In this article, we report the atomic-resolution prefusion structure of the spike protein from swine acute diarrhea syndrome coronavirus (SADS-CoV). SADS-CoV is a pathogenic alphacoronavirus that was responsible for a large-scale outbreak of fatal disease in pigs and that was reported to be capable of interspecies transmission. We describe the overall structure of the SADS-CoV spike protein and conducted a detailed analysis of its main structural elements. Our results and analyses are consistent with those of previous phylogenetic studies and suggest that the SADS-CoV spike protein is evolutionarily related to the spike proteins of betacoronaviruses, with a strong similarity in S1-NTDs and a marked divergence in S1-CTDs. Moreover, we discuss the possible immune evasion strategies used by the SADS-CoV spike protein. Our study provides insights into the structure and immune evasion strategies of the SADS-CoV spike protein and broadens the understanding of the evolutionary relationships between coronavirus spike proteins of different genera.
KEYWORDS: coronavirus, cryo-EM, protein structure-function, spike protein, virus entry
ABSTRACT
Coronaviruses (CoV) have caused a number of major epidemics in humans and animals, including the current pandemic of coronavirus disease 2019 (COVID-19), which has brought a renewed focus on the evolution and interspecies transmission of coronaviruses. Swine acute diarrhea syndrome coronavirus (SADS-CoV), which was recently identified in piglets in southern China, is an alphacoronavirus that originates from the same genus of horseshoe bats as severe acute respiratory syndrome CoV (SARS-CoV) and that was reported to be capable of infecting cells from a broad range of species, suggesting a considerable potential for interspecies transmission. Given the importance of the coronavirus spike (S) glycoprotein in host range determination and viral entry, we report a cryo-electron microscopy (cryo-EM) structure of the SADS-CoV S trimer in the prefusion conformation at a 3.55-Å resolution. Our structure reveals that the SADS-CoV S trimer assumes an intrasubunit quaternary packing mode in which the S1 subunit N-terminal domain (S1-NTD) and the S1 subunit C-terminal domain (S1-CTD) of the same protomer pack together by facing each other in the lying-down state. SADS-CoV S has several distinctive structural features that may facilitate immune escape, such as a relatively compact architecture of the S trimer and epitope masking by glycan shielding. Comparison of SADS-CoV S with the spike proteins of the other coronavirus genera suggested that the structural features of SADS-CoV S are evolutionarily related to those of the spike proteins of the other genera rather than to the spike protein of a typical alphacoronavirus. These data provide new insights into the evolutionary relationship between spike glycoproteins of SADS-CoV and those of other coronaviruses and extend our understanding of their structural and functional diversity.
IMPORTANCE In this article, we report the atomic-resolution prefusion structure of the spike protein from swine acute diarrhea syndrome coronavirus (SADS-CoV). SADS-CoV is a pathogenic alphacoronavirus that was responsible for a large-scale outbreak of fatal disease in pigs and that was reported to be capable of interspecies transmission. We describe the overall structure of the SADS-CoV spike protein and conducted a detailed analysis of its main structural elements. Our results and analyses are consistent with those of previous phylogenetic studies and suggest that the SADS-CoV spike protein is evolutionarily related to the spike proteins of betacoronaviruses, with a strong similarity in S1-NTDs and a marked divergence in S1-CTDs. Moreover, we discuss the possible immune evasion strategies used by the SADS-CoV spike protein. Our study provides insights into the structure and immune evasion strategies of the SADS-CoV spike protein and broadens the understanding of the evolutionary relationships between coronavirus spike proteins of different genera.
INTRODUCTION
Coronaviruses (CoV), named after the characteristic crown-like spike (S) proteins on their surface, are a group of enveloped positive-strand RNA viruses from the order Nidovirales. Four genera are recognized: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (1). Coronaviruses originate from bats (2–4) and have a tendency for interspecies transmission, which occurred frequently during CoV evolution and which has shaped their diversity (5). Known to infect mammals and birds, coronaviruses can afflict respiratory, hepatic, central nervous system, and gastrointestinal diseases with various degrees of severity (6, 7). In the past 2 decades, coronaviruses have become a serious threat to humans, causing major epidemics and pandemics, including severe acute respiratory syndrome (SARS) in 2003, Middle East Respiratory Syndrome (MERS) in 2012, and coronavirus disease 2019 (COVID-19) in 2020.
In addition to the threat that they pose to humans, coronaviruses, such as swine acute diarrhea syndrome coronavirus (SADS-CoV), infect domestic animals and bring substantial economic losses. SADS-CoV (also known as swine enteric alphacoronavirus [SeACoV] or porcine enteric alphacoronavirus [PEAV]) is an alphacoronavirus that appeared in southern China in August 2016 and caused outbreaks of severe watery diarrhea in newborn piglets with a mortality rate of up to 90% (3, 6, 8). SADS-CoV shares ∼95% sequence identity with the Rhinolophus bat coronavirus HKU2 and was recently reported to be capable of infecting cell lines of multiple species, including bats, mice, rats, hamsters, pigs, chickens, nonhuman primates, and humans (3, 9). These findings emphasize the significance of coronavirus spillover from bats to domestic animals and indicate that SADS-CoV displays considerable potential for interspecies transmission.
The entry of coronaviruses relies on a specific interaction between the S trimer on the virion surface and a host cell receptor (10). The S protein can be divided into three parts: a large ectodomain, a single-pass transmembrane anchor, and a small intracellular tail. The ectodomain comprises an N-terminal viral attachment and entry subunit, subunit S1 (approximately 700 amino acids), that forms a crown-like structure and a C-terminal membrane-fusion subunit, subunit S2 (approximately 600 amino acids) (11). The S1 subunit contains an N-terminal domain (S1-NTD) that plays a key role in attachment to host cell surface glycan and a C-terminal domain (S1-CTD) with the receptor-binding domain (RBD) responsible for specific binding to the host protein receptor. S1-CTDs are stabilized in an inactive lying-down conformation and expand into the active standing-up conformation once the S trimer engages the host receptor (7, 12–14). Host receptor engagement destabilizes the S trimer, exposing the cleavage site between the S1 and S2 subunits, which is then cleaved by a cathepsin, TMPRSS2, or another extracellular protease to initiate S2-mediated membrane fusion and viral entry (15, 16). The host receptor-binding S1 subunit, especially S1-NTD and S1-CTD/RBD, shows marked variation among coronaviruses and is the primary determinant of host tropism and transmission limits (10). Numerous coronaviruses, including human CoV 229E (HCoV-229E), bind to aminopeptidase N (APN) and MERS-CoV binds to dipeptidyl-peptidase 4 (DPP4), whereas severe acute respiratory syndrome coronavirus (SARS-CoV), HCoV-NL63, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) bind angiotensin-converting enzyme 2 (ACE2). Studies on SADS-CoV demonstrated that none of the known host protein receptors are crucial for its cell entry, suggesting the divergence of the SADS-CoV S protein from the S proteins of other coronaviruses (17). Therefore, structural characterization of the SADS-CoV S protein is necessary to deepen our understanding of its structural and functional divergence.
In this study, we determined by cryo-electron microscopy (cryo-EM) the structure of the SADS-CoV spike glycoprotein trimer at a 3.55-Å resolution. We analyzed the structure and compared it to the structures of spike proteins of representative coronaviruses from each genus. Particular focus was given to the structural features and conformations of SADS-CoV S1-NTD and S1-CTD, for which we delineated distinctions from and similarities to other spike proteins. Furthermore, based on our structural data, we discuss potential immune evasion strategies that the SADS-CoV spike protein may use. During our manuscript submission process, another structural study on the HKU2 and SADS-CoV spike glycoproteins by Yu et al. was published in Nature Communications (18). Similar to the present study, the authors systematically compared their structures with those of other known spike proteins and indicated the unique conformation of the connecting region after the fusion peptide in the S2 subunit of HKU2/SADS-CoV (18). The findings in our study will further broaden the understanding of the SADS-CoV spike protein and its evolutionary relationship with spike proteins of other coronaviruses.
RESULTS AND DISCUSSION
Overall structure of SADS-CoV spike protein.
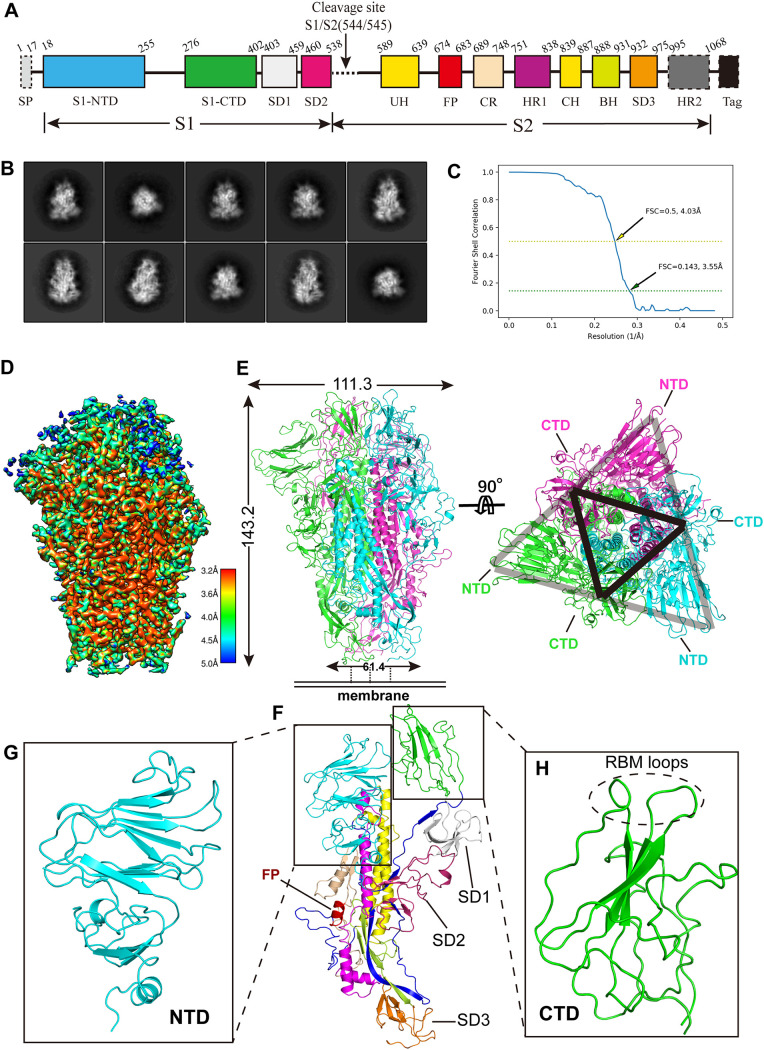
In order to investigate the structural features of SADS-CoV S, we sought to determine its cryo-EM structure. The ectodomain of SADS-CoV S was expressed in insect cells and purified from medium 3 days after infection with baculovirus. The general control protein GCN4 peptide was fused to the C-terminal end of S protein to promote trimerization (19) (Fig. 1A). The results of size exclusion chromatography indicated that the protein sample existed as a trimer in solution. Sample purity was determined by SDS-PAGE to be more than 95% (see Fig. S1A and B in the supplemental material). The protein displayed a high homogeneity in cryo-EM screening and was diluted to 0.63 mg ml−1 for data collection. Cryo-EM micrograph movies were collected on a Gatan K2 direct electron detector mounted on an FEI Titan Krios electron microscope (Fig. S2). Following reference-free two-dimensional (2D) classification, we determined a three-dimensional (3D) structure of the SADS-CoV S trimer at a 3.55-Å resolution using the gold standard Fourier shell correlation (FSC) threshold criterion of 0.143 (Fig. 1B and C and Table 1).
FIG 1.
The 3.55-Å cryo-EM structure of the SADS-CoV S trimer in the prefusion conformation. (A) Domain arrangement of the SADS-CoV S ectodomain. S1, receptor-binding subunit; S2, membrane-fusion subunit; SP, signal peptide; S1-NTD, N-terminal domain of S1; S1-CTD, C-terminal domain of S1; SD1 to SD3, subdomains 1 to 3, respectively; FP, fusion peptide; UH, upstream helix; CH, central helix; HR1 and HR2, heptad repeats 1 and 2, respectively; Tag, GCN4 trimerization tag followed by an 8×His tag. Regions indicated by dotted rectangles (signal peptide, HR2, and GCN4 tag and 8×His tag) were not included in the structural model. (B) Representative 2D class averages of SADS-CoV S trimer in different orientations. (C) Gold-standard FSC curve of SADS-CoV S. The resolution was determined to be 3.55 Å at an FSC threshold criterion of 0.143. The yellow dashed line indicates the cutoff FSC value of 0.5, which corresponds to a resolution of 4.03 Å. (D) Final cryo-EM density map of SADS-CoV S colored according to the local resolution. (E) (Left) Ribbon diagram of the SADS-CoV S trimer in the prefusion conformation. The three S subunits are in green, cyan, and magenta. (Right) Same as the left panel, but the SADS-CoV S trimer was rotated 90 degrees to show the top view of the trimer cap. The locations of the S1-CTDs of each protomer can be seen as a vertex of the black triangle. Each S1-CTD is sandwiched by two S1-NTDs, one from the same protomer and one from the adjacent protomer, thus forming one side of the gray triangle. (F) Ribbon diagram of a SADS-CoV S protomer. The structural elements are colored as described in panel A. (G and H) Structures of SADS-CoV S1-NTD (G) (cyan) and S1-CTD (H) (green). The putative RBM loops are indicated by a dashed circle.
TABLE 1.
Structural data collection and refinement statistics
| Parameter | Value for SADS-CoV S ectodomaina |
|---|---|
| Data collection statistics | |
| EM equipment | FEI Titan Krios |
| Voltage (kV) | 300 |
| Detector | K2 Summit |
| Pixel size (Å) | 1.04 |
| Electron dose (e−/Å2) | 60 |
| Defocus range (μm) | −1.8 to −2.5 |
| Reconstruction statistics | |
| Software | RELION (v3.0) |
| No. of particles | 35,000 |
| Symmetry | C3 |
| FSC threshold | 0.143 |
| Final resolution (Å) | 3.55 |
| Map-sharpening B factor (Å2) | −152 |
| Model-building software | Coot (v0.8.9) |
| Refinement statistics | |
| Software | Phenix-1.18rc1 |
| CC_maskb | 0.7888 |
| RMSD | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.222 |
| Validation statistics | |
| Clash score | 5.39 |
| Rotamer outliers (%) | 1.23 |
| Ramachandran plot (%) | |
| Favored | 91.26 |
| Allowed | 8.74 |
| Outliers | 0 |
EMDB accession number EMD-30071 and PDB accession number 6M39.
CC_mask, model-to-map correlation coefficient calculated in the map region around the model.
The resolved atomic structure of the prefusion SADS-CoV S ectodomain covered nearly all of the key structural elements (Fig. 1A), with the exception of residues 81 to 101 of S1-NTD and residues 999 to 1068 of HR2 (Fig. 1D; Fig. S3). Forty-five N-linked glycans (15 on each subunit) were found to be distributed across the surface of the SADS-CoV S trimer, with another 15 being predicted but not observed. Each S protein monomer was stabilized by 10 pairs of intramolecular disulfide bonds (Fig. S4). The SADS-CoV S trimer assumes a mushroom-like shape in which three S1 heads form a crown-like structure sitting on top of the trimeric S2 stalk (Fig. 1D and E; Movie S1). The trimer measures 143.2 Å in length from S1 to S2 and 111.3 Å and 61.4 Å in width at S1 and S2, respectively (Fig. 1E, left). The S1 subunit of SADS-CoV S can be divided into two major domains, S1-NTD and S1-CTD, which bind, respectively, glycan receptors and protein receptors and thus play a vital role in coronavirus entry, and two subdomains (SD1 and SD2), whose functions are unclear (Fig. 1F to H; Movie S1). The three S1-CTDs are located at the top center of the S trimer and are arranged as vertices of a small triangle, whereas the three S1-NTDs are located at the lower outer side of S1-CTDs and are arranged as a big triangle, so that each side of the triangle is formed by an S1-CTD sandwiched between S1-NTD of the same protomer and S1-NTD of the adjacent protomer (Fig. 1E, right). The protease cleavage site at the boundary of the S1 and S2 subunits is located between residues Val544 and Arg545. The upstream helix (UH) and central helix (CH) from the S2 subunit of each protomer collectively form a six-helix bundle in the core of the S trimer. Heptad repeat 1 (HR1), which comprises four helices interconnected by three loops, is located between the S1 and S2 subunits at the outer side of the S2 stalk, whereas a segment of HR2 is missing in the structure (Fig. 1A).
Structural evolution of SADS-CoV spike protein.
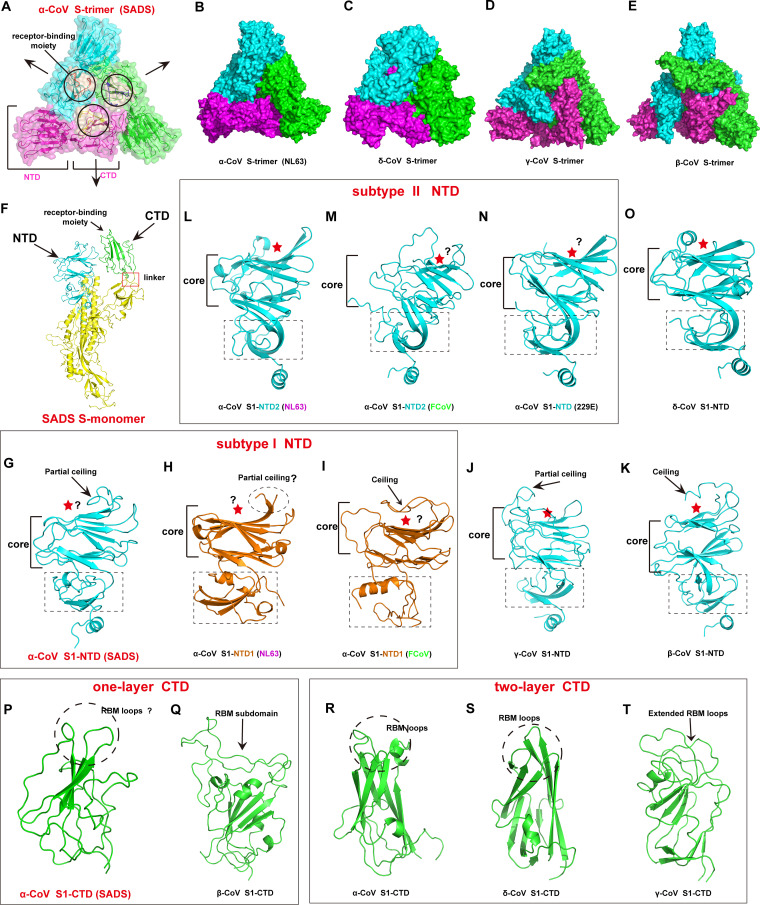
The spike proteins from all four genera of coronaviruses pack into a trimeric crown-like structure via one of two quaternary packing modes, i.e., the cross-subunit mode and the intrasubunit mode (20). Our structure revealed that SADS-CoV S uses the intrasubunit quaternary packing mode, in which the S1-NTD and S1-CTD of the same protomer are packed head-to-head. In such an architecture, the three S1-NTDs are located at the vertices of a big triangle and sandwich S1-CTDs to the center of each side of the triangle. As a result, the putative receptor-binding moieties, which are located on top of the S1-CTDs, are concentrated at the center top of the crown-like spike trimer (Fig. 2A). Thus, the geometry of SADS-CoV S resembles the prefusion structures of the spike proteins of other alpha- and deltacoronaviruses, and the structure of SADS-CoV S is more compact than the structures of the spike proteins of beta- and gammacoronaviruses that use the cross-subunit packing mode (Fig. 2A to E). Of note, although all alphacoronavirus spike proteins use the intrasubunit packing mode, they differ in the number of S1-NTDs within the S protomer. While the majority of known coronavirus spike proteins (including SADS-CoV, HKU2, and HCoV-229E spike proteins) contain a single NTD, certain alphacoronavirus spike proteins, such as the spike proteins of HCoV-NL63, feline CoV (FCoV), and porcine epidemic diarrhea virus (PEDV), contain two structurally distinct S1-NTDs named domain 0 and domain 1 (also classified as subtype I NTD [S1-NTD1] and subtype II NTD [S1-NTD2] by Yu et al. [18]) (Fig. 2F; Fig. S5A to C). Two evolutionary processes have been proposed to explain the existence of two NTDs: one study suggested that two NTDs in HCoV-NL63 are the result of gene duplication (21), whereas other studies suggested that two NTDs arose from the recombination of two separate primitive domains within the viral genome (1, 18, 19, 22).
FIG 2.
Structural evolution of coronavirus spike protein. (A) The architecture of the SADS-CoV S trimer. The three S protomers are shown in different colors (magenta, cyan, and green), and the receptor-binding moieties are colored wheat and indicated with black circles. The black arrows indicate the direction of S1-CTD expansion during the transition from an inactive state to an active state. (B to E) Architectural features of spike protein trimers from the four coronavirus genera. (B) S trimer of the alphacoronavirus HCoV-NL63 (PDB accession number 5SZS); (C) S trimer of the deltacoronavirus PdCoV (PDB accession number 6BFU); (D) S trimer of the gammacoronavirus infectious bronchitis virus (IBV) (PDB accession number 6CV0); (E) S trimer of the betacoronavirus SARS-CoV (PDB accession number 5X58). (F) Structure of the SADS-CoV S protomer. The red frame indicates the linker between CTD and SD1. NTD, CTD, and the receptor-binding moiety are labeled and indicated by black arrows. (G to O) Structural characteristics of spike protein S1-NTDs from the four coronavirus genera. The NTD core and subdomain (dotted rectangle) are labeled. A partial ceiling or ceiling is indicated with a black arrow, and the sugar-binding site or putative sugar-binding site is indicated by a red star. (G to I) Subtype I alphacoronavirus S1-NTDs. (G) SADS-CoV S1-NTD; (H) HCoV-NL63 S1-NTD1 (PDB accession number 5SZS); (I) FCoV S1-NTD1 (PDB accession number 6JX7). (J) S1-NTD of the gammacoronavirus IBV (PDB accession number 6CV0). (K) S1-NTD of the betacoronavirus SARS-CoV (PDB accession number 5X58). (L to N) Subtype II alphacoronavirus NTDs. (L) HCoV-NL63 S1-NTD2 (PDB accession number 5SZS); (M) FCoV S1-NTD1 (PDB accession number 6JX7); (N) HCoV-229E S1-NTD (PDB accession number 6U7H). (O) S1-NTD of the deltacoronavirus PdCoV (PDB accession number 6BFU). (P to T) Structural characteristics of the spike protein S1-CTDs from the four coronavirus genera. The RBM loops or putative RBM loops are indicated by dotted circles, whereas the extended RBM loops and RBM subdomain are indicated by black arrows. (P and Q) One-layer S1-CTDs. (P) SADS-CoV S1-CTD; (Q) S1-CTD of the betacoronavirus SARS-CoV (PDB accession number 5X58). (R to T) Two-layer S1-CTDs. (R) S1-CTD of the alphacoronavirus HCoV-NL63 (PDB accession number 5SZS); (S) S1-CTD of the deltacoronavirus PdCoV (PDB accession number 6BFU); (T) S1-CTD of the gammacoronavirus IBV (PDB accession number 6CV0).
Regardless of the packing mode, the S trimer needs to undergo conformational changes to engage in receptor binding. Apart from the S trimer structures in the receptor-binding inactive state, where all three S1-CTDs assume lying-down states (Fig. 2A to E), previous studies have also reported S trimer prefusion structures of the betacoronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2 in which S1-CTD assumes a standing-up state (7, 12, 14, 23, 24) (Fig. S5D). The standing-up state represents the active, unstable conformation of S1-CTDs required for host receptor binding (25). The S1-CTDs in our structure maintained a lying-down state, in which receptor binding was hindered by the partial concealment of the receptor-binding motif (RBM) and steric clashes between S1-NTD and the receptor (Fig. 2F). The linker between S1-CTD and the remainder of the S1 subdomain acts as a hinge to facilitate the conformational transition of S1-CTDs from the lying-down state to the standing-up state (Fig. 2F). Based on our structure, we speculated that, unlike betacoronavirus spike proteins, strong intermolecular interactions within the SADS-CoV S trimer may obstruct this conformational transition and the expansion of the S1-CTD/receptor-binding site into an open state before host receptor engagement (Fig. S6). To investigate the intrinsic mobility of the S1-CTD/RBD of the SADS-CoV S trimer in the apo form (i.e., to verify whether the S1-CTD/RBD can spontaneously expand to an open state), we carried out a 2,000-ns-long coarse-grained (CG) molecular dynamics (MD) simulation (26). A Martini CG model was initially generated using the lying-down state structure (Fig. S7). The simulation indicated that all three CTDs/RBDs were stabilized in a lying-down state similar to the cryo-EM structure determined in this study (Fig. S7), with the center-of-mass distances between two S1-CTDs/RBDs calculated from the structure and in the simulation differing by less than 5 Å. Although we did not observe an apparent transition of the S1-CTDs/RBDs from the lying-down state to the standing-up state during the simulations, we cannot rule out the possibility of the coexistence of the two states in the SADS-CoV S trimer apo form because such a large conformational transition may happen over a greater time scale (milliseconds to seconds), which is far beyond the time scale in the simulation performed. Therefore, to enhance sampling of the standing-up state, we ran a 100-ns-long atomistic steered MD (SMD) simulation in which the radii of gyration of all three CTDs/RBDs were mildly steered. SMD has been successfully used to investigate the conformational transition pathways, protein folding, and ligand binding. The SMD simulation indicated that one of the three CTDs/RBDs gradually opens up, while the other two CTDs/RBDs remain in the lying-down state (Fig. S7C). This suggests that the conformational transition between the lying-down state and the standing-up state of the CTDs/RBDs in the SADS-CoV S trimer follows a sequential pathway to eject each CTD/RBD one by one, which is in accordance with the findings of an earlier study (25). Thus, the results of the simulation may explain the homogeneity of the SADS-CoV S trimer observed in our 3D classification and the fact that different S1-CTD conformations are not detectable in our structural data (Fig. 1B).
Next, we analyzed the structures of the SADS-CoV spike protein S1-NTD and S1-CTD from an evolutionary perspective. The core structure of SADS-CoV S1-NTD consists of two six-stranded antiparallel β-sheet layers stacked together, which is a structure that is the same as the galectin fold adopted by human galectins and NTDs of spike proteins from other coronavirus genera (Fig. 2G to O). In addition to the core structure, SADS-CoV S1-NTD has a loop (residues 133 to 150) that resembles the partial ceiling of subtype I S1-NTDs (S1-NTD1s) of alphacoronaviruses (with the exception of FCoV S1-NTD1, which has a full ceiling) and gammacoronaviruses. The SADS-CoV S1-NTD loop is thus in contrast to the structure of subtype II S1-NTDs (S1-NTD2s) of alphacoronaviruses and deltacoronaviruses, which do not contain a ceiling-like structure, as well as the structure of betacoronaviruses S1-NTDs, which have a reinforced ceiling-like structure (Fig. 2G to O). Furthermore, the structure of the subdomain below the core domain of SADS-CoV S1-NTD is most comparable to that of subtype I S1-NTDs of alpha-, gamma-, and betacoronaviruses (Fig. 2G to K). Although the sugar-binding site in SADS-CoV S1-NTD has not been experimentally identified, the structural similarities to the S1-NTDs of other coronaviruses suggest that the sugar-binding site in SADS-CoV S1-NTD may also be located in the pocket formed between the core structure and the partial ceiling loop (Fig. 2G). Moreover, even though subtype I and II S1-NTDs of alphacoronaviruses have certain structural differences (18), their similar overall architectures and the local structure of their (putative) sugar-binding sites suggest that both subtypes of S1-NTDs can serve as the glycan attachment receptor-binding domain (Fig. 2G to O). While certain subtype II alphacoronavirus S1-NTDs, such as S1-NTD2 of HCoV-NL63, may require additional conformational changes to expose the putative sugar-binding site on the surface of the S trimer (Fig. S5A), the putative sugar-binding sites on the two S1-NTDs of FCoV and PEDV do not need to undergo conformational changes to become exposed (Fig. S5B and C). Given that the evolutionary spectrum of S1-NTDs is considered to follow the alpha-, delta-, gamma-, and betacoronavirus genus order, with alphacoronavirus S1-NTDs possibly being the most ancestral (20), our analysis is consistent with that of Yu et al. (18) and suggests the existence of two parallel pathways in the evolution of S1-NTDs. The ancestral alphacoronaviruses with these two pathways possessed a single S1-NTD, with one having the subtype I S1-NTD and the other having the subtype II S1-NTD (represented by SADS-CoV S1-NTD and HCoV-229E S1-NTD, respectively). Recombination of these two ancestral alphacoronaviruses probably gave rise to the alphacoronaviruses that contain two S1-NTDs (i.e., S1-NTD1 and S1-NTD2, such as HCoV-NL63, FCoV, and PEDV). The ancestral subtype I S1-NTD would evolve to subtype I S1-NTDs of alphacoronaviruses (e.g., SADS-CoV S1-NTD, HCoV-NL63 S1-NTD1, FCoV S1-NTD1, and PEDV S1-NTD1) and S1-NTDs of beta- and gammacoronaviruses, whereas the ancestral subtype II S1-NTD would evolve to subtype II S1-NTDs of alphacoronaviruses (e.g., HCoV-NL63 S1-NTD2, FCoV S1-NTD2, and PEDV S1-NTD2) as well as S1-NTDs of deltacoronaviruses.
Despite the dramatic differences in their primary sequences, SADS-CoV S1-CTD displays a structural topology generally similar to that of S1-CTDs of other coronaviruses (Fig. 2P to T). While the S1-CTDs of other alphacoronaviruses (e.g., HCoV-NL63, HCoV-229E, PEDV, transmissible gastroenteritis virus, and porcine respiratory coronavirus) and a deltacoronavirus (porcine deltacoronavirus [PdCoV]) adopt a compact core β-sandwich structure with two β-sheet layers (a two-layer CTD), the SADS-CoV S1-CTD core is more like those of betacoronavirus CTDs, which are loosely packed and which contain a single β-sheet layer and several α-helix and coil structures (one-layer CTD) (Fig. 2R and S); the S1-CTDs of gammacoronavirus are significantly different from the S1-CTDs of the other coronavirus genera, with a loosely packed two-β-sheet layer (Fig. 2T). The receptor-binding motif (RBM) of S1-CTD, located above the core, displays considerable structural divergence across the four coronavirus genera. The two-layer S1-CTD of alpha- and deltacoronaviruses has three short discontinuous loops that may represent a primitive RBM structure of the two-layer S1-CTD family, gammacoronaviruses have two reinforced loops, and betacoronaviruses have a single continuous insertion domain (Fig. 2P to T). The putative RBM of SADS-CoV consists of two loops, and structural comparison of the RBMs of other coronaviruses indicated the closest similarity to the RBMs of two-layer S1-CTD alpha- and deltacoronaviruses (Fig. 2P, R, and S), suggesting that the SADS-CoV S1-CTD may represent a primitive structure in the one-layer S1-CTD family. When these findings are taken together, we propose that SADS-CoV S1-CTD belongs to the one-layer S1-CTD family, which evolved in the betacoronaviruses after a recombination event between an alphacoronavirus S1-CTD core domain and a betacoronavirus-like RBM (8, 18, 27, 28). The unique two-layer S1-CTD of gammacoronavirus may have evolved independently from the two-layer S1-CTD family of alpha- and deltacoronaviruses (18) (Fig. 2P to T). Based on our structural analysis, we also speculate that, similar to other coronavirus spike proteins, SADS-CoV uses the two-RBM system, in which S1-NTD serves as the glycan attachment receptor-binding domain and S1-CTD functions as the protein receptor-binding domain.
Structural alignment of SADS-CoV, SARS-CoV, and SARS-CoV-2 spike proteins.
Previous whole-genome- and nucleocapsid (N) gene-based phylogenetic analyses of eight SADS-CoV strains suggested that all SADS-CoV sequences group with the bat coronavirus HKU2 and form a well-defined branch within the alphacoronaviruses (29). Interestingly, phylogenetic analysis of the alphacoronavirus S genes showed that they form two separate groups, α-CoV-1 and α-CoV-2. While most alphacoronavirus S genes clustered in the α-CoV-1 group, the S genes of SADS-CoV and HKU2 were assigned to the α-CoV-2 group, which clustered together with betacoronavirus S genes, indicating that HKU2 and SADS-CoV may have arisen from an ancient recombination event between an alphacoronavirus genomic backbone and a betacoronavirus spike gene (8, 29). Considering the implications for the evolutionary relationship on the gene sequence level, we decided to explore whether the evolutionary relationship between the spike proteins of SADS-CoV and betacoronaviruses can also be observed in protein structures. The results of a Dali server analysis (http://ekhidna2.biocenter.helsinki.fi/dali/) indicated that the SADS-CoV S trimer shares significant similarity with the spike proteins of betacoronaviruses, as reflected by generally high Z-scores and low root mean square deviation (RMSD) values (Table S1). Moreover, separate analyses of SADS-CoV S1-NTD and the S1-CTD on the Dali server showed that the SADS-CoV S1-NTD is largely related to the S1-NTDs of betacoronaviruses, whereas the SADS-CoV RBD bears no significant similarities to betacoronavirus RBDs (Tables S2 and S3). These results are consistent with those of the previous phylogenetic analyses and our evolutionary analyses and suggest that SADS-CoV S has a close evolutionary relationship with the spike proteins of betacoronaviruses, with a strong similarity in S1-NTDs and a marked divergence in S1-CTDs.
To further investigate the structural similarities and differences between the SADS-CoV S protein and the spike proteins of other betacoronaviruses in terms of receptor-binding specificity, we structurally compared the SADS-CoV spike protein S1-CTD to the S1-CTDs of SARS-CoV-2, SARS-CoV, and HCoV-NL63 S in complex with ACE2 (Fig. 3B to D) and the S1-CTDs of SARS-CoV-2 and SARS-CoV S in the apo form (Fig. 3E). The S1-CTDs of SADS-CoV, SARS-CoV-2, SARS-CoV, and HCoV-NL63 do not share significant sequence homology and belong to different coronavirus genera (Fig. 3A); they share a roughly identical organization and core folding; and SARS-CoV-2, SARS-CoV, and HCoV-NL63 use ACE2 as the receptor. SARS-CoV-2 and SARS-CoV use the additional insertion domain to bind ACE2, whereas HCoV-NL63 uses the RBM loops for the same purpose. However, although the SADS-CoV S1-CTD also possesses an RBM loop, it cannot be used to bind ACE2 (Fig. 3E). Altogether, our analysis implies that the receptor-binding specificity of coronavirus spike proteins is determined collectively by the sequence and local structure of the receptor-binding loops.
FIG 3.
Comparison of S1-CTD domains from SADS-CoV, SARS-CoV, HCoV-NL63, and SARS-CoV-2. (A) Sequence alignment of S1-CTDs from SADS-CoV, SARS-CoV, SARS-CoV-2, and HCoV-NL63. (B to D) Structures of SARS-CoV-2 S1-CTD (B), SARS-CoV S1-CTD (C), and NL63-CoV S1-CTD (D) in complex with ACE2. CTDs are in green, and ACE2 is in gray. (E) Structural alignment of SARS-CoV-2 (gray and red), SARS-CoV (gray and purple), and SADS-CoV S1-CTD (green). Red and purple ribbons indicate the ACE2-binding motifs (insertion domains) of SARS-CoV-2 and SARS-CoV, respectively.
Possible immune evasion strategies used by SADS-CoV S.
Located on the surface of coronaviruses, the spike proteins mediate viral entry into host cells but, at the same time, are subjected to pressure from the host immune system, which is why they developed strategies to evade immune detection (21). Similar to the S trimers of other alphacoronaviruses, the SADS-CoV S trimer exhibits a compact structure formed by intrasubunit packing (Fig. 2A and B), which maximally reduces its surface area exposed to the host immune system. Moreover, our structure of the SADS-CoV S trimer shows that the S1-NTDs and S1-CTDs of each protomer assume the lying-down state (i.e., the closed conformation), which may further reduce immune pressure (Fig. 2F). This closed conformation is also stabilized by a loop from S1-CTD (residues 321 to 328) that interacts with S1-NTD of the same protomer. Upon engaging the host receptor, S1-CTDs would need to switch into the standing-up state (the open conformation) to render the putative RBM loops accessible for host receptor binding (Fig. S6B). Thus, this closed conformation-to-open conformation mechanism can also minimize the exposure of the putative RBM loops to the immune system (1).
In addition to the subunit packing mode and the lying-down state, epitope masking by glycan shielding is another immune evasion strategy commonly used by coronavirus spike proteins (21). Our cryo-EM map shows that 45 N-linked glycans are spread across the surface of the SADS-CoV S trimer (Fig. 4A). Access to the putative RBMs of the SADS-CoV S trimer is blocked both by S1-NTDs and by N-linked glycosylation of residue Asp358, located on the RBM (Fig. 4B and C). Although the S trimer of another alphacoronavirus (HCoV-NL63) is covered by a more extensive glycan shield consisting of 34 N-linked oligosaccharides per protomer, SADS-CoV is likely to use similar strategies of glycan shielding (21). Furthermore, unlike the spike proteins of alpha- and deltacoronaviruses, whose S1-NTD sialic receptor-binding sites are concealed solely by surrounding glycans, the putative sialic receptor-binding sites on SADS-CoV S are shielded both by glycans and by the partial ceiling-like structure on top of the S1-NTD core, which is similar to the shielding of gamma- and betacoronavirus S1-NTDs (30). Taken together, SADS-CoV has several structural features that may facilitate immune evasion, including tight intrasubunit packing, the lying-down conformation of S1-CTDs, the partial ceiling-like structure above the S1-NTD core, and glycan shielding.
FIG 4.
N-linked glycan distribution on the surface of SADS-CoV S. (A) Distribution of the observed N-linked glycosylation sites (indicated by Ψ) on the primary structure diagram of SADS-CoV S. (B) Distribution of observed N-linked glycosylation sites on the three-dimensional structure of the SADS-CoV S trimer. Blue spheres indicate N-linked glycosylation sites. (C) Distribution of observed N-linked glycosylation sites in the S1 subunit of a SADS-CoV S protomer. Regions of the S1 subunit are colored as described in panel A, and N-linked glycosylation sites are indicated by blue spheres.
MATERIALS AND METHODS
Protein expression and purification.
The SADS-CoV spike glycoprotein (virus strain GDS04; GenBank accession no. ASK51717.1) gene was synthesized with optimized codons and inserted into a pFastBac vector (Life Technologies Inc.). A GCN4 trimerization tag (31–33), followed by a tobacco etch virus cleavage site and an 8×His tag, were fused to the C terminus of the S protein. The ectodomain of the SADS-CoV spike protein lacking the transmembrane anchor and intracellular tail (residues 18 to 1068) was expressed in the Bac-to-Bac insect cell system (Invitrogen). The cells were centrifuged at 4,000 × g to separate the supernatant from the cellular debris. The supernatant was then loaded onto an Ni-nitrilotriacetic acid (NTA) column (Invitrogen) for affinity purification. The spike protein was further purified using a Superose 6 HR10/300 column (GE Healthcare) that had been preequilibrated with buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, and 1 mM dithiothreitol (DTT). The purified protein samples were then concentrated with a centrifugal filter (Amicon Ultra) to approximately 1 mg/ml, divided into aliquots, flash-frozen in liquid nitrogen, and stored at −80°C until further use.
Cryo-EM sample preparation and data acquisition.
Purified S protein was diluted to 0.63 mg ml−1 with buffer containing 20 mM HEPES (pH 7.5), 100 mM NaCl, and 2 mM DTT. Four microliters of the sample was applied to a glow-discharged Quantifoil copper grid and vitrified by plunge freezing in liquid ethane, using a Vitrobot Mark system with a blotting time of 3 s. Data collection was performed at the Center for Biological Imaging, Institute of Biophysics, on a Titan Krios microscope operated at 300 kV and equipped with a field emission gun, a Gatan GIF Quantum energy filter, and a Gatan K2 Summit direct electron camera in superresolution mode. The calibrated magnification was ×130,000 in the ejection fraction transmission electron microscopy mode, corresponding to a pixel size of 1.04 Å. The automated software SerialEM was used to collect 1,000 movies at a defocus range of between 1.8 and 2.3 μm. Each exposure (with a 10-s exposure time) comprised 32 subframes, amounting to a total dose of 60 electrons Å−2.
Image processing.
Micrograph movie stacks were corrected for beam-induced motion using MotionCor2 software (34). The contrast transfer function parameters for each dose-weighted image were determined with the Gctf program (35). Particles were initially automatically picked with the Gautomatch program without the template and extracted with a 256-pixel-by-256-pixel box. A reference-free 2D-class average was determined using RELION software (36), and the well-resolved 2D averages were subjected to another iteration of particle automatic picking as a template with the Gautomatch program. After iterative 2D-class averaging in RELION software, particles with the best-resolved 2D averages were selected for initial model generation and 3D classification using RELION. The classes with identical detailed features were merged for further autorefinement with a sphere mask and postprocessed with a 3-pixel extension and a 3-pixel falloff around the entire molecule to produce the final density map with an overall resolution of 3.55 Å. The Chimera and PyMOL programs (https://pymol.org/) were used for graphical visualization (37).
Model building.
Ab initio modeling of the SADS-CoV spike glycoprotein was performed in Coot software (38), using the structure predictions calculated by the Phyre2 program (39). The partial structure was modeled using the EMBuilder program (40) and the reference model (PDB accession number 5X58). Map refinement was carried out using the Phenix.real_space_refine program (41) with secondary structure and Ramachandran restraints. Cryo-EM data collection, refinement, and validation statistics are listed in Table 1.
Data availability.
The cryo-EM structure of SADS-CoV S has been deposited in the Electron Microscopy Data Bank under accession number EMD-30071. Coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession number 6M39.
Supplementary Material
ACKNOWLEDGMENTS
S.O. was funded by the Special Open Fund of the Key Laboratory of Experimental Marine Biology, Chinese Academy of Sciences (grant number SKF2020NO1), the National Natural Science Foundation of China (grant number 31770948), and a high-level personnel introduction grant from Fujian Normal University (grant number Z0210509). P.Z. was funded by the National Natural Science Foundation of China (grant number 31425007), the Chinese Ministry of Science and Technology (grant number 2017YFA0504700), and the Strategic Priority Research Program of the Chinese Academy of Sciences (grant number XDB37010102). H.G. was funded by the National Natural Science Foundation of China (grant number 31900879) and the Natural Science Foundation of Fujian Province (grant number 2019J05064).
We declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Shang J, Zheng Y, Yang Y, Liu C, Geng Q, Tai W, Du L, Zhou Y, Zhang W, Li F. 2018. Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state. J Virol 92:e01556-17. doi: 10.1128/JVI.01556-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, Zheng XS, Li B, Li JM, Guo H, Pei GQ, An XP, Chen JW, Zhou L, Mai KJ, Wu ZX, Li D, Anderson DE, Zhang LB, Li SY, Mi ZQ, He TT, Cong F, Guo PJ, Huang R, Luo Y, Liu XL, Chen J, Huang Y, Sun Q, Zhang XL, Wang YY, Xing SZ, Chen YS, Sun Y, Li J, Daszak P, Wang LF, Shi ZL, Tong YG, Ma JY. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham RL, Baric RS. 2010. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol 84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. 2012. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song W, Gui M, Wang X, Xiang Y. 2018. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog 14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y, Tian X, Qin P, Wang B, Zhao P, Yang YL, Wang L, Wang D, Song Y, Zhang X, Huang YW. 2017. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet Microbiol 211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang YL, Qin P, Wang B, Liu Y, Xu GH, Peng L, Zhou J, Zhu SJ, Huang YW. 2019. Broad cross-species infection of cultured cells by bat HKU2-related swine acute diarrhea syndrome coronavirus and identification of its replication in murine dendritic cells in vivo highlight its potential for diverse interspecies transmission. J Virol 93:e01448-19. doi: 10.1128/JVI.01448-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fehr AR, Perlman S. 2015. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F. 2016. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, Corbett KS, Graham BS, McLellan JS, Ward AB. 2018. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep 8:15701. doi: 10.1038/s41598-018-36918-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y, Zhang X, Gao GF. 2017. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun 8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. 2010. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui J, Li F, Shi Z-L. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, Qiao S, Guo R, Wang X. 2020. Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution. Nat Commun 11:3070. doi: 10.1038/s41467-020-16876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrapp D, McLellan JS. 2019. The 3.1-angstrom cryo-electron microscopy structure of the porcine epidemic diarrhea virus spike protein in the prefusion conformation. J Virol 93:e00923-19. doi: 10.1128/JVI.00923-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang J, Zheng Y, Yang Y, Liu C, Geng Q, Luo C, Zhang W, Li F. 2018. Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins. PLoS Pathog 14:e1007009. doi: 10.1371/journal.ppat.1007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walls AC, Tortorici MA, Frenz B, Snijder J, Li W, Rey FA, DiMaio F, Bosch BJ, Veesler D. 2016. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol 23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang TJ, Chang YC, Ko TP, Draczkowski P, Chien YC, Chang YC, Wu KP, Khoo KH, Chang HW, Hsu SD. 2020. Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans. Proc Natl Acad Sci U S A 117:1438–1446. doi: 10.1073/pnas.1908898117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gui M, Song W, Zhou H, Xu J, Chen S, Xiang Y, Wang X. 2017. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res 27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder J, Quispe J, Cameroni E, Gopal R, Dai M, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D. 2019. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176:1026–1039.e1015. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, Cottrell CA, Becker MM, Wang L, Shi W, Kong WP, Andres EL, Kettenbach AN, Denison MR, Chappell JD, Graham BS, Ward AB, McLellan JS. 2017. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A 114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Jong DH, Singh G, Bennett WF, Arnarez C, Wassenaar TA, Schafer LV, Periole X, Tieleman DP, Marrink SJ. 2013. Improved parameters for the Martini coarse-grained protein force field. J Chem Theory Comput 9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 27.Fu X, Fang B, Liu Y, Cai M, Jun J, Ma J, Bu D, Wang L, Zhou P, Wang H, Zhang G. 2018. Newly emerged porcine enteric alphacoronavirus in southern China: identification, origin and evolutionary history analysis. Infect Genet Evol 62:179–187. doi: 10.1016/j.meegid.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau SK, Woo PC, Li KS, Huang Y, Wang M, Lam CS, Xu H, Guo R, Chan KH, Zheng BJ, Yuen KY. 2007. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology 367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou L, Sun Y, Lan T, Wu R, Chen J, Wu Z, Xie Q, Zhang X, Ma J. 2019. Retrospective detection and phylogenetic analysis of swine acute diarrhoea syndrome coronavirus in pigs in southern China. Transbound Emerg Dis 66:687–695. doi: 10.1111/tbed.13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng G, Xu L, Lin YL, Chen L, Pasquarella JR, Holmes KV, Li F. 2012. Crystal structure of bovine coronavirus spike protein lectin domain. J Biol Chem 287:41931–41938. doi: 10.1074/jbc.M112.418210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harbury PB, Zhang T, Kim PS, Alber T. 1993. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 32.Eckert DM, Malashkevich VN, Kim PS. 1998. Crystal structure of GCN4-pIQI, a trimeric coiled coil with buried polar residues. J Mol Biol 284:859–865. doi: 10.1006/jmbi.1998.2214. [DOI] [PubMed] [Google Scholar]
- 33.Walls AC, Tortorici MA, Bosch BJ, Frenz B, Rottier PJM, DiMaio F, Rey FA, Veesler D. 2016. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang K. 2016. Gctf: real-time CTF determination and correction. J Struct Biol 193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH. 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou N, Wang H, Wang J. 2017. EMBuilder: a template matching-based automatic model-building program for high-resolution cryo-electron microscopy maps. Sci Rep 7:2664. doi: 10.1038/s41598-017-02725-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM structure of SADS-CoV S has been deposited in the Electron Microscopy Data Bank under accession number EMD-30071. Coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession number 6M39.