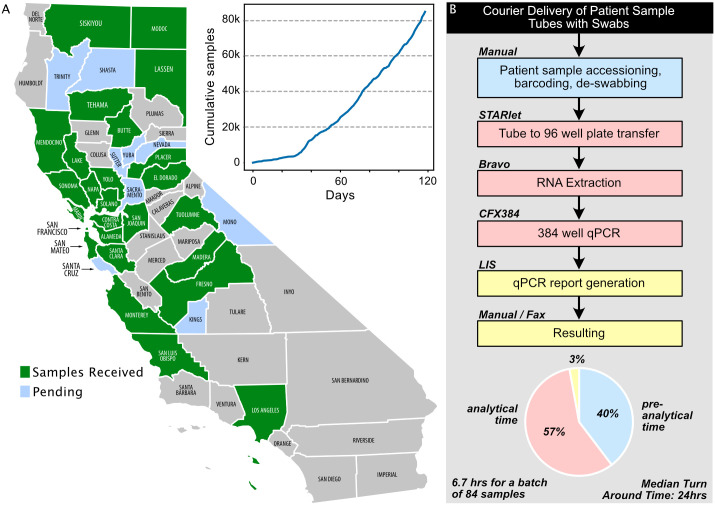
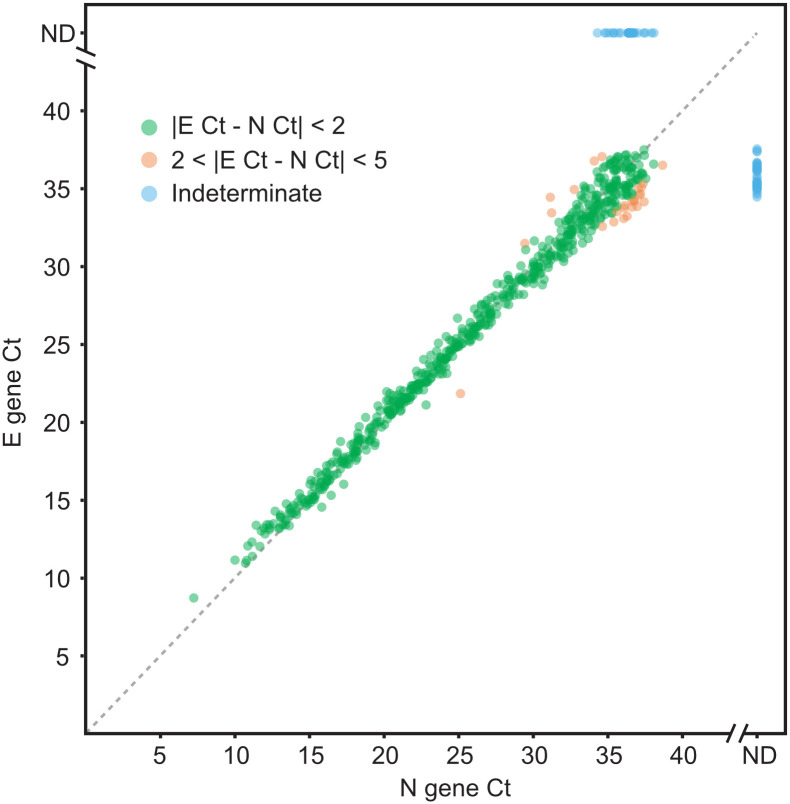
Emily D Crawford
Emily D Crawford
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
2University of California San Francisco, Department of Microbiology and Immunology, San Francisco, California, United States of America
1,2,
Irene Acosta
Irene Acosta
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Vida Ahyong
Vida Ahyong
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Erika C Anderson
Erika C Anderson
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Shaun Arevalo
Shaun Arevalo
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Daniel Asarnow
Daniel Asarnow
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Shannon Axelrod
Shannon Axelrod
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Patrick Ayscue
Patrick Ayscue
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Camillia S Azimi
Camillia S Azimi
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Caleigh M Azumaya
Caleigh M Azumaya
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Stefanie Bachl
Stefanie Bachl
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Iris Bachmutsky
Iris Bachmutsky
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Aparna Bhaduri
Aparna Bhaduri
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Jeremy Bancroft Brown
Jeremy Bancroft Brown
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Joshua Batson
Joshua Batson
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Astrid Behnert
Astrid Behnert
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Ryan M Boileau
Ryan M Boileau
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Saumya R Bollam
Saumya R Bollam
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Alain R Bonny
Alain R Bonny
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
David Booth
David Booth
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
Michael Jerico B Borja
Michael Jerico B Borja
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
David Brown
David Brown
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Bryan Buie
Bryan Buie
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Cassandra E Burnett
Cassandra E Burnett
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Lauren E Byrnes
Lauren E Byrnes
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Katelyn A Cabral
Katelyn A Cabral
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
5University of California San Francisco, Institute for Neurodegenerative Diseases, San Francisco, California, United States of America
3,5,
Joana P Cabrera
Joana P Cabrera
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Saharai Caldera
Saharai Caldera
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
6University of California San Francisco, Division of Infectious Disease, San Francisco, California, United States of America
1,6,
Gabriela Canales
Gabriela Canales
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Gloria R Castañeda
Gloria R Castañeda
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Agnes Protacio Chan
Agnes Protacio Chan
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Christopher R Chang
Christopher R Chang
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Arthur Charles-Orszag
Arthur Charles-Orszag
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
7Howard Hughes Medical Institute, Chevy Chase, Maryland, United States of America
3,7,
Carly Cheung
Carly Cheung
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Unseng Chio
Unseng Chio
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Eric D Chow
Eric D Chow
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
Y Rose Citron
Y Rose Citron
8University of California, Berkeley, California, United States of America
8,
Allison Cohen
Allison Cohen
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Lillian B Cohn
Lillian B Cohn
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
9University of California San Francisco, Department of Experimental Medicine, San Francisco, California, United States of America
1,9,
Charles Chiu
Charles Chiu
10University of California San Francisco, Department of Laboratory Medicine, San Francisco, California, United States of America
10,
Mitchel A Cole
Mitchel A Cole
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Daniel N Conrad
Daniel N Conrad
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Angela Constantino
Angela Constantino
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Andrew Cote
Andrew Cote
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Tre’Jon Crayton-Hall
Tre’Jon Crayton-Hall
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Spyros Darmanis
Spyros Darmanis
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Angela M Detweiler
Angela M Detweiler
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Rebekah L Dial
Rebekah L Dial
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
Shen Dong
Shen Dong
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Elias M Duarte
Elias M Duarte
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
David Dynerman
David Dynerman
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Rebecca Egger
Rebecca Egger
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Alison Fanton
Alison Fanton
8University of California, Berkeley, California, United States of America
8,
Stacey M Frumm
Stacey M Frumm
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Becky Xu Hua Fu
Becky Xu Hua Fu
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Valentina E Garcia
Valentina E Garcia
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
Julie Garcia
Julie Garcia
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Christina Gladkova
Christina Gladkova
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
7Howard Hughes Medical Institute, Chevy Chase, Maryland, United States of America
3,7,
Miriam Goldman
Miriam Goldman
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Rafael Gomez-Sjoberg
Rafael Gomez-Sjoberg
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
M Grace Gordon
M Grace Gordon
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
James C R Grove
James C R Grove
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Shweta Gupta
Shweta Gupta
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Alexis Haddjeri-Hopkins
Alexis Haddjeri-Hopkins
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Pierce Hadley
Pierce Hadley
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
5University of California San Francisco, Institute for Neurodegenerative Diseases, San Francisco, California, United States of America
3,5,
John Haliburton
John Haliburton
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Samantha L Hao
Samantha L Hao
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
George Hartoularos
George Hartoularos
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Nadia Herrera
Nadia Herrera
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Melissa Hilberg
Melissa Hilberg
10University of California San Francisco, Department of Laboratory Medicine, San Francisco, California, United States of America
10,
Kit Ying E Ho
Kit Ying E Ho
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Nicholas Hoppe
Nicholas Hoppe
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Shayan Hosseinzadeh
Shayan Hosseinzadeh
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Conor J Howard
Conor J Howard
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Jeffrey A Hussmann
Jeffrey A Hussmann
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Elizabeth Hwang
Elizabeth Hwang
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Danielle Ingebrigtsen
Danielle Ingebrigtsen
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Julia R Jackson
Julia R Jackson
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Ziad M Jowhar
Ziad M Jowhar
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Danielle Kain
Danielle Kain
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
James Y S Kim
James Y S Kim
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Amy Kistler
Amy Kistler
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Oriana Kreutzfeld
Oriana Kreutzfeld
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Jessie Kulsuptrakul
Jessie Kulsuptrakul
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Andrew F Kung
Andrew F Kung
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Charles Langelier
Charles Langelier
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
6University of California San Francisco, Division of Infectious Disease, San Francisco, California, United States of America
1,6,
Matthew T Laurie
Matthew T Laurie
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
Lena Lee
Lena Lee
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Kun Leng
Kun Leng
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Kristoffer E Leon
Kristoffer E Leon
11Gladstone Institute, San Francisco, California, United States of America
11,
Manuel D Leonetti
Manuel D Leonetti
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Sophia R Levan
Sophia R Levan
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Sam Li
Sam Li
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
Aileen W Li
Aileen W Li
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Jamin Liu
Jamin Liu
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
Heidi S Lubin
Heidi S Lubin
12eSix Development, Oakland, California, United States of America
12,
Amy Lyden
Amy Lyden
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Jennifer Mann
Jennifer Mann
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Sabrina Mann
Sabrina Mann
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Gorica Margulis
Gorica Margulis
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Diana M Marquez
Diana M Marquez
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Bryan P Marsh
Bryan P Marsh
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Calla Martyn
Calla Martyn
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Elizabeth E McCarthy
Elizabeth E McCarthy
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Aaron McGeever
Aaron McGeever
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Alexander F Merriman
Alexander F Merriman
11Gladstone Institute, San Francisco, California, United States of America
11,
Lauren K Meyer
Lauren K Meyer
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Steve Miller
Steve Miller
10University of California San Francisco, Department of Laboratory Medicine, San Francisco, California, United States of America
10,
Megan K Moore
Megan K Moore
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Cody T Mowery
Cody T Mowery
11Gladstone Institute, San Francisco, California, United States of America
11,
Tanzila Mukhtar
Tanzila Mukhtar
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Lusajo L Mwakibete
Lusajo L Mwakibete
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Noelle Narez
Noelle Narez
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Norma F Neff
Norma F Neff
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Lindsay A Osso
Lindsay A Osso
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Diter Oviedo
Diter Oviedo
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Suping Peng
Suping Peng
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Maira Phelps
Maira Phelps
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Kiet Phong
Kiet Phong
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Peter Picard
Peter Picard
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Lindsey M Pieper
Lindsey M Pieper
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Neha Pincha
Neha Pincha
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Angela Oliveira Pisco
Angela Oliveira Pisco
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Angela Pogson
Angela Pogson
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Sergei Pourmal
Sergei Pourmal
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
Robert R Puccinelli
Robert R Puccinelli
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Andreas S Puschnik
Andreas S Puschnik
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Elze Rackaityte
Elze Rackaityte
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
Preethi Raghavan
Preethi Raghavan
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Madhura Raghavan
Madhura Raghavan
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
James Reese
James Reese
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Joseph M Replogle
Joseph M Replogle
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Hanna Retallack
Hanna Retallack
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
Helen Reyes
Helen Reyes
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Donald Rose
Donald Rose
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Marci F Rosenberg
Marci F Rosenberg
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Estella Sanchez-Guerrero
Estella Sanchez-Guerrero
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Sydney M Sattler
Sydney M Sattler
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Laura Savy
Laura Savy
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Stephanie K See
Stephanie K See
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Kristin K Sellers
Kristin K Sellers
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Paula Hayakawa Serpa
Paula Hayakawa Serpa
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
6University of California San Francisco, Division of Infectious Disease, San Francisco, California, United States of America
1,6,
Maureen Sheehy
Maureen Sheehy
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Jonathan Sheu
Jonathan Sheu
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Sukrit Silas
Sukrit Silas
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Jessica A Streithorst
Jessica A Streithorst
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Jack Strickland
Jack Strickland
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Doug Stryke
Doug Stryke
10University of California San Francisco, Department of Laboratory Medicine, San Francisco, California, United States of America
10,
Sara Sunshine
Sara Sunshine
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
Peter Suslow
Peter Suslow
10University of California San Francisco, Department of Laboratory Medicine, San Francisco, California, United States of America
10,
Renaldo Sutanto
Renaldo Sutanto
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Serena Tamura
Serena Tamura
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Michelle Tan
Michelle Tan
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Jiongyi Tan
Jiongyi Tan
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Alice Tang
Alice Tang
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Cristina M Tato
Cristina M Tato
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Jack C Taylor
Jack C Taylor
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Iliana Tenvooren
Iliana Tenvooren
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Erin M Thompson
Erin M Thompson
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Edward C Thornborrow
Edward C Thornborrow
10University of California San Francisco, Department of Laboratory Medicine, San Francisco, California, United States of America
10,
Eric Tse
Eric Tse
13Joint Bioengineering Graduate Program, University of California, Berkeley, California, United States of America
13,
Tony Tung
Tony Tung
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Marc L Turner
Marc L Turner
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Victoria S Turner
Victoria S Turner
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Rigney E Turnham
Rigney E Turnham
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Mary J Turocy
Mary J Turocy
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Trisha V Vaidyanathan
Trisha V Vaidyanathan
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Ilia D Vainchtein
Ilia D Vainchtein
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Manu Vanaerschot
Manu Vanaerschot
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Sara E Vazquez
Sara E Vazquez
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
4,
Anica M Wandler
Anica M Wandler
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Anne Wapniarski
Anne Wapniarski
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
James T Webber
James T Webber
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Zara Y Weinberg
Zara Y Weinberg
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Alexandra Westbrook
Alexandra Westbrook
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Allison W Wong
Allison W Wong
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Emily Wong
Emily Wong
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Gajus Worthington
Gajus Worthington
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
1,
Fang Xie
Fang Xie
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Albert Xu
Albert Xu
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Terrina Yamamoto
Terrina Yamamoto
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Ying Yang
Ying Yang
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Fauna Yarza
Fauna Yarza
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Yefim Zaltsman
Yefim Zaltsman
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Tina Zheng
Tina Zheng
3University of California San Francisco, School of Medicine, San Francisco, California, United States of America
3,
Joseph L DeRisi
Joseph L DeRisi
1Chan Zuckerberg Biohub, San Francisco, California, United States of America
4University of California San Francisco, Department of Biochemistry and Biophysics, San Francisco, California, United States of America
1,4,*