Respiratory diseases caused by influenza viruses still pose a serious concern to global health, and neutralizing antibodies constitute a promising area of antiviral therapeutics. However, the potential application of antibodies is often hampered by the challenge in generating nonimmunogenic antibodies in large scale. In the present study, transchromosomic (Tc) cattle were used for the generation of nonimmunogenic monoclonal antibodies (MAbs), and characterization of such MAbs revealed one monoclonal antibody, 53C10, exhibiting a potent neutralization activity against H1N1 influenza viruses. Further characterization of the neutralization escape mutant generated using this MAb showed that three amino acid substitutions in the HA head domain contributed to the resistance. These findings emphasize the importance of Tc cattle in the production of nonimmunogenic MAbs and highlight the potential of MAb 53C10 in the therapeutic application against H1 influenza virus infection in humans.
KEYWORDS: epitope, influenza, monoclonal antibodies, neutralizing antibodies
ABSTRACT
Influenza remains a global health risk and challenge. Currently, neuraminidase (NA) inhibitors are extensively used to treat influenza, but their efficacy is compromised by the emergence of drug-resistant variants. Neutralizing antibodies targeting influenza A virus surface glycoproteins are critical components of influenza therapeutic agents and may provide alternative strategies to the existing countermeasures. However, the major hurdle for the extensive application of antibody therapies lies in the difficulty of generating nonimmunogenic antibodies in large quantities rapidly. Here, we report that one human monoclonal antibody (MAb), 53C10, isolated from transchromosomic (Tc) cattle exhibits potent neutralization and hemagglutination inhibition titers against different clades of H1N1 subtype influenza A viruses. In vitro selection of antibody escape mutants revealed that 53C10 recognizes a novel noncontinuous epitope in the hemagglutinin (HA) head domain involving three amino acid residues, glycine (G), serine (S), and glutamic acid (E) at positions 172, 207, and 212, respectively. The results of our experiments supported a critical role for substitution of arginine at position 207 (S207R) in mediating resistance to 53C10, while substitutions at either G172E or E212A did not alter antibody recognition and neutralization. The E212A mutation may provide structural stability for the epitope, while the substitution G172E probably compensates for loss of fitness introduced by S207R. Our results offer novel insights into the mechanism of action of MAb 53C10 and indicate its potential role in therapeutic treatment of H1 influenza virus infection in humans.
IMPORTANCE Respiratory diseases caused by influenza viruses still pose a serious concern to global health, and neutralizing antibodies constitute a promising area of antiviral therapeutics. However, the potential application of antibodies is often hampered by the challenge in generating nonimmunogenic antibodies in large scale. In the present study, transchromosomic (Tc) cattle were used for the generation of nonimmunogenic monoclonal antibodies (MAbs), and characterization of such MAbs revealed one monoclonal antibody, 53C10, exhibiting a potent neutralization activity against H1N1 influenza viruses. Further characterization of the neutralization escape mutant generated using this MAb showed that three amino acid substitutions in the HA head domain contributed to the resistance. These findings emphasize the importance of Tc cattle in the production of nonimmunogenic MAbs and highlight the potential of MAb 53C10 in the therapeutic application against H1 influenza virus infection in humans.
INTRODUCTION
Respiratory diseases caused by influenza viruses still pose a serious concern to global health. Considerable morbidity and mortality are caused by influenza virus; it is estimated that 290,000 to 650,000 deaths are associated with influenza every year worldwide (1). Vaccines against influenza viruses are available and have been in use since the 1940s (2). However, these vaccines vary in effectiveness, often due to differences between the circulating virus and the isolate chosen for inclusion in the vaccine (3).
Influenza viruses, belonging to the Orthomyxoviridae family, are divided into four genera: A, B, C, and D viruses (4, 5). Influenza A viruses are further divided into distinct subtypes based on the antigenic variation of two surface glycoproteins on the viral membrane: hemagglutinin (HA) and neuraminidase (NA). Eighteen HA and 11 NA subtypes have been detected in nature (6), and H1N1 influenza A viruses were the most prevalent strains circulating in humans in the United States during the 2019–2020 influenza season (7). Swine influenza virus may transmit to humans by direct contact. The H1N1 viruses circulating in the U.S. swine population are classified into several distinct clades according to antigenic differences in HA, including the alpha, beta, gamma, delta1, delta2, and pdm09 clades (8). This virus diversity may compromise vaccine efficacy, underscoring the critical role of broad neutralizing antibodies in combating influenza virus infections of humans.
Monoclonal antibody (MAb) therapy is a novel and promising technique against multiple pathogens, including influenza viruses (9–11), Ebola virus (12, 13), respiratory syncytial virus (14), and human immunodeficiency virus (15, 16). A major hurdle for extensive application of MAb therapies lies in the difficulty of generating nonimmunogenic antibodies in large quantities rapidly. MAbs are usually generated from animals or prepared from the plasma of human donors. However, MAb from a human source is hampered by supply shortages, and animal-derived MAbs suffer from severe side effects since they are often immunogenic in humans (17). Until now, multiple strategies have been employed to humanize animal-derived MAbs aiming to minimize foreign immunogenicity, such as complementarity-determining region (CDR) grafting and antibody resurfacing (18). It has been demonstrated that CDR grafting can lead to a substantial decrease in antibody affinity in many cases, whereas the comprehensive evaluation of resurfaced antibody immunogenicity is not currently available (19, 20).
To overcome these obstacles, a transchromosomic (Tc) cattle platform capable of expressing fully human, antigen-specific MAbs was developed (21). In these genetically engineered cattle, the immunoglobulin genes were knocked out and then replaced by the entire germ line loci of human immunoglobulin heavy chain, lambda chain, and kappa chain (21). Not only can this procedure produce human polyclonal antibodies in large quantities in a time-efficient manner, but also Tc cattle can be used to produce MAbs that recognize specific antigens of interest from a variety of viruses. This expands the potential application in passive immune therapies, allowing for both broad, polyclonal approaches and specific, monoclonal approaches to therapeutic treatment. Until now, Tc cattle have been employed to generate antibodies with high therapeutic efficacy in vivo against diverse viruses, such as Middle East respiratory syndrome coronavirus (MERS-CoV) (22), Ebola virus (23), hantavirus (24), Venezuelan equine encephalitis virus (25), and Zika virus (26). Therefore, the Tc cattle platform provides a feasible way to produce large amounts of fully human monoclonal antibodies rapidly and hence a valuable therapeutic tool to combat emerging virus outbreaks.
Here, we report a novel fully human monoclonal antibody (53C10) generated from Tc cattle that is capable of neutralizing multiple clades of H1N1 influenza A viruses in vitro. The results of our study collectively show that 53C10 binds with high affinity to a conformational epitope in the HA globular head domain that blocks virus entry. The characterization of this novel antibody presented here further suggests its role as a potential therapeutic agent to treat H1 influenza virus infections in humans.
RESULTS
Monoclonal antibody 53C10 binds to recombinant HA proteins from different clades of H1 viruses in ELISA.
To determine whether the 53C10 antibody interacted with the HA proteins from different clades of H1, enzyme-linked immunosorbent assay (ELISA) was performed using different clades of recombinant HA proteins produced by the baculovirus expression system. In ELISA, the antibody was diluted using 2-fold serial dilutions and the recombinant HA protein from influenza B virus (B/Florida/4/2006-Yamagata lineage) was used as a negative control. As shown in Fig. 1A, MAb 53C10 was able to recognize the representative HA proteins from the alpha and pdm09 clades with high affinity. Remarkably, antibody binding still could be detected when the antibody was diluted at 1:105. The HA proteins from only two H1 clades were tested in ELISA. We were unable to test reactivity against HA from other H1 clades due to the unavailability of representative recombinant HA proteins.
FIG 1.
53C10 neutralizes multiple H1 clades of influenza A virus. (A) Monoclonal antibody 53C10 recognizes HAs from the alpha clade and pdm09 clade. The baculovirus-expressed HAs of A/California/04/2009 and A/Iowa/15/1930 were subjected to ELISA with monoclonal antibody 53C10. 53C10 was 2-fold serially diluted. The baculovirus-expressed HA of strain B/Florida/4/2006 was used as negative control. (B) Neutralization activity of 53C10 against representative strains from different clades of H1 viruses. The online swine H1 clade classification tool (https://www.fludb.org) was employed to classify the virus strains. (C) Phylogenetic analysis of the H1 representative stains by maximum likelihood method in MEGA X. The phylogenetic tree with the highest log likelihood (−9917.90) was generated in the Jones-Taylor-Thornton (JTT) model with a bootstrapping of 1,000 replicates using the global swine H1 clade classification reference sequences (27). In the phylogenetic tree, strain name, isolated species and country, subtype, and H1 clade are shown. The H1 clades are labeled as alpha, beta, delta1, delta2, delta like, gamma, gamma2, alpha beta gamma2 like, gamma2 beta like, other, pdm09 (npdm), NA (not applicable), and ND (not determined) (www.fludb.org).
Monoclonal antibody 53C10 neutralizes different clades of H1 viruses in vitro.
To assess whether MAb 53C10 can neutralize various clades of H1 viruses, including alpha, beta, gamma, delta1, delta2 and pdm09, virus-neutralizing and hemagglutination inhibition (HI) assays were performed in vitro. The online swine H1 clade classification tool (https://www.fludb.org) was used to classify the virus strains (27). As summarized in Fig. 1B, 53C10 exhibited potent neutralizing capability against the representative strains from the pdm09, alpha, beta and gamma clades, with neutralizing titers up to 1:1,067, 1:533, 1:853, and 1:67 (average titers of three independent assays), respectively. In contrast, 53C10 failed to neutralize the representative viruses from the delta1 and delta2 clades. This may be due to the fact that the cluster involving the delta1 and delta2 clades is well separated phylogenetically and more divergent from the clusters involving the pdm09, alpha, beta, and gamma clades (Fig. 1C).
Next, the HI assay using different clades of H1 viruses was performed to characterize the cross-clade reactivity of 53C10. The HI assay showed results consistent with those of the virus neutralization assay. As shown in Fig. 1B, 53C10 displayed HI titers against representative viruses from the pdm09, alpha, beta, and gamma clades, at 1:640, 1:160, 1:320, and 1:20, respectively. Consistent with the neutralization assay results, 53C10 showed no detectable HI titer against the delta1 and delta2 clades. Together, our results suggest that the MAb 53C10 can bind to HA protein and as a result interfere with receptor binding and neutralize multiple clades of the H1 subtype.
Monoclonal antibody 53C10 recognizes a conformational epitope on the HA molecule.
The neutralization and HI assays suggest that the epitope of MAb 53C10 may overlap the receptor-binding site. To further investigate whether the epitope is comprised of continuous or noncontinuous amino acid residues, Western blotting using MAb 53C10 was performed using the baculovirus-expressed HA protein. Prior to Western blotting, recombinant HA (rHA) of A/California/04/2009 (CA/04, H1N1) was denatured at 95°C for 10 min. As shown in Fig. 2A, 53C10 antibody failed to react with CA/04 rHA at concentrations of 1, 4, 16, 64, 256, and 1,024 ng, whereas a linear epitope-targeted antibody, 63C7 (data not shown), was found to bind to the HA at a concentrations of 64, 256, and 1,024 ng (Fig. 2B), indicating that 53C10 likely recognizes a conformational epitope on the HA protein. It is worth noting that a fluorescence-activated cell sorting (FACS) assay performed using 53C10 and 63C7 antibodies on CA/04-infected Madin-Darby canine kidney (MDCK) cells demonstrated slightly higher percentages of HA-positive cells (77.6% and 86.9%, respectively) than the commercial nucleoprotein (NP) antibody (74.3%), which suggests comparable binding efficiencies of these antibodies (Fig. 2C). This finding further supports our position that 53C10 interacts with a nonlinear epitope.
FIG 2.
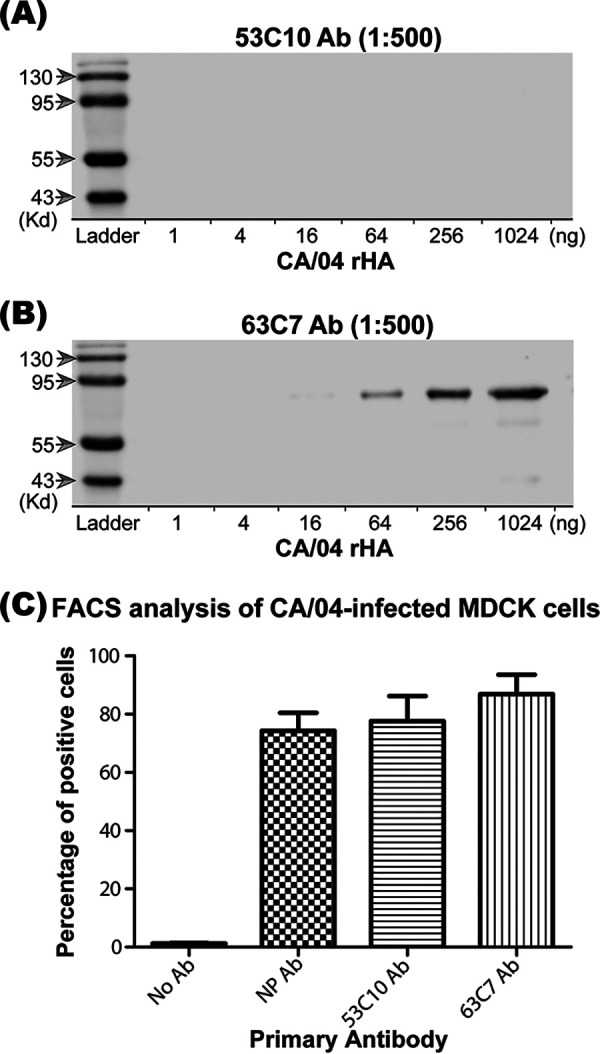
Monoclonal antibody 53C10 recognizes a conformational epitope on the HA molecule. The recognition of baculovirus-expressed CA/04 HA by antibody 53C10 (A) and antibody 63C7 (B) were evaluated by Western blotting (WB) as described in Materials and Methods. Below each lane is the amount of HA loaded. Molecular sizes are labeled on the left. Prior to WB, HA was denatured at 95°C for 10 min. (C) The binding of HA to 53C10 and 63C7 was determined in flow cytometry. MDCK cells were infected with CA/04. At 24 h postinfection, MDCK cells were evenly split into four tubes which were stained individually using either no antibody control, the antibody targeting NP protein (NP-Ab FR-667), 53C10 antibody, or 63C7 antibody. The percentage of cells bound by each antibody is shown.
In vitro selection and characterization of antibody escape mutants.
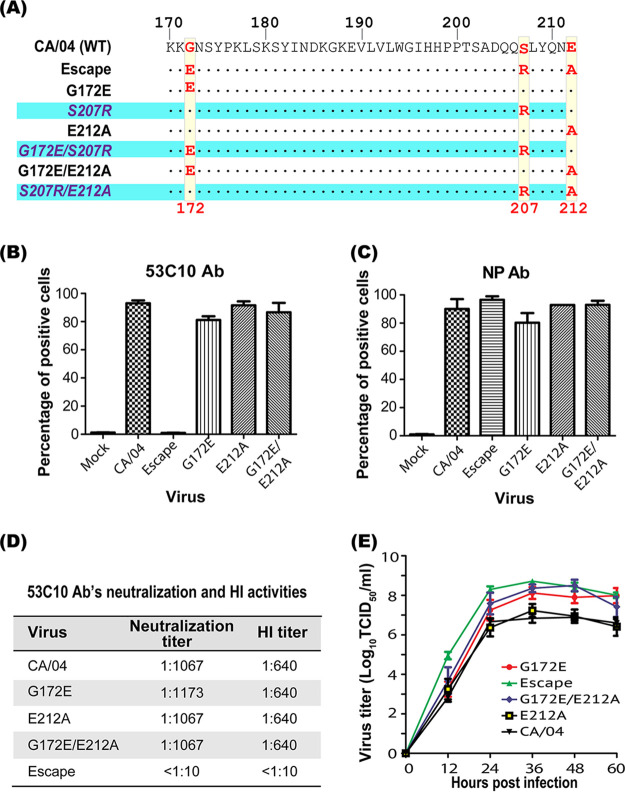
As 53C10 has a strong neutralization activity in both HI and virus neutralization assays, it may inhibit viral replication probably through blocking HA-mediated entry function. To further map the 53C10 epitope and to elucidate the critical amino acid residues mediating 53C10’s recognition, we selected antibody escape mutants using CA/04 in the presence of increasing concentrations of 53C10 antibody during the serial passage of the virus in MDCK cells. Specifically, the CA/04 wild type (WT) was propagated in the presence of 53C10 antibody. Antibody concentration was increased 2-fold in the subsequent propagations and the virus sent for MiSeq to determine the full-genome sequence after 10 passages for the identification of potential resistance-conferring mutations in all eight virus gene segments. Sequence analysis revealed that the escape mutant possessed three mutations in the HA segment after passage in the presence of the 53C10 antibody. Surprisingly, our next-generation sequencing (NGS) analyses detected no mutations in other segments compared to the respective control virus cultures propagated in the absence of 53C10 antibody. The three HA segment mutations observed in the escape mutant derived from passage 10 were G172E, S207R, and E212A (Fig. 3A), which were located in the antigenic site Sa, receptor-binding site (RBS), and antigenic site Sb, respectively (Fig. 3B) (28–31). The H1 residue numbering was initiated from the first methionine, as described previously (32, 33). Then, passages 1 to 9 were screened to identify the emergence of triple substitutions. The sequencing results revealed that the virus harbored triple substitutions since passage 3. To generate a pure population of the mutant harboring these three mutations, we introduced these mutations into our reverse genetic system (RGS)-HA plasmid for virus rescue. The escape mutant was successfully rescued and used for further characterization.
FIG 3.
Escape mutant harboring three substitutions completely escapes 53C10 recognition. (A) Alignment of the HA sequences of CA/04 and the survived antibody escape mutant. Three substitutions (G172E, S207R, and E212A) identified from the in vitro antibody selection assay are highlighted using boxes. (B) The structural locations of these three substitutions in the context of CA/04 HA (PDB accession number 3LZG) are shown. G172E, S207R, and E212A are in the antigenic site Sa, receptor-binding site (RBS), and antigenic site Sb, respectively. The influenza virus HA binding sialic acid is shown as sticks. This figure was generated using PyMOL.
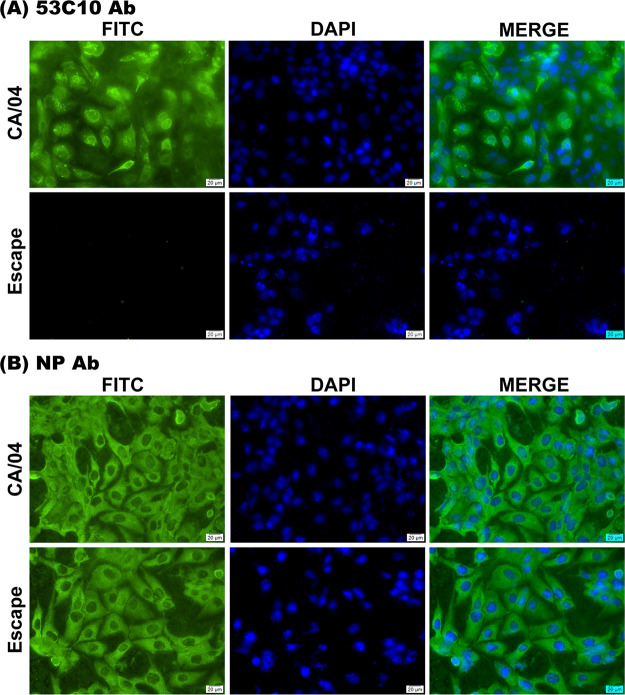
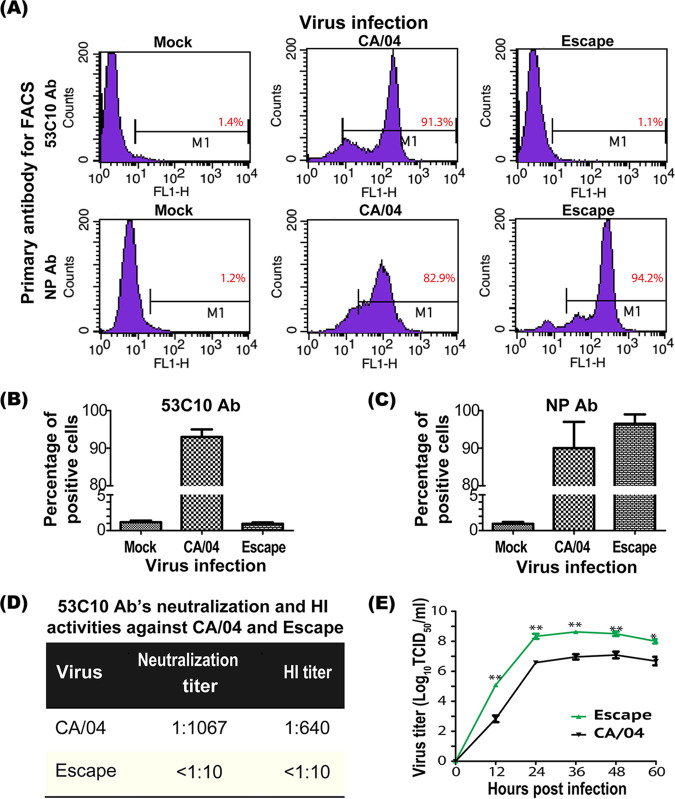
We performed immunofluorescence assay (IFA), flow cytometry, and virus neutralization assay to confirm antibody evasion using the RGS-rescued escape mutant. In IFA, escape mutant HA completely lost its binding to 53C10, as no immunofluorescent signals were detected (Fig. 4A). Notably, similar levels of MDCK cells were infected with the CA/04 WT and escape mutant as determined using anti-NP antibody (Fig. 4B). The remarkable reduction of 53C10’s recognition and neutralization for the mutant was also observed using flow cytometry, neutralization assay, and HI assay. As shown in Fig. 5A to C, the escape mutant showed a complete loss of reactivity with 53C10 antibody compared to the CA/04 WT in flow cytometry (Fig. 5A and B). Since no mutations in NP segment were observed in the escape mutant virus, the percentage of virus-infected MDCK cells could still be determined using anti-NP MAb (Fig. 5A and C). The proportions of CA/04 WT- and escape mutant-infected MDCK cells detected by NP-specific Ab were 82.89% and 94.22%, respectively, whereas the proportions of MDCK cells recognized by 53C10 were 91% and 1.1% (background level), respectively, suggesting that the escape mutant bearing these three mutations could completely escape 53C10 recognition. In addition, 53C10 exhibited no detectable neutralization or HI titers against the escape mutant (Fig. 5D). Together, these results show that these three substitutions in CA/04 HA abrogated the ability of 53C10 antibody to recognize and neutralize CA/04 WT virus.
FIG 4.
Binding comparison of the antibody 53C10 to CA/04 HA and escape mutant HA in IFA. The chamber slide-cultured MDCK cells were infected by the CA/04 WT and escape mutant. At 24 h postinfection, the cells were fixed with 80% acetone, followed by staining with the primary antibody 53C10 (upper images) and the antibody targeting NP protein (FR-667; lower images). Virus-infected cells were detected by staining with NP-specific monoclonal antibody. DAPI was used to stain the nucleus blue. Scale bar: 20 μm.
FIG 5.
Characterization of RGS-rescued escape mutant. (A) Quantitative comparison of the binding of antibody 53C10 to CA/04 HA and escape mutant HA using flow cytometry. Representative data of three independent experiments are presented. (B) MDCK cells were infected with the CA/04 WT and escape mutant and then stained by 53C10 to determine antibody binding. (C) Virus-infected cells were detected by staining with NP-specific monoclonal antibody FR-667. (D) Neutralization activity of 53C10 against the escape mutant and CA/04 WT. HI and neutralization titers against the Escape mutant were undetectable in the HI assay and virus neutralization assay. (E) Replication kinetics of the escape mutant and CA/04 WT in MDCK cells. MDCK cells were inoculated with respective viruses at an MOI of 0.01 and maintained at 37°C for 60 h. Supernatant samples were collected at 12, 24, 36, 48, and 60 h postinfection and titrated on MDCK cells by TCID50 infectivity assay.
To determine the impact of these three mutations on virus replication fitness, MDCK cells were inoculated with the escape mutant and CA/04 WT viruses at a multiplicity of infection (MOI) of 0.01. Supernatants (100 μl) were collected at 12, 24, 36, 48, and 60 h postinfection (h.p.i.) to determine viral infectivity. The replication kinetics show that the RGS-derived escape mutant replicated more efficiently than the CA/04 WT virus in MDCK cells (Fig. 5E).
Role of individual substitutions in 53C10 neutralization and virus replication fitness.
To assess the contribution of individual amino acid substitutions to 53C10 antibody evasion, we attempted to rescue viruses bearing each individual amino acid substitution and double amino acid substitutions in the context of the CA/04 HA backbone (Fig. 6A). We were unable to rescue mutants containing the S207R mutations after 6 independent attempts (Table 1). However, we successfully rescued three mutants that did not express the S207R substitution. Thus, it appears that position 207 plays a critical role in mediating receptor binding steps that initiate the infection cycle. Some substitutions at this position may disrupt receptor binding and as a result significantly impair virus replication fitness.
FIG 6.
Characterization of mutants with individual antibody-resistant mutations. (A) To study the contributions of individual mutations to 53C10 evasion, six mutants were designed. Single mutation and double mutations highlighted were designed to be introduced into the CA/04 WT virus. Dots indicate the identical residues observed when comparing mutant sequences with the CA/04 WT virus. (B and C) Quantitative comparison of the binding of antibody 53C10 to CA/04 HA and mutant HAs using flow cytometry. MDCK cells were infected with virus and then stained by 53C10 to determine antibody binding and NP-specific FR-667 antibody to quantify virus-infected cells. (D) Neutralization and HI titers against CA/04 WT and the RGS-rescued mutants with individual antibody-resistant mutations. (E) Replication comparison of CA/04 WT and the RGS-rescued mutants in MDCK cells. MDCK cells were inoculated with virus at an MOI of 0.01, and supernatant samples were collected at 12, 24, 36, 48, and 60 h postinfection followed by titration in MDCK cells by TCID50 assay.
TABLE 1.
Mutant rescue frequency from reverse genetic system
| Mutant | Rescue frequencya |
|---|---|
| CA/04 | 6/6 |
| Escape | 6/6 |
| G172E | 5/6 |
| S207R | 0/6 |
| E212A | 3/6 |
| G172E S207R | 0/6 |
| G172E E212A | 3/6 |
| S207R E212A | 0/6 |
Rescue frequency is shown as number of successful mutant rescues out of total attempts.
To further analyze the interaction of 53C10 Ab with the three rescued mutants, we performed flow cytometry, neutralization, and HI assays. The mutant viruses expressing G172E, E212A, and G172E/E212A reacted with 53C10 at a level similar to that of CA/04 WT virus, suggesting that residues 172G and 212E have no impact on 53C10 antibody recognition (Fig. 6B and C). Similarly, a neutralization assay showed that these mutants were neutralized by 53C10 similarly to the CA/04 WT, indicating that substitutions at positions 172 and 212 had no effect on 53C10 antibody recognition and neutralization. In addition, the G172E, E212A, and G172E/E212A mutants showed the same HI titer as the WT (Fig. 6D). Collectively, substitutions at positions 172 and 212 are not critical for antibody binding affinity and neutralization, underscoring the crucial role of position 207 in mediating 53C10 antibody recognition and neutralization of influenza A virus.
We next determined the replication fitness of these rescued mutants in MDCK cells. As shown in Fig. 6E, two mutants (the G172E and G172E/E212A mutants) displayed significant, 10-fold-higher titers than the CA/04 WT and E212A mutant (P = 0.029 at 36 h postinfection), suggesting that substitution at position 172 improves virus growth rate and, as a result, may compensate for the loss of fitness introduced by S207R. In contrast, the E212A substitution did not affect the viral growth rate, since the E212A mutant displayed growth kinetics similar to those of the CA/04 WT virus in MDCK cells.
Sequence conservation of 53C10 antibody recognition residues.
We analyzed sequence variations by retrieving the H1 sequences from the Influenza Research Database (IRD; https://www.fludb.org). Sequence logos (https://weblogo.berkeley.edu/) (34, 35)) were generated to visualize the conservation of 53C10 antibody recognition residues in each H1N1 clade (Fig. 7). In sequence logos, the proportion of each residue is indicated by the height of the amino acid symbol. The antibody-binding residues are highly conserved in all H1 clades except for delta1 and delta2 clades, which show a good correlation with our neutralization data. 53C10-resistant delta1 and delta2 strains differ significantly in these three residues from other 53C10-sensitive H1 clades, which may explain why the delta1 and delta2 clades cannot be recognized and neutralized by 53C10. For position 207, delta2 strains have an alanine (A), while other clades have a serine (S) or threonine (T) at this position. Based on our observations, substitution at T207 has no substantial effects on the recognition of HA by monoclonal antibody 53C10, which can be expected considering the similarity in side chains in the structures of serine and threonine amino acid residues. Interestingly, the HA from some delta1 strains harboring T207 could not be recognized by 53C10 in this study, which indicates a context-dependent recognition mechanism by the 53C10 antibody that is primarily driven by the three critical amino acid residues identified here. For position 172, glutamic acid (E) rather than glycine (G) was found in the alpha, beta, and gamma clades, which are sensitive to 53C10 antibody-mediated neutralization. As stated above, this substitution has no obvious impact on 53C10 activity. Conservation analysis also revealed that strains from delta1 and delta2 contain E, while the remaining clades maintain an alanine at position 212. Since A was observed in the escape mutant at position 212, and alpha, beta, gamma, and pdm09 clades harboring alanine were all recognized by 53C10, confirming that position 212 was not critical for 53C10 binding.
FIG 7.
Conservative analysis of three identified antibody resistant-related residues among different clades of H1 viruses. Sequence logos were employed to demonstrate the conservation of three antibody resistant-related residues (highlighted by arrows) in influenza virus strains from H1 clades. The proportion of individual residues in each clade is indicated by the height of the amino acid symbol. For conservative analysis, HA sequences from each clade were retrieved from the Influenza Research Database (IRD) and aligned for generating sequence logos (www.fludb.org).
DISCUSSION
Respiratory diseases caused by influenza viruses still raise serious concern to global health and lead to significant economic burden every year. NA inhibitors are extensively used to treat influenza, but their efficacy is compromised by the emergence of drug-resistant variants. Antibody-based treatment and prevention measures are gradually emerging as an attractive direction that can complement well the current influenza therapeutics. However, the clinical application of antibodies is hampered by their high immunogenicity in humans. In this study, we demonstrated that fully human monoclonal antibody 53C10 generated from Tc cattle recognizes a novel conformational epitope in the HA head and exhibits an impressive cross-clade (H1) neutralizing activity. Our results also revealed that a mutant carrying three amino acid substitutions in the HA head domain was capable of mediating total escape from 53C10 antibody recognition. Furthermore, results from our study indicated that only residue S207 is critical for 53C10 binding and neutralization activity, whereas G172E seems to compensate for the fitness cost introduced by the S207R mutation. While the role of E212A is inconclusive, our findings suggest that this mutation may hold a potential benefit to the overall viral fitness in combination with 172E and/or 207R. To our knowledge, the combination of these three mutations has not been previously documented for their contribution to antibody evasion.
The discontinuous epitope we identified in this study overlaps with one known epitope reported previously (36). In that study, the epitope-targeting antibodies could neutralize A/WSN RG/33 (H1), A/Aichi/2/68 (H3), and A/gull/Maryland/704/77 (H13), which are not neutralized by 53C10 antibody. This functional discrepancy supports our position that while these epitopes overlap, the epitope of 53C10 identified in our study is different from the one defined previously. We further demonstrate that G172E, S207R, and E212A substitutions are capable of mediating complete 53C10 antibody evasion. It was reported that residue 207 (also described as position 193, using numbering without inclusion of the signal peptide) is located in the 190 helix, which is one conserved region that mediates receptor binding (30, 31, 37). Antibody binding to this region inhibits viral replication by interfering with receptor binding and ultimately blocks virus entry. Therefore, S207 plays a critical role in virus replication, and direct binding to this residue can significantly block receptor binding. Substitutions in HA position 207 may reduce receptor binding avidity to escape from antibody recognition at the significant cost of viral fitness (38). The fact that we failed to rescue some viruses containing the S207R mutation highlights a critical role for this residue in virus entry and replication.
We demonstrated that two substitutions, G172E and E212A, had no substantial impact on 53C10 antibody recognition and neutralization but may compensate for the fitness costs of the S207R substitution. Mutant virus rescued from RGS harboring these two substitutions showed a higher growth rate than the wild-type virus in our replication kinetics experiment. Notably, the mutant carrying mutation E212A only showed replication kinetics similar to those of the CA/04 WT virus in MDCK cells. We speculate that this mutation may provide structural stability to the epitope recognized by 53C10. In sharp contrast to the E212A substitution, mutants carrying the G172E substitution alone displayed 10-fold-increased titers, suggesting that residue 172 is critical to compensate for the reduced fitness of the S207R substitution. Consistent with previous reports, the introduction of the G172E substitution (G155E in their study) can lead to a higher growth rate of A/New Jersey/76 in eggs (33). It was also reported that A/California/07/2009 containing the G172E mutation in HA replicated much better in eggs (39). Here, we demonstrated that the introduction of G172E substitution can increase CA/04 growth rate 10-fold in MDCK cells based on viral replication kinetics, and this 10-fold increase may restore the virus fitness that was reduced by the S207R mutation. Taken together, the results of our experiments pinpoint a critical role for HA position 172 in virus replication.
The selection of antibody escape mutants in vitro is commonly employed to identify conformational epitopes recognized by neutralizing antibody, and the amino acid mutations identified in escape mutants are considered to be residues constituting conformational epitopes. However, our findings suggest that some HA mutations may not engage directly in antibody binding but may rather be related to viral fitness or required to maintain the structural integrity critical for display of a functional epitope. Since HA globular head-binding, neutralizing antibody inhibits viral entry by binding to the RBS, substitutions in the RBS may enable the virus to evade neutralizing antibody pressure while also leading to reduced virus fitness or even a replication-incompetent nature. In this regard, it can be envisioned that an antibody escape mutant can acquire compensatory mutations to recover virus fitness at the cost of major resistance mutations (40–42). In this study, we confirmed that the compensatory mutations play no essential role in antibody recognition and neutralization; hence, these are more likely nonepitope mutations. Nonepitope mutations may emerge because these residues are highly tolerant to mutation and may also have an impact on proper presentation of conformational epitope structures. Therefore, future characterization of conformational epitopes by selection of escape mutants needs to distinguish compensatory mutations from the residues constituting the conformational epitope.
It would be important and interesting to investigate the mechanistic details of how these compensatory mutations restore virus fitness. Kosik et al. demonstrated that glycosylation might play a critical role in compensating for fitness loss of major antibody resistance mutations (38). Glycosylation may be responsible for the increased viral growth rate in this study, and residue 170 was reported to be involved in binding with glycans (43). Negatively charged glutamate at residues 170, 171, and 172 may present a better way to communicate with glycans, which as a result facilitates viral growth in MDCK cells (39).
Neutralizing antibody may facilitate the emergence of antibody-resistant mutants, as has been observed for multiple pathogens, such as influenza virus, Zika virus (44), HIV (45, 46), respiratory syncytial virus (47), hepatitis C virus (48, 49), and porcine reproductive and respiratory syndrome virus (50). Influenza virus HAs have high plasticity and undergo frequent antigenic drifts to evade humoral immunity (51). The high antigenic drift rate in influenza viruses may be due to the selective pressure exerted by neutralizing antibodies. In fact, selection by neutralizing antibodies is proposed as one primary force that drives the evolution of human influenza virus HA (52). Most influenza virus-specific neutralization antibodies target the HA globular head and block viral entry. Influenza virus can evade antibody pressure by acquiring one or several mutations that can either eliminate or significantly reduce antibody binding affinity (53). Due to the presence of highly conserved epitopes on the HA stem, HA stem-targeting antibodies are commonly shown to have broader binding efficiency than HA head-targeting antibodies. However, resistance mutations could also occur to escape HA stem-targeting antibody by disrupting antibody binding or enhancing membrane fusion (53). The accumulation of these mutations leads to antigenic drift, or antigenic variation, which may substantially reduce a vaccine’s effectiveness (54, 55). The antigenic drift/variation poses a major health concern and makes influenza prevention and control more challenging. It is worth noting that the combination of multiple antibodies or the mixture of antibody and influenza drugs may serve as an effective way to prevent the emergence of escape mutants. Prabakaran et al. reported that the combination of two antibodies recognizing different epitopes can prevent the formation of escape mutants in mice and enhance therapeutic efficacy against H5N1 infection (56). Shen et al. also demonstrated that the combination of antibody C7G6-IgM and oseltamivir could confer protective efficacy better than that of either agent administered alone (57). Although there is no evidence to show that oseltamivir can inhibit the replication of antibody-resistant mutants, it is tempting to speculate that variants resistant to one treatment may still be inhibited by the other treatment since these treatments inhibit viral replication through distinct mechanisms.
There are several limitations to our study. First, the characterization of 53C10 antibody’s protective efficacy in vivo is necessary for future studies. Second, it is worthwhile to select for and characterize antibody-resistant mutants in an animal model, as use of an animal model or cell lines may result in distinct mutation profiles. Third, the conformational epitope recognized by 53C10 still needs to be mapped precisely. Selection of antibody escape mutants only shows one critical residue mediating 53C10 binding; the molecular nature and composition of this novel conformational epitope remain unclear. Further characterization of the conformational epitope may provide insights with the rational design of effective antibodies to combat influenza epidemics.
In summary, we report here that a human monoclonal antibody (53C10) generated from Tc cattle is broadly active against diverse clades of viruses within the H1 subtype. By in vitro selection of antibody escape mutants, we identified a novel discontinuous epitope that overlaps with the RBS of HA protein. Further characterization demonstrated that two escape substitutions do not affect antibody binding; rather, they compensate for the loss of fitness introduced by the escape mutation that is critical for antibody evasion. In this regard, our observations broaden our understanding of antibody escape mechanisms in their description that some mutations are critical for disruption of antibody binding sites, while other mutations are essential either for compensating the loss of fitness or for maintaining structural integrity of an epitope. Further characterization of 53C10 antibody’s mechanism of action and its antiviral activity in animal models may offer useful information for the development of future antibody-based therapeutic agents against influenza A virus infections in humans.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby canine kidney (MDCK) cells and human embryonic kidney cells (293T) were ordered from the ATCC. These cells were passaged and grown in Dulbecco’s modified Eagle’s medium (DMEM; HyClone) with the addition of 100 U/ml of penicillin-streptomycin (Gibco) and 10% fetal bovine serum (FBS; HyClone) at 37°C with 5% CO2. A/California/04/2009 (CA/04) was acquired from BEI Resources. Swine influenza A viruses A/swine/Nebraska/A01399492/2013 (NE9492; GenBank accession number KF712462), A/swine/Oklahoma/A01134903/2011 (OK4903; GenBank accession number CY114784), A/swine/Minnesota/A01349281/2013 (MN9281; GenBank accession number KC844197), and A/swine/Illinois/A01047079/2010 (IL7079; GenBank accession number CY114631) were ordered from the U.S. Department of Agriculture (USDA). A/Swine/Minnesota/02053/2008 (MN02053; GenBank accession number CY082650) was isolated by Ben Hause. Viruses were grown in virus growth medium (VGM) containing1 μg/ml of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Thermo Fisher Scientific) and 0.3% bovine serum albumin (BSA; Sigma-Aldrich).
Recombinant H1 proteins.
The baculovirus-expressed recombinant H1 proteins of A/California/04/2009 (NR-15749) and B/Florida/4/2006 (NR-15748) were obtained from BEI Resources. The recombinant H1 protein of A/swine/Iowa/15/1930 (FR-699) was provided by International Reagent Resources (IRR).
Vaccination of Tc cattle.
Tc cattle were produced as previously described (21). Briefly, Tc cattle are homozygous for triple knockouts in the endogenous bovine immunoglobulin genes (IGHM−/− IGHML1−/− IGL−/−) and carry a human artificial chromosome (HAC) vector with an IgG1 production bias, labeled as isKcHACD.
One of the Tc cattle was immunized with a licensed 2012–2013 trivalent seasonal influenza vaccine (Fluzone TIV; Sanofi-Pasteur) at a total of 1 mg of HA/dose formulated with SAB’s proprietary adjuvant formulation (SAB-adj-2) for the first vaccination (V1) to the fourth vaccination (V4) and then boosted with recombinant HA1 (H1N1) and HA1 (H3N2) at 2 mg of HA1 each formulated with SAB’s proprietary adjuvant formulation (SAB-adj-1) for the fifth vaccination (V5) and the sixth vaccination (V6). The vaccinations were done at 3- to 4-week intervals.
Generation of fully human monoclonal antibodies from the Tc cattle.
Superficial cervical lymph node was surgically retrieved from the immunized Tc cattle on day 3 post-V6 booster. The lymphocytes isolated from superficial cervical lymph node were then be used for cell fusion with mouse myeloma cell line SP 2/0 for hybridoma generation. On day 10 after cell fusion, hybridoma screening with HA1 (H1N1)- and HA1 (H3N2)-specific human IgG ELISAs were performed.
Due to the very low efficiency of conventional cell-based subcloning, a molecular cloning approach to obtain antigen-specific clones was applied. HA1 (H1N1) high-positivity clones from hybridoma screening were subjected to RNA isolation followed by molecular cloning of VDJ/VJ sequences into the expression vector. The antigen specificity of recombinant human IgG from each positive clone was verified using cell culture supernatant of 293F cells transfected with the expression vectors. Recombinant human IgG MAbs, purified from a larger-scale cell culture containing the verified expression vectors, were used for in vitro characterization.
Twenty-seven monoclonal antibodies isolated from Tc cattle were screened using the standard virus neutralization assay to determine the broadly neutralizing antibodies. The selection criterion is based on the breadth of antibody’s neutralizing activity. Among these antibodies, 53C10 MAb was found to be capable of neutralizing multiple clades of H1 influenza viruses. Some monoclonal antibodies were clade specific, while other antibodies possessed insignificant neutralizing activity. The concentration of each antibody used for in vitro characterization was 1 mg/ml.
Selection of in vitro escape mutants against monoclonal antibody 53C10.
The 53C10-resistant mutant was selected by propagating CA/04 virus in the presence of an increasing concentration of 53C10 antibody. In brief, MDCK cells were infected with CA/04 virus and incubated for 1 h at 37°C with 5% CO2. Then the medium was replaced with the virus growth medium (VGM) with 53C10 and plates were incubated at 37°C until cytopathic effect (CPE) was apparent. The virus cultural supernatants were then collected for the next passage. The concentration of 53C10 was doubled after each passage, and the selection process ended after 10 passages. Virus was propagated in the absence of 53C10 antibody in parallel to detect any passage-related mutations. All viruses from passage 10 were deep sequenced to identify the amino acid mutations in all segments.
Viral whole-genome sequencing.
The whole genomes of passage 10 viruses were sequenced by the Illumina MiSeq instrument as reported previously (5). Briefly, viral RNA was extracted using TRAzol and then amplified by a universal influenza A virus amplification kit (Life Technologies). The Nextera XT DNA library preparation kit and MiSeq reagent nano kit v2 (Illumina) were used for library preparation. DNA libraries were loaded into a MiSeq reagent cartridge for sequencing.
ELISA.
ELISA was performed as previously described (58). Briefly, high-binding and flat-bottom 96-well plates (Thermo Fisher Scientific) were coated with recombinant HA protein for 18 h at 4°C. After being washed with PBST (phosphate-buffered saline [PBS] with 0.05% Tween 20; Sigma-Aldrich) six times, the plates were blocked with 300 μl/well of blocking buffer (PBST with 0.2% bovine skin gelatin; Sigma-Aldrich) for 2 h at 37°C. After six washes, the plates were incubated with antibody 53C10 (2-fold serially diluted in PBST with 0.01% gelatin) for 1 h, followed by an additional six washes. A total of 100 μl of horseradish peroxidase (HRP)-conjugated goat anti-human IgG (Bethyl) at a 1:100,000 dilution was added to the plates and incubated for 30 min at 37°C. A total of 100 μl/well of tetramethylbenzidine (TMB; SeraCare) was added as the substrate, and the plates were incubated in the dark for 10 min followed by the addition of 4 M sulfuric acid to stop the reaction. The signal was detected using an ELISA microplate reader (BioTek) at 450 nm.
RGS.
Eight bidirectional reverse genetic system (RGS) plasmids of A/New York/1682/2009 were used to rescue the virus as described previously (59). The HA segment of CA/04 virus was amplified by reverse transcription-PCR (RT-PCR) and then cloned into the pHW2000 vector to generate HA plasmid for “7 + 1” virus rescue. A single point mutation or double mutations in the HA gene were introduced using specific primers. The mutated plasmids were sequenced before virus rescue. The RGS-rescued viruses were sequenced to confirm the absence of undesired mutations.
Indirect immunofluorescence assay (IFA).
MDCK cells cultured in a chamber slide (Lab-Tek) were infected with the virus. At 24 h postinfection (h.p.i.), cells were fixed with 80% acetone for 20 min at room temperature and blocked with blocking buffer (PBST with 1% BSA) for an additional 1 h at 37°C. The first antibody, 53C10, was added at a 1:400 dilution to each of the wells, and then the slides were incubated at 37°C for 1 h. Following three washes with PBS, goat anti-human fluorescein-conjugated secondary antibody (Bethyl) was added at a 1:1,000 dilution, followed by incubation at 37°C for 45 min. After the chamber was washed three times, 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) was added and incubated for 20 min at room temperature before data collection and analysis.
Fluorescence-activated cell sorting (FACS).
MDCK cells were infected with the virus. At 24 h.p.i., the supernatant was discarded and MDCK cells were collected, followed by washing with PBS. Permeable buffer (BD Biosciences) was added to the cells and incubated for 1 h at 4°C. Cells were collected by centrifugation at 2,000 rpm for 5 min and evenly split into two tubes before immunostaining. The anti-NP antibody (FR-667; International Reagent Resource) generated from mouse was diluted at 1:250 and added to one tube to detect the percentage of virus-infected cells, while 53C10 antibody was diluted at 1:500 and added to the other tube. After a 1-h incubation at room temperature, the first antibody was removed, followed by three washes with PBS. Bound antibodies were detected with goat anti-mouse fluorescein isothiocyanate (FITC; Thermo Fisher Scientific) and goat anti-human FITC (Bethyl) at 37°C for 45 min. Following washes with PBS, the cells were suspended and fixed in 1% paraformaldehyde (PFA; Polysciences). Flow cytometry was carried out on a FACSCalibur (BD Biosciences) and analyzed by Cell Quest software (BD Biosciences). A total of 20,000 events were analyzed in each sample.
Western blotting.
Recombinant HA proteins expressed from baculovirus were heated at 95°C for 10 min and then subjected to 12% SDS-PAGE. The gel was transferred to a nitrocellulose membrane (Thermo Fisher Scientific) at 100 V, followed by 2 h of blocking with 5% nonfat milk at room temperature. The membrane was incubated with primary antibody overnight at 4°C, followed by incubation with goat anti-human secondary antibody (Li-Cor) for 1 h at room temperature. The membrane was washed three times with PBST (PBS and 0.1% Tween 20) after each incubation. Following the final washes with PBST, antibody binding was detected using the Odyssey infrared gel imaging system (Li-Cor).
Virus neutralization assay.
Virus neutralization assays were performed as previously described (58, 60, 61), with slight modifications. In brief, the 50% tissue culture infectious dose (TCID50) of viruses used in this assay was determined using MDCK cells, followed by the dilution of viruses to 100 TCID50s/100 μl with VGM. MAb 53C10 was treated with receptor-destroying enzyme (RDE; Denka Seiken) at 37°C for 18 h, heat inactivated at 56°C, and 2-fold serially diluted in PBS in a U-bottom 96-well plate (Thermo Fisher Scientific). Then 60 μl of virus at 100 TCID50s/100 μl was added to each 53C10 dilution (60 μl), followed by a 1-h incubation at 37°C in 5% CO2. After the incubation, 100 μl of virus-53C10 mixture was transferred to PBS-washed MDCK cells, followed by additional incubation for 4 days. Viral neutralization was evaluated with HA assay involving 1% turkey erythrocytes (Lampire Biological Products). Back titrations, cell-only control, and negative antibody control were performed to confirm assay reliability.
HI assay.
Hemagglutination inhibition (HI) assay was carried out in V-bottom 96-well plates as previously described (62). Briefly, RDE-treated 53C10 was 2-fold serially diluted and incubated with an equal volume of 4 HA units of H1 virus for 30 min at room temperature, and then 1% turkey erythrocytes were added. The HI titer was determined as the highest antibody dilution that exhibits complete inhibition of hemagglutination.
Virus replication kinetics.
Monolayer MDCK cells were infected with influenza virus at an MOI of 0.01. At 1 h postinfection, cells were washed three times with PBS to remove unbound virus before the addition of VGM. At 6, 12, 18, and 24 h.p.i., 100 μl of supernatant was collected and kept at –80°C before titration on MDCK cells.
Structural locations of the identified mutations in CA/04 HA.
The 3D location of three mutant residues was generated by PyMOL software in the context of the crystal structure of CA/04 hemagglutinin (PDB accession number 3LZG).
Statistics analysis.
Data are shown as means ± standard errors. Statistical significance evaluation was assessed by Student’s t test and is indicated in figures as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
ACKNOWLEDGMENTS
We thank the members of Wang and Li laboratories for reading and critiquing the manuscript and for providing cell culture media and reagents used in this study. We also thank the NIAID Biodefense and Emerging Infections Research Resources Repository (BEI Resources) and International Reagent Resources (IRR) for providing recombinant HA proteins.
The study was supported in part by the South Dakota Agricultural Experiment Station (3AH-673), National Science Foundation/EPSCoR Cooperative Agreement IIA-1355423, the South Dakota Research and Innovation Center, and BioSNTR. This research was also in part funded by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences (NIGMS) of the NIH (P20GM103443 to V.C.H.), with additional support from the Division of Basic Biomedical Sciences at USD (to V.C.H.).
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Hua Wu, Christoph L. Bausch, and Eddie J. Sullivan have a financial interest in SAB Biotherapeutics. However, this interest has not influenced the results and conclusions of this paper.
REFERENCES
- 1.Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, Pereira da Silva S, Aungkulanon S, Buchholz U, Widdowson M-A, Bresee JS, Azziz-Baumgartner E, Cheng P-Y, Dawood F, Foppa I, Olsen S, Haber M, Jeffers C, MacIntyre CR, Newall AT, Wood JG, Kundi M, Popow-Kraupp T, Ahmed M, Rahman M, Marinho F, Sotomayor Proschle CV, Vergara Mallegas N, Luzhao F, Sa L, Barbosa-Ramírez J, Sanchez DM, Gomez LA, Vargas XB, Acosta Herrera A, Llanés MJ, Global Seasonal Influenza-associated Mortality Collaborator Network. 2018. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barberis I, Myles P, Ault SK, Bragazzi NL, Martini M. 2016. History and evolution of influenza control through vaccination: from the first monovalent vaccine to universal vaccines. J Prev Med Hyg 57:E115–E120. [PMC free article] [PubMed] [Google Scholar]
- 3.Osterholm MT, Kelley NS, Sommer A, Belongia EA. 2012. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis 12:36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 4.Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, Webby RJ, Simonson RR, Li F. 2013. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog 9:e1003176. doi: 10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, Wang D, Nelson EA, Li F. 2014. Characterization of a novel influenza virus in cattle and swine: proposal for a new genus in the Orthomyxoviridae family. mBio 5:e00031-14. doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New World bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CDC. 2020. Weekly U.S. influenza surveillance report, key updates for week 7, ending February 15, 2020. https://www.cdc.gov/flu/weekly/index.htm.
- 8.Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL. 2013. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir Viruses 7(Suppl 4):42–51. doi: 10.1111/irv.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chai N, Swem LR, Park S, Nakamura G, Chiang N, Estevez A, Fong R, Kamen L, Kho E, Reichelt M, Lin Z, Chiu H, Skippington E, Modrusan Z, Stinson J, Xu M, Lupardus P, Ciferri C, Tan MW. 2017. A broadly protective therapeutic antibody against influenza B virus with two mechanisms of action. Nat Commun 8:14234. doi: 10.1038/ncomms14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjamin E, Wang W, McAuliffe JM, Palmer-Hill FJ, Kallewaard NL, Chen Z, Suzich JA, Blair WS, Jin H, Zhu Q. 2014. A broadly neutralizing human monoclonal antibody directed against a novel conserved epitope on the influenza virus H3 hemagglutinin globular head. J Virol 88:6743–6750. doi: 10.1128/JVI.03562-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasugi M, Kubota-Koketsu R, Yamashita A, Kawashita N, Du A, Sasaki T, Nishimura M, Misaki R, Kuhara M, Boonsathorn N, Fujiyama K, Okuno Y, Nakaya T, Ikuta K. 2013. Human monoclonal antibodies broadly neutralizing against influenza B virus. PLoS Pathog 9:e1003150. doi: 10.1371/journal.ppat.1003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta M, Mahanty S, Bray M, Ahmed R, Rollin PE. 2001. Passive transfer of antibodies protects immunocompetent and immunodeficient mice against lethal Ebola virus infection without complete inhibition of viral replication. J Virol 75:4649–4654. doi: 10.1128/JVI.75.10.4649-4654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flyak AI, Kuzmina N, Murin CD, Bryan C, Davidson E, Gilchuk P, Gulka CP, Ilinykh PA, Shen X, Huang K, Ramanathan P, Turner H, Fusco ML, Lampley R, Kose N, King H, Sapparapu G, Doranz BJ, Ksiazek TG, Wright DW, Saphire EO, Ward AB, Bukreyev A, Crowe JE Jr. 2018. Broadly neutralizing antibodies from human survivors target a conserved site in the Ebola virus glycoprotein HR2-MPER region. Nat Microbiol 3:670–677. doi: 10.1038/s41564-018-0157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, Pfarr DS, Losonsky GA, Kiener PA. 2008. Immunoprophylaxis of RSV infection: advancing from RSV-IGIV to palivizumab and motavizumab. Curr Top Microbiol Immunol 317:103–123. doi: 10.1007/978-3-540-72146-8_4. [DOI] [PubMed] [Google Scholar]
- 15.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 16.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol GPI, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H, Fan Z, Brandsrud M, Meng Q, Bobbitt M, Regouski M, Stott R, Sweat A, Crabtree J, Hogan RJ, Tripp RA, Wang Z, Polejaeva IA, Sullivan EJ. 2019. Generation of H7N9-specific human polyclonal antibodies from a transchromosomic goat (caprine) system. Sci Rep 9:366. doi: 10.1038/s41598-018-36961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Safdari Y, Farajnia S, Asgharzadeh M, Khalili M. 2013. Antibody humanization methods—a review and update. Biotechnol Genet Eng Rev 29:175–186. doi: 10.1080/02648725.2013.801235. [DOI] [PubMed] [Google Scholar]
- 19.Pavlinkova G, Colcher D, Booth BJ, Goel A, Wittel UA, Batra SK. 2001. Effects of humanization and gene shuffling on immunogenicity and antigen binding of anti-TAG-72 single-chain Fvs. Int J Cancer 94:717–726. doi: 10.1002/ijc.1523. [DOI] [PubMed] [Google Scholar]
- 20.Almagro JC, Fransson J. 2008. Humanization of antibodies. Front Biosci 13:1619–1633. [DOI] [PubMed] [Google Scholar]
- 21.Matsushita H, Sano A, Wu H, Jiao JA, Kasinathan P, Sullivan EJ, Wang Z, Kuroiwa Y. 2014. Triple immunoglobulin gene knockout transchromosomic cattle: bovine lambda cluster deletion and its effect on fully human polyclonal antibody production. PLoS One 9:e90383. doi: 10.1371/journal.pone.0090383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luke T, Wu H, Zhao J, Channappanavar R, Coleman CM, Jiao JA, Matsushita H, Liu Y, Postnikova EN, Ork BL, Glenn G, Flyer D, Defang G, Raviprakash K, Kochel T, Wang J, Nie W, Smith G, Hensley LE, Olinger GG, Kuhn JH, Holbrook MR, Johnson RF, Perlman S, Sullivan E, Frieman MB. 2016. Human polyclonal immunoglobulin G from transchromosomic bovines inhibits MERS-CoV in vivo. Sci Transl Med 8:326ra21. doi: 10.1126/scitranslmed.aaf1061. [DOI] [PubMed] [Google Scholar]
- 23.Dye JM, Wu H, Hooper JW, Khurana S, Kuehne AI, Coyle EM, Ortiz RA, Fuentes S, Herbert AS, Golding H, Bakken RA, Brannan JM, Kwilas SA, Sullivan EJ, Luke TC, Smith G, Glenn G, Li W, Ye L, Yang C, Compans RW, Tripp RA, Jiao JA. 2016. Production of potent fully human polyclonal antibodies against Ebola Zaire virus in transchromosomal cattle. Sci Rep 6:24897. doi: 10.1038/srep24897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hooper JW, Brocato RL, Kwilas SA, Hammerbeck CD, Josleyn MD, Royals M, Ballantyne J, Wu H, Jiao JA, Matsushita H, Sullivan EJ. 2014. DNA vaccine-derived human IgG produced in transchromosomal bovines protect in lethal models of hantavirus pulmonary syndrome. Sci Transl Med 6:264ra162. doi: 10.1126/scitranslmed.3010082. [DOI] [PubMed] [Google Scholar]
- 25.Gardner CL, Sun C, Luke T, Raviprakash K, Wu H, Jiao JA, Sullivan E, Reed DS, Ryman KD, Klimstra WB. 2017. Antibody preparations from human transchromosomic cows exhibit prophylactic and therapeutic efficacy against Venezuelan equine encephalitis virus. J Virol 91:e00226-17. doi: 10.1128/JVI.00226-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddharthan V, Miao J, Van Wettere AJ, Li R, Wu H, Sullivan E, Jiao J, Hooper JW, Safronetz D, Morrey JD, Julander JG, Wang Z. 2019. Human polyclonal antibodies produced from transchromosomal bovine provides prophylactic and therapeutic protections against Zika virus infection in STAT2 KO Syrian hamsters. Viruses 11:92. doi: 10.3390/v11020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, Swenson SL, Simon G, Saito T, Berhane Y, Ciacci-Zanella J, Pereda A, Davis CT, Donis RO, Webby RJ, Vincent AL. 2016. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza A viruses. mSphere 1:e00275-16. doi: 10.1128/mSphere.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE Jr, Wilson IA. 2010. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sriwilaijaroen N, Suzuki Y. 2012. Molecular basis of the structure and function of H1 hemagglutinin of influenza virus. Proc Jpn Acad Ser B 88:226–249. doi: 10.2183/pjab.88.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunthaboot N, Rungrotmongkol T, Malaisree M, Kaiyawet N, Decha P, Sompornpisut P, Poovorawan Y, Hannongbua S. 2010. Evolution of human receptor binding affinity of H1N1 hemagglutinins from 1918 to 2009 pandemic influenza A virus. J Chem Inf Model 50:1410–1417. doi: 10.1021/ci100038g. [DOI] [PubMed] [Google Scholar]
- 31.Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, Wiley DC, Skehel JJ. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 32.Matsuzaki Y, Sugawara K, Nakauchi M, Takahashi Y, Onodera T, Tsunetsugu-Yokota Y, Matsumura T, Ato M, Kobayashi K, Shimotai Y, Mizuta K, Hongo S, Tashiro M, Nobusawa E. 2014. Epitope mapping of the hemagglutinin molecule of A/(H1N1)pdm09 influenza virus by using monoclonal antibody escape mutants. J Virol 88:12364–12373. doi: 10.1128/JVI.01381-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Job ER, Deng YM, Barfod KK, Tate MD, Caldwell N, Reddiex S, Maurer-Stroh S, Brooks AG, Reading PC. 2013. Addition of glycosylation to influenza A virus hemagglutinin modulates antibody-mediated recognition of H1N1 2009 pandemic viruses. J Immunol 190:2169–2177. doi: 10.4049/jimmunol.1202433. [DOI] [PubMed] [Google Scholar]
- 34.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, Ito K, Takada A. 2009. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog 5:e1000350. doi: 10.1371/journal.ppat.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell RJ, Stevens DJ, Haire LF, Gamblin SJ, Skehel JJ. 2006. Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconj J 23:85–92. doi: 10.1007/s10719-006-5440-1. [DOI] [PubMed] [Google Scholar]
- 38.Kosik I, Ince WL, Gentles LE, Oler AJ, Kosikova M, Angel M, Magadan JG, Xie H, Brooke CB, Yewdell JW. 2018. Influenza A virus hemagglutinin glycosylation compensates for antibody escape fitness costs. PLoS Pathog 14:e1006796. doi: 10.1371/journal.ppat.1006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z, Wang W, Zhou H, Suguitan AL Jr, Shambaugh C, Kim L, Zhao J, Kemble G, Jin H. 2010. Generation of live attenuated novel influenza virus A/California/7/09 (H1N1) vaccines with high yield in embryonated chicken eggs. J Virol 84:44–51. doi: 10.1128/JVI.02106-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rimmelzwaan GF, Berkhoff EG, Nieuwkoop NJ, Smith DJ, Fouchier RA, Osterhaus AD. 2005. Full restoration of viral fitness by multiple compensatory co-mutations in the nucleoprotein of influenza A virus cytotoxic T-lymphocyte escape mutants. J Gen Virol 86:1801–1805. doi: 10.1099/vir.0.80867-0. [DOI] [PubMed] [Google Scholar]
- 41.Seki S, Matano T. 2011. CTL escape and viral fitness in HIV/SIV infection. Front Microbiol 2:267. doi: 10.3389/fmicb.2011.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez-Merino V, Farrow MA, Brewster F, Somasundaran M, Luzuriaga K. 2008. Identification and characterization of HIV-1 CD8+ T cell escape variants with impaired fitness. J Infect Dis 197:300–308. doi: 10.1086/524845. [DOI] [PubMed] [Google Scholar]
- 43.Soundararajan V, Tharakaraman K, Raman R, Raguram S, Shriver Z, Sasisekharan V, Sasisekharan R. 2009. Extrapolating from sequence—the 2009 H1N1 ‘swine’ influenza virus. Nat Biotechnol 27:510–513. doi: 10.1038/nbt0609-510. [DOI] [PubMed] [Google Scholar]
- 44.Collins MH, Tu HA, Gimblet-Ochieng C, Liou GA, Jadi RS, Metz SW, Thomas A, McElvany BD, Davidson E, Doranz BJ, Reyes Y, Bowman NM, Becker-Dreps S, Bucardo F, Lazear HM, Diehl SA, de Silva AM. 2019. Human antibody response to Zika targets type-specific quaternary structure epitopes. JCI Insight 4:e124588. doi: 10.1172/jci.insight.124588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou P, Wang H, Fang M, Li Y, Wang H, Shi S, Li Z, Wu J, Han X, Shi X, Shang H, Zhou T, Zhang L. 2019. Broadly resistant HIV-1 against CD4-binding site neutralizing antibodies. PLoS Pathog 15:e1007819. doi: 10.1371/journal.ppat.1007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKeating JA, Gow J, Goudsmit J, Pearl LH, Mulder C, Weiss RA. 1989. Characterization of HIV-1 neutralization escape mutants. AIDS 3:777–784. doi: 10.1097/00002030-198912000-00001. [DOI] [PubMed] [Google Scholar]
- 47.Crowe JE, Firestone CY, Crim R, Beeler JA, Coelingh KL, Barbas CF, Burton DR, Chanock RM, Murphy BR. 1998. Monoclonal antibody-resistant mutants selected with a respiratory syncytial virus-neutralizing human antibody fab fragment (Fab 19) define a unique epitope on the fusion (F) glycoprotein. Virology 252:373–375. doi: 10.1006/viro.1998.9462. [DOI] [PubMed] [Google Scholar]
- 48.Velazquez-Moctezuma R, Galli A, Law M, Bukh J, Prentoe J. 2019. Hepatitis C virus escape studies of human antibody AR3A reveal a high barrier to resistance and novel insights on viral antibody evasion mechanisms. J Virol 93:e01909-18. doi: 10.1128/JVI.01909-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brimacombe CL, Grove J, Meredith LW, Hu K, Syder AJ, Flores MV, Timpe JM, Krieger SE, Baumert TF, Tellinghuisen TL, Wong-Staal F, Balfe P, McKeating JA. 2011. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J Virol 85:596–605. doi: 10.1128/JVI.01592-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Costers S, Vanhee M, Van Breedam W, Van Doorsselaere J, Geldhof M, Nauwynck HJ. 2010. GP4-specific neutralizing antibodies might be a driving force in PRRSV evolution. Virus Res 154:104–113. doi: 10.1016/j.virusres.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 51.Thyagarajan B, Bloom JD. 2014. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. Elife 3:e03300. doi: 10.7554/eLife.03300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guarnaccia T, Carolan LA, Maurer-Stroh S, Lee RT, Job E, Reading PC, Petrie S, McCaw JM, McVernon J, Hurt AC, Kelso A, Mosse J, Barr IG, Laurie KL. 2013. Antigenic drift of the pandemic 2009 A(H1N1) influenza virus in a ferret model. PLoS Pathog 9:e1003354. doi: 10.1371/journal.ppat.1003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chai N, Swem LR, Reichelt M, Chen-Harris H, Luis E, Park S, Fouts A, Lupardus P, Wu TD, Li O, McBride J, Lawrence M, Xu M, Tan MW. 2016. Two escape mechanisms of influenza A virus to a broadly neutralizing stalk-binding antibody. PLoS Pathog 12:e1005702. doi: 10.1371/journal.ppat.1005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao J, Couzens L, Burke DF, Wan H, Wilson P, Memoli MJ, Xu X, Harvey R, Wrammert J, Ahmed R, Taubenberger JK, Smith DJ, Fouchier RAM, Eichelberger MC. 2019. Antigenic drift of the influenza A(H1N1)pdm09 virus neuraminidase results in reduced effectiveness of A/California/7/2009 (H1N1pdm09)-specific antibodies. mBio 10:e00307-19. doi: 10.1128/mBio.00307-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burton DR, Poignard P, Stanfield RL, Wilson IA. 2012. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prabakaran M, Prabhu N, He F, Hongliang Q, Ho HT, Qiang J, Meng T, Goutama M, Kwang J. 2009. Combination therapy using chimeric monoclonal antibodies protects mice from lethal H5N1 infection and prevents formation of escape mutants. PLoS One 4:e5672. doi: 10.1371/journal.pone.0005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen C, Zhang M, Chen Y, Zhang L, Wang G, Chen J, Chen S, Li Z, Wei F, Chen J, Yang K, Guo S, Wang Y, Zheng Q, Yu H, Luo W, Zhang J, Chen H, Chen Y, Xia N. 2019. An IgM antibody targeting the receptor binding site of influenza B blocks viral infection with great breadth and potency. Theranostics 9:210–231. doi: 10.7150/thno.28434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Huang B, Thomas M, Sreenivasan CC, Sheng Z, Yu J, Hause BM, Wang D, Francis DH, Kaushik RS, Li F. 2018. Detailed mapping of the linear B cell epitopes of the hemagglutinin (HA) protein of swine influenza virus. Virology 522:131–137. doi: 10.1016/j.virol.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza A viruses. J Virol 83:10309–10313. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ran Z, Shen H, Lang Y, Kolb EA, Turan N, Zhu L, Ma J, Bawa B, Liu Q, Liu H, Quast M, Sexton G, Krammer F, Hause BM, Christopher-Hennings J, Nelson EA, Richt J, Li F, Ma W. 2015. Domestic pigs are susceptible to infection with influenza B viruses. J Virol 89:4818–4826. doi: 10.1128/JVI.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yasuhara A, Yamayoshi S, Ito M, Kiso M, Yamada S, Kawaoka Y. 2018. Isolation and characterization of human monoclonal antibodies that recognize the influenza A(H1N1)pdm09 virus hemagglutinin receptor-binding site and rarely yield escape mutant viruses. Front Microbiol 9:2660. doi: 10.3389/fmicb.2018.02660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hause BM, Stine DL, Sheng Z, Wang Z, Chakravarty S, Simonson RR, Li F. 2012. Migration of the swine influenza virus delta-cluster hemagglutinin N-linked glycosylation site from N142 to N144 results in loss of antibody cross-reactivity. Clin Vaccine Immunol 19:1457–1464. doi: 10.1128/CVI.00096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]