Epstein-Barr virus (EBV), as the first human tumor virus, infects more than 90% of the human population worldwide and is associated with numerous human cancers. Exploring EBV-mediated transcription regulatory networks is critical to understand viral-associated lymphomagenesis. However, the detailed mechanism is not fully explored. Now we describe the regulatory profiles of the E2F-Rb-HDAC complex together with EBV latent antigens, and we found that EBV latent antigens cooperatively facilitate KLF14 expression by antagonizing this multisubunit repressor complex in EBV-positive cells. This provides potential therapeutic targets for the treatment of EBV-associated cancers.
KEYWORDS: E2F-Rb-HDAC, Epstein-Barr virus, KLF14, latent infection
ABSTRACT
Epstein-Barr virus (EBV) was discovered as the first human tumor virus more than 50 years ago. EBV infects more than 90% of the human population worldwide and is associated with numerous hematologic malignancies and epithelial malignancies. EBV establishes latent infection in B cells, which is the typical program seen in lymphomagenesis. Understanding EBV-mediated transcription regulatory networks is one of the current challenges that will uncover new insights into the mechanism of viral-mediated lymphomagenesis. Here, we describe the regulatory profiles of several cellular factors (E2F6, E2F1, Rb, HDAC1, and HDAC2) together with EBV latent nuclear antigens using next-generation sequencing (NGS) analysis. Our results show that the E2F-Rb-HDAC complex exhibits similar distributions in genomic regions of EBV-positive cells and is associated with oncogenic super-enhancers involving long-range regulatory regions. Furthermore, EBV latent antigens cooperatively hijack this complex to bind at KLFs gene loci and facilitate KLF14 gene expression in lymphoblastoid cell lines (LCLs). These results demonstrate that EBV latent antigens can function as master regulators of this multisubunit repressor complex (E2F-Rb-HDAC) to reverse its suppressive activities and facilitate downstream gene expression that can contribute to viral-induced lymphomagenesis. These results provide novel insights into targets for the development of new therapeutic interventions for treating EBV-associated lymphomas.
IMPORTANCE Epstein-Barr virus (EBV), as the first human tumor virus, infects more than 90% of the human population worldwide and is associated with numerous human cancers. Exploring EBV-mediated transcription regulatory networks is critical to understand viral-associated lymphomagenesis. However, the detailed mechanism is not fully explored. Now we describe the regulatory profiles of the E2F-Rb-HDAC complex together with EBV latent antigens, and we found that EBV latent antigens cooperatively facilitate KLF14 expression by antagonizing this multisubunit repressor complex in EBV-positive cells. This provides potential therapeutic targets for the treatment of EBV-associated cancers.
INTRODUCTION
The E2F family is traditionally divided into several subgroups as activators (E2F1 to E2F3) or suppressors (E2F4 to E2F8) based on their transcriptional properties in vivo (1). E2Fs have also been described for their critical roles in regulating cell proliferation and cell cycle (2). These convergent studies revealed that E2Fs were the functional target of the first identified tumor repressor retinoblastoma (Rb) (3, 4). Importantly, the E2F/Rb pathway is also critical in regulation of cell growth and development of cancer (5, 6). Rb can block E2F transcription activation by binding to its transactivation domain (7, 8). Furthermore, Rb can interact with E2Fs and HDACs (histone deacetylase), simultaneously. Thus, the E2F/Rb complex suppresses transcription through the recruitment of HDACs, which contain an LXCXE motif required for their interaction with the domain B of the Rb protein (9–11). Recruitment of the E2F-Rb-HDAC complex can regulate chromatin structure by modifying histone acetylation and further inhibit transcription activity (11–13). Additionally, adenovirus E1A protein, simian virus 40 (SV40) large T antigen, and human papillomavirus-16 E7 oncoprotein bind the Rb pocket domain and inactivate its function to facilitate cell transformation (14–17). These results suggest that the cooperation of these cellular factors is important for the regulation of downstream signaling targets. For example, DNMT1, a predominant mammalian DNA methyltransferase, can bind the E2F-Rb-HDAC complex to repress the transcription activity of E2F-responsive promoters (18).
Epstein-Barr virus (EBV) is the first known human tumor virus and has been studied for more than 50 years (19–21). EBV infects more than 90% of the population worldwide and is associated with numerous diseases, including Burkitt’s lymphoma (BL), Hodgkin lymphoma (HL), diffuse large B-cell lymphoma (DLBCL), nasopharyngeal carcinoma (NPC), and gastric carcinoma (GC) (22, 23). EBV is still the most efficient transforming virus capable of immortalizing human primary B lymphocytes in vitro (24). Two types of infection can be predominantly established in EBV-infected cells, latency in primary B lymphocytes and lytic infection in epithelial cells (22, 25). During latent infection, different latent programs are defined by the expression programs of the viral genome, which results in a specific repertoire of EBV proteins (24). This lifelong and persistent EBV infection in the infected host plays a critical role in driving EBV-associated tumorigenesis. EBV latent antigens can induce lymphomagenesis in B cells by dysregulating the transcription network and, thus, protein expression at multiple levels (24). Additional studies focus on the complicated transcription regulatory networks in EBV-induced B cell transformation in vitro (19). Super-enhancers are clusters of transcriptional enhancers bound by multiple transcription factors that are critical for cell identity or cell diseases (26). EBV super-enhancers with higher H3K27ac signals increase the expression of both MYC and BCL2 genes to promote cell proliferation (27). EBV nuclear antigens (EBNAs) are essential for the organization of MYC super-enhancer-associated chromatin loops, which further increases MYC expression (28, 29). However, revealing the effects of chromosome modification on transcriptional regulatory networks, as well as EBV-induced B-cell transformation, does not provide the entire picture. How EBV oncogenes regulate the human three-dimensional (3D) genome and reprogram the cellular functions is still mostly an unexplored area.
As a classical signaling pathway, the E2F-Rb-HDAC complex can be controlled and utilized by EBV to facilitate its tumorigenic activities. For example, EBV immediate early protein BRLF1 also recruits the E2F family members to activate the EBV DNA polymerase promoter (30). Interestingly, another EBV immediate early protein, BZLF1, also induces E2F1 expression and other cell cycle-associated genes (31). LMP1 decreases p27 transcription by recruiting the repressive E2F4 protein to E2F sites within the p27 promoter (32). E2F1 transcription factor could also activate the EBV BamHI-F promoter (Fp) activity by antagonizing EBNA1-mediated transcription inhibition (33). Our previous studies showed that EBNA3C can interact with the Rb protein and mediates its degradation through an SCF (Skp1-Cul1-F-box-protein) ubiquitin ligase Skp2 (34, 35). EBNA3C mediates E2F1 downregulation and E2F6 upregulation to facilitate cell cycle progression (36, 37). Also, HDACs are strictly regulated and target multiple genes by modulating chromosome structure during EBV infection (38, 39). For instance, the myocyte enhancer-binding factor 2 (MEF2) recruits class II HDACs to the BZLF1 gene promoter and changes the status of chromatin acetylation during latent infection (40). Furthermore, the histone deacetylase inhibitor romidepsin exerts strong antitumor activity via reducing LMP1 and c-Myc expression in EBV-positive DLBCL (41). Treatment with the HDAC inhibitor sodium butyrate regulates the switch from lytic to latent infection, which increases STAT3 expression (42). Therefore, HDAC inhibitors induce EBV lytic-phase gene expression and can be potently utilized for treatment of EBV-associated lymphomas (43). However, whether EBV antigens target the E2F-Rb-HDAC complex to regulate organization of oncogenic super-enhancers is yet to be fully resolved.
Here, we investigated the binding characteristics of cellular factors E2F6, E2F1, Rb, HDAC1, and HDAC2 on the human genome to explore the role of EBV-mediated transcriptional regulatory networks in controlling the organization of super-enhancers. We found that EBV latent antigens cooperated with this repressor complex E2F-Rb-HDAC and promoted the expression of two new targets (KLF10 and KLF14) in EBV-positive cells. These results provide novel insights into the molecular mechanism and a deeper understanding of EBV-associated regulation which can be harnessed for development of novel treatment strategies.
RESULTS
The E2F, Rb, and HDAC exhibit similar distribution at annotated genomic regions in lymphoblastoid cell lines (LCLs).
E2F-Rb-HDAC is a functional transcription regulatory module that modulates the expression of several specific target genes in human cancers (5, 44). Both E2F6 and E2F1/Rb can act as tumor repressors to inhibit gene expression through the recruitment of histone deacetylases 1 (HDAC1) or histone deacetylases 2 (HDAC2) (9–11, 45–47). Our previous studies demonstrated that EBV latent antigens interact with these molecules to regulate their expression and functional activities (34, 36–38). How these viral antigens modulate the downstream target genes through cooperative binding of the E2F-Rb-HDAC complex, and whether viral antigens can reverse their inhibition of expression of specific targets are not fully understood in EBV-infected cells.
To explore the detailed regulatory mechanisms of E2F-Rb-HDAC in the background of EBV, we first performed high-throughput chromatin immunoprecipitation sequencing (ChIP-Seq) analysis of the EBV-transformed lymphoblastoid cell line (LCL1) to identify the genomic regions enriched for these transcription regulatory factors, which included E2F6, E2F1, Rb, HDAC1, and HDAC2 (Fig. 1). The results from the ChIP-Seq analysis demonstrated that E2F6 and E2F1 as well as Rb primarily bound to noncoding DNA regions of the human genome, particularly introns and intergenic regions, while another 3% to 5% were regions which include transcription start sites and promoters associated with regulation of coding sequences (Fig. 1A). Similarly, HDAC 1 and 2 were enriched at introns and intergenic regions (Fig. 1A). E2F6 was shown to be enriched at approximately 3-fold the number of mutually exclusive binding sites compared to E2F1; however, the number of common binding sites shared by these factors in LCLs were much greater than the single sites (Fig. 1B). Additionally, HDAC1 and HDAC2 may indirectly bind the genomic sites through their association with E2Fs or Rb protein. Interestingly, HDAC2 is associated with approximately 90-fold more mutually exclusive binding sites than HDAC1 in LCL1 cells, although they have overlapping sites. These results suggest that HDAC2 has a prominent and multifunctional role in EBV-mediated lymphomagenesis (Fig. 1C).
FIG 1.
The E2F, Rb, and HDAC exhibit similar distribution at annotated genomic regions in LCLs. (A) Distribution of E2F6, E2F1, Rb, HDAC1, and HDAC2 occupancy at annotated genomic regions in EBV-transformed LCLs. TTS, transcription termination site; Exon, coding region. (B and C) Venn diagrams showing the overlap of E2F6 and E2F1 (B) and HDAC1 and HDAC2 (C) peaks in LCLs. The numbers indicate the binding sites of these cellular factors. (D) Overlap of E2F6, E2F1, Rb, HDAC1, and HDAC2 binding sites in LCLs. The numbers demonstrate the overlapping peaks of the indicated factors.
To further investigate the binding characteristics, we determined the overlap of these five cellular factors that are located at enriched regions on the human genome. Our results showed that approximately 20,073 sites were bound similarly for E2F6, E2F1, Rb, HDAC1, and HDAC2 (Fig. 1D). Therefore, E2F-Rb-HDAC is a critical transcriptional functional complex, the parts of which are distributed similarly to each other owning many common binding sites in EBV-transformed LCL cells, which implies that they are tightly coordinated in the regulation of EBV latency.
The E2F-Rb-HDAC complex can function as a super-enhancer in LCLs.
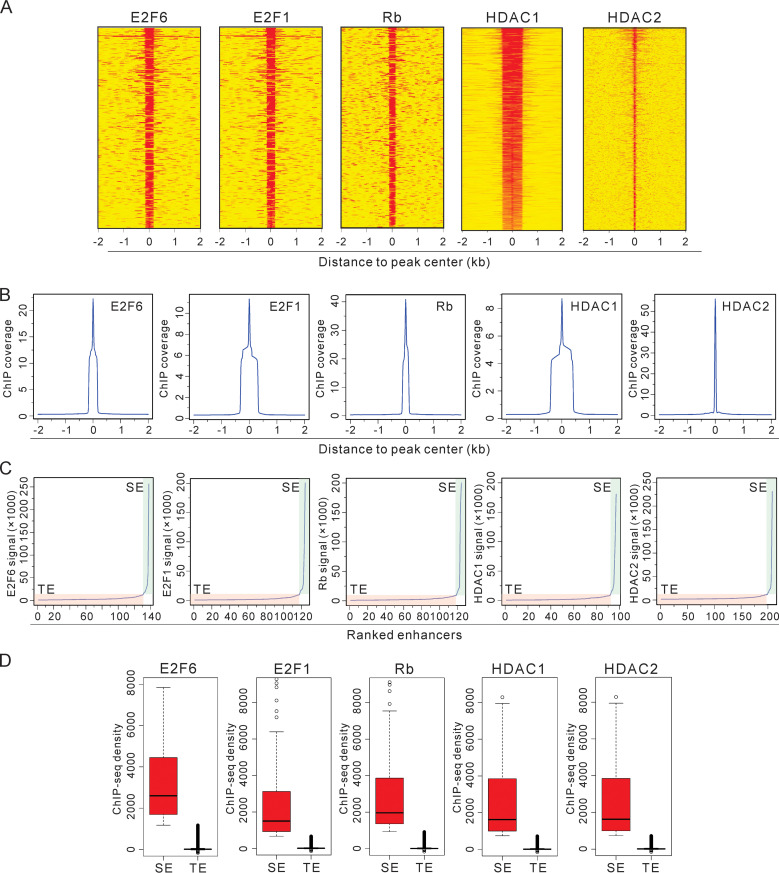
The E2F-Rb-HDAC signaling pathway that is linked to many functional activities can transcriptionally regulate its downstream targets (9, 10, 47). Following our determination of the patterns of their binding sites or regions in EBV-positive LCL cells, we found that E2F6, Rb, and HDAC2 could bind to very small regions of 100 to 200 bp, while E2F1 and HDAC1 bound to a relatively wider region which encompasses approximately 400 bp in size (Fig. 2, A and B). These protein complexes can only bind to a small region and have universal characteristics of binding to human genomic regions as a single binding factor. However, they may function in long-range regulatory regions when generating a multicomponent complex and coordinating with other proteins to perform a specific function in gene regulation. To detect whether E2F-Rb-HDAC can cooperate as a regulatory complex in LCL1 cells, super-enhancer analysis using the HOMER program was performed to identify their associated enhancers (48, 49). The results from our analyses identified super-enhancers (SE) as well as typical enhancers (TE) that were defined by the slope threshold of 1 (Fig. 2C). Furthermore, these five cellular transcription factors are more enriched at the super-enhancer sites than the typical enhancer sites in EBV-transformed LCLs (Fig. 2D), suggesting that the functions of these proteins are consistent with regulation of transcriptional networks from long-range enhancers, although they may need the assistance of additional recruiters.
FIG 2.
The E2F-Rb-HDAC complex can function as a super-enhancer in LCLs. (A and B) Heatmap view (A) and anchor plots (B) of E2F6, E2F1, Rb, HDAC1, and HDAC2 binding intensity at annotated human genome in LCLs. ChIP-Seq signals of the indicated cellular factors were created in a ±2-kb window. (C) The rank order of E2F6, E2F1, Rb, HDAC1, and HDAC2 ChIP-Seq signals for their enhancers with the module of “finding super enhancers” in the HOMER program. The typical enhancers (TE) and super enhancers (SE) are highlighted with light orange or blue, respectively. (D) Boxplots of E2F6, E2F1, Rb, HDAC1, and HDAC2 ChIP-Seq signal density at super-enhancers and typical enhancers. SE, super enhancers; TE, typical enhancers.
The E2F-Rb-HDAC complex regulates shared downstream signaling pathways.
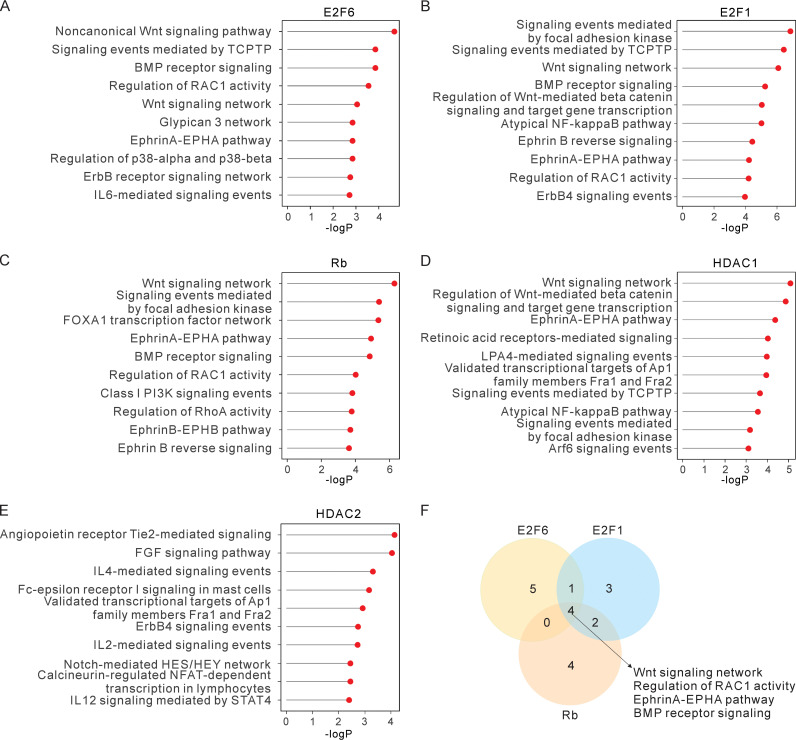
To further investigate the cooperation of E2F-Rb-HDAC, we analyzed their regulated downstream signaling pathways with gene ontology analysis based on their binding regions. The results demonstrated that they are potentially involved in several common pathways (Fig. 3A to E). These cellular factors can target many common binding sites as shown in Results (Fig. 1D). More specifically, both E2F6 and E2F1/Rb are linked to the Wnt signaling network, regulation of RAC1 activity, the EphrinA-EPHA pathway, and BMP receptor signaling (Fig. 3F), which are tightly associated with EBV infection (50–54). Similarly, HDAC1, but not HDAC2, is also associated with the Wnt signaling network, suggesting that E2F-Rb interactions are more likely to recruit HDAC1 for the related regulation (Fig. 3D). Besides, the shared signaling pathway of HDAC1 and HDAC2 in LCLs is related to AP1 family members Fra1 and Fra2 (Fig. 3D and E), which are also involved in EBV infection (55). Therefore, these results suggest that multiple common transcription regulatory patterns linked to these cellular factors are associated with EBV-induced B-cell lymphomas.
FIG 3.
The E2F-Rb-HDAC complex regulates shared downstream signaling pathways. (A to E) Gene ontology analysis is conducted to annotate enriched peaks or regions with the ChIP-Seq data of E2F6 (A), E2F1 (B), Rb (C), HDAC1 (D), and HDAC2 (E) and shows the regulated signaling pathways by these cellular factors. GO annotation of the top 10 enriched signaling pathways was shown according to the indicated P value. (F) Venn diagram demonstrating the overlapped signaling pathways among E2F6, E2F1, and Rb in LCLs. Four shared signaling pathways of these three transcription factors (E2F6, E2F1, and Rb) were highlighted.
EBV regulates the binding of E2F6 and E2F1/Rb at KLF gene regions.
E2F6 and E2F1 are the critical transcription factors in the E2F-Rb-HDAC complex that can directly bind to genomic DNA (2, 56, 57). To further explore the enriched genomic regions bound by these cellular factors, motif analysis was performed to identify their shared targets in EBV-positive LCLs. Among these binding motifs, we found that these five cellular transcription regulators could bind several common target genes, including KLF10 and KLF14, which were two of the most significant targets as determined by P value (Fig. 4A and B). Therefore, these results demonstrated that the E2F-Rb-HDAC complex which contains E2F6 or E2F1/Rb, and HDAC1/HDAC2 showed several consistent patterns, such as binding sites, the length of enrichment regions, the associated downstream pathways, and multiple targeted genes.
FIG 4.
EBV regulates the binding of E2F6 and E2F1/Rb at KLF gene regions. (A) Anchor plots show the indicated cellular factor (E2F6, E2F1, Rb, HDAC1, and HDAC2) binding sites and targeted genes. ChIP-Seq signals of the cellular factors on the described targets were created in a ±500-bp window. (B) Motif analysis by HOMER presents the enriched binding motifs of E2F6 and E2F1 in LCLs. (C and D) ChIP experiments show the enrichment of E2F6, E2F1, and Rb at the coding region of KLF10 (C) or KLF14 (D) in BJAB and LCL1 cells. Results are the mean ± standard error of the triplicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
KLF10 and KLF14 are two members of the Krüppel-like family of transcription factors (KLFs) (58). KLF10, also named TGF-β inducible early gene-1 (TIEG-1), is rapidly induced by TGF-β1 (59). Furthermore, TGF-β1 can be induced by EBV infection and this induction leads to activation of EBV lytic infection (60, 61). This suggests that KLF10 may be upregulated during EBV infection following the induction of TGF-β1. The functions of KLF14 and its potential roles in EBV-infected cells are not well-known. One study showed that KLF14 was necessary for the suppression of centrosome amplification and tumorigenesis (62). To verify the regulation of the E2F-Rb-HDAC complex on KLF10 and KLF14 genes, we performed chromatin immunoprecipitation (ChIP) assays to detect whether E2F6 or E2F1/Rb could bind to KLF10 and KLF14 loci in EBV-negative BJAB and EBV-positive LCL1 cells. Our results demonstrated that both E2F6 and E2F1/Rb could bind to both KLF10 and KLF14 coding regions. These three cellular factors (E2F6, E2F1, and Rb) were more enriched at the KLF10 gene in LCL1 cells compared to that seen in BJAB cells (Fig. 4C). Additionally, E2F6 and E2F1 showed more enrichment at the KLF14 gene in EBV-positive LCL1 cells than in EBV-negative BJAB cells, but Rb showed the opposite enrichment in these cell lines (Fig. 4D). The results suggested that the regulation of KLF10 and KLF14 may be mediated by distinct mechanisms in LCL1 cells. In particular, KLF10 expression is more dependent on Rb than KLF14 expression, which can be modulated by EBV latent antigens through directly targeting E2F6 or E2F1 without the assistance of Rb (36, 37). Therefore, these findings demonstrated not only that these proteins could bind to KLF10 and KLF14 genes in the B-cell background, but also that EBV latent antigens may coordinate with E2F6 or E2F/Rb to regulate the downstream genes which include KLF10 and KLF14.
EBV latent antigens are responsible for KLF14 upregulation.
In EBV latently infected B cells, the E2F-Rb-HDAC multifunctional repressor complex involved in crucial regulatory pathways controlled by EBV latent antigens (34, 36, 37, 63, 64). To determine whether KLFs are associated with EBV infection, we first explored the expression of KLF10 and KLF14 in EBV-transformed GM12878 cells from public data sets (65, 66). GM12878 is another EBV-transformed lymphoblastoid cell line that has been widely used in the International HapMap Project and the Encyclopedia of DNA Elements (ENCODE) Project (65, 66). By searching the histone modifications of the KLF10 or KLF14 gene in the ENCODE project, we found that the upstream regions of KLF10 or KLF14 coding sequences were highly enriched by H3K4me1, H3K4me2, and H3K4me3 modifications, which were related to activated promoters or enhancers (67) (Fig. 5A). Additionally, the RNA-Seq data indicated the peaks of KLF10 or KLF14 gene in EBV-transformed GM12878 cells (Fig. 5A). These results showed that KLF10 and KLF14 genes are associated with high expression in EBV-positive cells.
FIG 5.
EBV latent antigens are responsible for KLF14 upregulation. (A) RNA-Seq from the ENCODE data set shows the mRNA expression of KLF10 or KlF14 in EBV-transformed GM12878 cells, while ChIP-Seq from the ENCODE data set shows the enrichment of histone modifications (H3K4me1, H3K4me, H3K4me3) at the KLF10 or KLF14 gene in GM12878 cells. (B) Real-time PCR analysis shows the level of KLF14 mRNA expression in BJAB (EBV-negative cell line), Raji (EBV-positive cell line expressing only EBNA1 antigen), BL41/B95.8 and LCL1 (EBV-positive cell lines expressing all of the EBV latent antigens) cells. Results are the mean ± standard error of the triplicates. **, P < 0.01; ***, P < 0.001. (C) Real-time PCR analysis shows KLF14 mRNA expression in EBV-positive cell lines with latency I program (SavI, KemI, MutuI) or latency III program (SavIII, KemIII, MutuIII). Results are the mean ± standard error of the triplicates. ***, P < 0.001; ****, P < 0.0001.
The expression and functional role of KLF14 in EBV-infected cells have not yet been elucidated. To determine KLF14 expression in the presence of EBV infection, quantitative real-time PCR analyses were conducted with different EBV-negative or EBV-positive B cell lines. The results showed that KLF14 expression was slightly downregulated in EBV-positive Raji cells compared to EBV-negative BJAB cells (Fig. 5B). The viral-encoded EBNA1 antigen is the most prominent latent antigen expressed in Raji cells, which has a deletion in the EBNA3C gene and lacks EBNA3C (68). This suggested that EBNA1 may function as a repressor of KLF14 mRNA expression in EBV latently infected B cells. Interestingly, two other EBV-positive cell lines (BL41/B95.8 and LCL1) expressed more KLF14 mRNA than the BJAB cell line (Fig. 5B). All EBV latent antigens were expressed in these two cell lines, therefore, this strongly suggests that EBV latent antigens, except for EBNA1 can significantly promote the upregulation of KLF14 mRNA expression in EBV-positive cells. To further validate our results, we detected KLF14 mRNA expression in other EBV latently infected cell lines, which were matched in latency I program (SavI, KemI, MutuI) only expressing EBNA1 antigen or in latency III program (SavIII, KemIII, MutuIII) expressing all EBV latent antigens. The results clearly showed that KLF14 mRNA was upregulated in EBV latency III cells compared to EBV latency I cells, which indicated that the EBV latent antigens besides EBNA1 played a critical role in upregulating KLF14 mRNA expression (Fig. 5C).
EBV latent antigens cooperate with the E2F-Rb-HDAC complex to target genes.
In addition to KLF10 and KLF14 genes, we further detected the enrichment of the E2F-Rb-HDAC complex on other gene regions of crucial cellular factors in EBV-transformed LCL cells. These results identified the binding regions of the EBV latent antigens (EBNA1, EBNA2, EBNA3C, EBNA-LP) and the E2F-Rb-HDAC complex on BCL2 and MIR155HG loci in LCLs (Fig. 6A and B). These two genes are representative key players that are induced in EBV-mediated lymphomagenesis (69, 70). More interestingly, although these viral antigens and cellular factors could bind to the coding regions of BCL2 and MIR155HG, their binding regions were different although adjacent. Previous studies also showed these loci were highly active through interaction with other regions or transcription factors using RNAPII ChIA-PET and CTCF ChIA-PET data in the context of EBV infection (27, 28). Our findings further demonstrate that the E2F-Rb-HDAC complex may also involve complicated regulation as an important component of super-enhancers. In addition, the lack of EBNA1 enrichment on these genes indicated that it may not associate with the E2F-Rb-HDAC complex, which was consistent with our previous conclusion on KLF14 expression in EBV-infected cells (Fig. 5B and C). These results indicate that EBV latent antigens can modulate the E2F-Rb-HDAC complex by regulating E2F6 or E2F1/Rb to further target the downstream gene expression pathways (Fig. 6C).
FIG 6.
EBV latent antigens cooperate with the E2F-Rb-HDAC complex to target genes. (A and B) ChIP-Seq analysis shows the snapshot signal maps of representative targets BCL2 (A) and MIR155HG (B) by the E2F-Rb-HDAC complex and EBV latent antigens. The peaks or binding regions are highlighted. (C) Models of human-EBV regulatory interactions in LCLs. E2F6 or E2F1/Rb cooperates with HDAC1 or HDAC2 and binds to the target enhancers as a complex to modulate the downstream gene expression (e.g., KLF10/KLF14, BCL2, MIR155HG), but EBV latent antigens (EBNA2, EBNA3C, and EBNA-LP) can reverse the suppressive activities and facilitate gene expression in EBV-transformed LCLs.
DISCUSSION
EBV is the first human tumor virus that was identified more than 50 years ago. EBV is best known for its ability to establish latent infection and transforming normal B-lymphocytes in the infected host. However, it is still a huge challenge to conditionally control the transformation process and fully describe the critical processes during EBV-induced tumorigenesis. A dysregulated transcriptional network is one of the hallmarks of EBV-induced transformation (71). With the rapid development of high-throughput sequencing methods, our current studies now provide a more comprehensive view of EBV antigens associated with cellular transcription regulatory networks (27, 28). Using chromatin immunoprecipitation and sequencing (ChIP-Seq), we identified the binding sites of several crucial cellular factors (E2F6, E2F1, Rb, HDAC, and HDAC2) and explored the patterns of EBV induced regulation through long-range enhancers. The shared characteristics of their binding sites suggest analogous functions of these cellular factors. E2F1 is the transcriptional activator of E2F family members, while E2F6 is a repressor whose functions may be associated with the recruitment of the polycomb transcriptional repressor complex (72). Both E2F1 and E2F6 can bind to the E2F site within the promoters of numerous genes, which have also been shown in our results. Together with Rb, E2F1 binds to specific sites or regions of the human genome and recruits histone deacetylases (HDACs) or histone acetyltransferases (HATs) to modify chromatin structure (44, 73, 74). E2F6 can function in an Rb-independent pathway (1, 57). Thus, their similar mechanisms, in turn, are supported by their similar distribution on the human genome. In this study, we showed that KLF10 and KL14 were two new targets of the E2F-Rb-HDAC complex, which can be modulated by multiple EBV latent antigens. Our ChIP experiments showed that these cellular factors bound to the coding regions of KLF10 and KLF14 loci, but whether they can directly target their promoter regions need further investigation. More importantly, it is still not clear how EBV latent antigens upregulate KLF10 and KLF14 expression by modulating the E2F-Rb-HDAC complex and what are the key functions of KLFs in EBV-mediated lymphomagenesis.
Although E2F-Rb-HDAC could function as a complex to regulate the expression of target genes, the dynamic components of this functional structure are still unanswered. Rb interacts with other cellular proteins to remodel chromatin structure, including hBRM, BRG1, and SUV39H1 (75). To be specific, hBRM and BRG1 are the human homologs of SWI2/SNF2 that are the ATPases in yeast (76). These ATPases are involved in ATP-dependent nucleosome remodeling complexes to remodel chromatin structure and further control transcriptional repression and activation (13, 77). Rb also forms a repressor with HDACs and hSWI/SNF complex to suppress cyclin E and cyclin A expression (13). Moreover, Rb can inhibit E2F1 transcription activity by recruiting hBRM (78). The fact that Rb interacts with hBRM and E2F1 simultaneously suggests that the potential regulatory complex may cooperate at promoters with E2F binding sites (78). Loss of Rb function induces a p53-dependent apoptotic pathway, which may release E2F expression as well as ARF activation (79, 80). Besides Rb and hBRM proteins, p300/CBP and pCAF also act as a coactivator of E2F1 (81). Therefore, it is a huge challenge to completely understand how these cellular factors cooperate and organize on the human genome. The complex cooperation is likely to be critical for revealing the transcriptional regulatory networks in oncogenesis. Here, we concentrated on several key transcription factors (E2F6, E2F1, Rb, HDAC1, and HDAC2) to explore their genome-wide functional characteristics by using high-throughput sequencing strategies. The ChIP-Seq results showed that BCL2 and MIR155HG were two crucial targets of the E2F-Rb-HDAC complex. Previous studies also indicated the BCL2 and MIR155HG loci were highly active and linked to EBV super-enhancers by cooperating with multiple transcription factors (27, 28). These results suggested that the E2F-Rb-HDAC complex together with EBV latent antigens were also involved in complex transcriptional regulation, but how this complex interacted with other transcription factors in the presence of EBV latent antigen is still a mystery.
The transcriptional regulatory network of human cells involves the modification of chromatin structure. In EBV-infected cells, how the latent antigens reorganize these complexes and reprogram transcription networks to facilitate tumorigenesis is also a complicated topic. Fortunately, many recent studies have implicated these interaction networks or models between EBV latent antigens and cellular factors by utilizing high-throughput genome sequence analysis (19). The novel proximity-based labeling techniques, such as BioID, APEX, and TurboID, have been developed as powerful approaches to capture the weak and transient interactions and map various scales of protein-protein interaction networks (82–84). How EBV latent antigens regulate host transcriptional networks to mediate lymphomagenesis will be further investigated with a combination of technical platforms for the development of novel treatments. Nevertheless, this study has demonstrated the potential functions of E2F-Rb-HDAC, which involve super-enhancers, and has now identified two new KLF targets that can be promoted by EBV latent antigens and may be potential therapeutic targets for treating EBV-associated lymphomas.
MATERIALS AND METHODS
Cells and antibodies.
EBV-transformed immortalized LCL1 (lymphoblastoid cell line) cells were generated in our laboratory and were grown in RPMI 1640 medium (HyClone, Logan, UT) supplemented with 5% fetal bovine serum (FBS), 25 U/ml penicillin, 50 μg/ml streptomycin, and 2 mM l-glutamine (85). Two Burkitt’s lymphoma cell lines (BJAB and Raji), EBV converted BL41/B95.8 lymphoma cell line, and other EBV-positive cell lines (SavI, SavIII; KemI, KemIII; MutuI, MutuIII) were also cultured with the above-described RPMI 1640 medium.
The following antibodies were used for ChIP-Seq experiments: rabbit anti-E2F1 (sc-193X; Santa Cruz), rabbit anti-E2F6 (sc-22823X; Santa Cruz), mouse anti-Rb (G3-245; BD Pharmingen), rabbit anti-HDAC1 (ab7028; Abcam), rabbit anti-HDAC2 (ab7029; Abcam), normal mouse IgG (sc-2025; Santa Cruz), and normal rabbit IgG (sc-2027; Santa Cruz).
Chromatin immunoprecipitation (ChIP).
The method has been described in the previous paper (86). Briefly, 30 million BJAB or LCL1 cells were cross-linked with 1% formaldehyde and harvested for sonication. Then cell lysates including sheared genomic DNA were immunoprecipitated with the indicated antibody (anti-E2F1, anti-E2F6, anti-Rb, anti-HDAC1, and anti-HDAC2) or normal IgG to collect the DNA fragments that can bind with the proteins of interest. The immune complexes were incubated with the salmon sperm DNA/protein A agarose slurry. Then the collected DNA was purified and subjected to real-time PCR analysis. The primers used for KLF10 were 5′-CCTTCCAGCCTCCATATTC-3′ and 5′-CAACACAGGTAGCACAGAT-3′; those used for KLF14 were 5′-GACTTGTAATAGGCTTTGGTG-3′ and 5′-GGAGGAGGTCTGTCACAC-3′.
ChIP-Seq assay.
The DNA library was prepared with a TruSeq ChIP library preparation kit (IP-202-1012; Illumina, San Diego, CA). Then the purified DNAs were sequenced using the Illumina HiSeq platform by the Genome Technology Access Center (GTAC) at the Washington University in St. Louis. These ChIP-Seq data are available at the NCBI Gene Expression Omnibus (GEO) with accession number GSE148165.
ChIP-Seq analysis.
ChIP-Seq reads were mapped to the hg19 genome using Bowtie (87). Peak calling, the annotations of enriched regions, motif discovery, and pathway analysis were performed with HOMER (Hypergeometric Optimization of Motif EnRichment; http://homer.ucsd.edu/homer/) (48) or CLC Genomics Workbench version 12.0 (CLC Bio, Qiagen, USA). Pathway analysis was also performed using HOMER or Ingenuity Pathway Analysis (IPA; Qiagen, USA). The analyzed data were visualized using the R program (https://www.r-project.org/) and the Integrative Genomics Viewer (IGV) (88).
Quantitative real-time PCR.
Total RNAs from the cells were extracted with TRIzol reagent (Invitrogen, Inc., Carlsbad, CA), treated with DNase I (Invitrogen, Inc., Carlsbad, CA), and reverse-transcribed with a Superscript II reverse transcriptase kit (Invitrogen, Inc., Carlsbad, CA). Then quantitative real-time PCR analysis was performed with the SYBR green real-time master mix (MJ Research, Inc., Waltham, MA) according to the manufacturer’s protocol. The primers used for KL14 were 5′-AAGCCTATTACAAGTCGTC-3′ and 5′-TAAACTTCTTGTCGCAGTC-3′. GAPDH was set as an internal control as previously described (86). These assays were conducted in triplicate.
Statistical analysis.
GraphPad Prism software version 6.01 was used for statistical analysis. The mean values with standard deviation (SD) were presented in this study, and the significance of differences was calculated with a 2-tailed Student’s t test. The P value of <0.05 was considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Data availability.
The EBV latent antigen-associated ChIP-Seq data in EBV-positive cells were downloaded from NCBI GEO data sets with accession numbers GSE73887 (EBNA1), GSE29498 (EBNA2), GSE52632 (EBNA3C), and GSE49338 (EBNA-LP). These RNA-Seq data and the histone marker-related ChIP-Seq data in GM12878 (EBV-transformed lymphoblastoid cell line) were analyzed and visualized with the WashU Epigenome Browser (https://epigenomegateway.wustl.edu/).
ACKNOWLEDGMENTS
We express our gratitude to Elliott Kieff (Harvard Medical School, Boston, MA), Paul M. Lieberman (The Wistar Institute, Philadelphia, PA), and Yan Yuan (University of Pennsylvania, Philadelphia, PA) for kindly providing cell lines and reagents.
This work was supported by the National Cancer Institute at the National Institutes of Health public health service grants P30-CA016520, R01-CA171979, P01-CA174439, and R01-CA177423 to E.S.R. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Chen HZ, Tsai SY, Leone G. 2009. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer 9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attwooll C, Lazzerini Denchi E, Helin K. 2004. The E2F family: specific functions and overlapping interests. EMBO J 23:4709–4716. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nevins JR. 1992. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science 258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 4.Dyson N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev 12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 5.Nevins JR. 2001. The Rb/E2F pathway and cancer. Hum Mol Genet 10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 6.Sengupta S, Henry RW. 2015. Regulation of the retinoblastoma-E2F pathway by the ubiquitin-proteasome system. Biochim Biophys Acta 1849:1289–1297. doi: 10.1016/j.bbagrm.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Flemington EK, Speck SH, Kaelin WG Jr. 1993. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci U S A 90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helin K, Harlow E, Fattaey A. 1993. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol 13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo RX, Postigo AA, Dean DC. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 10.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 11.Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 12.Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, Te Riele H, Dynlacht BD. 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev 16:933–947. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 14.Dyson N, Howley PM, Munger K, Harlow E. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 15.Whyte P, Buchkovich KJ, Horowitz JM, Friend SH, Raybuck M, Weinberg RA, Harlow E. 1988. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature 334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 16.DeCaprio JA, Ludlow JW, Figge J, Shew JY, Huang CM, Lee WH, Marsilio E, Paucha E, Livingston DM. 1988. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell 54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 17.Lee JO, Russo AA, Pavletich NP. 1998. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature 391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 18.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet 25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 19.Pei Y, Lewis AE, Robertson ES. 2017. Current progress in EBV-associated B-cell lymphomas. Adv Exp Med Biol 1018:57–74. doi: 10.1007/978-981-10-5765-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jha H, Pei Y, Robertson E. 2016. Epstein Barr virus: diseases linked to infection and transformation. Front Microbiol 7:1602. doi: 10.3389/fmicb.2016.01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epstein MA, Achong BG, Barr YM. 1964. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet 1:702–703. doi: 10.1016/S0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- 22.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. 2012. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol 13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 23.Kutok JL, Wang F. 2006. Spectrum of Epstein-Barr virus-associated diseases. Annu Rev Pathol 1:375–404. doi: 10.1146/annurev.pathol.1.110304.100209. [DOI] [PubMed] [Google Scholar]
- 24.Mesri EA, Feitelson MA, Munger K. 2014. Human viral oncogenesis: a cancer hallmarks analysis. Cell Host Microbe 15:266–282. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munz C. 2019. Latency and lytic replication in Epstein-Barr virus-associated oncogenesis. Nat Rev Microbiol 17:691–700. doi: 10.1038/s41579-019-0249-7. [DOI] [PubMed] [Google Scholar]
- 26.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H, Schmidt SC, Jiang S, Willox B, Bernhardt K, Liang J, Johannsen EC, Kharchenko P, Gewurz BE, Kieff E, Zhao B. 2015. Epstein-Barr virus oncoprotein super-enhancers control B cell growth. Cell Host Microbe 17:205–216. doi: 10.1016/j.chom.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang S, Zhou H, Liang J, Gerdt C, Wang C, Ke L, Schmidt SCS, Narita Y, Ma Y, Wang S, Colson T, Gewurz B, Li G, Kieff E, Zhao B. 2017. The Epstein-Barr virus regulome in lymphoblastoid cells. Cell Host Microbe 22:561–573.e564. doi: 10.1016/j.chom.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood CD, Veenstra H, Khasnis S, Gunnell A, Webb HM, Shannon-Lowe C, Andrews S, Osborne CS, West MJ. 2016. MYC activation and BCL2L11 silencing by a tumour virus through the large-scale reconfiguration of enhancer-promoter hubs. Elife 5:e18270. doi: 10.7554/eLife.18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C, Sista ND, Pagano JS. 1996. Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J Virol 70:2545–2555. doi: 10.1128/JVI.70.4.2545-2555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mauser A, Holley-Guthrie E, Zanation A, Yarborough W, Kaufmann W, Klingelhutz A, Seaman WT, Kenney S. 2002. The Epstein-Barr virus immediate-early protein BZLF1 induces expression of E2F-1 and other proteins involved in cell cycle progression in primary keratinocytes and gastric carcinoma cells. J Virol 76:12543–12552. doi: 10.1128/jvi.76.24.12543-12552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everly DN Jr, Mainou BA, Raab-Traub N. 2009. Transcriptional downregulation of p27KIP1 through regulation of E2F function during LMP1-mediated transformation. J Virol 83:12671–12679. doi: 10.1128/JVI.01422-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sung NS, Wilson J, Davenport M, Sista ND, Pagano JS. 1994. Reciprocal regulation of the Epstein-Barr virus BamHI-F promoter by EBNA-1 and an E2F transcription factor. Mol Cell Biol 14:7144–7152. doi: 10.1128/mcb.14.11.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knight JS, Sharma N, Robertson ES. 2005. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci U S A 102:18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szekely L, Selivanova G, Magnusson KP, Klein G, Wiman KG. 1993. EBNA-5, an Epstein-Barr virus-encoded nuclear antigen, binds to the retinoblastoma and p53 proteins. Proc Natl Acad Sci U S A 90:5455–5459. doi: 10.1073/pnas.90.12.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pei Y, Banerjee S, Sun Z, Jha HC, Saha A, Robertson ES. 2016. EBV nuclear antigen 3C mediates regulation of E2F6 to inhibit E2F1 transcription and promote cell proliferation. PLoS Pathog 12:e1005844. doi: 10.1371/journal.ppat.1005844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saha A, Lu J, Morizur L, Upadhyay SK, Aj MP, Robertson ES. 2012. E2F1 mediated apoptosis induced by the DNA damage response is blocked by EBV nuclear antigen 3C in lymphoblastoid cells. PLoS Pathog 8:e1002573. doi: 10.1371/journal.ppat.1002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saha A, Jha HC, Upadhyay SK, Robertson ES. 2015. Epigenetic silencing of tumor suppressor genes during in vitro Epstein-Barr virus infection. Proc Natl Acad Sci U S A 112:E5199–E5207. doi: 10.1073/pnas.1503806112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ropero S, Esteller M. 2007. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol 1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gruffat H, Manet E, Sergeant A. 2002. MEF2-mediated recruitment of class II HDAC at the EBV immediate early gene BZLF1 links latency and chromatin remodeling. EMBO Rep 3:141–146. doi: 10.1093/embo-reports/kvf031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin DY, Kim A, Kang HJ, Park S, Kim DW, Lee SS. 2015. Histone deacetylase inhibitor romidepsin induces efficient tumor cell lysis via selective down-regulation of LMP1 and c-myc expression in EBV-positive diffuse large B-cell lymphoma. Cancer Lett 364:89–97. doi: 10.1016/j.canlet.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 42.Daigle D, Megyola C, El-Guindy A, Gradoville L, Tuck D, Miller G, Bhaduri-McIntosh S. 2010. Upregulation of STAT3 marks Burkitt lymphoma cells refractory to Epstein-Barr virus lytic cycle induction by HDAC inhibitors. J Virol 84:993–1004. doi: 10.1128/JVI.01745-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh SK, Perrine SP, Williams RM, Faller DV. 2012. Histone deacetylase inhibitors are potent inducers of gene expression in latent EBV and sensitize lymphoma cells to nucleoside antiviral agents. Blood 119:1008–1017. doi: 10.1182/blood-2011-06-362434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harbour JW, Dean DC. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev 14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 45.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. 2000. Regulation of E2F1 activity by acetylation. EMBO J 19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Woodford N, Xia X, Hamburger AW. 2003. Repression of E2F1-mediated transcription by the ErbB3 binding protein Ebp1 involves histone deacetylases. Nucleic Acids Res 31:2168–2177. doi: 10.1093/nar/gkg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caretti G, Salsi V, Vecchi C, Imbriano C, Mantovani R. 2003. Dynamic recruitment of NF-Y and histone acetyltransferases on cell-cycle promoters. J Biol Chem 278:30435–30440. doi: 10.1074/jbc.M304606200. [DOI] [PubMed] [Google Scholar]
- 48.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morrison JA, Klingelhutz AJ, Raab-Traub N. 2003. Epstein-Barr virus latent membrane protein 2A activates beta-catenin signaling in epithelial cells. J Virol 77:12276–12284. doi: 10.1128/jvi.77.22.12276-12284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao B, Mar JC, Maruo S, Lee S, Gewurz BE, Johannsen E, Holton K, Rubio R, Takada K, Quackenbush J, Kieff E. 2011. Epstein-Barr virus nuclear antigen 3C regulated genes in lymphoblastoid cell lines. Proc Natl Acad Sci U S A 108:337–342. doi: 10.1073/pnas.1017419108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin Q, Wang X, Fewell C, Cameron J, Zhu H, Baddoo M, Lin Z, Flemington EK. 2010. MicroRNA miR-155 inhibits bone morphogenetic protein (BMP) signaling and BMP-mediated Epstein-Barr virus reactivation. J Virol 84:6318–6327. doi: 10.1128/JVI.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J, Sathiyamoorthy K, Zhang X, Schaller S, Perez White BE, Jardetzky TS, Longnecker R. 2018. Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat Microbiol 3:172–180. doi: 10.1038/s41564-017-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang H, Li Y, Wang HB, Zhang A, Chen ML, Fang ZX, Dong XD, Li SB, Du Y, Xiong D, He JY, Li MZ, Liu YM, Zhou AJ, Zhong Q, Zeng YX, Kieff E, Zhang Z, Gewurz BE, Zhao B, Zeng MS. 2018. Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat Microbiol 3:1–8. doi: 10.1038/s41564-017-0080-8. [DOI] [PubMed] [Google Scholar]
- 55.Lan YY, Hsiao JR, Chang KC, Chang JS, Chen CW, Lai HC, Wu SY, Yeh TH, Chang FH, Lin WH, Su IJ, Chang Y. 2012. Epstein-Barr virus latent membrane protein 2A promotes invasion of nasopharyngeal carcinoma cells through ERK/Fra-1-mediated induction of matrix metalloproteinase 9. J Virol 86:6656–6667. doi: 10.1128/JVI.00174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dimova DK, Dyson NJ. 2005. The E2F transcriptional network: old acquaintances with new faces. Oncogene 24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 57.Trimarchi JM, Lees JA. 2002. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 58.McConnell BB, Yang VW. 2010. Mammalian Kruppel-like factors in health and diseases. Physiol Rev 90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Subramaniam M, Hawse JR, Rajamannan NM, Ingle JN, Spelsberg TC. 2010. Functional role of KLF10 in multiple disease processes. Biofactors 36:8–18. doi: 10.1002/biof.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang CL, Chen JL, Hsu YP, Ou JT, Chang YS. 2002. Epstein-Barr virus BZLF1 gene is activated by transforming growth factor-beta through cooperativity of Smads and c-Jun/c-Fos proteins. J Biol Chem 277:23345–23357. doi: 10.1074/jbc.M107420200. [DOI] [PubMed] [Google Scholar]
- 61.Foster AE, Dotti G, Lu A, Khalil M, Brenner MK, Heslop HE, Rooney CM, Bollard CM. 2008. Antitumor activity of EBV-specific T lymphocytes transduced with a dominant negative TGF-beta receptor. J Immunother 31:500–505. doi: 10.1097/CJI.0b013e318177092b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan G, Sun L, Shan P, Zhang X, Huan J, Zhang X, Li D, Wang T, Wei T, Zhang X, Gu X, Yao L, Xuan Y, Hou Z, Cui Y, Cao L, Li X, Zhang S, Wang C. 2015. Loss of KLF14 triggers centrosome amplification and tumorigenesis. Nat Commun 6:8450. doi: 10.1038/ncomms9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zacny VL, Wilson J, Pagano JS. 1998. The Epstein-Barr virus immediate-early gene product, BRLF1, interacts with the retinoblastoma protein during the viral lytic cycle. J Virol 72:8043–8051. doi: 10.1128/JVI.72.10.8043-8051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ohtani N, Brennan P, Gaubatz S, Sanij E, Hertzog P, Wolvetang E, Ghysdael J, Rowe M, Hara E. 2003. Epstein-Barr virus LMP1 blocks p16INK4a-RB pathway by promoting nuclear export of E2F4/5. J Cell Biol 162:173–183. doi: 10.1083/jcb.200302085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, Onate KC, Graham K, Miyasato SR, Dreszer TR, Strattan JS, Jolanki O, Tanaka FY, Cherry JM. 2018. The Encyclopedia of DNA Elements (ENCODE): data portal update. Nucleic Acids Res 46:D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sims RJ III, Nishioka K, Reinberg D. 2003. Histone lysine methylation: a signature for chromatin function. Trends Genet 19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 68.Robertson ES, Ooka T, Kieff ED. 1996. Epstein-Barr virus vectors for gene delivery to B lymphocytes. Proc Natl Acad Sci U S A 93:11334–11340. doi: 10.1073/pnas.93.21.11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Henderson S, Rowe M, Gregory C, Croom-Carter D, Wang F, Longnecker R, Kieff E, Rickinson A. 1991. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell 65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- 70.Elton TS, Selemon H, Elton SM, Parinandi NL. 2013. Regulation of the MIR155 host gene in physiological and pathological processes. Gene 532:1–12. doi: 10.1016/j.gene.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 71.Bradner JE, Hnisz D, Young RA. 2017. Transcriptional addiction in cancer. Cell 168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trimarchi JM, Fairchild B, Wen J, Lees JA. 2001. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc Natl Acad Sci U S A 98:1519–1524. doi: 10.1073/pnas.041597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang HS, Dean DC. 2001. Rb-mediated chromatin structure regulation and transcriptional repression. Oncogene 20:3134–3138. doi: 10.1038/sj.onc.1204338. [DOI] [PubMed] [Google Scholar]
- 74.Macaluso M, Montanari M, Giordano A. 2006. Rb family proteins as modulators of gene expression and new aspects regarding the interaction with chromatin remodeling enzymes. Oncogene 25:5263–5267. doi: 10.1038/sj.onc.1209680. [DOI] [PubMed] [Google Scholar]
- 75.Giacinti C, Giordano A. 2006. RB and cell cycle progression. Oncogene 25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- 76.Phelan ML, Sif S, Narlikar GJ, Kingston RE. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell 3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 77.Tyler JK, Kadonaga JT. 1999. The “dark side” of chromatin remodeling: repressive effects on transcription. Cell 99:443–446. doi: 10.1016/s0092-8674(00)81530-5. [DOI] [PubMed] [Google Scholar]
- 78.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. 1997. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci U S A 94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, Cordon-Cardo C, DePinho RA. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell 92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 80.Morgenbesser SD, Williams BO, Jacks T, DePinho RA. 1994. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature 371:72–74. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- 81.Trouche D, Cook A, Kouzarides T. 1996. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res 24:4139–4145. doi: 10.1093/nar/24.21.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roux KJ, Kim DI, Raida M, Burke B. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. 2013. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY. 2018. Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cotter MA II, Robertson ES. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol Cell Biol 20:5722–5735. doi: 10.1128/mcb.20.15.5722-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pei Y, Banerjee S, Jha HC, Sun Z, Robertson ES. 2017. An essential EBV latent antigen 3C binds Bcl6 for targeted degradation and cell proliferation. PLoS Pathog 13:e1006500. doi: 10.1371/journal.ppat.1006500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The EBV latent antigen-associated ChIP-Seq data in EBV-positive cells were downloaded from NCBI GEO data sets with accession numbers GSE73887 (EBNA1), GSE29498 (EBNA2), GSE52632 (EBNA3C), and GSE49338 (EBNA-LP). These RNA-Seq data and the histone marker-related ChIP-Seq data in GM12878 (EBV-transformed lymphoblastoid cell line) were analyzed and visualized with the WashU Epigenome Browser (https://epigenomegateway.wustl.edu/).