Abstract
FimA is an important virulence factor of Porphyromonas gingivalis (P. gingivalis). According to its DNA sequence, the fimA genotype of P. gingivalis can be divided into six categories (I, Ib, Ⅱ, III, IV, V). The fimA gene may be a key factor in the diversity of virulence found in P. gingivalis. Moreover, the role fimA plays in the pathogenesis of P. gingivalis is closely associated with periodontitis, making it an important factor of study for disease prevention and treatment. In this study, the prevalence of fimA genotypes of P. gingivalis in patients with periodontal diseases was evaluated by meta-analysis. The Embase and PubMed databases were searched for articles from 1999 to 2019 using the following search terms: Porphyromonas gingivalis or P. gingivalis; periodontitis or chronic periodontal disease; fimA or fimA genotype. The reference lists of relevant published articles were searched manually. A total of 17 studies were included in this report. A statistical software package (Stata, version 11.0/mp, StataCorp) was utilized to calculate and analyze the P. gingivalis fimA genotypes for each combined incidence estimate. The pooled rates of fimA Ⅰ, fimA Ib, fimA Ⅱ, fimA Ⅲ, fimA Ⅳ and fimA Ⅴ genotypes of P. gingivalis were 8.4% (95% CI: 5.7–11.1), 11.7% (95% CI: 7.4–16), 42.9% (95% CI: 34.2–51.7), 6.5% (95% CI: 5.1–7.9), 17.8% (95% CI: 9.0–26.5), and 3.2% (95% CI: 1.6–4.9), respectively. This study showed that the fimA Ⅱ and fimA Ⅳ genotypes of P. gingivalis are highly present in patients with periodontal disease. Therefore, these two genotypes may be related to the pathogenesis and progress of periodontal disease, one of the main risk factors of periodontitis.
Introduction
Periodontitis is the chronic inflammation of periodontal tissue caused by an imbalance between local bacterial infection and host immune response, which leads to the formation of periodontal pockets. Periodontitis is characterized by the progressive loss of gum and alveolar bone absorption. Without treatment, this chronic inflammation can lead to the loss of affected teeth [1].
The major microorganisms associated with chronic periodontal disease are Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, Treponemadenticola, Prevotella nigrescens, and Fusobacterium nucleatum [2]. The most common bacterium related to periodontitis is Porphyromonas gingivalis (P. gingivalis), a gram-negative anaerobe. P. gingivalis colonizes in the mouth and is closely linked to chronic periodontitis [3–5].
Many virulent factors contribute to P. gingivalis’ ability to affect the gums; proteases (collagenases, gingipains, hemolysin, trypsin), lipopolysaccharides, its capsule, fimbriae, and hemagglutinins all work in parallel to colonize the host tissue [6–8]. However, studies point to fimbriae as the leading virulence factor of P. gingivalis [9, 10]. According to the nucleotide sequence differences of the fimA gene, which encodes the fimA protein subunit, the fimA gene is divided into six genotypes: I, Ib, Ⅱ, III, IV, and V [11]. Many animal experiments and clinical studies have shown that P. gingivalis with different fimA genotypes resulted in different virulent characteristics; genotypes I, III, and V are non-toxic, while fimA Ⅱ, Ib and IV are highly toxic [11–13].
Due to the high prevalence of periodontitis and the myriad of complications that patients face, determining the risk factors that influence the incidences of this disease is crucial to prevention and management strategies. FimA genotypes of P. gingivalis have been extensively researched and reports have indicated that fimA genotypes are closely connected with periodontitis. Therefore, it is of important to investigate the prevalence of fimA genotypes of P. gingivalis as they may provide a novel approach in the prevention and treatment of periodontitis. Accordingly, a meta-analysis is important for identifying the prevalence of fimA genotypes in periodontal patients [3, 5, 7]. Using a meta-analysis vs a systematic review has the advantages of a larger sample size and higher resolution by combining different study statistics, and is also a comprehensive, accurate, and effective way of understanding the subject under investigation [14]. In our study, articles obtained from Embase and PubMed databases from 1999 to 2019 were analyzed using a statistical software package. The aim of this study is to provide a comprehensive summary of the measures to explore the prevalence of fimA genotypes of P. gingivalis in patients with periodontal disease.
Materials and methods
Literature search
The Embase and PubMed databases were searched from 1999 to January 2019 using the following search terms: Porphyromonas gingivalis or P. gingivalis; periodontitis or chronic periodontal disease; fimA or fimA genotype. The reference lists of relevant published articles were searched manually.
Study selection
167 articles with the keywords were included in the initial list and 150 unrelated studies were excluded. In addition, all studies reporting a fimA genotype of P. gingivalis in chronic periodontal disease were reviewed. Studies were included only if they met the following criteria: (a) studies were written in English or Chinese; (b) studies were from between 1999 and 2019; (c) studies reported the rate of fimA genotypes of P. gingivalis among chronic periodontal disease. Studies were excluded using the following criteria: (a) studies were reviews, case reports, or comments; (b) studies were non-human studies; (c) studies were duplicates.
Data extraction and methodological assessment
Relevant information was extracted, including the year of publication, first author's name, sample size, total number of cases, and number of positive numbers. Adjusted point estimation with 95% confidence intervals was required for quality assessment (Table 1). Data extraction was completed by two authors. Disagreements were resolved by a third reviewer.
Table 1. Summary of data from selected articles on the fimA genotypes of P. gingivalis found in periodontitis.
| First author, published year | Language | Quality assessment score | Sample size | FimA genotype | |||||
|---|---|---|---|---|---|---|---|---|---|
| I | Ib | II | III | IV | V | ||||
| Amano A,1999 [7] | English | 6 | 73 | 4 | Nd | 43 | 5 | 9 | 0 |
| Amano A,2000 [15] | English | 6 | 121 | 8 | Nd | 80 | 7 | 35 | 21 |
| Beikler T,2003 [16] | English | 7 | 102 | 26 | 0 | 39 | 5 | 19 | 4 |
| Guo YH,2005 [17] | Chinese | 7 | 89 | 3 | 0 | 22 | 6 | 25 | 16 |
| Zhao L,2007 [12] | English | 7 | 94 | 8 | 9 | 23 | 6 | 20 | 3 |
| Yang BT,2016 [18] | Chinese | 8 | 24 | 0 | 3 | 10 | 3 | 4 | 3 |
| Enersen M,2008 [19] | English | 7 | 82 | 3 | 4 | 28 | 8 | 17 | 1 |
| Missailidis CG,2004 [20] | English | 7 | 135 | 6 | 33 | 53 | 9 | 7 | 0 |
| Teixeira SR, 2009 [21] | English | 5 | 152 | Nd | Nd | 47 | Nd | 117 | Nd |
| Davila-Perez C, 2007 [22] | English | 8 | 25 | 3 | 5 | 4 | 1 | 1 | 0 |
| Pérez-Chaparro PJ,2009 [5] | English | 7 | 30 | 2 | 5 | 16 | 3 | 4 | 0 |
| Sandra Moreno, 2015 [23] | English | 7 | 26 | 3 | 2 | 15 | 2 | 0 | 0 |
| Feng X,2013 [24] | English | 7 | 55 | 9 | 11 | 22 | 3 | 3 | 0 |
| Ji-Hoi Moon,2013 [3] | English | 7 | 277 | 36 | 49 | 191 | 19 | 43 | 14 |
| Miriam Puig-Silla,2012 [25] | English | 7 | 33 | 4 | 4 | 13 | 3 | 5 | 0 |
| Asano H,2003 [26] | English | 5 | 32 | 3 | Nd | 15 | 2 | 5 | 2 |
| Feng X,2014 [27] | English | 7 | 39 | 4 | 9 | 20 | 2 | 1 | 0 |
*NT = not tested.
The authors assessed data quality using an appropriate quality assessment tool [28]. We assessed each article’s quality using eight criteria: (1) definition of the target population; (2) representative probability sampling; (3) sample characteristics matching the population; (4) adequate response rates; (5) standardized data collection methods; (6) reliable survey measures; (7) effective survey measures; (8) appropriate statistical methods. The criteria score was either 0 or 1, which stood for "non conformity" or "conformity", respectively. The total score for each study was between 0 and 8.
Statistical analysis
We used a statistical software package (Stata, version 11.0/mp, StataCorp) to analyze fimA genotypes of P. gingivalis for each combined incidence estimate. The comprehensive prevalence estimate and 95% confidence interval (CI) were determined based on the random or fixed effect model. Considering the possibility of heterogeneity between the studies, the Q-test (P<0.10 was considered as significant heterogeneity) and I2 test (values 25, 50, 75%) were used to represent low, medium, and high heterogeneity, respectively. Due to high heterogeneity (P<0.10), the random effect model was used for statistical analysis. Furthermore, publication bias was assessed by Egger’s and Begger’s tests (P<0.05 represents significant publication bias).
Results
Study selection and characteristics of eligible studies
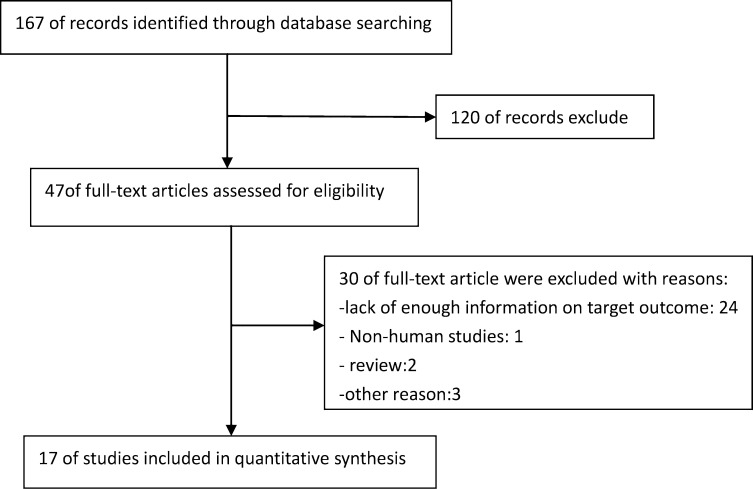
A total of 167 studies were originally selected through the search process. These articles were identified based on abstracts, titles, and full texts, and after the exclusion process a total of 17 papers were selected for our study. The total sample size was 1389 individuals, including samples from different countries. The search process is demonstrated in Fig 1.
Fig 1. Flow diagram of studies search.
Results of the meta-analysis
In the 17 included studies, the sample size was 1389 periodontal patients from articles published between 1999 and 2016. Studies were divided into two cohorts (1999–2010 and 2011–2016) for group analysis. To begin there were four main early genotypes: fimA Ⅰ, fimA Ⅱ, fimA Ⅲ, fimA Ⅳ and fimA Ⅴ. Over time, the fimA Ib subtype appeared (Table 1).
Correlation of fimA genotypes and periodontitis
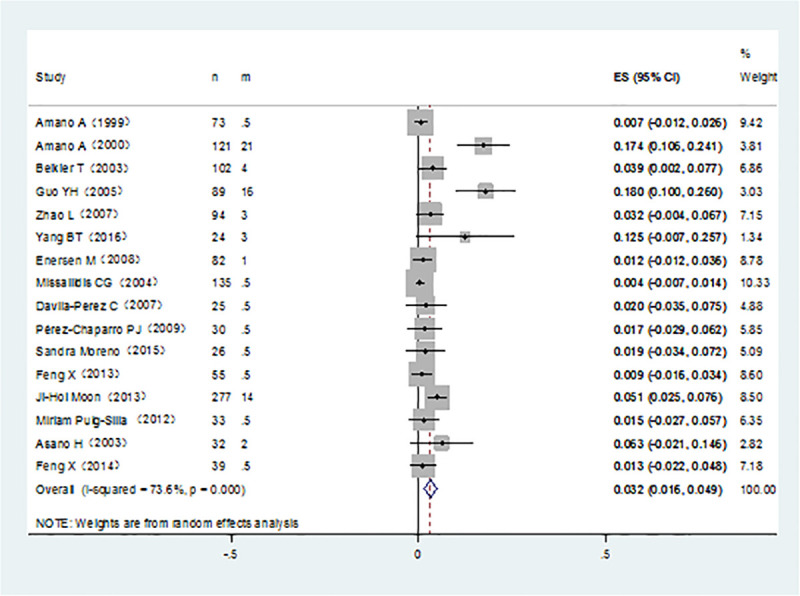
It was noticed that P. gingivalis with various fimA genotypes were present from chronic periodontitis patients. The pooled rates of fimAⅠ, fimA Ib, fimA Ⅱ, fimA Ⅲ, fimA Ⅳ, and fimA ⅤP. gingivalis were 8.4% (95% CI:5.7–11.1; Fig 2), 11.7% (95% CI: 7.4–16; Fig 3), 42.9% (95% CI: 34.2–51.7; Fig 4), 6.5% (95% CI: 5.1–7.9; Fig 5), 17.8%(95% CI: 9.0–26.5; Fig 6), and 3.2% (95% CI: 1.6–4.9; Fig 7) respectively. As we can see in Figs 3 and 6, the prevalence of fimA Ⅱ and fimA Ⅳ P. gingivalis were statistically significant compared to others. In general, the fimA Ⅱ and fimA Ⅳ genotypes contributed to a elevated risk of periodontitis.
Fig 2. Forest plot of the prevalence of the fimA I genotype of P. gingivalis in periodontitis.
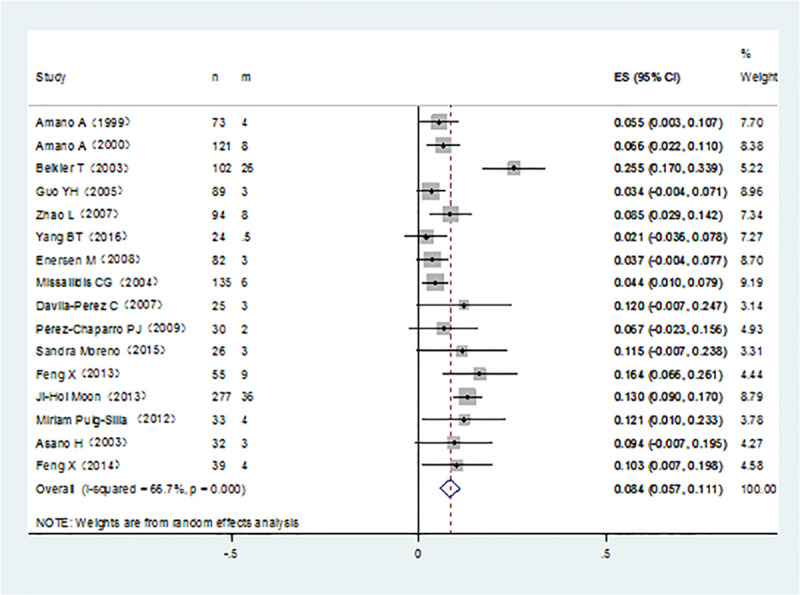
Fig 3. Forest plot of the prevalence of the fimA Ib genotype of P. gingivalis in periodontitis.
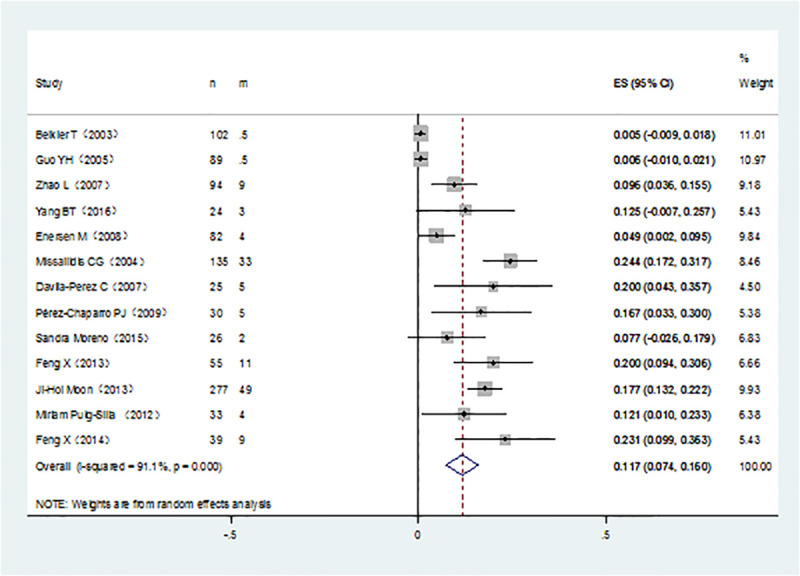
Fig 4. Forest plot of the prevalence of the fimA II genotype of P. gingivalis in periodontitis.
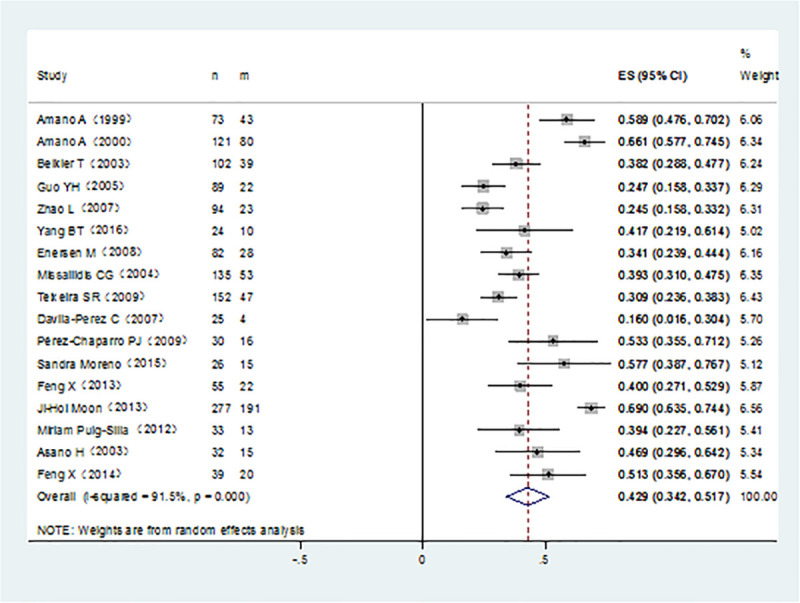
Fig 5. Forest plot of the prevalence of the fimA III genotype of P. gingivalis in periodontitis.
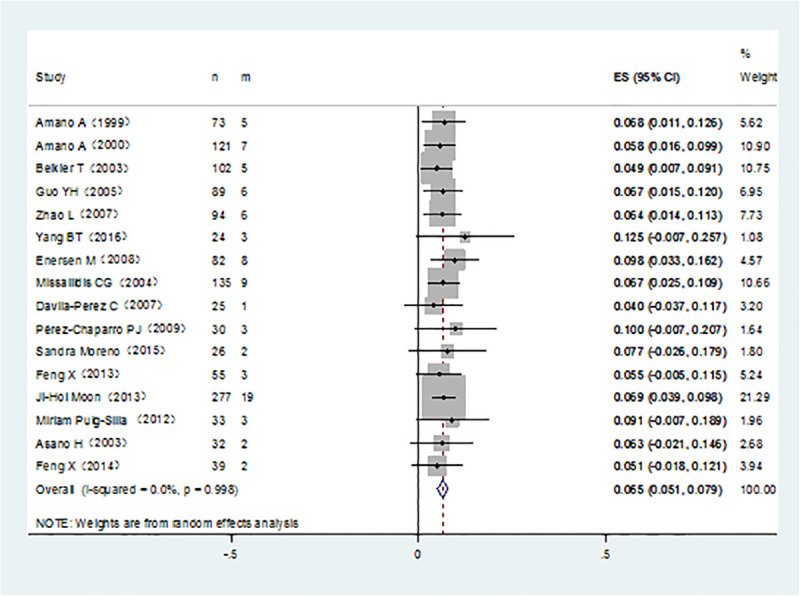
Fig 6. Forest plot of the prevalence of the fimA IV genotype of P. gingivalis in periodontitis.
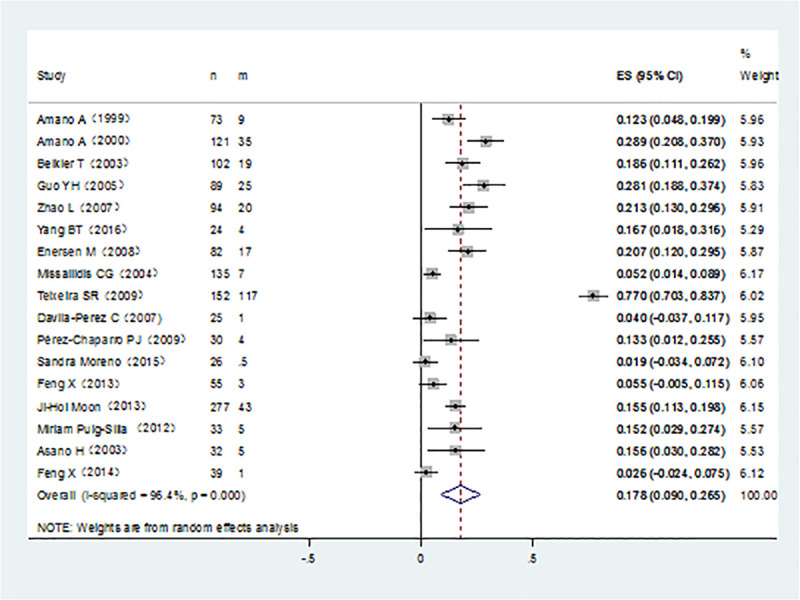
Fig 7. Forest plot of the prevalence of the fimA V genotype of P. gingivalis in periodontitis.
Publication bias
When the Begg’s funnel plots of the prevalence of the fimA Ⅰ, Ib, Ⅲ, and Ⅴ genotypes of P. gingivalis in chronic periodontal patients were examined, no sign of publication bias was observed. In fact, most studies were located in the funnel plots, and therefore the results of all related studies were included in the analysis (P = 0.005) (Fig 8).
Fig 8. Begg’s funnel plot of the prevalence of the fimA II genotype of P. gingivalis in periodontitis.
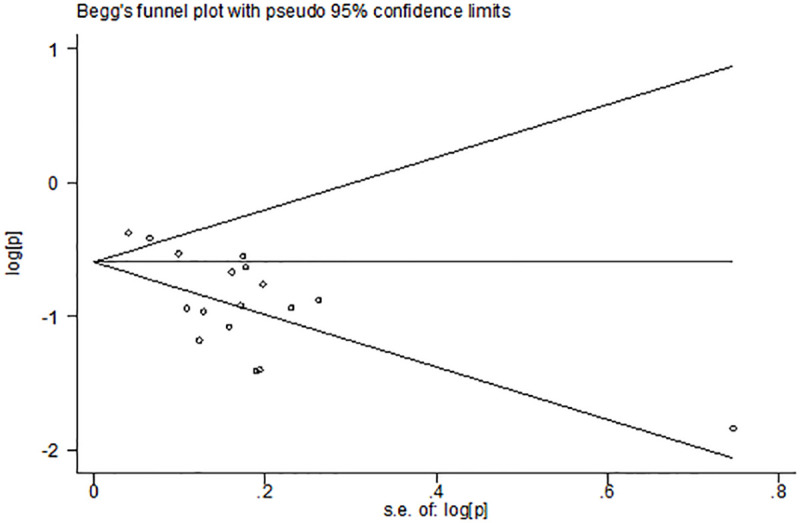
When examining the Begg’s funnel plot of the prevalence of the fimA Ⅱ genotype of P. gingivalis in chronic periodontal patients, signs of publication bias were observed (P = 0.005) (Fig 8).
Discussion
Previous studies indicated that fimA genotypes of P. gingivalis are closely connected with periodontitis. In this meta-analysis, we examined the presence of P. gingivalis with various fimA genotypes in chronic periodontitis patients. We found that the fimA Ⅱ and fimA Ⅳ genotypes of P. gingivalis were the two most predominant genotypes [7, 12, 15–19, 21, 24–27]. There are several plausible reasons that may explain why the fimA Ⅱ and fimA Ⅳ genotypes were more predominant in this study. Periodontitis is a chronic inflammatory disease that leads to persistent, low-level, systemic inflammation with elevated levels of circulating inflammatory markers [29]. Various studies have indicated that the P. gingivalis fimA protein could initiate an immune inflammatory response through a variety of receptors, signal pathways, and cytokines, which enable periodontal tissue cells to express proteolytic enzymes, such as matrix metalloproteinase (MMP). These proteolytic enzymes participate in the destruction of periodontal connective tissue and alveolar bone absorption, leading to bone defects [30–35]. The fimA Ⅱ genotype had a greater ability to adhere and invade epithelial cells in mouse models [36] and in vitro studies showed that fimA Ⅱ could induce higher levels of IL-6 and IL-β than fimA Ⅰ [37]. Compared with fimA Ⅰ, the fimA Ⅳ genotype was highly invasive, and this invasiveness was increased in gingival fibroblasts if there was the substituting of allele fimA Ⅳ for allele fimA Ⅰ [38].In addition, fimA Ⅱ and fimA Ⅳ genotypes of P. gingivalis could induce the expression and secretion of MMP-8 and MMP-9 in neutrophils in vivo and in vitro [39, 40]. On the contrary, some studies have implicated that the incidence of fimA Ⅰ and fimA Ib genotypes of P. gingivalis are also high [3, 16, 22–23]. A previous study indicated that the fimA Ⅰ genotype was significantly associated with healthy periodontal people [15]. Further investigation found that fimA Ib had a high sequence homology with fimA Ⅰ, but their distribution profiles were different [41]. Statistical analysis in the same study showed a significant relationship between the fimA Ib genotype and periodontitis [41]. The variations of fimA play a major definitive factor in the virulence and pathogenicity of P. gingivalis, in relation to the specific proteases, capsulation, and other factors that influence pathogenicity [42–44]. Presently, there is no other meta-analysis similar to this research in terms of ability to compare results. To the best of our knowledge, we identified the fimA genotypes of P. gingivalis, which is one of the main etiological factors of chronic periodontal disease. Exploration of the part these genotypes play in this disease can lead to the development of new treatment and prevention methods in the future. In recent years, the research on reducing the pathogenicity of P. gingivalis and preventing periodontal disease through the inhibition of fimA protein function has attracted much attention. Researchers have cloned an IgG monoclonal antibody from purified fimA protein and produced the monoclonal antibody using rice suspension [45]. These results suggest that the specific antibodies produced can be used for passive immunization to prevent P. gingivalis induced periodontal disease, but further research is needed. Recent studies have shown that photo-activated disinfection mediated by photosensitizer toluidine blue has a significant inhibitory effect on the formation of P. gingivalis biofilm. Because of this, the fimA gene is a suitable target for interactions with photo-activated disinfection [46, 47]. There were several limitations in this study that must be addressed. First, publication bias is an inevitable problem in a meta-analysis process, negative trials are sometimes less likely. Considering that our data were extracted from studies written in English and Chinese, which should reduce publication bias to some extent. Second, we were unable to avoid some influential factors, such as smoking, alcohol, age, and gender on account of inadequate data These factors may influence the prevalence of fimA genotypes of P. gingivalis and incidence of chronic periodontitis by affecting the ability of P. gingivalis to invade the gingival tissue and influence the malignant process. Third, the demographic features of the populations in these studies could have an impact on the results. Finally, some studies that examined the prevalence of fimA genotypes of P. gingivalis in periodontitis were not available.
In conclusion, the fimA gene is P. gingivalis' main virulence factor, it is important to identify the different fimA genotypes and their prevalence in chronic periodontitis. This study showed that the fimA Ⅱ and fimA Ⅳ genotypes were present at increased levels in patients with periodontal disease. Therefore, these two genotypes may be related to the pathogenesis and progression of periodontal disease and can be considered as the main potential risk factors of periodontal disease. These results indicate that the pathogenicity of fimA genotypes of P. gingivalis needs to be further studied.
Supporting information
(DOC)
Acknowledgments
The authors wish to thank Professor Dongmeng Qian and Dr. Qin Zheng for their help in revising the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors acknowledge the funding support of Qingdao Key Health Discipline Development Fund 2020-2022 to HW. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kato A, Imai K, Ochiai K, Ogata Y. Higher prevalence of Epstein-Barr virus DNA in deeper periodontal pockets of chronic periodontitis in Japanese patients. PLoS One. 2013; 8(8):e71990 10.1371/journal.pone.0071990 eCollection 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrell LN, Papapanou PN. Analytical epidemiology of periodontitis. J Clin Periodontol. 2005;32(Suppl 6):132–58. 10.1111/j.1600-051X.2005.00799.x . [DOI] [PubMed] [Google Scholar]
- 3.Moon JH, Herr Y, Lee HW, Shin SI, Kim C, Amano A, et al. Genotype analysis of Porphyromonas gingivalis fimA in Korean adults using new primers. J Med Microbiol. 2013;62(Pt 9):1290–1294. 10.1099/jmm.0.054247-0 . [DOI] [PubMed] [Google Scholar]
- 4.Bostanci N, Belibasakis GN. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012;333(1):1–9. 10.1111/j.1574-6968.2012.02579.x . [DOI] [PubMed] [Google Scholar]
- 5.Pérez-Chaparro PJ, Lafaurie GI, Gracieux P, Meuric V, Tamanai-Shacoori Z, Castellanos JE, et al. Distribution of Porphyromonas gingivalis fimA genotypes in isolates from subgingival plaque and blood sample during bacteremia. Biomedica. 2009;29(2):298–306. . [PubMed] [Google Scholar]
- 6.Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000. 1999;20:168–238. 10.1111/j.1600-0757.1999.tb00162.x . [DOI] [PubMed] [Google Scholar]
- 7.Amano A, Nakagawa I, Kataoka K, Morisaki I, Hamada S. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J Clin Microbiol.1999; 37(5):1426–30. 10.1128/JCM.37.5.1426-1430.1999 ; PMCID: PMC84792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inaba H, Nakano K, Kato T, Nomura R, Kawai S, Kuboniwa M, et al. Heterogenic virulence and related factors among clinical isolates of Porphyromonas gingivalis with type II fimbriae. Oral Microbiol Immunol. 2008;23(1):29–35. 10.1111/j.1399-302X.2007.00386.x . [DOI] [PubMed] [Google Scholar]
- 9.Nagano K, Hasegawa Y, Yoshida Y, Yoshimura F. A Major Fimbrilin Variant of Mfa1 Fimbriae in Porphyromonas gingivalis. J Dent Res. 2015;94(8):1143–8. 10.1177/0022034515588275 . [DOI] [PubMed] [Google Scholar]
- 10.Hasegawa Y, Iijima Y, Persson K, Nagano K, Yoshida Y, Lamont RJ, et al. Role of Mfa5 in Expression of Mfa1 Fimbriae in Porphyromonas gingivalis. J Dent Res. 2016;95(11):1291–7. 10.1177/0022034516655083 ; PMCID: PMC5076756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enersen M, Nakano K, Amano A. Porphyromonas gingivalis fimbriae. J Oral Microbiol. 2013;5 10.3402/jom.v5i0.20265 ; PMCID: PMC3647041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao L, Wu YF, Meng S, Yang H, OuYang YL, Zhou XD. Prevalence of fimA genotypes of Porphyromonas gingivalis and periodontal health status in Chinese adults.J Periodontal Res.2007;42(6):511–7. 10.1111/j.1600-0765.2007.00975.x . [DOI] [PubMed] [Google Scholar]
- 13.Krishnan M, Krishnan P, Chandrasekaran SC. Detection of Porphyromonas gingivalis fimA Type I Genotype in Gingivitis by Real-Time PCR-A Pilot Study. J Clin Diagn Res. 2016;10(6):ZC32–5. 10.7860/JCDR/2016/17938.7979 ; PMCID: PMC4963766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steiner M. Postnatal depression: a few simple questions. Fam Pract. 2002;19(5):469–70. 10.1093/fampra/19.5.469 . [DOI] [PubMed] [Google Scholar]
- 15.Amano A, Kuboniwa M, Nakagawa I, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res.2000;79(9):1664–8. 10.1177/00220345000790090501 . [DOI] [PubMed] [Google Scholar]
- 16.Beikler T, Peters U, Prajaneh S, Prior K, Ehmke B, Flemmig TF. Prevalence of Porphyromonas gingivalis fimA genotypes in Caucasians. Eur J Oral Sci. 2003;111(5):390–4. 10.1034/j.1600-0722.2003.00065.x . [DOI] [PubMed] [Google Scholar]
- 17.Guo YH, Wu YF, Liu TJ, Xiao XR, Zhou B, Zhou XP. The distribution of fimA genotype of Porphyromonas gingivalis in chronic periodontitis patients. Hua Xi Kou Qiang Yi Xue Za Zhi. 2005;23(2):99–102. . [PubMed] [Google Scholar]
- 18.Yang BT, Xu JL, He L, Meng HX, Xu L. Porphyromonas gingivalis FimA genotype distribution among periodontitis patients with type 2 diabetes. Zhonghua Kou Qiang Yi Xue Za Zhi. 2016;51(1):20–4. 10.3760/cma.j.issn.1002-0098.2016.01.006 . [DOI] [PubMed] [Google Scholar]
- 19.Enersen M, Olsen I, Kvalheim Ø, Caugant DA. fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis. J Clin Microbiol. 2008;46(1):31–42. 10.1128/JCM.00986-07 ; PMCID: PMC2224254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Missailidis CG, Umeda JE, Ota-Tsuzuki C, Anzai D, Mayer MP. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol Immunol. 2004;19(4):224–9. 10.1111/j.1399-302X.2004.00140.x . [DOI] [PubMed] [Google Scholar]
- 21.Teixeira SR, Mattarazo F, Feres M, Figueiredo LC, de Faveri M, Simionato MR, et al. Quantification of Porphyromonas gingivalis and fimA genotypes in smoker chronic periodontitis. J Clin Periodontol. 2009;36(6):482–7. 10.1111/j.1600-051X.2009.01411.x . [DOI] [PubMed] [Google Scholar]
- 22.Davila-Perez C, Amano A, Alpuche-Solis AG, Patiño-Marin N, Pontigo-Loyola AP, Hamada S, et al. Distribution of genotypes of Porphyromonas gingivalis in type 2 diabetic patients with periodontitis in Mexico. J Clin Periodontol. 2007;34(1):25–30. 10.1111/j.1600-051X.2006.01011.x . [DOI] [PubMed] [Google Scholar]
- 23.Moreno S, Jaramillo A, Parra B, Botero JE, Contreras A. Porphyromonas gingivalis Fim-A genotype distribution among Colombians. Colomb Med (Cali). 2015;46(3):122–7. ; PMCID: PMC4640434. [PMC free article] [PubMed] [Google Scholar]
- 24.Feng X, Zhang L, Xu L, Meng H, Lu R, Chen Z, et al. Detection of eight periodontal microorganisms and distribution of Porphyromonas gingivalis fimA genotypes in Chinese patients with aggressive periodontitis. J Periodontol. 2014;85(1):150–9. 10.1902/jop.2013.120677 . [DOI] [PubMed] [Google Scholar]
- 25.Puig-Silla M, Dasí-Fernández F, Montiel-Company JM, Almerich-Silla JM. Prevalence of fimA genotypes of Porphyromonas gingivalis and other periodontal bacteria in a Spanish population with chronic periodontitis. Med Oral Patol Oral Cir Bucal. 2012;17(6):e1047–53. 10.4317/medoral.17009 ; PMCID: PMC3505701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asano H, Ishihara K, Nakagawa T, Yamada S, Okuda K. Relationship between transmission of Porphyromonas gingivalis and fimA type in spouses. J Periodontol. 2003;74(9):1355–60. 10.1902/jop.2003.74.9.1355 . [DOI] [PubMed] [Google Scholar]
- 27.Feng X, Zhu L, Xu L, Meng H, Zhang L, Ren X, et al. Distribution of 8 periodontal microorganisms in family members of Chinese patients with aggressive periodontitis. Arch Oral Biol. 2015;60(3):400–7. 10.1016/j.archoralbio.2014.11.015 . [DOI] [PubMed] [Google Scholar]
- 28.Boyle MH. Guidelines for evaluating prevalence studies. Evidence-Based Mental Health. 1998;1:37–9. 10.1136/ebmh.1.2.37 [DOI] [Google Scholar]
- 29.Moutsopoulos NM, Madianos PN. Low-grade inflammation in chronic infectious diseases: paradigm of periodontal infections. Ann N Y Acad Sci. 2006;1088:251–64. 10.1196/annals.1366.032 . [DOI] [PubMed] [Google Scholar]
- 30.Eskan MA, Hajishengallis G, Kinane DF. Differential activation of human gingival epithelial cells and monocytes by Porphyromonas gingivalis fimbriae. Infect Immun. 2007;75(2):892–8. 10.1128/IAI.01604-06 ; PMCID: PMC1828485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hajishengallis G, Shakhatreh MA, Wang M, Liang S. Complement receptor 3 blockade promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J Immunol. 2007;179(4):2359–67. 10.4049/jimmunol.179.4.2359 . [DOI] [PubMed] [Google Scholar]
- 32.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3(2):133–46. 10.1038/nri1001 . [DOI] [PubMed] [Google Scholar]
- 33.Hajishengallis G, Wang M, Liang S, Triantafilou M, Triantafilou K. Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proc Natl Acad Sci U S A. 2008;105(36):13532–7. 10.1073/pnas.0803852105 ; PMCID: PMC2533224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoki Y, Tabeta K, Murakami Y, Yoshimura F, Yamazaki K. Analysis of immunostimulatory activity of Porphyromonas gingivalis fimbriae conferred by Toll-like receptor 2. Biochem Biophys Res Commun. 2010;398(1):86–91. 10.1016/j.bbrc.2010.06.040 . [DOI] [PubMed] [Google Scholar]
- 35.Nozoe K, Sanui T, Takeshita M, Fukuda T, Haraguchi A, Aida Y, et al. Innate immune-stimulatory activity of Porphyromonas gingivalis fimbriae is eliminated by phase separation using Triton X-114. J Immunol Methods. 2017;441:31–38. 10.1016/j.jim.2016.11.012 . [DOI] [PubMed] [Google Scholar]
- 36.Kato T, Kawai S, Nakano K, Inaba H, Kuboniwa M, Nakagawa I, et al. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell Microbiol. 2007;9(3):753–65. 10.1111/j.1462-5822.2006.00825.x . [DOI] [PubMed] [Google Scholar]
- 37.Gao L, Xu Y, Meng S, Wu Y, Huang H, Su R, et al. Identification of the putative specific pathogenic genes of Porphyromonas gingivalis with type II fimbriae. DNA Cell Biol. 2012; 31(6):1027–37. 10.1089/dna.2011.1487 . [DOI] [PubMed] [Google Scholar]
- 38.Kerr JE, Abramian JR, Dao DH, Rigney TW, Fritz J, Pham T, et al. Genetic exchange of fimbrial alleles exemplifies the adaptive virulence strategy of Porphyromonas gingivalis. PLoS One. 2014;9(3):e91696 10.1371/journal.pone.0091696 ; PMCID: PMC3953592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouyang YL, Wu YF, Zhao L, Xiao XR, Zhang JY, et al. Matrix metalloproteinase 8 and 9 regulations of polymorphonuclear leukocytes stimulated by Porphyromonas gingivalis with different fimA genotypes. Hua Xi Kou Qiang Yi Xue Za Zhi. 2009; 27(2):206–9. . [PubMed] [Google Scholar]
- 40.Zhao L, Yang H, Wu YF, Ouyang YL, Meng S. Matrix metalloproteinases regulations of human gingival fibroblasts by Porphyromonas gingivalis with different fimA genotypes. Zhonghua Kou Qiang Yi Xue Za Zhi. 2008;43(12):727–31. . [PubMed] [Google Scholar]
- 41.Nakagawa I, Amano A, Ohara-Nemoto Y, Endoh N, Morisaki I, Kimura S, et al. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J Periodontal Res. 2002;37(6):425–32. 10.1034/j.1600-0765.2002.01637.x . [DOI] [PubMed] [Google Scholar]
- 42.van Winkelhoff AJ, Appelmelk BJ, Kippuw N, de Graaff J. K-antigens in Porphyromonas gingivalis are associated with virulence. Oral Microbiol Immunol. 1993; 8(5):259–65. 10.1111/j.1399-302x.1993.tb00571.x . [DOI] [PubMed] [Google Scholar]
- 43.Kontani M, Kimura S, Nakagawa I, Hamada S. Adherence of Porphyromonas gingivalis to matrix proteins via a fimbrial cryptic receptor exposed by its own arginine-specific protease. Mol Microbiol.1997;24(6):1179–87. 10.1046/j.1365-2958.1997.4321788.x . [DOI] [PubMed] [Google Scholar]
- 44.Kuboniwa M, Amano A, Shizukuishi S, Nakagawa I, Hamada S. Specific antibodies to Porphyromonas gingivalis Lys-gingipain by DNA vaccination inhibit bacterial binding to hemoglobin and protect mice from infection. Infect Immun. 2001;69(5):2972–9. 10.1128/IAI.69.5.2972-2979.2001 ; PMCID: PMC98250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim BG, Kim S H, Kim N S, et al. Production of monoclonal antibody against FimA protein from Porphyromonas gingivalis in rice cell suspension culture. Plant Cell Tiss Organ Cult. 2014;118:293–304. 10.1007/s11240-014-0481-9 [DOI] [Google Scholar]
- 46.Pourhajibagher M, Chiniforush N, Raoofian R, Ghorbanzadeh R, Shahabi S, Bahador A. Effects of sub-lethal doses of photo-activated disinfection against Porphyromonas gingivalis for pharmaceutical treatment of periodontal-endodontic lesions. Photodiagnosis Photodyn Ther.2016;16:50–53. 10.1016/j.pdpdt.2016.08.013 . [DOI] [PubMed] [Google Scholar]
- 47.Pourhajibagher M, Bahador A. Evaluation of the crystal structure of a fimbrillin (FimA) from Porphyromonas gingivalis as a therapeutic target for photo-activated disinfection with toluidine blue O. Photodiagnosis Photodyn Ther. 2017;17:98–102. 10.1016/j.pdpdt.2016.11.007 . [DOI] [PubMed] [Google Scholar]