Epstein-Barr virus (EBV) is a ubiquitous human pathogen and associated with various human diseases. EBV undergoes latency and lytic replication stages in its life cycle. The transition into the lytic replication stage, at which virus is produced, is mainly regulated by the viral gene product, Zta. Therefore, the regulation of Zta function becomes a central issue regarding viral biology and pathogenesis. Known modifications of Zta include phosphorylation and sumoylation. Here, we report the role of ubiquitination in regulating Zta function. We found that Zta is subjected to ubiquitination in both EBV-infected and EBV-negative cells. The ubiquitin modification targets 4 lysine residues on Zta, leading to both mono- and polyubiquitination of Zta. Ubiquitination of Zta affects the protein’s stability and likely contributes to the progression of viral lytic replication. The function and fate of Zta may be determined by the specific lysine residue being modified.
KEYWORDS: Epstein-Barr virus, Zta, lytic replication, reactivation, ubiquitination
ABSTRACT
The Epstein-Barr virus (EBV) immediate early transactivator Zta plays a key role in regulating the transition from latency to the lytic replication stages of EBV infection. Regulation of Zta is known to be controlled through a number of transcriptional and posttranscriptional events. Here, we show that Zta is targeted for ubiquitin modification and that this can occur in EBV-negative and in EBV-infected cells. Genetic studies show critical roles for both an amino-terminal region of Zta and the basic DNA binding domain of Zta in regulating Zta ubiquitination. Pulse-chase experiments demonstrate that the bulk population of Zta is relatively stable but that at least a subset of ubiquitinated Zta molecules are targeted for degradation in the cell. Mutation of four out of a total of nine lysine residues in Zta largely abrogates its ubiquitination, indicating that these are primary ubiquitination target sites. A Zta mutant carrying mutations at these four lysine residues (lysine 12, lysine 188, lysine 207, and lysine 219) cannot induce latently infected cells to produce and/or release infectious virions. Nevertheless, this mutant can induce early gene expression, suggesting a possible defect at the level of viral replication or later in the lytic cascade. As far as we know, this is the first study that has investigated the targeting of Zta by ubiquitination or its role in Zta function.
IMPORTANCE Epstein-Barr virus (EBV) is a ubiquitous human pathogen and associated with various human diseases. EBV undergoes latency and lytic replication stages in its life cycle. The transition into the lytic replication stage, at which virus is produced, is mainly regulated by the viral gene product, Zta. Therefore, the regulation of Zta function becomes a central issue regarding viral biology and pathogenesis. Known modifications of Zta include phosphorylation and sumoylation. Here, we report the role of ubiquitination in regulating Zta function. We found that Zta is subjected to ubiquitination in both EBV-infected and EBV-negative cells. The ubiquitin modification targets 4 lysine residues on Zta, leading to both mono- and polyubiquitination of Zta. Ubiquitination of Zta affects the protein’s stability and likely contributes to the progression of viral lytic replication. The function and fate of Zta may be determined by the specific lysine residue being modified.
INTRODUCTION
Epstein-Barr virus (EBV) is a DNA tumor virus that infects more than 90% of the adult population worldwide (1, 2). It is involved in the development of a number of human nonmalignant and malignant diseases (1–6), including infectious mononucleosis, Burkitt’s lymphoma (BL), Hodgkin’s disease, nasopharyngeal carcinoma (NPC), gastric carcinoma, lymphoproliferative diseases in immunosuppressed patients, and non-small cell lung cancers. Like other herpesviruses, EBV has latent and lytic gene expression programs that support distinct biological functions. Following initial infection, EBV is maintained in the host indefinitely in part by establishment of a latent infection in which little or no viral gene expression occurs. Upon receiving appropriate induction signals, latently infected cells undergo virus reactivation. The two earliest genes expressed following reactivation are the lytic transactivators Zta (also referred to as BZLF1, ZEBRA, and EB1) and Rta (also referred to as BRLF1 and R). Expression of these viral transactivators induces early and late gene expression, ultimately resulting in the production of infectious virions.
Zta is an immediate early gene that is a key mediator of the transition from latency to productive lytic replication. Zta contains a nonacidic amino terminal transactivation domain and a basic leucine zipper (bZIP)-type DNA binding/dimerization domain located near its carboxyl terminus (7, 8). The Zta activation domain induces transcription in part through stabilizing the association of basal transcription factors such as TATA binding protein (TBP) and TFIIA with the “TATA” promoter element as well as stabilizing the interaction between TBP-associated factors (TAFs) and the “TATA” promoter elements (9–11). The Zta activation domain also binds and recruits the coactivators CREB binding protein (CBP) (12, 13) and its close relative, p300 (Z. Lin, unpublished data). Both of these coactivators contain intrinsic histone acetyltransferase activity, which enhances promoter activity in part by facilitating a more open chromatin environment (14, 15).
In addition to transactivation of viral lytic genes, Zta also plays an essential role in lytic genomic replication as an origin binding protein (16, 17). The lytic origin of replication (oriLyt) contains seven ZREs, and binding of Zta to oriLyt is necessary for the activation of lytic replication (16–22). The DNA binding/dimerization domain of Zta is required for its replication function in addition to a subregion of the activation domain (amino acids 11 to 25) that facilitates recruitment of a helicase-primase complex (19, 23). The replication function of this amino-terminal region is separable from Zta’s transactivation functions since nonconservative mutation of amino acids 12 and 13 abrogates Zta’s replication activity but does not alter its ability to induce transcription (19).
Besides Zta’s key roles in the activation of lytic EBV transcription and lytic genome replication, it also has significant influences on host cells, including the manipulation of cellular gene expression (24–31), the cell cycle (32–38), cellular immunity (28, 29, 39–43), the DNA damage response (44–46), and subcellular structures (47–49). It is currently uncertain whether all Zta molecules in a reactive cell universally carry out all of Zta’s various functions or whether posttranslational modifications tag subpopulations of Zta to support certain functions. Nevertheless, there is evidence for the latter. For example, phosphorylation of serine 173 by casein kinase 2 (CK2) appears to play a role in temporally regulating the function of Zta to allow Zta to activate early genes without activating certain late genes (50). When serine 173 is phosphorylated, Zta inhibits the ability of Rta to activate expression of some late genes (50). Further investigations into the functional consequences of the various Zta posttranslation modifications will likely reveal myriad ways in which these modifications temporally and spatially control the various functions of Zta.
Zta is known to be subjected to posttranslational modifications, including phosphorylation at a number of serine and threonine residues (50–56), as well as sumoylation of lysine 12 (47, 57). Here, we report that Zta is subjected to posttranslational modification by ubiquitination. Ubiquitin is a highly conserved polypeptide with ubiquitous expression in higher eukaryotic organisms. Ubiquitin conjugation occurs through a biochemical cascade executed by a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-ligase (E3). At the end of the reaction, ubiquitin molecules are covalently linked through their C-terminal carboxyl group to an ε-amino group of lysine residues in the target protein (58–61).
Ubiquitination regulates target protein activity in both a proteolytic and nonproteolytic manner (60). In the proteolytic pathway, the ubiquitinated protein is directed to and degraded in the proteasome system. In the nonproteolytic pathway, ubiquitination can alter a protein’s subcellular localization or interaction with cofactors. After ubiquitination, whether the target undergoes degradation largely depends on the specific type of ubiquitin modification that it received and the extent of ubiquitin polymerization. For instance, polyubiquitin chains with four or more ubiquitin molecules formed through the lysine 48 residue of ubiquitin function mainly as a degradation signal that directs the target to the proteasome (62–64). In contrast, ubiquitin chains of fewer than four molecules, or polyubiquitin chains linked through other lysine residues such as lysine 11 or lysine 63, signal the regulation of targets independently of degradation (60, 64–75). Frequently, factors controlled by the ubiquitin pathway are regulated by both the proteolytic and nonproteolytic types of ubiquitination. The ubiquitin-mediated regulation of c-Myc is a well-studied example (66, 76–78). The F box E3 ligase, Skp2, binds to a conserved element (MycboxII) in the amino terminus of c-Myc which is essential for transformation and transcriptional regulation (78), and Skp2-mediated ubiquitination contributes to the proteasomal degradation of c-Myc (78). A homologous-to-E6-AP carboxy terminus domain (HECT) ubiquitin ligase, HECT-H9, regulates c-Myc’s function through ubiquitination but in a nonproteolytic manner (66). HECT-H9 has been shown to ubiquitinate c-Myc in a carboxyl-terminal region that is required for recruitment of the coactivator p300 (66). This site-specific ubiquitination of c-Myc is believed to regulate the switch between transcription activation and repression (66). Here, we provide evidence that modification of Zta by ubiquitin is likely to be similarly complex and that it regulates Zta’s function in both a nonproteolytic and proteolytic manner during the lytic cycle.
RESULTS
Zta is ubiquitinated in vivo.
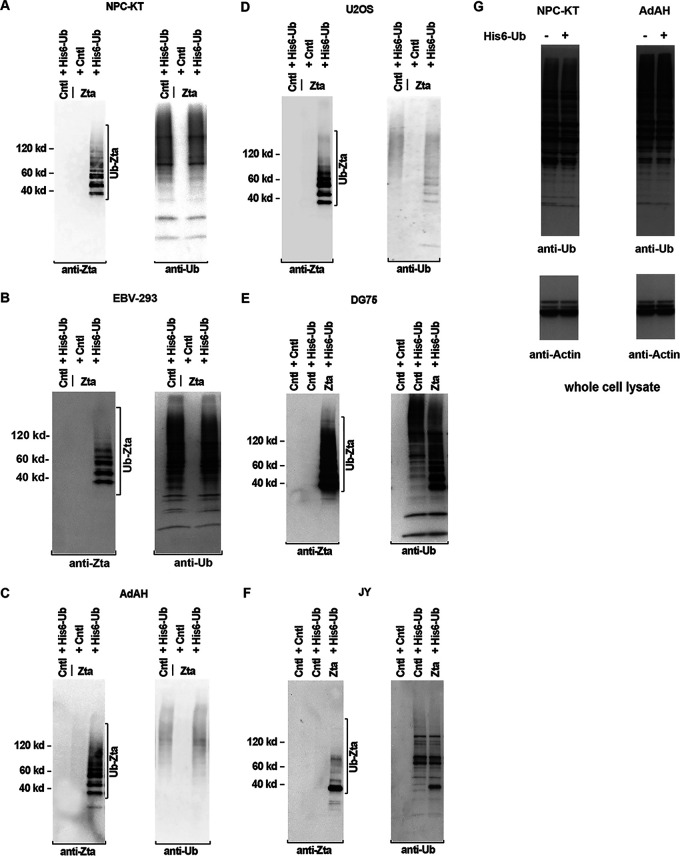
To examine whether Zta is subject to ubiquitination, nickel-nitrilotriacetic acid (Ni-NTA)-histidine pulldown-based ubiquitination assays were performed. With this method, ubiquitin-conjugated proteins can be captured under denaturing conditions. Denaturing conditions make this assay more specific than coimmunoprecipitation assays, in which the protein of interest can potentially be pulled down as an artifact through its association with a binding partner that is ubiquitinated. A control or a Zta expression vector was cotransfected with either a control (hemagglutinin-tagged ubiquitin [HA-Ub]) or a histidine-tagged ubiquitin expression vector (His6-Ub) into the EBV-positive nasopharyngeal carcinoma (NPC) cell line NPC-KT or the EBV-positive epithelial cell line EBV-293 (the proteasome inhibitor lactacystin was added to NPC-KT cells for the last 16 h of culture). Forty hours after transfection, cells were harvested and lysed under denaturing conditions in a buffer containing 6 M guanidinium hydrochloride. Histidine-tagged ubiquitin conjugates were purified using a nickel-NTA affinity resin, and eluates were subjected to Western blot analysis using an anti-Zta antibody. As shown in Fig. 1A and B, multiple ubiquitin-conjugated forms of Zta were observed in cells cotransfected with the Zta and His6-Ub expression vectors but not in cells transfected with His6-Ub expression vector alone or with the Zta plus control HA-Ub vectors.
FIG 1.
Zta is ubiquitinated in epithelial and B cells independently of viral factors. NPC-KT (A), EBV-293 (B), AdAH (C), U2OS (D), DG75 (E), and JY (F) cells were transfected with the indicated plasmids as described in Materials and Methods. (Lactacystin [10 μM] was added to NPC-KT, AdAH, and U2OS cells for the last 16 h of culture.) Cells were lysed under denaturing conditions and extracts were purified over nickel-NTA beads. The purified material was assayed by Western blot analysis employing anti-Zta and anti-ubiquitin antibodies (left and right, respectively). The whole-cell lysates of nontransfected NPC-KT and AdAH cells as well as cells that were transfected with the His6-Ub expression vector were analyzed with anti-ubiquitin and anti-actin antibodies (G).
Transfection of Zta expression vectors into EBV-positive cells typically results in expression of other lytic genes. To test whether EBV latency gene products or Zta-induced viral lytic gene products are required to facilitate Zta ubiquitination, a ubiquitination experiment was performed in the EBV-negative NPC line AdAH and the osteosarcoma cell line U2OS. Zta ubiquitination was observed in these cell lines (Fig. 1C and D), indicating that although EBV-encoded proteins may regulate Zta ubiquitination in some fashion, ubiquitination of Zta can be mediated entirely through cellular regulatory proteins.
Since EBV also infects B lymphocytes in vivo, ubiquitination of Zta in the EBV-negative Burkitt’s lymphoma cell line DG75 and the EBV-positive lymphoblastoid cell line JY was assessed. For these experiments, cells were transfected by electroporation and no lactacystin was added prior to harvesting. Zta ubiquitination was observed in both of these cell lines (Fig. 1E and F). Together, these results indicate that Zta is subjected to ubiquitin modification in both epithelial cells and B lymphocytes and that this can occur in the absence of other viral gene products.
To ensure that the detection of ubiquitinated Zta species in the experiments outlined above was not due to an artificially high level of overall ubiquitin levels in cells transfected with the His6-Ub expression vector, we assessed the total ubiquitin levels in nontransfected cells and cells transfected with His6-Ub expression vector. This analysis was carried out in two cell lines (NPC-KT and AdAH) in which high transfection efficiencies are typically obtained (80% and 75% transfection efficiencies, respectively). As shown in Fig. 1G, transfection of the His6-Ub expression vector did not notably alter the overall levels of ubiquitin. This is in line with previous observations that eukaryotic cells typically express very high levels of endogenous ubiquitin (79–82).
Endogenous Zta is ubiquitinated following reactivation.
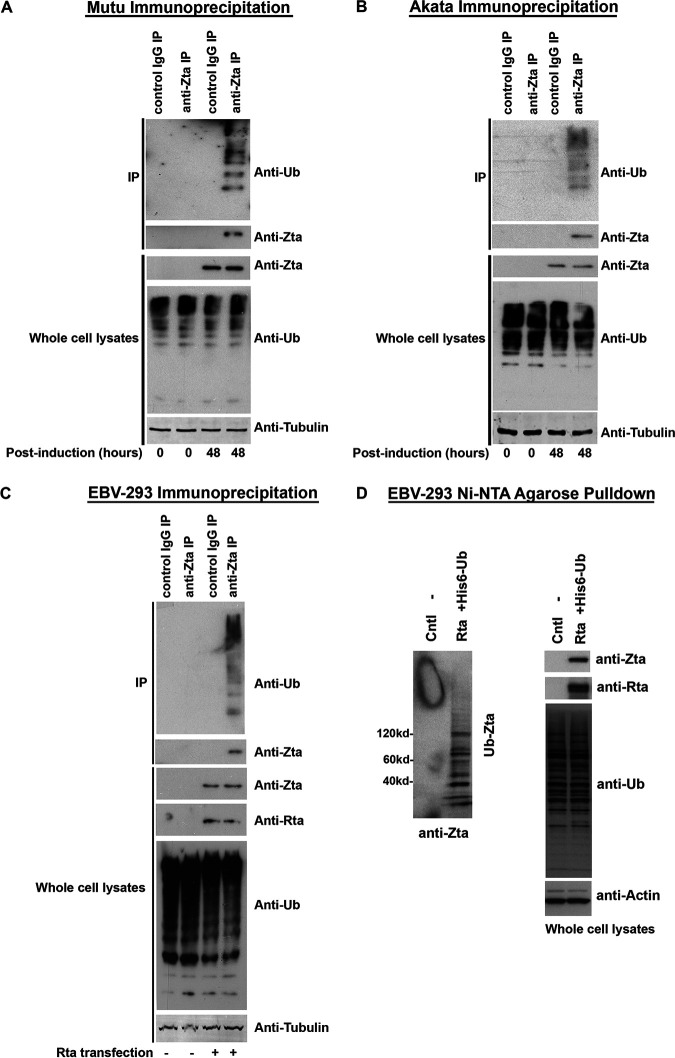
Next, we set out to examine whether endogenously expressed Zta is targeted for ubiquitination following activation of the lytic cycle. The lytic cycles in EBV-positive Burkitt’s lymphoma (BL) cells were induced by cross-linking of the surface B-cell receptors (BCR) with anti-immunoglobulin. EBV-positive Mutu and Akata BL cells were treated with either an anti-human IgM antibody for Mutu cells or an anti-human IgG antibody for Akata cells and at 0 h and 48 h postinduction, and the cells were lysed and immunoprecipitated with either a control antibody or an anti-Zta antibody. Immunoprecipitates were analyzed for the presence of ubiquitin and Zta by Western blot analysis. As shown in Fig. 2A for Mutu cells and Fig. 2B for Akata cells, ubiquitinated Zta species were readily detected at 48 h postinduction.
FIG 2.
Endogenously expressed Zta is ubiquitinated following activation of the lytic cycle in Mutu cells, Akata cells, and EBV-293 cells. (A) EBV lytic reactivation was induced in EBV-positive Mutu cells by anti-human IgM treatment. At 0 h and 48 h postinduction, cell extracts were prepared and immunoprecipitated with either a control IgG antibody or an anti-Zta antibody. Immunoprecipitates were analyzed for ubiquitin and Zta by Western blot analysis. The levels of Zta, ubiquitin, and α/β tubulin in the whole-cell lysates were also analyzed by Western blot analysis. (B) EBV lytic reactivation was induced in EBV-positive Akata cells by anti-human IgG treatment. At 0 h and 48 h postinduction, cell extracts were prepared and immunoprecipitated with either a control IgG antibody or an anti-Zta antibody. Immunoprecipitates were analyzed for ubiquitin and Zta by Western blot analysis. The levels of Zta, ubiquitin, and α/β tubulin in the whole-cell lysates were also analyzed by Western blot analysis. (C) EBV lytic reactivation was induced in EBV-293 cells by transfection of an EBV Rta expression vector. At 48 h postinduction, cell extracts were prepared and immunoprecipitated with either a control IgG antibody or an anti-Zta antibody. Immunoprecipitates were analyzed for ubiquitin and Zta by Western blot analysis. The levels of Zta, Rta, ubiquitin, and α/β tubulin in the whole-cell lysates were also analyzed by Western blot analysis. (D) EBV lytic reactivation was induced in EBV-293 cells by transfection of an EBV Rta expression vector. After 48 h, cells were lysed under denaturing conditions and extracts were purified over nickel-NTA beads. The purified material was assayed by Western blot analysis employing an anti-Zta antibody. Expression levels of Zta, Rta, ubiquitin, and actin were determined by Western blot analysis of whole-cell lysates.
To further address whether endogenously expressed Zta is ubiquitinated following reactivation in EBV-infected epithelial cells, viral reactivation was induced by transfecting EBV-293 cells with an EBV immediate early Rta gene expression or its control vectors. At 48 h posttransfection/induction, the cells were lysed and immunoprecipitated with either a control antibody or an anti-Zta antibody. Immunoprecipitates were analyzed for the presence of ubiquitin and Zta by Western blot analysis. As shown in Fig. 2C, ubiquitinated Zta species were readily detected at 48 h postinduction. Meanwhile, EBV-293 cells were transfected with an Rta expression vector with or without a His6-Ub expression vector. Histidine-tagged ubiquitin conjugates were purified using a nickel-NTA affinity resin, and eluates were subjected to Western blot analysis using an anti-Zta antibody. As shown in Fig. 2D, endogenous Zta is ubiquitinated following Rta-mediated reactivation.
Amino-terminal sequences of Zta influence ubiquitination of Zta.
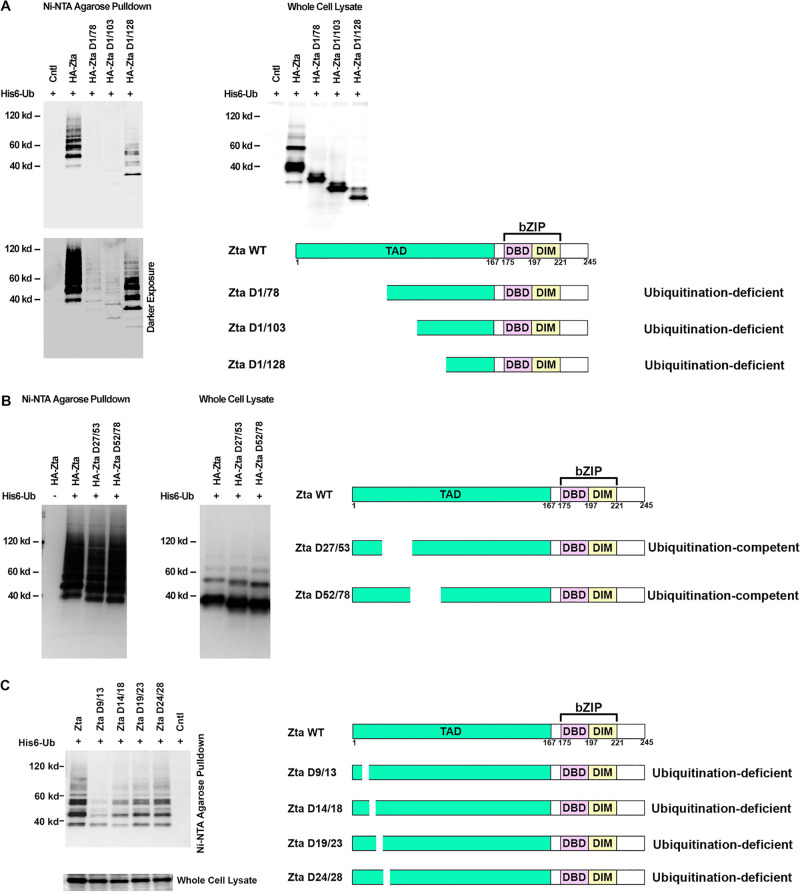
To identify motifs of Zta that influence the protein’s ubiquitination, a series of amino-terminal deletion constructs were assessed for ubiquitination. For ease of interpretation, these experiments were performed in the EBV-negative cell line AdAH, which avoids possible complications related to heterodimerization with endogenously expressed wild-type Zta. Deletion of the first 78 amino acids or the first 103 amino acids caused a substantial decrease in overall Zta ubiquitination although clear evidence of ubiquitination was observed in a darker/longer exposure (Fig. 3A). Unexpectedly, deletion of the next 25 amino acids from the amino terminus resulted in an increase in the level of Zta ubiquitination. This suggests that there is a negative regulatory element in the region between amino acids 103 and 129, and deletion of this negative regulatory element results in an increase in ubiquitination. Together, our data indicate that sequences between residues 1 and 79 are required for maximal ubiquitination and that there is a negative regulatory element between residues 103 and 129.
FIG 3.
The Zta transactivation domain regulates Zta ubiquitination. EBV-negative AdAH cells were transfected with or without 5 μg of a ubiquitin (His6-Ub) expression vector as well as 5 μg of the indicated Zta, Zta mutant, or control expression plasmids. Cells were lysed under denaturing conditions and extracts were purified over nickel-NTA beads. The purified material was assayed by Western blot analysis employing either an anti-HA antibody (A and B) or an anti-Zta antibody (C). Expression levels of Zta and its mutants were determined by Western blot analysis of whole-cell lysates. Darker exposure corresponds to longer exposure time. Shown is a schematic illustration of functional domains in Zta. Deletion regions are marked for each of the Zta mutants. Abbreviations: TAD, transactivation domain; DBD, DNA-binding domain; DIM, dimerization domain; bZIP, basic leucine zipper-like domain.
To further investigate sequences between amino acids 1 and 79 that are crucial for high-level ubiquitination, internal deletion mutants spanning most of this region were tested. Deletion of amino acids 27 to 53 and deletion of amino acids 52 to 78 did not influence the level of Zta ubiquitination (Fig. 3B). In contrast, analysis of a series of 5 amino acid deletion mutants spanning amino acids 9 through 28 showed an influence of these sequences on the level of Zta ubiquitination. Deletion of amino acids 9 to 13 (Zta D9/13) showed the most substantial impact on Zta ubiquitination, and deletion of amino acids 14 to 18 had a significant but more moderate impact on Zta ubiquitination (Fig. 3C). Importantly, within the deleted sequence in the Zta D9/13 mutant is a lysine residue (lysine 12). This raises the possibility that the decrease in ubiquitination with this mutant was due, at least in part, to a loss of a ubiquitination site. Nevertheless, deletion of the adjacent downstream sequence also appears to have a negative influence on overall ubiquitination, suggesting a possible regulatory role of this sequence or a possible role in facilitating specific lysine recognition.
Regulatory role of carboxyl-terminal domains of Zta in facilitating ubiquitination.
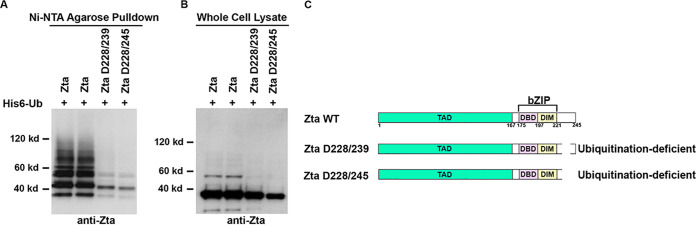
Despite the strong negative influence of deleting amino acids 1 to 78 on Zta ubiquitination, the residual ubiquitination observed with Zta D1/78 and Zta D1/103, and the more significant level of ubiquitination of the Zta D1/128 mutant, indicates the presence of additional downstream sequences that may help facilitate binding of a ubiquitin ligase(s). To address this issue, mutants within the basic DNA binding domain and sequences between the coiled-coil interface and the carboxyl terminus were assessed for their influence on Zta ubiquitination. As shown in Fig. 4, deletion of amino acids 228 to 239 or 228 to 245 has a negative impact on Zta ubiquitination. These sequences therefore appear to contribute to full Zta ubiquitination. Since this region may play a role in stabilizing the coiled-coil dimerization interface (83–85), we cannot discriminate whether the observed diminution of Zta ubiquitination is due to a regulatory role of this region in mediating ubiquitination or whether it is due to less stable dimerization which secondarily influences the binding of an E3 ligase.
FIG 4.
Deletion of the carboxyl-terminal tail of Zta inhibits Zta ubiquitination. EBV-negative AdAH cells were transfected with 5 μg of a ubiquitin (His6-Ub) expression vector and 5 μg of the indicated Zta or Zta mutant expression plasmids. Cells were lysed under denaturing conditions and extracts were purified over nickel-NTA beads. The purified material was assayed by Western blot analysis employing an anti-Zta antibody (A). Expression levels of Zta and its mutants were determined by Western blot analysis of whole-cell lysates (B). Shown is a schematic illustration of functional domains in Zta (C). Deletion regions are marked for each of Zta mutants.
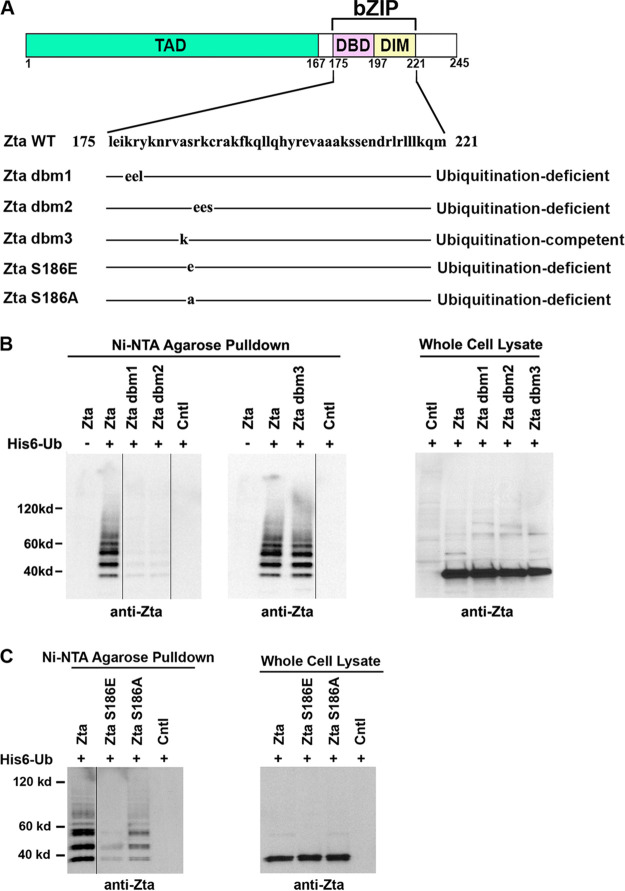
In addition to facilitating sequence-specific DNA binding, the basic region of Zta is a nuclear localization signal (86, 87). Degradation of nuclear survivin was shown previously to require a basic domain that facilitates subnuclear localization of survivin to the nucleolus, where it is ubiquitinated and degraded (88). A role for Zta’s basic region in supporting Zta ubiquitination was therefore investigated by analyzing a panel of site-directed mutants. Two 3-amino-acid mutants, Zta dbm1 and Zta dbm2, which localize to the nucleus but are defective for DNA binding (89), showed substantially lower levels of ubiquitination (Fig. 5B). The inability of these mutants to be ubiquitinated is not likely due specifically to an inability to binding DNA since a third mutant, Zta dbm3, which was shown previously to be defective for DNA binding (87), was fully ubiquitinated (Fig. 5B).
FIG 5.
Genetic analysis of the Zta basic/DNA binding domain in mediating Zta ubiquitination. (A) Schematic diagram of Zta basic leucine zipper-like (bZIP) domain mutants. (B and C) EBV-negative AdAH cells were transfected with or without 5 μg of a ubiquitin (His6-Ub) expression vector and 5 μg of the indicated Zta, Zta mutant, or control expression plasmids. Cells were lysed under denaturing conditions and extracts were purified over nickel-NTA beads. The purified material was assayed by Western blot analysis employing an anti-Zta antibody. Expression levels of Zta and its mutants were determined by Western blot analysis of whole-cell lysates. The images were spliced to unify the Western blot format.
It is also notable that both the Zta dbm1 and Zta dbm2 mutations alter lysine residues, which are potential targets of ubiquitination, and mutation of these residues may result in a loss of the target sites. However, as shown below, mutation of lysine 178 (mutated in Zta dbm1) to an arginine did not alter the ability of Zta to become ubiquitinated and mutation of lysine 188 (mutated in Zta dbm2) had a relatively minor impact on Zta ubiquitination. This suggests that the basic region of Zta plays a role in regulating overall ubiquitination outside of simply providing potential ubiquitin target residues. This conclusion is further supported by the finding that mutation of a nonlysine residue, serine 186, to glutamic acid substantially inhibited overall Zta ubiquitination (Fig. 5C). A more moderate mutation of serine 186 to alanine showed an intermediate effect on Zta ubiquitination. Together, our data support the idea that the basic domain of Zta plays a key positive regulatory role in Zta ubiquitination.
Role of ubiquitination in Zta degradation.
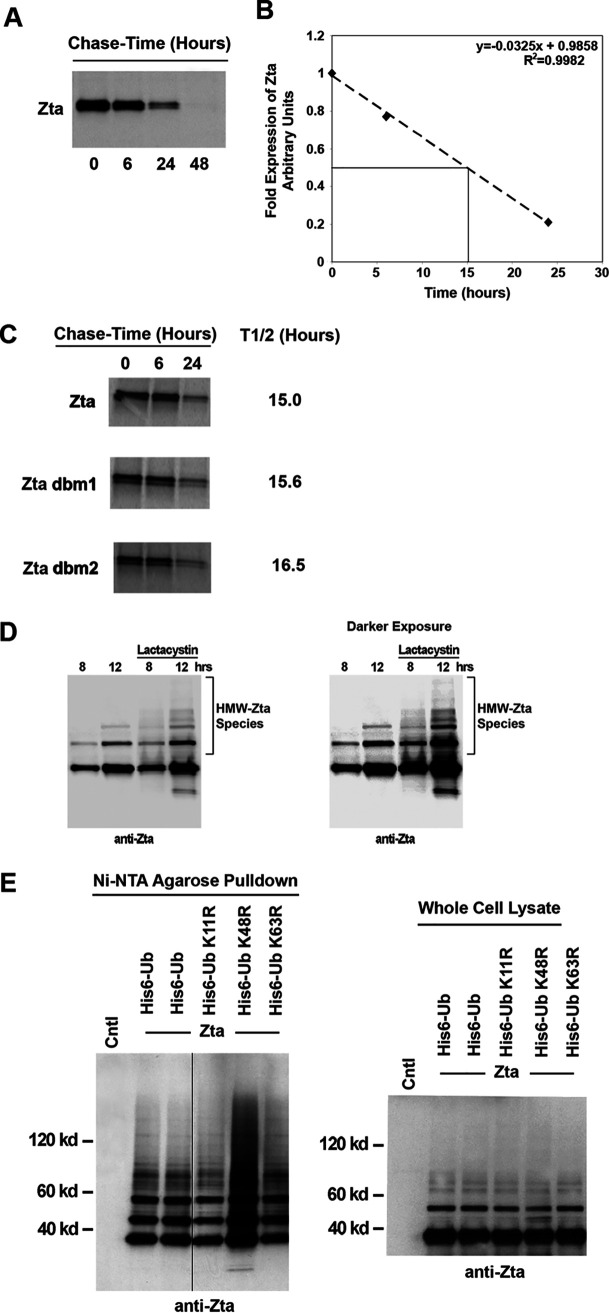
Polyubiquitination of proteins can serve as a signal that targets proteins for degradation by the proteasome system. Nevertheless, ubiquitination also plays a number of roles in modulating protein function independently of degradation, including facilitating subcellular localization. To initially test the role of Zta ubiquitination in facilitating Zta degradation, [35S]methionine pulse-chase experiments were carried out to determine the half-life of wild-type Zta compared to those of Zta mutants that display lower levels of ubiquitination. An analysis of wild-type Zta showed that it is a fairly stable protein, with a half-life of approximately 15 h (Fig. 6A and B). Analysis of Zta dbm1 and Zta dbm2, which are poorly ubiquitinated, showed slightly longer half-lives, 15.6 h and 16.5 h, respectively (Fig. 6C).
FIG 6.
Analysis of ubiquitination-dependent degradation of Zta. (A) EBV-negative U2OS osteosarcoma cells were transfected with a Zta expression plasmid and split into four different plates the following day. Two days after transfection, cells were pulsed with [35S]methionine and chased for the indicated times. Extracts were prepared under denaturing conditions and immunoprecipitated with an anti-Zta MAb. Samples were analyzed by SDS-PAGE and autoradiography. (B) The experiment shown in panel A was analyzed using a PhosphorImager to quantitate the levels of Zta immunoprecipitated at the different chase time points. The abundance at each time point was calculated relative to the abundance at 0 h. The line was generated by linear regression of the data in panel A. (C) U2OS cells were transiently transfected with either wild-type Zta or mutant expression plasmids and pulse-chase analysis was performed. (D) NPC-KT cells were transfected with Zta expression vector and treated for the indicated periods of time with 10 μM lactacystin or a vehicle. Aliquots from cell extracts were immunoblotted with the anti-Zta monoclonal antibody. A species of high-molecular-weight Zta (HMW-Zta) were readily detected at 12 h posttreatment with lactacystin but not with the vehicle. (E) NPC-KT cells were transfected with 5 μg of either wild-type ubiquitin (His6-Myc-Ub) or its mutant expression vectors plus 5 μg of the Zta expression plasmid or its control vector. Cells were lysed under denaturing conditions and extracts were purified over nickel-NTA beads. The purified material was assayed by Western blot analysis employing an anti-Zta antibody. Expression levels of Zta and its mutants were determined by Western blot analysis of whole-cell lysates. The image was spliced to unify the Western blot format.
Since pulse-chase experiments showed that the bulk of Zta has a fairly long half-life and since the half-lives of Zta dbm1 and Zta dbm2 were only marginally longer, this indicates that at a minimum, the bulk of the Zta molecules in a cell are not subjected to ubiquitin-dependent degradation. Nevertheless, the slightly longer half-life observed for Zta dbm1 and Zta dbm2 raised the possibility that a subset of Zta molecules in the cell are subjected to ubiquitin-dependent degradation (perhaps following certain posttranslational modification events that signal degradation-type-ubiquitination). This issue was therefore investigated further by assessing Zta ubiquitination following treatment of cells with the proteasome inhibitor lactacystin. As shown in Fig. 6D, an increase in higher-molecular-weight species is observed in cells treated with lactacystin, suggesting that at least some of the ubiquitinated Zta protein is subjected to proteasome-dependent degradation.
Polyubiquitination can occur through conjugation to different lysine residues on ubiquitin peptides. The most common polyubiquitination targets on ubiquitin are lysine residues 11, 48, and 63 (90, 91). Importantly, polyubiquitin chains conjugated at lysine 48 typically mediate proteasomal degradation, while conjugation at lysine residue 63 or 11 signals other types of regulation of the recipient protein. To further investigate whether a subset of ubiquitinated Zta molecules are subjected to degradation, mutant His6-Ub expression vectors containing lysine-to-arginine mutations at each of these lysine residues were cotransfected with a Zta expression vector and Zta polyubiquitination was assessed. Notably, mutation of ubiquitin lysine 48 resulted in a substantial increase in polyubiquitinated Zta species (Fig. 6E). This suggests that Zta can be polyubiquitinated through other residues besides lysine 48. The increase in polyubiquitinated species observed with the lysine 48 mutant also suggests that Zta is normally ubiquitinated through lysine 48 polyubiquitin chains and that this leads to degradation of Zta. It is also notable that mutation of lysine 11 results in an alteration of the polyubiquitinated Zta pattern, suggesting that at least a portion of Zta is polyubiquitinated through a lysine 11 mechanism. Together, these results indicate that a portion of Zta is targeted for proteasomal degradation and that a portion of ubiquitinated Zta is regulated in a nonproteasomally related pathway.
Zta is ubiquitinated at multiple lysine residues.
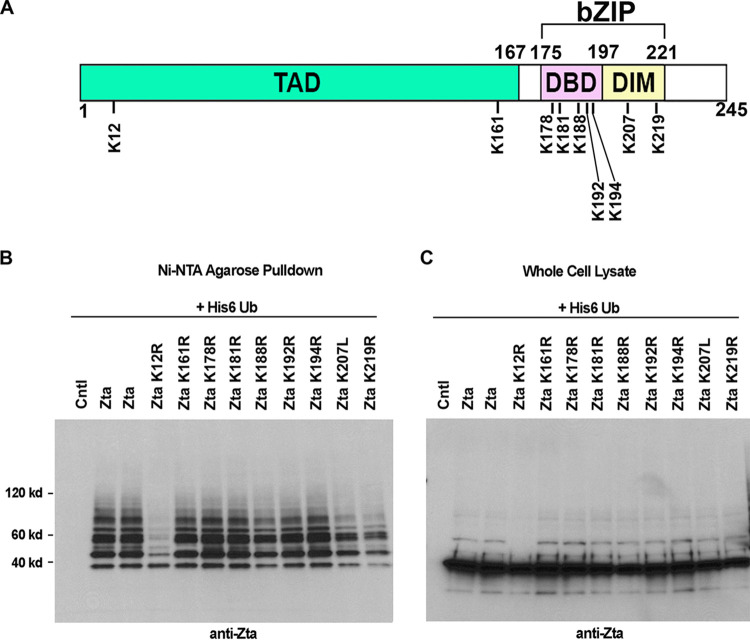
Zta contains 9 lysine residues which are candidates for ubiquitin conjugation (Fig. 7A). To identify residues that are targeted for ubiquitination, lysine-to-arginine (in most cases) or lysine-to-leucine mutants were generated for each lysine residue of Zta. Wild-type or lysine mutant Zta expression vectors were then analyzed for ubiquitination in AdAH cells (Fig. 7B). The most pronounced impact on ubiquitination was observed with the lysine 12 mutant, for which there was a substantial loss of all but the fastest migrating ubiquitinated species (Fig. 7B). This suggests that lysine 12 is critical for high-level ubiquitination and is a likely target site for polyubiquitination.
FIG 7.
Analysis of single-lysine-residue Zta mutants. (A) Schematic diagram of Zta lysine residues. (B) EBV-negative AdAH cells were transfected with 5 μg of a ubiquitin (His6-Ub) expression vector and 5 μg of the indicated Zta, Zta mutant, or control expression plasmids. Cells were lysed under denaturing conditions and extracts were purified over nickel-NTA beads. The purified material was assayed by Western blot analysis employing anti-Zta antibodies. (C) Comparable expression levels of Zta and its mutants were confirmed by Western blot analysis of whole-cell lysates.
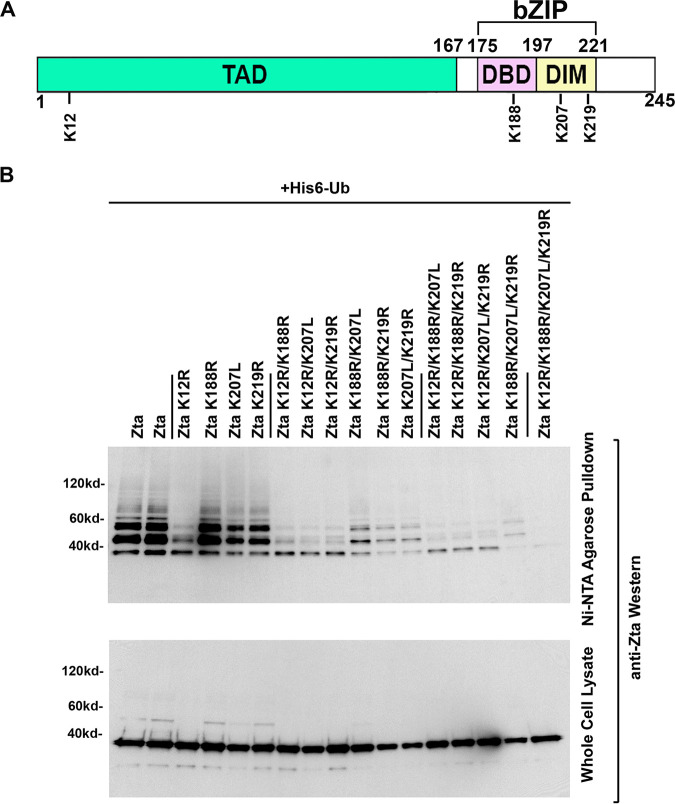
Despite the decrease in ubiquitination observed with the lysine 12 mutant, the polyubiquitinated species that remained and the lack of apparent change in the intensity of the fastest-migrating ubiquitinated species suggest that Zta can also be ubiquitin conjugated at other lysine residues. Careful inspection of the data in Fig. 7B revealed subtle decreases in Zta ubiquitination with the lysine 188, 207, and 219 mutants. To further address the possible contribution of these residues to the overall ubiquitination of Zta, double, triple, and quadruple mutants were generated at lysine residues 12, 188, 207, and 219. This analysis revealed progressive decreases in Zta ubiquitination with the double, triple, and quadruple mutants and little ubiquitination observed with the quadruple mutant (Fig. 8). This result supports the conclusion that each of these lysine residues contributed to the total observed Zta ubiquitination.
FIG 8.
Analysis of multiple-lysine-residue Zta mutants. (A) Schematic diagram of selected Zta lysine residues. (B) EBV-negative AdAH cells were transfected with 5 μg of a ubiquitin (His6-Ub) expression vector and 5 μg of the indicated Zta or Zta mutant expression plasmids. Cells were lysed under denaturing conditions and extracts were purified over nickel-NTA beads. The purified material was assayed by Western blot analysis employing anti-Zta antibodies. Expression levels of Zta and its mutants were determined by Western blot analysis of whole-cell lysates.
Mutation of ubiquitin target sites on Zta results in a loss of progression through the lytic cycle.
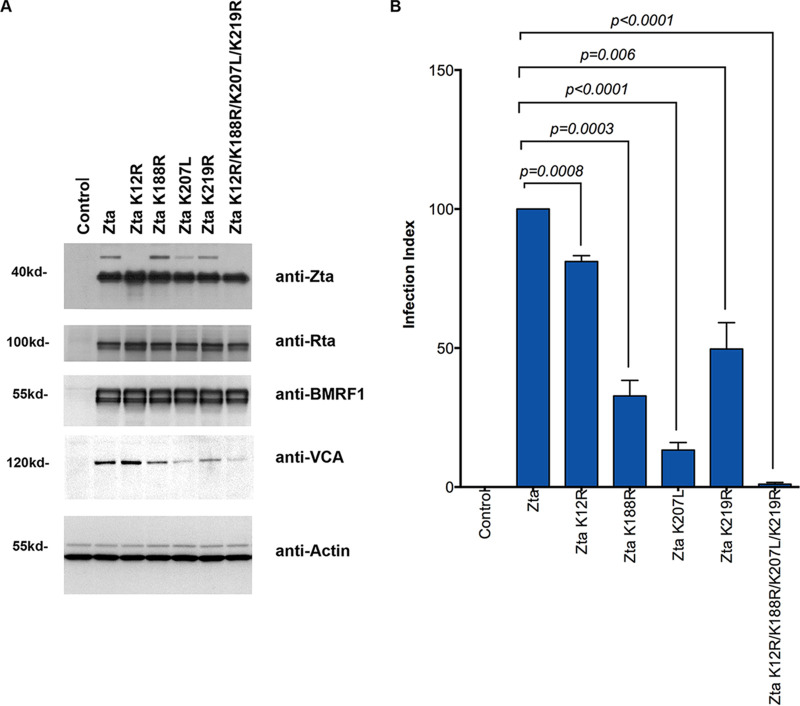
The experiments outlined above indicate that a subpopulation of ubiquitinated Zta is targeted for degradation. To evaluate whether ubiquitination also affects Zta function in a nonproteolytic manner, Zta lysine mutants showing decreased ubiquitin modification were analyzed for the ability to induce lytic reactivation in EBV (Zta knockout)-infected 293 cells [EBV(Z K/O)-293 cells]. None of the mutants showed any detectable influence on induction of the immediate early/early markers Rta and BMRF1 (Fig. 9A). Nevertheless, the single lysine mutants K188R, K207L, and K219R as well as the quadruple mutant showed defects in the ability to induce the late viral capsid antigen (VCA) gene (Fig. 9A).
FIG 9.
Loss of ubiquitination leads to deficit progression of Zta-mediated lytic replication. (A) EBV (Zta knockout)-infected 293 cells [EBV(Z K/O)-293] were transfected with wild-type or mutant Zta expression vectors or a control vector. Forty-eight hours posttransfection, cells were harvested and analyzed with each antibody shown. (B) EBV(Z K/O)-293 cells were transfected with wild-type or mutant Zta expression vectors or a control vector. Viral supernatants were harvested 5 days posttransfection and used to infect Raji cells. Since the virus from EBV(Z K/O)-293 cells contains a GFP expression cassette, the number of infected Raji cells was counted by FACS analysis. The infection index represents the percentage of GFP-positive cells (infected cells) relative to the number of GFP-positive cells observed following infection with supernatants from wild-type-Zta-transfected EBV(Z K/O)-293 cells. The averages and standard errors from the results of 3 independent infection assays are shown. Significance for differences between experimental groups were determined using Student’s t test using Prism 6 software, and P values of <0.05 were considered significant.
To further evaluate progression through the lytic cycle, the ubiquitin target site mutants were assessed for the ability to mediate production of infectious virus. EBV(Z K/O)-293 cells were transfected with wild-type or mutant Zta expression vectors. Viral supernatants were harvested 5 days later and used to infect Raji cells. Since the virus from EBV(Z K/O)-293 cells contains a green fluorescent protein (GFP) expression cassette, the number of infected Raji cells was counted by fluorescence-activated cell sorting (FACS) analysis. As shown in Fig. 9B, each of the single-ubiquitin-target-site mutants produced significantly fewer infected cells than wild-type Zta. Strikingly, although the quadruple mutant appears to be fully capable of inducing Rta and BMRF1, this mutant produced no detectable infectious virus (P < 0.05 compared to each of the wild-type and single-mutant Zta). These results indicate that ubiquitination of Zta is not required for the early stages of reactivation but ubiquitination of Zta is required for the late stages of viral production.
DISCUSSION
Multiple lines of evidence have indicated that ubiquitination signaling may play important roles in the regulation of EBV lytic replication. For instance, the SUMO-targeted ubiquitin ligase RNF4 targets EBV immediate early gene Rta for ubiquitination and thus influences the lytic progression of EBV (92). Recently, Tikhmyanova and colleagues showed that treatment with the proteasome inhibitor MG132 leads to the accumulation of Zta in EBV-positive lymphoma cells, indicating that ubiquitin-mediated proteolysis may regulate Zta protein stability and EBV lytic reactivation (93). Here, we provide new evidence that modification of Zta by ubiquitin is likely a complex process and that it regulates Zta’s function in both a nonproteolytic and proteolytic manner during the lytic cycle.
As shown in Fig. 6, we found that Zta is a relatively stable protein and the bulk of Zta molecules in the cell are probably not subjected to ubiquitin-dependent degradation. This indicates that ubiquitination of Zta probably does not function to facilitate transient expression of the overall population of Zta molecules, as is the case for many cellular early response genes like c-Fos or c-Myc. This is in line with the known requirement of Zta at multiple stages throughout the lytic cascade, a property that sets it apart from the globally transient requirements of cellular immediate early genes like c-Fos. Nevertheless, our studies indicate that a subpopulation of Zta molecules are likely modified in a lysine 48 polyubiquitin manner and are in fact degraded. It could be argued that this level of degradation is an inherent and perhaps inconsequential by-product of inefficiently regulated Zta ubiquitination whereby a low level of K48 modification and Zta degradation can be tolerated. On the other hand, there is accumulating evidence that certain modifications of Zta induce a transition in the function of targeted Zta molecules from early promoter activation to late lytic functions (50). Zta molecules that are modified in a way that facilitates functions that are no longer needed and/or desirable could be subjected to lysine 48 polyubiquitination/degradation as part of a mechanism to transition the functions of Zta throughout the lytic cascade.
In addition to being subjected to polyubiquitination, Zta is monoubiquitinated at single and possibly multiple lysine residues. Based on a wide array of studies in other systems, monoubiquitination typically plays a role in altering protein function. Further, in addition to the lysine 48 polyubiquitination, Zta is likely polyubiquitinated by nondegradative ubiquitin chain conjugation such as through lysine 11 (or lysine 63) of ubiquitin. These forms of Zta modification are also likely to play nondegradative roles in governing Zta function, such as regulating subnuclear localization or protein-protein interactions.
Except for lysine 12, all of the lysine residues in Zta are located in or near the DNA binding and dimerization domains. In addition to possible roles in mediating subnuclear localization and changes in association with binding partners, it is likely that ubiquitination of one or more of these residues has substantial influences on DNA binding and/or dimerization. Ubiquitination of residues such as lysine 188 is likely to be highly disruptive for DNA binding, and ubiquitination of lysine residue 207 or 219 may have an impact on dimerization/DNA binding. The DNA binding-dependent functions of Zta, then, would require that at least lysine 188 not be ubiquitinated. Consistent with this idea, our results indicate that ubiquitination is not required for some of Zta’s early transactivation functions that are mediated by DNA binding.
EBV has previously been shown to encode at least three deubiquitinases (DUBs), two of which (BSLF1 and BXLF1) are early proteins (94); one of these (BSLF1) is required for viral DNA replication (16). A compelling speculation is that viral (or cellular) encoded DUBs downmodulate ubiquitination of Zta during this phase of the lytic cycle or that downmodulation occurs in subcellular compartments where Zta is engaged in DNA binding functions (such as transcriptional activation or DNA replication).
Our genetic studies identified residues within the DNA binding domain that are critical for facilitating Zta ubiquitination. Therefore, an additional component of the model discussed above is the possible exclusion of certain ubiquitin ligases from interacting with Zta that is bound to DNA. This would be predicted to help sustain Zta DNA binding functions in these settings that would otherwise be inhibited by ubiquitination of basic and/or downstream lysine target residues.
We have identified multiple regions of Zta that control the level and type of Zta ubiquitination. Based on studies from other systems, it is clear that ubiquitination of proteins is typically a highly regulated process controlled by an array of regulatory factors and mechanisms. Phosphorylation is a critical regulator of ubiquitin ligase recruitment (76, 77, 95–100), and it will be interesting to carry out a more detailed analysis of putative phosphor-acceptor sites on Zta to identify possible phosphorylation events that regulate Zta ubiquitination. Our initial results show that mutation of serine 186, which is a protein kinase C (PKC) phosphorylation site, to glutamic acid has a substantial negative influence on Zta ubiquitination. Phosphorylation of this serine residue may therefore have an impact on Zta ubiquitination by altering the binding of a ubiquitin ligase. Phosphorylation of serine 186 was shown previously to enhance the ability of Zta to bind DNA (51). This phosphorylation event therefore appears to favor Zta’s functions that are associated with DNA binding. If phosphorylation of serine 186 were to impede the binding of a ubiquitin ligase, this too, would inhibit ubiquitin conjugation to residues such as lysine 188, thereby favoring DNA binding.
Although disruption of DNA binding/dimerization by ubiquitination of lysines 188, 207, and 219 is a reasonable hypothesis, it is also possible that modification at these sites (particularly residues 207 and 219) could have other consequences on Zta function, such as possibly stabilizing the dimerization structure. Alternatively, since the Zta dimerization domain is involved in interactions with a number of cellular proteins, ubiquitin moieties at position 207 or 219 could enhance or destabilize protein-protein interactions with cellular factors. Such interactions could play a role in facilitating late Zta functions and/or late-stage subcellular localization. This may explain the late-stage defect observed with the Zta ubiquitin target site mutants. It is worth noting that although we predict that ubiquitination of lysine 188 disrupts the DNA binding activity of Zta, there may be unknown late-stage functions of Zta that do not require DNA binding. These activities may be facilitated in part by ubiquitin targeting of lysine 188 (as well as lysine 207 or 219).
Previous studies have shown that Zta lysine 12 is also modified by a related peptide, SUMO-1 (47, 57), which is known to control protein localization and turnover. SUMO-1 modification of Zta was shown to decrease its ability to activate certain lytic cycle promoters but enhance expression of the lytic gene BMRF1 (57). Since SUMO-1 and ubiquitin modification of lysine 12 is likely competitive, modification of Zta molecules by SUMO-1 and ubiquitin conjugation probably have distinct influences on Zta function. This may be a means of tagging different populations of Zta for different functions or outcomes during progression through the lytic cycle in a mutually exclusive manner. Notably, mutation of lysine 12 to arginine has a more pronounced effect on polyubiquitination than on monoubiquitination (for example, see Fig. 7), raising the possibility that modification at this residue may be primarily degradative. SUMO-1 modification of this lysine residue would therefore be expected to be protective against Zta degradation.
Consistent with the hypothesis that ubiquitination of lysine 12 primarily serves a degradative function as opposed to a positive role in late-stage replication, mutation of lysine 12 showed a milder effect on virus production than mutation of lysine 188, 207, or 219 (Fig. 9B). Further, in contrast to the lysine 188, 207, and 219 mutations, the K12R mutation did not diminish induction of VCA expression (Fig. 9A). It should be pointed out, however, that interpretation of the functional consequences of a lysine 12 mutant must be done with caution since this mutant is defective for both SUMO-1 and ubiquitin modification.
Our results show that maximal ubiquitination of Zta requires sequences between residues 1 and 79, a region actively involved in recruiting a number of Zta-interacting factors, including the cellular factor p300. Interestingly, p300 has been reported to have intrinsic ubiquitin ligase activity which may serve as an E3 ligase in catalyzing Zta ubiquitination (101). Besides p300, our genetic studies aimed at identifying regions of Zta that facilitate regulation of Zta ubiquitination also support the possibility that there are more than one ubiquitin ligase that control ubiquitination of Zta. There is ample precedent for ubiquitin targeting of individual proteins being mediated by multiple distinct ubiquitin ligases (66, 76–78, 101–107). For example, at least 5 different ubiquitin ligases have been shown to be involved in ubiquitinating p53 under different circumstances (101–107). Further studies will be required to identify the full repertoire of ubiquitin ligases that bind to Zta, to determine their respective roles in mediating modification of specific residues on Zta, and to determine their roles in regulating the function of Zta (or associated proteins).
An unanticipated result from our genetic studies is the reconstitution of Zta ubiquitination when sequences between amino acids 103 and 129 were deleted (Fig. 3). This suggests that this region plays a role in recruiting a negative regulator of Zta ubiquitination. The binding of enzymes with deubiquitination activity is a common feature that is apparently involved in further refining the regulation of proteins targeted for control by ubiquitin modification (108–113). For example, the deubiquitinating enzyme HAUSP interacts with and stabilizes p53 through a deubiquitination mechanism (108, 112, 113). c-Myc is deubiquitinated by the ubiquitin-specific protease USP28, and intriguingly, USP28 is actually recruited to c-Myc by the ubiquitin ligase FBW7 (110). Although there are other possible explanations for the negative influence of amino acids 104 to 128 on Zta ubiquitination, there is a reasonable possibility that this domain contributes to the interaction with a deubiquitinating enzyme, as is observed with other transcription factors. The binding of a deubiquitinase to Zta would help maintain the level of Zta polyubiquitination in certain populations of Zta below the threshold of 4-unit polyubiquitin chains required for proteasome targeting. As mentioned above, it could also confer temporally and/or spatially dependent deubiquitination of Zta to facilitate particular Zta functions during progression of the lytic cascade.
In addition to playing a role in modifying the function of Zta, the recruitment of ubiquitin ligases and the possible recruitment of deubiquitinating enzymes (through amino acids 104 to 128, for example) may also play a role in altering the function of viral and cellular factors that associate with Zta, such as components of the lytic replication complex or the cellular factors NF-κB and p53 (23, 45, 46, 114, 115). Sato et al. (116) demonstrated that Zta can facilitate proteasomally dependent degradation of p53 during lytic replication. It is likely, then, that the complex nature of Zta’s interactions with ubiquitin ligases and possibly DUBs and the possible modulation of ubiquitin targeting by phosphorylation of serine 186 may play a role in regulating not only Zta functions but also the functions of Zta binding partners during progression of the lytic cycle. Our study provides a basis to explore the effects of these interactions on the functions of Zta as well as their influence on other cellular and viral binding partners.
Our data indicate that disruption of Zta ubiquitination may halt EBV lytic progression and lead to an abortive replication, which raises the possibility of managing EBV infection and its associated diseases by pharmaceutically targeting the ubiquitination system. Indeed, some ubiquitination-modulating drugs have shown potential to treat certain types of cancers, including lymphomas (117, 118). It will be worth exploring if any such drugs can target Zta ubiquitination and disrupt the EBV life cycle and thus ultimately eliminate EBV-associated cancers.
MATERIALS AND METHODS
Cell culture.
The EBV-positive Akata cell line (type I latency) was established from an EBV-positive Burkitt’s lymphoma from a Japanese patient and expresses surface IgG and has a t(8:14) chromosome translocation (119). The EBV-positive Mutu (Mutu I) cell line (type I latency) was derived from an EBV-positive Burkitt’s lymphoma biopsy specimen from a Kenyan patient and exhibits surface IgM (μK+) expression and a typical t(8:14) chromosome translocation (119). JY is an EBV-positive lymphoblastoid cell line (type III latency). DG75 is an EBV-negative Burkitt’s lymphoma cell line. Raji is an EBV-positive Burkitt’s lymphoma cell line. NPC-KT is an EBV-positive hybrid cell line which was generated by the fusion of EBV genome-positive NPC epithelial explant culture cells with AdAH cells (120). AdAH is an EBV-negative human nasopharyngeal adenoid epithelial cell line (121). U2OS is an EBV-negative osteosarcoma cell line. EBV-293 is an EBV-positive human embryonic kidney 293 cell line (122). EBV(Z K/O)-293 was established by infecting human embryonic kidney 293 cells with an EBV genome with a Zta deletion (123). All cell lines were maintained in either Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific; catalog no. SH30243.02) for adherent cells or RPMI 1640 (Thermo Fisher Scientific; catalog no.SH30027.02) for suspension cells, supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific; catalog no. 10437-028), penicillin, streptomycin, and glutamine. Cells were grown at 37°C in a humidified 5% CO2-containing atmosphere. Hygromycin (100 μg/ml; Thermo Fisher Scientific; catalog no. 10687010) was added to 293 stable cell cultures.
Transfection.
Transient-transfection experiments were performed by using either a modified version of the calcium phosphate precipitation procedure for adherent cells (38) or the Amaxa electroporation method for suspension cells (124). For the calcium phosphate precipitation procedure, 1 × 106 cells were plated onto 100-mm-diameter tissue culture dishes. The following day, the medium was replaced with 8 ml of fresh supplemented DMEM; 4 h later, 0.5 ml of 1× HEPES-buffered saline (0.5% HEPES, 0.8% NaCl, 0.1% dextrose, 0.01% anhydrous Na2HPO4, 0.37% KCl [pH 7.10]) was mixed with a total of 30 μg of plasmid DNA (effector plasmids were added in the amounts indicated in the figure legends, and carrier DNA [pUHD10] was added to make a total of 30 μg). Thirty microliters of 2.5 M CaCl2 was added, and samples were mixed immediately. Precipitates were allowed to form at room temperature for 20 min before being added dropwise to cells. Cells were incubated at 37°C with 5% CO2 for 16 h before the medium was replaced with 10 ml of fresh supplemented DMEM. For the Amaxa electroporation method, cells were placed in antibiotic-free RPMI 1640 2 days before electroporation. For each transfection, 2 × 106 cells in 100 μl of Nucleofector solution R (Lonza; catalog no. VCA-1001) were electroporated with a total of 5 μg of plasmid DNA (containing both effector and carrier DNA [pUHD10]) and transferred to a six-well plate with 1.5 ml of medium per well. After 24 h, 1.5 ml of fresh RPMI 1640 was added to each well. Cells were incubated for another 24 h before harvesting.
In vivo ubiquitination assay.
Cells transfected with His6-Ub or the control vector were washed twice with 1× phosphate-buffered saline (PBS) and lysed in 1 ml of buffer A (6 M guanidinium hydrochloride, 0.1 M Na2HPO4/NaH2PO4 [pH 8.0], 10 mM imidazole) per 100-mm tissue culture dish at 4°C. The lysate was subsequently sonicated for 30 s to reduce viscosity and then mixed on a rotator with 50 μl (settled volume) of nickel-NTA-agarose beads (Qiagen; catalog no.1018244) for 3 h at 4°C. The beads were washed three times with 1 ml of buffer A, twice with 1 ml of buffer A diluted in 25 mM Tris-HCl (pH 6.8)/20 mM imidazole (1:4), and twice with 1 ml of 25 mM Tris-HCl (pH 6.8)/20 mM imidazole. All the above-named buffers were supplemented with protease inhibitors (protease inhibitor cocktail; Sigma; catalog no. P8849). Purified proteins were eluted by boiling the beads in 50 μl of 2× Laemmli sample buffer (Sigma; catalog no. S3401) supplemented with 200 mM imidazole at 95°C for 15 min and analyzed by Western blot analysis.
Lytic cycle induction.
EBV-positive Mutu cells (119) and EBV-positive Akata cells (119) were grown to near saturation, and the day before induction, a 50% volume of fresh RPMI 1640 supplemented with 10% FBS and penicillin/streptomycin was added. The following day, cells were spun down and resuspended in an equal volume of fresh-warmed RPMI 1640 supplemented with 10% FBS and penicillin/streptomycin, containing either 10 μg/ml of anti-human IgM (Sigma; catalog no. I0759) for Mutu cells or 10 μg/ml of anti-human IgG (Sigma; catalog no. I5260) for Akata cells. Cells were harvested at 0 and 48 h posttreatment and subjected to the immunoprecipitation analysis. To induce the lytic cycle in EBV-293 cells, cells were transfected with 5 μg of Rta expression plasmid or its control vector. Forty-eight hours posttransfection, cells were harvested for the immunoprecipitation analysis or in vivo ubiquitination assay.
Immunoprecipitation.
Cells were suspended in 500 μl of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 0.1% SDS) supplemented with protease inhibitors and incubated for 1 h at 4°C. Lysates were then centrifuged at 13,000 rpm for 30 min at 4°C, and the supernatants were transferred to new tubes. Cell lysates were precleared with 20 μl of protein G-agarose beads (Pierce; catalog no. 20421) for 1 h. At the same time, 10 μl of the anti-Zta antibody (Santa Cruz; catalog no. sc-53904) or a control antibody (Millipore; catalog no. 12-371) was incubated with 20 μl of protein G-agarose beads in 500 μl of RIPA buffer for 1 h at 4°C and then washed three times with RIPA buffer. The lysates were then centrifuged briefly, and the supernatants were transferred to the antibody-agarose complexes and incubated overnight on a rotator at 4°C. The next day, the beads were washed three times with RIPA buffer with 15 min of incubation (at room temperature) for each wash. Twenty microliters of 2× Laemmli sample buffer was then added to the agarose bead complexes, and samples were boiled for 15 min at 95°C before analysis by Western blotting.
Pulse-chase analysis.
Human osteosarcoma U2OS cells were transiently transfected with the desired plasmids and evenly split into different plates the following day. Twenty-four hours after transfection, cells were methionine starved for 30 min and then pulsed for 60 min with [35S]methionine (PerkinElmer; catalog no. NEG772). Cells were then washed twice with 1× PBS and chased for the desired periods of time with fresh supplemented DMEM. Cells were washed twice with 1× PBS, trypsinized, and washed twice again with 1× PBS. Next, cells were lysed in 0.5 ml of 25 mM Tris-HCl (pH 6.8)/1.5% SDS per 100-mm tissue culture dish or T25 tissue culture flask and boiled at 95°C for 15 min. Samples were then diluted in 8 volumes of EBC/BSA buffer (50 mM Tris-HCl, 180 mM NaCl, 0.5% Nonidet P-40, 0.5% bovine serum albumin [BSA]) supplemented with protease inhibitors (protease inhibitor cocktail; Sigma; catalog no. P8340). Extracts were precleared for 30 min at 4°C with protein G-agarose beads (Roche) and then incubated with anti-Zta monoclonal antibody (MAb) (Argene; catalog no. 11-007) overnight at 4°C. Forty microliters of a 50% (wt/vol) slurry of protein G-agarose beads was then added, and the mixture was rocked for 60 min at 4°C and washed four times in EBC/BSA buffer, plus once in 50 mM Tris-HCl (pH 6.8). Immunobeads were boiled in 2× Laemmli sample buffer (Sigma; catalog no. S3401) at 95°C for 10 min, and the samples were then run on an SDS-PAGE gel. Image acquisition and quantitation were carried out using a Typhoon Trio+ instrument (Amersham Bioscience).
Western blot analysis.
After a single 1× PBS wash, a fraction of harvested cells was separated for Western blot analysis. Cells were immediately suspended in 100 μl of 1× Laemmli sample buffer and boiled for 15 min at 95°C to shear the genomic DNA. Protein concentrations of the whole-cell extracts were measured with the Bio-Rad protein assay kit according to the manufacturer’s instructions. Equal amounts of protein were subjected to SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Whatman). The blots were blocked for 60 min in Tris-buffered saline containing 5% nonfat powdered milk and 1% FBS and then incubated with the primary antibody (in blocking buffer) overnight at 4°C. The blots were washed three times with 1× TBST (140 mM NaCl, 3 mM KCl, 25 mM Tris-HCl [pH 7.4], 0.1% Tween 20) (each wash was carried out for approximately 15 min). To detect the chemiluminescence signals, the blots were then incubated with horseradish peroxidase-conjugated secondary antibody (Bio-Rad) in blocking buffer for 1 h at room temperature. Blots were washed as described above and analyzed with an enhanced chemiluminescence detection system (Perkin-Elmer) according to the manufacturer’s recommendations, and filters were exposed to either Fuji Super RX films (Fujifilm) or Kodak image station 4000 mm (Kodak, NY). To detect the infrared fluorescence signal, an Odyssey infrared imaging system (Li-Cor Biosciences) was used with a secondary antibody (IRDye 680RD goat anti-rabbit antibody; Li-Cor; catalog no. 925-68071). The following primary antibodies were used for Western blot analysis: anti-Zta MAb (Argene; catalog no. 11-007), anti-Zta MAb (Santa Cruz; catalog no. sc-53904), anti-HA.11 MAb (Covance; catalog no. MMS-101R), anti-ubiquitin polyclonal antibody (PAb) (Covance; catalog no. PRB-268C), anti-ubiquitin PAb (Santa Cruz; catalog no. sc-9133), anti-ubiquitin PAb (Cell Signaling Technology; catalog no. 3933S), anti-Rta MAb (Argene; catalog no. 11-008), anti-BMRF1 MAb (Capricon; catalog no. EBV-018-48180), anti-VCA MAb (Chemicon; catalog no. MAB8184), anti-actin PAb (Santa Cruz; catalog no. sc-1615), anti-α/β-tubulin PAb (Cell Signaling Technology; catalog no. 2148).
Infection assay.
EBV(Z K/O)-293 cells were transfected with 5 μg of plasmids encoding either wild-type or mutant Zta plus 25 μg of the carrier plasmid pUHD10 by the calcium phosphate method. Five days after transfection, the supernatants were collected and filtered (0.45-mm pore size). Three-milliliter aliquots of supernatant were mixed with 2 × 105 Raji cells. Seventy-two hours after infection, cells were harvested and analyzed for GFP fluorescence by FACS (BD).
Plasmid construction.
Zta or HA-Zta proteins were expressed from pBS(SVp/e) plasmids containing the simian virus 40 (SV40) early promoter and enhancer upstream from the genomic BZLF1 gene or an HA tag-fused genomic BZLF1 gene fragment. The Zta deletion mutants HA-Zta 79–245, HA-Zta 104–245, HA-Zta 129–245, HA-Zta D27/53, and HA-Zta D52/78 were made by first introducing restriction sites (with a Bio-Rad Muta-Gene kit) at the indicated loci and closing down the plasmids to delete the respective coding sequences. The Zta deletion mutants Zta D9/13, Zta D14/18, Zta D19/23, Zta D24/28, Zta D228/239, and Zta D228/245 were generated from pBS(SVp/e)-Zta by site-directed mutagenesis with the Bio-Rad Muta-Gene kit. All of the Zta lysine mutants (mono-, double-, triple-, and quadruple-lysine mutants) were generated by replacing lysine codons with arginine codons (or a leucine codon, for K207) in the plasmid, pBS(SVp/e)-Zta, by successive PCR-based site-directed mutagenesis (QuikChange mutagenesis kit; Stratagene; catalog no. 200521) according to the manufacturer’s instructions. Zta dbm1, Zta dbm2, and Zta dbm3 expression vectors were described elsewhere (89). Zta S186E and Zta S186A were generated by site-directed mutagenesis using a Bio-Rad Muta-Gene kit. All mutants were screened and verified by sequencing. His6-ubiquitin plasmid and its control vector (HA-ubiquitin plasmid) are described elsewhere (125). His6-Myc-ubiquitin plasmid and its mutants (His6-Myc-Ub-K11R, -K63R, and -K48R) were generous gifts from Martin Eilers (IMT, Germany). CMV-BRLF1 expression plasmid and its control vector were generous gifts from Shannon Kenney (University of Wisconsin, Madison, WI).
ACKNOWLEDGMENTS
This work was supported by a National Institutes of Health COBRE grant (P20 GM121288), a U.S.-Japan Cooperative Medical Sciences Program Collaborative Award from CRDF Global, a Tulane School of Medicine faculty research pilot grant, and a Carol Lavin Bernick faculty grant to Z.L., as well as by the Japan Society for the Promotion of Science (grant number JP17K4567), the Takeda Science Foundation, and a Shiseido Female Researcher Science Grant to A.N.
We thank Martin Eilers (IMT, Germany), Shannon Kenney (University of Wisconsin, Madison, WI), and Henri-Jacques Delecluse (DKFZ, Germany) for kindly providing us with the reagents.
REFERENCES
- 1.Rickinson AB, Kieff E. 2007. Epstein-Barr virus, p 2655–2700. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Young LS, Yap LF, Murray PG. 2016. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer 16:789–802. doi: 10.1038/nrc.2016.92. [DOI] [PubMed] [Google Scholar]
- 3.de Sanjose S, Bosch R, Schouten T, Verkuijlen S, Nieters A, Foretova L, Maynadie M, Cocco PL, Staines A, Becker N, Brennan P, Benavente Y, Boffetta P, Meijer CJ, Middeldorp JM. 2007. Epstein-Barr virus infection and risk of lymphoma: immunoblot analysis of antibody responses against EBV-related proteins in a large series of lymphoma subjects and matched controls. Int J Cancer 121:1806–1812. doi: 10.1002/ijc.22857. [DOI] [PubMed] [Google Scholar]
- 4.Kheir F, Zhao M, Strong MJ, Yu Y, Nanbo A, Flemington EK, Morris GF, Reiss K, Li L, Lin Z. 2019. Detection of Epstein-Barr virus infection in non-small cell lung cancer. Cancers (Basel) 11:759. doi: 10.3390/cancers11060759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strong MJ, O’Grady T, Lin Z, Xu G, Baddoo M, Parsons C, Zhang K, Taylor CM, Flemington EK. 2013. Epstein-Barr virus and human herpesvirus 6 detection in a non-Hodgkin’s diffuse large B-cell lymphoma cohort by using RNA sequencing. J Virol 87:13059–13062. doi: 10.1128/JVI.02380-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strong MJ, Xu G, Coco J, Baribault C, Vinay DS, Lacey MR, Strong AL, Lehman TA, Seddon MB, Lin Z, Concha M, Baddoo M, Ferris M, Swan KF, Sullivan DE, Burow ME, Taylor CM, Flemington EK. 2013. Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy. PLoS Pathog 9:e1003341. doi: 10.1371/journal.ppat.1003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrell PJ, Rowe DT, Rooney CM, Kouzarides T. 1989. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J 8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieberman PM, Berk AJ. 1990. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J Virol 64:2560–2568. doi: 10.1128/JVI.64.6.2560-2568.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman PM, Berk AJ. 1991. The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev 5:2441–2454. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- 10.Lieberman PM, Berk AJ. 1994. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA–promoter DNA complex formation. Genes Dev 8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 11.Lieberman PM, Ozer J, Gursel DB. 1997. Requirement for transcription factor IIA (TFIIA)-TFIID recruitment by an activator depends on promoter structure and template competition. Mol Cell Biol 17:6624–6632. doi: 10.1128/mcb.17.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adamson AL, Kenney S. 1999. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein. J Virol 73:6551–6558. doi: 10.1128/JVI.73.8.6551-6558.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zerby D, Chen CJ, Poon E, Lee D, Shiekhattar R, Lieberman PM. 1999. The amino-terminal C/H1 domain of CREB binding protein mediates zta transcriptional activation of latent Epstein-Barr virus. Mol Cell Biol 19:1617–1626. doi: 10.1128/mcb.19.3.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CJ, Deng Z, Kim AY, Blobel GA, Lieberman PM. 2001. Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol Cell Biol 21:476–487. doi: 10.1128/MCB.21.2.476-487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng Z, Chen CJ, Chamberlin M, Lu F, Blobel GA, Speicher D, Cirillo LA, Zaret KS, Lieberman PM. 2003. The CBP bromodomain and nucleosome targeting are required for Zta-directed nucleosome acetylation and transcription activation. Mol Cell Biol 23:2633–2644. doi: 10.1128/mcb.23.8.2633-2644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fixman ED, Hayward GS, Hayward SD. 1992. trans-Acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol 66:5030–5039. doi: 10.1128/JVI.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fixman ED, Hayward GS, Hayward SD. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol 69:2998–3006. doi: 10.1128/JVI.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammerschmidt W, Sugden B. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 19.Sarisky RT, Gao Z, Lieberman PM, Fixman ED, Hayward GS, Hayward SD. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol 70:8340–8347. doi: 10.1128/JVI.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schepers A, Pich D, Hammerschmidt W. 1993. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein-Barr virus. EMBO J 12:3921–3929. doi: 10.1002/j.1460-2075.1993.tb06070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schepers A, Pich D, Mankertz J, Hammerschmidt W. 1993. cis-Acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J Virol 67:4237–4245. doi: 10.1128/JVI.67.7.4237-4245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schepers A, Pich D, Hammerschmidt W. 1996. Activation of oriLyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology 220:367–376. doi: 10.1006/viro.1996.0325. [DOI] [PubMed] [Google Scholar]
- 23.Gao Z, Krithivas A, Finan JE, Semmes OJ, Zhou S, Wang Y, Hayward SD. 1998. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication proteins. J Virol 72:8559–8567. doi: 10.1128/JVI.72.11.8559-8567.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flemington E, Speck SH. 1990. Epstein-Barr virus BZLF1 trans activator induces the promoter of a cellular cognate gene, c-fos. J Virol 64:4549–4552. doi: 10.1128/JVI.64.9.4549-4552.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshizaki T, Sato H, Murono S, Pagano JS, Furukawa M. 1999. Matrix metalloproteinase 9 is induced by the Epstein-Barr virus BZLF1 transactivator. Clin Exp Metastasis 17:431–436. doi: 10.1023/a:1006699003525. [DOI] [PubMed] [Google Scholar]
- 26.Lu J, Chua HH, Chen SY, Chen JY, Tsai CH. 2003. Regulation of matrix metalloproteinase-1 by Epstein-Barr virus proteins. Cancer Res 63:256–262. [PubMed] [Google Scholar]
- 27.Lu J, Chen SY, Chua HH, Liu YS, Huang YT, Chang Y, Chen JY, Sheen TS, Tsai CH. 2000. Upregulation of tyrosine kinase TKT by the Epstein-Barr virus transactivator Zta. J Virol 74:7391–7399. doi: 10.1128/jvi.74.16.7391-7399.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cayrol C, Flemington EK. 1995. Identification of cellular target genes of the Epstein-Barr virus transactivator Zta: activation of transforming growth factor beta igh3 (TGF-beta igh3) and TGF-beta 1. J Virol 69:4206–4212. doi: 10.1128/JVI.69.7.4206-4212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahot S, Sergeant A, Drouet E, Gruffat H. 2003. A novel function for the Epstein-Barr virus transcription factor EB1/Zta: induction of transcription of the hIL-10 gene. J Gen Virol 84:965–974. doi: 10.1099/vir.0.18845-0. [DOI] [PubMed] [Google Scholar]
- 30.Chang Y, Lee HH, Chen YT, Lu J, Wu SY, Chen CW, Takada K, Tsai CH. 2006. Induction of the early growth response 1 gene by Epstein-Barr virus lytic transactivator Zta. J Virol 80:7748–7755. doi: 10.1128/JVI.02608-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones RJ, Seaman WT, Feng WH, Barlow E, Dickerson S, Delecluse HJ, Kenney SC. 2007. Roles of lytic viral infection and IL-6 in early versus late passage lymphoblastoid cell lines and EBV-associated lymphoproliferative disease. Int J Cancer 121:1274–1281. doi: 10.1002/ijc.22839. [DOI] [PubMed] [Google Scholar]
- 32.Cayrol C, Flemington E. 1996. G0/G1 growth arrest mediated by a region encompassing the basic leucine zipper (bZIP) domain of the Epstein-Barr virus transactivator Zta. J Biol Chem 271:31799–31802. doi: 10.1074/jbc.271.50.31799. [DOI] [PubMed] [Google Scholar]
- 33.Cayrol C, Flemington EK. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J 15:2748–2759. doi: 10.1002/j.1460-2075.1996.tb00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez A, Armstrong M, Dwyer D, Flemington E. 1999. Genetic dissection of cell growth arrest functions mediated by the Epstein-Barr virus lytic gene product, Zta. J Virol 73:9029–9038. doi: 10.1128/JVI.73.11.9029-9038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez A, Jung EJ, Yin Q, Cayrol C, Flemington EK. 2001. Role of c-myc regulation in Zta-mediated induction of the cyclin-dependent kinase inhibitors p21 and p27 and cell growth arrest. Virology 284:159–169. doi: 10.1006/viro.2001.0923. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez A, Jung EJ, Flemington EK. 2001. Cell cycle analysis of Epstein-Barr virus-infected cells following treatment with lytic cycle-inducing agents. J Virol 75:4482–4489. doi: 10.1128/JVI.75.10.4482-4489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu FY, Chen H, Wang SE, ApRhys CM, Liao G, Fujimuro M, Farrell CJ, Huang J, Hayward SD, Hayward GS. 2003. CCAAT/enhancer binding protein alpha interacts with ZTA and mediates ZTA-induced p21(CIP-1) accumulation and G(1) cell cycle arrest during the Epstein-Barr virus lytic cycle. J Virol 77:1481–1500. doi: 10.1128/jvi.77.2.1481-1500.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Z, Yin Q, Flemington E. 2004. Identification of a negative regulatory element in the Epstein-Barr virus Zta transactivation domain that is regulated by the cell cycle control factors c-Myc and E2F1. J Virol 78:11962–11971. doi: 10.1128/JVI.78.21.11962-11971.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrison TE, Mauser A, Wong A, Ting JP, Kenney SC. 2001. Inhibition of IFN-gamma signaling by an Epstein-Barr virus immediate-early protein. Immunity 15:787–799. doi: 10.1016/s1074-7613(01)00226-6. [DOI] [PubMed] [Google Scholar]
- 40.Hahn AM, Huye LE, Ning S, Webster-Cyriaque J, Pagano JS. 2005. Interferon regulatory factor 7 is negatively regulated by the Epstein-Barr virus immediate-early gene, BZLF-1. J Virol 79:10040–10052. doi: 10.1128/JVI.79.15.10040-10052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keating S, Prince S, Jones M, Rowe M. 2002. The lytic cycle of Epstein-Barr virus is associated with decreased expression of cell surface major histocompatibility complex class I and class II molecules. J Virol 76:8179–8188. doi: 10.1128/jvi.76.16.8179-8188.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison TE, Mauser A, Klingelhutz A, Kenney SC. 2004. Epstein-Barr virus immediate-early protein BZLF1 inhibits tumor necrosis factor alpha-induced signaling and apoptosis by downregulating tumor necrosis factor receptor 1. J Virol 78:544–549. doi: 10.1128/jvi.78.1.544-549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guenther JF, Cameron JE, Nguyen HT, Wang Y, Sullivan DE, Shan B, Lasky JA, Flemington EK, Morris GF. 2010. Modulation of lung inflammation by the Epstein-Barr virus protein Zta. Am J Physiol Lung Cell Mol Physiol 299:L771–L784. doi: 10.1152/ajplung.00408.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inman GJ, Binne UK, Parker GA, Farrell PJ, Allday MJ. 2001. Activators of the Epstein-Barr virus lytic program concomitantly induce apoptosis, but lytic gene expression protects from cell death. J Virol 75:2400–2410. doi: 10.1128/JVI.75.5.2400-2410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mauser A, Saito S, Appella E, Anderson CW, Seaman WT, Kenney S. 2002. The Epstein-Barr virus immediate-early protein BZLF1 regulates p53 function through multiple mechanisms. J Virol 76:12503–12512. doi: 10.1128/jvi.76.24.12503-12512.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Q, Gutsch D, Kenney S. 1994. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol Cell Biol 14:1929–1938. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adamson AL, Kenney S. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J Virol 75:2388–2399. doi: 10.1128/JVI.75.5.2388-2399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LaJeunesse DR, Brooks K, Adamson AL. 2005. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 alter mitochondrial morphology during lytic replication. Biochem Biophys Res Commun 333:438–442. doi: 10.1016/j.bbrc.2005.05.120. [DOI] [PubMed] [Google Scholar]
- 49.Adamson AL. 2005. Epstein-Barr virus BZLF1 protein binds to mitotic chromosomes. J Virol 79:7899–7904. doi: 10.1128/JVI.79.12.7899-7904.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El-Guindy AS, Miller G. 2004. Phosphorylation of Epstein-Barr virus ZEBRA protein at its casein kinase 2 sites mediates its ability to repress activation of a viral lytic cycle late gene by Rta. J Virol 78:7634–7644. doi: 10.1128/JVI.78.14.7634-7644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baumann M, Mischak H, Dammeier S, Kolch W, Gires O, Pich D, Zeidler R, Delecluse HJ, Hammerschmidt W. 1998. Activation of the Epstein-Barr virus transcription factor BZLF1 by 12-O-tetradecanoylphorbol-13-acetate-induced phosphorylation. J Virol 72:8105–8114. doi: 10.1128/JVI.72.10.8105-8114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asai R, Kato A, Kato K, Kanamori-Koyama M, Sugimoto K, Sairenji T, Nishiyama Y, Kawaguchi Y. 2006. Epstein-Barr virus protein kinase BGLF4 is a virion tegument protein that dissociates from virions in a phosphorylation-dependent process and phosphorylates the viral immediate-early protein BZLF1. J Virol 80:5125–5134. doi: 10.1128/JVI.02674-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El-Guindy A, Heston L, Delecluse HJ, Miller G. 2007. Phosphoacceptor site S173 in the regulatory domain of Epstein-Barr virus ZEBRA protein is required for lytic DNA replication but not for activation of viral early genes. J Virol 81:3303–3316. doi: 10.1128/JVI.02445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El-Guindy AS, Paek SY, Countryman J, Miller G. 2006. Identification of constitutive phosphorylation sites on the Epstein-Barr virus ZEBRA protein. J Biol Chem 281:3085–3095. doi: 10.1074/jbc.M506076200. [DOI] [PubMed] [Google Scholar]
- 55.Daibata M, Humphreys RE, Sairenji T. 1992. Phosphorylation of the Epstein-Barr virus BZLF1 immediate-early gene product ZEBRA. Virology 188:916–920. doi: 10.1016/0042-6822(92)90553-2. [DOI] [PubMed] [Google Scholar]
- 56.Kolman JL, Taylor N, Marshak DR, Miller G. 1993. Serine-173 of the Epstein-Barr virus ZEBRA protein is required for DNA binding and is a target for casein kinase II phosphorylation. Proc Natl Acad Sci U S A 90:10115–10119. doi: 10.1073/pnas.90.21.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adamson AL. 2005. Effects of SUMO-1 upon Epstein-Barr virus BZLF1 function and BMRF1 expression. Biochem Biophys Res Commun 336:22–28. doi: 10.1016/j.bbrc.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 58.Hershko A, Ciechanover A, Varshavsky A. 2000. The ubiquitin system. Nat Med 6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 59.Pickart CM. 2001. Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 60.Weissman AM. 2001. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 61.Hochstrasser M. 2000. Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol 2:E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 62.Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V. 1994. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol 14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 64.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J 19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bres V, Kiernan RE, Linares LK, Chable-Bessia C, Plechakova O, Treand C, Emiliani S, Peloponese JM, Jeang KT, Coux O, Scheffner M, Benkirane M. 2003. A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat Cell Biol 5:754–761. doi: 10.1038/ncb1023. [DOI] [PubMed] [Google Scholar]
- 66.Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, Kogel U, Scheffner M, Helin K, Eilers M. 2005. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 67.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. 2000. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351–361. doi: 10.1016/S0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 68.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 69.Pickart CM. 2002. DNA repair: right on target with ubiquitin. Nature 419:120–121. doi: 10.1038/419120a. [DOI] [PubMed] [Google Scholar]
- 70.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. 2001. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 71.Haglund K, Dikic I. 2005. Ubiquitylation and cell signaling. EMBO J 24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hofmann RM, Pickart CM. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 73.Spence J, Sadis S, Haas AL, Finley D. 1995. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol 15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. 2000. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- 75.Galan JM, Haguenauer-Tsapis R. 1997. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J 16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Welcker M, Orian A, Jin J, Grim JE, Grim JA, Harper JW, Eisenman RN, Clurman BE. 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A 101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J 23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, Soderberg O, Kerppola TK, Larsson LG. 2003. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell 11:1189–1200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 79.Glickman MH, Ciechanover A. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 80.Hershko A, Ciechanover A. 1998. The ubiquitin system. Annu Rev Biochem 67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 81.Guerreiro P, Rodrigues-Pousada C. 1996. Characterization of a polyubiquitin gene in T. thermophila and of ubiquitin gene expression during sexual reproduction and under stress conditions. Gene 182:183–188. doi: 10.1016/S0378-1119(96)00551-3. [DOI] [PubMed] [Google Scholar]
- 82.Weber G. 2007. Molecular mechanisms of cancer, p 309–335. Springer, Dordrecht, the Netherlands. [Google Scholar]
- 83.Petosa C, Morand P, Baudin F, Moulin M, Artero JB, Muller CW. 2006. Structural basis of lytic cycle activation by the Epstein-Barr virus ZEBRA protein. Mol Cell 21:565–572. doi: 10.1016/j.molcel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 84.Schelcher C, Al Mehairi S, Verrall E, Hope Q, Flower K, Bromley B, Woolfson DN, West MJ, Sinclair AJ. 2007. Atypical bZIP domain of viral transcription factor contributes to stability of dimer formation and transcriptional function. J Virol 81:7149–7155. doi: 10.1128/JVI.00215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hicks MR, Al-Mehairi SS, Sinclair AJ. 2003. The zipper region of Epstein-Barr virus bZIP transcription factor Zta is necessary but not sufficient to direct DNA binding. J Virol 77:8173–8177. doi: 10.1128/jvi.77.14.8173-8177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mikaelian I, Drouet E, Marechal V, Denoyel G, Nicolas JC, Sergeant A. 1993. The DNA-binding domain of two bZIP transcription factors, the Epstein-Barr virus switch gene product EB1 and Jun, is a bipartite nuclear targeting sequence. J Virol 67:734–742. doi: 10.1128/JVI.67.2.734-742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giot JF, Mikaelian I, Buisson M, Manet E, Joab I, Nicolas JC, Sergeant A. 1991. Transcriptional interference between the EBV transcription factors EB1 and R: both DNA-binding and activation domains of EB1 are required. Nucleic Acids Res 19:1251–1258. doi: 10.1093/nar/19.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Song Z, Wu M. 2005. Identification of a novel nucleolar localization signal and a degradation signal in Survivin-deltaEx3: a potential link between nucleolus and protein degradation. Oncogene 24:2723–2734. doi: 10.1038/sj.onc.1208097. [DOI] [PubMed] [Google Scholar]
- 89.Flemington EK, Lytle JP, Cayrol C, Borras AM, Speck SH. 1994. DNA-binding-defective mutants of the Epstein-Barr virus lytic switch activator Zta transactivate with altered specificities. Mol Cell Biol 14:3041–3052. doi: 10.1128/mcb.14.5.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu P, Peng J. 2006. Dissecting the ubiquitin pathway by mass spectrometry. Biochim Biophys Acta 1764:1940–1947. doi: 10.1016/j.bbapap.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. 2003. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 92.Yang YC, Yoshikai Y, Hsu SW, Saitoh H, Chang LK. 2013. Role of RNF4 in the ubiquitination of Rta of Epstein-Barr virus. J Biol Chem 288:12866–12879. doi: 10.1074/jbc.M112.413393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tikhmyanova N, Tutton S, Martin KA, Lu F, Kossenkov AV, Paparoidamis N, Kenney S, Salvino JM, Lieberman PM. 2017. Small molecule perturbation of the CAND1-Cullin1-ubiquitin cycle stabilizes p53 and triggers Epstein-Barr virus reactivation. PLoS Pathog 13:e1006517. doi: 10.1371/journal.ppat.1006517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sompallae R, Gastaldello S, Hildebrand S, Zinin N, Hassink G, Lindsten K, Haas J, Persson B, Masucci MG. 2008. Epstein-Barr virus encodes three bona fide ubiquitin-specific proteases. J Virol 82:10477–10486. doi: 10.1128/JVI.01113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rao N, Dodge I, Band H. 2002. The Cbl family of ubiquitin ligases: critical negative regulators of tyrosine kinase signaling in the immune system. J Leukoc Biol 71:753–763. [PubMed] [Google Scholar]
- 96.Ungureanu D, Saharinen P, Junttila I, Hilton DJ, Silvennoinen O. 2002. Regulation of Jak2 through the ubiquitin-proteasome pathway involves phosphorylation of Jak2 on Y1007 and interaction with SOCS-1. Mol Cell Biol 22:3316–3326. doi: 10.1128/mcb.22.10.3316-3326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG Jr. 2005. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 98.Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD. 1999. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci U S A 96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shieh SY, Ikeda M, Taya Y, Prives C. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 100.Fuchs SY, Adler V, Pincus MR, Ronai Z. 1998. MEKK1/JNK signaling stabilizes and activates p53. Proc Natl Acad Sci U S A 95:10541–10546. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, Nakatani Y, Livingston DM. 2003. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 102.Harris SL, Levine AJ. 2005. The p53 pathway: positive and negative feedback loops. Oncogene 24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 103.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. 2003. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 104.Rajendra R, Malegaonkar D, Pungaliya P, Marshall H, Rasheed Z, Brownell J, Liu LF, Lutzker S, Saleem A, Rubin EH. 2004. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J Biol Chem 279:36440–36444. doi: 10.1074/jbc.C400300200. [DOI] [PubMed] [Google Scholar]
- 105.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 106.Alarcon-Vargas D, Ronai Z. 2002. p53-Mdm2—the affair that never ends. Carcinogenesis 23:541–547. doi: 10.1093/carcin/23.4.541. [DOI] [PubMed] [Google Scholar]
- 107.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O’Rourke K, Koeppen H, Dixit VM. 2004. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 108.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 109.Wilkinson KD. 2000. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol 11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 110.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. 2007. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol 9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 111.Li Z, Wang D, Messing EM, Wu G. 2005. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep 6:373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lim SK, Shin JM, Kim YS, Baek KH. 2004. Identification and characterization of murine mHAUSP encoding a deubiquitinating enzyme that regulates the status of p53 ubiquitination. Int J Oncol 24:357–364. [PubMed] [Google Scholar]
- 113.Li M, Brooks CL, Kon N, Gu W. 2004. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 114.Morrison TE, Kenney SC. 2004. BZLF1, an Epstein-Barr virus immediate-early protein, induces p65 nuclear translocation while inhibiting p65 transcriptional function. Virology 328:219–232. doi: 10.1016/j.virol.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 115.Gutsch DE, Holley-Guthrie EA, Zhang Q, Stein B, Blanar MA, Baldwin AS, Kenney SC. 1994. The bZIP transactivator of Epstein-Barr virus, BZLF1, functionally and physically interacts with the p65 subunit of NF-kappa B. Mol Cell Biol 14:1939–1948. doi: 10.1128/mcb.14.3.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sato Y, Shirata N, Kudoh A, Iwahori S, Nakayama S, Murata T, Isomura H, Nishiyama Y, Tsurumi T. 2009. Expression of Epstein-Barr virus BZLF1 immediate-early protein induces p53 degradation independent of MDM2, leading to repression of p53-mediated transcription. Virology 388:204–211. doi: 10.1016/j.virol.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 117.Morrow JK, Lin HK, Sun SC, Zhang S. 2015. Targeting ubiquitination for cancer therapies. Future Med Chem 7:2333–2350. doi: 10.4155/fmc.15.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang Y, Staudt LM. 2015. Protein ubiquitination in lymphoid malignancies. Immunol Rev 263:240–256. doi: 10.1111/imr.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin Z, Wang X, Strong MJ, Concha M, Baddoo M, Xu G, Baribault C, Fewell C, Hulme W, Hedges D, Taylor CM, Flemington EK. 2013. Whole-genome sequencing of the Akata and Mutu Epstein-Barr virus strains. J Virol 87:1172–1182. doi: 10.1128/JVI.02517-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Takimoto T, Kamide M, Umeda R. 1984. Establishment of Epstein-Barr virus (EBV)-associated nuclear antigen (EBNA)-positive nasopharyngeal carcinoma hybrid cell line (NPC-KT). Arch Otorhinolaryngol 239:87–92. doi: 10.1007/BF00454266. [DOI] [PubMed] [Google Scholar]
- 121.Takimoto T, Furukawa M, Hatano M, Umeda R. 1984. Epstein-Barr virus nuclear antigen-positive nasopharyngeal hybrid cells. Ann Otol Rhinol Laryngol 93:166–169. doi: 10.1177/000348948409300213. [DOI] [PubMed] [Google Scholar]
- 122.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci U S A 95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Feederle R, Kost M, Baumann M, Janz A, Drouet E, Hammerschmidt W, Delecluse HJ. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J 19:3080–3089. doi: 10.1093/emboj/19.12.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cao S, Moss W, O’Grady T, Concha M, Strong MJ, Wang X, Yu Y, Baddoo M, Zhang K, Fewell C, Lin Z, Dong Y, Flemington EK. 2015. New noncoding lytic transcripts derived from the Epstein-Barr virus latency origin of replication, oriP, are hyperedited, bind the paraspeckle protein, NONO/p54nrb, and support viral lytic transcription. J Virol 89:7120–7132. doi: 10.1128/JVI.00608-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Campanero MR, Flemington EK. 1997. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc Natl Acad Sci U S A 94:2221–2226. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]