Yellow fever is a mosquito-borne acute hemorrhagic disease caused by yellow fever virus (YFV). The mechanisms responsible for its pathogenesis remain largely unknown, although increased inflammation has been linked to worsened outcome. YFV targets the liver, where it primarily infects hepatocytes. We found that two RNA-sensing proteins, RIG-I and PKR, participate in the induction of proinflammatory mediators in human hepatocytes infected with YFV. We show that YFV infection promotes the formation of cytoplasmic structures, termed stress granules, in a PKR- but not RIG-I-dependent manner. While stress granules were previously postulated to be essential platforms for immune activation, we found that they are not required for the production of proinflammatory mediators upon YFV infection. Collectively, our work uncovered molecular events triggered by the replication of YFV, which could prove instrumental in clarifying the pathogenesis of the disease, with possible repercussions for disease management.
KEYWORDS: cytokines, flavivirus, innate immunity, interferons, liver inflammation, pattern recognition receptors, stress granules, yellow fever virus
ABSTRACT
Yellow fever virus (YFV) is an RNA virus primarily targeting the liver. Severe YF cases are responsible for hemorrhagic fever, plausibly precipitated by excessive proinflammatory cytokine response. Pathogen recognition receptors (PRRs), such as the cytoplasmic retinoic acid inducible gene I (RIG-I)-like receptors (RLRs), and the viral RNA sensor protein kinase R (PKR), are known to initiate a proinflammatory response upon recognition of viral genomes. Here, we sought to reveal the main determinants responsible for the acute cytokine expression occurring in human hepatocytes following YFV infection. Using a RIG-I-defective human hepatoma cell line, we found that RIG-I largely contributes to cytokine secretion upon YFV infection. In infected RIG-I-proficient hepatoma cells, RIG-I was localized in stress granules. These granules are large aggregates of stalled translation preinitiation complexes known to concentrate RLRs and PKR and are so far recognized as hubs orchestrating RNA virus sensing. Stable knockdown of PKR in hepatoma cells revealed that PKR contributes to both stress granule formation and cytokine induction upon YFV infection. However, stress granule disruption did not affect the cytokine response to YFV infection, as assessed by small interfering RNA (siRNA)-knockdown-mediated inhibition of stress granule assembly. Finally, no viral RNA was detected in stress granules using a fluorescence in situ hybridization approach coupled with immunofluorescence. Our findings suggest that both RIG-I and PKR mediate proinflammatory cytokine induction in YFV-infected hepatocytes, in a stress granule-independent manner. Therefore, by showing the uncoupling of the cytokine response from the stress granule formation, our model challenges the current view in which stress granules are required for the mounting of the acute antiviral response.
IMPORTANCE Yellow fever is a mosquito-borne acute hemorrhagic disease caused by yellow fever virus (YFV). The mechanisms responsible for its pathogenesis remain largely unknown, although increased inflammation has been linked to worsened outcome. YFV targets the liver, where it primarily infects hepatocytes. We found that two RNA-sensing proteins, RIG-I and PKR, participate in the induction of proinflammatory mediators in human hepatocytes infected with YFV. We show that YFV infection promotes the formation of cytoplasmic structures, termed stress granules, in a PKR- but not RIG-I-dependent manner. While stress granules were previously postulated to be essential platforms for immune activation, we found that they are not required for the production of proinflammatory mediators upon YFV infection. Collectively, our work uncovered molecular events triggered by the replication of YFV, which could prove instrumental in clarifying the pathogenesis of the disease, with possible repercussions for disease management.
INTRODUCTION
Flaviviruses are a group of more than 70 enveloped RNA viruses that can cause serious diseases in humans and animals (1, 2). They have provided some of the most important examples of emerging or resurging diseases of global significance. Most flaviviruses, such as dengue virus (DENV), yellow fever virus (YFV), and Zika virus (ZIKV), are arthropod-borne viruses transmitted to vertebrate hosts by mosquitoes or ticks (1). YFV is the prototype and eponymous virus of the Flavivirus genus. It is a small (40- to 60-nm), enveloped virus harboring a single positive-strand RNA genome of 11 kb. The genome encodes a polyprotein that is co- and posttranslationally cleaved into three structural proteins, namely, the capsid (C), membrane precursor (prM), and envelope (E), and seven nonstructural (NS) proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5). The C, prM, and E proteins are incorporated into the virions, while NS proteins are found only in infected cells (3). NS proteins coordinate RNA replication and viral assembly and modulate innate immune responses, while the structural proteins constitute the virion.
YFV is endemic in the tropical regions of sub-Saharan Africa and South America. The reference strain Asibi was isolated in 1927 in West Africa from the blood of a human patient. The vaccine strain 17D was developed empirically in the 1930s by passaging the Asibi strain in rhesus macaques and in mouse and chicken embryonic tissues (4). 17D is one of the most effective vaccines ever generated. It has been used safely and effectively on more than 600 million individuals over the past 70 years (4). However, due to poor vaccine coverage and vaccine shortages, the virus continues to cause disease in areas of endemicity, as illustrated by recent outbreaks in Angola and Brazil (5, 6).
Yellow fever (YF) pathogenesis is viscerotropic in humans, with viral replication in the liver being critical to the establishment of the disease (3). Severe YF is responsible for multisystem organ failure and viral hemorrhagic fever, resulting in up to 50% fatality. Similar to the case for Ebola hemorrhagic fever, cytokine dysregulation is thought to result in endothelial damage, disseminated intravascular coagulation, and circulatory shock in the terminal stage of the disease. Viral hemorrhagic fever is considered an illness precipitated by an excessive proinflammatory cytokine response (“cytokine storm”) (7, 8). Almost every cell type in the body can produce proinflammatory cytokines in response to various stimuli, such as a viral infection (9). The pattern of cytokine expression in response to infection with pathogenic strains of YFV has been described in both humans and rhesus macaques. Studies performed on patients during a YF outbreak in the Republic of Guinea in 2000 showed that excessive production of proinflammatory cytokines, such as interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α), correlates with high viremia and severe disease outcome (10). The levels of these two cytokines were statistically higher in patients with fatal hemorrhagic fever than in patients who survived. Similar findings were obtained with rhesus macaques infected with the strain Dakar-HD1279 (11). The animals died of hemorrhagic fever within 5 days. These studies also suggest that proinflammatory cytokines might be produced by injured organs, especially the liver, and not by cells circulating in the blood (11).
Upon viral infection, the cytokine response is initiated by the recognition of viral genomes and viral replication intermediates by pathogen recognition receptors (PRRs) (12), such as the ubiquitously expressed cytoplasmic retinoic acid inducible gene I (RIG-I)-like receptors (RLRs). Three RLRs have been identified in vertebrates: melanoma differentiation-associated gene 5 (MDA5), laboratory of genetics and physiology 2 (LGP2), and RIG-I (13). Upon binding to viral nucleic acids, MDA5 and RIG-I undergo a conformational change, which allows interaction with the adaptor protein mitochondrial antiviral signaling protein (MAVS) via their caspase-associated recruitment domains (CARDs) and subsequent recruitment of a signaling complex which comprises protein kinases such as TBK1, IκB kinase α (IKKα), IKKβ, and IKKε (14). These events lead to the activation and nuclear translocation of the transcription factors interferon regulatory factor 3 (IRF3) and NF-κB, which stimulate the rapid expression of proinflammatory cytokines and type I interferons (IFNs).
RIG-I plays an important role in initiating the IFN-mediated antiviral response against DENV and ZIKV (15–18). Induction of cytokines upon infection with the vaccine strain of YFV is also mediated by RIG-I in plasmacytoid dendritic cells (19). Whether RIG-I contributes to cytokine production in cells infected with wild-type YFV strains is not known.
The subcellular localization where RIG-I interacts with its ligands is not well described. Upon infection with influenza A virus lacking its nonstructural protein 1 (IAVΔNS1), Newcastle disease virus, or Sendai virus (20–23), RIG-I accumulates in cytoplasmic foci. Disruption of these foci, which have been identified as stress granules, reduced the abundance of IFNB mRNA in IAVΔNS1-infected cells (20). Thus, stress granules may be important for RIG-I signaling and could serve as a platform for viral RNA sensing. Moreover, other viral nucleic sensors, such as MDA5 (24), LGP2 (25), and cyclic GMP-AMP synthase (cGAS) (26), are also concentrated in these stress granules. Finally, numerous viruses interfere with stress granule formation, via, for instance, the cleavage of one of their core components by a viral protease. Thus, accumulation of viral sensors in stress granules and viral counteraction mechanisms suggest that these granules act as hubs that coordinate foreign nucleic acid sensing (27, 28).
Stress granules are nonmembranous cytoplasmic structures that concentrate translationally stalled mRNAs and proteins (29). Their assembly is triggered by sensing distinct types of stress, including viral infection, by one of four kinases, resulting in phosphorylation of the eukaryotic initiation factor 2α (eIF2α). These kinases are as follows: the heme-regulated inhibitor (HRI), which is activated by heat shock or oxidative stress (e.g., by reactive oxygen species [ROS] produced upon treatment with sodium arsenite); general control nondepressible protein 2 (GCN2), which reacts to UV-induced damage or amino acid deprivation; double-stranded RNA (dsRNA)-sensing protein kinase R (PKR); and PKR-like endoplasmic reticulum (ER) kinase (PERK), which is activated upon ER stress (29). Assembly of stress granules not only arrests the host translation machinery but also heavily disturbs viral translation, which is further evidence for their antiviral nature (27, 28, 30). Here, we investigated the role of RIG-I, PKR, and stress granules in the induction of inflammatory cytokines upon infection of hepatoma cells with YFV.
RESULTS
The Asibi strain of YFV triggers the production of proinflammatory cytokines in hepatoma cells through RIG-I.
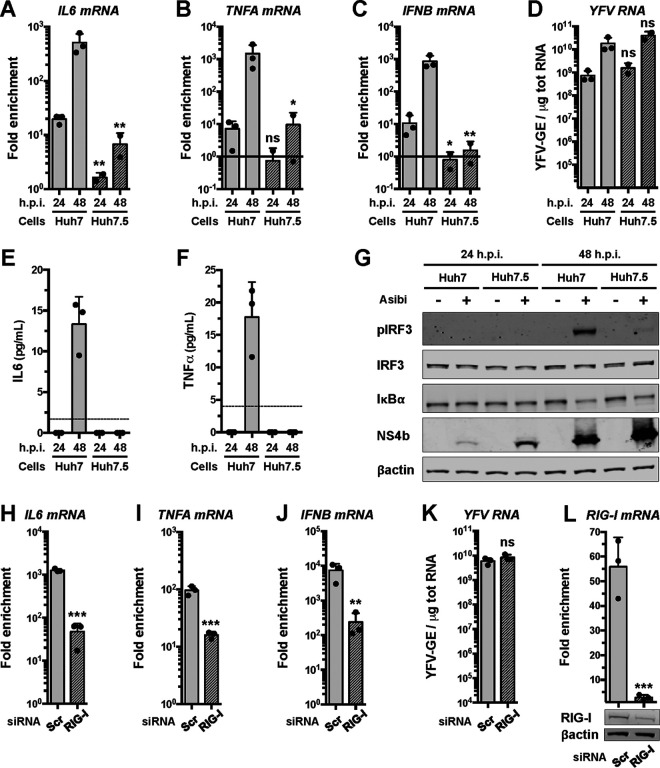
We examined whether the YFV reference strain Asibi could trigger the expression of proinflammatory cytokines in human hepatoma cells through the RIG-I signaling pathway. Huh7 and Huh7.5 cells were infected with the Asibi strain for 24 or 48 h. Huh7.5 cells, which derive from Huh7 cells, express an inactive form of RIG-I that possesses a mutation (T55I) in its first CARD (31). Reverse transcription-quantitative PCR (RT-qPCR) analysis revealed that the abundances of TNFA, IL-6, and IFNB mRNAs in Huh7 cells increased over time upon infection, while being significantly lower in Huh7.5 cells at 48 h postinfection (hpi) (Fig. 1A to C). We noticed that Huh7.5 cells infected for 48 h produced approximately 3-fold more viral RNA than Huh7 cells infected for the same amount of time (Fig. 1D). Although it was not statistically significant, this modest effect was reproducible (Fig. 1D) and suggests that absence of RIG-I signaling favors viral replication. Huh7 cells secreted around 10 to 20 pg/ml of IL-6 and TNF-α protein after 48 h of infection but not after 24 hpi (Fig. 1E, F). In contrast, the amount of IL-6 and TNF-α protein secreted by cells expressing the inactive form of RIG-I was below the detection limit of the ELISA (Fig. 1E and F). These data suggest that Asibi infection triggers the induction and secretion of proinflammatory cytokines in a RIG-I-dependent manner in human hepatoma cells.
FIG 1.
The Asibi strain of YFV triggers the production of proinflammatory cytokines in hepatoma cells through RIG-I. Huh7 and Huh7.5 cells were left uninfected or infected with Asibi at an MOI of 1 for 24 or 48 h. (A to C) The relative amounts of IL-6 (A), TNFA (B), and IFNB (C) mRNAs were determined by RT-qPCR analysis and normalized to GAPDH mRNA and noninfected samples. (D) The relative amounts of cell-associated viral RNA were determined by RT-qPCR analysis and expressed as genome equivalents (YFV-GE) per μg of total cellular RNA. (E and F) Culture media of the indicated cells were analyzed by ELISA to determine the amounts of secreted IL-6 (E) and TNF-α (F). The data are the means ± SD from two (Huh7.5) or three (Huh7) experiments. The dashed lines indicate the limits of detection of the IL-6 and TNF-α ELISAs (2 and 4 pg/ml, respectively). Statistical analysis compared Huh7.5 samples to Huh7 samples. (G) Whole-cell lysates of the indicated cells were analyzed by Western blotting with antibodies against the indicated proteins. (H to J) The relative amounts of IL-6 (H), TNFA (I), and IFNB (J) mRNAs were determined by RT-qPCR analysis in cells transfected with RIG-I-specific siRNAs or control siRNAs (Scr). The samples were normalized to GAPDH mRNAs. (K) The relative amounts of cell-associated viral RNA were determined by RT-qPCR analysis and expressed as genome equivalents (YFV-GE) per μg of total cellular RNA. (L) The relative amounts of RIG-I mRNAs were determined by RT-qPCR analysis in cells transfected with RIG-I-specific siRNAs or control siRNAs (Scr). The samples were normalized to GAPDH mRNAs. Bottom, the abundances of RIG-I and actin were determined by Western blotting in cells that were transfected with control or RIG-I-specific siRNAs. Whole-cell lysates were analyzed 36 h later. The RT-qPCR and ELISA data are the means ± SD from two or three independent experiments, as indicated by the dots on the graphs. ns, nonsignificant; *, P < 0.05; **, P < 0.01. Western blots are representative of two independent experiments.
By Western blotting analysis, we found that IRF3 was activated in Huh7 cells infected with Asibi for 48 h but not in noninfected cells (Fig. 1G). IRF3 was not phosphorylated in Huh7.5 cells infected with Asibi for 48 h (Fig. 1G). IκBα is a negative regulator of the NF-κB pathway; its degradation triggers the nuclear translocation of the NF-κB subunits p65 and p50. IκBα was degraded in Huh7 cells infected for 48 h and, less so, in Huh.7.5 cells (Fig. 1G). More of the viral NS4b protein was produced in Huh7.5 cells than in Huh7 cells after 24 and 48 h of Asibi infection (Fig. 1G), which is consistent with the RT-qPCR analysis (Fig. 1D). These experiments indicate that both IRF3 and NF-κB are activated during Asibi infection of human hepatoma cells and that RIG-I contributes to IRF3 activation.
To further validate the role of RIG-I in activating a cytokine response in Asibi-infected hepatoma cells, we performed gene-silencing experiments using RIG-I specific small interfering RNAs (siRNAs). At 24 hours posttransfection, Huh7 cells were infected with Asibi for 48 h. The abundances of IL-6, TNFA, and IFNB mRNAs were significantly lower in cells transfected with RIG-I-specific siRNAs than in cells transfected with scrambled siRNA (Fig. 1H to K). Transfection of Huh7 cells with siRNAs targeting RIG-I reduced the abundance of RIG-I mRNAs by more than 90% in comparison to cells transfected with control siRNAs (Fig. 1L). Consistently, Western blot analysis showed that the abundance of RIG-I was reduced in RIG-I siRNA-transfected cells in comparison to cells transfected with control siRNAs (Fig. 1L). These data further demonstrate that RIG-I contributes to the induction of cytokines upon Asibi infection.
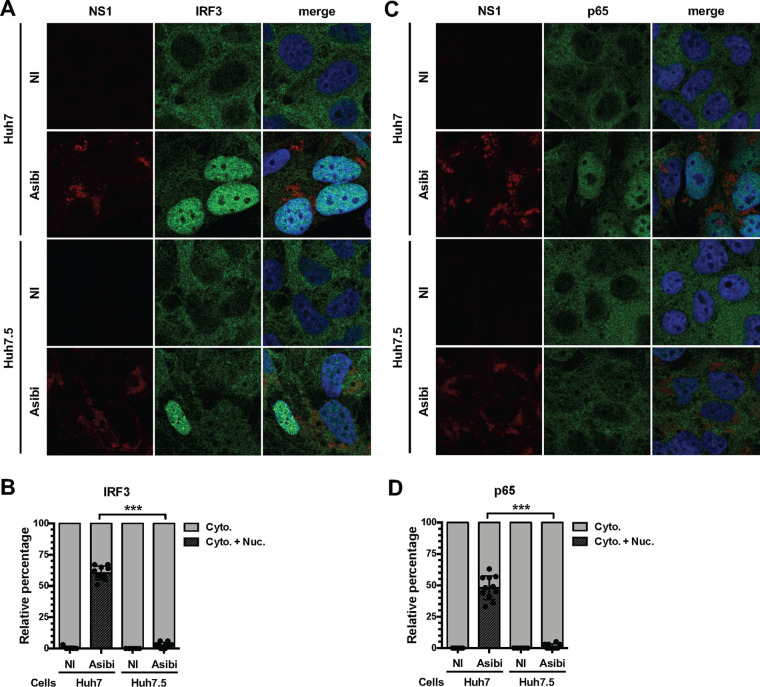
Immunofluorescence analysis revealed that IRF3 was detectable exclusively in the cytosol of nearly all uninfected Huh7 cells, whereas it was found in nuclei of around 60% of cells positive for the viral protein NS1 (Fig. 2A and B). In infected Huh7.5 cells, the number of nuclei positive for IRF3 was observed to be around 3% (Fig. 2A and B). Similar quantification showed that no uninfected cells exhibited nuclei positive for the NF-κB subunit p65, whereas around 50% of infected Huh7 cells showed nuclei positive for p65 (Fig. 2C and D). In Huh7.5 cells positive for NS1, the number of nuclei positive for p65 was estimated to be around 1% (Fig. 2C and D).
FIG 2.
RIG-I contributes to IRF3 and NF-κB activation in hepatoma cells infected with the Asibi strain of YFV. Huh7 or Huh7.5 cells were left uninfected (NI) or infected with YFV-Asibi for 48 h at an MOI of 20. Cells were then stained with antibodies recognizing NS1 (red), IRF3 (A and B), or p65 (C and D) (green), as well as with NucBlue (blue). Ten field containing around 30 cells were scored for nuclear translocation of IRF3 or p65. ns, nonsignificant; ***, P < 0.001.
Together, these results suggest that both the NF-κB and IRF3 pathways are activated upon Asibi infection of human hepatoma cells and that their initiation is dependent on RIG-I.
RIG-I localizes in stress granules in human and monkey cells infected with YFV.
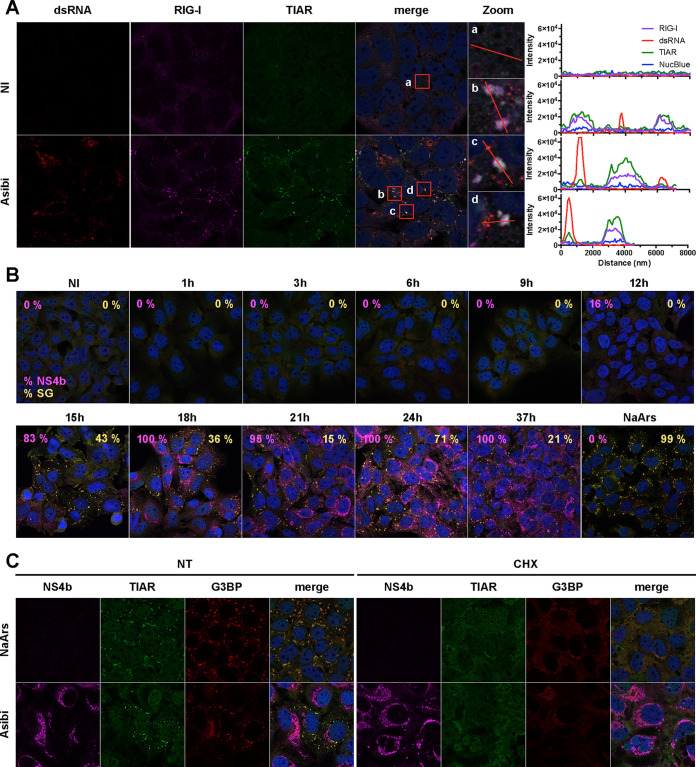
Having determined that RIG-I is important for mediating a cytokine response in Huh7 cells infected with Asibi (Fig. 1 and 2), we then investigated its subcellular localization. The distribution of the RIG-I signal was found to be weak and diffuse in the cytosol of uninfected Huh7 cells (Fig. 3A), which is consistent with data reported for unstimulated HeLa cells (20). After infection with Asibi for 24 h, RIG-I accumulated in cytosolic foci (Fig. 3A). Since RIG-I was previously shown to localize in stress granules in HeLa cells infected with several types of viruses (20–23), the Huh7 cells were also stained with the stress granule marker TIA-1-related protein (TIAR). In noninfected cells, the TIAR signal was weak and diffused in the cytoplasm and in the nuclei of the cells. In infected cells, TIAR was redistributed into cytoplasmic foci (Fig. 3A). The RIG-I structures present in infected cells colocalized with TIAR foci (Fig. 3A, see zoom images for details), suggesting that RIG-I is recruited to stress granules upon infection. The intensity profiles of individual foci confirmed that the RIG-I signal and the TIAR signal overlapped (Fig. 3A, right panels). Infected cells were identified with antibodies recognizing dsRNA structures, including probable viral replicative intermediates. These analysis revealed that dsRNAs seemed to be excluded from stress granules (Fig. 3A).
FIG 3.
RIG-I localizes to stress granules in Huh7 cells infected with YFV. (A) Huh7 cells were left uninfected (NI) or infected with Asibi at an MOI of 20 for 24 h. Cells were then stained with dsRNA (red), RIG-I (purple), or TIAR (green) antibodies and NucBlue (blue). (B) Huh7 cells were left uninfected (NI), treated with 0.5 mM NaArs for 30 min, or infected with Asibi at an MOI of 20 for the indicated time. Cells were then stained with NS4b (purple), G3BP (red), or TIAR (green) antibodies and NucBlue (blue). Percentages of NS4b-positive and G3BP-positive cells were estimated and are shown in purple and yellow, respectively. (C) Huh7 cells were treated with 0.5 mM NaArs for 30 min or infected with YFV-Asibi at an MOI of 20 for 48 h. Cells were then left untreated (NT) or treated with 100 μg/ml cycloheximide (CHX) for 1 h and stained with antibodies against NS4b (purple), TIAR (green), and G3BP (red), as well as with NucBlue (blue).
To follow the formation of stress granules in infected cells, the localization of TIAR and RAS GTPase-activating protein SH3 domain-binding proteins 1 and 2 (G3BP), which are canonical stress granule markers (29), was examined in Huh7 cells infected with Asibi over time (Fig. 3B). Infected cells were identified by the presence of the viral protein NS4b (Fig. 3B). TIAR and G3BP were diffuse in noninfected cells. They remained diffuse until 12 hpi, when 16% of cells were determined to be positive for NS4b. Fifteen hours after infection, around 80% of cells were infected and around 40% of the total number of cells exhibited TIAR- and G3BP-positive foci (Fig. 3B). At 24 hpi, all cells were infected and around 70% of them were positive for TIAR- and G3BP-containing foci. At 37 hpi, all cells remained infected and around 20% of them exhibited TIAR- and G3BP positive foci. These foci resembled the ones detected in Huh7 cells after a 30-min treatment with sodium arsenite (NaArs), a chemical commonly used for induction of oxidative stress in eukaryotic cells (Fig. 3B). These results indicate that upon Asibi infection, TIAR- and G3BP-positive foci appear between 12 and 15 hpi, peak around 24 hpi, and persist in around 20% of cells until later times. Moreover, these data suggest that the formation of the stress granules occurs throughout infection with Asibi.
To further validate the nature of the TIAR- and G3BP-positive foci observed in infected cells, we assessed their sensitivity to cycloheximide. Cycloheximide is a compound that inhibits stress granule formation and fosters their disassembly by stabilizing mRNAs on polysomes (32). Huh7 cells treated with NaArs for 30 min and subsequently incubated with cycloheximide for 1 h or left untreated were used as positive controls (Fig. 3C). Alternatively, cells were infected with Asibi for 48 h and then treated, or not, with cycloheximide for 1 h. The TIAR and the G3BP signals overlapped, both in cells infected by Asibi for 48 h and in NaArs-treated cells (Fig. 3C). Cycloheximide treatment led to a complete loss of TIAR- and G3BP-positive foci, both in NaArs-treated cells and in Asibi-infected cells (Fig. 3C, compare nontreated and treated cells). These results suggest that, under both conditions, most mRNAs trapped in these cytoplasmic foci can be released within 1 h and undergo some translation on polysomes. Most importantly, these experiments confirmed that the cytoplasmic foci containing TIAR, G3BP, and RIG-I identified in infected cells also contained mRNAs and, as such, can be defined as canonical stress granules.
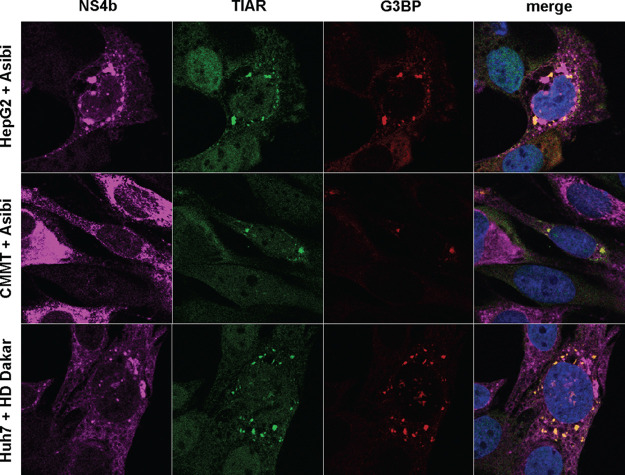
Next, we monitored the formation of stress granules upon Asibi infection in cell models different from Huh7. Like Huh7 cells, HepG2 cells are hepatocyte-derived cells and are thus relevant for infection with YFV, a hepatotropic virus. TIAR- and G3BP-positive foci were detected in HepG2 cells infected for 48 h (Fig. 4). Stress granules were also detected in CMMT cells, a Macaca mulatta epithelial cell line, upon 24 h of infection with Asibi (Fig. 4). Macaques are natural hosts of YFV during the jungle cycle of transmission (33). Similarly, TIAR- and G3BP-positive foci were observed in Huh7 cells infected with the clinical isolate HD1279 (HD Dakar) for 24 h (Fig. 4). These data indicate that infection with two YFV strains triggers the formation of stress granules in several mammalian cell lines, suggesting that the stress response is neither viral strain nor cell type specific.
FIG 4.
YFV infection induces the formation of stress granules in several mammalian cells. HepG2 or CMMT cells were infected with Asibi at an MOI of 20 for, respectively, 48 and 24 h. Huh7 cells were infected with YFV-HD Dakar at an MOI of 10 for 24 h. Cells were then stained with NS4b (purple), TIAR (green), or G3BP (red) antibodies and NucBlue (blue).
PKR contributes to stress granule formation and innate response in Asibi-infected Huh7 cells.
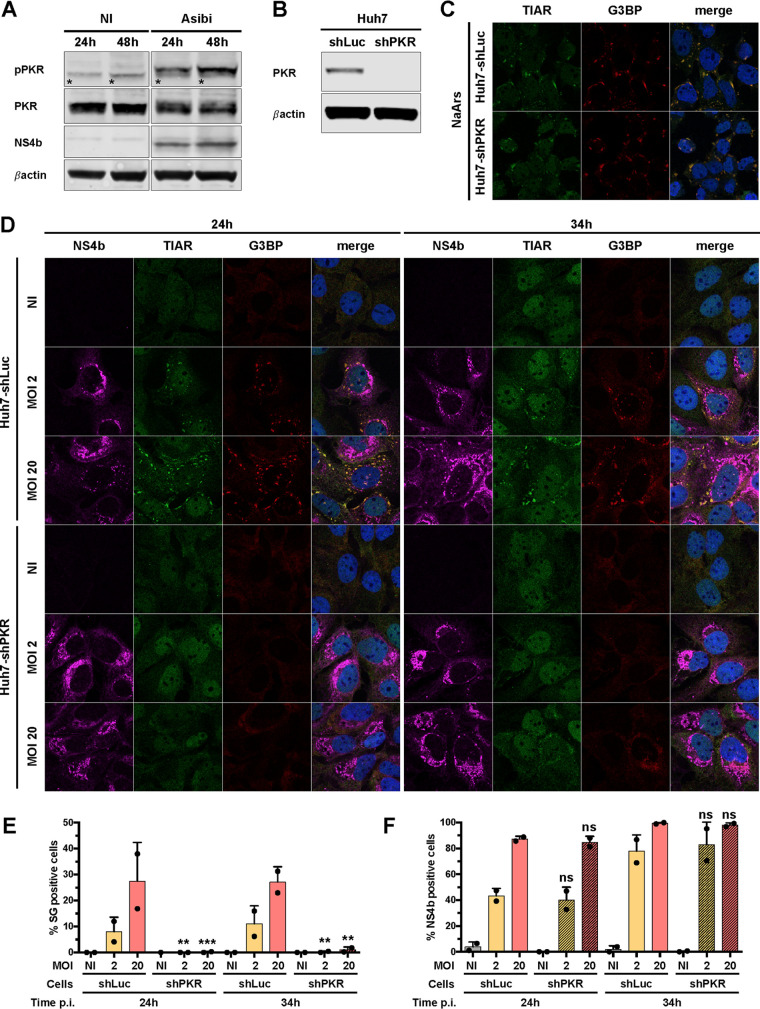
PKR is known to be activated upon infection with several flaviviruses, including ZIKV, WNV and Japanese encephalitis virus (JEV) (34–36), and to play an important role in stress granule nucleation through its ability to phosphorylate the eukaryotic initiation factor eIF2α (37). We first assessed whether PKR was activated upon Asibi infection of Huh7 cells. Western blot analysis showed that PKR was phosphorylated in Huh7 cells infected for 24 and 48 h but not in noninfected cells (Fig. 5A). Thus, PKR is activated upon Asibi infection of Huh7 cells. To evaluate the role of PKR in the formation of stress granules in Asibi-infected human hepatic cells, we generated a Huh7 short hairpin PKR (shPKR) cell line by transduction with lentiviruses expressing short hairpin RNAs (shRNAs) specific for PKR. While PKR expression was detectable in Huh7 cells transduced with lentivirus expressing control shRNA targeting luciferase (Huh7-shLuc cells), it was not detectable in Huh7-shPKR cells (Fig. 5B). As a control experiment, we treated the cells with NaArs, which induces stress granule formation via HRI-mediated eIF2α phosphorylation (38). TIAR- and G3BP-positive stress granules were detected in both Huh7-shLuc and Huh7-shPKR cells treated with NaArs for 30 min (Fig. 5C). This confirms that the absence of PKR does not disrupt the formation of NaArs-dependent stress granule induction.
FIG 5.
PKR contributes to stress granule formation in Asibi-infected Huh7 cells. (A) Huh7 cells were left uninfected (NI) or infected with Asibi at an MOI of 20 for 24 or 48 h. Whole-cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. Samples were loaded on the same gel. *, unspecific bands. (B) Whole-cell lysates of Huh7-shLuc and Huh7-shPKR cells were analyzed by Western blotting with antibodies against the indicated proteins. (C) Huh7-shLuc and Huh7-shPKR cells were treated with 0.5 mM NaArs for 30 min and stained with TIAR (green) and G3BP (red) antibodies, as well as with NucBlue (blue). (D) Huh7 cells were left uninfected (NI) or infected with YFV-Asibi at an MOI of 2 or 20 for 24 or 34 h. Cells were then stained with NS4b (purple), TIAR (green), or G3BP (red) antibodies and NucBlue (blue). Images are representative of two independent experiments. (E and F) Percentages of G3BP-positive (E) or NS4b-positive (F) cells were estimated by analyzing at least 200 cells per condition. Statistical analysis compared Huh7-shPKR cells to Huh7-shLuc cells. ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Automated microscopic quantification revealed that around 10% of Huh7-shLuc cells infected at a multiplicity of infection (MOI) of 2 for 24 or 34 h contained at least two stress granules (Fig. 5D and E). At an MOI of 20, around 25% of Huh7-shLuc cells were positive for stress granules at 24 and 34 hpi (Fig. 5D and E). Neither TIAR- nor G3BP-positive foci were detected in Huh7-shPKR cells infected at an MOI of 2 for 24 or 34 h. At an MOI of 20, a low percentage (around 1%) of Huh7-shPKR cells exhibited detectable stress granules (Fig. 5D and E). These stress granules possibly formed in a PKR-independent manner, via the activation of other members of the eIF2α kinase family. Alternatively, a minute quantity of PKR still being expressed in shPKR cells may have initiated the formation of these granules. Similar proportions of Huh7-shLuc and Huh7-shPKR cells were found positive for NS4b at 24 or 34 hpi at the two tested MOIs (Fig. 5F). Taking the results together, we conclude that PKR plays a key role in stress granule formation in Asibi-infected human hepatic cells and that the absence of stress granules has no effect on viral infection.
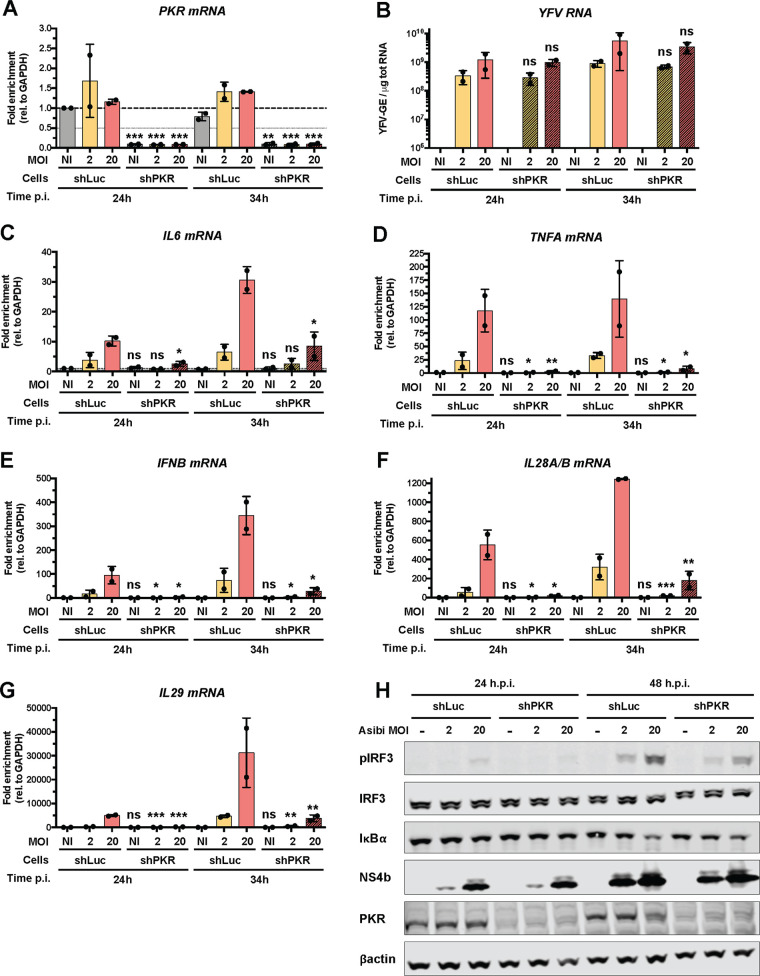
Since no stress granules were detected in cells depleted of PKR under three of four conditions tested (Fig. 5E), we used Huh7-shPKR cells to determine whether absence of granules affected cytokine induction. Consistent with our Western blot data (Fig. 5B), RT-qPCR analysis showed that PKR mRNA abundance was reduced by more than 90% in Huh7-shPKR cells in comparison to Huh7-shLuc cells (Fig. 6A). These RT-qPCR analyses also showed that the viral RNA yield in Huh7-shPKR cells was not markedly different from that in Huh7-shLuc cells at 24 h or 34 h after infection at an MOI of either 2 or 20 (Fig. 6B), which is consistent with our microscopic analysis (Fig. 5F). IL-6 mRNA abundance was significantly lower in Huh7-shPKR cells than in Huh7-shLuc cells when cells were infected at an MOI of 20 (Fig. 6C). The abundances of TNFA and IFNB mRNAs were significantly lower in Huh7-shPKR cells than in infected Huh7-shLuc cells, independently of the MOI and the time of infection (Fig. 6D and E). The abundances of IL28A/B and IL-29 mRNAs were also analyzed (Fig. 6F and G). They were significantly lower in Huh7-shPKR cells than in Huh7-shLuc cells under all conditions tested (Fig. 6F and G). Thus, induction of cytokines, including IFN-β and type III IFN, is affected by PKR knockdown, further demonstrating the contribution of PKR to the antiviral response against YFV in Huh7 cells. This is consistent with a previous report showing that PKR is involved in the induction of the IFN through the activation of NF-κB (39).
FIG 6.
PKR contributes to innate response in Asibi-infected Huh7 cells. Huh7-shLuc or -shPKR cells were left uninfected (NI) or infected with Asibi at an MOI of 2 or 20 for 24 or 34 h. (A and C to G) The relative amounts of PKR (A), IL-6 (C), TNFA (D), IFNB (E), IL28A/B (F), and IL-29 (G) mRNAs were determined by qPCR analysis and normalized to that of GAPDH mRNA and noninfected shLuc samples. (B) The relative amounts of cell-associated viral RNA were determined by RT-qPCR analysis and expressed as genome equivalents (YFV-GE) per μg of total cellular RNA. Means ± SD are shown. One-way ANOVA on log-transformed data with Tukey-corrected multiple-comparison tests were performed. shPKR samples were compared to shLuc samples. ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (H) Whole-cell lysates of the indicated cells were analyzed by Western blotting with antibodies against the indicated proteins. The Western blot is representative of two independent experiments.
By Western blotting analysis, we found that IRF3 was activated in Huh7-shLuc cells infected with Asibi at an MOI of 2 and 20 for 48 h but not in noninfected cells (Fig. 6H), which is consistent with our previous analysis (Fig. 1G). The level of IRF3 phosphorylation was higher at an MOI of 20 than at an MOI of 2, despite the total protein abundance being similar under both conditions (Fig. 6H). The level of IRF3 phosphorylation was reduced in Huh7-shPKR cells infected with Asibi at the same MOI (Fig. 6H), despite the cells being infected at similar levels as indicated by the comparable amount of the detected viral protein NS4b. IκBα was degraded in Huh7-shLuc cells infected for 48 h at an MOI of 20 (Fig. 6H), which is in agreement with data shown in Fig. 1G. The degradation of IκBα was less pronounced in Huh7-shPKR cells than in Huh7-shLuc cells infected under the same conditions. These experiments indicate that PKR contributes to IRF3 activation during Asibi infection of human hepatoma cells.
Taken together, our data show that at least two nucleic acid sensors, namely, RIG-I (Fig. 1 and 2) and PKR (Fig. 5 and 6), contribute to the antiviral response against Asibi in Huh7 cells. Our data also revealed that PKR contributes to both stress granule formation and induction of cytokines, including IFN-β and IFN type III, in Huh7 cells infected with Asibi. However, since PKR does not require its kinase activity to stimulate NF-κB (39, 40), it might contribute to the cytokine induction in Asibi-infected cells in an eIF2α (and thus stress granule)-independent manner.
Induction of proinflammatory cytokines is not affected by the inhibition of stress granules.
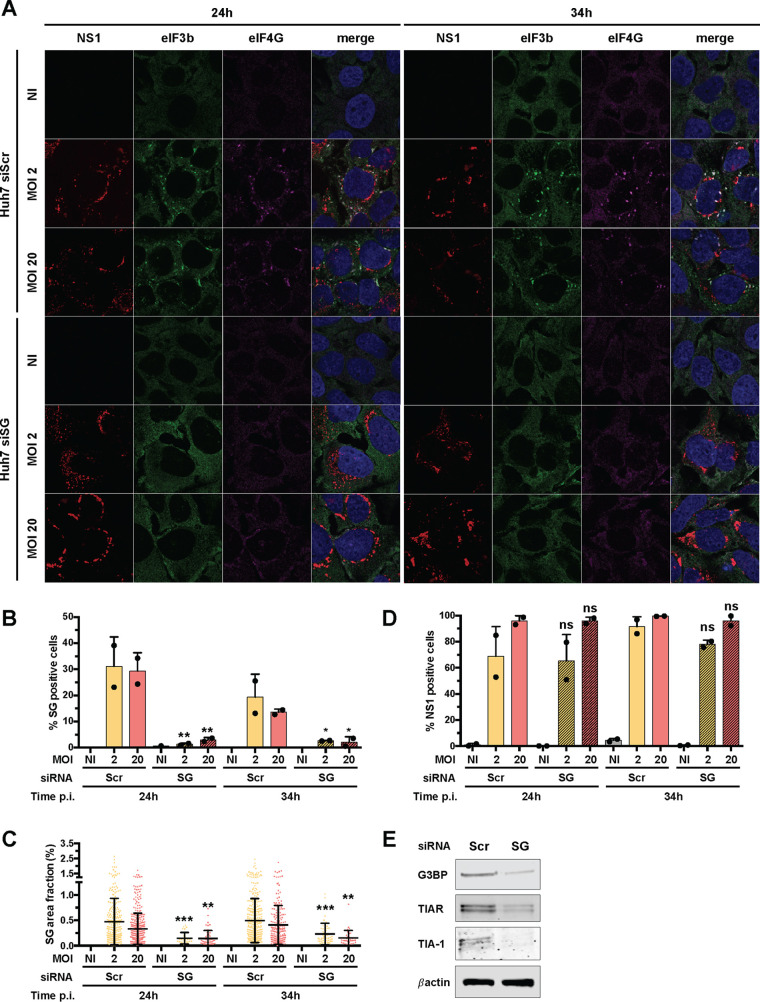
To further investigate the putative link between stress granules and the induction of antiviral cytokines, we used an siRNA knockdown approach to inhibit stress granule assembly. We tested numerous combinations of siRNAs targeting core granule components in infected cells. The most efficient strategy consisted of transfecting Huh7 cells twice with four pools of siRNAs targeting TIAR, TIA-1, G3BP1, and G3BP2 (siSG). In these cells, we monitored stress granule formation using antibodies against eIF3b and eIF4G, two core components of stress granules. Around 30% of Huh7 cells transfected with scrambled control siRNAs and infected at an MOI of 2 or 20 for 24 h contained at least two eIF4G- and eIF3b-positive foci (Fig. 7A and B). When cells were transfected with the 4 siRNA pools and infected under the same conditions and for the same amount of time, fewer than 5% of cells exhibited stress granules (Fig. 7A and B). The impact of the siRNA against TIAR, TIA-1, G3BP1, and G3BP2 on the number of cells positive for stress granules following 34 h of infection was also significant at the two tested MOIs (Fig. 7B). Under the four tested conditions, the surface area occupied by stress granules per cell was significantly reduced in cells transfected with the mixture of four siRNA pools compared to the controls (Fig. 7C). Transfection with the 4 siRNA pools had no effect on viral infection compared to transfection with control siRNAs (Fig. 7D). Western blot analysis showed that the abundances of TIAR, TIA-1, and G3BP1/2 proteins were reduced in siSG cells compared to control cells (Fig. 7E). These experiments validate our silencing approach as an efficient way to reduce the abundances of TIAR, TIA-1, and G3BP1/2 and significantly decrease both the number of cells positive for stress granules and the surface occupied by stress granules per cell.
FIG 7.
Knockdown of G3BP1, G3BP2, TIAR, and TIA1 inhibits stress granule formation. (A) Huh7 cells were transfected with scramble siRNA (siScr) or with siRNA targeting G3BP1, G3BP2, TIAR, and TIA1 (siSG) at 48 and 24 h prior to Asibi infection. Transfected cells were left uninfected (NI), treated with 0.5 mM NaArs for 30 min (NaArs), or infected with Asibi at an MOI of 2 or 20 for 24 or 34 h. Cells were then stained with NS1 (purple), eIF3b (green), or eIF4G (red) antibodies and NucBlue (blue). Images are representative of two independent confocal microscopy analyses. (B to D) Percentages of eIF4G- and eIF3b-positive (B) or NS1-positive (D) cells were estimated, and the stress granule area was determined for each cell as the sum of the whole area occupied by stress granules divided by the total cell surface (C). Around 200 cells per independent experiment were scored. Statistical analysis compared Huh7-siSG cells to Huh7-siScr cells. ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (E) The abundances of G3BP1/2, TIAR, TIA1, and actin were determined by Western blotting in Huh7 cells, which were transfected with control siRNAs or with siSG. Whole-cell lysates were analyzed with antibodies against the indicated proteins. Western blots are representative of two independent experiments.
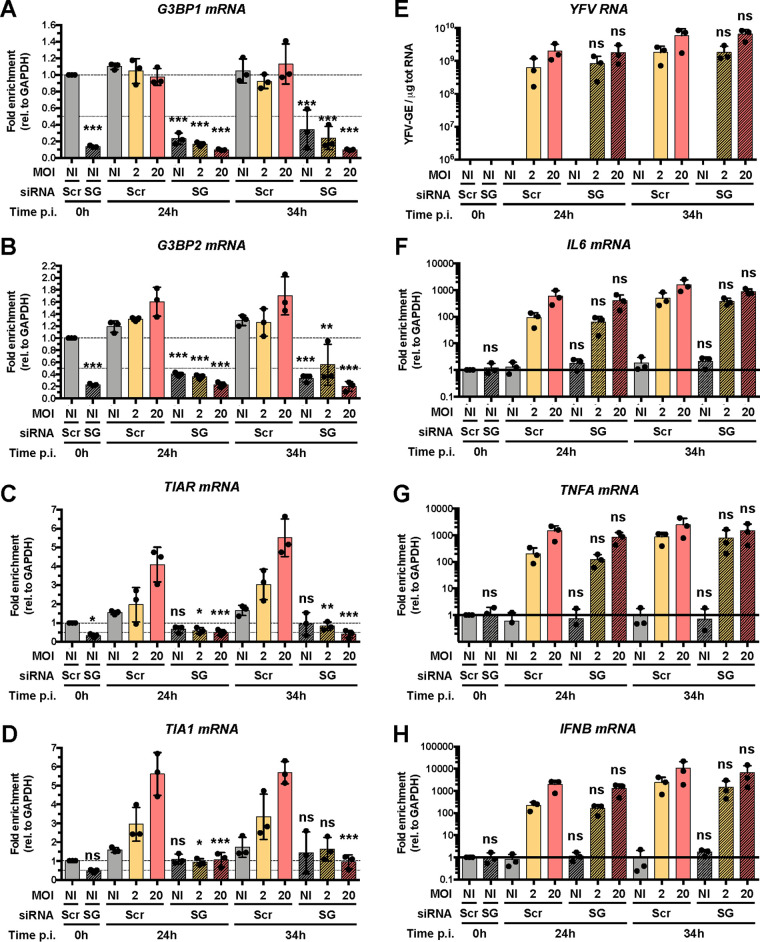
RT-qPCR analysis showed that TIAR, TIA-1, G3BP1, and G3BP2 mRNA abundances were significantly reduced in cells transfected with the 4 siRNA pools compared to cells transfected with control siRNAs (Fig. 8A to D), which is consistent with our Western blot analysis (Fig. 7E). Of note, TIAR and TIA-1 expression was induced 3- to 4-fold by infection, in an MOI-dependent response. This may be a consequence of virus-induced stress. Transfection of the four siRNA pools had no effect on TNFA, IL-6, and IFNB mRNA abundances or on viral RNA yield (Fig. 8E to H). Together, these data show that significant reduction of stress granule numbers and size did not suppress the cytokine response to Asibi infection. Thus, in our model, stress granules do not contribute to the initiation of the antiviral response.
FIG 8.
Inhibition of stress granule formation has a limited effect on the antiviral response. Huh7 cells were transfected with scramble siRNA (siScr) or with four pools of siRNA targeting G3BP1, G3BP2, TIAR, and TIA1 (siSG) at 48 and 24 h prior to infection. Transfected cells were left uninfected (NI) or infected with Asibi at an MOI of 2 or 20. Cells were harvested prior to infection (0 h) and at 24 and 34 h postinfection. (A to D and F to H) The relative amounts of G3BP1 (A), G3BP2 (B), TIAR (C), TIA1 (D), IL-6 (F), TNFA (G), and IFNB (H) mRNAs were determined by qPCR analysis and normalized to that of GAPDH mRNA and noninfected shScr samples. (E) The relative amounts of cell-associated viral RNA were determined by RT-qPCR analysis and expressed as genome equivalents (YFV-GE) per μg of total cellular RNA. Means ± SD are shown. One-way ANOVA on log-transformed data with Tukey-corrected multiple-comparison tests were performed. Samples treated with siSG RNA were compared to siScr samples. ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
RIG-I signaling is not involved in stress granule assembly.
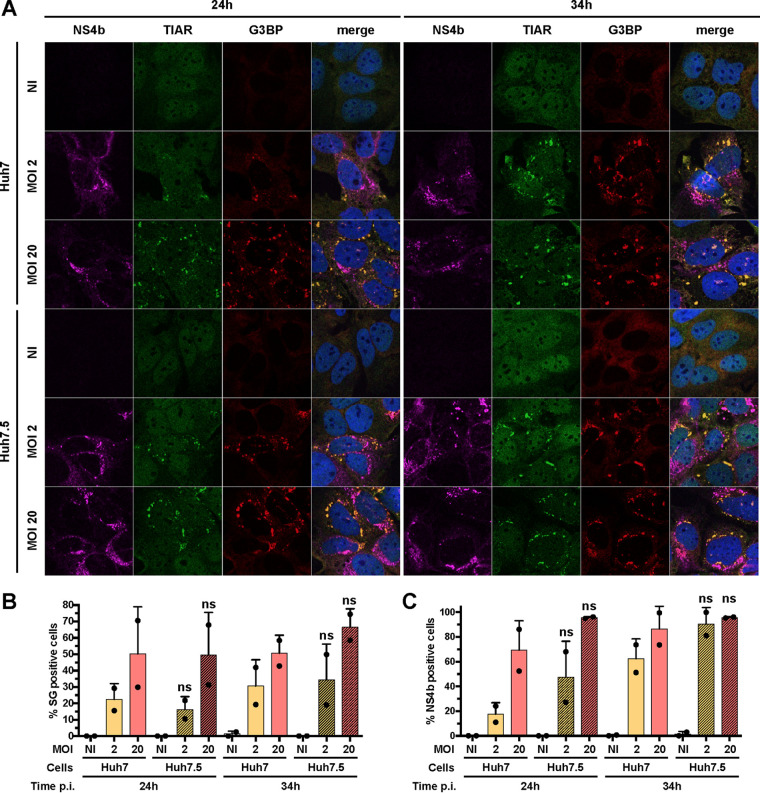
In Huh7 cells, RIG-I signaling is crucial for cytokine induction and secretion upon Asibi infection (Fig. 1 and 2). Therefore, we wondered whether RIG-I signaling would participate in stress granule assembly and/or maintenance. We quantified the proportion of Huh7 and Huh7.5 cells that were positive for stress granules at 24 and 34 hpi following infection at two MOIs. Similar proportions of Huh7 and Huh7.5 cells were found positive for stress granule markers under all tested conditions (Fig. 9A and B). No difference in the number of cells positive for NS4b was observed between Huh7 and Huh7.5 cells (Fig. 9C). We conclude that the absence of RIG-I signaling had no effect on the number of granules that assemble in response to infection. It is thus unlikely that RIG-I mediates a feed-forward loop to maintain stress granule assembly.
FIG 9.
RIG-I signaling is not involved in stress granule assembly. (A) Huh7 and Huh7.5 cells were left uninfected (NI), treated with 0.5 mM NaArs for 30 min (NaArs), or infected with Asibi at an MOI of 2 or 20 for 24 or 34 h. Cells were then stained with NS4b (purple), TIAR (green), or G3BP (red) antibodies and NucBlue (blue). Images are representative of two independent confocal microscopy analyses. (B and C) Around 200 cells per independent experiment were scored. Percentages of G3BP-positive (B) or NS4b-positive (C) cells were estimated. Statistical analysis compared Huh7.5 samples to Huh7 samples. ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
YFV RNA is excluded from stress granules.
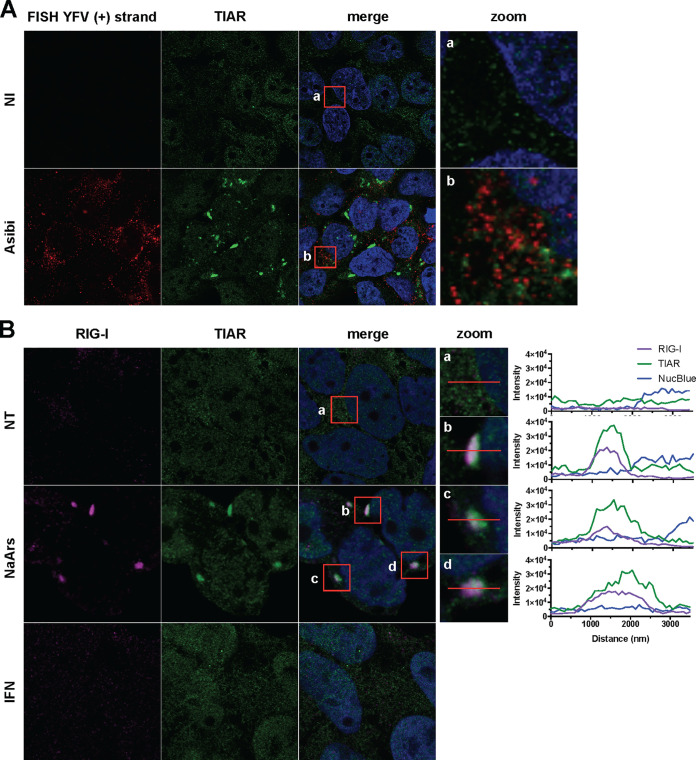
Our previous analysis showed that neither dsRNA (Fig. 3A) nor the viral proteins NS4b and NS1 (Fig. 3B and C, 4, 5D, 7A, and 9A) seem to be recruited to stress granules. To further investigate the putative association between stress granules containing RIG-I (Fig. 3A) and viral RNA, we used an RNA fluorescence in situ hybridization (FISH) approach coupled with immunofluorescence and confocal microscopy. The Asibi-specific FISH probe produced no signal in noninfected Huh7 control cells (Fig. 10A) and a bright punctate signal in the cytoplasm of infected cells (Fig. 10A), confirming the specificity of the probe (41). Asibi RNA was never found to be associated with TIAR (Fig. 10A). This was unexpected since RIG-I signaling is key to cytokine induction and production (Fig. 1 and 2) and massively concentrates in stress granules in Asibi-infected cells (Fig. 3A). These results suggest that an interaction between viral RNA and RIG-I might not happen in stress granules in Asibi-infected Huh7 cells. A minute quantity of RIG-I might, however, interact with viral RNA in ER-derived replication sites and initiate a stress granule-independent IFN response.
FIG 10.
YFV RNA is excluded from stress granules, and IFN-α treatment is not sufficient to induce stress granule formation. (A) Huh7 cells were left uninfected (NI) or infected with Asibi at an MOI of 20 for 24 h. Cells were then stained with TIAR (green) and NucBlue (blue) prior to be processed for FISH using a probe specific for YFV plus-strand RNA (red). (B) Huh7 cells were left untreated (NT) or were treated with 0.5 mM NaArs for 30 min or with 1,000 U/ml IFN-α2a for 8 h. Cells were then stained with RIG-I (purple) and TIAR (green) antibodies, as well as with NucBlue (blue).
In line with this, RIG-I was recruited to TIAR-positive granules in Huh7 cells treated with NaArs (Fig. 10B), which is consistent with previous observations on NaArs-treated HeLa cells (20). These data further indicate that RIG-I recruitment to stress granules can occur in the absence of viral RNA. Finally, we observed that in IFN-α-treated Huh7 cells, the RIG-I signal was weak and diffuse in the cytosol, similar to the signal in uninfected cells (Fig. 10B). Thus, IFN-α treatment is not sufficient to trigger the formation of RIG-I-containing granules.
DISCUSSION
The identity of the PRR(s) responsible for the initiation of a proinflammatory response upon direct infection of human hepatocytes with wild-type strains of YFV has not been uncovered thus far. In this study, we first identified RIG-I and PKR as main actors responsible for the induction and production of proinflammatory cytokines upon infection of human hepatocytes with the Asibi strain of YFV. We demonstrated the crucial involvement of RIG-I in this process by directly comparing the dysfunctional RIG-I-expressing human hepatoma cell line Huh7.5 with its RIG-I-competent counterpart (Huh7 cells).
The level of IL-6 and TNF-α secreted in the first 48 h of infection by Huh7.5 cells upon Asibi infection was under the limit of detection, while both cytokines were produced in Huh7 cells under the same experimental conditions. Gene-silencing experiments performed with siRNAs specific for RIG-I further demonstrated that RIG-I contributes to the induction of TNFA, IL-6, and IFNB mRNAs upon Asibi infection. These results are in accordance with our previous studies showing that human plasmacytoid dendritic cells infected with the YFV vaccine strain 17D induce an IFN response in a RIG-I-dependent manner (19). Other members of the RLR family may contribute to YFV sensing in other cell types or at later time of infection. Such distinct temporal activity of RIG-I and MDA5 during infection with the related West Nile virus has been reported in mice (42). Members of the Toll-like receptor (TLR) family, such as TLR3 and TLR7, may also contribute to cytokine induction upon infection. TLR3 has been shown to inhibit DENV and ZIKV replication in 293 cells and skin fibroblasts, respectively (43, 44). TLR7 signaling has been shown to participate in the immune response against DENV and ZIKV infection in primary myeloid cells (45, 46). Minor contributions of MDA5 and/or TLRs could explain the activation of IκBα in Huh7.5 cells upon Asibi infection.
In Huh7 cells silenced for PKR and infected with Asibi, cytokine expression was, under most of the tested conditions, below the detection limit. This suggests that PKR, like RIG-I, largely contributes to the initiation of the antiviral response. This is consistent with the previously reported role of PKR in the production of IFN and other proinflammatory cytokines upon viral infection, possibly via its interaction with the IKK complex and subsequent activation of NF-κB (39, 47). Moreover, PKR was previously described as an important effector protein against infection with WNV in mice (36).
The fact that abolishing RIG-I signaling or silencing PKR drastically reduces the proinflammatory response in Asibi-infected cells shows the nonredundant nature of these two dsRNA sensors in maintaining cytokine production. We speculate that RIG-I and PKR cooperate to induce an anti-YFV response. Indeed, PKR interacts with RIG-I and MDA5 in HEK293T cells transfected with FLAG-tagged versions of RIG-I or MDA5 and infected with a mutant vaccinia virus lacking a gene known to inhibit PKR (48). However, in this context, PKR was required for only MDA5-mediated, not RIG-I-mediated, IFN production (48). Consistent with the hypothesis that PKR and RIG-I cooperate to orchestrate the antiviral response, PKR is known to interact with several members of the TRAF family, including TRAF2 and TRAF6 (49, 50), two proteins involved in MAVS signaling. Finally, MAVS, which acts downstream of RIG-I and MDA5, interacts with PKR (51) and is essential for PKR-induced IFN production (48).
Stress granules have been demonstrated to contain and concentrate viral nucleic sensors; hence, they are generally regarded as a functional platform for ligand recognition and subsequent induction of an antiviral response. We report here that Asibi replication triggers the formation of stress granules in two human hepatoma cell lines and epithelial monkey cells. This is consistent with previous studies showing that infection with the vaccine strain 17D induces the formation of stress granules in human dendritic cells and hamster kidney cells (52). Thus, formation of stress granules seems to be a general feature of flavivirus infection (53–56). However, unlike WNV, DENV, ZIKV, and the related tick-borne encephalitis virus (34, 53–58), YFV does not seem to efficiently antagonize stress granule biogenesis. YFV may not express proteins that inhibit granule formation. Alternatively, stress granules may not have an antiviral function against YFV, and thus the virus has no pressure to evolve an evasion strategy.
Stress granules were not detected in Huh7 cells depleted of PKR upon 24 h of Asibi infection, strongly suggesting a PKR-dependent granule establishment early in infection. Of note, RIG-I signaling was not involved in stress granule assembly, indicating that while the two sensors are both critically implicated in the proinflammatory cytokine production, only PKR is able to activate stress granule formation. At 34 h, very few cells silenced for PKR exhibited stress granules. Small quantities of remaining PKR, below the detection limit of Western blot and qPCR analysis, may be responsible for the formation of these foci. Alternatively, they may form subsequent to the phosphorylation of eIF2α by one of the other three eIF2α kinases. YFV replication and assembly, which take place within ER membranes (3), likely cause an ER stress and might thus trigger the activation of the ER-resident kinase PERK. Since GCN2 is phosphorylated in mouse and human dendritic cells upon infection with 17D (52), this kinase could also potentially contribute to granule formation in Asibi-infected cells.
Similar to what was reported in the context of infection with numerous RNA viruses (27, 28, 59), we showed that RIG-I accumulates in stress granules upon Asibi infection. PKR, MDA5, LGP2, and the DNA sensor cGAS were also shown to concentrate in stress granules in cells infected with a large panel of RNA and DNA viruses (20, 22–26). Surprisingly, we were unable to detect either Asibi RNA or dsRNA structures in stress granules of infected cells. Moreover, the stress granule proteins TIAR and G3BP were not recruited to Asibi replication complexes identified by NS4b or NS1. Thus, no interaction between stress granules and viral RNAs was detected in our model. Similarly, in cells infected with orthohantaviruses, no viral RNA was found associated with stress granules (60). Conversely, viral RNA was detected in stress granules induced by infection with IAVΔNS1 (20), and stress granule components were recruited to ZIKV, DENV, and WNV replication complexes (53, 54).
The lack of detectable association between stress granules and viral RNA implies that RIG-I and presumably PKR do not interact with large amount of YFV RNA. We propose that the large accumulation of RLRs in stress granules masks the detection of small quantities of PKR and RIG-I which are recruited to perinuclear ER-derived replication sites, where viral RNA sensing may occur. The absence of viral RNA in RIG-I-containing stress granules also suggests that RIG-I may be bound to translationally stalled mRNAs that accumulate in these structures. Recent advances in RLR purification methods coupled to RNA-sequencing approaches have indeed revealed the existence of cellular RNAs bound to RIG-I (61, 62). These immunostimulatory cellular RNAs are generated by virally induced cellular dysregulations, including cellular RNA misprocessing, unshielding by RNA-binding proteins, and RNA mislocalization (12, 63). During viral infection, RLRs may first be activated by viral RNA generated during viral replication, and once cellular disturbance occurs with accumulation of viral proteins, RLRs may be activated by endogenous RNAs that accumulate in stress granules. These interactions may perpetuate and/or amplify the RLR-mediated antiviral response.
Importantly, inhibition of stress granule formation did not affect the proinflammatory cytokine expression by infected hepatocytes, indicating that the antiviral response and stress granule formation are independent events in the early phases of YFV infection. Whether the formation of stress granules and their composition affect later stages of infection remains to be established.
Collectively, our data strongly suggest that in cells infected with YFV, proinflammatory cytokine induction is mediated by RIG-I and PKR, independently of their localization in stress granules. Thus, contrary to the widely believed consensus, we demonstrated that stress granule formation can be uncoupled from the initiation of a proinflammatory response upon viral infection.
MATERIALS AND METHODS
Cell lines, viruses, antibodies, and reagents.
Huh7 human hepatocellular carcinoma cells (kindly provided by A. Martin, Institut Pasteur Paris) and Huh7.5 cells (64) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) containing GlutaMAX I and sodium pyruvate (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Dominique Dutscher) and 1% penicillin and streptomycin (10,000 U/ml; Thermo Fisher). HepG2 cells (kindly provided by C. Neuveut, Institut Pasteur Paris) were maintained in DMEM-F12 complemented with 10% FBS, 3.5 × 10−7 M hydrocortisone, and 5 μg/ml insulin. Human embryonic kidney (HEK) 293FT cells (American Type Culture Collection [ATCC] CRL-1573), Macaca mulatta mammary gland epithelial CMMT cells (kindly provided by O. Schwartz, Institut Pasteur Paris), and African green monkey kidney epithelial Vero cells (ATCC) were maintained in DMEM with 10% FBS.
The Asibi strain, which is the YFV reference strain, was provided by the Biological Resource Center of Institut Pasteur. The YFV-DAK strain (YFV-Dakar HD1279) was provided by the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) through the University of Texas Medical Branch at Galveston, TX, USA. Virus stocks were propagated on Vero cells. Viruses were concentrated by polyethylene glycol 6000 precipitation and purified by centrifugation in a discontinuous gradient of sucrose. Infectious virus titers were determined by plaque assays on Vero cells. Cells were infected with 10-fold serial dilutions of viral stocks and incubated for 4 days in DMEM containing 2% FBS and carboxymethyl cellulose. Cells were then washed with phosphate-buffered saline (PBS), fixed with 3% formaldehyde crystal violet solution for 20 min at room temperature, and rinsed with water, and plaques were counted. Cell infections were carried out at the indicated MOIs. The viral inoculum was replaced with fresh culture medium at 2 h postinfection.
The following antibodies were used: phosphorylated IRF3 S386 (clone EPR2346; Abcam); IRF3 (FL-425; Santa Cruz); NF-κB p65 (D14E12; Cell Signaling); IκBα (L35A5; Cell Signaling); β-actin (clone AC-74; Sigma-Aldrich); phosphorylated PKR T451 (EPR2152Y; Abcam); dsRNA J2 (English & Scientific Consulting Kft); G3BP (BD Biosciences); TIAR (C-18; Santa Cruz); eIF4G (sc‐11373; Santa Cruz); eIF3b (N-20, sc‐16377); YFV-NS4b (a kind gift from C. M. Rice, Rockefeller University); Alexa Fluor 488 donkey anti-goat IgG (H+L), Alexa Fluor 594 donkey anti-mouse IgG (H+L), and Alexa Fluor 647 donkey anti-rabbit IgG (H+L) (Life Technologies); anti-mouse 680 (Li-Cor Bioscience); and anti-rabbit 800 (Thermo Fisher Scientific). Both RIG-I and PKR antibodies were previously described (20, 50).
Sodium arsenite (NaArs) (Sigma) and cycloheximide (Sigma) were diluted in water and used at the indicated concentration. IFN-α2a (SRE0013; Sigma) was used at a final concentration of 1,000 U/ml.
Generation of shRNA-expressing Huh7 cells by lentiviral transduction.
pALPS-shPKR1 and pALPS-shPKR3 were generated by cloning the following sequences in pALPS (65): TGC TGT TGA CAG TGA GCG CCA GCA GAT ACA TCA GAG ATA ATA GTG AAG CCA CAG ATG TAT TAT CTC TGA TGT ATC TGC TGA TGC CTA CTG CCT CGG A and TGC TGT TGA CAG TGA GCG ACA GAA GAA AAG ACT AAC TGT ATA GTG AAG CCA CAG ATG TAT ACA GTT AGT CTT TTC TTC TGG TGC CTA CTG CCT CGG A. HEK293FT cells were transfected with codon-optimized pCMV-VSV-G and pCMVR8.74 (both plasmids were given by P. Charneau, Institut Pasteur) and lentiviral vectors (pALPS-shPKR1/3 or pALPS-shLuc) at a mass ratio of 1:4:4. Huh7 cells were transduced with supernatant from transfected HEK293FT cells and selected with 10 μg/μl puromycin (Sigma-Aldrich) 4 days later.
siRNA transfection.
Huh7 cells were transfected using Lipofectamine RNAiMax (Life Technologies) with 30 nM nontargeting scrambled negative-control (siScr) or G3BP1, G3BP2, and TIAR small interfering RNAs (siRNAs). Alternatively, cells were transfected with 40 nM siRNA-TIA1 or 20 nM siRNA-RIG-I. siRNAs were obtained from Dharmacon (ON-TARGETplus SMARTpool). Transfected cells were harvested or used for infection assays at 48 h after transfection.
ELISA.
The amount of TNF-α or IL-6 in cell culture supernatants was quantified by enzyme-linked immunosorbent assay (ELISA) using the human TNF-α or IL-6 ELISA kit (eBioscience), according to the manufacturer’s instructions. The detection limits were 4 pg/ml for TNF-α and 2 pg/ml for IL-6.
Western blot analysis.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (Sigma) containing a protease and phosphatase inhibitor mixture (Roche). Cell lysates were normalized for protein content and boiled in NuPAGE lithium dodecyl sulfate (LDS) sample buffer (Thermo Fisher Scientific) under nonreducing conditions, and the proteins were separated by SDS-PAGE (NuPAGE 4 to 12% Bis-Tris gel; Life Technologies). Proteins were transferred to nitrocellulose membranes (Bio-Rad) using a Trans-Blot Turbo Transfer system (Bio-Rad). After blocking with PBS–0.1% Tween 20 (PBST) containing 5% milk or bovine serum albumin (BSA) for 1 h at room temperature, the membrane was incubated overnight at 4°C with primary antibodies diluted in a blocking buffer. Finally, the membrane was incubated for 45 min at room temperature with secondary antibodies (anti-rabbit/mouse IgG [H+L] DyLight 680/800) diluted in a blocking buffer, washed, and scanned using an Odyssey CLx infrared imaging system (Li-Cor Bioscience).
RNA extraction and RT-qPCR analysis.
Total RNAs were extracted from cell lysates using the NucleoSpin RNA II kit (Macherey-Nagel) following the manufacturer’s protocol and were eluted in nuclease-free water. First-strand cDNA synthesis was performed with the RevertAid H Minus Moloney murine leukemia virus (M-MuLV) reverse transcriptase (Thermo Fisher Scientific). Quantitative real-time PCR was performed on a real-time PCR system (Quant Studio 6 Flex; Applied Biosystems) with SYBR green PCR master mix (Life Technologies). Data were analyzed with the ΔΔCT method, with all samples normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase). All experiments were performed in technical triplicate. The primers used in this study are as follows: GAPDH, GGT CGG AG TCA ACG GAT TTG and ACT CCA CGA CGT ACT CAG CG; YF-NS3, GCG TAA GGC TGG AAA GAG TG and CTT CCT CCC TTC ATC CAC AA; TNFA, GCC CAT GTT GTA GCA AAC CC and GGA GGT TGA CCT TGG TCT GG; IL-6, TGC AAT AAC CAC CCC TGA CC and GTG CCC ATG CTA CAT TTG CC; IFNB, AAG CAA TTG TCC AGT CCC A and TGC ATT ACC TGA AGG CCA AG; IL28A/B, TCC AGA ACC TTC AGC GTC AG and AGG GCC AAA GAT GCC TTA GA; IL-29, TCA GCT TGA GTG ACT CTT CCA AGG and GCC ACA TTG GCA GGT TCA AAT CTC; PKR, CAG AAT TGA CGG AAA GAC TTA CG and CTC TCA AGA GAA TCA TCA CTG GT; G3BP1, CAG TTA TAT AGA CCG GCG GC and TAT GTC CAA ACC TAC GCG GC; G3BP2, CAA TCT CAG CCA CCT CGT GT and CCA CAA CGT TTC CAA AAC TCA T; TIAR, CGT CTG GGT TAA CAG ATC AGC and CAT GGG CTG CAC TTT CAT GG; and TIA1, AAT CCC GTG CAA CAG CAG GA and AGG CGG TTG CAC TCC ATA AT). Viral genome equivalent concentrations (genome equivalents per milliliter) were determined by extrapolation from a standard curve generated from serial dilutions of the plasmid encoding the YF-R.luc2A-RP (66).
Immunofluorescence, FISH, and confocal analysis.
Cells were fixed with 4% paraformaldehyde (PFA) (Sigma-Aldrich) for 30 min at room temperature, permeabilized with 0.5% Triton X-100 in PBS, and then blocked with PBS containing 0.05% Tween and 5% BSA before incubation with the indicated primary antibodies. Antibody labeling was revealed with Alexa Fluor 488-, 594-, or 647-conjugated antibodies. Coverslips were mounted on slides using ProLong Gold Antifade reagent with NucBlue solution (Invitrogen). Images were acquired with a Zeiss LSM 700 inverted confocal microscope. Fluorescence in situ hybridization (FISH) was performed after immunostaining. The plus RNA strands of YFV were detected following the manufacturer’s protocol (ViewRNA ISH Cell Assays) using probe sets designed by Affymetrix. The Alexa Fluor 546-conjugated plus-strand probe set targets a region between nucleotides 4567 and 5539 of the YFV genome. Intensity plot data were generated using Zen 2 Lite (Zeiss) and plotted using GraphPad Prism 6.
Stress granule quantification.
Nucleus and stress granule quantifications were performed using CellProfiler (v3.0.0) with a custom pipeline (available on request). Briefly, (i) nuclei were detected using the NucBlue signal, (ii) cellular borders were estimated using the TIAR or eiF3b signal, and (iii) stress granule borders were estimated using the G3BP, TIAR, eIF4G, or eIF3b signal. Further analyses were performed using R and RStudio (67). Cells were considered positive for stress granules if they exhibited at least two of them.
Statistical analysis.
Data are presented as means ± standard deviations (SD) and were analyzed using GraphPad Prism 6. Statistical analysis of percent values or fold enrichment values were performed on logit or log-transformed values, respectively. Statistical analysis was performed with the two-tailed unpaired t test with Welch correction for pairwise comparisons or by one- or two-way analysis of variance (ANOVA) with Tukey’s or Dunnett’s multiple-comparison test when comparing more than two sets of values. Each experiment was performed at least twice, unless otherwise stated. The statistical significance of differences is indicated as follows: *, P < 0.05, **, P < 0.01; ***, P < 0.001; and ns, not significant.
ACKNOWLEDGMENTS
We thank Charlie Rice for generously providing the anti-YFV-NS4b antibodies, Richard Kuhn for the YFV replicon construct, Marie Flamand for anti-NS1 antibodies (17A12), Christine Neuveut for the HepG2 cells, Olivier Schwartz for the CMMT cells, Annette Martin for Huh7 cells, Pamela Schnupf for eIF3b and eIF4G antibodies, Pierre Charneau for the plasmids pCMV-VSV-G and pCMVR8.74, Andrea Cimarelli for the pALPS plasmid, Olivier Disson for immunohistochemistry advice, and Léa Richard for cloning shPKR into pALPS. We are grateful to the members of our laboratory for helpful discussions and technical advice. We thank Audrey Salles and Julien Fernandes (UTechS Photonic Bioimaging Platform, Institut Pasteur) for their help in confocal acquisition. We are grateful to Stéphane Rigaud (Image Analysis Hub, Institut Pasteur) for his advice on analyzing confocal images and to Vincent Guillemot (Bioinformatics and Biostatistics Hub, Institut Pasteur) for his help with statistical analysis. Finally, we thank Stephanie Straubel for critical readings of the manuscript.
REFERENCES
- 1.Gould EA, Solomon T. 2008. Pathogenic flaviviruses. Lancet 371:500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- 2.Gubler DJ, Kuno G, Markoff L. 2007. Flaviviruses, p 1153–1252. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Douam F, Ploss A. 2018. Yellow fever virus: knowledge gaps impeding the fight against an old foe. Trends Microbiol 26:913–928. doi: 10.1016/j.tim.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pulendran B. 2009. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol 9:741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 5.Barrett AD. 2016. Yellow fever in Angola and beyond—the problem of vaccine supply and demand. N Engl J Med 375:301–303. doi: 10.1056/NEJMp1606997. [DOI] [PubMed] [Google Scholar]
- 6.Barrett ADT. 2018. The reemergence of yellow fever. Science 361:847–848. doi: 10.1126/science.aau8225. [DOI] [PubMed] [Google Scholar]
- 7.Messaoudi I, Basler CF. 2015. Immunological features underlying viral hemorrhagic fevers. Curr Opin Immunol 36:38–46. doi: 10.1016/j.coi.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paessler S, Walker DH. 2013. Pathogenesis of the viral hemorrhagic fevers. Annu Rev Pathol 8:411–440. doi: 10.1146/annurev-pathol-020712-164041. [DOI] [PubMed] [Google Scholar]
- 9.Chen IY, Ichinohe T. 2015. Response of host inflammasomes to viral infection. Trends Microbiol 23:55–63. doi: 10.1016/j.tim.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 10.ter Meulen J, Sakho M, Koulemou K, Magassouba N, Bah A, Preiser W, Daffis S, Klewitz C, Bae HG, Niedrig M, Zeller H, Heinzel-Gutenbrunner M, Koivogui L, Kaufmann A. 2004. Activation of the cytokine network and unfavorable outcome in patients with yellow fever. J Infect Dis 190:1821–1827. doi: 10.1086/425016. [DOI] [PubMed] [Google Scholar]
- 11.Engelmann F, Josset L, Girke T, Park B, Barron A, Dewane J, Hammarlund E, Lewis A, Axthelm MK, Slifka MK, Messaoudi I. 2014. Pathophysiologic and transcriptomic analyses of viscerotropic yellow fever in a rhesus macaque model. PLoS Negl Trop Dis 8:e3295. doi: 10.1371/journal.pntd.0003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Streicher F, Jouvenet N. 2019. Stimulation of innate immunity by host and viral RNAs. Trends Immunol 40:1134–1148. doi: 10.1016/j.it.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Chow KT, Gale M, Loo Y-M. 2018. RIG-I and other RNA sensors in antiviral immunity. Annu Rev Immunol 36:667–694. doi: 10.1146/annurev-immunol-042617-053309. [DOI] [PubMed] [Google Scholar]
- 14.Nan Y, Nan G, Zhang YJ. 2014. Interferon induction by RNA viruses and antagonism by viral pathogens. Viruses 6:4999–5027. doi: 10.3390/v6124999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chazal M, Beauclair G, Gracias S, Najburg V, Simon-Loriere E, Tangy F, Komarova AV, Jouvenet N. 2018. RIG-I recognizes the 5′ region of dengue and Zika virus genomes. Cell Rep 24:320–328. doi: 10.1016/j.celrep.2018.06.047. [DOI] [PubMed] [Google Scholar]
- 16.Sprokholt JK, Kaptein TM, van Hamme JL, Overmars RJ, Gringhuis SI, Geijtenbeek TBH. 2017. RIG-I-like receptor triggering by dengue virus drives dendritic cell immune activation and TH1 differentiation. J Immunol 198:4764–4771. doi: 10.4049/jimmunol.1602121. [DOI] [PubMed] [Google Scholar]
- 17.Sprokholt JK, Kaptein TM, van Hamme JL, Overmars RJ, Gringhuis SI, Geijtenbeek TBH. 2017. RIG-I-like receptor activation by dengue virus drives follicular T helper cell formation and antibody production. PLoS Pathog 13:e1006738. doi: 10.1371/journal.ppat.1006738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hertzog J, Junior AGD, Rigby RE, Donald CL, Mayer A, Sezgin E, Song C, Jin B, Hublitz P, Eggeling C, Kohl A, Rehwinkel J. 2018. Infection with a Brazilian isolate of Zika virus generates RIG-I stimulatory RNA and the viral NS5 protein blocks type I IFN induction and signaling. Eur J Immunol 48:1120–1136. doi: 10.1002/eji.201847483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruni D, Chazal M, Sinigaglia L, Chauveau L, Schwartz O, Despres P, Jouvenet N. 2015. Viral entry route determines how human plasmacytoid dendritic cells produce type I interferons. Sci Signal 8:ra25. doi: 10.1126/scisignal.aaa1552. [DOI] [PubMed] [Google Scholar]
- 20.Onomoto K, Jogi M, Yoo J-S, Narita R, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, Matsumiya T, Namiki H, Yoneyama M, Fujita T. 2012. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One 7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh S-W, Onomoto K, Wakimoto M, Onoguchi K, Ishidate F, Fujiwara T, Yoneyama M, Kato H, Fujita T. 2016. Leader-containing uncapped viral transcript activates RIG-I in antiviral stress granules. PLoS Pathog 12:e1005444. doi: 10.1371/journal.ppat.1005444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo J-S, Takahasi K, Ng CS, Ouda R, Onomoto K, Yoneyama M, Lai JC, Lattmann S, Nagamine Y, Matsui T, Iwabuchi K, Kato H, Fujita T. 2014. DHX36 enhances RIG-I signaling by facilitating PKR-mediated antiviral stress granule formation. PLoS Pathog 10:e1004012. doi: 10.1371/journal.ppat.1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sánchez-Aparicio MT, Ayllón J, Leo-Macias A, Wolff T, García-Sastre A. 2017. Subcellular localizations of RIG-I, TRIM25, and MAVS complexes. J Virol 91:e01155-16. doi: 10.1128/JVI.01155-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langereis MA, Feng Q, van Kuppeveld FJ. 2013. MDA5 localizes to stress granules, but this localization is not required for the induction of type I interferon. J Virol 87:6314–6325. doi: 10.1128/JVI.03213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narita R, Takahasi K, Murakami E, Hirano E, Yamamoto SP, Yoneyama M, Kato H, Fujita T. 2014. A novel function of human pumilio proteins in cytoplasmic sensing of viral infection. PLoS Pathog 10:e1004417. doi: 10.1371/journal.ppat.1004417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu S, Sun H, Yin L, Li J, Mei S, Xu F, Wu C, Liu X, Zhao F, Zhang D, Huang Y, Ren L, Cen S, Wang J, Liang C, Guo F. 2019. PKR-dependent cytosolic cGAS foci are necessary for intracellular DNA sensing. Sci Signal 12:eaav7934. doi: 10.1126/scisignal.aav7934. [DOI] [PubMed] [Google Scholar]
- 27.Onomoto K, Yoneyama M, Fung G, Kato H, Fujita T. 2014. Antiviral innate immunity and stress granule responses. Trends Immunol 35:420–428. doi: 10.1016/j.it.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCormick C, Khaperskyy DA. 2017. Translation inhibition and stress granules in the antiviral immune response. Nat Rev Immunol 17:647–660. doi: 10.1038/nri.2017.63. [DOI] [PubMed] [Google Scholar]
- 29.Ivanov P, Kedersha N, Anderson P. 2019. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol 11:a032813. doi: 10.1101/cshperspect.a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reineke LC, Lloyd RE. 2013. Diversion of stress granules and P-bodies during viral infection. Virology 436:255–267. doi: 10.1016/j.virol.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sumpter R, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. 2000. Dynamic shuttling of Tia-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monath TP. 2005. Yellow fever vaccine. Expert Rev Vaccines 4:553–574. doi: 10.1586/14760584.4.4.553. [DOI] [PubMed] [Google Scholar]
- 34.Hou S, Kumar A, Xu Z, Airo AM, Stryapunina I, Wong CP, Branton W, Tchesnokov E, Götte M, Power C, Hobman TC. 2017. Zika virus hijacks stress granule proteins and modulates the host stress response. J Virol 91:e00474-17. doi: 10.1128/JVI.00474-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tu Y-C, Yu C-Y, Liang J-J, Lin E, Liao C-L, Lin Y-L. 2012. Blocking double-stranded RNA-activated protein kinase PKR by Japanese encephalitis virus nonstructural protein 2A. J Virol 86:10347–10358. doi: 10.1128/JVI.00525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samuel MA, Whitby K, Keller BC, Marri A, Barchet W, Williams BRG, Silverman RH, Gale M, Diamond MS. 2006. PKR and RNase L contribute to protection against lethal West Nile virus infection by controlling early viral spread in the periphery and replication in neurons. J Virol 80:7009–7019. doi: 10.1128/JVI.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dabo S, Meurs EF. 2012. dsRNA-dependent protein kinase PKR and its role in stress, signaling and HCV infection. Viruses 4:2598–2635. doi: 10.3390/v4112598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu L, Han AP, Chen JJ. 2001. Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol Cell Biol 21:7971–7980. doi: 10.1128/MCB.21.23.7971-7980.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonnet MC, Weil R, Dam E, Hovanessian AG, Meurs EF. 2000. PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol Cell Biol 20:4532–4542. doi: 10.1128/mcb.20.13.4532-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonnet MC, Daurat C, Ottone C, Meurs EF. 2006. The N-terminus of PKR is responsible for the activation of the NF-κB signaling pathway by interacting with the IKK complex. Cell Signal 18:1865–1875. doi: 10.1016/j.cellsig.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Sinigaglia L, Gracias S, Decembre E, Fritz M, Bruni D, Smith N, Herbeuval JP, Martin A, Dreux M, Tangy F, Jouvenet N. 2018. Immature particles and capsid-free viral RNA produced by yellow fever virus-infected cells stimulate plasmacytoid dendritic cells to secrete interferons. Sci Rep 8:10889. doi: 10.1038/s41598-018-29235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M. 2013. The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol 87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai YT, Chang SY, Lee CN, Kao CL. 2009. Human TLR3 recognizes dengue virus and modulates viral replication in vitro. Cell Microbiol 11:604–615. doi: 10.1111/j.1462-5822.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- 44.Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, Perera-Lecoin M, Surasombatpattana P, Talignani L, Thomas F, Cao-Lormeau VM, Choumet V, Briant L, Despres P, Amara A, Yssel H, Misse D. 2015. Biology of Zika virus infection in human skin cells. J Virol 89:8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang JP, Liu P, Latz E, Golenbock DT, Finberg RW, Libraty DH. 2006. Flavivirus activation of plasmacytoid dendritic cells delineates key elements of TLR7 signaling beyond endosomal recognition. J Immunol 177:7114–7121. doi: 10.4049/jimmunol.177.10.7114. [DOI] [PubMed] [Google Scholar]
- 46.Vanwalscappel B, Tada T, Landau NR. 2018. Toll-like receptor agonist R848 blocks Zika virus replication in human monocytes by inducing the antiviral protein viperin. Virology 522:199–208. doi: 10.1016/j.virol.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zamanian-Daryoush M, Mogensen TH, DiDonato JA, Williams BR. 2000. NF-kappaB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-kappaB-inducing kinase and IkappaB kinase. Mol Cell Biol 20:1278–1290. doi: 10.1128/mcb.20.4.1278-1290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pham AM, Santa Maria FG, Lahiri T, Friedman E, Marié IJ, Levy DE. 2016. PKR transduces MDA5-dependent signals for type I IFN induction. PLoS Pathog 12:e1005489. doi: 10.1371/journal.ppat.1005489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gil J, García MA, Gomez-Puertas P, Guerra S, Rullas J, Nakano H, Alcamí J, Esteban M. 2004. TRAF family proteins link PKR with NF-κB activation. Mol Cell Biol 24:4502–4512. doi: 10.1128/mcb.24.10.4502-4512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arnaud N, Dabo S, Akazawa D, Fukasawa M, Shinkai-Ouchi F, Hugon J, Wakita T, Meurs EF. 2011. Hepatitis C virus reveals a novel early control in acute immune response. PLoS Pathog 7:e1002289. doi: 10.1371/journal.ppat.1002289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang P, Li Y, Xia J, He J, Pu J, Xie J, Wu S, Feng L, Huang X, Zhang P. 2014. IPS-1 plays an essential role in dsRNA-induced stress granule formation by interacting with PKR and promoting its activation. J Cell Sci 127:2471–2482. doi: 10.1242/jcs.139626. [DOI] [PubMed] [Google Scholar]
- 52.Ravindran R, Khan N, Nakaya HI, Li S, Loebbermann J, Maddur MS, Park Y, Jones DP, Chappert P, Davoust J, Weiss DS, Virgin HW, Ron D, Pulendran B. 2014. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science 343:313–317. doi: 10.1126/science.1246829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonenfant G, Williams N, Netzband R, Schwarz MC, Evans MJ, Pager CT. 2019. Zika virus subverts stress granules to promote and restrict viral gene expression. J Virol 93:e00520-19. doi: 10.1128/JVI.00520-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emara MM, Brinton MA. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci U S A 104:9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albornoz A, Carletti T, Corazza G, Marcello A. 2014. The stress granule component TIA-1 binds tick-borne encephalitis virus RNA and is recruited to perinuclear sites of viral replication to inhibit viral translation. J Virol 88:6611–6622. doi: 10.1128/JVI.03736-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roth H, Magg V, Uch F, Mutz P, Klein P, Haneke K, Lohmann V, Bartenschlager R, Fackler OT, Locker N, Stoecklin G, Ruggieri A. 2017. Flavivirus infection uncouples translation suppression from cellular stress responses. mBio 8:e00488-17. doi: 10.1128/mBio.00488-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Basu M, Courtney SC, Brinton MA. 2017. Arsenite-induced stress granule formation is inhibited by elevated levels of reduced glutathione in West Nile virus-infected cells. PLoS Pathog 13:e1006240. doi: 10.1371/journal.ppat.1006240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amorim R, Temzi A, Griffin BD, Mouland AJ. 2017. Zika virus inhibits eIF2α-dependent stress granule assembly. PLoS Negl Trop Dis 11:e0005775. doi: 10.1371/journal.pntd.0005775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Q, Sharma NR, Zheng Z-M, Chen M. 2019. Viral regulation of RNA granules in infected cells. Virol Sin 34:175–191. doi: 10.1007/s12250-019-00122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Christ W, Tynell J, Klingström J. 2019. Puumala and Andes orthohantaviruses cause transient protein kinase R-dependent formation of stress granules. J Virol 94:e01168-19. doi: 10.1128/JVI.01168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiang JJ, Sparrer KMJ, van Gent M, Lässig C, Huang T, Osterrieder N, Hopfner K-P, Gack MU. 2018. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat Immunol 19:53–62. doi: 10.1038/s41590-017-0005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Y, Ye X, Dunker W, Song Y, Karijolich J. 2018. RIG-I like receptor sensing of host RNAs facilitates the cell-intrinsic immune response to KSHV infection. Nat Commun 9:4841. doi: 10.1038/s41467-018-07314-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rehwinkel J, Gack MU. 2020. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol 20:537–515. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76:13001–13014. doi: 10.1128/jvi.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grutter MG, Luban J. 2011. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones CT, Patkar CG, Kuhn RJ. 2005. Construction and applications of yellow fever virus replicons. Virology 331:247–259. doi: 10.1016/j.virol.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 67.RStudio Team. 2016. RStudio: integrated development environment for R. RStudio, Boston, MA. [Google Scholar]