Abstract
Antibiotic-resistant Enterococcus (ARE) are among leading causes of nosocomial infections worldwide. Enterococcus spp. are ubiquitous in sewage, which can contaminate surface waters via many pathways, providing a route of exposure for humans. This review focuses on ARE in marine and estuarine habitats, including marine animals. Phylogenetic confirmation of the genus Enterococcus and intermediate or full resistance to clinically relevant antibiotics were inclusion criteria. The proportion of resistant isolates varied greatly among antibiotics, for example, 24.2% for ampicillin and 2.4% for vancomycin. The water column contained the highest proportion of ARE observations (18.8%), followed by animal feces and tissues (14.8%), sediment (9.4%), and sand (2.0%). The proportion of multidrug-resistant isolates was the greatest in animal tissue and fecal samples, followed by water and sediments. This review indicates that clinically relevant ARE are present in marine/estuarine habitats and that animals may be important reservoirs.
Keywords: antibiotic resistance, enterococci, marine, estuarine, review
Introduction
Antibiotic-resistant bacteria and their genes are among the most recalcitrant issues in infectious disease. Pathogens once thought nearly vanquished, such as Mycobacterium tuberculosis and Neisseria gonorrohea, are reemerging in antibiotic-resistant strains that are difficult to treat, whereas others formerly considered relatively unimportant opportunists such as Enterococcus species have acquired genes to combat multiple antibiotics, even those of last resort such as vancomycin (VRE) [1,2]. As a result, ARE are among the leading causes of nosocomial infections [3] and have been recently listed as a serious threat to public health [4]. Enterococcus species, including the most clinically important members of the genus, Enterococcus faecalis and Enterococcus faecium, are normal flora of the human gastrointestinal tract, and are thus ubiquitous in domestic sewage [5,6]. Sewage can contaminate surface and groundwater via many pathways, including inadequate treatment of sewage, spills due to infrastructure malfunction or overload, and land or lagoon disposal of animal waste [7,8], providing a route of exposure for humans who ingest or otherwise come in contact with contaminated water [8].
Recent reports indicate that antibiotics are over-prescribed worldwide [9–11]. For example, in the United States nearly 300 million antibiotic prescriptions annually (equivalent to more than 800 prescriptions per 1000 individuals) were prescribed for outpatient care alone [11]. An estimated 30% of those antibiotic prescriptions were unnecessary or inappropriate because they were prescribed for respiratory conditions for which they were not indicated (e.g. viral infections, bronchitis, asthma and allergy, nonsuppurative otitis media) or the antibiotic prescribed did not adhere to treatment guidelines (e.g. β-lactams are first-line therapy for common bacterial infections yet azithromycin is the most commonly prescribed antibiotic) [10,11]. Table 1 provides a summary of the most commonly prescribed antibiotic classes (along with the example antibiotics for each) used in the treatment of infections caused by various gram-positive organisms, including Enterococcus.
Table 1.
Most prescribed/used antibiotic classes for infections caused by Enterococcus species.
| Classification | Examples | Usage | Activity spectrum | Comments | References |
|---|---|---|---|---|---|
| Glycopeptides | Vancomycin teicoplanin telavancin | Serious infections of blood, watery diarrhea, skin infections, endocarditis, bone and joint infections, and meningitis | C. difficile, E. faecalis, S. aureus | Prescriptions worldwide | [39] |
| Quinolones | Ciprofloxacin lomefloxacin norfloxacin ofloxacin | Treat conditions associated with bacteria including lower respiratory tract infections, skin infections and urinary tract infections. | Streptococcus spp., P. aeruginosa, S. aureus | Widely used in human, animal husbandry and veterinary medicine | [40] |
| Macrolides | Erythromycin azithromycin clarithromycin | Used to treat infections of the respiratory tract, whooping cough, diphtheria syphilis, ear, intestine, gynecological, urinary tract, and skin infections. | Enterococcus spp., M. pneumoniae, V. trachomatis, L. pneumophila, C. jejuni, B. pertussis | The most commonly prescribed antibiotics worldwide used in human and veterinary medicine | [41] |
| Sulfonamides | Sulfisoxazole sulfadiazine | Treat urinary tract infections and parasitic disease | S. typhi, Serratia, Enterobacter, Pneumocystis spp. and some protozoa, such as Toxoplasma and Plasmodia | Widely used in clinical and veterinary medicine to treat bacterial and protozoal infections | [42] |
| Tetracyclines | Tetracycline | Treat many different bacterial infections of the skin, intestines, respiratory tract, urinary tract, genitals, lymph nodes, and other body systems. | Gram-positive bacilli, Clostridium and Nocardia spp. and gram-negative bacteria such as, Chlamydia, Mycoplasmas, Rickettsia, Vibrio, Helicobacter spp. and protozoan parasites. | Tetracyclines retain important roles in both human and veterinary medicine globally | [43] |
| Lipopeptides | Daptomycin | Complicated skin infection and MRSA | E. faecalis, E. faecium, S. aureus, | Used in clinical and veterinary medicine | [44] |
| Aminoglycosides | Rifampicin gentamicin amikacin neomycin | Treat several types of bacterial infections, including tuberculosis, leprosy and other bacterial infections. | Gram-positive cocci (except for some enterococci), H. influenzae, N. gonorrhoeae, N. meningitidis, Legionella spp., L. monocytogenes, M. tuberculosis | Used in clinical and veterinary medicine worldwide | [45] |
| Lincosamides | Lincomycin, clindamycin, pirlimycin | Used to treat severe bacterial infections | Streptococcus, Staphylococcus and Mycoplasma spp. | Rarely used due to toxicity and adverse effects. Used for patients that are allergic to penicillin. First-choice use antibiotic in veterinary. | [46] |
| Beta-lactams | Penicillin, amoxicillin, cephalosporins, carbapenems, aminopenicillins | Treatment of bacterial infections caused by susceptible organisms. | Streptococcus spp., S. aureus, L. monocytogenes, Enterococcus spp., P. mirabilis, E. coli | Used in clinical and veterinary medicine worldwide | [47] |
| Oxazolidinones | Linezolid, eperzolid | Treatment of pneumonia, skin infections, and infections that are resistant to other antibiotics | E. faecium, S. aureus, S. agalactiae, S pneumoniae, S. pyogenes | Used in clinical and veterinary medicine | [48] |
This review focuses on antibiotic resistance in Enterococcus in marine and estuarine ecosystems. Currently Enterococcus species are the only fecal indicator bacteria recommended for monitoring the sanitary quality of marine and brackish waters in the U.S [5]. Furthermore, recent review articles have summarized available literature on antibiotic-resistant bacteria and their genes in wastewater and freshwaters [12,13], but marine environments have received less attention. This review focuses on antibiotic-resistant Enterococcus species isolated from marine and estuarine ecosystems; specifically those that displayed intermediate or full resistance to antibiotics used in human medicine or animal husbandry [14,15](Table 2). Habitats sampled within these ecosystems included water, sediment, sand, and marine animals (e.g. feces and tissues). Many publications on antibiotic-resistant bacteria in the environment include isolates with low-level resistance in the data and/or do not confirm the identity of isolates. Such practices can exaggerate the potential relevance of findings to human health, and therefore we used resistance level and phylogenetic confirmation as criteria for inclusion in the study.
Table 2.
Study characteristics and antibiotic resistance tested among the studies.
| Reference | Location | Habitat | Antibiotic included in primary isolation* | Antibiotics tested | Antibiotic resistance reported |
|---|---|---|---|---|---|
| [34] | Italy | Sediments | None | Ampicillin, chloramphenicol, ciprofloxacin, erythromycin, quinupristin/dalfopristin, streptomycin, tetracycline, vancomycin | Chloramphenicol, ciprofloxacin, erythromycin, quinupristin/dalfopristin, tetracycline, |
| [22] | Italy | Water and sediment | Tetracycline (10 mg/L) or erythromycin (20 mg/L) or ampicillin (20 mg/L), or linezolid (10 mg/L), or levofloxacin (20 mg/L), or chloramphenicol (40 mg/L) or gentamicin (250 mg/L) or streptomycin (1000 mg/L) | Ampicillin, chloramphenicol, erythromycin, gentamicin, levofloxacin, linezolid, streptomycin, tetracycline | Ampicillin, erythromycin, tetracycline |
| [21] | Italy | Sediments | None | Ampicillin, erythromycin, gentamicin, tetracycline | None |
| [16] | Portugal | Gilthead seabream | Vancomycin (4 mg/L) | Ampicillin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, quinupristin/dalfopristin, streptomycin, teicoplanin, tetracycline, vancomycin | Ampicillin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, quinupristin/dalfopristin, streptomycin, teicoplanin, tetracycline, vancomycin, |
| [32] | Poland | Water and sediment | None | Ampicillin, ciprofloxacin, daptomycin, erythromycin, gentamicin, levofloxacin, linezolid, quinupristin/dalfopristin, streptomycin, teicoplanin, tetracycline, vancomycin | Ampicillin, ciprofloxacin, erythromycin, levofloxacin, gentamicin, quinupristin/dalfopristin, streptomycin, tetracycline |
| [25] | Portugal | Echinoderms | None | Ampicillin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, quinupristin/dalfopristin, streptomycin, teicoplanin, tetracycline, vancomycin | Ampicillin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, quinupristin/dalfopristin, teicoplanin, tetracycline, vancomycin |
| [26] | India | Water | None | Ampicillin, clindamycin, erythromycin, penicillin, tetracycline | Ampicillin |
| [33] | Italy | Sediment | None | Ampicillin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, linezolid, tetracycline, vancomycin | Ampicillin, erythromycin, gentamicin, tetracycline |
| [17] | Portugal | Gilthead seabream | None | Ampicillin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, quinupristin/dalfopristin, streptomycin, teicoplanin, tetracycline, vancomycin | Ampicillin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, quinupristin/dalfopristin, streptomycin, tetracycline |
| [23] | Venezuela | Water | None | Ampicillin, ampicillin/sulbactam, penicillin, sulfamethoxazole | Ampicillin, ampicillin/sulbactam, penicillin, sulfamethoxazole |
| [29] | Brazil | Sea turtles, sea birds, mammals (Minke and, Humpback whale, Risso’s dolphin) | None | Ampicillin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, nitrofurantoin, norfloxacin, rifampicin, streptomycin, tetracycline, vancomycin | Ciprofloxacin, erythromycin, norfloxacin, rifampicin, tetracycline |
| [20] | Malaysia | Water and sediment | None | Ampicillin, chloramphenicol, erythromycin, tetracycline | Ampicillin, chloramphenicol, erythromycin, tetracycline |
| [37] | Italy | Sand | Tetracycline (10 mg/L) or erythromycin (20 mg/L) or ampicillin (20 mg/L), or linezolid (10 mg/L), or levofloxacin (20 mg/L), or chloramphenicol (40 mg/L) or gentamicin (250 mg/L) or streptomycin (1000 mg/L) | Ampicillin, erythromycin, gentamicin, tetracycline, vancomycin | Erythromycin, tetracycline, vancomycin |
| [24] | United States | Water | Ampicillin (50 mg/L), or cefotaxime (25 mg/L), or ciprofloxacin (0.1 mg/L), or erythromycin (20 mg/L), or streptomycin (50 mg/L), or sulfamethoxazole (250 mg/L), or tetracycline (18 mg/L) | Ampicillin, ciprofloxacin, erythromycin, streptomycin, sulfamethoxazole, tetracycline | Ampicillin, ciprofloxacin, erythromycin, streptomycin, sulfamethoxazole, tetracycline |
| [28] | Australia | Silver gull | Vancomycin (32 mg/L) | Ampicillin, chloramphenicol, ciprofloxacin, erythromycin, gentamicin, nitrofurantoin, quinupristin/dalfopristin, rifampicin, teicoplanin, tetracycline, tigecycline, vancomycin | Ampicillin, erythromycin, gentamicin, rifampicin, teicoplanin, vancomycin |
| [19] | Italy | Venus clam | None | Ampicillin, daptomycin, erythromycin, gentamicin, levofloxacin, linezolid, quinupristin/dalfopristin, streptomycin, teicoplanin, tetracycline, vancomycin | Ampicillin, daptomycin, erythromycin, gentamicin, levofloxacin, linezolid, quinupristin/dalfopristin, streptomycin, tetracycline |
| [27] | Italy | Water | None | Ampicillin, clindamycin, erythromycin, gentamicin, tetracycline, vancomycin | Ampicillin, clindamycin, erythromycin, gentamicin, tetracycline, vancomycin |
| [31] | Puerto Rico | Water and sand | Vancomycin (6 mg/L) | Vancomycin | Vancomycin |
| [36] | Puerto Rico | Water and sand | Tetracycline (16 mg/L) or vancomycin (20 mg/L) | Vancomycin | Vancomycin |
Denotes the type and concentration of antibiotics used to select for resistant Enterococcus spp.
Literature Search Criteria
We searched PubMed (https://www.ncbi.nlm.nih.gov/pubmed/) and Web of Science (https://clarivate.com/webofsciencegroup/solutions/web-of-science/) data-bases for articles published between 2009 and 2019 and containing the following keywords: enterocc* AND antibiotic resistan* (OR antibiotic susceptible OR antimicrobial resistan*) AND marine (OR sea OR salt OR estuarine OR brackish). Three hundred ten unique articles were first examined to verify that studies measured antibiotic-resistant Enterococcus spp. in marine or estuarine habitats. Two hundred eighty-eight articles were eliminated due to at least one of the following reasons: (1) studies were carried out in wastewater and/or hospital setting, (2) food items were tested, and (3) the antimicrobial effect of extracts/chemicals was tested. The remaining 22 articles [16–37] were further examined to ensure that (1) confirmation of genus and/or species was carried out using either biochemical (e.g. API20Strep, matrix-assisted laser desorption ionization time of flight mass spectrometry) or phylogenetic (e.g. DNA sequencing, quantitative PCR or [q]PCR) methods and (2) antibiotics with clinical application for Enterococcus species were used at recommended concentrations and isolates displayed either intermediate or full resistance, as described in Performance Standards for Antimicrobial Susceptibility Testing Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) [14,15] or as evidenced by presence of appropriate genes (e.g. vanA, vanB). Some publications did not discriminate between intermediate and full resistance; therefore, we have grouped them into a single category. Three publications were removed because of failure to confirm genus/species [35] or use of inappropriate antibiotics and/or antibiotic concentrations [18,30]. Within the remaining 19 articles, certain antibiotics tested were either inappropriate for Enterococcus (e.g. clindamycin) or were tested at concentrations insufficient to isolate organisms with intermediate or full resistance [17,20,23–26,31,33], and these partial data sets were also removed from our analysis.
Characteristics of included studies
A summary of studies containing location, habitat (estuarine/marine waters and/or sediments and animal tissue and/or fecal samples) and antibiotics tested is provided in Table 2. Study locations were distributed worldwide, including Australia, Europe, North America, Southeast Asia, South America, and India (Table 2). The primary isolation method (e.g. selective-differential media supplemented with antibiotics or not) is also included. Seven of 19 studies (36.8%) relied on media supplemented with antibiotics for isolation of bacteria. Utilization of media supplemented with antibiotics can bias the results by increasing the likelihood of selecting the resistant organisms and can therefore affect the calculated resistance frequency. Although we did not separate studies based on isolation method when calculating the proportion of resistant isolates, we note this issue particularly in the case where small sample size (number of studies) results in a high proportion of resistant isolates.
In many studies isolates were tested against multiple antibiotics, yielding a total of 9099 observations on 1769 unique isolates. Because many studies did not report results on an isolate-by-isolate basis, comparisons across antibiotics are reported on a per-observation basis. When the proportion of isolates resistant to a given antibiotic is reported, the calculation is simply the number of resistant to the given antibiotic divided by the total number tested against the antibiotic. Although many studies tested multiple antibiotics against multiple isolates, the multiple antibiotics were not necessarily tested against a single isolate to generate multidrug-resistant (MDR) data. The MDR comparisons were performed for studies that explicitly tested multiple antibiotics on single, confirmed Enterococcus isolates.
Studies most frequently tested Enterococcus for resistance to ampicillin, followed by erythromycin, tetracycline, vancomycin, chloramphenicol, gentamicin, streptomycin, and ciprofloxacin (Table 2). Less than 50% of studies tested resistance to quinupristin-dalfopristin, teicoplanin, linezolid, levofloxacin, nitrofurantoin, rifampicin, sulfamethoxazole-trimethoprim (Table 2). Resistance to daptomycin, kanamycin, norfloxacin, penicillin, and tigecycline was tested in only one study each (Table 2). At least one study reported resistance to ampicillin, erythromycin, tetracycline, gentamicin, ciprofloxacin, quinupristin-dalfopristin, streptomycin, vancomycin, chloramphenicol, levofloxacin, rifampicin, sulfamethoxazole-trimethoprim, daptomycin, norfloxacin and penicillin (Table 2), whereas resistance to linezolid, nitrofurantoin, and tigecycline was not observed.
Relationship of habitat and Enterococcus species to antibiotic resistance
The proportion of antibiotic-resistant Enterococcus isolates varied widely by habitat (Table 3). For example, of the 1133 isolates tested for ampicillin susceptibility across all habitats, 24.2% were resistant, but the proportion of Enterococcus isolated from water that was resistant to ampicillin was 37.6%. A smaller proportion of isolates from animal fecal and tissue samples (17.4%), as well as sediment samples (9.7%) were ampicillin resistant. A greater proportion of Enterococcus isolates were resistant to erythromycin (29.9%) compared to ampicillin, and followed a different pattern of habitat distribution, as isolates from animal samples were most frequently resistant (37.4%), followed closely by isolates from water (36.6%), while sediment isolates were least frequently resistant (9.7%). One publication [23] reported data from water and oysters so that the source of isolates could not be distinguished, and therefore they were not accounted for in Table 3.
Table 3.
Proportion of antibiotic-resistant Enterococcus spp. (%) by habitat and total isolates tested against each antibiotic.
| Number of isolates tested and percent resistant (%) | |||||
|---|---|---|---|---|---|
| Water | Sediment | Sand | Animals | All habitats | |
| Ampicillin | 495 (37.6%) | 299 (9.70%) | 5 (0%) | 334 (17.4%) | 1133 (24.2%) |
| Chloramphenicol | 150 (0.67%) | 149 (7.39%) | N/Aa | 432 (0.93%) | 731 (2.2%) |
| Ciprofloxacin | 81 (38.3%) | 98 (8.16%) | N/A | 288 (31.3%) | 467 (27.6%) |
| Daptomycin | 74 (0%) | N/A | N/A | 46 (43.5%) | 120 (16.7%) |
| Erythromycin | 495 (36.6%) | 299 (9.70%) | 5 (80%) | 334 (37.4%) | 1133 (29.9%) |
| Gentamicin | 272 (20.6%) | 214 (0.93%) | 5 (0%) | 334 (2.69%) | 825 (8.1%) |
| Levofloxacin | 120 (3.33%) | 14 (0%) | N/A | 46 (21.7%) | 180 (7.8%) |
| Linezolid | 120 (0%) | 64 (0%) | N/A | 48 (0%) | 232 (0%) |
| Nitrofurantoin | N/A | N/A | N/A | 64 (0%) | 64 (0%) |
| Norfloxacin | N/A | N/A | N/A | 62 (12.9%) | 62 (12.9%) |
| Penicillin | 112 (0%) | N/A | N/A | N/A | 112 (0%) |
| Quinupristin-dalfopristin | 66 (54.5%) | 48 (22.9%) | N/A | 272 (17.3%) | 386 (24.4%) |
| Rifampicin | N/A | N/A | N/A | 64 (60.9%) | 64 (60.9%) |
| Streptomycin | 127 (6.30%) | 62 (9.68%) | N/A | 334 (2.99%) | 523 (4.6%) |
| Sulfamethoxazole | 7 (100%) | N/A | N/A | N/A | 7 (100%) |
| Teicoplanin | 74 (0%) | N/A | N/A | 272 (3.68%) | 346 (2.9%) |
| Tetracycline | 495 (13.1%) | 299 (19.4%) | 5 (100%) | 334 (26.9%) | 1133 (19.2%) |
| Tigecycline | N/A | N/A | N/A | 2 (0%) | 2 (0%) |
| Vancomycin | 479 (4.35%) | 98 (0%) | 421 (0%) | 334 (3.29%) | 1332 (2.4%) |
N/A: No isolates obtained from that combination of habitat and antibiotic
Vancomycin resistance was much less common than resistance to ampicillin or erythromycin (2.5% of all isolates tested); but was observed most frequently in isolates from water (4.4%) followed by animal samples (3.3%). No vancomycin-resistant Enterococcus were isolated from sediment (n = 98) or sand (n = 421). One study tested for vancomycin resistance in water and sand, finding 3.1% to be resistant, but the proportion of isolates from each habitat was not specified [31]. Considering all isolates (n = 1769) tested against all antibiotics (n = 9099 observations), isolates from water were most frequently resistant (18.8%) followed by those from animals (14.8%), sediment (9.4%), and finally sand (2.0%). It should be noted that 95% of observations on sand isolates were for vancomycin resistance, which was rare in all habitats, thus the frequency of antibiotic resistance in sand is artificially low. The observed frequency of resistance in various habitats is influenced by the fact that studies varied in terms of habitats sampled and antibiotics tested, for example, the mean number of antibiotics used in water habitats was 5.1, that in sediment 7.3, that in animals 10.7, and that in sand 2.7.
Eight different Enterococcus spp. were isolated across all studies including E. avium, E. casseliflavus, E. dispar, E. durans, E. faecalis, E. faecium, E. gallinarum, and E. hirae, Figure 1 shows a compilation of antibiotic resistance by species across all studies. A bar denoting a particular species indicates that resistance to that antibiotic was observed in at least one study. E. faecium and E. faecalis displayed resistance to 14 and 13 antibiotics, respectively, in at least one study, and sulfamethoxazole was the only antibiotic to which all E. faecium and E. faecalis were sensitive (Figure 1). E. casseliflavus and E. durans isolates displayed resistance to 9 antibiotics each, but each species was susceptible to a different group of antibiotics. E. casseliflavus was susceptible to daptomycin, penicillin, rifampicin, streptomycin, sulfamethoxazole and teicoplanin, while E. durans was sensitive to ciprofloxacin, daptomycin, gentamicin, levofloxacin, rifampicin, and streptomycin. E. gallinarum and E. hirae were resistant to eight and nine antibiotics each, E. dispar to five, and E. avium to three antibiotics (Figure 1). All eight Enterococcus species displayed resistance to erythromycin, followed by tetracycline (seven species), then ampicillin, gentamicin, quinupristin-dalfopristin and vancomycin (six species each). E. avium and E. dispar were the only species that were completely susceptible to ampicillin and quinupristin-dalfopristin, while E. durans and E. hirae were the only species completely susceptible to gentamicin (Figure 1).
Figure 1.
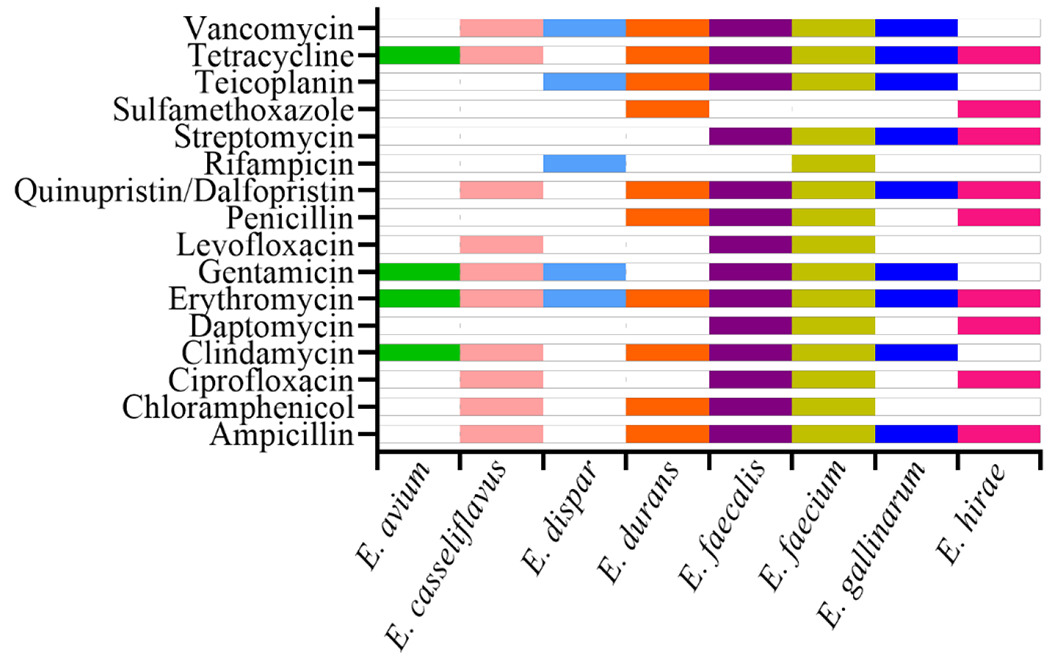
Antibiotic resistance observations in various Enterococcus spp. across all studies. A bar denotes resistance to a particular antibiotic observed in at least one isolate of a given species. Antibiotics to which no resistance was observed in any species are omitted.
MDR enterococci
We considered resistance to three or more antibiotics to represent multidrug resistance. Of 17 studies that tested individual isolates against at least three antibiotics, ten reported a total of 102 MDR Enterococcus isolates [16,17,19,20,24,25,27–29,34]. Within these ten studies, only one study reported resistance to either eight [17] or seven antibiotics [16], with one [24] and three studies [24,25,34] reporting resistance to six and five antibiotics, respectively. Resistance to four or three antibiotics was reported more frequently, in six [16,19,24,25,28,29] and seven [16,17,19,20,25,29,34] studies, respectively. Multidrug resistance profiles most commonly included resistance to erythromycin and tetracycline, followed by ampicillin, ciprofloxacin, and quinupristin-dalfopristin.
The great majority of the 102 confirmed MDR Enterococcus isolates (83.3%) came from animal fecal and/or tissues sample [16,17,19,25,28,29], followed by water (8.8%) [20,24,27]. The proportion of MDR isolates among all Enterococcus isolated from animal samples was 9.9%, while that from the water was 3.1%. The proportion of MDR isolates from sediments were described in only one study, but comprised 7.8% of all MDR identified and 6.5% of all Enterococcus isolated from this habitat [34]. The comparatively high frequency of MDR isolates in animal samples was probably influenced by the comparatively high number of antibiotics used in the studies in which animal samples were tested. Of the 102 isolates, speciation was performed on 98, and the majority (59.2%) were identified as E. faecium, followed by E. faecalis (27.6%). MDR E. gallinarum and E. hirae were detected less frequently (4.1% and 5.1%, respectively), with only a singular MDR isolate each of E. casseliflavus, E. dispar and E. durans. Even though isolates from water were most frequently resistant to at least one antibiotic, animal samples from seabirds, whales and clams were the most common source of MDR strains (and in particular E. faecium).
Conclusions
Our review of the relevant literature indicates that ARE are widespread in marine environments, but their observed distribution across habitats varied widely. The proportion of ARE observations was the greatest in water column, followed by animals (fecal and tissue samples) and sediment/sand and was not consistent across antibiotics, for example, the proportion of ampicillin resistant isolates in water samples (37.6%) was more than twice that of any other habitat. Among different Enterococcus species, E. faecium and E. faecalis were resistant to the greatest number of different antibiotics. Erythromycin was the only antibiotic to which each species showed resistance in at least one study. Resistance to vancomycin was observed infrequently, and it was mainly associated with isolates from the water column and animals. Similarly, MDR isolates were detected most frequently in marine animals, suggesting they may be an important source and/or reservoir of these strains in these environments.
While we were able to synthesize the available information on ARE in marine habitats to draw some preliminary conclusions, our review also identified many knowledge gaps which should be addressed in future studies to aid in more accurate risk assessment for exposure to ARE in aquatic environments. The most common issue identified resides with study design and methodological considerations. No selective-differential medium is infallible, and all produce a certain percentage of false-positive or false-negative results, even widely used formulations that are a part of standard methods such as mEI [38]. Confirmation of the phylogenetic identity of ARE isolates is of paramount importance due to the fact that the frequency of false-positive isolates can be much higher when antibiotics are used in the primary isolation medium, and to the public health implications of the topic. Another important methodological consideration is the antibiotic tested and the resistance level reported. Standardized protocols, such as CLSI [14] and EUCAST [15], provide clinically relevant antibiotics for many different genera, as well as suggestions for interpretation of the resulting data. Enterococcus spp. antibiotic resistance has three tiers (sensitive, intermediate, and full resistance) with the fully resistant phenotype carrying the most public health implications. Despite the availability of these guidelines, many studies used either inappropriate antibiotics, tested below the intermediate resistance level, or failed to discriminate between intermediate and full-level resistance. Addressing these shortcomings by standardizing methodological approaches to studying antibiotic-resistant bacteria in the environment will enable better management of ARE and other antibiotic-resistant bacteria and allow for more accurate risk assessment.
Acknowledgments
Disclaimer
The United States Environmental Protection Agency through its Office of Research and Development funded and managed the research described here. It has been subjected to Agency’s administrative review and approved for publication. The views expressed in this article are those of the author(s) and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
References:
- 1.* Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJ, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:4e00534–00513. [DOI] [PMC free article] [PubMed] [Google Scholar]; Examines the effect of various factors, including human behavior, on emergence of multi-drug resistant Enterococcus faecium strain
- 2.Tenover FC, McDonald LC. 2005. Vancomycin-resistant staphylococci and enterococci: epidemiology and control. Curr Opin Infect Dis 18:300–305. [DOI] [PubMed] [Google Scholar]
- 3.Weiner-Lastinger LM, Abner S, Edwards JR, Kallen AJ, Karlsson M, Magill SS, Pollock D, See I, Soe MM, Walters MS, Dudeck MA. 2020. Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network, 2015–2017. Infection Control and Hospital Epidemiology 41:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2019. Antibiotic Resistance Threats in the United States. U.S. Department of Health and Human Services, Atlanta, GA. [Google Scholar]
- 5.** Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. 2012. Enterococci in the Environment. Microbiology and Molecular Biology Reviews 76:685–706. [DOI] [PMC free article] [PubMed] [Google Scholar]; An extensive review of enterococci ecology, including responses to environmental stressors and descriptions of environmental reservoirs of Enterococcus spp.
- 6.Harwood VJ, Levine AD, Scott TM, Chivukula V, Lukasik J, Farrah SR, Rose JB. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Applied and Environmental Microbiology 71:3163–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.* Korajkic A, Wanjugi P, Brooks L, Cao Y, Harwood VJ. 2019. Persistence and Decay of Fecal Microbiota in Aquatic Habitats. Microbiol Mol Biol Rev 83. [DOI] [PMC free article] [PubMed] [Google Scholar]; An in-depth review of the effect of various abiotic and abiotic factors on decay of fecal microbiota, including Enterococccus spp.
- 8.** Burgmann H, Frigon D, W HG, C MM, Pruden A, Singer AC, B FS, Zhang T. 2018. Water and sanitation: an essential battlefront in the war on antimicrobial resistance. FEMS Microbiol Ecol 94. [DOI] [PubMed] [Google Scholar]; Provides a multi-level framework for limiting the spread of antimicrobial resistance.
- 9.World Health Organization. 2018. WHO Report on Surveillance of Antibiotic Consumption: 2016–2018 early implementation. Organization WH, Geneva. [Google Scholar]
- 10.Fleming-Dutra KE, Hersh AL, Shapiro DJ, Bartoces M, Enns EA, File TM Jr., Finkelstein JA, Gerber JS, Hyun DY, Linder JA, Lynfield R, Margolis DJ, May LS, Merenstein D, Metlay JP, Newland JG, Piccirillo JF, Roberts RM, Sanchez GV, Suda KJ, Thomas A, Woo TM, Zetts RM, Hicks LA. 2016. Prevalence of Inappropriate Antibiotic Prescriptions Among US Ambulatory Care Visits, 2010–2011. JAMA 315:1864–1873. [DOI] [PubMed] [Google Scholar]
- 11.Hicks LA, Bartoces MG, Roberts RM, Suda KJ, Hunkler RJ, Taylor TH Jr., Schrag SJ. 2015. US outpatient antibiotic prescribing variation according to geography, patient population, and provider specialty in 2011. Clin Infect Dis 60:1308–1316. [DOI] [PubMed] [Google Scholar]
- 12.Pazda M, Kumirska J, Stepnowski P, Mulkiewicz E. 2019. Antibiotic resistance genes identified in wastewater treatment plant systems - A review. Sci Total Environ 697:134023. [DOI] [PubMed] [Google Scholar]
- 13.Nnadozie CF, Odume ON. 2019. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ Pollut 254:113067. [DOI] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. 2016. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA. [Google Scholar]
- 15.European Committee on Antimicrobial Susceptibility Testing. 2019. Breakpoint tables for interpretation of MICs and zone diameteres. Version 9.0. [Google Scholar]
- 16.Barros J, Andrade M, Radhouani H, Lopez M, Igrejas G, Poeta P, Torres C. 2012. Detection of vanA-containing Enterococcus species in faecal microbiota of gilthead seabream (Sparus aurata). Microbes Environ 27:509–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.* Barros J, Igrejas G, Andrade M, Radhouani H, Lopez M, Torres C, Poeta P. 2011. Gilthead seabream (Sparus aurata) carrying antibiotic resistant enterococci. A potential bioindicator of marine contamination? Marine Pollution Bulletin 62:1245–1248. [DOI] [PubMed] [Google Scholar]; Reported mult-drug resistant enterococci isolates that displayed resistance to eight different antibiotics- more than what was reported in any other studies.
- 18.Bourouni OC, El Bour M, Calo-Mata P, Barros-Velazquez J. 2012. Antimicrobial Resistance and Potential Probiotic Application of Enterococcus spp. in Sea Bass and Sea Bream Aquaculture. Antibiotic Resistant Bacteria - a Continuous Challenge in the New Millennium doi:Book_Doi 10.5772/1058:513-530. [DOI] [Google Scholar]
- 19.Citterio B, Pasquaroli S, Mangiaterra G, Vignaroli C, Di Sante L, Leoni F, Chierichetti S, Ottaviani D, Rocchi M, Biavasco F. 2017. Venus clam (Chamelea gallina): A reservoir of multidrug-resistant enterococci. Food Control 82:184–189. [Google Scholar]
- 20.Dada AC, Ahmad A, Usup G, Heng LY. 2013. Speciation and antimicrobial resistance of enterococci isolated from recreational beaches in Malaysia. Environmental Monitoring and Assessment 185:1583–1599. [DOI] [PubMed] [Google Scholar]
- 21.Di Cesare A, Luna GM, Vignaroli C, Pasquaroli S, Tota S, Paroncini P, Biavasco F. 2013. Aquaculture can promote the presence and spread of antibiotic-resistant enterococci in marine sediments. PLoS One 8:e62838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Cesare A, Vignaroli C, Luna GM, Pasquaroli S, Biavasco F. 2012. Antibiotic-resistant enterococci in seawater and sediments from a coastal fish farm. Microb Drug Resist 18:502–509. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Delgado M, Suarez P. 2009. Multiple antibiotic resistances of enteric bacteria isolated from recreational coastal waters and oysters of the Caribbean Sea. Annals of Microbiology 59:409–414. [Google Scholar]
- 24.Kawecki S, Kuleck G, Dorsey JH, Leary C, Lum M. 2017. The prevalence of antibiotic-resistant bacteria (ARB) in waters of the Lower Ballona Creek Watershed, Los Angeles County, California. Environmental Monitoring and Assessment 189. [DOI] [PubMed] [Google Scholar]
- 25.Marinho C, Silva N, Pombo S, Santos T, Monteiro R, Goncalves A, Micael J, Rodrigues P, Costa AC, Igrejas G, Poeta P. 2013. Echinoderms from Azores islands: An unexpected source of antibiotic resistant Enterococcus spp. and Escherichia coli isolates. Marine Pollution Bulletin 69:122–127. [DOI] [PubMed] [Google Scholar]
- 26.Meena B, Anburajan L, Sathish T, Raghavan RV, Jha DK, Venkateshwaran P, Das AK, Dheenan PS, Vinithkumar NV, Dharani G, Kirubagaran R. 2015. Enterococcus species diversity and molecular characterization of biomarker genes in Enterococcus faecalis in Port Blair Bay, Andaman and Nicobar Islands, India. Marine Pollution Bulletin 94:217–227. [DOI] [PubMed] [Google Scholar]
- 27.Monticelli LS, Decembrini F, Bergamasco A, Caruso G. 2019. Water quality assessment of transitional and coastal marine Sicilian waters (Italy): Ecological and epidemiological significance of multiple antimicrobial resistant Enterococcus spp. Estuarine Coastal and Shelf Science 217:173–184. [Google Scholar]
- 28.Oravcova V, Svec P, Literak I. 2017. Vancomycin-resistant enterococci with vanA and vanB genes in Australian gulls. Environ Microbiol Rep 9:316–318. [DOI] [PubMed] [Google Scholar]
- 29.Prichula J, Pereira RI, Wachholz GR, Cardoso LA, Tolfo NC, Santestevan NA, Medeiros AW, Tavares M, Frazzon J, d’Azevedo PA, Frazzon AP. 2016. Resistance to antimicrobial agents among enterococci isolated from fecal samples of wild marine species in the southern coast of Brazil. Mar Pollut Bull 105:51–57. [DOI] [PubMed] [Google Scholar]
- 30.Rince A, Baliere C, Hervio-Heath D, Cozien J, Lozach S, Parnaudeau S, Le Guyader FS, Le Hello S, Giard JC, Sauvageot N, Benachour A, Strubbia S, Gourmelon M. 2018. Occurrence of Bacterial Pathogens and Human Noroviruses in Shellfish-Harvesting Areas and Their Catchments in France. Frontiers in Microbiology 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts MC, Soge OO, Giardino MA, Mazengia E, Ma G, Meschke JS. 2009. Vancomycin-resistant Enterococcus spp. in marine environments from the West Coast of the USA. J Appl Microbiol 107:300–307. [DOI] [PubMed] [Google Scholar]
- 32.Sadowy E, Luczkiewicz A. 2014. Drug-resistant and hospital-associated Enterococcus faecium from wastewater, riverine estuary and anthropogenically impacted marine catchment basin. BMC Microbiol 14:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vignaroli C, Luna GM, Pasquaroli S, Di Cesare A, Petruzzella R, Paroncini P, Biavasco F. 2013. Epidemic Escherichia coli ST131 and Enterococcus faecium ST17 in Coastal Marine Sediments from an Italian Beach. Environmental Science & Technology 47:13772–13780. [DOI] [PubMed] [Google Scholar]
- 34.Vignaroli C, Pasquaroli S, Citterio B, Di Cesare A, Mangiaterra G, Fattorini D, Biavasco F. 2018. Antibiotic and heavy metal resistance in enterococci from coastal marine sediment. Environ Pollut 237:406–413. [DOI] [PubMed] [Google Scholar]
- 35.Vignesh S, Muthukumar K, James RA. 2012. Antibiotic resistant pathogens versus human impacts: a study from three eco-regions of the Chennai coast, southern India. Mar Pollut Bull 64:790–800. [DOI] [PubMed] [Google Scholar]
- 36.Santiago-Rodriguez TM, Rivera JI, Coradin M, Toranzos GA. 2013. Antibiotic-resistance and virulence genes in Enterococcus isolated from tropical recreational waters. J Water Health 11:387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Cesare A, Pasquaroli S, Vignaroli C, Paroncini P, Luna GM, Manso E, Biavasco F. 2014. The marine environment as a reservoir of enterococci carrying resistance and virulence genes strongly associated with clinical strains. Environ Microbiol Rep 6:184–190. [DOI] [PubMed] [Google Scholar]
- 38.U.S. Environmental Protection Agency: Method 1600: enterococci in water by membrane filtration using membrane Enterococcus indoxyl-beta-D-glucoside agar (mEI). Washington, D.C: U.S. Environmental Protection Agency; 2006. [Google Scholar]
- 39.Binda E, Marinelli F, Marcone GL. 2014. Old and New Glycopeptide Antibiotics: Action and Resistance. Antibiotics (Basel) 3:572–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siporin C 1989. The evolution of fluorinated quinolones: pharmacology, microbiological activity, clinical uses, and toxicities. Annu Rev Microbiol 43:601–627. [DOI] [PubMed] [Google Scholar]
- 41.Dinos GP. 2017. The macrolide antibiotic renaissance. Br J Pharmacol 174:2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdul Qadir M, Ahmed M, Aslam H, Waseem S, Imtiaz Shafiq M. 2015. Amidine Sulfonamides and Benzene Sulfonamides: Synthesis and Their Biological Evaluation. Journal of Chemistry 2015:524056. [Google Scholar]
- 43.Grossman TH. 2016. Tetracycline Antibiotics and Resistance. Cold Spring Harb Perspect Med 6:a025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bell JM, Turnidge JD, Sader HS, Jones RN. 2010. Antimicrobial activity and spectrum of daptomycin: results from the surveillance program in Australia and New Zealand (2008). Pathology 42:470–473. [DOI] [PubMed] [Google Scholar]
- 45.Krause KM, Serio AW, Kane TR, Connolly LE. 2016. Aminoglycosides: An Overview. Cold Spring Harb Perspect Med 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rezanka T, Spizek J, Sigler K. 2007. Medicinal Use of Lincosamides and Microbial Resistance to Them. Anti-Infective Agents in Medicinal Chemistry 6:133–144. [Google Scholar]
- 47.Kristich CJ, Rice LB, Arias CA. 2014. Enterococcal Infection-Treatment and Antibiotic Resistance In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: From Commensals to Leading Causes of Drug Resistant Infection, Boston. [PubMed] [Google Scholar]
- 48.Ager S, Gould K. 2012. Clinical update on linezolid in the treatment of Gram-positive bacterial infections. Infect Drug Resist 5:87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]


