Abstract
Colorectal cancer remains one of the most common cancers in men and women, and it accounts for a large proportion of cancer-related deaths worldwide. Tripartite motif (TRIM) proteins are a novel class of “single protein RING finger” E3 ubiquitin ligases, which have been shown to be involved in many cancers. The aim of this study was to investigate the potential role of TRIM24 in human colorectal cancer. By using a lentivirus-mediated RNA interference system, we first explored the effect of TRIM24 knockdown on HCT116 cell proliferation and colony formation. Moreover, flow cytometry analysis was used to examine its effects on cell cycle distribution and apoptosis. Our data showed that knockdown of TRIM24 expression in HCT116 cells significantly decreased cell growth due to the induction of apoptosis. Hence, the present study provides evidence that TRIM24 functions as an oncogene in colorectal carcinogenesis.
Key words: TRIM24, Proliferation, Apoptosis, Colorectal cancer, Lentivirus
INTRODUCTION
Increasing scientific and clinical evidence indicates that the deregulation of ubiquitin-mediated degradation of oncogene or antioncogene may be involved in carcinogenesis. Tripartite motif (TRIM) proteins are defined as a novel class of the RING type E3 ubiquitin ligases, which function as scaffold proteins that mediate the interaction between E2 ubiquitin-conjugating enzyme and the substrate (1). TRIM proteins generally prefer the D and E classes of E2 enzymes, including E2D1 (UBCH5A), E2D2 (UBCH5B), E2D3 (UBCH5C), E2E1 (UBCH6), E2E2, and E2E3 (UBCH9). Recently, the biological functions and implications of TRIM proteins in cancers have been reviewed in detail, showing that some of them are involved in cancer cell growth, apoptosis, differentiation, and transcriptional regulation (2). For instance, TRIM29 is highly expressed in gastric cancer tissues, and its expression is strongly associated with tumor size, extent of invasion, and lymph node metastasis (3). TRIM25, a target gene product of ERα, is predominantly expressed in breast cancers and is essential for estrogen-dependent breast cancer growth (4,5). Moreover, TRIM24, together with three other TRIM family members (TRIM19, TRIM33, and TRIM27) has been shown to acquire oncogenic activity upon chromosomal translocations (6–9).
TRIM24, originally named as transcriptional intermediary factor (TIF) 1α, is a ligand-dependent mediator that interacts with and regulates the transactivation of multiple nuclear receptors (10). Studies have shown that the inactivation of TRIM24 gene function confers oncogenic activity in hepatocellular carcinoma (11–13). Also, TRIM24 is found to be overexpressed and associated with poor prognosis in breast cancers and head and neck squamous cell carcinoma (HNSCC) (14,15). During these processes, TRIM24 may interact with different nuclear receptors, including retinoic acid receptor-α (RARα), estrogen (ER), and progesterone (PR) receptors via its nuclear receptor interaction domain (NRID) (12,15). Besides, in prostate cancers, TRIM24 is able to regulate cancer cell growth via binding to bromodomain containing 7 (BRD7) (16). However, little is known regarding the biological role of TRIM24 in colorectal cancer development. It should be noted that TRIM24 is a key regulator of p53 protein since it ubiquitylates and mediates the degradation of p53 via its RING domain (17). Given the prevalence and important role of p53 deregulation in the development of various cancers, we may infer that TRIM24 is involved in colorectal cancer cell growth and apoptosis.
Hence, in the present investigation, we applied a lentivirus-mediated RNA interference (RNAi) system to specifically knock down the expression of TRIM24 in HCT116 colorectal cancer cells. The effects of TRIM24 knockdown on cancer cell growth, cell cycle, and apoptosis were studied.
MATERIALS AND METHODS
Cell Culture
Human colorectal cancer HCT116 cells were purchased from The Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in MCCOYS’5A Medium (Sigma, St. Louis, MO, USA) containing 10% fetal bovine serum (FBS). Cells were maintained at 37°C with 5% CO2.
Lentivirus Construction
The lentivirus-mediated silencing system targeting TRIM24 gene (NM_003852.3) was constructed as follows. First, short hairpin RNA (shRNA) sequence (5′-GCAGCAGTACAGCATTACTTTGGTACCAAAGTAATGCTGTACTGCTGC-3′) targeting TRIM24 gene was constructed into pFH-L vector (Shanghai Hollybio, China). Scrambled control shRNA sequence (5′-CTAGCCCGG CCAAGGAAGTGCAATTGCATACTCGAGTATGCAATTGCACTTCCTTGGTTTTTTGTTAAT-3′) was designed and constructed as well. Then, to generate reconstructed lentiviruses, the pFH-L vectors containing TRIM24-shRNA or control-shRNA were cotransfected into 293T cells with pVSVG-I and pCMVΔR8.92 (Shanghai Hollybio, China). After 72 h, the lentiviruses were harvested by centrifugation and purification, and the two plasmids were named as Lv-shTRIM24 and Lv-shCon. For lentivirus infection, the HCT116 cells were infected with either lentivirus (MOI = 20) for 72 h, and then the cells were examined by fluorescence microscopy to monitor the transfection efficiency. Quantitative real-time PCR (QRT-PCR) and Western blot analysis were used to monitor the knockdown efficiency of Lv-shTRIM24.
RNA Isolation, Reverse Transcription, and Quantitative Real-Time PCR
HCT116 cells were subjected to three treatments (Lv-shTRIM24, Lv-shCon, and Con). After 5 days, cells were collected for RNA isolation by using the Trizol reagent (Invitrogen). For reverse transcription, 2 μg total RNA was reverse transcribed by using the M-MLV Reverse Transcriptase Kit (Promega Corp., Madison, WI, USA). Then two pairs of primers were used in QRT-PCR: actin-F, 5′-GTG GAC ATC CGC AAA GAC-3′, actin-R, 5′-AAA GGG TGT AAC GCA ACT A-3′; TRIM24-F, 5′-ACG GTC CAG TCA CCA AAT TC-3′, TRIM24-R, 5′-CGG TGG TCC ACT GTT TTC TT-3′. The PCR reaction system was 20 µl volume for each tube [2 × SYBR premix ex taq 10 µl, forward and reverse primers (2.5 µM) 0.8 µl, cDNA 5 µl, ddH2O 4.2 µl]. The cycle condition for PCR was described as follows: (1) initial denaturation at 95°C for 1 min; (2) 40 cycles of denaturation 95°C for 5 s and annealing extension at 60°C for 20 s. Fluorescence was analyzed by using the BioRad Connet Real-Time PCR platform. To analyze the mRNA expression of TRIM24, the comparative threshold cycle (ct) method (2−ΔΔCt) was used, and the level of actin mRNA was used as control.
Western Blot Analysis
The lentiviruses were transduced into HCT116 cells for 5 days. Then cells in all three groups (Lv-shTRIM24, Lv-shCon, and Con) were subjected to protein extraction with cell lysis buffer (10 mM Tris-HCl, pH 7.4; 1 mM EDTA; 0.1% Triton X-100; and 0.1% SDS) by centrifugation. Before Western blot analysis, total protein concentration was examined by BCA protein assay (Pierce, Rockford, IL, USA). For electrophoresis, 30-µg cell extracts from three groups were mixed with sample buffer, respectively, separated by 10% SDS-PAGE gel, and wet-transferred to a polyvinylidene fluoride (PVDF) membrane. For immunoblotting, the specific primary antibodies against TRIM24 (14208-1-AP, Sigma, St. Louis, MO, USA) and GAPDH (Sc-32233, Santa Cruz, CA, USA) were incubated with the membrane overnight at 4°C. Goat anti-rabbit secondary antibody (SC-2054, Santa Cruz) was then used for 2-h incubation at room temperature. Immunoblots were visualized by using an enhanced chemiluminescence (ECL) kit (PE LifeScience).
MTT Proliferation Assay
The proliferative ability of HCT116 cells in three groups (Lv-shTRIM24, Lv-shCon, and Con) was determined by methylthiazoletetrazolium (MTT) assay. In detail, cells were subjected to the above three treatments for 96 h. Then the cells were reseeded in 96-well plates (2,500 cells/well). At indicated time points, the plates were incubated in MTT solution (5 mg/ml) for 4 h at 37°C. When MTT solution was removed, the acidic isopropanol solution (10% SDS, 5% isopropanol, and 0.01 mol/L HCl) was added to the samples. The absorbance of the plates was read at 595 nm. Experiments were performed in triplicate.
Colony Formation Assay
The colony formation ability of HCT116 cells in three groups (Lv-shTRIM24, Lv-shCon, and Con) was determined by colony formation assay. In detail, cells were subjected to the above three treatments for 96 h. Then the cells were reseeded in six-well plates (400 cells/well) and allowed to form natural colonies for 9 days. At the indicated time point, the plates were observed and photographed with a digital camera. For crystal purple staining, the plates were washed by PBS buffer, fixed by paraformaldehyde, and stained with crystal purple. The number of colonies (>50 cells/colony) was counted and statistically analyzed.
Cell Cycle Analysis by Flow Cytometry
Cell cycle distribution (sub-G1, G0/G1, S, or G2/M phase) was characterized by different DNA contents. For cell cycle analysis, propidium iodide (PI) (Sigma, St. Louis, MO, USA) was used for staining, and the DNA content of each sample was detected by using flow cytometry. Briefly, cells synchronized by serum starvation were subjected to three different treatments (Lv-shTRIM24, Lv-shCon, and Con). After 72 h of incubation, cells were washed and reseeded in 6 cm dishes at a density of 80,000 cells/dish before detection. After 24 h, the cells were trypsinized, washed with ice-cold PBS, fixed with 70% ethanol, and stained with PI/RNase/PBS (100 μg/ml PI and 10 μg/ml RNase A) buffer in the dark (37°C, 30 min). Samples in all three groups were determined by a FACs caliber II sorter and Cell Quest FACS system (BD Biosciences, San Diego, CA, USA), and the percentages of cells in all phases were statistically analyzed.
Apoptosis Analysis by Flow Cytometry
Cells were subjected to three different treatments (Lv-shTRIM24, Lv-shCon, and Con). After 72 h of incubation, cells were washed and reseeded in 6-cm dishes at a density of 80,000 cells/dish before detection. After 24 h, the cells were collected and subjected to Annexin V-APC/7-AAD double staining according to the manufacturer’s instructions (KeyGen Biotech, Nanjing, China).
Statistical Analysis
All data were shown as mean ± standard deviation of at least three independent experiments. The Student’s t test was used for statistical analysis. A value of p < 0.05 was considered significant.
RESULTS
Inhibition of TRIM24 Expression by Lentivirus Mediated RNA Interference
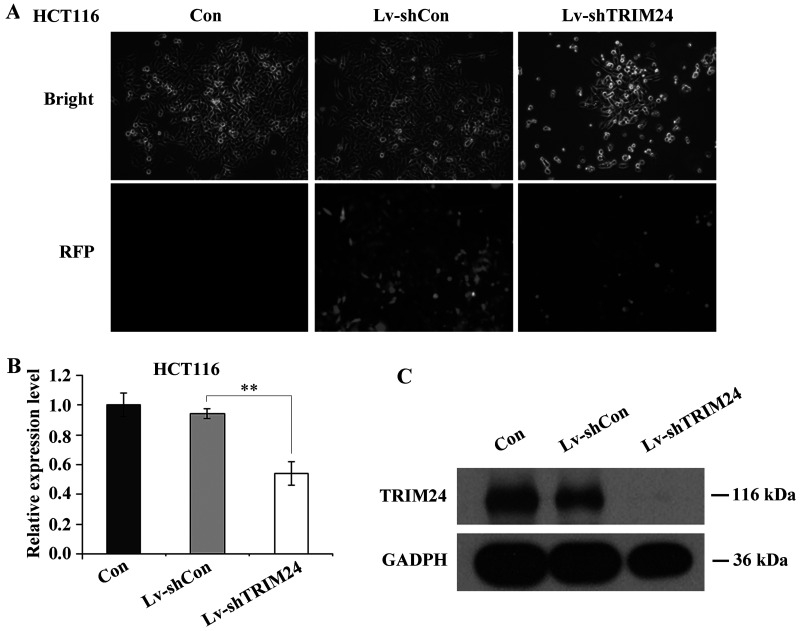
First, the lentiviruses containing TRIM24-shRNA and control-shRNA were reconstructed and generated. For RNA interference (RNAi), human colorectal cancer cells HCT116 were subjected to three groups of treatments (Lv-shTRIM24, Lv-shCon, and Con). After 72 h, cells in all three groups were examined via fluorescence microscopy to monitor the infection efficiency. As shown in Figure 1A, over 90% HCT116 cells expressed red fluorescence protein (RFP), indicating successful infection with either lentivirus. Then all cells were harvested for the following QRT-PCR and Western blot analysis to confirm the knockdown efficiency of TRIM24. As Figure 1B and C reveal, the mRNA and protein levels of TRIM24 were both remarkably decreased by Lv-shTRIM24 treatment, compared to the Lv-shCon and Con groups.
Figure 1.
Lentivirus-mediated silencing of TRIM24 in HCT116 colorectal cancer cells. (A) Fluorescence photomicrographs of HCT116 cells infected with either lentivirus. Pictures were taken 72 h after infection at a magnification of 100×. (B) QRT-PCR analysis of TRIM24 knockdown efficiency in HCT116 cells. The actin gene was used as control. (C) Western blot analysis of TRIM24 knockdown efficiency in HCT116 cells. GAPDH protein was used as control. **p < 0.01 compared to Lv-shCon.
Effect of TRIM24 Knockdown on Colorectal Cancer Cell Growth
To evaluate the biological role of TRIM24 in colorectal cancer cell growth, an MTT assay was applied. As shown in Figure 2A, the growth curve in the Lv-shTRIM24 group was much lower than that in the Lv-shCon group, while there was no difference between the Lv-shCon and Con groups. Specifically, on day 4, the cell proliferation in the Lv-shTRIM24 group (OD: 0.4818 ± 0.012) was significantly reduced compared with the Lv-shCon (OD: 0.8766 ± 0.049) and Con (OD: 0.9088 ± 0.030) groups. The growth inhibition was more obvious on day 5 (p < 0.001). This indicates that suppression of TRIM24 gene expression via Lv-shTRIM24 had a strong inhibitory effect on HCT116 cell proliferation.
Figure 2.
TRIM24 knockdown decreased HCT116 cell growth. (A) HCT116 cells were subjected to three treatments (Lv-shTRIM24, Lv-shCon, and Con). Cell proliferation assay was carried by MTT, and the absorbance of plates was recorded at 595 nm. (B) HCT116 cells in three groups were allowed to form natural colonies. At the indicated time point, cells were stained with crystal violet and observed by light microscopy and fluorescent microscopy. (C) Colonies larger than 50 cells were counted and analyzed in three groups. ***p < 0.001 compared to Lv-shCon.
Moreover, to explore the relative long-term effect of TRIM24 knockdown, the colony formation ability of HCT116 cells was also studied. As shown in Figure 2B, Lv-shTRIM24 treatment could strongly decrease both the size of single colony and the number of colonies formed in HCT116 cells. Compared to the Lv-shCon (122 ± 6) and Con (134 ± 9) groups, the colony numbers in the Lv-shTRIM24 group (11 ± 2) were markedly reduced (Fig. 2C), indicating that TRIM24 may have an oncogenic effect on HCT116 cell growth.
Effect of TRIM24 Knockdown on Colorectal Cancer Cell Cycle Distribution and Apoptosis
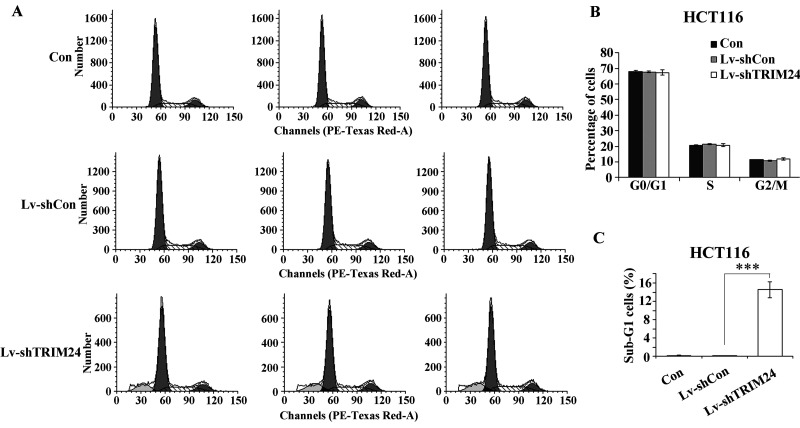
To find out whether TRIM24 mediates colorectal cancer cell growth through direct regulation of cell cycle, we then examined the cell cycle distribution in all three groups by flow cytometry analysis. In HCT116 cells, we did not observe any significant changes in the percentage of cells in G0/G1, S, or G2/M phase (Fig. 3A and B), suggesting that TRIM24 may not be directly involved in cell cycle control.
Figure 3.
Effect of TRIM24 knockdown on cell cycle distribution. (A) Flow cytometry histograms of HCT116 cells following lentivirus infection in all three groups (Lv-shTRIM24, Lv-shCon, and Con). (B) Quantification of cell cycle distribution in G0/G1, S, or G2/M phases in HCT116 cells by FACS. (C) Quantification of sub-G1 population in HCT116 cells by FACS. ***p < 0.001 compared to Lv-shCon.
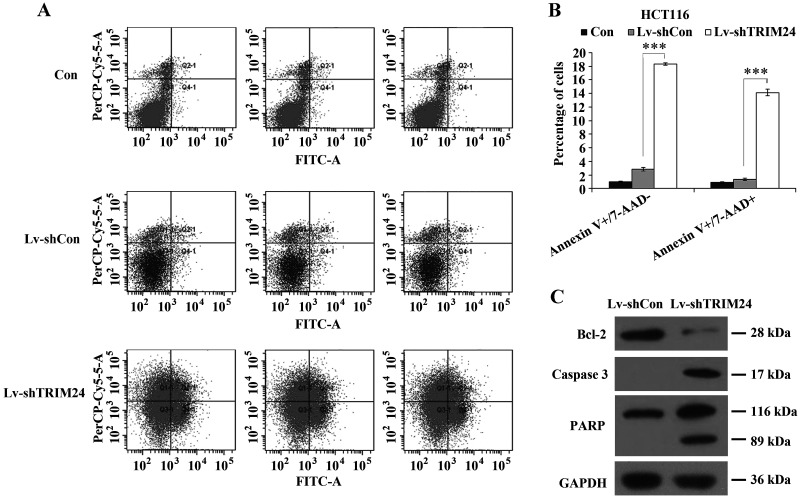
Meanwhile, the apoptotic cells with degraded DNA are represented in sub-G1 peaks as previously described (18). Our data showed that TRIM24 knockdown strongly increased the percentage of cells in the sub-G1 phase (Fig. 3C), which is a strong indication that TRIM24 may regulate cell apoptosis. To answer this question, we then applied Annexin V-APC/7-AAD double staining in HCT116 cells following lentivirus infection. Annexin V-APC versus 7-AAD plots from the gated cells showed the populations corresponding to viable and nonapoptotic (Annexin V−/7-AAD−), early (Annexin V+/7-AAD−), and late (Annexin V+/7-AAD+) apoptotic cells. Knockdown of TRIM24 augmented apoptotic cells (early apoptosis and late apoptosis) by nearly 10-fold, compared to controls (Fig. 4A and B). Furthermore, the expression alterations of apoptosis markers were detected in HCT116 cells, including Bcl-2, caspase 3, and PARP. Western blot showed that depletion of TRIM24 resulted in an obvious decrease in Bcl-2 expression and a remarkable increase in caspase 3 and PARP expression (Fig. 4C). These results suggested that knockdown of TRIM24 in HCT116 cells induced cell apoptosis attributed to downregulation of Bcl-2 and upregulation of caspase 3 and PARP. Hence, the above data reveal that TRIM24 inhibition induced a strong proapoptotic effect in human colorectal cancer HCT116 cells.
Figure 4.
Effect of TRIM24 knockdown on cell apoptosis. (A) Cytogram of Annexin V-APC binding versus 7-AAD uptake in HCT116 cells following lentivirus infection in all three groups (Lv-shTRIM24, Lv-shCon, and Con). (B) Quantification of apoptotic cells in HCT116 cells by FACS. Annexin V−/7-AAD−: viable and nonapoptotic cells, Annexin V-APC+/7-AAD−: early apoptotic cells; Annexin V-APC+/7-AAD+: late apoptotic cells. (C) Western blot analysis of Bcl-2, caspase 3, and PARP expression in HCT116 cells after TRIM24 knockdown. GAPDH protein was used as control.
DISCUSSION
Colorectal cancer is the second most common cancer in women and the third in men. Worldwide, it accounts for more than 1 million new cases and around 600,000 deaths each year (19,20). Ubiquitin–proteasome system has shown to be activated in many malignancies including colorectal cancers (21). The selective degradation of the ubiquitin−proteasome system for a target protein relies on the interaction between E3 ubiquitin ligase and E2 ubiquitin-conjugating enzyme (22). A number of E3 ubiquitin ligases have been shown to play a role in colorectal cancer development, including Hsc70-interacting protein (CHIP, a U-box containing E3 ubiquitin ligase), Cullin 4B (CUL4B), the S-phase kinase-associated protein 2 (SKP2), and MDM2 (23–26). TRIM proteins represent a novel class of “single protein RING finger” E3 ubiquitin ligases containing a B box domain and a coiled coil (27). Actually, the oncogenic effect of TRIM24 on cancer cell growth has been proved in several studies in various cancers, including breast cancer, prostate cancer, HNSCC, and hepatocellular carcinoma (11–16). However, the involvement of TRIM in colorectal cancer remains obscure.
In order to clarify the potential role of TRIM24 in colorectal cancer development, we knocked down the expression of TRIM24 and investigated its effect on HCT116 colorectal cancer cells in vitro. Our data showed that suppression of TRIM24 markedly decreased cell proliferation and colony formation ability. However, we did not observe any significant changes in the distribution of HCT116 cells in G0/G1, S, and G2/M phases, indicating that TRIM24 was not involved in cell cycle control. Notably, flow cytometry analysis by Annexin V-APC/7-AAD staining revealed that knockdown of TRIM24 in HCT116 cells augmented apoptotic cells (early apoptosis and late apoptosis). Furthermore, Western blot showed that knockdown of TRIM24 obviously downregulated the expression of Bcl-2, which belongs to the Bcl-2 family of antiapoptotic proteins (28), and upregulated the expression levels of caspase 3 and cleaved PARP that are hallmarks of apoptosis (29,30), suggesting that TRIM24 knockdown could suppress the growth of colorectal cancer cells through inducing cell apoptosis. Our data were in accordance with a previous study showing that TRIM24 silencing strongly inhibited the proliferation of HNSCC cells due to the induction of apoptosis (15). Since TRIM24 is a ligand-dependent mediator, further investigation is needed to evaluate whether TRIM24 regulates any nuclear receptors in colorectal cancer cells.
Taken together, our data strongly indicate that TRIM24 has an oncogenic effect on colorectal cancer, and it may serve as a promising therapeutic target for the treatment of colorectal cancer. Further studies will be helpful to clarify the mechanisms by which TRIM24 contributes to colorectal carcinogenesis.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (Grant No. 81272677) and the Natural Science Foundation of Ningbo (Grant No. 2012A610204 and 2014A610225).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Hatakeyama S. TRIM proteins and cancer. Nat. Rev. Cancer 11: 792–804; 2011. [DOI] [PubMed] [Google Scholar]
- 2. Cambiaghi V.; Giuliani V.; Lombardi S.; Marinelli C.; Toffalorio F.; et al. et al. TRIM proteins in cancer. Adv. Exp. Med. Biol. 770: 77–91; 2012. [DOI] [PubMed] [Google Scholar]
- 3. Kosaka Y.; Inoue H.; Ohmachi T.; Yokoe T.; Matsumoto T.; et al. Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph node metastasis in gastric cancer. Ann. Surg. Oncol. 14: 2543–2549; 2007. [DOI] [PubMed] [Google Scholar]
- 4. Urano T.; Saito T.; Tsukui T.; Fujita M.; Hosoi T.; et al. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature 417: 871–875; 2002. [DOI] [PubMed] [Google Scholar]
- 5. Zhao K. W.; Sikriwal D.; Dong X.; Guo P.; Sun X.; et al. Oestrogen causes degradation of KLF5 by inducing the E3 ubiquitin ligase EFP in ER-positive breast cancer cells. Biochem. J. 437: 323–333; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de The H.; Lavau C.; Marchio A.; Chomienne C.; Degos L.; et al. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 66: 675–684; 1991. [DOI] [PubMed] [Google Scholar]
- 7. Takahashi M.; Ritz J.; Cooper G. M. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell 42: 581–588; 1985. [DOI] [PubMed] [Google Scholar]
- 8. Le Douarin B.; Zechel C.; Garnier J. M.; Lutz Y.; Tora L.; et al. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 14: 2020–2033; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Drilon A.; Wang L.; Hasanovic A.; Suehara Y.; Lipson D.; et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 3: 630–635; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Le Douarin B.; Nielsen A. L.; Garnier J. M.; Ichinose H.; Jeanmougin F.; et al. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 15: 6701–6715; 1996. [PMC free article] [PubMed] [Google Scholar]
- 11. Khetchoumian K.; Teletin M.; Tisserand J.; Mark M.; Herquel B.; et al. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat. Genet. 39: 1500–1506; 2007. [DOI] [PubMed] [Google Scholar]
- 12. Herquel B.; Ouararhni K.; Khetchoumian K.; Ignat M.; Teletin M.; et al. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 108: 8212–8217; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khetchoumian K.; Teletin M.; Tisserand J.; Herquel B.; Ouararhni K.; et al. Trim24 (Tif1 alpha): An essential ‘brake’ for retinoic acid-induced transcription to prevent liver cancer. Cell Cycle 7: 3647–3652; 2008. [DOI] [PubMed] [Google Scholar]
- 14. Chambon M.; Orsetti B.; Berthe M. L.; Bascoul-Mollevi C.; Rodriguez C.; et al. Prognostic significance of TRIM24/TIF-1alpha gene expression in breast cancer. Am. J. Pathol. 178: 1461–1469; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cui Z.; Cao W.; Li J.; Song X.; Mao L.; et al. TRIM24 overexpression is common in locally advanced head and neck squamous cell carcinoma and correlates with aggressive malignant phenotypes. PLoS One 8: e63887; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kikuchi M.; Okumura F.; Tsukiyama T.; Watanabe M.; Miyajima N.; et al. TRIM24 mediates ligand-dependent activation of androgen receptor and is repressed by a bromodomain-containing protein, BRD7, in prostate cancer cells. Biochim. Biophys. Acta 1793: 1828–1836; 2009. [DOI] [PubMed] [Google Scholar]
- 17. Allton K.; Jain A. K.; Herz H. M.; Tsai W. W.; Jung S. Y.; et al. Trim24 targets endogenous p53 for degradation. Proc. Natl. Acad. Sci. USA 106: 11612–11616; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Telford W. G.; King L. E.; Fraker P. J. Comparative evaluation of several DNA binding dyes in the detection of apoptosis-associated chromatin degradation by flow cytometry. Cytometry 13: 137–143; 1992. [DOI] [PubMed] [Google Scholar]
- 19. Hong T. S.; Clark J. W.; Haigis K. M. Cancers of the colon and rectum: Identical or fraternal twins? Cancer Discov. 2: 117–121; 2012. [DOI] [PubMed] [Google Scholar]
- 20. Jemal A.; Center M. M.; DeSantis C.; Ward E. M. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol. Biomarkers Prev. 19: 1893–1907; 2010. [DOI] [PubMed] [Google Scholar]
- 21. Shafiee S. M.; Seghatoleslam A.; Nikseresht M.; Hosseini S. V.; Alizadeh-Naeeni M.; et al. UBE2Q1 expression in human colorectal tumors and cell lines. Mol. Biol. Rep. 40: 7045–7051; 2013. [DOI] [PubMed] [Google Scholar]
- 22. Ardley H. C.; Robinson P. A. E3 ubiquitin ligases. Essays Biochem. 41: 15–30; 2005. [DOI] [PubMed] [Google Scholar]
- 23. Wang Y.; Ren F.; Wang Y.; Feng Y.; Wang D.; et al. CHIP/Stub1 functions as a tumor suppressor and represses NF-kappaB-mediated signaling in colorectal cancer. Carcinogenesis 35:983–991; 2014. [DOI] [PubMed] [Google Scholar]
- 24. Jiang T.; Tang H. M.; Wu Z. H.; Chen J.; Lu S.; et al. Cullin 4B is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Med. Oncol. 30: 534; 2013. [DOI] [PubMed] [Google Scholar]
- 25. Tian Y. F.; Chen T. J.; Lin C. Y.; Chen L. T.; Lin L. C.; et al. SKP2 overexpression is associated with a poor prognosis of rectal cancer treated with chemoradiotherapy and represents a therapeutic target with high potential. Tumour Biol. 34: 1107–1117; 2013. [DOI] [PubMed] [Google Scholar]
- 26. Tuna G.; Kucukhuseyin O.; Arikan S.; Kaytan Saglam E.; Guler E.; et al. Do CDKN2 p16 540 C>G, CDKN2 p16 580 C>T, and MDM2 SNP309 T>G gene variants act on colorectal cancer development or progression? DNA. Cell. Biol. 32: 400–408; 2013. [DOI] [PubMed] [Google Scholar]
- 27. Meroni G.; Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 27: 1147–1157; 2005. [DOI] [PubMed] [Google Scholar]
- 28. Tsujimoto Y. Bcl-2 family of proteins: Life-or-death switch in mitochondria. Biosci. Rep. 22: 47–58; 2002. [DOI] [PubMed] [Google Scholar]
- 29. Bressenot A.; Marchal S.; Bezdetnaya L.; Garrier J.; Guillemin F.; et al. Assessment of apoptosis by immunohistochemistry to active caspase-3, active caspase-7, or cleaved PARP in monolayer cells and spheroid and subcutaneous xenografts of human carcinoma. J. Histochem. Cytochem. 57: 289–300; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luthi A. U.; Martin S. J. The CASBAH: A searchable database of caspase substrates. Cell. Death Differ. 14: 641–650; 2007. [DOI] [PubMed] [Google Scholar]