Abstract
The immune response is orchestrated by a variety of immune cells. The function of each cell is determined by the collective signals from various immunoreceptors, whose expression and activity depend on the developmental stages of the cell and its environmental context. Recent studies have highlighted the presence of mechanical force on several immunoreceptor–ligand pairs and the important role of force in regulating their interaction and function. In this Perspective, we use the T cell antigen receptor as an example with which to review the current understanding of the mechanosensing properties of immunoreceptors. We discuss the types of forces that immunoreceptors may encounter and the effects of force on ligand bonding, conformational change and the triggering of immunoreceptors, as well as the effects of force on the downstream signal transduction, cell-fate decisions and effector function of immune cells.
Immune cells experience myriad forces as they traffic through the body’s organs and tissues via the blood and lymphatic circulation to carry out immunosurveillance and effector functions. The action of forces in vivo can be readily inferred from the observable deformations of immune cells, neighboring cells or the extracellular matrix (ECM)1. In vitro studies have shown that endogenous forces are applied on the T cell antigen receptor (TCR)2–7 and B cell antigen receptor (BCR)8–12 during lymphocyte activation. Furthermore, exogenously applied forces modulate the kinetics of the dissociation of ligand from the TCR13–19, pre-TCR20,21, BCR22–25 and FcγRIIA (the receptor for the Fc fragment of immunoglobulin G (IgG))26, induce conformational changes in bonds between the TCR or pre-TCR and complexes of peptide and major histocompatibility complex (pMHC)14,19,21, trigger TCR signaling15,27–31 and potentiate the killing of target cells by cytotoxic T cells32. Moreover, lymphocytes use force to amplify antigen discrimination14–16,21,33 and respond to changes in substrate stiffness29,34–40. Forces exerted on specific immunoreceptor–ligand bonds are transmitted across the cell membrane and potentially induce mechanotransduction. As mechanobiology intersects with immunology, interest in exploring how immune cells sense, respond to and adapt to their mechanical environment is rapidly growing. In this Perspective, we review recent advances and suggest future directions for the emerging field of mechanoimmunology41–58, using the TCR as a prototypical immunoreceptor.
Immunoreceptors experience mechanical forces
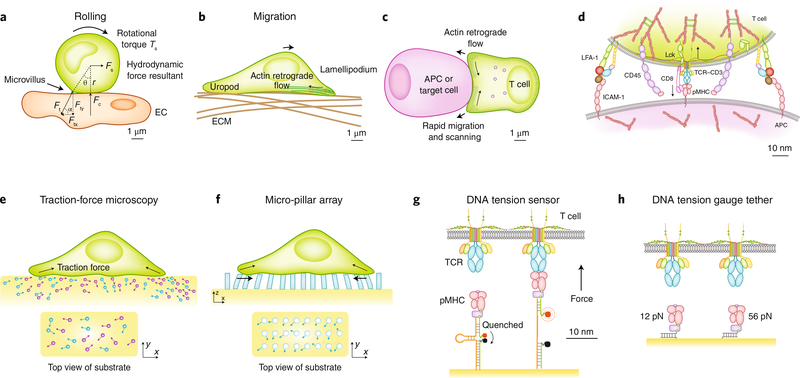
Mechanical forces on immunoreceptors emerge as an immune cell adheres to (Fig. 1a) and/or migrates on (Fig. 1b) another cell or the ECM and forms an immunological synapse with an antigen-presenting cell (APC) (Fig. 1c). For example, Fcγ receptors mediate the binding of neutrophils and monocytes to IgG immunocomplexes deposited on vascular endothelium26,59 (Fig. 1a). Mechanical analysis shows that the receptor–ligand complex bears a tensile force along its long axis with an inclined angle to the interface between the immune cell and endothelium. This force is needed to balance the normal force from the endothelium and the lateral force and rotational torque imposed by blood flow42,60. A T cell uses its TCRs to search for antigens during migration, a movement that is powered by cell-generated forces on integrins (Fig. 1b). Once engaged with pMHC complexes, the TCRs must resist relative displacements between the moving cell and the stationary substrate surface that result in forces on TCRs. During the formation of immunological synapses, TCRs are driven by actin retrograde flow for spatial reorganization61,62; this flow results in forces on the TCR–pMHC bonds that prevent the TCRs from moving away from the stationary pMHC complexes5–7,30 (Fig. 1c). On a finer scale, TCRs form bonds with pMHC complexes against steric repulsion from molecules of larger ectodomains such as integrin αLβ2 (LFA-1) and the phosphatase CD45, which are also present at the interfacial junction between the T cell and the APC (Fig. 1d). The cross-junctional gap must be pulled closer for the TCRs and pMHC complexes to reach each other, which results in tension in their bonds along the normal direction of the interface.
Fig. 1 |. Immune cells are subjected to external forces from, and apply endogenous forces to, their surroundings, which can be measured by biophysical techniques.
a, A lymphocyte rolling on the endothelium experiences forces. Blood flow applies a tangential force Fs to the cell center and a rotational torque Ts, which are supported by the tether force Ft at the rear with an inclined angle. The tangential component Ftx balances Fs. The normal component Fty generates a torque Fty × r × sinθ to balance Ts and the torque of the tangential force Ftx × r × cosθ. Fty also induces a reaction Fc from the endothelium. b,c, During migration (b) and formation of the immunological synapse (c), a lymphocyte can apply endogenous forces, generated by actomyosin contraction and actin retrograde flow, on the ECM (b) or the APC (c). d, The TCR and CD8, localized at the tip of the microvillus of a T cell interacting with an APC, bind pMHC in the center, while LFA-1 binds to ICAM-1 at the periphery. CD45 is uniformly distributed. e–g, TFM (e), mPADs (f) and molecular tension probes (g) measure the endogenous force generated by cells and applied on receptors. Force is measured from the displacements of beads embedded in an elastic substrate to which the cell adheres (TFM), the deflections of elastic pillars to which the cell attaches (mPAD) and the unquenched fluorescence (molecular tension probe). Cellular-level forces on many receptors are measured by TFM and mPAD, which reflect only the tangential force components, because only lateral bead displacements and pillar deflections can be visualized by the microscope’s top view. By comparison, molecular tension probes measure tension along the long axis of the receptors. h, Tension gauge tethers fail when force exceeds a designed threshold, which thus limits the level of tension that the cell can apply on its receptors.
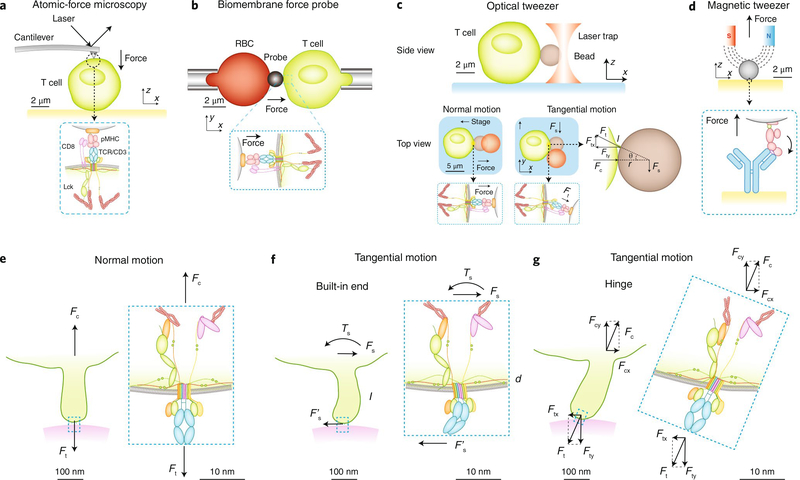
Although no technique available at present allows the in vivo measurement of forces on immunoreceptors, the lines of evidence noted above suggest that immunoreceptors probably experience forces in vivo; these may come from outside the cell as collateral effects of intermembrane expulsion of large molecules, from the relative displacement between the cell membrane and the substrate surface, or from inside the cell as a result of motor activity and cytoskeletal rearrangement. Furthermore, such forces have been directly observed in vitro through the use of traction-force microscopy (TFM)2,7,8 (Fig. 1e), micro-pillar array detectors (mPADs)3,4 (Fig. 1f) and molecular tension probes5,6,17 (Fig. 1g). Moreover, the requirement for above-threshold forces for certain biological functions has been demonstrated through the use of tension gauge tethers63 (Fig. 1h). In addition, for the study of mechanical impact in a controlled manner, given forces are applied on immunoreceptors to induce biophysical and biological effects through the use of single-molecule force techniques such as atomic-force microscopy22–24,30 (Fig. 2a), biomembrane force probes13,15–20,64 (Fig. 2b), optical tweezers14,21,27,31,65 (Fig. 2c) and magnetic tweezers19 (Fig. 2d). Regardless of its exogenous or endogenous origin, the same level of force acts on the immunoreceptor, as required by equilibrium.
Fig. 2 |. Single-molecule force techniques for applying forces to immuneoreceptors on immune cells.
a–d, Atomic-force microscopy (a), biomembrane force probes (b), optical tweezers (c) and magnetic tweezers (d) apply force to single receptors on cells and analyze the effects on receptor–ligand interactions, protein conformational changes and cellular functions. Atomic-force microscopy (a), biomembrane force probes (b) and magnetic tweezers (d) tend to apply force normal to the bead–cell (or substrate) interface by pulling them apart. Optical tweezers (c) apply a force Fs by moving the cell normally or tangentially relative to the bead–cell interface, pulling the receptor along the normal direction or an inclined direction with a force Ft. Ft is equal to Fs in the normal case but is larger than Fs in the tangential case. The mechanical analysis in the tangential case is similar to that in Fig. 1a. The inclined angle, determined by the relative dimensions of the microvillus length l and the bead radius r by the equation Ft = Fs/sin{cos−1[1/(1 + l/r)]}, is needed to generate both the normal force component Fty and tangential force component Ftx to balance the forces (Fc and Fs) and torques (Fty × r × sinθ and – Ftx × r × cosθ) on the bead. e, Normal motion of the T cell away from the substrate stretches the microvillus and pulls the TCR upward with a cell force Fc against the bond force Ft. f, Tangential motion of the T cell parallel to the substrate surface bends the microvillus and the TCR, assuming the root of the microvillus and the membrane anchor of the TCR can support bending moments (i.e., behave as a built-in end). g, If the microvillus root or the TCR membrane anchor, or both, behave(s) as a hinge incapable of supporting bending moment, the microvillus and/or the TCR would tilt to adjust its (their) orientation(s) to bear tension along its (their) long axis (axes).
Studies using TFM2,7,8 and mPADs3,4 have mapped whole-cell traction-force distributions through the TCRs, BCRs and associated molecules in vitro. However, relating such traction maps to the forces sensed by individual receptors remains challenging, due to the unknown spatial distribution of the receptors, such as clustering, which probably varies depending on the specific kind of receptor, cell type and physiological setting. Force is a vector with both magnitude and direction, and it can also generate rotational or bending moment, which is equal to force multiplied by the length of the lever arm. However, the two-dimensional versions of TFM2,7,8 and mPADs3,4 are able to measure only the force component tangential to the substrate surface, not the component normal to it, because the microscope’s top view can observe only tangential bead displacements and lateral pillar deflections, not normal bead displacements and axial pillar extension or compression (Fig. 1e,f). Therefore, although TFM and mPADs have provided ample evidence that cells pull on their immunoreceptors in a time-dependent fashion, the directionality of mechanical force and the moment that it may generate at the molecular scale have remained unclear.
Determining force direction and moment at the molecular scale requires information about how the lymphocyte’s membrane protrusions (called ‘microvilli’66) contact the substrate surface and how the microvilli and immunoreceptors support applied bending moment from the lateral force. When viewed by super-resolution fluorescence microscopy67,68, some TCRs are concentrated on the microvillus tips, which contact the substrate surface at variable orientations. At the cellular level, cell-generated forces on many TCRs may collectively result in a substantial tangential component at the contact interface, as seen with TFM2,7,8 (Fig. 1e) and mPADs3,4 (Fig. 1f). At the molecular scale, however, the TCR–pMHC bond experiences tension probably along the long axis, as visualized by molecular tension probes5,6,17 (Fig. 1g).
The suggestion above is based on mechanical analysis of how microvilli and immunoreceptors support force and moment. Such analysis requires models whose validity has not been established experimentally. Structurally, microvilli and TCR–pMHC bonds may be modeled as flexible structures because of their slender shapes, which seem to be better suited to support tension (Fig. 2e) than bending (Fig. 2f). On one end of the spectrum, the TCR anchor on the cell membrane can be modeled as a hinge (Fig. 2g) unable to support a bending moment, as suggested by the seemingly flexible connecting peptides in the proximal region of the transmembrane helices of the αβ TCR, TCR invariant chain CD3 molecules and MHC, all of which seem to lack rotational restrictions. On the other end of the spectrum, the opposite of a hinge is a ‘built-in’ end (Fig. 2f), which is able to support bending fully. Rigorous proving or ruling out of either model requires information about transmembrane and intramolecular interactions within the TCR–CD3 complex69 and interactions of the complex with the cytoskeleton70–72, which is currently unavailable; the reality may lie in between the two extremes. However, if either the flexible-structure model or the hinge model applies, the orientation of the TCR–pMHC long axis would be tilted to align with the axial tension (Fig. 2g).
The hinge model described above has been supported experimentally. T cells have been found to pull through the TCR and the co-receptor CD8 on pMHC complexes that were either adsorbed on glass surfaces, which support lateral force5, or reconstituted on supported lipid bilayers, which do not support lateral force6. In contrast, platelets can pull via integrins on ligands adsorbed on glass surfaces, but not those reconstituted on supported lipid bilayers73. Comparison of these results suggests that integrins need lateral force to mediate cell–cell interactions, but the TCR does not.
Dissociation kinetics and conformational changes
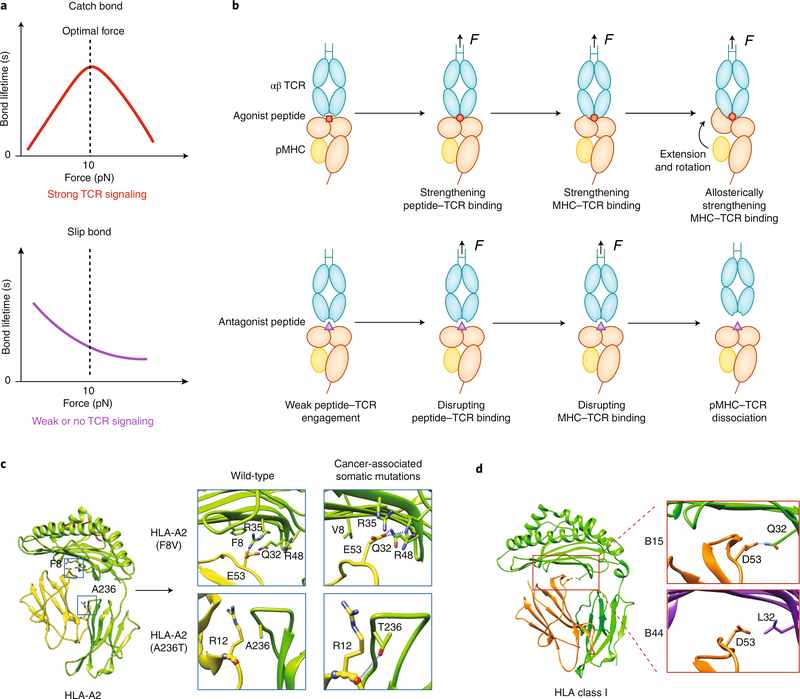
Studies using single-molecule force techniques (Fig. 2a–c) have revealed mechanical regulation of protein–protein dissociation kinetics, which can be classified into several types of dynamic bonds. A bond that shows accelerated dissociation with increasing force is called a ‘slip bond’. However, in many systems, a force lower than a critical level can strengthen the bond by slowing the dissociation; this is called a ‘catch bond’. When the force exceeds the critical level, the catch bond is overpowered and transitions to a slip bond42,47,74. Most catch bonds are found in systems that have structural and/or force-supporting roles in their functions, such as adhesive and cytoskeletal proteins22–25,42,74. For immunoreceptors, antibodies form slip bonds with antigen through the Fab fragment22–25 but form catch bonds with FcγRIIA though the Fc26. TCRs form catch bonds with agonist peptide–MHC complexes (Fig. 3a) but form slip bonds with weak agonist peptide–MHC or antagonist peptide–MHC complexes15 (Fig. 3a). The association of bond type with ligand potency has been confirmed in multiple mouse and human systems14–19. In addition to the ability of TCRs to form bimolecular catch bonds with agonist pMHC complexes14–16,18,19, TCRs and CD8 molecules can act together to form trimolecular catch bonds with negative-selection pMHC complexes, which distinguishes these negative-selection ligands from positive-selection ligands17. This type of catch bond is called a ‘dynamic catch’75 and emerges from the synergistic TCR–pMHC–CD8 interaction, rather than being intrinsic to either bimolecular arm of the trimolecular complex, because it can form even when both the TCR–pMHC interaction and pMHC–CD8 interaction are slip bonds.
Fig. 3 |. Force-regulated pMHC conformations determine TCR–MHC dynamic bonds.
a, When a TCR forms a catch bond with an agonistic pMHC (top), increasing force prolongs the bond lifetime at low forces, but the bond changes to a slip bond at high forces. When a TCR forms a slip bond with antagonistic pMHC (bottom), increasing force monotonically shortens the bond lifetime. b, A TCR catch bond with agonistic pMHC is initiated by force-enhanced engagement between the TCR’s CDR3 loops and the peptide’s ‘hotspot’ residues, followed by a force-induced betterment of the complementarity of the MHC–TCR interface (top). Force also induces separation of β2m from the MHC heavy chain, which leads to MHC extension and rotation of the α1α2 domains toward the TCR; this further strengthens the TCR–MHC engagement allosterically. For a weak ligand, force is unable to induce the conformational changes noted above in the pMHC, which leads to slip bonds (bottom). c,d, Cancer-associated somatic mutations (c) or genotype variations (d) encoding mutant HLA molecules may also affect the susceptibility of MHC to conformational changes under force. Examples of the former are HLA-A2 F8V and A236, which form more hydrogen bonds between β2m and the MHC heavy chains (c). As an example of the latter, HLA-B15 has more hydrogen bonds between β2m and the MHC heavy chain than does HLA-B44 (d). Diagrams in Fig. 3c,d are modified from ref. 19.
Many proteins utilize conformational changes to accomplish their function. Whereas protein conformational change is often regulated biochemically, such as (un)binding and/or (de)phosphorylation by another protein, it can also be modulated mechanically. Force acted on a TCR–pMHC or pre-TCR–pMHC bond can induce a sudden increase in the length of the complex, which correlates with the ligand potency14,21, suggestive of its biological relevance. A study using a combined approach of molecular-dynamics simulations, single-molecule magnetic tweezers and biomembrane-force-probe experiments and conformation-locking mutagenesis has indicated that the length increase results mainly from a conformational change in the pMHC, with a possible minor contribution from the TCR’s connecting peptide motif, and suggested a mechanistic model for the TCR–pMHC catch bond on the basis of such conformational changes19.
The mechano-chemical coupling mechanism noted above is similar to the sliding–rebinding mechanisms observed in several catch-bond systems76–78. In this model, the catch bond (Fig. 3a) is initiated by a normal pulling to strengthen the network of atomiclevel interactions between the TCR CDR loops and the agonist peptide ‘hotspot’ residues, which in turn strengthens the interaction of the TCR with the MHC (Fig. 3b). The increased force uncouples the α-chain–β2-microglobulin (β2m) interdomain interaction, which results in an extension of 5–10 nm in the MHC19 that further stabilizes the TCR–pMHC bond allosterically and enables the two molecules to slide against each other. Sliding promotes the formation of new interactions after force ruptures the preexisting interactions. In contrast, for non-activating ‘self’ peptides, force only weakens the TCR–pMHC bond without inducing the aforementioned conformational changes (Fig. 3b), which results in slip bonds (Fig. 3a). Thus, three layers of ‘mechanical signal amplification’ are proposed for the TCR to discriminate self from non-self (Fig. 3b). Distinct from the ‘shear-induced sliding’ model, in which force is applied perpendicular to the TCR long axis18, in the mechano-chemical coupling model, sliding is a consequence of the conformational change in the MHC induced by axial pulling and has been observed only in simulations with agonist pMHC19 (Fig. 3b, top). Future studies should test whether this model applies to MHC class II systems.
Somatic mutations or polymorphism of MHC may also affect its conformational changes under force and thereby affect the TCR’s dynamic bonds and antigen recognition. Cancer-associated somatic mutations encode mutant HLA-A molecules that suppress the TCR catch bond probably by limiting MHC’s conformational changes through enhancement of the α-chain–β2m interdomain coupling (Fig. 3c). Similarly, the α-chain–β2m coupling seems stronger for HLA-B15 than for HLA-B44 (Fig. 3d); this potentially results in fewer mechanically induced conformational changes, which might impair the TCR catch bond with agonist pMHC or neoantigen peptide–MHC complexes. This may explain why HLA-B15+ patients are less responsive to checkpoint blockade immunotherapy than are HLA-B44+ patients79.
Immunoreceptors as force sensors
The idea that the TCR may be an anisotropic mechanosensor was suggested a decade ago on the basis of a pioneering experiment in which optical tweezers were used to trap a bead bearing either antibody to CD3 or pMHC to apply sinusoidal force to the TCR (or TCR and CD8)27. When the bead was driven to move tangentially (but not normally) relative to the bead–cell interface (Fig. 2c), intracellular calcium was induced, which indicated conversion of the mechanical stimulation on the TCR into a biochemical signal. By comparison, a biomembrane force probe used to pull on the TCR (Fig. 2b) induced calcium by constant force normal to the bead–cell interface15. An optical-tweezers study found that calcium could be induced by a step displacement of the bead relative to the cell along the direction either tangential or normal to the bead–cell interface, but much denser pMHC coating was required in the latter case than the former case31. These data suggest that T cells can sense force on the TCR. However, the question of which force direction is most relevant to TCR mechanosensing remains incompletely addressed.
We note that in the aforementioned optical-tweezers experiments, for evaluation of the anisotropic, or force direction, effect, external forces were applied at the cellular level27,31. Relating those forces to the molecular-level force on the TCR requires a mechanical analysis whereby all forces and moments are balanced (Fig. 2c). A tangential force applied by optical tweezers to the bead center is balanced by (the tangential component of) the TCR force on the opposite direction, but not along the same line. The distance between the two parallel lines of force action provides a lever arm to generate a moment that would drive rotation. Because optical tweezers cannot apply torques to balance this moment, the bead would roll on the cell surface until it reached an equilibrium configuration (Fig. 2c), with the TCR bond on the tip of a stretched and tilted microvillus repositioned from the center to the rear of the bead–cell contact to provide an inclined tether force (Fig. 2g). The end result would be a tension along the long axis of the TCR bond, rather than a lateral force perpendicular to that axis. Therefore, the TCR bond (and the microvillus on which the TCR is localized) would adjust its orientation to bear tension along its long axis, which would translate a ‘lateral force’ at the cellular level to an ‘axial force’ at the molecular level.
Beyond magnitude and direction, several aspects of the force waveform, including the pattern, duration and frequency of the force, may be sensed by the TCR to generate different responses. In one study discussed above, calcium signal was induced by clamped normal forces (which were increased to a preset level and held constant until dissociation) but not by ramped normal forces (which were increased until rupture)15. The inability of ramped forces to induce calcium (despite their much larger magnitude) when only the TCR was pulled15 was overcome by permitting the pMHC to pull both the TCR and the CD829. Although more studies are needed for full characterization of the effects of force waveforms on TCR triggering, the existing studies suggest that the TCR does not respond to force in an all-or-none fashion; instead, it can interpret the force waveform and translate the complex input into distinct signals and thus function as a mechanosensor15,27,29,31.
Immunoreceptors as rigidity sensors
The mechanical environment of a cell includes not only the forces acting on it but also the stiffness of the substrate in which it resides. Not only can lymphocytes sense substrate stiffness through integrins80, similar to tissue cells, but also T cells29,34–36 and B cells37–39 can do so via their antigen receptors, responding with changes in their activation and development. Stiffness is the ease with which the substrate is deformed by force. Thus, rigidity sensing is a form of mechanosensing different from force sensing. The cell must be able not only to apply force to the substrate but also to gauge the degree of the resistance to the applied force. Thus, antigen receptor–mediated rigidity sensing represents an example in which immune cells use force to gather antigen-dependent information about their mechanical environment and to inform biological decisions.
Substrate stiffness is thought to be gauged by a molecular assembly that acts as a ‘clutch’ to transmit force from the cytoskeleton to the ECM. The proposed clutch is controlled by the dynamics of various bonds in the force transmission pathways81. For integrinmediated sensing of rigidity by tissue cells, combined experimental and modeling studies suggest that force transmission and transduction involve talin and vinculin, which bear intracellular forces82,83. A model called the ‘actin–talin–integrin–ligand clutch’ proposes that two force-dependent events, unfolding of talin and unbinding of the integrin–ligand bond, are related in such a way that talin would unfold on a stiff substrate but not on a soft substrate to expose cryptic binding sites for the recruitment of vinculin84. The force-dependent dynamics of other intracellular bonds, such as the integrin’s cytoplasmic tail with talin, talin with vinculin, and vinculin with the actin cytoskeleton, may also have important roles in this.
It might be useful to compare relevant aspects of rigidity sensing by integrins with such sensing by antigen receptors. Integrins can transmit forces and transduce signals across the cell membrane bidirectionally, known as ‘outside-in’ and ‘inside-out’ signaling. In both directions, talin is thought to bear forces and relay signals. On the other hand, antigen receptors are known for signal reception. Previously, only the biochemical aspect of the signal received from the antigen was considered. More recently, the mechanical aspect has been recognized. Regardless, both aspects permit only outside-in flow of information unidirectionally. Also, how the TCR and BCR interact directly and/or indirectly with the actomyosin cytoskeleton, which is a key node in the force-transmission pathway, is still poorly understood70–72. To this end, an outside-in–inside-out signaling loop of the TCR mechanotransduction apparatus17 may serve as a ‘clutch’ mechanism for sensing of rigidity by the T cell. The signaling kinase Lck and/or CD3 may have a role similar to that of talin, and the signaling kinase ZAP-70 and/or Lck may have a role similar to that of vinculin, which would make them part of the rigidity-sensing apparatus along with the TCR, CD3 and CD8.
Effect of force on TCR triggering and antigen discrimination
Considering the TCR as a mechanosensor can provide a new angle with which to investigate the central yet unresolved question of how the physical TCR–pMHC binding events are transduced across the membrane into peptide-specific biochemical signals. The existing models can be divided into roughly two categories, depending on whether conformational changes in the TCR–CD3 complex are required. For the first category, studies of force-free TCR–pMHC structures have revealed changes in the pMHC binding sites that may potentially induce allosteric changes in the TCR constant region85–88. Those observations alone cannot explain TCR triggering, due to the subtle effects of the changes and the lack of a mechanism for coupling TCR conformational changes to differential signaling of CD3. TCR–pMHC binding was proposed to release the cytoplasmic tails of CD3 from the inner leaflet of the plasma membrane via dynamic allostery, which would allow phosphorylation of the exposed immunoreceptor tyrosine-based activation motifs by Lck and/or the kinase Fyn89,90. The details of how the intracellular structures rearrange in response to extracellular ligand binding distal from the membrane have not been explained, and they cannot be elucidated by mere comparison of non-ligand-bound TCR structures and pMHC-bound TCR structures in force-free conditions85–88. Force may provide a natural mechanism for segmenting the bipartite helix in the transmembrane domain of the TCR α-subunit to alter its association with the CD3 subunits65, and/or to induce apposition of the CD3ζζ subunits that are spread before TCR–pMHC binding91; this may dislodge the CD3 tails from the plasma membrane, with more-potent ligands engaging for a longer time and transmitting more-durable forces and thus exposing CD3 for longer periods and with a greater potential to generate stronger signals.
In the second category, the kinetic segregation model does not require TCR–CD3 conformational changes, as it suggests that the TCR may be triggered by the segregation of phosphatases (such as CD45) from the TCR and kinases (such as Lck) and may thereby tilt the local phosphorylation equilibrium92,93. Although this is not explicitly considered, both the ‘kinetic’ and ‘segregation’ elements of the kinetic segregation model employ force as a critical factor. Force may apply on the TCR–pMHC bond during segregation passively, as the reaction to keep the two membranes together against the repulsion of the larger molecules; actively, through actin dynamics; or both. Force elicits catch bonds and slip bonds to prolong or shorten, respectively, the lifetime of TCR bonds with antigenic and non-cognate ligands14–16, which provides a kinetic mechanism for differential segregation and signaling. Indeed, catch bonds, but not slip bonds, were shown to cause exclusion of CD45 from the contact area between a giant unilamellar vesicle and the glass-supported bilayer, which served as a synthetic biology system with which to test the kinetic segregation model18.
Two previous models of T cell activation have integrated TCR–pMHC binding kinetics. The serial engagement model suggests that a pMHC may compensate for its low affinity and fast off rate by forming serial bonds with multiple TCRs94. The kinetic proofreading model proposes that a T cell may be activated by TCR bonds with strong ligands that outlive a threshold, but not by bonds with weak ligands that are shorter lived than the threshold95. Both models and their modifications96 have been used as conceptual frameworks for theoretical analyses of TCR triggering, although the basis of these analyses — the TCR–pMHC binding kinetic parameters — was limited to their force-free values97. Kinetic measurements under force have opened a new avenue for re-examining these theoretical analyses, because the force-free off rate represents only a single point in a full bond lifetime curve that may increase (catch) or decrease (slip) with force42,74. For example, the TCR–pMHC catch-bond or slip-bond pattern corresponds to the T cell signaling pattern, despite the fact that other binding parameters, most notably affinity, do not18. A durable force on a single TCR can induce intracellular calcium, which indicates that serial engagement is not required15,31. However, the calcium signal strength correlates with the accumulation of sequential stimulations by multiple intermittent brief forces, each of which acts on a single TCR15,29. These data support a kinetic proofreading mechanism in which a threshold is set for certain amount of signaling intermediates rather than certain phosphorylation states of a single TCR, and in which the TCR triggering does not require but is enhanced by the accumulation of serial (preferentially long-lived) bonds with pMHC.
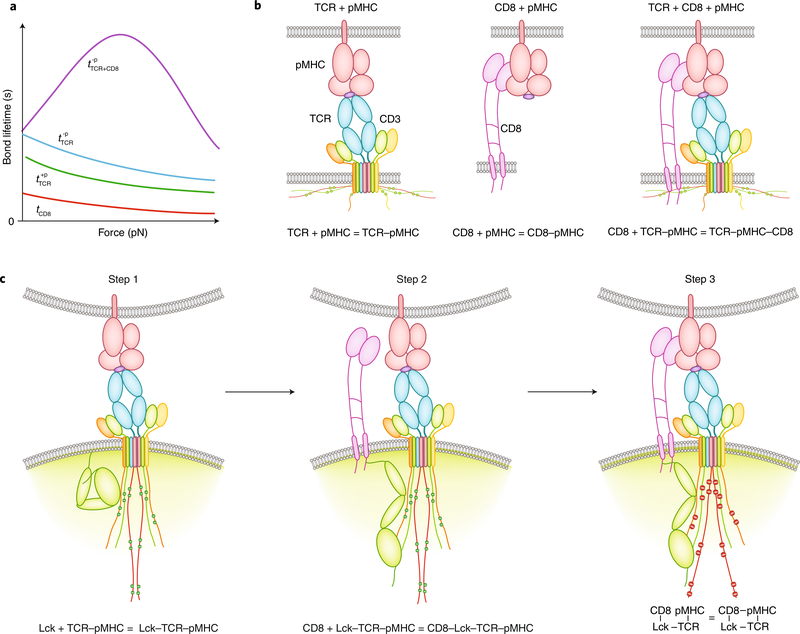
Trimolecular catch bonds (dynamic catch) amplify the ability of thymocytes to discriminate negative-selection ligands from positive-selection ligands17. Across the negative-selection border, both negative-selection ligands and positive-selection ligands form biomolecular slip bonds with the TCR. However, the slightly longer lived TCR bonds with the negative-selection ligands may generate signals to induce the cooperative binding of CD8 to the pMHC pre-engaged with the TCR to form trimolecular catch bonds. By comparison, the TCR bonds with positive-selection ligands have shorter lifetimes, are unable to induce binding of CD8 to the pMHC pre-engaged with the TCR, and remain as bimolecular slip bonds (Fig. 4a). Although only three ectodomain interactions were measured in these studies (Fig. 4b), experiments designed to modulate intracellular domain interactions suggest a three-step minimal model for the dynamic catch formation: first, initial binding of a negative-selection ligand by the TCR triggers binding of TCR–CD3 to Lck and its phosphorylation by Lck; second, CD8 is recruited to phosphorylated TCR–CD3 via Lck; and third, CD8 binds to pMHC cooperatively to stabilize the trimeric complex (Fig. 4c). Thus, the TCR’s recognition process includes not only the physical interactions at the extracellular binding interfaces of TCR, CD8 and pMHC but also the TCR-proximal signaling events and the adaptor property of Lck. Therefore, the dynamic catch represents a form of ‘inside-out’ signaling for the TCR. Its manifestation requires mechanical force, which acts not only on the ectodomains of TCR, CD8 and pMHC but also potentially on intracellular proteins; this may alter their binding, conformation and enzymatic activity. Future work should test the possible involvement of other intracellular proteins, such as ZAP-70.
Fig. 4 |. TCR mechanotransduction via dynamic catch.
a, Lifetime-versus-force curves of various bonds. Dynamic catch is a catch bond that results from the binding of CD8 to a negative-selection pMHC (–p) pre-engaged by a TCR (; purple). This is an emergent property from cooperative binding rather than a property intrinsic to the two arms of the trimeric complex, because both TCR–pMHC bimolecular interactions (; cyan) and pMHC–CD8 bimolecular interactions (tCD8; red) behave as slip bonds. Interaction of the TCR with a positive-selection pHMC (+p) also forms a slip bond (; green) with a lifetime slightly shorter than that of the bond with a negative-selection pMHC. However, it does not induce dynamic catch formation. b, Up to three extracellular interactions participate in the binding of a thymocyte to pMHC, which involve the TCR (left), CD8 (middle), or both (right). c, Minimal model for dynamic catch formation. For elucidation of the inner workings of the dynamic catch, intracellular interactions are shown here, which include the binding of Lck to TCR–CD3 pre-bound by pMHC and the phosphorylation of TCR–CD3 by Lck (Step 1; left), the recruitment of CD8 to phosphorylated TCR–CD3 by Lck (Step 2; middle), and the binding of CD8 to pMHC cooperatively to stabilize the trimeric complex (Step 3; right). During the recognition of antigen by the TCR and signal initiation, mechanical forces may apply to both extracellular interactions and intracellular interactions, which may elicit catch bonds or slip bonds, induce protein conformational change and regulate enzymatic activity.
Feedback loops have been introduced into TCR signaling models to bestow specificity, because the intrinsic off rate does not seem enough98. In contrast to the published feedback amplification mechanisms that involve molecules such as the kinase ERK and phosphatase SHP-199 or the costimulatory molecule CD28 in the respective pathways100, the signaling-dependent dynamic catch involves mechanotransduction. Positive feedback is employed to further amplify differential binding to positive-selection ligands versus negative-selection ligands. Different TCR–pMHC bond lifetimes would transduce different signals, which might induce different Lck-mediated intracellular interactions between TCR and CD8 and consequently bring them to different degrees of proximity to allow or prevent TCR–pMHC–CD8 intercellular interaction and thereby amplify the different durations of TCR engagement with different pMHC complexes17. In the case of negative-selection ligands, but not positive-selection ligands, not only can cooperative biophysical interaction result from cooperative biochemical signaling, but the latter can also be amplified by the former, as prolongation of the time that the TCR senses the antigen can be translated into signaling enhancement. Thus, the two cooperative mechanisms may be self-reinforcing as well as cross-reinforcing, which provides two levels of positive feedback and synergy for amplifying the TCR’s discrimination of different pMHC complexes.
Effect of force on TCR sensitivity and specificity
Considering the effects of force also sheds lights onto the question of how the TCR is able to correctly identify a minute amount of antigenic peptide–MHC (sensitivity) without responding to over-whelming amounts of self peptide–MHC complexes (specificity). Because antigen recognition is achieved via the TCR–pMHC interaction, it is generally thought that the key to having both high sensitivity and high specificity is hidden in the physical chemistry of this interaction. A common approach to finding this key is to characterize a particular property using a panel of TCR–pMHC interactions and examine the correlation (or the lack thereof) between the measured values and the biological responses induced by these interactions. Several binding parameters, most commonly binding affinity or dissociation off rate, correlate with the biological responses to the pMHC stimulations15–17,101–109. Sensitivity is thought to associate with the on rate and/or the affinity of the TCR–pMHC interaction, as the higher the on rate and the affinity, the faster and more bonds form for given TCR and pMHC densities. Three-dimensional measurements using soluble proteins have suggested that TCR–pMHC interactions have lower affinities than those of antibody–antigen interactions, which raises the question of how such low-affinity interactions could provide highly sensitive antigen recognition. Two-dimensional measurements of the T cell surface have shown that the affinities of interactions of TCRs with the antigenic peptide–MHC complexes, which can be further enhanced by cooperative binding with CD8, are comparable to those of the binding of the high-affinity integrin LFA-1 to its ligand ICAM-1, the dominant adhesion receptor–ligand interaction in the immunological synapse102,103,107.
However, specificity cannot be explained by difference in affinity and on rate alone, because their low values can be compensated for by high pMHC densities on the APC. Instead, specificity is thought to be governed by the off rate of the TCR–pMHC dissociation. Specificity analysis typically employs kinetic proofreading as a foundational concept, based on the duration or lifetime of the TCR–pMHC bond95,96,110. One difficulty in using TCR–pMHC off rates for the analysis of TCR specificity is the small differences between the off rates of agonists and those of antagonists when they are measured at zero force. Force greatly amplifies such differences, owing to the distinct catch-bond versus slip-bond types of the two ligands14–16.
Considering the effects of force also suggests modifications to the current frameworks used to evaluate sensitivity and specificity, which often plot the activation likelihood against either the ligand number or a kinetic parameter97. Given that the APC presents multiple antigenic peptides, the T cell could layer information from multiple interactions during contact with the APC. A model that combines kinetic proofreading and serial engagement to balance the sensitivity and specificity was used to mimic this multi-step decision-making process17. In the absence of mechanical force or CD8 interactions, the thymocyte lacks the ability to discriminate between the two pMHC complexes across the negative-selection border accurately. However, force and CD8 engagement add accuracy, which increases with the number of TCR bonding events; this allows the thymocyte to be negatively selected on a vast majority of encounters with the negative-selection ligand (sensitivity) and on only a negligible fraction of encounters with the positive-selection ligand (specificity).
The analysis described above suggests that the accumulation of multiple small differences in bond lifetime through biochemical and biomechanical feedbacks (for example, dynamic catch) is able to enhance sensitivity and specificity so that the thymocyte can respond appropriately when the information is layered sequentially. From a mechanistic standpoint, a TCR’s repetitive encounters with agonist pMHC could be processed by the downstream signaling network and could shift the phosphorylation and organizational states of the signaling intermediates in a cumulative way, at a rate proportional to the lifetime of the TCR bond in each encounter. If sufficient signal is accrued over a spatially and temporally defined threshold, cellular activation will occur. In this way, mechanical force may have several roles, from modulating conformational states of the TCR–CD3 complex to prolonging the lifetime of antigenic interactions, to triggering the TCR, and to integrating cumulative signals for T cell activation.
Future work
While future studies will continue to grow the list of immunoreceptors identified as mechanosensors, more work will provide the structural and/or organizational basis of their mechanotransduction and develop robust phonotypical mechanokinetic ‘markers’ (for example, catch or slip bond, long or short lifetime, high or low force) for immunoreceptors. The activation of immune cells is not determined by the antigenic stimulations alone. A variety of co-stimulatory and inhibitory receptors also deliver signals to be integrated with the antigen-receptor signals at multiple levels. Ion channels, in particular mechanosensitive channels such as Piezo1111, may also contribute to this. Spatially, different receptors may be organized in different membrane microdomains that dynamically merge or segregate in response to antigen stimulation, as suggested by imaging studies of molecular microclusters and protein islands112–116. The reorganization of these molecules is largely due to the active restructuring of the local cytoskeletal network to which they are coupled, which also apply forces on them7,61,62. It would be interesting to determine how force regulates the function of individual immunoreceptor species, as well as the interplay among them. It is possible that one receptor species can cross-modulate another by tuning the force applied via the shared cytoskeletal network. For example, signals from inhibitory receptors may disrupt the formation of a stable synapse by suppressing TCR signaling that triggers actin polymerization for active force generation117. This mechanically based communication may be able to propagate quickly and globally across the cell and thereby relax the requirement for the colocalization of two receptors when only the diffusion of downstream signaling intermediates is considered. Therefore, the eventual cell functions may not only be determined by the biochemical cascades weaved into the immunoreceptor networks but could also be regulated by the information ‘encoded’ in and transmitted by force. In decoding this information, experiments that use toolkits such as FRET-base tension sensors82,118,119 will shed light on the key players and details of the characteristics of the force-transmission and force-transduction pathways.
New insights gained from studies of immunoreceptor-mediated mechanosensing may have translational applications. The finding that TCR–pMHC catch bonds are suppressed by cancer-associated somatic mutations to the genes encoding HLA molecules suggests that force effects should be considered to counter the impairment of the TCR’s recognition of tumor antigen. Moreover, the presence of catch bonds may be a signature of cancer neoantigens, so such information may be used to predict neoantigens for the development of vaccines directed against cancer. As another example, chimeric antigen receptors (CARs) use antibody derivatives as the ligand-binding domain and the cytoplasmic signaling motifs of CD3 and CD28 as the signaling module120. Despite the promising applications of CARs, details of the molecular mechanism underlying CAR triggering remain elusive, which prevents CARs from achieving optimal design for the improvement of clinical efficacy. Because CARs probably share part of the TCR-triggering mechanisms, it may be important to incorporate force in the design process to achieve both durable binding and more-efficient triggering.
Acknowledgements
Supported by grants from the US National Institutes of Health (U01CA214354, R01AI124680, R01GM122489 and R21Al135753 to C.Z.), from the National Basic Research Program of China (2015CB910800 to W.C. and 2014CB910202 to J.L.) and from the National Science Foundation of China (31470900 and 31522021 to W.C. and 11672317 and 31222022 to J.L.).
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mempel TR, Henrickson SE & Von Andrian UH T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature 427, 154–159 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Hui KL, Balagopalan L, Samelson LE & Upadhyaya A Cytoskeletal forces during signaling activation in Jurkat T-cells. Mol. Biol. Cell 26, 685–695 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashour KT et al. CD28 and CD3 have complementary roles in T-cell traction forces. Proc. Natl Acad. Sci. USA 111, 2241–2246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashour KT et al. Cross talk between CD3 and CD28 is spatially modulated by protein lateral mobility. Mol. Cell. Biol. 34, 955–964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y et al. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc. Natl Acad. Sci. USA 113, 5610–5615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma VP et al. Ratiometric tension probes for mapping receptor forces and clustering at intermembrane junctions. Nano Lett. 16, 4552–4559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colin-York H et al. Cytoskeletal control of antigen-dependent T cell activation. Cell Rep. 26, 3369–3379.e3365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J et al. Profiling the origin, dynamics, and function of traction force in B cell activation. Sci. Signal. 11, eaai9192 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Wan Z et al. The activation of IgM- or isotype-switched IgG- and IgE-BCR exhibits distinct mechanical force sensitivity and threshold. eLife 4, e06925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nowosad CR, Spillane KM & Tolar P Germinal center B cells recognize antigen through a specialized immune synapse architecture. Nat. Immunol. 17, 870–877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spillane KM & Tolar P B cell antigen extraction is regulated by physical properties of antigen-presenting cells. J. Cell Biol. 216, 217–230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan Z et al. PI(4,5)P2 determines the threshold of mechanical force-induced B cell activation. J. Cell Biol. 217, 2565–2582 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu B et al. The cellular environment regulates in situ kinetics of T-cell receptor interaction with peptide major histocompatibility complex. Eur. J. Immunol. 45, 2099–2110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das DK et al. Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc. Natl Acad. Sci. USA 112, 1517–1522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B, Chen W, Evavold BD & Zhu C Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 157, 357–368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong J et al. Force-regulated in situ TCR-peptide-bound MHC class ii kinetics determine functions of CD4+ T cells. J. Immunol. 195, 3557–3564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong J et al. A TCR mechanotransduction signaling loop induces negative selection in the thymus. Nat. Immunol. 19, 1379–1390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sibener LV et al. Isolation of a structural mechanism for uncoupling T cell receptor signaling from peptide-MHC binding. Cell 174, 672–687.e627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu P et al. Mechano-regulation of peptide-MHC class I conformations determines TCR antigen recognition. Mol. Cell 73, 1015–1027.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallis RJ et al. Pre-TCR ligand binding impacts thymocyte development before αβTCR expression. Proc. Natl Acad. Sci. USA 112, 8373–8378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das DK et al. Pre-T cell receptors (pre-TCRs) leverage Vβ complementarity determining regions (CDRs) and hydrophobic patch in mechanosensing thymic self-ligands. J. Biol. Chem. 291, 25292–25305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall BT et al. Direct observation of catch bonds involving cell-adhesion molecules. Nature 423, 190–193 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Sarangapani KK et al. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J. Biol. Chem. 279, 2291–2298 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Kong F, García AJ, Mould AP, Humphries MJ & Zhu C Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 185, 1275–1284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen W, Lou J & Zhu C Forcing switch from short- to intermediate- and long-lived states of the αA domain generates LFA-1/ICAM-1 catch bonds. J. Biol. Chem. 285, 35967–35978 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishi H et al. Neutrophil FcγRIIA promotes IgG-mediated glomerular neutrophil capture via Abl/Src kinases. J. Clin. Invest. 127, 3810–3826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim ST et al. The αβ T cell receptor is an anisotropic mechanosensor. J. Biol. Chem. 284, 31028–31037 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y-C et al. Cutting edge: mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. J. Immunol. 184, 5959–5963 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Pryshchep S, Zarnitsyna VI, Hong J, Evavold BD & Zhu C Accumulation of serial forces on TCR and CD8 frequently applied by agonist antigenic peptides embedded in MHC molecules triggers calcium in T cells. J. Immunol. 193, 68–76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu KH & Butte MJ T cell activation requires force generation. J. Cell Biol. 213, 535–542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng Y et al. Mechanosensing drives acuity of αβ T-cell recognition. Proc. Natl Acad. Sci. USA 114, E8204–E8213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basu R et al. Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell 165, 100–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Natkanski E et al. B cells use mechanical energy to discriminate antigen affinities. Science 340, 1587–1590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Judokusumo E, Tabdanov E, Kumari S, Dustin ML & Kam LC Mechanosensing in T lymphocyte activation. Biophys. J. 102, L5–L7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitakis M et al. Different TCR-induced T lymphocyte responses are potentiated by stiffness with variable sensitivity. eLife 6, e23190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wahl A et al. Biphasic mechanosensitivity of T cell receptor-mediated spreading of lymphocytes. Proc. Natl Acad. Sci. USA 116, 5908–5913 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan Z et al. B cell activation is regulated by the stiffness properties of the substrate presenting the antigens. J. Immunol. 190, 4661–4675 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Zeng Y et al. Substrate stiffness regulates B-cell activation, proliferation, class switch, and T-cell-independent antibody responses in vivo. Eur. J. Immunol. 45, 1621–1634 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Shaheen S et al. Substrate stiffness governs the initiation of B cell activation by the concerted signaling of PKCβ and focal adhesion kinase. eLife 6, e23060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Connor RS et al. Substrate rigidity regulates human T cell activation and proliferation. J. Immunol. 189, 1330–1339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim ST et al. TCR mechanobiology: torques and tunable structures linked to early T cell signaling. Front. Immunol. 3, 76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen W & Zhu C Mechanical regulation of T-cell functions. Immunol. Rev. 256, 160–176 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brazin KN et al. Structural features of the αβTCR mechanotransduction apparatus that promote pMHC discrimination. Front. Immunol. 6, 441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reinherz EL & Wang JH Codification of bidentate pMHC interaction with TCR and its co-receptor. Trends Immunol. 36, 300–306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hivroz C & Saitakis M Biophysical aspects of T lymphocyte activation at the immune synapse. Front. Immunol. 7, 46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Basu R & Huse M Mechanical communication at the immunological synapse. Trends Cell Biol. 27, 241–254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y, Ju L, Rushdi M, Ge C & Zhu C Receptor-mediated cell mechanosensing. Mol. Biol. Cell 28, 3134–3155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huse M Mechanical forces in the immune system. Nat. Rev. Immunol. 17, 679–690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tolar P Cytoskeletal control of B cell responses to antigens. Nat. Rev. Immunol. 17, 621–634 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Upadhyaya A Mechanosensing in the immune response. Semin. Cell Dev. Biol. 71, 137–145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng Y, Reinherz EL & Lang MJ αβ T cell receptor mechanosensing forces out serial engagement. Trends Immunol. 39, 596–609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spillane KM & Tolar P Mechanics of antigen extraction in the B cell synapse. Mol. Immunol. 101, 319–328 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Pageon SV, Govendir MA, Kempe D & Biro M Mechanoimmunology: molecular-scale forces govern immune cell functions. Mol. Biol. Cell 29, 1919–1926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossy J, Laufer JM & Legler DF Role of mechanotransduction and tension in T cell function. Front. Immunol. 9, 2638 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim JK, Shin YJ, Ha LJ, Kim DH & Kim DH Unraveling the mechanobiology of the immune system. Adv. Healthc. Mater. 8, e1801332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu C, Chen Y & Ju L Dynamic bonds and their roles in mechanosensing. Curr. Opin. Chem. Biol. (in the press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aramesh M, Stoycheva D, Raaz L & Klotzsch E Engineering T-cell activation for immunotherapy by mechanical forces. Curr. Opin. Biomed. Eng. 10, 134–141 (2019). [Google Scholar]
- 58.Wan Z, Shaheen S, Chau A, Zeng Y & Liu W Imaging: gear up for mechano-immunology. Cell. Immunol. 10.1016/j.cellimm.2019.103926 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Saggu G et al. Cis interaction between sialylated FcγRIIA and the αI-domain of Mac-1 limits antibody-mediated neutrophil recruitment. Nat. Commun. 9, 5058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McEver RP & Zhu C Rolling cell adhesion. Annu. Rev. Cell Dev. Biol. 26, 363–396 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murugesan S et al. Formin-generated actomyosin arcs propel T cell receptor microcluster movement at the immune synapse. J. Cell Biol. 215, 383–399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hong J, Murugesan S, Betzig E & Hammer JA Contractile actomyosin arcs promote the activation of primary mouse T cells in a ligand-dependent manner. PLoS One 12, e0183174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luca VC et al. Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science 355, 1320–1324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kolawole EM, Andargachew R, Liu B, Jacobs JR & Evavold BD 2D kinetic analysis of TCR and CD8 coreceptor for LCMV GP33 epitopes. Front. Immunol. 9, 2348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brazin KN et al. The T cell antigen receptor α transmembrane domain coordinates triggering through regulation of bilayer immersion and CD3 subunit associations. Immunity 49, 829–841.e826 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Majstoravich S et al. Lymphocyte microvilli are dynamic, actin-dependent structures that do not require Wiskott-Aldrich syndrome protein (WASp) for their morphology. Blood 104, 1396–1403 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Cai E et al. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science 356, eaal3118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jung Y et al. Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc. Natl Acad. Sci. USA 113, E5916–E5924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wucherpfennig KW, Gagnon E, Call MJ, Huseby ES & Call ME Structural biology of the T-cell receptor: insights into receptor assembly, ligand recognition, and initiation of signaling. Cold Spring Harb. Perspect. Biol. 2, a005140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barda-Saad M et al. Dynamic molecular interactions linking the T cell antigen receptor to the actin cytoskeleton. Nat. Immunol. 6, 80–89 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Klieger Y et al. Unique ζ-chain motifs mediate a direct TCR-actin linkage critical for immunological synapse formation and T-cell activation. Eur. J. Immunol. 44, 58–68 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Comrie WA & Burkhardt JK Action and traction: cytoskeletal control of receptor triggering at the immunological synapse. Front. Immunol. 7, 68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y et al. Platelet integrins exhibit anisotropic mechanosensing and harness piconewton forces to mediate platelet aggregation. Proc. Natl Acad. Sci. USA 115, 325–330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu B, Chen W & Zhu C Molecular force spectroscopy on cells. Annu. Rev. Phys. Chem. 66, 427–451 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Fiore VF, Ju L, Chen Y, Zhu C & Barker TH Dynamic catch of a Thy-1-α5β1+syndecan-4 trimolecular complex. Nat. Commun. 5, 4886 (2014). [DOI] [PubMed] [Google Scholar]
- 76.Lou J & Zhu C A structure-based sliding-rebinding mechanism for catch bonds. Biophys. J. 92, 1471–1485 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yago T et al. Platelet glycoprotein Ibα forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J. Clin. Invest. 118, 3195–3207 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee CY et al. Actin depolymerization under force is governed by lysine 113:glutamic acid 195-mediated catch-slip bonds. Proc. Natl Acad. Sci. USA 110, 5022–5027 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chowell D et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359, 582–587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roy NH, MacKay JL, Robertson TF, Hammer DA & Burkhardt JK Crk adaptor proteins mediate actin-dependent T cell migration and mechanosensing induced by the integrin LFA-1. Sci. Signal. 11, eaat3178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Elosegui-Artola A, Trepat X & Roca-Cusachs P Control of mechanotransduction by molecular clutch dynamics. Trends Cell Biol. 28, 356–367 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Grashoff C et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ringer P et al. Multiplexing molecular tension sensors reveals piconewton force gradient across talin-1. Nat. Methods 14, 1090–1096 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Elosegui-Artola A et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 18, 540–548 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Beddoe T et al. Antigen ligation triggers a conformational change within the constant domain of the αβ T cell receptor. Immunity 30, 777–788 (2009). [DOI] [PubMed] [Google Scholar]
- 86.Krogsgaard M et al. Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation. Mol. Cell 12, 1367–1378 (2003). [DOI] [PubMed] [Google Scholar]
- 87.Lee JK et al. T cell cross-reactivity and conformational changes during TCR engagement. J. Exp. Med. 200, 1455–1466 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reiser JB et al. A T cell receptor CDR3β loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex. Immunity 16, 345–354 (2002). [DOI] [PubMed] [Google Scholar]
- 89.van der Merwe PA & Dushek O Mechanisms for T cell receptor triggering. Nat. Rev. Immunol. 11, 47–55 (2011). [DOI] [PubMed] [Google Scholar]
- 90.Kuhns MS & Davis MM TCR signaling emerges from the sum of many parts. Front. Immunol. 3, 159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee MS et al. A mechanical switch couples T cell receptor triggering to the cytoplasmic juxtamembrane regions of CD3ζζ. Immunity 43, 227–239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang VT et al. Initiation of T cell signaling by CD45 segregation at ‘close contacts’. Nat. Immunol. 17, 574–582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.James JR & Vale RD Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature 487, 64–69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Valitutti S, Müller S, Cella M, Padovan E & Lanzavecchia A Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature 375, 148–151 (1995). [DOI] [PubMed] [Google Scholar]
- 95.McKeithan TW Kinetic proofreading in T-cell receptor signal transduction. Proc. Natl Acad. Sci. USA 92, 5042–5046 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dushek O, Das R & Coombs D A role for rebinding in rapid and reliable T cell responses to antigen. PLOS Comput. Biol. 5, e1000578 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lever M, Maini PK, van der Merwe PA & Dushek O Phenotypic models of T cell activation. Nat. Rev. Immunol. 14, 619–629 (2014). [DOI] [PubMed] [Google Scholar]
- 98.Siller-Farfán JA & Dushek O Molecular mechanisms of T cell sensitivity to antigen. Immunol. Rev. 285, 194–205 (2018). [DOI] [PubMed] [Google Scholar]
- 99.Stefanová ŠtefanováI. et al. TCR ligand discrimination is enforced by competing ERK positive and SHP-1 negative feedback pathways. Nat. Immunol. 4, 248–254 (2003). [DOI] [PubMed] [Google Scholar]
- 100.Yang W et al. Dynamic regulation of CD28 conformation and signaling by charged lipids and ions. Nat. Struct. Mol. Biol. 24, 1081–1092 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Aleksic M et al. Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity 32, 163–174 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang J et al. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature 464, 932–936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jiang N et al. Two-stage cooperative T cell receptor-peptide major histocompatibility complex-CD8 trimolecular interactions amplify antigen discrimination. Immunity 34, 13–23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Adams JJ et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity 35, 681–693 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zarnitsyna V & Zhu C T cell triggering: insights from 2D kinetics analysis of molecular interactions. Phys. Biol. 9, 045005 (2012). [DOI] [PubMed] [Google Scholar]
- 106.Zhu C, Jiang N, Huang J, Zarnitsyna VI & Evavold BD Insights from in situ analysis of TCR-pMHC recognition: response of an interaction network. Immunol. Rev. 251, 49–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu B et al. 2D TCR-pMHC-CD8 kinetics determines T-cell responses in a self-antigen-specific TCR system. Eur. J. Immunol. 44, 239–250 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.O’Donoghue GP, Pielak RM, Smoligovets AA, Lin JJ & Groves JT Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. eLife 2, e00778 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pielak RM et al. Early T cell receptor signals globally modulate ligand:receptor affinities during antigen discrimination. Proc. Natl Acad. Sci. USA 114, 12190–12195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Altan-Bonnet G, Germain RN & Modeling T Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 3, e356 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu CSC et al. Cutting edge: Piezo1 mechanosensors optimize human T cell activation. J. Immunol. 200, 1255–1260 (2018). [DOI] [PubMed] [Google Scholar]
- 112.Lillemeier BF et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 90–96 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yokosuka T et al. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat. Immunol. 6, 1253–1262 (2005). [DOI] [PubMed] [Google Scholar]
- 114.Yokosuka T et al. Spatiotemporal regulation of T cell costimulation by TCR-CD28 microclusters and protein kinase C θ translocation. Immunity 29, 589–601 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yokosuka T et al. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J. Exp. Med. 209, 1201–1217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pageon SV et al. Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl Acad. Sci. USA 113, E5454–E5463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brunner-Weinzierl MC & Rudd CE CTLA-4 and PD-1 control of T-cell motility and migration: implications for tumor immunotherapy. Front. Immunol. 9, 2737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.LaCroix AS, Lynch AD, Berginski ME & Hoffman BD Tunable molecular tension sensors reveal extension-based control of vinculin loading. eLife 7, e33927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jurchenko C & Salaita KS Lighting up the force: investigating mechanisms of mechanotransduction using fluorescent tension probes. Mol. Cell. Biol. 35, 2570–2582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.June CH, O’Connor RS, Kawalekar OU, Ghassemi S & Milone MC CAR T cell immunotherapy for human cancer. Science 359, 1361–1365 (2018). [DOI] [PubMed] [Google Scholar]