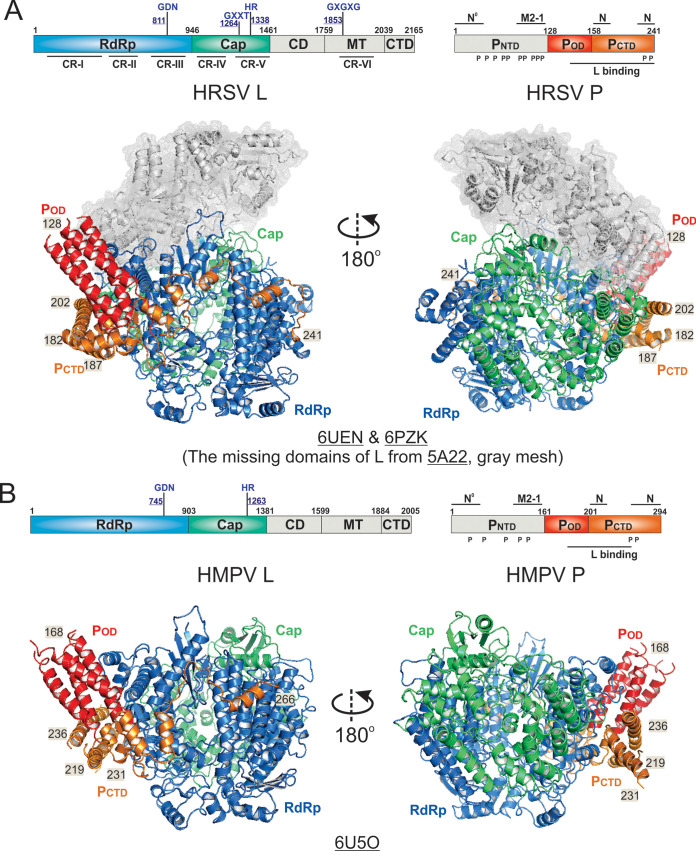
Multiple cryo-EM structures of the Pneumoviridae polymerases have also been reported in recent months, including a 3.2-Å (PDB: 6PZK) and a 3.67-Å (PDB: 6UEN) resolution structures of the HRSV polymerase and a 3.7-Å resolution structure of the HMPV polymerase (PDB: 6U5O) (61–63). Two structures of the HRSV polymerase are nearly identical, with an RMSD of 1.48 Å (61, 63) (Fig. 4A). The structures reveal that the RdRp (10 to 945) and Cap (946 to 1461) domains of the 2,165-residue L interact with the POD (128 to 157) and PCTD (158 to 241) of a tetramer of the 241-residue P. Interestingly, although full-length L and P were used to reconstitute the HRSV polymerases, the EM densities of MT domain and structural CD and CTD domains of the L and the PNTD are missing in 3-dimensional (3D) reconstructions (61, 63) (Fig. 4A, missing domains are shown in gray). The integrity of proteins was confirmed by mass spectrometry. The missing EM densities suggest that the intrinsic flexibility of those domains (61) and POD and PCTD are not sufficient to lock those domains of L into a homogenous conformation. Interestingly, four protomers of the tetrameric HRSV POD and PCTD adopt distinct conformation, and each of the promoters uses different ranges of residues, namely, 128 to 182, 128 to 187, 128 to 202, and 128 to 241, to interact with distinct regions of HRSV L (Fig. 4A). A further comparison of structures reveals slightly different intermolecular arrangements among L and tetrameric P, suggesting the plasticity of the L:P interface for structural rearrangements during RNA synthesis (61).
FIG 4.
The cryo-EM structures of the Pneumoviridae polymerases. (A) Linear domain representation of the L and P proteins of the human respiratory syncytial virus (HRSV) polymerase. The cartoon view of the 3.67-Å (PDB: 6UEN) and 3.2-Å (PDB: 6PZK) cryo-EM structures of HRSV polymerase complexes. The missing domains compared with the VSV L are shown in the gray meshes. (B) Linear domain representation of the L and P proteins of the human metapneumovirus (HMPV) polymerase. The cartoon view of the 3.7-Å (PDB: 6U5O) cryo-EM structure of the HMPV polymerase is shown. The domain colorings are the same as Fig. 2. The terminal residue numbers of the modeled POD and PCTD are indicated. The PDB accession codes are underlined.
The structure of the HMPV polymerase (PDB: 6U5O) shares a highly similar architecture to that of the HRSV polymerase, which contains the RdRp (8 to 902) and Cap (903 to 1380) domains of the 2,005-residue HMPV L and POD (168 to 193) and PCTD (194 to 266) of a tetramer of the 294-residue HMPV P (62). The RMSD between the HRSV and HMPV L is 1.49 Å. The HMPV polymerase also lacks the MT and other structural domains (CD and CTD) of L and PNTD in the 3D reconstructions (62) (Fig. 4B). Similarly, each of the four protomers of the tetrameric HMPV POD and PCTD adopts a distinct conformation and uses different ranges of residues, namely, 168 to 219, 168 to 231, 168 to 236, and 168 to 266, to interact with HMPV L (Fig. 4B).
There are high sequence identities between the HRSV and HMPV L and P, namely, 49.12%, and 37.18%, respectively. As expected, HRSV and HMPV polymerases share highly similar architectures between them, including the priming loop. Surprisingly, the priming loop in the Cap domain of the Pneumoviridae L shows a substantial shift and ∼37 Å away from the active sites of the RdRp domain, suggesting that L adopts an elongation state in the structures (61–63). Despite the similarities, there are several noticeable differences between the structures of HRSV and HMPV polymerases, as follows: (i) HRSV L contains an insertion (134 to 176) compared with that of HMPV L; (ii) HRSV L has a missing connecting helix (660 to 691), but the equivalent connecting helix of HMPV L can be partially modeled; (iii) one protomer of the HRSV P tetramers shows a different arrangement compared with its counterpart protomer of the HMPV P. Those slight differences between the two genera Metapneumovirus and Orthopneumovirus are likely due to genus-specific features of the RNA synthesis machine, and more detailed comparisons can be found in reference 128.
STRUCTURES OF THE PARAMYXOVIRIDAE POLYMERASES
Cryo-EM structures of the Paramyxoviridae polymerases have also been reported, including 4.38-Å (PDB: 6V85) and 4.63-Å (PDB: 6V86) resolution structures of the HPIV polymerase at two similar stable conformations (64). In the structure, all five domains of the 2,255-residue HPIV L are visible, including RdRp (1 to 912), Cap (913 to 1397), CD (1398 to 1730), MT (1731 to 2060), and CTD (2061 to 2255), but also, two domains of a tetramer of the 392-residue HPIV P, POD (198 to 271) and PCTD (also called PXD; 346 to 392), are present to interact with the HPIV L (Fig. 5A). Interestingly, although all five domains of HPIV L are presented, the CTD adopts a significant domain switch compared with that of the Rhabdoviridae L (Fig. 5B). The two conformations of the HPIV polymerase (L:P) are highly similar, with slightly different orientations of the CD-MT-CTD module with respect to RdRp and Cap (Fig. 5B, right panel). Furthermore, in contrast to Pneumoviridae P, only one protomer of PCTD EM-density is visible in Paramyxoviridae P, suggesting the versatile roles of P in RNA synthesis. It is noticeable that the tetrameric Paramyxoviridae POD is much longer than that of Rhabdoviridae and Pneumoviridae POD, highlighting the potential mechanistic differences among those families.
FIG 5.
The cryo-EM structure of the Paramyxoviridae polymerase. (A) Linear domain representation of the L and P proteins of the human parainfluenza virus (HPIV) polymerase. The side view of the ribbon diagram of the 4.3-Å (PDB: 6V85) cryo-EM structure of the HPIV polymerase complex. (B) The top view of the superimposed VSV L and HPIV L shows the domain switch of the CD-MT-CTD module. The superimposition is based on the RdRp (surface view), and CD, MT, and CTD are shown as the ribbon diagram. The domain colorings are the same as Fig. 2. The VSV L is shown in the left panel (box), and the HPIV L is shown in the right panel. The HPIV L (PDB: 6V85) is colored the same as A, and another stable conformation of the HPIV L (PDB: 6V86) is colored in gray. Note the significant location switch of CTD, facing down (VSV) versus facing up (HPIV L). The PDB accession codes are underlined.
STRUCTURAL SIMILARITIES AND DIFFERENCES AMONG THE MONONEGAVIRALES POLYMERASES
The L proteins of Rhabdoviridae, Pneumoviridae, and Paramyxoviridae have similar lengths (2,000 to 2,300 residues) and share a similar architecture. Indeed, the RdRp domains of Mononegavirales L share a standard right-hand thumb-palm-finger ring-like configuration of RNA and DNA polymerases. Comprehensive comparisons of the RNA/DNA polymerases and viral polymerases have been extensively reviewed elsewhere (129–136). The structural superimpositions of the motifs, namely, fingers, palm, thumb, and structural support, of the RdRp domains of the Mononegavirales L, are shown in blue, red, green, and gray, respectively. The active sites (GDN) of the RdRp domains are shown as magenta spheres (Fig. 6A to E). For comparison, we also showed the structural motifs of representative RdRps of reovirus (ReoV) and influenza B (FluB) (Fig. 6F and G), of which both are further discussed in the model section.
FIG 6.
Structural comparison of the RNA-dependent RNA polymerization (RdRp) domain. (A to E) The ribbon representations of the RdRp domain of the Rhabdoviridae (VSV and RABV), Pneumoviridae (HRSV and HMPV), and Paramyxoviridae (HPIV) L in conventional orientation. The structural motifs finger, palm, thumb, and support region are in blue, red, green, and gray, respectively. The tri-residues (GDN) of the RdRp active sites at a β-hairpin tip of the palm motif are shown in magenta spheres. (F and G) Similarities of the Mononegavirales RdRp domain to other viral polymerases. Structures of the polymerases of reovirus λ3 (ReoV; PDB: 1MUK) and influenza B (FluB; PDB: 4WRT) are shown as the same orientation and coloring scheme as in A. The PDB accession codes are underlined.
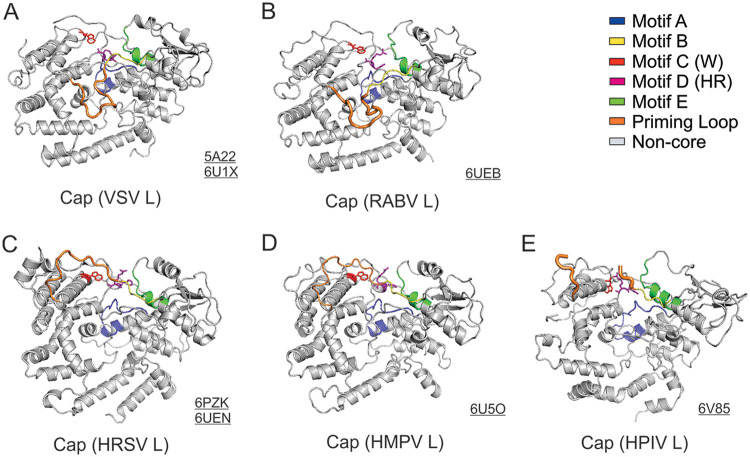
The previous studies highlighted the conserved structural motifs A to E of the Cap domain of L (33, 56, 137). Unlike the capping in the host cells, the capping reaction of the Mononegavirales L forms a covalent protein:RNA intermediate linkage between the 5′ of the RNA transcript and the active site H residue (motif D), followed by the attack by a guanosine nucleotide. The motifs A to E of the Cap domain of the Mononegavirales L are shown as a ribbon diagram in blue, yellow, red, magenta, and green, respectively. Those motifs are centered around the motif D (HR) active site. The proposed priming loops (orange) are next to the motif B (yellow) but exhibit a dramatic conformational rearrangement (Fig. 7).
FIG 7.
Structural comparison of the Cap domain. The motifs A to E of the Cap domain of the Rhabdoviridae (VSV and RABV), Pneumoviridae (HRSV and HMPV), and Paramyxoviridae (HPIV) L are shown as ribbon diagrams in blue, yellow, red, magenta, and green, respectively. Those motifs are centered around the active site motif D (HR). The proposed priming loop (orange) is next to motif B. The PDB accession codes are underlined.
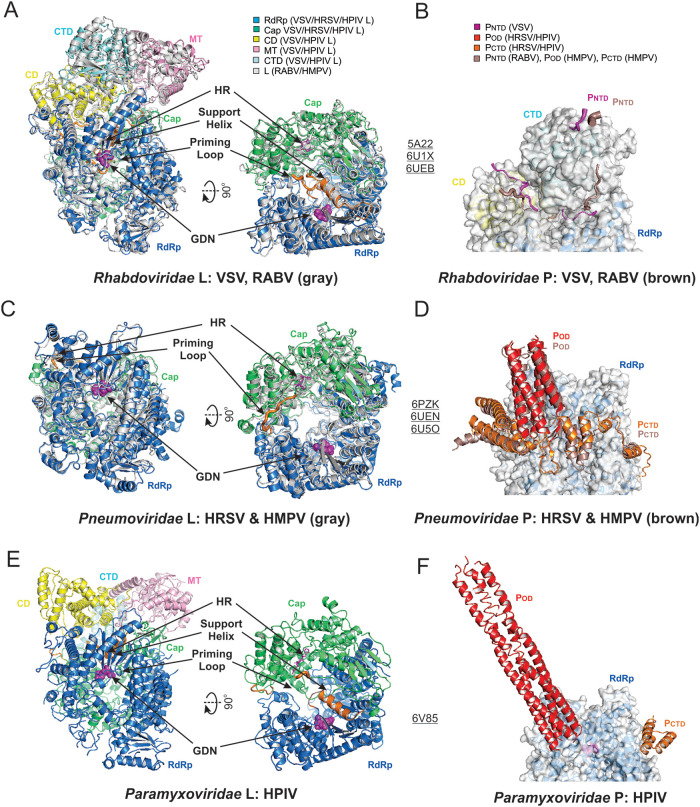
Despite the high similarities, there are several significant differences between the known structures of Mononegavirales polymerases. (i) All five domains (RdRp, Cap, CD, MT, and CTD) of the Rhabdoviridae and Paramyxoviridae L compared with only two domains (RdRp and Cap) of the Pneumoviridae L are visible in the cryo-EM structures. (ii) P forms dimers in Rhabdoviridae but tetramers in Pneumoviridae and Paramyxoviridae. It is thought that P displays distinct structural features due to low sequence identity and different oligomerization states. Interestingly, different domains of P interact with L in the reported structures. In Rhabdoviridae, only the PNTD interacts with mostly CD and CTD and part of RdRp of L (Fig. 8B). However, in Pneumoviridae and Paramyxoviridae, the POD and PCTD interact with the RdRp domain of L (59–63) (Fig. 8D and 8F). Compared with the oligomeric P shown in Pneumoviridae and Paramyxoviridae, the lack of the POD in Rhabdoviridae resulted in a monomeric P binding to L. (iii) The priming loop and the supporting helix of L (Fig. 8, colored in orange) adopt three different conformations, as follows: in Rhabdoviridae (VSV and RABV), the priming loop together with a supporting helix in the RdRp domain project into the GDN active sites (Fig. 8A) of the RdRp domain and close off a channel toward the Cap domain; in Pneumoviridae (HRSV and HMPV), the supporting helix is (partially) disordered, and the priming loop retracts from the RdRp active sites (Fig. 8C) and opens the channel connecting to the Cap domain; and in Paramyxoviridae (HPIV), the supporting helix is visible (similar as Rhabdoviridae), but the priming loop with a disordered tip is projected away from the RdRp active sites (similar as Pneumoviridae) (Fig. 8E).
FIG 8.
Structural comparisons of the Mononegavirales RNA polymerases. The active sites of the RdRp and Cap domains of L, GDN, and HR are shown in magenta spheres and sticks, respectively. The priming loops and supporting helix are colored in orange. (A) The structural superimposition of the Rhabdoviridae L. The VSV L is colored the same as Fig. 3A, and the RABV L is colored in gray. (B) The structural superimposition of the Rhabdoviridae P. The VSV P is colored in magenta the same as Fig. 3A, and the RABV P is colored in brown. Only the interacting domains RdRp, CD, and CTD of L are shown as surface. (C) The structural superimposition of the Pneumoviridae L. The HRSV L is colored the same as Fig. 4A, and the HMPV L is colored in gray. Note that the supporting helix is missing. (D) The superimposition of the Pneumoviridae P. The HRSV P is colored the same as Fig. 4A, and the HMPV P is colored in brown. Only the interacting domain RdRp of L is shown as surface. (E) The structural representation of the Paramyxoviridae L. The HPIV L is colored the same as Fig. 5A. (F) The location of the Paramyxoviridae P. The HPIV P is colored the same as Fig. 5A. Only the interacting domain RdRp of L is shown as surface. The PDB accession codes are underlined.
MECHANISMS AND MODELS OF MONOMEGAVIRALES RNA SYNTHESIS
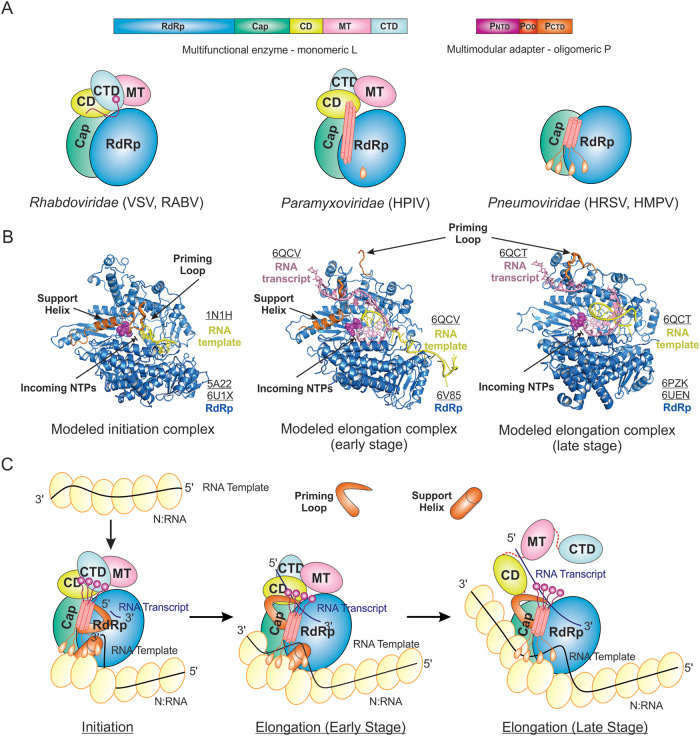
Collectively, the structures of the Mononegavirales polymerases discussed here reveal multiple distinct conformational arrangements of the L and P proteins, as shown in the cartoon diagrams (Fig. 9A). The comparison analyses suggest potential RNA synthesis mechanisms of Mononegavirales, switching of initiation, and elongation associated with priming loop and supporting helix rearrangements (59–63). Based on the structural similarities and differences among the Mononegavirales polymerases, we hypothesize that (i) the polymerases of the Rhabdoviridae (VSV and RABV) are likely at the initiation stage of genome replication, and (ii) the polymerases of Pneumoviridae (HRSV and HMPV) and Paramyxoviridae (HPIV) are at different phases, possibly late phase and early phase, of the elongation stages of transcription, respectively.
FIG 9.
Structural models of the Mononegavirales RNA synthesis. (A) The cartoon diagrams of recently reported structures of the Rhabdoviridae (VSV and RABV), Paramyxoviridae (HPIV), and Pneumoviridae (HRSV and HMPV) polymerases. The same color scheme as Fig. 2. (B) The modeled initiation and elongation complexes. The RdRp domain of the L proteins of Rhabdoviridae (VSV), Paramyxoviridae (HPIV), and Pneumoviridae (HRSV) with modeled RNA template from reovirus λ3 polymerase (PDB: 1N1H), FluB polymerase (PDB: 6QCV), and FluB polymerase (PDB: 6QCT), respectively. The same color scheme for the RdRp domain of Mononegavirales L. The priming loop (from the Cap domain) and the support helix (from the RdRp domain) are colored in orange. The modeled RNA template and RNA transcript are shown in yellow and pink, respectively. (C) The proposed cartoon models of the initiation and elongation stages on the nucleoprotein (N) encapsidated N:RNA (NC) template. Initiation, the priming loop and support helix are at the close approximate of the GDN active site of the RdRp domain of L; elongation (early stage), the priming loop is away from but the support helix stays at the close approximate to the active site of the RdRp domain of L; elongation (late stage), the priming loop is away from the active site of the RdRp domain of L, the support helix is missing, and the CD, MT, and CTD domains of L are disordered and linked by dashed lines. The nucleoprotein (N) protein is shown as the yellow oval. The RNA template, RNA transcript, and the flexible linker are shown in the black, blue, and red lines, respectively. The priming loop and support helix are shown as the thick orange bar and cylinder, respectively. The PDB accession codes are underlined.
To better understand the RNA synthesis mechanism by the Mononegavirales polymerases, we superimposed other viral polymerase complexes in the initiation and elongation stages. For the initiation, the superimposition of the reovirus (ReoV) λ3 initiation complex reveals in the presence of the RNA template (yellow), the initiating nucleotide stacks with a Trp (W1167 in VSV L and W1180 in RABV L) residue of the priming loop, which is also similar to the Y630 in hepatitis C virus (NS5B) (59, 60, 138, 139) (Fig. 9B, left panel). The mutation of this Trp residue severely affects the genome or antigenome end initiation but not internal initiation or capping (140). For the elongation, the polymerases require the retraction of the priming loop and possibly the support helix to pave the way to accommodate the product. Indeed, the fully retracted priming loop configurations are observed in both Pneumoviridae (HRSV and HMPV) and Paramyxoviridae (HPIV). The superimpositions of the influenza B (FluB) elongation complexes at early and later stages reveal that the RNA transcripts (pink) have sufficient space to extend and pass through a continuous tunnel when the priming loop is entirely retracted (141) (Fig. 9B, middle and right panels). The remaining support helix in Paramyxoviridae (HPIV) results in a partially extruded tunnel, where the missing support helix in Pneumoviridae (HRSV and HMPV) leads to a fully open tunnel, which is ideal for highly processive transcription.
As highlighted above, the NC is the cognate RNA template for Mononegavirales RNA synthesis. Based on the structures of Mononegavirales RNA polymerases, we propose the models of the initiation and early and late stage elongation of RNA synthesis, as shown in cartoon diagrams (Fig. 9C). The template RNA (black line) are coated by N at all times except when passing through the active sites of the RdRp domain of L. (i) At the initiation stage, the priming loop of the Cap domain is at the close approximate of the active site of the RdRp domain of L, and a short RNA transcript (blue line) is synthesized (Fig. 9C, left panel). (ii) At the early elongation stage, the priming loop is away from but the support helix stays at the close approximate to the active site of the RdRp domain of L (Fig. 9C, middle panel). (iii) At the late elongation stage, the priming loop of the Cap domain of L is away from the active site of the RdRp domain of L, and the CD, MT, and CTD domains of L are flexible when the RNA transcript (blue line) is being extended (Fig. 9C, right panel).
CONCLUSIONS
Many Mononegavirales are significant human pathogens, imposing a tremendous public threat and health care burden. However, no effective vaccines and antiviral therapeutics against many Mononegavirales are currently available (18–21, 23, 29, 142–148). Viral polymerases have been attractive and major antiviral therapeutic targets, as seen in multiple drug discovery successes in various viral pathogens, including HIV-1, hepatitis C virus (HCV), and hepatitis B virus (HBV) (149–157). Drug design and target search heavily rely on an accurate understanding of the structure and functions of the target molecules. Therefore, various viral polymerases have been extensively investigated for their structures and functions (129, 130). To understand the mechanistic insights of Mononegavirales RNA synthesis, the precise composition and structure of the Mononegavirales polymerases, how the different activities of the L protein influence one another, and how the cofactor regulates RNA synthesis need to be elucidated.
The structures of the Mononegavirales polymerases discussed here, including the L protein in complex with its cofactor P protein of VSV, RABV, HRSV, HMPV, and HPIV, reveal three conformations poised for initiation and elongation of RNA synthesis (59–63). The potential channels and the relative locations of multiple catalytic sites of L suggest that L coordinates a distinct capping and methyltransferase reaction with priming for de novo initiation of transcription. Transcription and replication might have different priming configurations and potential different product exit sites. The high similarity between L and P of the Mononegavirales polymerases provides a structural basis for the development of antiviral drugs that inhibit the RNA synthesis in transcription or replication.
This difference might also explain why L shows different architecture in three different families. PNTD is speculated to lock the CD, MT, and CTD domains into a closed conformation, which represents that L is poised for initiation at the 3′ end of the genome or antigenome and ready for RNA synthesis. The interactions between multiple domains of L and the PNTD reveal how P induces a compact, closed, and initiation-compatible state of L and how P positions the RNA template and the putative RNA product exit channel.
Several interesting questions arise by comparing and analyzing the known structures of the Mononegavirales polymerases. First, although the mass spectrometry data indicated that the Pneumoviridae L proteins used in structural studies are intact, the mystery of the missing MT domain and structural domains of L remains. Where do the MT and structural domains (CD and CTD) go? How do we capture the snapshots of their intermediates? Second, the known structures of the Mononegavirales polymerases are protein only without RNA present in the complex. However, those polymerases are in different initiation and elongation-compatible stages. Why do the priming loop and the supporting helix of L adopt different conformations in the protein-only complex? Third, the tetrameric P has a large interaction surface between POD and L in Pneumoviridae and Paramyxoviridae. Given that P is a dimer in Rhabdoviridae but a tetramer in Pneumoviridae and Paramyxoviridae, is it possible that the dimeric P in Rhabdoviridae may not form a tight complex with L with large interfaces? This may explain why the HRSV, HMPV, and HPIV L need to be coexpressed in the presence of P, but not VSV L, which can be expressed and purified alone.
From an evolutionary perspective, Mononegavirales have evolved to utilize a single multifunctional enzyme to transcribe individual genes (make, cap, and methylate the mRNAs) and replicate the entire genome without capping and methylation. This may be due to reduced evolutionary pressure; typically, this multifaceted process is sensitive to cell state and signaling inputs. These viruses have evolved to drive this process efficiently forward using minimal components. In eukaryotes, RNA transcription (copying the genetic information) is a delicate and complicated process involving many molecular machines, such as DNA-dependent RNA polymerases, capping enzymes, and methyltransferases. For example, the eukaryotic counterparts of the RdRp, Cap, and MT domains of the multifunctional enzyme L are (i) RNA polymerase II and polyadenylate polymerase, (ii) RNA triphosphatase and guanylyltransferase, and (iii) RNA methyltransferase, respectively (158–167). Additionally, Mononegavirales L also mimics the replication of the entire genome by accessing the N protein-coated RNA genome, similar to eukaryotic counterparts of DNA polymerases on the histone-assembled DNA genome (168–170).
The structural similarity of the Mononegavirales polymerases agrees with the relatively high sequence conservation. Nonetheless, the structural differences also highlight the virus- or genus-specific features. Collectively, the structures of the Mononegavirales polymerases provide significant advances into understanding the molecular architectures, interrelationship, the inhibitors, and the evolutionary implications of the Mononegavirales polymerases. Other polymerases from measles, mumps, Nipah virus, and Hendra virus in Paramyxoviridae and Ebola virus and Marburg virus in Filoviridae need to be determined for us to fully understand the similarities and differences of the polymerases in Mononegavirales. Furthermore, structures of Mononegavirales polymerases in complex with RNA templates, RNA products, or inhibitors are desired to appreciate the specific protein:RNA interactions and druggable sites.
FIGURE PREPARATION
All the figures presenting the structural models were generated using PyMOL (171).
ACKNOWLEDGMENTS
The research programs in the Liang laboratory at Emory are supported by the U.S. National Institute of General Medical Sciences (NIGMS), National Institutes of Health (NIH), under award number R01GM130950 and by the Research Start-Up Fund at Emory University School of Medicine.
I acknowledge the members of the Liang laboratory for helpful support and critical discussion.
I declare that I have no competing interests.
REFERENCES
- 1.Lamb RA. 2013. Mononegavirales In Knipe DM, Howley PM (ed), Fields virology, 6th ed, Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Whelan SP, Barr JN, Wertz GW. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol 283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- 3.Conzelmann KK. 1998. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu Rev Genet 32:123–162. doi: 10.1146/annurev.genet.32.1.123. [DOI] [PubMed] [Google Scholar]
- 4.Amarasinghe GK, Ayllón MA, Bào Y, Basler CF, Bavari S, Blasdell KR, Briese T, Brown PA, Bukreyev A, Balkema-Buschmann A, Buchholz UJ, Chabi-Jesus C, Chandran K, Chiapponi C, Crozier I, de Swart RL, Dietzgen RG, Dolnik O, Drexler JF, Dürrwald R, Dundon WG, Duprex WP, Dye JM, Easton AJ, Fooks AR, Formenty PBH, Fouchier RAM, Freitas-Astúa J, Griffiths A, Hewson R, Horie M, Hyndman TH, Jiāng D, Kitajima EW, Kobinger GP, Kondō H, Kurath G, Kuzmin IV, Lamb RA, Lavazza A, Lee B, Lelli D, Leroy EM, Lǐ J, Maes P, Marzano S-YL, Moreno A, Mühlberger E, Netesov SV, Nowotny N, Nylund A, et al. 2019. Taxonomy of the order Mononegavirales: update 2019. Arch Virol 164:1967–1980. doi: 10.1007/s00705-019-04247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Afonso CL, Amarasinghe GK, Bányai K, Bào Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand F-X, Briese T, Bukreyev A, Calisher CH, Chandran K, Chéng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Dürrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astúa J, Formenty P, Fouchier RAM, Fù Y, Ghedin E, Goodin MM, Hewson R, Horie M, Hyndman TH, Jiāng D, Kitajima EW, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lenardon S, Leroy EM, Li C-X, Lin X-D, Liú L, Longdon B, Marton S, Maisner A, Mühlberger E, Netesov SV, Nowotny N, et al. 2016. Taxonomy of the order Mononegavirales: update 2016. Arch Virol 161:2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hume AJ, Muhlberger E. 2019. Distinct genome replication and transcription strategies within the growing filovirus family. J Mol Biol 431:4290–4320. doi: 10.1016/j.jmb.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell CJ, Simoes EAF, Hurwitz JL. 2018. Vaccines for the paramyxoviruses and pneumoviruses: successes, candidates, and hurdles. Viral Immunol 31:133–141. doi: 10.1089/vim.2017.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffiths C, Drews SJ, Marchant DJ. 2017. Respiratory syncytial virus: infection, detection, and new options for prevention and treatment. Clin Microbiol Rev 30:277–319. doi: 10.1128/CMR.00010-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laksono BM, de Vries RD, McQuaid S, Duprex WP, de Swart RL. 2016. Measles virus host invasion and pathogenesis. Viruses 8:210. doi: 10.3390/v8080210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubin S, Eckhaus M, Rennick LJ, Bamford CG, Duprex WP. 2015. Molecular biology, pathogenesis and pathology of mumps virus. J Pathol 235:242–252. doi: 10.1002/path.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuster JE, Williams JV. 2014. Human Metapneumovirus. Microbiol Spectr 2:10.1128/microbiolspec.AID-0020-2014. doi: 10.1128/microbiolspec.AID-0020-2014. [DOI] [PubMed] [Google Scholar]
- 12.NIAID. 2018. NIAID Emerging infectious diseases/pathogens. www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens.
- 13.Singh RK, Dhama K, Chakraborty S, Tiwari R, Natesan S, Khandia R, Munjal A, Vora KS, Latheef SK, Karthik K, Singh Malik Y, Singh R, Chaicumpa W, Mourya DT. 2019. Nipah virus: epidemiology, pathology, immunobiology and advances in diagnosis, vaccine designing and control strategies—a comprehensive review. Vet Q 39:26–55. doi: 10.1080/01652176.2019.1580827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shifflett K, Marzi A. 2019. Marburg virus pathogenesis—differences and similarities in humans and animal models. Virol J 16:165. doi: 10.1186/s12985-019-1272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baseler L, Chertow DS, Johnson KM, Feldmann H, Morens DM. 2017. The pathogenesis of Ebola virus disease. Annu Rev Pathol 12:387–418. doi: 10.1146/annurev-pathol-052016-100506. [DOI] [PubMed] [Google Scholar]
- 16.Davis BM, Rall GF, Schnell MJ. 2015. Everything you always wanted to know about rabies virus (but were afraid to ask). Annu Rev Virol 2:451–471. doi: 10.1146/annurev-virology-100114-055157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Middleton D. 2014. Hendra virus. Vet Clin North Am Equine Pract 30:579–589. doi: 10.1016/j.cveq.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suschak JJ, Schmaljohn CS. 2019. Vaccines against Ebola virus and Marburg virus: recent advances and promising candidates. Hum Vaccin Immunother 15:2359–2377. doi: 10.1080/21645515.2019.1651140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denis M, Knezevic I, Wilde H, Hemachudha T, Briggs D, Knopf L. 2019. An overview of the immunogenicity and effectiveness of current human rabies vaccines administered by intradermal route. Vaccine 37:A99–A106. doi: 10.1016/j.vaccine.2018.11.072. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds P, Marzi A. 2017. Ebola and Marburg virus vaccines. Virus Genes 53:501–515. doi: 10.1007/s11262-017-1455-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satterfield BA, Dawes BE, Milligan GN. 2016. Status of vaccine research and development of vaccines for Nipah virus. Vaccine 34:2971–2975. doi: 10.1016/j.vaccine.2015.12.075. [DOI] [PubMed] [Google Scholar]
- 22.Neuzil KM. 2016. Progress toward a respiratory syncytial virus vaccine. Clin Vaccine Immunol 23:186–188. doi: 10.1128/CVI.00037-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broder CC, Xu K, Nikolov DB, Zhu Z, Dimitrov DS, Middleton D, Pallister J, Geisbert TW, Bossart KN, Wang LF. 2013. A treatment for and vaccine against the deadly Hendra and Nipah viruses. Antiviral Res 100:8–13. doi: 10.1016/j.antiviral.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivera CA, Gomez RS, Diaz RA, Cespedes PF, Espinoza JA, Gonzalez PA, Riedel CA, Bueno SM, Kalergis AM. 2015. Novel therapies and vaccines against the human respiratory syncytial virus. Expert Opin Invest Drugs 24:1613–1630. doi: 10.1517/13543784.2015.1099626. [DOI] [PubMed] [Google Scholar]
- 25.Cagno V, Andreozzi P, D'Alicarnasso M, Jacob Silva P, Mueller M, Galloux M, Le Goffic R, Jones ST, Vallino M, Hodek J, Weber J, Sen S, Janecek ER, Bekdemir A, Sanavio B, Martinelli C, Donalisio M, Rameix Welti MA, Eleouet JF, Han Y, Kaiser L, Vukovic L, Tapparel C, Kral P, Krol S, Lembo D, Stellacci F. 2018. Broad-spectrum non-toxic antiviral nanoparticles with a virucidal inhibition mechanism. Nat Mater 17:195–203. doi: 10.1038/nmat5053. [DOI] [PubMed] [Google Scholar]
- 26.Fearns R, Deval J. 2016. New antiviral approaches for respiratory syncytial virus and other mononegaviruses: inhibiting the RNA polymerase. Antiviral Res 134:63–76. doi: 10.1016/j.antiviral.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Cui R, Wang Y, Wang L, Li G, Lan K, Altmeyer R, Zou G. 2016. Cyclopiazonic acid, an inhibitor of calcium-dependent ATPases with antiviral activity against human respiratory syncytial virus. Antiviral Res 132:38–45. doi: 10.1016/j.antiviral.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Velkov T, Carbone V, Akter J, Sivanesan S, Li J, Beddoe T, Marsh GA. 2014. The RNA-dependent-RNA polymerase, an emerging antiviral drug target for the Hendra virus. Curr Drug Targets 15:103–113. doi: 10.2174/1389450114888131204163210. [DOI] [PubMed] [Google Scholar]
- 29.Elshabrawy HA, Fan J, Haddad CS, Ratia K, Broder CC, Caffrey M, Prabhakar BS. 2014. Identification of a broad-spectrum antiviral small molecule against severe acute respiratory syndrome coronavirus and Ebola, Hendra, and Nipah viruses by using a novel high-throughput screening assay. J Virol 88:4353–4365. doi: 10.1128/JVI.03050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bharat TA, Noda T, Riches JD, Kraehling V, Kolesnikova L, Becker S, Kawaoka Y, Briggs JA. 2012. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc Natl Acad Sci U S A 109:4275–4280. doi: 10.1073/pnas.1120453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liljeroos L, Huiskonen JT, Ora A, Susi P, Butcher SJ. 2011. Electron cryotomography of measles virus reveals how matrix protein coats the ribonucleocapsid within intact virions. Proc Natl Acad Sci U S A 108:18085–18090. doi: 10.1073/pnas.1105770108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge P, Tsao J, Schein S, Green TJ, Luo M, Zhou ZH. 2010. Cryo-EM model of the bullet-shaped vesicular stomatitis virus. Science 327:689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogino T, Green TJ. 2019. RNA synthesis and capping by non-segmented negative strand RNA viral polymerases: lessons from a prototypic virus. Front Microbiol 10:1490. doi: 10.3389/fmicb.2019.01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fearns R, Plemper RK. 2017. Polymerases of paramyxoviruses and pneumoviruses. Virus Res 234:87–102. doi: 10.1016/j.virusres.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins PL, Fearns R, Graham BS. 2013. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr Top Microbiol Immunol 372:3–38. doi: 10.1007/978-3-642-38919-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnheiter H, Davis NL, Wertz G, Schubert M, Lazzarini RA. 1985. Role of the nucleocapsid protein in regulating vesicular stomatitis virus RNA synthesis. Cell 41:259–267. doi: 10.1016/0092-8674(85)90079-0. [DOI] [PubMed] [Google Scholar]
- 37.Mazumder B, Barik S. 1994. Requirement of casein kinase II-mediated phosphorylation for the transcriptional activity of human respiratory syncytial viral phosphoprotein P: transdominant negative phenotype of phosphorylation-defective P mutants. Virology 205:104–111. doi: 10.1006/viro.1994.1624. [DOI] [PubMed] [Google Scholar]
- 38.Grosfeld H, Hill MG, Collins PL. 1995. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J Virol 69:5677–5686. doi: 10.1128/JVI.69.9.5677-5686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marriott AC, Wilson SD, Randhawa JS, Easton AJ. 1999. A single amino acid substitution in the phosphoprotein of respiratory syncytial virus confers thermosensitivity in a reconstituted RNA polymerase system. J Virol 73:5162–5165. doi: 10.1128/JVI.73.6.5162-5165.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horikami SM, Curran J, Kolakofsky D, Moyer SA. 1992. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J Virol 66:4901–4908. doi: 10.1128/JVI.66.8.4901-4908.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cowton VM, McGivern DR, Fearns R. 2006. Unravelling the complexities of respiratory syncytial virus RNA synthesis. J Gen Virol 87:1805–1821. doi: 10.1099/vir.0.81786-0. [DOI] [PubMed] [Google Scholar]
- 42.Qanungo KR, Shaji D, Mathur M, Banerjee AK. 2004. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc Natl Acad Sci U S A 101:5952–5957. doi: 10.1073/pnas.0401449101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munday DC, Wu W, Smith N, Fix J, Noton SL, Galloux M, Touzelet O, Armstrong SD, Dawson JM, Aljabr W, Easton AJ, Rameix-Welti MA, de Oliveira AP, Simabuco FM, Ventura AM, Hughes DJ, Barr JN, Fearns R, Digard P, Eleouet JF, Hiscox JA. 2015. Interactome analysis of the human respiratory syncytial virus RNA polymerase complex identifies protein chaperones as important cofactors that promote L-protein stability and RNA synthesis. J Virol 89:917–930. doi: 10.1128/JVI.01783-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fearns R, Collins PL. 1999. Role of the M2–1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J Virol 73:5852–5864. doi: 10.1128/JVI.73.7.5852-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutherland KA, Collins PL, Peeples ME. 2001. Synergistic effects of gene-end signal mutations and the M2–1 protein on transcription termination by respiratory syncytial virus. Virology 288:295–307. doi: 10.1006/viro.2001.1105. [DOI] [PubMed] [Google Scholar]
- 46.Mason SW, Aberg E, Lawetz C, DeLong R, Whitehead P, Liuzzi M. 2003. Interaction between human respiratory syncytial virus (RSV) M2–1 and P proteins is required for reconstitution of M2–1-dependent RSV minigenome activity. J Virol 77:10670–10676. doi: 10.1128/jvi.77.19.10670-10676.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoenen T, Jung S, Herwig A, Groseth A, Becker S. 2010. Both matrix proteins of Ebola virus contribute to the regulation of viral genome replication and transcription. Virology 403:56–66. doi: 10.1016/j.virol.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Kirchdoerfer RN, Moyer CL, Abelson DM, Saphire EO. 2016. The Ebola virus VP30-NP interaction is a regulator of viral RNA synthesis. PLoS Pathog 12:e1005937. doi: 10.1371/journal.ppat.1005937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferron F, Longhi S, Henrissat B, Canard B. 2002. Viral RNA-polymerases—a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem Sci 27:222–224. doi: 10.1016/s0968-0004(02)02091-1. [DOI] [PubMed] [Google Scholar]
- 50.Sleat DE, Banerjee AK. 1993. Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J Virol 67:1334–1339. doi: 10.1128/JVI.67.3.1334-1339.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grdzelishvili VZ, Smallwood S, Tower D, Hall RL, Hunt DM, Moyer SA. 2005. A single amino acid change in the L-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J Virol 79:7327–7337. doi: 10.1128/JVI.79.12.7327-7337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hercyk N, Horikami SM, Moyer SA. 1988. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology 163:222–225. doi: 10.1016/0042-6822(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 53.Ogino T, Banerjee AK. 2007. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell 25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 54.Li J, Fontaine-Rodriguez EC, Whelan SP. 2005. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J Virol 79:13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J, Wang JT, Whelan SP. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc Natl Acad Sci U S A 103:8493–8498. doi: 10.1073/pnas.0509821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Rahmeh A, Morelli M, Whelan SP. 2008. A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J Virol 82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hunt DM, Mehta R, Hutchinson KL. 1988. The L protein of vesicular stomatitis virus modulates the response of the polyadenylic acid polymerase to S-adenosylhomocysteine. J Gen Virol 69:2555–2561. doi: 10.1099/0022-1317-69-10-2555. [DOI] [PubMed] [Google Scholar]
- 58.Morin B, Kranzusch PJ, Rahmeh AA, Whelan SP. 2013. The polymerase of negative-stranded RNA viruses. Curr Opin Virol 3:103–110. doi: 10.1016/j.coviro.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jenni S, Bloyet LM, Diaz-Avalos R, Liang B, Whelan SPJ, Grigorieff N, Harrison SC. 2020. Structure of the vesicular stomatitis virus L protein in complex with its phosphoprotein cofactor. Cell Rep 30:53–60.e5. doi: 10.1016/j.celrep.2019.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horwitz JA, Jenni S, Harrison SC, Whelan SPJ. 2020. Structure of a rabies virus polymerase complex from electron cryo-microscopy. Proc Natl Acad Sci U S A 117:2099–2107. doi: 10.1073/pnas.1918809117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cao D, Gao Y, Roesler C, Rice S, D'Cunha P, Zhuang L, Slack J, Domke M, Antonova A, Romanelli S, Keating S, Forero G, Juneja P, Liang B. 2020. Cryo-EM structure of the respiratory syncytial virus RNA polymerase. Nat Commun 11:368. doi: 10.1038/s41467-019-14246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan J, Qian X, Lattmann S, El Sahili A, Yeo TH, Jia H, Cressey T, Ludeke B, Noton S, Kalocsay M, Fearns R, Lescar J. 2020. Structure of the human metapneumovirus polymerase phosphoprotein complex. Nature 577:275–279. doi: 10.1038/s41586-019-1759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gilman MSA, Liu C, Fung A, Behera I, Jordan P, Rigaux P, Ysebaert N, Tcherniuk S, Sourimant J, Eleouet JF, Sutto-Ortiz P, Decroly E, Roymans D, Jin Z, McLellan JS. 2019. Structure of the respiratory syncytial virus polymerase complex. Cell 179:193–204.e14. doi: 10.1016/j.cell.2019.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abdella R, Aggarwal M, Okura T, Lamb RA, He Y. 2020. Structure of a paramyxovirus polymerase complex reveals a unique methyltransferase-CTD conformation. Proc Natl Acad Sci U S A 117:4931–4941. doi: 10.1073/pnas.1919837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kao CC, Singh P, Ecker DJ. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251–260. doi: 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- 66.Malet H, Masse N, Selisko B, Romette JL, Alvarez K, Guillemot JC, Tolou H, Yap TL, Vasudevan S, Lescar J, Canard B. 2008. The flavivirus polymerase as a target for drug discovery. Antiviral Res 80:23–35. doi: 10.1016/j.antiviral.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 67.Kuo L, Fearns R, Collins PL. 1997. Analysis of the gene start and gene end signals of human respiratory syncytial virus: quasi-templated initiation at position 1 of the encoded mRNA. J Virol 71:4944–4953. doi: 10.1128/JVI.71.7.4944-4953.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuo L, Grosfeld H, Cristina J, Hill MG, Collins PL. 1996. Effects of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J Virol 70:6892–6901. doi: 10.1128/JVI.70.10.6892-6901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jordan PC, Liu C, Raynaud P, Lo MK, Spiropoulou CF, Symons JA, Beigelman L, Deval J. 2018. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog 14:e1006889. doi: 10.1371/journal.ppat.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mink MA, Stec DS, Collins PL. 1991. Nucleotide sequences of the 3' leader and 5' trailer regions of human respiratory syncytial virus genomic RNA. Virology 185:615–624. doi: 10.1016/0042-6822(91)90532-g. [DOI] [PubMed] [Google Scholar]
- 71.Iverson LE, Rose JK. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 72.Piedra FA, Qiu X, Teng MN, Avadhanula V, Machado AA, Kim DK, Hixson J, Bahl J, Piedra PA. 2020. Non-gradient and genotype-dependent patterns of RSV gene expression. PLoS One 15:e0227558. doi: 10.1371/journal.pone.0227558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pagan I, Holmes EC, Simon-Loriere E. 2012. Level of gene expression is a major determinant of protein evolution in the viral order Mononegavirales. J Virol 86:5253–5263. doi: 10.1128/JVI.06050-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fearns R, Collins PL, Peeples ME. 2000. Functional analysis of the genomic and antigenomic promoters of human respiratory syncytial virus. J Virol 74:6006–6014. doi: 10.1128/jvi.74.13.6006-6014.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fearns R, Peeples ME, Collins PL. 1997. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology 236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 76.Noton SL, Tremaglio CZ, Fearns R. 2019. Killing two birds with one stone: how the respiratory syncytial virus polymerase initiates transcription and replication. PLoS Pathog 15:e1007548. doi: 10.1371/journal.ppat.1007548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poch O, Blumberg BM, Bougueleret L, Tordo N. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol 71:1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- 78.Rahmeh AA, Li J, Kranzusch PJ, Whelan SP. 2009. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J Virol 83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tekes G, Rahmeh AA, Whelan SP. 2011. A freeze frame view of vesicular stomatitis virus transcription defines a minimal length of RNA for 5′ processing. PLoS Pathog 7:e1002073. doi: 10.1371/journal.ppat.1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang JT, McElvain LE, Whelan SP. 2007. Vesicular stomatitis virus mRNA capping machinery requires specific cis-acting signals in the RNA. J Virol 81:11499–11506. doi: 10.1128/JVI.01057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li J, Chorba JS, Whelan SP. 2007. Vesicular stomatitis viruses resistant to the methylase inhibitor sinefungin upregulate RNA synthesis and reveal mutations that affect mRNA cap methylation. J Virol 81:4104–4115. doi: 10.1128/JVI.02681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li J, Rahmeh A, Brusic V, Whelan SP. 2009. Opposing effects of inhibiting cap addition and cap methylation on polyadenylation during vesicular stomatitis virus mRNA synthesis. J Virol 83:1930–1940. doi: 10.1128/JVI.02162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gerard FC, Ribeiro Ede A Jr, Albertini AA, Gutsche I, Zaccai G, Ruigrok RW, Jamin M. 2007. Unphosphorylated rhabdoviridae phosphoproteins form elongated dimers in solution. Biochemistry 46:10328–10338. doi: 10.1021/bi7007799. [DOI] [PubMed] [Google Scholar]
- 84.Das SC, Pattnaik AK. 2005. Role of the hypervariable hinge region of phosphoprotein P of vesicular stomatitis virus in viral RNA synthesis and assembly of infectious virus particles. J Virol 79:8101–8112. doi: 10.1128/JVI.79.13.8101-8112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takacs AM, Barik S, Das T, Banerjee AK. 1992. Phosphorylation of specific serine residues within the acidic domain of the phosphoprotein of vesicular stomatitis virus regulates transcription in vitro. J Virol 66:5842–5848. doi: 10.1128/JVI.66.10.5842-5848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ivanov I, Crépin T, Jamin M, Ruigrok RW. 2010. Structure of the dimerization domain of the rabies virus phosphoprotein. J Virol 84:3707–3710. doi: 10.1128/JVI.02557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ding H, Green TJ, Lu S, Luo M. 2006. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J Virol 80:2808–2814. doi: 10.1128/JVI.80.6.2808-2814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bruhn JF, Kirchdoerfer RN, Urata SM, Li S, Tickle IJ, Bricogne G, Saphire EO. 2017. Crystal structure of the Marburg virus VP35 oligomerization domain. J Virol 91:e01085-16. doi: 10.1128/JVI.01085-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zinzula L, Nagy I, Orsini M, Weyher-Stingl E, Bracher A, Baumeister W. 2019. Structures of Ebola and Reston virus VP35 oligomerization domains and comparative biophysical characterization in all Ebolavirus species. Structure 27:39–54.e6. doi: 10.1016/j.str.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 90.Bruhn JF, Barnett KC, Bibby J, Thomas JM, Keegan RM, Rigden DJ, Bornholdt ZA, Saphire EO. 2014. Crystal structure of the nipah virus phosphoprotein tetramerization domain. J Virol 88:758–762. doi: 10.1128/JVI.02294-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Communie G, Crepin T, Maurin D, Jensen MR, Blackledge M, Ruigrok RW. 2013. Structure of the tetramerization domain of measles virus phosphoprotein. J Virol 87:7166–7169. doi: 10.1128/JVI.00487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cox R, Green TJ, Purushotham S, Deivanayagam C, Bedwell GJ, Prevelige PE, Luo M. 2013. Structural and functional characterization of the mumps virus phosphoprotein. J Virol 87:7558–7568. doi: 10.1128/JVI.00653-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tarbouriech N, Curran J, Ruigrok RW, Burmeister WP. 2000. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat Struct Biol 7:777–781. doi: 10.1038/79013. [DOI] [PubMed] [Google Scholar]
- 94.Leyrat C, Renner M, Harlos K, Grimes JM. 2013. Solution and crystallographic structures of the central region of the phosphoprotein from human metapneumovirus. PLoS One 8:e80371. doi: 10.1371/journal.pone.0080371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gerard FC, Ribeiro Ede A Jr, Leyrat C, Ivanov I, Blondel D, Longhi S, Ruigrok RW, Jamin M. 2009. Modular organization of rabies virus phosphoprotein. J Mol Biol 388:978–996. doi: 10.1016/j.jmb.2009.03.061. [DOI] [PubMed] [Google Scholar]
- 96.Habchi J, Mamelli L, Darbon H, Longhi S. 2010. Structural disorder within Henipavirus nucleoprotein and phosphoprotein: from predictions to experimental assessment. PLoS One 5:e11684. doi: 10.1371/journal.pone.0011684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Karlin D, Belshaw R. 2012. Detecting remote sequence homology in disordered proteins: discovery of conserved motifs in the N-termini of Mononegavirales phosphoproteins. PLoS One 7:e31719. doi: 10.1371/journal.pone.0031719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kirchdoerfer RN, Abelson DM, Li S, Wood MR, Saphire EO. 2015. Assembly of the Ebola virus nucleoprotein from a chaperoned VP35 complex. Cell Rep 12:140–149. doi: 10.1016/j.celrep.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leung DW, Borek D, Luthra P, Binning JM, Anantpadma M, Liu G, Harvey IB, Su Z, Endlich-Frazier A, Pan J, Shabman RS, Chiu W, Davey RA, Otwinowski Z, Basler CF, Amarasinghe GK. 2015. An intrinsically disordered peptide from Ebola virus VP35 controls viral RNA synthesis by modulating nucleoprotein-RNA interactions. Cell Rep 11:376–389. doi: 10.1016/j.celrep.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Raux H, Flamand A, Blondel D. 2000. Interaction of the rabies virus P protein with the LC8 dynein light chain. J Virol 74:10212–10216. doi: 10.1128/jvi.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Leung DW, Ginder ND, Fulton DB, Nix J, Basler CF, Honzatko RB, Amarasinghe GK. 2009. Structure of the Ebola VP35 interferon inhibitory domain. Proc Natl Acad Sci U S A 106:411–416. doi: 10.1073/pnas.0807854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barik S, Banerjee AK. 1992. Phosphorylation by cellular casein kinase II is essential for transcriptional activity of vesicular stomatitis virus phosphoprotein P. Proc Natl Acad Sci U S A 89:6570–6574. doi: 10.1073/pnas.89.14.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Briggs K, Wang L, Nagashima K, Zengel J, Tripp RA, He B. 2020. Regulation of mumps virus replication and transcription by kinase RPS6KB1. J Virol 94:e00387-20. doi: 10.1128/JVI.00387-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huntley CC, De BP, Banerjee AK. 1997. Phosphorylation of Sendai virus phosphoprotein by cellular protein kinase C zeta. J Biol Chem 272:16578–16584. doi: 10.1074/jbc.272.26.16578. [DOI] [PubMed] [Google Scholar]
- 105.Lenard J. 1999. Host cell protein kinases in nonsegmented negative-strand virus (mononegavirales) infection. Pharmacol Ther 83:39–48. doi: 10.1016/s0163-7258(99)00016-9. [DOI] [PubMed] [Google Scholar]
- 106.Schwemmle M, De B, Shi L, Banerjee A, Lipkin WI. 1997. Borna disease virus P-protein is phosphorylated by protein kinase Cepsilon and casein kinase II. J Biol Chem 272:21818–21823. doi: 10.1074/jbc.272.35.21818. [DOI] [PubMed] [Google Scholar]
- 107.Sun M, Fuentes SM, Timani K, Sun D, Murphy C, Lin Y, August A, Teng MN, He B. 2008. Akt plays a critical role in replication of nonsegmented negative-stranded RNA viruses. J Virol 82:105–114. doi: 10.1128/JVI.01520-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Emerson SU, Yu Y. 1975. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol 15:1348–1356. doi: 10.1128/JVI.15.6.1348-1356.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rahmeh AA, Morin B, Schenk AD, Liang B, Heinrich BS, Brusic V, Walz T, Whelan SP. 2012. Critical phosphoprotein elements that regulate polymerase architecture and function in vesicular stomatitis virus. Proc Natl Acad Sci U S A 109:14628–14633. doi: 10.1073/pnas.1209147109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sourimant J, Rameix-Welti MA, Gaillard AL, Chevret D, Galloux M, Gault E, Eleouet JF. 2015. Fine mapping and characterization of the L-polymerase-binding domain of the respiratory syncytial virus phosphoprotein. J Virol 89:4421–4433. doi: 10.1128/JVI.03619-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ribeiro Ede A Jr, Leyrat C, Gerard FC, Albertini AA, Falk C, Ruigrok RW, Jamin M. 2009. Binding of rabies virus polymerase cofactor to recombinant circular nucleoprotein-RNA complexes. J Mol Biol 394:558–575. doi: 10.1016/j.jmb.2009.09.042. [DOI] [PubMed] [Google Scholar]
- 112.Green TJ, Luo M. 2009. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc Natl Acad Sci U S A 106:11713–11718. doi: 10.1073/pnas.0903228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Galloux M, Tarus B, Blazevic I, Fix J, Duquerroy S, Eléouët JF. 2012. Characterization of a viral phosphoprotein binding site on the surface of the respiratory syncytial nucleoprotein. J Virol 86:8375–8387. doi: 10.1128/JVI.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Galloux M, Gabiane G, Sourimant J, Richard CA, England P, Moudjou M, Aumont-Nicaise M, Fix J, Rameix-Welti MA, Eléouët JF. 2015. Identification and characterization of the binding site of the respiratory syncytial virus phosphoprotein to RNA-free nucleoprotein. J Virol 89:3484–3496. doi: 10.1128/JVI.03666-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yabukarski F, Leyrat C, Martinez N, Communie G, Ivanov I, Ribeiro EA Jr, Buisson M, Gerard FC, Bourhis JM, Jensen MR, Bernado P, Blackledge M, Jamin M. 2016. Ensemble structure of the highly flexible complex formed between vesicular stomatitis virus unassembled nucleoprotein and its phosphoprotein chaperone. J Mol Biol 428:2671–2694. doi: 10.1016/j.jmb.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 116.Renner M, Bertinelli M, Leyrat C, Paesen GC, Saraiva de Oliveira LF, Huiskonen JT, Grimes JM. 2016. Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein. Elife 5:e12627. doi: 10.7554/eLife.12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guryanov SG, Liljeroos L, Kasaragod P, Kajander T, Butcher SJ. 2015. Crystal structure of the measles virus nucleoprotein core in complex with an N-terminal region of phosphoprotein. J Virol 90:2849–2857. doi: 10.1128/JVI.02865-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Leyrat C, Yabukarski F, Tarbouriech N, Ribeiro EA Jr, Jensen MR, Blackledge M, Ruigrok RW, Jamin M. 2011. Structure of the vesicular stomatitis virus N(0)-P complex. PLoS Pathog 7:e1002248. doi: 10.1371/journal.ppat.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Selvaraj M, Yegambaram K, Todd E, Richard CA, Dods RL, Pangratiou GM, Trinh CH, Moul SL, Murphy JC, Mankouri J, Eleouet JF, Barr JN, Edwards TA. 2018. The structure of the human respiratory syncytial virus M2–1 protein bound to the interaction domain of the phosphoprotein p defines the orientation of the complex. mBio 9:e01554-18. doi: 10.1128/mBio.01554-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tanner SJ, Ariza A, Richard CA, Kyle HF, Dods RL, Blondot ML, Wu W, Trincao J, Trinh CH, Hiscox JA, Carroll MW, Silman NJ, Eleouet JF, Edwards TA, Barr JN. 2014. Crystal structure of the essential transcription antiterminator M2–1 protein of human respiratory syncytial virus and implications of its phosphorylation. Proc Natl Acad Sci U S A 111:1580–1585. doi: 10.1073/pnas.1317262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Leyrat C, Renner M, Harlos K, Huiskonen JT, Grimes JM. 2014. Drastic changes in conformational dynamics of the antiterminator M2–1 regulate transcription efficiency in Pneumovirinae. Elife 3:e02674. doi: 10.7554/eLife.02674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Watanabe S, Noda T, Halfmann P, Jasenosky L, Kawaoka Y. 2007. Ebola virus (EBOV) VP24 inhibits transcription and replication of the EBOV genome. J Infect Dis 196:S284–S290. doi: 10.1086/520582. [DOI] [PubMed] [Google Scholar]
- 123.Grimes JM, Hall DR, Ashton AW, Evans G, Owen RL, Wagner A, McAuley KE, von Delft F, Orville AM, Sorensen T, Walsh MA, Ginn HM, Stuart DI. 2018. Where is crystallography going? Acta Crystallogr D Struct Biol 74:152–166. doi: 10.1107/S2059798317016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cheng Y. 2018. Single-particle cryo-EM—how did it get here and where will it go. Science 361:876–880. doi: 10.1126/science.aat4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SPJ. 2015. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qiu S, Ogino M, Luo M, Ogino T, Green TJ. 2016. Structure and function of the N-terminal domain of the vesicular stomatitis virus RNA polymerase. J Virol 90:715–724. doi: 10.1128/JVI.02317-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Paesen GC, Collet A, Sallamand C, Debart F, Vasseur JJ, Canard B, Decroly E, Grimes JM. 2015. X-ray structure and activities of an essential Mononegavirales L-protein domain. Nat Commun 6:8749. doi: 10.1038/ncomms9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cao D, Liang B. 2020. Cryo-electron microscopy structures of the Pneumoviridae polymerases. Viral Immunol doi: 10.1089/vim.2020.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Peersen OB. 2019. A comprehensive superposition of viral polymerase structures. Viruses 11:745. doi: 10.3390/v11080745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sauguet L. 2019. The extended “two-barrel” polymerases superfamily: structure, function and evolution. J Mol Biol 431:4167–4183. doi: 10.1016/j.jmb.2019.05.017. [DOI] [PubMed] [Google Scholar]
- 131.Ferrero D, Ferrer-Orta C, Verdaguer N. 2018. Viral RNA-dependent RNA polymerases: a structural overview. Subcell Biochem 88:39–71. doi: 10.1007/978-981-10-8456-0_3. [DOI] [PubMed] [Google Scholar]
- 132.Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N. 2006. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol 16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 133.Venkataraman S, Prasad B, Selvarajan R. 2018. RNA dependent RNA polymerases: insights from structure, function and evolution. Viruses 10:76. doi: 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wu J, Liu W, Gong P. 2015. A structural overview of RNA-dependent RNA polymerases from the Flaviviridae family. Int J Mol Sci 16:12943–12957. doi: 10.3390/ijms160612943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Te Velthuis AJW. 2014. Common and unique features of viral RNA-dependent polymerases. Cell Mol Life Sci 71:4403–4420. doi: 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Choi KH, Rossmann MG. 2009. RNA-dependent RNA polymerases from Flaviviridae. Curr Opin Struct Biol 19:746–751. doi: 10.1016/j.sbi.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 137.Neubauer J, Ogino M, Green TJ, Ogino T. 2016. Signature motifs of GDP polyribonucleotidyltransferase, a non-segmented negative strand RNA viral mRNA capping enzyme, domain in the L protein are required for covalent enzyme-pRNA intermediate formation. Nucleic Acids Res 44:330–341. doi: 10.1093/nar/gkv1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tao Y, Farsetta DL, Nibert ML, Harrison SC. 2002. RNA synthesis in a cage–structural studies of reovirus polymerase lambda3. Cell 111:733–745. doi: 10.1016/S0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 139.Appleby TC, Perry JK, Murakami E, Barauskas O, Feng J, Cho A, Fox D III, Wetmore DR, McGrath ME, Ray AS, Sofia MJ, Swaminathan S, Edwards TE. 2015. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science 347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 140.Ogino M, Gupta N, Green TJ, Ogino T. 2019. A dual-functional priming-capping loop of rhabdoviral RNA polymerases directs terminal de novo initiation and capping intermediate formation. Nucleic Acids Res 47:299–309. doi: 10.1093/nar/gky1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kouba T, Drncova P, Cusack S. 2019. Structural snapshots of actively transcribing influenza polymerase. Nat Struct Mol Biol 26:460–470. doi: 10.1038/s41594-019-0232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dhama K, Karthik K, Khandia R, Chakraborty S, Munjal A, Latheef SK, Kumar D, Ramakrishnan MA, Malik YS, Singh R, Malik SVS, Singh RK, Chaicumpa W. 2018. Advances in designing and developing vaccines, drugs, and therapies to counter Ebola virus. Front Immunol 9:1803. doi: 10.3389/fimmu.2018.01803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Latorre V, Mattenberger F, Geller R. 2018. Chaperoning the Mononegavirales: current knowledge and future directions. Viruses 10:699. doi: 10.3390/v10120699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lu DY, Wu HY, Yarla NS, Lu TR, Xu B, Jian D. 2019. Ebola therapeutic study and future directions. Infect Disord Drug Targets 19:17–29. doi: 10.2174/1871526518666180813160348. [DOI] [PubMed] [Google Scholar]
- 145.Pfaller CK, Cattaneo R, Schnell MJ. 2015. Reverse genetics of Mononegavirales: how they work, new vaccines, and new cancer therapeutics. Virology 479–480:331–344. doi: 10.1016/j.virol.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Venkatraman N, Silman D, Folegatti PM, Hill AVS. 2018. Vaccines against Ebola virus. Vaccine 36:5454–5459. doi: 10.1016/j.vaccine.2017.07.054. [DOI] [PubMed] [Google Scholar]
- 147.World Health Organization. 2018. Rabies vaccines: WHO position paper, April 2018—recommendations. Vaccine 36:5500–5503. doi: 10.1016/j.vaccine.2018.06.061. [DOI] [PubMed] [Google Scholar]
- 148.Wu XX, Yao HP, Wu NP, Gao HN, Wu HB, Jin CZ, Lu XY, Xie TS, Li LJ. 2015. Ebolavirus vaccines: progress in the fight against ebola virus disease. Cell Physiol Biochem 37:1641–1658. doi: 10.1159/000438531. [DOI] [PubMed] [Google Scholar]
- 149.Spyrou E, Smith CI, Ghany MG. 2020. Hepatitis B: current status of therapy and future therapies. Gastroenterol Clin North Am 49:215–238. doi: 10.1016/j.gtc.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Milani A, Basimi P, Agi E, Bolhassani A. 2020. Pharmaceutical approaches for treatment of hepatitis C virus. Curr Pharm Des doi: 10.2174/1381612826666200509233215. [DOI] [PubMed] [Google Scholar]
- 151.Martinello M, Bajis S, Dore GJ. 2020. Progress toward hepatitis C virus elimination: therapy and implementation. Gastroenterol Clin North Am 49:253–277. doi: 10.1016/j.gtc.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 152.Chaudhuri S, Symons JA, Deval J. 2018. Innovation and trends in the development and approval of antiviral medicines: 1987–2017 and beyond. Antiviral Res 155:76–88. doi: 10.1016/j.antiviral.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Akram M, Tahir IM, Shah SMA, Mahmood Z, Altaf A, Ahmad K, Munir N, Daniyal M, Nasir S, Mehboob H. 2018. Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: a systematic review. Phytother Res 32:811–822. doi: 10.1002/ptr.6024. [DOI] [PubMed] [Google Scholar]
- 154.De Clercq E, Li G. 2016. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev 29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kohli A, Shaffer A, Sherman A, Kottilil S. 2014. Treatment of hepatitis C: a systematic review. JAMA 312:631–640. doi: 10.1001/jama.2014.7085. [DOI] [PubMed] [Google Scholar]
- 156.Arts EJ, Hazuda DJ. 2012. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med 2:a007161. doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.De Clercq E. 2009. Another ten stories in antiviral drug discovery (part C): “old” and “new” antivirals, strategies, and perspectives. Med Res Rev 29:611–645. doi: 10.1002/med.20153. [DOI] [PubMed] [Google Scholar]
- 158.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 159.Cramer P, Bushnell DA, Kornberg RD. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 160.Balbo PB, Bohm A. 2007. Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis. Structure 15:1117–1131. doi: 10.1016/j.str.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bienroth S, Keller W, Wahle E. 1993. Assembly of a processive messenger RNA polyadenylation complex. EMBO J 12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cowling VH. 2009. Regulation of mRNA cap methylation. Biochem J 425:295–302. doi: 10.1042/BJ20091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. 2004. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci U S A 101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim H-J, Jeong S-H, Heo J-H, Jeong S-J, Kim S-T, Youn H-D, Han J-W, Lee H-W, Cho E-J. 2004. mRNA capping enzyme activity is coupled to an early transcription elongation. Mol Cell Biol 24:6184–6193. doi: 10.1128/MCB.24.14.6184-6193.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fabrega C, Hausmann S, Shen V, Shuman S, Lima CD. 2004. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol Cell 13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- 166.Proudfoot NJ, Furger A, Dye MJ. 2002. Integrating mRNA processing with transcription. Cell 108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 167.Ho CK, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. 1998. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 168.Kang S, Kang MS, Ryu E, Myung K. 2018. Eukaryotic DNA replication: orchestrated action of multi-subunit protein complexes. Mutat Res 809:58–69. doi: 10.1016/j.mrfmmm.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 169.Parker MW, Botchan MR, Berger JM. 2017. Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit Rev Biochem Mol Biol 52:107–144. doi: 10.1080/10409238.2016.1274717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Alabert C, Jasencakova Z, Groth A. 2017. Chromatin replication and histone dynamics. Adv Exp Med Biol 1042:311–333. doi: 10.1007/978-981-10-6955-0_15. [DOI] [PubMed] [Google Scholar]
- 171.Rigsby RE, Parker AB. 2016. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem Mol Biol Educ 44:433–437. doi: 10.1002/bmb.20966. [DOI] [PubMed] [Google Scholar]