The COVID-19 pandemic has already led to more than 700,000 deaths and innumerable changes to daily life worldwide. Along with development of a vaccine, identification of effective antivirals to treat infected patients is of the highest importance. However, rapid drug discovery requires efficient methods to identify novel compounds that can inhibit the virus. In this work, we present a method for identifying inhibitors of the SARS-CoV-2 main protease, 3CLpro. This reporter-based assay allows for antiviral drug screening in human cell culture at biosafety level 2 (BSL2) with high-throughput compatible sample processing and analysis. This assay may help identify novel antivirals to control the COVID-19 pandemic.
KEYWORDS: antivirals, coronavirus, FlipGFP, protease, screening
ABSTRACT
In late 2019, a human coronavirus, now known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged, likely from a zoonotic reservoir. This virus causes COVID-19, has infected millions of people, and has led to hundreds of thousands of deaths across the globe. While the best interventions to control and ultimately stop the pandemic are prophylactic vaccines, antiviral therapeutics are important to limit morbidity and mortality in those already infected. At this time, only one FDA-approved anti-SARS-CoV-2 antiviral drug, remdesivir, is available, and unfortunately, its efficacy appears to be limited. Thus, the identification of new and efficacious antivirals is of the highest importance. In order to facilitate rapid drug discovery, flexible, sensitive, and high-throughput screening methods are required. With respect to drug targets, most attention is focused on either the viral RNA-dependent RNA polymerase or the main viral protease, 3CLpro. 3CLpro is an attractive target for antiviral therapeutics, as it is essential for processing newly translated viral proteins and the viral life cycle cannot be completed without protease activity. In this work, we report a new assay to identify inhibitors of 3CLpro. Our reporter is based on a green fluorescent protein (GFP)-derived protein that fluoresces only after cleavage by 3CLpro. This experimentally optimized reporter assay allows for antiviral drug screening in human cell culture at biosafety level 2 (BSL2) with high-throughput compatible protocols. Using this screening approach in combination with existing drug libraries may lead to the rapid identification of novel antivirals to suppress SARS-CoV-2 replication and spread.
IMPORTANCE The COVID-19 pandemic has already led to more than 700,000 deaths and innumerable changes to daily life worldwide. Along with development of a vaccine, identification of effective antivirals to treat infected patients is of the highest importance. However, rapid drug discovery requires efficient methods to identify novel compounds that can inhibit the virus. In this work, we present a method for identifying inhibitors of the SARS-CoV-2 main protease, 3CLpro. This reporter-based assay allows for antiviral drug screening in human cell culture at biosafety level 2 (BSL2) with high-throughput compatible sample processing and analysis. This assay may help identify novel antivirals to control the COVID-19 pandemic.
INTRODUCTION
In December 2019, a novel human coronavirus (hCoV) was identified in the Hubei Province of China (1–3). The virus, now known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causes the transmissible and pathogenic disease COVID-19 (4). COVID-19 has become a global pandemic and infected over 8 million people and caused ∼800,000 deaths to date (5). Current efforts to control COVID-19 are largely focused on behavioral modifications such as social distancing and the use of masks (6). These approaches attempt to slow the spread of the virus, but meaningful control of the virus will ultimately be the result of a combination of efficacious vaccines and antiviral therapeutics (7).
Antiviral therapeutics aim to disrupt the replication cycle and reduce viral load in infected individuals. Therapeutic development efforts have led to a number of candidate antiviral compounds focused mainly on two essential viral enzymes, the RNA-dependent RNA polymerase (RdRp) and the main viral protease. Remdesivir (GS-5734), recently FDA approved as an antiviral for SARS-CoV-2, targets the polymerase to suppress hCoV replication by inducing termination of RNA polymerization (8); however, the benefits of this drug in clinical trials and early use appear limited (9). Another nucleoside analogue, β-d-N4-hydroxycytidine (NHC; EIDD-1931), also inhibits SARS-CoV-2 polymerase activity, likely via inducing lethal mutagenesis of the viral genome (10). In addition to the RdRp, the viral proteases, which are critical to liberate individual viral proteins from the polyprotein produced by initial genome translation, present another attractive drug target. For SARS-CoV-2, lopinavir/ritonavir, a protease inhibitor combination, is shown to interact with the main coronavirus protease, known as 3CLpro or Mpro (11); however, early clinical trial results with these compounds have shown no significant benefits to SARS-CoV-2 patients (12). More recently, structure-based design has enabled the rapid development of new antivirals targeting the SARS-CoV-2 protease 3CLpro (13–15). At this time, these newly designed compounds are in the early stages of testing. Thus, the discovery of additional effective SARS-CoV-2 antiviral drugs remains of high importance. The identification (and subsequent improvement) of novel drugs targeting SARS-CoV-2 will require robust and high-throughput screening approaches.
Here, we report the development and validation of a fluorescent reporter optimized to detect SARS-CoV-2 3CLpro activity. This assay is performed in human cell culture and does not require biosafety level 3 (BSL3) containment. Our reporter is based on FlipGFP, which fluoresces only after protease-mediated activation (16). We generated and tested three reporter constructs with distinct cleavage target sequences for activation by the SARS-CoV-2 3CLpro. We also show that the reporter with the best signal-to-noise ratio for SARS-CoV-2 is also activable by other coronavirus 3CLpro proteins across subgroups (Betacoronavirus, Alphacoronavirus, and Gammacoronavirus) and host species (human, rodent, and bird). Finally, we used this reporter to test the inhibition of SARS-CoV-2 3CLpro with a known coronavirus 3CLpro inhibitor, GC376 (17), and then validated the correlation between reporter inhibition and inhibition of SARS-CoV-2 replication. These experiments together demonstrate the utility of this approach for the identification of novel antiviral drugs that target the SAR-CoV-2 main protease, 3CLpro.
RESULTS
Generation of a fluorescent SARS-CoV-2 3CLpro activity reporter.
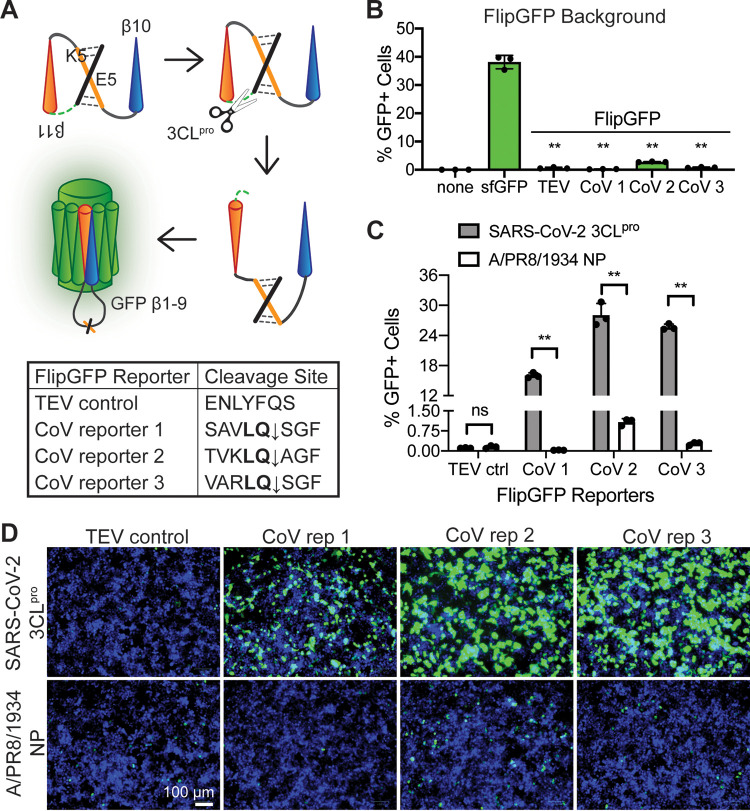
In order to develop a fluorescent reporter responsive to the SARS-CoV-2 main protease, we started with the FlipGFP protein (16). FlipGFP is used to detect protease activity by expressing the green fluorescent protein (GFP) 10th and 11th beta-strands, β10-11, separately from, and in a conformation incompatible with, the rest of the GFP beta-barrel, β1-9 (Fig. 1A, top). A linker containing a cleavage site holds the two GFP beta-strands, β10-11, in an inactive, parallel conformation. When the appropriate protease is present, the linker containing the cleavage site is cut. This cleavage allows GFP β11 to reorient such that GFP β10-11 are antiparallel and able to fit into GFP β1-9, inducing fluorescence ∼100-fold over background (16).
FIG 1.
A FlipGFP protease reporter with coronavirus cleavage sites fluoresces after SARS-CoV-2 3CLpro expression. (A) Diagram of the FlipGFP protease reporter (16) with coronavirus cleavage sequences. FlipGFP splits GFP into β1-9 and β10-11, with β11 held in parallel to β10 by heterodimerized coiled coils E5/K5 and a linker sequence containing a coronavirus cleavage site. The CoV main protease, 3CLpro, cuts at the cleavage site, allowing β11 to “flip” antiparallel to β10, enabling self-assembly of the complete GFP beta-barrel and resulting in detectable fluorescence. The pan-coronavirus 3CLpro consensus sequence, LQ, is in bold. (B) Quantification of fluorescence from 293T cells 48 h after transfection with each FlipGFP reporter or superfolder GFP (sfGFP) individually and without a protease. Statistical analysis is relative to sfGFP. (C) Quantification of fluorescence from 293T cells 48 h after transfection with each FlipGFP reporter and either the SARS-CoV-2 3CLpro or an influenza virus protein (A/PR8/1834 NP). Statistical analysis is relative to NP control. (D) Images corresponding to panel C. Green, cleaved FlipGFP; blue, nuclei. Data are shown as means ± SDs (n = 3). P values were calculated using unpaired, two-tailed Student’s t tests (*, P < 0.05; **, P < 0.001; ns, not significant). Experiments were performed twice.
SARS-CoV-2 generates two proteases that cleave the viral polyprotein, a papain-like protease (PLpro) and a chymotrypsin-like protease (3CLpro). 3CLpro, also known as the main protease or Mpro, is the more conserved viral protease, with only 5 amino acid changes between SARS/SARS-like CoVs and SARS-CoV-2, compared to the 102 differences found in PLpro (18). 3CLpro cleaves at a consensus sequence, LQ↓, which is highly conserved across the coronavirus family (19). Additionally, 3CLpro has been shown to effectively cleave luciferase-based protease biosensors (20, 21) and fluorescence resonance energy transfer (FRET)-based probes (13, 17, 22–28). With the aim of generating a protease reporter compatible with SARS-CoV-2 and other present and future coronaviruses to support viral inhibitor screening, we selected CoV 3CLpro as our protease target.
Although CoV 3CLpro requires the minimal consensus sequence LQ↓ for cleavage, the cleavage site context influences cleavage efficiency (29). Among the CoV polyprotein cleavage sites, 3CLpro targeted sites surrounding the 3CLpro sequence itself are generally cleaved most efficiently (30). Additionally, different CoVs have distinctive optimal cleavage site sequences (24). In order to develop an efficiently cleaved CoV 3CLpro reporter, we tested three different cleavage sequences predicted to be highly compatible with the SARS-CoV-2 3CLpro (Fig. 1A, bottom). CoV reporter 1 contains the conserved nsp4-5 cleavage site present in the SARS-CoV and SARS-CoV-2 polyproteins (31). CoV reporter 2 contains an optimized cleavage sequence for the SARS-CoV 3CLpro (32). CoV reporter 3 contains an optimized sequence shown to be highly cleaved by many CoV family members (24). As a negative control, we included a construct harboring the tobacco etch virus (TEV) protease cleavage site, as in the initial FlipGFP construct (16). To observe whether these FlipGFP constructs background fluoresced without CoV 3CLpro activity, we transfected cells with each reporter or a superfolder GFP (sfGFP) expression plasmid. Compared to sfGFP, which is properly folded and fluorescing, the inactive FlipGFP constructs produced significantly lower fluorescence, from a 10- to 100-fold reduction depending on the reporter (Fig. 1B; see also Fig. S2 in the supplemental material).
Our goal was to identify a construct that showed minimal background fluorescence while still being efficiently cleaved by SARS-CoV-2 3CLpro, allowing strong fluorescence for detection via microscopy, plate reader, or flow cytometry. To test our 3CLpro reporters, we cotransfected each reporter with a SARS-CoV-2 3CLpro expression plasmid. At 48 h posttransfection, we could detect GFP-positive cells with each of the three CoV reporters transfected with the SARS-CoV-2 3CLpro (Fig. 1C and D). In contrast, with transfection of a negative control, nucleoprotein from an H1N1 influenza virus (A/PR8/1934 NP), we did not detect any signal above background levels of fluorescence. Further, the reporter containing the TEV cleavage site was not activated by SARS-CoV-2 3CLpro. Fluorescent signal across the treatment conditions demonstrated that while all three CoV reporters showed significant induction of GFP signal when coexpressed with the SARS-CoV-2 3CLpro, CoV reporter 2 had substantial background and the level of induction with CoV reporter 1 reached only half that of the other two CoV reporters. With a 100-fold change in fluorescence and minimal background, we selected CoV reporter 3 for further testing (Fig. 1C and D).
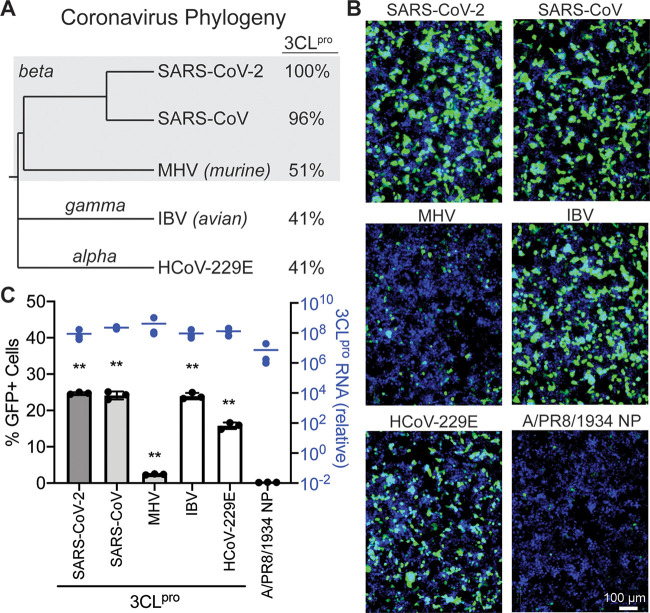
Many CoV 3CLpro proteins activate the FlipGFP CoV 3CLpro reporter.
CoV 3CLpro proteins are reasonably conserved across coronavirus groups (Fig. 2A) (33). Further, CoV reporter 3 was based on an optimized cleavage sequence for CoV 3CLpros from each coronavirus group (24). To test whether this protease reporter was compatible with a variety of CoV 3CLpro proteins, we expressed CoV reporter 3 with four other coronavirus proteases from different groups (Alphacoronavirus, Betacoronavirus, and Gammacoronavirus) and host species (human, mouse, and bird). At 48 h after transfection, all CoV 3CLpros induced visible fluorescence compared to the control influenza virus nucleoprotein with CoV reporter 3 (Fig. 2B). Expression of the CoV 3CLpro or PR8 NP constructs was quantified using quantitative PCR (qPCR) (Fig. 2C). Quantification with a plate reader demonstrated that SARS-CoV (Betacoronavirus, human) and avian infectious bronchitis virus (IBV; Gammacoronavirus, avian) resulted in levels of fluorescence similar to that with SARS-CoV-2 (Betacoronavirus, human) (Fig. 2C). Murine hepatitis virus (MHV; Betacoronavirus, murine) and human coronavirus 229E (HCoV-229E; Alphacoronavirus, human) were less compatible with CoV reporter 3, while still producing 12- and 80-fold changes in fluorescence, respectively, over background (Fig. 2C). These experiments show that our FlipGFP 3CLpro reporter is generally compatible with many CoV 3CLpro proteins across coronavirus groups and host species, potentially enabling protease inhibitor screening for a variety of CoVs in addition to SARS-CoV-2.
FIG 2.
Conservation of coronavirus 3CLpro activity enables CoV protease reporter compatibility with many coronaviruses. (A) Phylogenetic tree of five coronaviruses, SARS-CoV-2, SARS-CoV, murine hepatitis virus (MHV), avian infectious bronchitis virus (IBV), and HCoV-229E, generated based on the polyprotein ORF1ab using NCBI Virus (34). These viruses span three coronavirus groups: Alphacoronavirus, Betacoronavirus, and Gammacoronavirus. 3CLpro protein sequence identities are compared to the SARS-CoV-2 3CLpro. (B) Microscopy of 293T cells 48 h after transfection with CoV reporter 3 and coronavirus 3CLpro proteins or an influenza virus protein (A/PR8/1834 NP). Green, cleaved FlipGFP; blue, nuclei. (C) In black is quantification of the data in panel B. In blue are results of qPCR of CoV 3CLpro or PR8 NP RNA levels relative to untransfected cells. Data are shown as means ± SDs (n = 3); statistical analysis is relative to the NP control. P values were calculated using unpaired, two-tailed Student’s t tests (*, P < 0.05; **, P < 0.001). Experiments were performed twice.
Development of a FlipGFP CoV 3CLpro reporter-based assay for protease inhibitor screening in human cells.
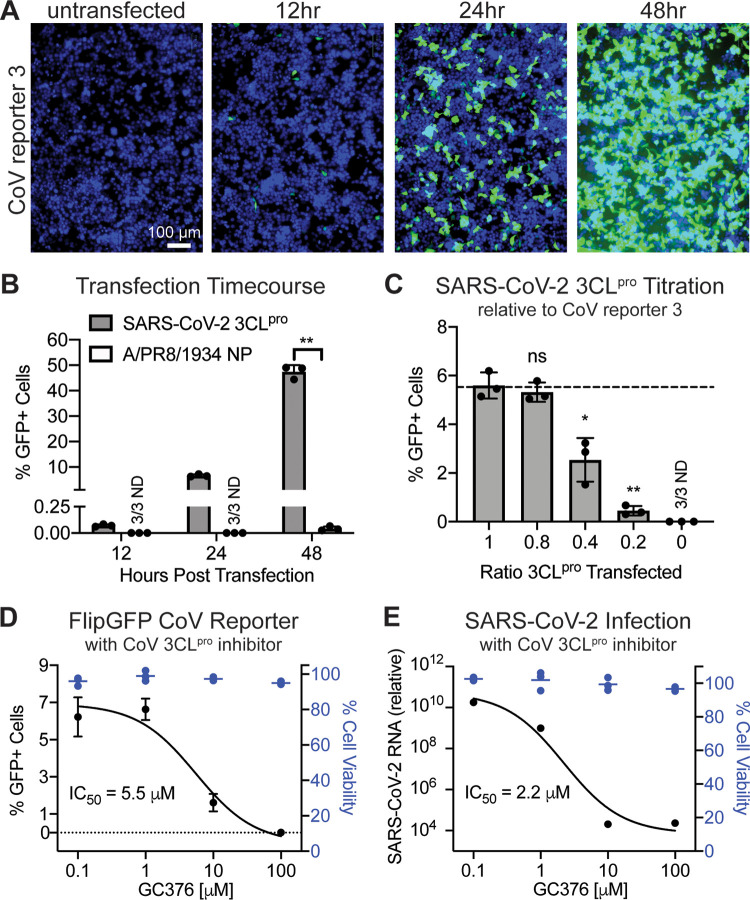
To develop an assay for protease inhibitor screening using our CoV reporter 3, we first needed to optimize the experimental conditions. We performed a transfection time course with SARS-CoV-2 3CLpro to determine an early, appropriate time point for sample collection (Fig. 3A). At 12 h posttransfection, only a few GFP fluorescing cells were visible and fluorescent signal was just above background (Fig. 3B). At 24 h posttransfection, green cells were visible without appreciable background signaling (Fig. 3B). At 48 h postinfection, most cells produced a high GFP signal, with some background fluorescence detectable (Fig. 3B). We therefore selected the 24-h posttransfection time point. To increase the sensitivity of our assay, we titrated the level of SARS-CoV-2 3CLpro transfected with CoV reporter 3; our goal was to maximize activation of the reporter while minimizing the amount of protease available in the cell. We transfected cells with five ratios of reporter to protease: 1:1, 1:0.8, 1:0.4, 1:0.2, and 1:0. At 24 h posttransfection, we observed significant decreases in reporter activation at reporter-to-protease ratios of 1:0.4 and 1:0.2 (Fig. 3C). However, a 1:0.8 reporter-to-protease ratio resulted in no significant loss of fluorescence compared to that with a 1:1 ratio (Fig. 3C). Based on these experiments together, we selected a 1:0.8 reporter-to-protease ratio for transfection and a 24-h posttransfection endpoint as the optimal conditions for our protease inhibitor assay using FlipGFP 3CLpro reporter 3.
FIG 3.
Inhibition of the SARS-CoV-2 3CLpro by the protease inhibitor GC376 is measurable with the fluorescent CoV protease reporter. (A) Microscopy of 293T cells before or 12, 24, and 48 h after transfection with CoV reporter 3 and SARS-CoV-2 3CLpro. Green, cleaved FlipGFP; blue, nuclei. (B) Quantification of data in panel A. Data are shown as means ± SDs (n = 3); statistical analysis is relative to the NP control. P values were calculated using unpaired, two-tailed Student’s t tests (*, P < 0.05; **, P < 0.001). (C) Quantification of 293T cells 24 h after transfection with CoV reporter 3 and SARS-CoV-2 3CLpro, with decreasing levels of 3CLpro. Data are shown as means ± SDs (n = 3); statistical analysis is relative to a 1:1 ratio of reporter to protease. P values were calculated using unpaired, two-tailed Student’s t tests (*, P < 0.05; **, P < 0.001). (D) In black is quantification of 293T cells 24 h after transfection with CoV reporter 3 and SARS-CoV-2 3CLpro and treatment with the pan-coronavirus protease inhibitor GC376. Data are shown as means ± SDs with nonlinear fit curve (n = 3). In blue is calculation of cell viability relative to vehicle-only (DMSO) samples. (E) In black are results of RT-qPCR of VeroE6 cells 24 h after infection with SARS-CoV-2 at an MOI of 0.01 and treatment with the pan-coronavirus protease inhibitor GC376. Data are shown as means ± SDs with nonlinear fit curve (n = 4). In blue is calculation of cell viability relative to vehicle-only (DMSO) samples. Data are shown as means ± SDs (n = 3). Experiments were performed twice.
Finally, we wanted to verify that our assay could detect drug inhibition of the SARS-CoV-2 3CLpro with a known inhibitor. Therefore, we selected a recognized pan-coronavirus 3CLpro inhibitor, GC376, to test our assay (17). Four concentrations of GC376, that did not significantly impact cell viability compared to vehicle alone, were applied to cells at the time of transfection with CoV reporter 3 and SARS-CoV-2 3CLpro. As expected, reporter activity levels were maintained at the lower protease inhibitor concentrations, while fluorescence was reduced at the higher concentrations of GC376 (Fig. 3D). Thus, our assay successfully detected inhibition of SARS-CoV-3 3CLpro by the protease inhibitor GC376. However, it is also important to verify that inhibition of our reporter is strongly correlated with inhibition of SARS-CoV-2. We infected VeroE6 cells with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.01 before applying protease inhibitor at the same four concentrations as tested with the protease reporter. At 24 h postinfection, we collected RNA and performed reverse transcription (RT)-qPCR to detect SARS-CoV-2 RNA; similar to the case with the reporter, viral RNA levels were suppressed in a dose-dependent manner (Fig. 3E). Our observed inhibition of the virus is consistent with reports of inhibition of SARS-CoV-2 by GC376 in the literature (22, 23). Together, these experiments demonstrate feasibility of using our FlipGFP CoV 3CLpro reporter assay to identify protease-targeting inhibitors of SARS-CoV-2.
DISCUSSION
Our goal for this study was to develop a cell-based assay to screen for novel SARS-CoV-2 antiviral drugs at BSL2; to our knowledge, no such assay optimized for SARS-CoV-2 currently exists. Therefore, we generated a reporter requiring a coronavirus protease, 3CLpro, for activation of a GFP fluorescent signal. We showed that this reporter is responsive to the SARS-CoV-2 3CLpro, in addition to many different coronavirus 3CLpro proteins. After optimizing screening conditions, we demonstrated that our reporter was sensitive to treatment with a known coronavirus protease inhibitor, GC376. These experiments illustrate the utility of our approach to identify, and subsequently optimize, novel protease inhibitors of SARS-CoV-2.
To meet the demands of virus research during the SARS-CoV-2 pandemic, reporter assays need to be flexible and high-throughput. Our reporter is activated with expression of a single CoV protein, 3CLpro, allowing for SARS-CoV-2 drug testing at BSL2. Additionally, the reporter is compatible with many CoV 3CLpro proteins, supporting rapid testing of inhibitors against a variety of coronaviruses, present or future, and without synthesis of protease substrates or purification of viral proteins (13, 17, 22–28). Further, as our assay is performed in living cells, our system enables the discovery of protease inhibitors while simultaneously evaluating effects on cellular viability. Our assay is scalable, and the analysis requires only a basic fluorescent plate reader, supporting high-throughput screening. In addition to applications in drug discovery pipelines, this assay could be deployed to determine targets of antivirals identified via viral screening.
Reporter assays, including ours, also have limitations. Our reporter utilizes CoV 3CLpro expressed alone; during a CoV infection, the protease is only one of many viral proteins present, and any inhibitors that may affect cross-viral protein interactions would be missed. Additionally, CoV infection induces significant cellular membrane rearrangements that transfection of the protease alone does not. Thus, the subcellular access of therapeutic compounds to the viral protease may fail to be reflected in our assay, and the effects of an identified protease inhibitor could significantly differ when applied to authentic viral infection. Finally, although this plasmid-based expression presents many advantages, it also necessitates further screen hit testing in the context of coronavirus infection. Although the correlation between our reporter and inhibition of viral infection was appreciable with the drug GC376, testing of more inhibitors is required to make generalizable correlations between our reporter assay and viral infection readouts.
To have the greatest impact on the COVID-19 pandemic, an effective SARS-CoV-2 antiviral needs to be identified as early as possible. Countries around the world have taken drastic and necessary steps to limit the spread of virus, yet infection rates continue to rise in some. An antiviral treatment is unlikely to stop the spread of infection, but it is likely to limit the mortality associated with SARS-CoV-2 infection. It is our hope that this reporter assay facilitates the identification of SARS-CoV-2 protease inhibitor candidates to be rapidly optimized and translated to clinical use.
MATERIALS AND METHODS
Cell culture.
All cells were obtained from the ATCC and grown at 37°C in 5% CO2. 293T cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 5% fetal bovine serum (FBS), GlutaMAX, and penicillin-streptomycin. VeroE6 cells were grown in minimum essential medium (MEM) supplemented with 10% fetal bovine serum, pyruvate, nonessential amino acids (NEAA), and penicillin-streptomycin. For transfection, plates were polylysine treated and seeded with 293Ts. Twenty-four hours later, plasmid DNA, Opti-MEM, and TransIT-LT (Mirus) were combined using pipetting and incubated at room temperature for 20 min before being added to cells by droplet.
Plasmids.
Constructs (excluding CoV reporters 1, 2, and 3) were cloned into the pLEX expression vector using the BamHI and NotI restriction sites and DNA assembly (New England BioLabs [NEB]). Coronavirus 3CLpro expression plasmids (SARS-CoV-2, SARS-CoV, MHV, IBV, and HCoV-229E) were generated using codon-optimized gBlocks (IDT). The TEV control FlipGFP (Addgene; number 124429) was designed to include a silent NheI restriction site ahead of the TEV cleavage sequence and generated using a gBlock (IDT) (Fig. S1) with primers FlipGFP For and FlipGFP Rev (Table S1) for insertion into BamHI- and NotI-digested pLEX. The coronavirus 3CLpro FlipGFP reporters (reporters 1, 2, and 3) were generated using forward primers containing the cleavage sequences (CoV rep 1 For, CoV rep 2 For, and CoV rep 3 For) along with the FlipGFP Rev primer (Table S1) and assembled into NheI- and NotI-digested TEV control FlipGFP plasmid (Table S1). DNA was transformed into NEB 5-alpha high-efficiency competent cells (NEB). Insert size was verified with PCR, and purified plasmids were sequenced using Sanger sequencing.
Imaging and quantification.
Cells were fixed with 2% paraformaldehyde (PFA) at room temperature for 20 min before incubation in 1:10,000 Hoechst 33342 (Life Technologies) in phosphate-buffered saline (PBS) at 4°C overnight. Images were obtained using the ZOE fluorescent cell imager (Bio-Rad). Quantification was performed using the CellInsight CX5 platform (Thermo Scientific).
CoV 3CLpro and PR8 NP qPCR.
At 48 h after transfection of 293Ts, RNA was prepped according to the RNeasy 96 kit (Qiagen). One-step RT-PCR was performed using primers targeting the respective CoV 3CLpro (SARS-CoV-2, SARS-CoV, MHV, IBV, or 229E) or PR8 NP and a housekeeping gene (18S) with the iTaq universal SYBR green one-step kit (Bio-Rad) on an Applied Biosystems QuantStudio3 instrument.
Cytotoxicity assays.
At 24 h before treatment, plates were polylysine treated and seeded with 293T or VeroE6 cells. The next day, medium was replaced with complete medium containing GC376 (MedKoo) at the desired concentrations using a constant level of a vehicle (dimethyl sulfoxide [DMSO]). After 24 h of treatment, cells were collected according the CellTiter-GLO (Promega) protocol and luminescence levels assessed using a luminometer.
SARS-CoV-2 infections and viral qPCR.
At 24 h before infection, plates were polylysine treated and seeded with VeroE6 cells. The next day, the cells were washed with PBS before infection with SARS-CoV-2 isolate USA-WA1/2020 from BEI Resources in 2% FBS-MEM infection medium at an MOI of 0.01 for 1 h. Virus was removed and cells were then placed in infection media containing GC376 (MedKoo) at the desired concentrations using a constant level of a vehicle (DMSO). At 24 h postinfection, cells were collected in TRIzol (Invitrogen), followed by RNA isolation. One-step RT-qPCR was performed with primers targeting the SARS-CoV-2 N region (BEI) using the EXPRESS one-step Superscript qRT-PCR kit (Thermo Fisher) on an Applied Biosystems QuantStudio3 instrument. RNA was normalized using an endogenous 18S rRNA primer/probe set (Applied Biosystems).
Supplementary Material
ACKNOWLEDGMENTS
N.S.H. is partially funded by the Defense Advanced Research Projects Agency’s (DARPA) PReemptive Expression of Protective Alleles and Response Elements (PREPARE) program (cooperative agreement number HR00111920008).
The views, opinions, and/or findings expressed are those of the authors and should not be interpreted as representing the official views or policies of the U.S. government.
The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-related coronavirus 2, isolate USA-WA1/2020, NR-52281.
Biocontainment work was performed in the Duke Regional Biocontainment Laboratory, which received partial support for construction from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (UC6-AI058607).
We thank Clare Smith for help establishing SARS-CoV-2 infection assays at BSL3 and Laura Froggatt for designing the FlipGFP diagram.
Duke University may file for intellectual property protection for the technology described in this report.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. 2020. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, Xing F, Liu J, Yip CC-Y, Poon RW-S, Tsoi H-W, Lo SK-F, Chan K-H, Poon VK-M, Chan W-M, Ip JD, Cai J-P, Cheng VC-C, Chen H, Hui CK-M, Yuen K-Y. 2020. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johns Hopkins Coronavirus Resource Center. 2020. COVID-19 map. https://coronavirus.jhu.edu/map.html.
- 6.Chu DK, Akl EA, Duda S, Solo K, Yaacoub S, Schünemann HJ, Akl EA, El-Harakeh A, Bognanni A, Lotfi T, Loeb M, Hajizadeh A, Bak A, Izcovich A, Cuello-Garcia CA, Chen C, Harris DJ, Borowiack E, Chamseddine F, Schünemann F, Morgano GP, Schünemann GEUM, Chen G, Zhao H, Neumann I, Chan J, Khabsa J, Hneiny L, Harrison L, Smith M, Rizk N, Rossi PG, AbiHanna P, El-Khoury R, Stalteri R, Baldeh T, Piggott T, Zhang Y, Saad Z, Khamis A, Reinap M, Duda S, Solo K, Yaacoub S, Schünemann HJ, COVID-19 Systematic Urgent Review Group Effort (SURGE) study authors. 1 June 2020. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet doi: 10.1016/S0140-6736(20)31142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tse LV, Meganck RM, Graham RL, Baric RS. 2020. The current and future state of vaccines, antivirals and gene therapies against emerging coronaviruses. Front Microbiol 11:658. doi: 10.3389/fmicb.2020.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I, Bannister R, Park Y, Babusis D, Mo C, Mackman RL, Spahn JE, Palmiotti CA, Siegel D, Ray AS, Cihlar T, Jordan R, Denison MR, Baric RS. 2017. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, Fu S, Gao L, Cheng Z, Lu Q, Hu Y, Luo G, Wang K, Lu Y, Li H, Wang S, Ruan S, Yang C, Mei C, Wang Y, Ding D, Wu F, Tang X, Ye X, Ye Y, Liu B, Yang J, Yin W, Wang A, Fan G, Zhou F, Liu Z, Gu X, Xu J, Shang L, Zhang Y, Cao L, Guo T, Wan Y, Qin H, Jiang Y, Jaki T, Hayden FG, Horby PW, Cao B, Wang C. 2020. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, Leist SR, Schäfer A, Dinnon KH, Stevens LJ, Chappell JD, Lu X, Hughes TM, George AS, Hill CS, Montgomery SA, Brown AJ, Bluemling GR, Natchus MG, Saindane M, Kolykhalov AA, Painter G, Harcourt J, Tamin A, Thornburg NJ, Swanstrom R, Denison MR, Baric RS. 2020. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med 12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nutho B, Mahalapbutr P, Hengphasatporn K, Pattaranggoon NC, Simanon N, Shigeta Y, Hannongbua S, Rungrotmongkol T. 2020. Why are lopinavir and ritonavir effective against the newly emerged coronavirus 2019? Atomistic insights into the inhibitory mechanisms. Biochemistry 59:1769–1779. doi: 10.1021/acs.biochem.0c00160. [DOI] [PubMed] [Google Scholar]
- 12.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, Ruan L, Song B, Cai Y, Wei M, Li X, Xia J, Chen N, Xiang J, Yu T, Bai T, Xie X, Zhang L, Li C, Yuan Y, Chen H, Li H, Huang H, Tu S, Gong F, Liu Y, Wei Y, Dong C, Zhou F, Gu X, Xu J, Liu Z, Zhang Y, Li H, Shang L, Wang K, Li K, Zhou X, Dong X, Qu Z, Lu S, Hu X, Ruan S, Luo S, Wu J, Peng L, Cheng F, Pan L, Zou J, Jia C, et al. 2020. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N Engl J Med 382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai W, Zhang B, Su H, Li J, Zhao Y, Xie X, Jin Z, Liu F, Li C, Li Y, Bai F, Wang H, Cheng X, Cen X, Hu S, Yang X, Wang J, Liu X, Xiao G, Jiang H, Rao Z, Zhang L-K, Xu Y, Yang H, Liu H, Yang H, Liu H. 2020. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. 2020. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y, Yu J, Wang L, Yang K, Liu F, Jiang R, Yang X, You T, Liu X, Yang X, Bai F, Liu H, Liu X, Guddat LW, Xu W, Xiao G, Qin C, Shi Z, Jiang H, Rao Z, Yang H. 2020. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Q, Schepis A, Huang H, Yang J, Ma W, Torra J, Zhang SQ, Yang L, Wu H, Nonell S, Dong Z, Kornberg TB, Coughlin SR, Shu X. 2019. Designing a green fluorogenic protease reporter by flipping a beta strand of GFP for imaging apoptosis in animals. J Am Chem Soc 141:4526–4530. doi: 10.1021/jacs.8b13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y, Lovell S, Tiew K-C, Mandadapu SR, Alliston KR, Battaile KP, Groutas WC, Chang K-O. 2012. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J Virol 86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T. 2020. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiemer L, Lund O, Brunak S, Blom N. 2004. Coronavirus 3CLpro proteinase cleavage sites: possible relevance to SARS virus pathology. BMC Bioinformatics 5:72. doi: 10.1186/1471-2105-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J, Fang L, Yang Z, Xu S, Lv M, Sun Z, Chen J, Wang D, Gao J, Xiao S. 2019. Identification of novel proteolytically inactive mutations in coronavirus 3C-like protease using a combined approach. FASEB J 33:14575–14587. doi: 10.1096/fj.201901624RR. [DOI] [PubMed] [Google Scholar]
- 21.Kilianski A, Mielech AM, Deng X, Baker SC. 2013. Assessing activity and inhibition of Middle East respiratory syndrome coronavirus papain-like and 3C-like proteases using luciferase-based biosensors. J Virol 87:11955–11962. doi: 10.1128/JVI.02105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma C, Sacco MD, Hurst B, Townsend JA, Hu Y, Szeto T, Zhang X, Tarbet B, Marty MT, Chen Y, Wang J. 2020. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res 30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vuong W, Khan MB, Fischer C, Arutyunova E, Lamer T, Shields J, Saffran HA, McKay RT, van Belkum MJ, Joyce MA, Young HS, Tyrrell DL, Vederas JC, Lemieux MJ. 2020. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat Commun 11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuck C-P, Chow H-F, Wan DC-C, Wong K-B. 2011. Profiling of substrate specificities of 3C-like proteases from group 1, 2a, 2b, and 3 coronaviruses. PLoS One 6:e27228. doi: 10.1371/journal.pone.0027228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng X, StJohn SE, Osswald HL, O’Brien A, Banach BS, Sleeman K, Ghosh AK, Mesecar AD, Baker SC. 2014. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J Virol 88:11886–11898. doi: 10.1128/JVI.01528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanchard JE, Elowe NH, Huitema C, Fortin PD, Cechetto JD, Eltis LD, Brown ED. 2004. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem Biol 11:1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y, Liu H, Galasiti Kankanamalage AC, Weerasekara S, Hua DH, Groutas WC, Chang K-O, Pedersen NC. 2016. Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog 12:e1005531. doi: 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jo S, Kim S, Shin DH, Kim M-S. 2020. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem 35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegyi A, Ziebuhr J. 2002. Conservation of substrate specificities among coronavirus main proteases. J Gen Virol 83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- 30.Fan K, Wei P, Feng Q, Chen S, Huang C, Ma L, Lai B, Pei J, Liu Y, Chen J, Lai L. 2004. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J Biol Chem 279:1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen YW, Yiu C-PB, Wong K-Y. 2020. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CLpro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Res 9:129. doi: 10.12688/f1000research.22457.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan K, Ma L, Han X, Liang H, Wei P, Liu Y, Lai L. 2005. The substrate specificity of SARS coronavirus 3C-like proteinase. Biochem Biophys Res Commun 329:934–940. doi: 10.1016/j.bbrc.2005.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stobart CC, Sexton NR, Munjal H, Lu X, Molland KL, Tomar S, Mesecar AD, Denison MR. 2013. Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J Virol 87:12611–12618. doi: 10.1128/JVI.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brister JR, Ako-Adjei D, Bao Y, Blinkova O. 2015. NCBI viral genomes resource. Nucleic Acids Res 43:D571–D577. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.