Abstract
Background:
Some 20 y ago, scientific and regulatory communities identified the potential of omics sciences (genomics, transcriptomics, proteomics, metabolomics) to improve chemical risk assessment through development of toxicogenomics. Recognizing that regulators adopt new scientific methods cautiously given accountability to diverse stakeholders, the scope and pace of adoption of toxicogenomics tools and data have nonetheless not met the ambitious, early expectations of omics proponents.
Objective:
Our objective was, therefore, to inventory, investigate, and derive insights into drivers of and obstacles to adoption of toxicogenomics in chemical risk assessment. By invoking established social science frameworks conceptualizing innovation adoption, we also aimed to develop recommendations for proponents of toxicogenomics and other new approach methodologies (NAMs).
Methods:
We report findings from an analysis of 56 scientific and regulatory publications from 1998 through 2017 that address the adoption of toxicogenomics for chemical risk assessment. From this purposeful sample of toxicogenomics discourse, we identified major categories of drivers of and obstacles to adoption of toxicogenomics tools and data sets. We then mapped these categories onto social science frameworks for conceptualizing innovation adoption to generate actionable insights for proponents of toxicogenomics.
Discussion:
We identify the most salient drivers and obstacles. From 1998 through 2017, adoption of toxicogenomics was understood to be helped by drivers such as those we labeled Superior scientific understanding, New applications, and Reduced cost & increased efficiency but hindered by obstacles such as those we labeled Insufficient validation, Complexity of interpretation, and Lack of standardization. Leveraging social science frameworks, we find that arguments for adoption that draw on the most salient drivers, which emphasize superior and novel functionality of omics as rationales, overlook potential adopters’ key concerns: simplicity of use and compatibility with existing practices. We also identify two perspectives—innovation-centric and adopter-centric—on omics adoption and explain how overreliance on the former may be undermining efforts to promote toxicogenomics. https://doi.org/10.1289/EHP6500
Introduction
Toxicogenomics is part of a new generation of scientific techniques in toxicology and ecotoxicology referred to as new approach methodologies (NAMs) (ECHA 2016a). The Society of Environmental Toxicology and Chemistry (SETAC 2019) defines toxicogenomics as “the study of the relationship between the genome and the adverse biological effects of external agents,” and highlights its connection with other omics disciplines, including genomics [“the study of the genome, which is the entire set of genes (DNA) in an organism”], transcriptomics [“the study of all messenger genes (RNA) transcripts under specific conditions, which is the transcriptome”], proteomics (“the study of proteins, which are important components of organisms”), and metabolomics (“the study of the metabolome, which are the molecules involved in metabolism including sugars, lipids, and amino and nucleic acids”). Despite impressive advances, the scope and pace of adoption of new toxicogenomics tools and data sets in chemical risk assessment have, generally, not met the ambitious expectations of their proponents (Birnbaum 2013; Cote et al. 2016; Leung 2018). In particular, regulatory uptake has been slow despite notable improvements and new applications (Grondin et al. 2018; Li et al. 2019; Tice et al. 2013), as well as considerable national and international efforts (Arnold 2015; ECHA 2016a; ICCVAM 2018; Kavlock et al. 2018).
A range of factors that serve as drivers of (or, conversely, barriers or obstacles to) adoption of toxicogenomics and other NAMs have been posited. For example, Zeiger (1999) foresaw adoption as driven by the tools’ ability to predict apical outcomes of interest, cost effectiveness, and contributions to reduced animal use, among other factors. Some research invokes structured frameworks to bring conceptual order to drivers and obstacles: Ankley et al. (2006) discuss obstacles using the practical categories of “methods and capabilities,” “research needs,” and “implementation challenges,” and address drivers in terms of “potential applications” and “regulatory challenges”; Balbus (2005) employs an empirically derived dichotomy between “scientific” and “sociopolitical” factors; Sauer et al. (2017) present “legal,” “regulatory,” “scientific,” and “technical” challenges to adoption; Vachon et al. (2017) distinguish “individual” from “organizational” factors inhibiting adoption; and Zaunbrecher et al. (2017) distinguish “scientific or technical” barriers to adoption from “social/legal/institutional” ones. These are important contributions, but the frameworks invoked are not directly informed by theories of innovation adoption. In fact, to date we are not aware of efforts to comprehensively inventory both drivers of and obstacles to toxicogenomics and NAM adoption and to examine them in light of established social science frameworks for conceptualizing the adoption of innovations.
More than 20 y after the potential of the omics fields of biology to accelerate and improve the assessment of chemical risks to human health and the environment was recognized (Fielden and Zacharewski 2001; Iannaccone 2001; Olden et al. 2001; Schmidt 2002) and more than 10 y after publication of the U.S. National Research Council report Toxicity Testing in the 21st Century: A Vision and a Strategy, referred to as TTC21 (NRC 2007), answering the following questions remains crucial: a) What are the drivers of and obstacles to adoption of toxicogenomics in chemical risk assessment? b) Which drivers and obstacles are more and less salient in terms of attention paid to them? and c) How could proponents of toxicogenomics better leverage drivers and overcome obstacles? We describe our approach to answering these questions in the “Methods” section herein, and we derive “Recommendations for policy and practice” in our Discussion (see Figure 1 for an overview).
Figure 1.
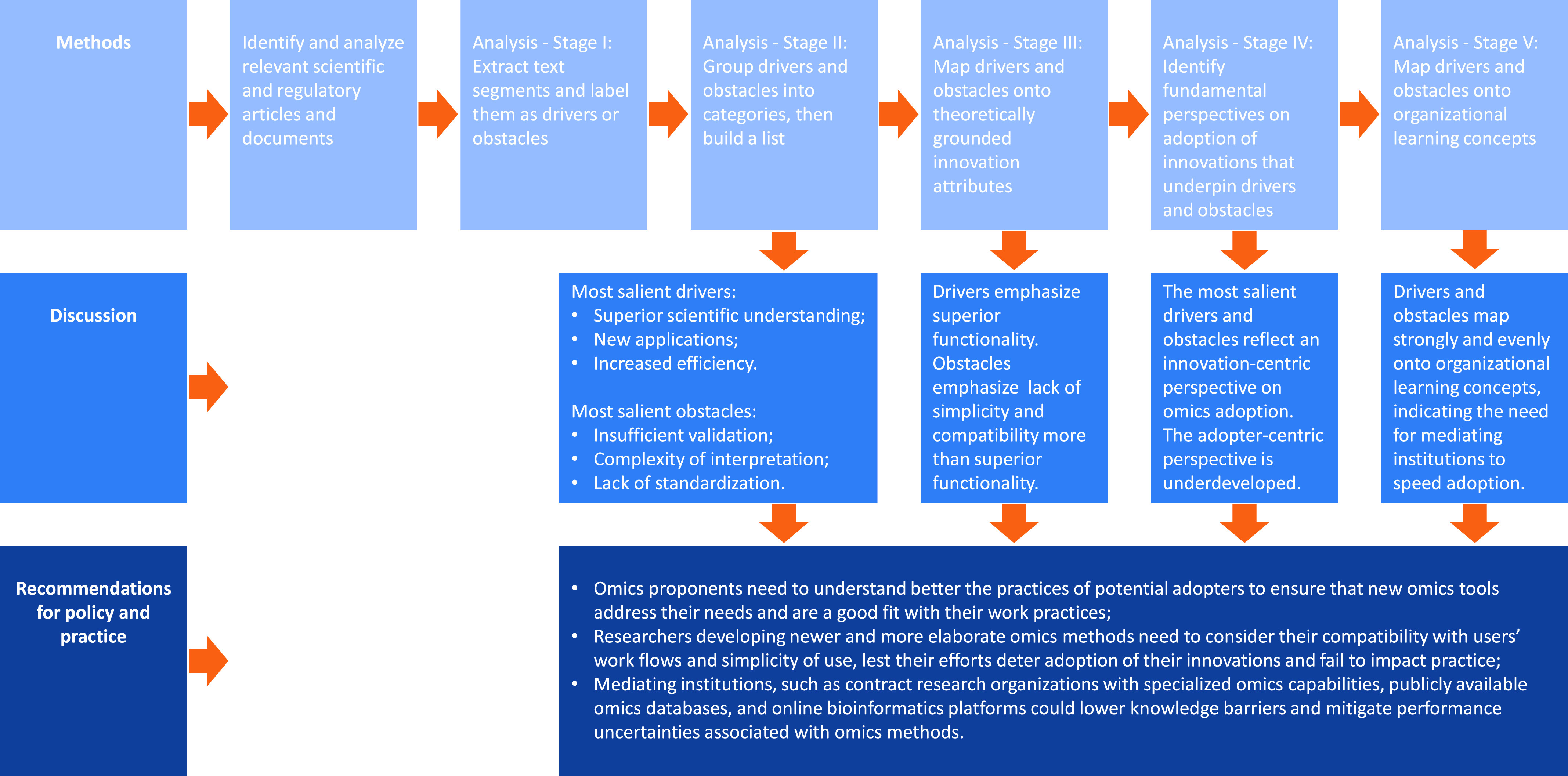
Overview of approach to constructing social science insights into adoption of toxicogenomics for chemical risk assessment.
Methods
Our research methods followed guidelines for qualitative research from the Equator Network, namely, O’Brien et al.’s (2014) standards for reporting qualitative research (SRQR); Tong et al.’s (2007, 2012) consolidated criteria for reporting qualitative studies (COREQ) and enhancing transparency in reporting the synthesis of qualitative research (ENTREQ); Clark’s (2003) Relevance, Appropriateness, Transparency, and Soundness (RATS) criteria; and Malterud’s (2001) qualitative research standards (see Table S1).
Data Collection
We used an expert-driven approach to identify relevant studies. We consulted 20 toxicologists from academia, regulatory agencies, and industry who helped us identify relevant sources: peer-reviewed journals (Table S2), books (Table S3), and authoritative sources (Table S4) with searchable websites and online document repositories [e.g., the Society of Toxicology (SOT), the European Chemicals Agency (ECHA)] that potentially contained discussions of what helped or hindered the adoption of toxicogenomics in chemical risk assessment. We then conducted a targeted review of these sources and generated a purposeful sample of toxicogenomics discourse focused on drivers of and obstacles to adoption of toxicogenomics in chemical risk assessment. We retained 56 documents (listed in Table S5) for subsequent coding.
Data Analysis
In Stage I, to capture the original ideas and language of authors, we extracted 300 verbatim text segments associated with drivers and obstacles from the 56 sources making up the corpus of texts, using broad operationalizations of drivers and obstacles, i.e., positive or negative influences of any kind on adoption. A researcher not involved in extraction reviewed text segments to verify that they indeed addressed drivers or obstacles. (See Table 1 for examples and Table S1 for the complete list.)
Table 1.
Examples of text segments and their coding as “drivers” or “obstacles.”
| Coding | Sample text segments |
|---|---|
| Drivers (positive influences on adoption) | With more tools available and importantly more experienced practitioners of the art of interpretation forthcoming it is most likely that environmental science will increasingly experience the application of genomic tools in chemical assessments (ECETOC 2007 p. 19). |
| Industry, government, and academic institutions all are engaged in developing and applying omic data. The strongest driver behind the development of these technologies is the pharmaceutical industry, which is confident that these techniques will accelerate drug discovery and toxicity testing (Balbus and Environmental Defense 2005 p. 12). | |
| Obstacles (negative influences on adoption) | Without a clearly defined approach to categorize in vitro effects as beneficial, adverse, or irrelevant (normal variation), there is the concern that pathway perturbation results will not be credible as a risk assessment tool for the regulatory community (Andersen and Krewski 2010 p. 19). |
| Data sharing and providing adequate informatics support to retrieve and utilise available data, including those from NAMs [new approach methodologies], is a key challenge to supporting their use for regulatory purposes (ECHA 2016 p. 13). |
Note: Underlined phrases in this table are the summary labels for each driver which are used in the rest of this article for ease of reference.
In Stage II, we coded text segments using “open coding,” a qualitative method that identifies major information categories in data (Creswell 2013). We assigned each segment to one or more driver or obstacle categories. After the initial coding, we merged overlapping categories and adjusted labels to ensure that the final categories were analytically distinct and labeled according to original ideas in corpus sources. To establish the relative salience of drivers and obstacles identified in our sample of toxicogenomics discourse, we counted the number of corpus sources that mentioned each one, so the maximum possible “score” for any driver or obstacle was 56. Because the data set had, on average, 5.4 text segments per source, this approach prevented giving undue weight to sources that repeated the same point. The resulting 11 drivers (D1–D11) and 12 obstacles (O1–O12) are presented, defined, cross-referenced to sources, and detailed below.
Because the analysis spanned 20 y, we revisited the data to see whether there were notable differences over time. The first, median, and final year of mention for each driver and obstacle were noted (Table S6). From the resulting consistent distribution of mentions for empirically derived drivers and obstacles, especially the most salient ones, we concluded that the data adequately represent toxicogenomics discourse over the period 1998–2017.
In subsequent stages, we deployed social science frameworks widely used for theorizing innovation adoption but heretofore not used to study toxicogenomics. In Stage III, we drew upon the work of Rogers (1962, 2003), who posits five fundamental drivers of adoption, articulated in terms of innovation attributes (relative advantage, compatibility, simplicity, trialability, and observability). We mapped each empirically derived driver and obstacle onto these attributes by establishing the presence or absence of semantic correspondence. Although we recognized that this approach unavoidably involves interpretation and subjectivity, our analysis allowed a coarse-grained view of the distribution of drivers and obstacles across Rogers’ attributes, and in turn, by noting patterns in how specific attributes relate to more or less salient drivers and obstacles, allowed development of recommendations for proponents of toxicogenomics.
In Stage IV, we noted that, whereas Rogers’ five attributes focus on the innovation and de-emphasize features of the adopting system, some of our empirically derived drivers and obstacles refer directly to the adopting system. We labeled the perspective emphasizing innovation attributes as the main cause behind (non)adoption “innovation-centric.” It suggests that proponents of an innovation should focus efforts on the innovation itself to improve it sufficiently to interest potential adopters. We labeled the perspective emphasizing features of the adopting system as the main cause behind (non)adoption “adopter-centric.” It suggests that proponents of an innovation should focus efforts on potential adopters to make them more inclined as well as better prepared and able to adopt it. We then categorized empirically derived drivers and obstacles as innovation- or adopter-centric using semantic correspondence. This analysis made it possible to observe the distribution of drivers and obstacles across the two perspectives and to make inferences from the relative balance/imbalance between them.
In Stage V, we mapped drivers and obstacles onto key concepts from Paul Attewell’s work, which relates technology adoption to organizational learning (Attewell 1992; Bhaskarabhatla 2016; Compagni et al. 2015; Cusumano et al. 2015). Attewell (1992) notes that, for “simple” innovations, information flows explain innovation adoption, because organizational ties suffice to inform potential adopters about an innovation’s existence, relative advantage, simplicity, and compatibility. However, information flows are insufficient to spur adoption of “complex” innovations, for which Attewell posits two generic obstacles to adoption: knowledge barriers (e.g., lack of know-how; few opportunities for “hands-on” learning by doing; limited transferability of technical expertise) and resulting performance uncertainties. Two generic drivers of adoption help overcome these generic obstacles: skill development (i.e., individuals cultivate hands-on understanding of the technology within their specific context) and organizational learning (i.e., organizations consolidate this situated understanding into organizational practices). In such situations, mediating institutions, such as service bureaus and consultants, can “progressively lower [knowledge] barriers, and make it easier for firms to adopt and use the technology without extensive in-house expertise” (Attewell 1992, p. 1). In other words, mediating institutions can substitute for skill development and organizational learning early in the adoption process and facilitate them over time. Reframing toxicogenomics as a complex innovation, we mapped empirically derived drivers and obstacles onto Attewell’s generic drivers and obstacles, guided by semantic correspondence, which allowed us to draw conclusions about how mediating institutions could facilitate adoption of toxicogenomics.
Discussion
Drivers of and Obstacles to Adoption of Toxicogenomics
Stages I and II of analysis of toxicogenomics discourse from 1998 through 2017 yielded 11 drivers and 12 obstacles (See Tables 2 and 3). (Here, we note the possibility that our corpus inadvertently excluded texts containing relevant drivers and obstacles, a limitation of our approach. However, given the number of sources supporting our most salient drivers and obstacles, we believe they approximate well the content of toxicogenomics discourse during the period 1998–2017.) With one exception (D8: 2007), the final years of mention of the nine most salient drivers fell within the period 2015–2017. More strikingly, with two exceptions (O7: 2011, O9: 2016), the final year of mention of the nine most salient obstacles was 2017. These findings indicate that our drivers and obstacles, which are labeled with overarching concepts to capture a range of more finely grained issues, remained important concerns for the toxicogenomics community in 2017, notwithstanding significant progress made on specific, more finely grained issues (e.g., standardization, as discussed below).
Table 2.
Drivers cross-referenced to sources.
| Drivers | Sources | First, median, and last year of mention |
|---|---|---|
| D1) Superior scientific understanding: Omics methods allow for a better understanding of health and ecological effects from exposures to chemicals. |
(26 sources) (Andersen and Krewski 2010; Ankley et al. 2006; Bergeson 2008; ECETOC 2007; ECHA 2016b; Fent and Sumpter 2011; Fielden and Zacharewski 2001; Gershon 2002; Grodsky 2007; Hartung 2011; Hattis 2009; IPCS 2003; Kramer and Kolaja 2002; Krewski et al. 2009; Lühe et al. 2005; NRC 2007, 2012; NTP 2004; OECD 2005, 2010; Olden et al. 2001; Orphanides 2003; SOT 2015; Tralau et al. 2015; Trosko and Upham 2010; Tsuji and Garry 2009) | First year: 2001 Median year: 2007 Last year: 2016 |
| D2) New applications: Omics methods offer new applications in human and ecological toxicology. |
(20 sources) (Ankley et al. 2006; Bergeson 2008; Chen et al. 2012; ECHA 2016b; Fent and Sumpter 2011; Fielden and Zacharewski 2001; Hartung 2011; Hattis 2009; IPCS 2003; Krewski et al. 2009; Lühe et al. 2005; NASEM 2017; NRC 2007; NTP 2004; OECD 2005; Olden and Wilson 2000; Olden et al. 2001; Orphanides 2003; Tralau et al. 2015; Trosko and Upham 2010) | First year: 2000 Median year: 2007 Last year: 2017 |
| D3) Reduced cost & increased efficiency: Omics methods will reduce the cost and time, increase the efficiency and scale of testing. |
(16 sources) (Balbus and Environmental Defense 2005; Boverhof and Zacharewski 2006; Chen et al. 2012; ECETOC 2007; ECHA 2016b; Government of Canada 2016; Iannaccone 2001; NRC 2007; OECD 2010; Olden and Wilson 2000; Olden et al. 2001; Orphanides 2003; Sauer et al. 2017; Tralau et al. 2015; Tsuji and Garry 2009; Wittwehr et al. 2017) | First year: 2000 Median year: 2007 Last year: 2017 |
| D4) Scientific and technological advances: Scientific and technological advances enhance the capacity of omics methods to improve scientific understanding and generate new applications in toxicology. |
(13 sources) (Chen et al. 2012; ECETOC 2007; Frueh 2006; Gershon 2002; Grodsky 2007; Hartung 2011; Iannaccone 2001; NRC 2007, 2012; OECD 2005; SOT 2015; Tralau et al. 2015; Tsuji and Garry 2009) | First year: 2001 Median year: 2007 Last year: 2015 |
| D5) Belief in the potential of omics: Field actors express their belief in the potential and promise of omics methods. |
(12 sources) (ECETOC 2007; ECVAM and ICCVAM 2003; Fielden and Zacharewski 2001; Gershon 2002; IPCS 2003; Lühe et al. 2005; NRC 2007; OECD 2005; Olden and Wilson 2000; Orphanides 2003; Tralau et al. 2015; Tsuji and Garry 2009) | First year: 2000 Median year: 2003 Last year: 2015 |
| D6) Stakeholder commitment & investment: Government and industry are committed to, and invest in, the development of omics methods. |
(9 sources) (Balbus and Environmental Defense 2005; Boverhof and Zacharewski 2006; Chen et al. 2012; ECHA 2016b; Hartung 2011; NRC 2007; OECD 2010; Olden and Wilson 2000; Wilks et al. 2015) | First year: 2000 Median year: 2010 Last year: 2016 |
| D7) Reduced animal use: Omics methods are expected to reduce animal use in toxicity testing. |
(6 sources) (ECVAM and ICCVAM 2003; Hartung 2011; OECD 2010; Olden et al. 2001; Tralau et al. 2015; Tsuji and Garry 2009) | First year: 2001 Median year: 2009 Last year: 2015 |
| D8) Numerous untested chemicals: The high number of untested incumbent and new chemicals creates pressure for adopting new testing approaches. |
(5 sources) (Ankley et al. 2006; Lane and Pray 2002; NRC 2007; NTP 2004; Olden et al. 2001) | First year: 2001 Median year: 2004 Last year: 2007 |
| D9) Enabling laws & regulations: New laws and regulations convey directions that foster omics methods. |
(4 sources) (Ankley et al. 2006; Balbus and Environmental Defense 2005; ECHA 2016b; Hartung 2011) | First year: 2005 Median year: 2006 Last year: 2016 |
| D10) Accessibility of capabilities & resources: Omics tools and expertise are becoming more available. |
(2 sources) (Bergeson 2008; ECETOC 2007) | First year: 2007 Last year: 2008 |
| D11) International collaboration & harmonization: International collaboration supports the harmonization of omics methods. |
(1 source) (SOT 2015) | First and last year: 2015 |
Note: Underlined phrases in this table are the summary labels for each driver which are used in the rest of this article for ease of reference.
Table 3.
Obstacles cross-referenced to sources.
| Obstacles | Sources | First, median and last year of mention |
|---|---|---|
| O1) Insufficient validation: Omics methods are insufficiently validated, especially for regulatory uses. |
(33 sources) (Andersen and Krewski 2010; Ankley et al. 2006; Balbus and Environmental Defense 2005; Bergeson 2008; Dunn and Kolaja 2003; ECETOC 2007; ECHA 2016b; Fent and Sumpter 2011; Frueh 2006; Government of Canada 2016; Grodsky 2007; Hattis 2009; IPCS 2003; Krewski et al. 2009; Lühe et al. 2005; Malloy et al. 2017; NASEM 2017; Nature Publishing Group 2006; NRC 2007; NTP 2004; OECD 2005; Olden et al. 2001; Pettit et al. 2010; Sauer et al. 2017; Tralau et al. 2015; Tsuji and Garry 2009; Vachon et al. 2017; van Leeuwen 2007; Wakefield 2003; Willett et al. 2014; Wittwehr et al. 2017; Zaunbrecher et al. 2017; Zeiger 1999) | First year: 1999 Median year: 2008 Last year: 2017 |
| O2) Complexity of interpretation: Interpretation of omics data is complex and needs clearer links to biological impacts. |
(22 sources) (Andersen and Krewski 2010; Ankley et al. 2006; Bahamonde et al. 2016; Balbus and Environmental Defense 2005; ECETOC 2007; Fent and Sumpter 2011; Frueh 2006; Government of Canada 2016; IPCS 2003; Krewski et al. 2009; NASEM 2017; NRC 2005, 2007; NTP 2004; OECD 2005; Olden and Wilson 2000; Olden et al. 2001; Pennie et al. 2004; Pettit et al. 2010; Tsuji and Garry 2009; Villeneuve et al. 2012; Wakefield 2003) | First year: 2000 Median year: 2006 Last year: 2017 |
| O3) Lack of standardization: Omics methods need standardization. |
(20 sources) (Andersen and Krewski 2010; Ankley et al. 2006; Balbus and Environmental Defense 2005; Bergeson 2008; ECVAM and ICCVAM 2003; Fent and Sumpter 2011; Frueh 2006; Gant 2016; Government of Canada 2016; Malloy et al. 2017; NASEM 2017; Nature Publishing Group 2006; NRC 2005, 2007; NTP 2004; OECD 2005; Pettit et al. 2010; Sauer et al. 2017; Taylor et al. 2007; Tralau et al. 2015) | First year: 2003 Median year: 2007 Last year: 2017 |
| O4) Lack of expertise: Field actors lack expertise and need training. |
(12 sources) (Ankley et al. 2006; Balbus 2005; Balbus and Environmental Defense 2005; ECHA 2016b; ECVAM and ICCVAM 2003; Fent and Sumpter 2011; OECD 2005; Olden et al. 2001; Sauer et al. 2017; Tralau et al. 2015; Vachon et al. 2017; Zeiger 1999) | First year: 1999 Median year: 2005 Last year: 2017 |
| O5) Difficulty of coordination: The efforts of field actors lack coordination. Technical and IP rights issues with data sharing hinder coordination. |
(10 sources) (Balbus and Environmental Defense 2005; Bergeson 2008; ECHA 2016b; Hartung 2009; Krewski et al. 2009; Malloy et al. 2017; OECD 2005; Pettit et al. 2010; SOT 2015; Tralau et al. 2015) | First year: 2005 Median year: 2009 Last year: 2017 |
| O6) Resistance to change: Field actors resist the use and adoption of omics methods. |
(8 sources) (Ankley et al. 2006; Balbus 2005; Balbus and Environmental Defense 2005; Grodsky 2007; Hartung 2009; NRC 2007; Pettit et al. 2010; Zaunbrecher et al. 2017) | First year: 2005 Median year: 2007 Last year: 2017 |
| O7) High level of required investment: Using omics requires significant investment. |
(7 sources) (Ankley et al. 2006; Balbus and Environmental Defense 2005; Dunn and Kolaja 2003; Fent and Sumpter 2011; Hattis 2009; NRC 2007; OECD 2005) | First year: 2003 Median year: 2006 Last year: 2011 |
| O8) Lack of organizational support: There is a lack of funding, resources, and organizational support for omics methods. |
(6 sources) (Ankley et al. 2006; Krewski et al. 2009; NRC 2007; OECD 2005; Pettit et al. 2010; Vachon et al. 2017) | First year: 2005 Median year: 2007 Last year: 2017 |
| O9) Uncertain economic benefits: The economic benefits of omics are uncertain. |
(4 sources) (Balbus and Environmental Defense 2005; ECHA 2016b; Hattis 2009; Pettit et al. 2010) | First year: 2005 Median year: 2009 Last year: 2016 |
| O10) Inadequacy for some applications: There are some areas of toxicology where omics cannot be applied. |
(4 sources) (Andersen and Krewski 2010; Malloy et al. 2017; NRC 2007; Pettit et al. 2010) | First year: 2007 Median year: 2010 Last year: 2017 |
| O11) Concerns about litigation: Actors are concerned with litigation from retrospective analysis. |
(2 sources) (Freeman 2004; Pettit et al. 2010) | First year: 2004 Last year: 2010 |
| O12) Frustrated expectations: Omics created unrealistic expectations which are not fulfilled. |
(2 sources) (Bergeson 2008; Dunn and Kolaja 2003) | First year: 2003 Last year: 2008 |
Note: Underlined phrases in this table are the summary labels for each obstacle which are used in the rest of this article for ease of reference.
We identified several types of potential adopters/users of toxicogenomics: government agencies and regulators (ECVAM and ICCVAM 2003; OECD 2005, 2010; Tralau et al. 2015); businesses (Kramer and Kolaja 2002; Lühe et al. 2005; Orphanides 2003); academia (Bahamonde et al. 2016; Boverhof and Zacharewski 2006; Grodsky 2007; Hartung 2009; Hattis 2009; Trosko and Upham 2010); and cross-sector collaborations (Andersen and Krewski 2010; Malloy et al. 2017; Taylor et al. 2007). These groups are considered together as “potential adopters” in this commentary. Notably, the nature of what precisely was to be adopted varied. Sources discussed toxicogenomics as a suite of approaches, tests, and methods (e.g., Hartung 2011; SOT 2015; Zaunbrecher et al. 2017); a set of technologies (e.g., ECETOC 2007; NRC, 2005; Sauer et al. 2017); and data or data sets produced by particular approaches, tests, methods, and technologies (e.g., Bergeson 2008; Freeman 2004; Gant 2016). Distinguishing whether the specific toxicogenomics innovations being adopted were knowledge, practices, physical artifacts or data was therefore challenging. Given a tradition in social science of conceptualizing technologies as a nexus of knowledge, artifacts, and practices (Garud and Rappa 1994), we elided distinctions and present findings below with reference to “toxicogenomics” writ large and/or “toxicogenomics tools and data sets.” The rest of this section describes the five most salient drivers and obstacles, explaining how they were understood during the period 1998–2017 to help or hinder the adoption of toxicogenomics in chemical risk assessment. We labeled drivers and obstacles according to their relative salience, from those mentioned in the most to the fewest number of sources in our corpus (D1, D2, … D11; and O1, O2, … O12).
Drivers.
The first driver, Superior scientific understanding (D1), reflects the view that omics methods allow a detailed scientific understanding of health and ecological effects from chemical exposure (Olden et al. 2001). Sources mentioning this driver suggested that the more comprehensive toxicogenomics knowledge of chemical hazards becomes, the more users will adopt toxicogenomics to improve risk assessment (Table 2). This driver includes the discovery of new toxicity pathways and mechanisms of action (Fent and Sumpter 2011), refinements of risk models (Andersen and Krewski 2010), optimization of assays for specific chemicals (Ankley et al. 2006), more accurate health risk assessments (Tsuji and Garry 2009), and enhanced public health and regulatory decisions (ECETOC 2007; ECHA 2016b; Tralau et al. 2015).
The second driver refers to New applications (D2) afforded by omics in human and ecological toxicology (ECHA 2016b; Olden et al. 2001). Sources mentioning this driver argued that access to previously unavailable tests motivates the adoption of toxicogenomics. New applications include identifying biomarkers of exposure (IPCS 2003), determining species-specific toxicity or mixture toxicity, assessing low-dose effects, examining endocrine effects, investigating nanotoxicology (Tralau et al. 2015), generating toxicity data to identify the best analogs to chemicals of concern (NASEM 2017), and removing new substances with unsuitable safety margins early in the testing process (Chen et al. 2012).
The third driver, Reduced cost & increased efficiency (D3), captures the notion that omics have been expected to reduce testing cost and time, thus increasing testing efficiency (Iannaccone 2001). Sources highlighted alternative methods’ potential for time and cost savings in comparison with conventional approaches (ECHA 2016b; OECD 2010; Tsuji and Garry 2009) and noted that costs for using some new technologies were already reasonable (Sauer et al. 2017). Continued improvement of the economics of toxicogenomics, it was argued, could lead to greater toxicological coverage of the chemical universe (NRC 2007), especially if omics-based methods are applied widely, generating efficiencies of scale (Sauer et al. 2017).
The fourth driver highlights how Scientific and technological advances (D4) have been viewed to expand the capacities and scope of omics methods in toxicology, resulting in more New applications (D2) and fueling the uptake of omics. Advances in molecular technology, proteomics, metabolomics, bioinformatics, and modeling improve testing efficiency and efficacy (Iannaccone 2001), understanding of mechanistic toxicology (Tralau et al. 2015), alternative methods validation (Hartung 2011) and, more broadly, the capacity to deal with important issues in human and ecological toxicology (Chen et al. 2012), according to toxicogenomics discourse during the period 1998–2017.
The fifth driver, Belief in the potential of omics (D5), refers to confidence in the promise of omics methods to generate Superior scientific understanding (D1) and New applications (D2). During the period 1998–2017, it manifested itself in assertions that omics would transform risk assessment in medical science (ECVAM and ICCVAM 2003) and ecotoxicology (OECD 2005), with specific claims regarding better classification of chemicals and drugs based on transcriptomic profiling (Fielden and Zacharewski 2001), improved specificity of chemical risk assessment (IPCS 2003), and increased speed and efficiency of toxicity testing (NRC 2007; Tralau et al. 2015).
Obstacles.
The first obstacle, Insufficient validation (O1), reflects the claim made in numerous sources during the period 1998–2017 that omics methods lack adequate validation, especially for regulatory uses (Table 3). Different validity requirements across specific uses and user needs [e.g., regulatory vs. other contexts (Malloy et al. 2017; Zeiger 1999)] may amplify this concern. Ultimately, lack of appropriate validation discourages actors from using omics (Sauer et al. 2017). Concerns during the past 20 y include the likelihood of false positives (Andersen and Krewski 2010; Balbus and Environmental Defense 2005; Villeneuve et al. 2012), difficulty in distinguishing chemically induced from normal gene expression (Balbus and Environmental Defense 2005), insufficient knowledge of data quality (ECHA 2016b; Fent and Sumpter 2011; Vachon et al. 2017), as well as limited biological understanding of omics data (Pettit et al. 2010). Some sources claimed that toxicogenomics need further scientific justification (Frueh 2006; Wakefield 2003; Zaunbrecher et al. 2017). Complicating matters, the validation of omics and other novel methods has been a moving target due to their rapid evolution (Balbus and Environmental Defense 2005; NASEM 2017; NRC 2007). Further, validation has been constructed as requiring lengthy, expensive, and technically and logistically demanding efforts (NRC 2007; Olden et al. 2001; Tralau et al. 2015). Other validation challenges discussed during the period 1998–2017 include the perceived requirement to compare data from alternative methods with data from incumbent methods that are still considered the “gold standard” (Sauer et al. 2017; Wittwehr et al. 2017) despite shortcomings (Andersen and Krewski 2010); and the constraints of a necessarily multistakeholder approach to validation (Bergeson 2008).
The second obstacle, Complexity of interpretation (O2), reflects the complexity of omics data analysis (Ankley et al. 2006; ECETOC 2007; Fent and Sumpter 2011), which has been understood to bring uncertainty to data interpretation and make some actors reluctant to use omics. During the period 1998–2017, interpretation challenges came primarily from limited knowledge of gene sequences and annotations (Fent and Sumpter 2011; OECD 2005; Pennie et al. 2004); lack of a rigorous, established and harmonized interpretation framework, including baseline data (ECETOC 2007; Fent and Sumpter 2011; Frueh 2006; NASEM 2017; NRC 2007; NTP 2004); and uncertainty in extrapolations from gene expression to outcomes in cells, organisms, and populations (Fent and Sumpter 2011; NTP 2004; Olden et al. 2001). Challenges also arose from the time required for data analysis and interpretation (ECETOC 2007; Pettit et al. 2010), and the need to integrate data from separate disciplines (NASEM 2017). Big data have further complicated interpretation through significant computational requirements, multiple online data sources, and the considerable investments that were needed to generate and analyze data sets, especially early in the period from 1998 through 2017 (Balbus and Environmental Defense 2005; ECETOC 2007).
Many sources from 1998 through 2017 called for more standardization of omics assays, data evaluation, and reporting (NASEM 2017) to establish a simpler interpretation framework that would also be compatible with users’ routines (Table 3). Accordingly, the third obstacle was labeled Lack of standardization (O3). Even recent research identified lack of standardization as a crucial reason risk assessors continue to balk at using omics data (Sauer et al. 2017). Researchers have argued that standardization is essential for credible and consistent processing of omics data in regulatory risk assessment (ECVAM and ICCVAM 2003; Pettit et al. 2010; Sauer et al. 2017), and for characterizing new risk assessment approaches (Government of Canada 2016). Standardization, it has been argued, can help address issues such as low compliance with data standards (Pettit et al. 2010) and variability in results across methods and laboratories (Fent and Sumpter 2011; Frueh 2006; Nature Publishing Group 2006). Notwithstanding the benefits that might flow from standardization, proponents of toxicogenomics confronted persistent hurdles to standardization that compounded their challenges (Balbus and Environmental Defense 2005). These hurdles included the high levels of expertise across numerous, diverse actors needed to achieve standardization (Tralau et al. 2015), norms of storing experimental data in unconnected silos (Malloy et al. 2017), and rapid technological progress (ECVAM and ICCVAM 2003).
The Lack of expertise (O4) obstacle highlights that addressing the Complexity of interpretation (O2) of omics data requires expertise in several domains, including experimentation, data gathering, data analysis, result interpretation, reporting, and decision making, which often require training (ECVAM and ICCVAM 2003; Fent and Sumpter 2011; Vachon et al. 2017). There has been concern about the lack of expertise in the regulatory community (ECVAM and ICCVAM 2003) and broader public (Balbus and Environmental Defense 2005). Limited training has been understood to engender costly recruitment of experts to interpret omics data, difficult integration of omics knowledge into existing paradigms (Balbus 2005), low individual acceptance of omics methods (Zeiger 1999), and poor regulatory uptake of omics (Sauer et al. 2017). Training and education have therefore been considered necessary to develop technical expertise (ECHA 2016b; OECD 2005) and familiarity (Sauer et al. 2017; Zeiger 1999).
The fifth obstacle, Difficulty of coordination (O5), refers to the need for diverse actors to align their efforts around toxicogenomics (ECETOC 2007; Zaunbrecher et al. 2017) and the associated challenges. Insufficient coordination has been understood to impede integration of omics into chemical risk assessment (OECD 2005). Coordination relates explicitly to Lack of standardization (O3), because concerted efforts help harmonize laboratory procedures (OECD 2005), validation processes (SOT 2015), and (inter)national regulatory guidelines on alternative methods (SOT 2015). Coordination also helps legal, regulatory, ethical, and policy communities develop stable frameworks for the use of omics data in regulatory and legal settings (Bergeson 2008). During the period 1998–2017, coordination challenges included the absence of harmonized and publicly available tools for data analysis (Ankley et al. 2006; ECHA 2016a; ECVAM and ICCVAM 2003), barriers to data sharing (Balbus and Environmental Defense 2005; ECHA 2016b), scarce infrastructure to bridge different types of information and expertise (IPCS 2003), and the need for extensive cross-sector collaboration (Krewski et al. 2009).
Given contrasts between empirically derived drivers and obstacles [e.g., D3 indicates that Reduced cost & increased efficiency drive adoption, whereas O7 (High level of required investment) and O9 (Uncertain economic benefits) counter this claim with economics-based rationales for nonadoption] we note that there is no consensus in our corpus (Tables 2 and 3). Nonetheless, clear patterns were uncovered in how authors have talked about what helps and what hinders the adoption of toxicogenomics for chemical risk assessment.
Drivers, Obstacles, and Adoption of Innovations
The social science literature on innovation adoption permits anchoring empirically derived drivers and obstacles into theoretical frameworks that can provide guidance for proponents of toxicogenomics. In particular, Everett Rogers’ highly cited Diffusion of Innovations (Rogers 1962, 2003) theorizes five innovation attributes that facilitate the adoption of a given innovation (Table 4). Guided by semantic correspondence, we mapped drivers and obstacles onto Rogers’ (1962, 2003) innovation attributes. For example, International collaboration & harmonization (D11) refers to efforts to align omics practices across jurisdictions and to create a shared basis for omics data interpretation. Harmonization is unrelated to the perception that omics methods are superior to incumbent technologies (i.e., relative advantage) or to adopters’ ability to experiment directly or vicariously with omics (i.e., trialability and observability). Rather, it refers to ensuring consistency and ease of use of omics tools and data across jurisdictions, so we mapped D11 onto compatibility and simplicity.
Table 4.
Five innovation attributes that facilitate and accelerate adoption.
| Innovation attribute | Definition (from Rogers 2003) |
|---|---|
| Relative advantage | The extent to which an innovation has superior functionality relative to cost, as compared to incumbent technologies. |
| Compatibility | The extent to which an innovation is consistent with potential adopters’ values, past experiences, and needs. |
| Simplicity | The extent to which an innovation is easy to understand and use. |
| Trialability | The extent to which an innovation can be experimented with by potential adopters. |
| Observability | The extent to which an innovation’s benefits can be clearly seen by later adopters when early adopters use it. |
The majority of empirically derived drivers—the eight most salient ones—map onto relative advantage, indicating a widespread assumption during the period 1998–2017 that the superior functionality of toxicogenomics tools in comparison with legacy methods would drive adoption (Table 5). Our mapping also indicates some attention to ensuring that toxicogenomics tools and data sets have the requisite compatibility with skills and routines of potential adopters, and the requisite simplicity to be understood and used easily by them, to drive adoption. Finally, our mapping highlights that trialability and observability received little attention in discussions of what drives adoption of toxicogenomics, during the period 1998–2017.
Table 5.
Mapping drivers and obstacles onto innovation attributes that facilitate and accelerate adoption.
| Innovation attributes | Drivers | Obstacles |
|---|---|---|
| Relative advantage | D1 - Superior scientific understanding D2 - New applications D3 - Reduced cost & increased efficiency D4 - Scientific and technological advances D5 - Belief in the potential of omics D6 - Stakeholder commitment & investment D7 - Reduced animal use D8 - Numerous untested chemicals |
O1 - Insufficient validation O2 - Complexity of interpretation O5 - Difficulty of coordination O7 - High level of required investment O8 - Lack of organizational support O9 - Uncertain economic benefits O10 - Inadequacy for some applications |
| Compatibility | D6 - Stakeholder commitment & investment D9 - Enabling laws & regulations D10 - Accessibility of capabilities & resources D11 - International collaboration & harmonization |
O1 - Insufficient validation O2 - Complexity of interpretation O3 - Lack of standardization O4 - Lack of expertise O5 - Difficulty of coordination O8 - Lack of organizational support |
| Simplicity | D6 - Stakeholder commitment & investment D10 - Accessibility of capabilities & resources D11 - International collaboration & harmonization |
O2 - Complexity of interpretation O3 - Lack of standardization O4 - Lack of expertise O5 - Difficulty of coordination O8 - Lack of organizational support |
| Trialability | D6 - Stakeholder commitment & investment D10 - Accessibility of capabilities & resources |
O1 - Insufficient validation O5 - Difficulty of coordination O8 - Lack of organizational support |
| Observability | D6 - Stakeholder commitment & investment D10 - Accessibility of capabilities & resources |
O1 - Insufficient validation O5 - Difficulty of coordination O8 - Lack of organizational support |
Trialability and observability received slightly more consideration in discussions of obstacles in comparison with drivers; i.e., low trialability and low observability hindered adoption: Two out of the five most salient obstacles map onto these attributes, suggesting that adopters lacked opportunities for direct and proxy experimentation with omics tools and data sets (Table 5). Three of the five most salient obstacles map onto relative advantage, indicating that the performance of omics relative to legacy methods was also a concern. Four of the five most salient obstacles map onto simplicity, indicating that difficulties in understanding and using omics tools and data sets were key barriers to adoption. Finally, all of the five most salient obstacles map onto compatibility, suggesting that poor fit of toxicogenomics with risk assessors’ routines was an important barrier to their adoption during the period 1998–2017.
Recommendations for policy and practice.
The difference in focus we observe between drivers and obstacles suggests how toxicogenomics’ proponents may redirect their efforts to ensure attention to all innovation attributes. Arguments that invoke the most salient drivers, thus highlighting relative advantage, are unlikely to convince potential adopters concerned with compatibility and simplicity. Further, the apparent lack of attention to trialability and observability in the discourse on drivers and obstacles could aggravate compatibility issues; if potential adopters cannot experiment directly or vicariously with toxicogenomics tools and data sets, it is unlikely they can evaluate the compatibility of these tools and data with their work. Our findings corroborate previous studies of toxicogenomics that have highlighted their unfamiliarity, a need for more knowledge about them, and resistance to change as important obstacles to their adoption (Balbus 2005; Vachon et al. 2017) but additionally cast these studies in new light by mobilizing social science theorizing of innovation adoption.
Our findings also provide theoretical support for initiatives aiming to increase trialability and observability of emerging tools and data sets, such as the U.S. Food and Drug Administration’s provision of a “safe harbor” for industry to submit genomic data voluntarily for learning purposes, with reassurances that regulatory decisions would not leverage premature data (see Goodsaid et al. 2010). Also, we acknowledge that the need for toxicogenomics researchers to make their tools and data sets more readily and easily usable by regulators and other end users has been identified in recent research (Farmahin et al. 2017; Harrill et al. 2019; Thomas et al. 2019). Our commentary complements existing work by pointing to potential solutions, including the expansion of the priorities of developers of toxicogenomics tools beyond further scientific advances that yield superior functionality and novel uses. Specifically, ensuring that tools and data sets are not overly complex to use (simplicity) and not unnecessarily disruptive of established risk assessment practices (compatibility) would speed their adoption.
We propose that one way to increase the simplicity and compatibility of toxicogenomics tools and data is by addressing the Lack of standardization obstacle (O3), i.e., by developing standardized approaches to guide the development of tools, associated data sets, and reports that are simple to use and compatible with users’ workflows (see mapping in Table 5). Standardization has been difficult because the technology for evaluating gene expression has changed repeatedly since 1998 (NASEM 2017; NRC 2007), even if researchers have long recognized that consistent data sets built on a stable platform represent a necessary foundation for adoption of toxicogenomics by regulators (Frueh 2006). In fact, academic and regulatory scientists have made important progress toward this goal. As part of the U.S. National Institutes of Health’s Library of Integrated Network-Based Cellular Signature (LINCS) initiative, bioinformaticians identified 978 “landmark” genes that when perturbed could represent changes across the human transcriptome (Subramanian et al. 2017); whereas Thomas et al. (2019) describe the U.S. Environmental Protection Agency’s plan to develop a similar database of sentinel genes for toxicological applications, an approach now used by multiple stakeholders. For example, researchers at the U.S. National Toxicology Program (NTP) realized the S1500+ gene set based on an analysis of available Affymetrix Human Whole Genome Microarrays (HG-U133plus2) (Mav et al. 2018); an academic team built the Toxicogenomics-1000 (T1000) gene set based on analyses of in vivo and in vitro data from human and rat studies from the Toxicogenomics Project-Genomics Assisted Toxicity Evaluation System (Open TG-GATEs) database (Soufan et al. 2019); and a group of industry researchers used the Connectivity Map (CMap) concept in a read-across study that spanned 186 chemicals and 19 cell lines (de Abrew et al. 2019). In particular, researchers are seeking to standardize the reporting of omics data through various initiatives, e.g., development of a generic Transcriptomics Reporting Framework (TRF) that includes Reference Baseline Analysis (RBA) (Gant et al. 2017); the MEtabolomics standaRds Initiative in Toxicology (MERIT) (Viant et al. 2019); and incorporation of omics data into the Adverse Outcome Pathway (AOP) framework (Brockmeier et al. 2017). Our analysis underlines the importance of these and similar efforts, because an innovation’s simplicity and compatibility bear critically on innovation adoption.
Adoption of Innovations: Innovation-Centric vs. Adopter-Centric Perspectives
In Stage IV in the “Methods” section, we introduced the analytical distinction between an innovation-centric and an adopter-centric perspective on the adoption of innovations (Figure 1). Following semantic correspondence, we mapped each empirically derived driver and obstacle onto one of these perspectives. For example, cost and efficiency are attributes of omics methods, and it falls on omics proponents to refine omics methods in ways that improve these attributes. Therefore, we linked Reduced cost & increased efficiency (D3) to the innovation-centric perspective. Conversely, Stakeholder commitment & investment (D6) refers to a feature of the adopting system; the likelihood of adoption increases when potential adopters allocate resources to toxicogenomics training and other efforts that increase organizations’ “absorptive capacity” (Cohen and Levinthal 1990, p. 128). We thus linked D6 to the adopter-centric perspective.
Drivers from 1998 through 2017 are distributed almost evenly between the innovation-centric and adopter-centric perspectives (Table 6), but the four most salient ones reflect an innovation-centric perspective. This finding suggests that attributes of omics innovations received more attention than features of the adopting system in discourse about drivers of adoption of toxicogenomics. Similarly, the three most salient obstacles are innovation-centric, which suggests more attention was paid to attributes of omics innovations than to features of the adopting system in discourse about obstacles to adoption of toxicogenomics. This finding is mitigated somewhat, however, by most obstacles mapping to the adopter-centric perspective, including two of the top five, which suggests a heightened sensitivity to adopters in discussions of what hindered adoption, in comparison with what helped it during the period 1998–2017.
Table 6.
Relating drivers and obstacles to innovation- and adopter-centric perspectives on adoption of innovations.
| Perspective | Drivers | Obstacles |
|---|---|---|
| Innovation-centric | D1 - Superior scientific understanding D2 - New applications D3 - Reduced cost and increased efficiency D4 - Scientific and technological advances D7 - Reduced animal use D10 - Accessibility of capabilities and resources |
O1 - Insufficient validation O2 - Complexity of interpretation O3 - Lack of standardization O9 - Uncertain economic benefits O10 - Inadequacy for some applications |
| Adopter-centric | D5 - Belief in the potential of omics D6 - Stakeholder commitment and investment D8 - Numerous untested chemicals D9 - Enabling laws and regulations D11 - International collaboration & harmonization |
O4 - Lack of expertise O5 - Difficulty of coordination O6 - Resistance to change O7 - High level of required investment O8 - Lack of organizational support O11 - Concerns about litigation O12 - Frustrated expectations |
Recommendations for policy and practice.
These findings indicate that the innovation-centric perspective was more prevalent than the adopter-centric one in discourse about the (non)adoption of toxicogenomics during the period 1998–2017. Given recent expressions of concern about the pace of adoption (Bergeson 2008; LaLone et al. 2017; Leung 2018), this prevalence may in itself represent an obstacle. It appears that research and development of omics tools has heretofore focused more on the tools and associated data abstracted from context than on characterizing the adopting system to develop a clearer “situated” view of the “tools in use.” We propose that a more balanced approach—one that places more emphasis on the adopter-centric perspective and, therefore, users—could facilitate and speed the adoption of toxicogenomics and other NAMs.
Adoption of Innovations and Organizational Learning
Reframing toxicogenomics as a complex innovation, we mapped empirically derived drivers and obstacles onto Attewell’s (1992) generic drivers and obstacles, guided by semantic correspondence. This analysis yielded interesting insights (Table 7). Except for Inadequacy for some applications (O10), empirically derived obstacles map richly and positively onto Attewell’s two generic obstacles as well as richly and negatively onto his two generic drivers. For example, potential adopters’ Lack of expertise (O4) clearly increases knowledge barriers and performance uncertainties, but undermines and slows skill development and organizational learning; the more expertise is lacking, the more skill development and organizational learning are required, and the more difficult they are.
Table 7.
Mapping drivers and obstacles onto concepts from Attewell’s (1992) framework for understanding the adoption of complex innovations.
| Concept | Drivers | Obstacles |
|---|---|---|
| Knowledge barriers (generic obstacle) | D1 - Superior scientific understanding () D2 - New applications () D4 - Scientific and technological advances () D6 - Stakeholder commitment & investment () D10 - Accessibility of capabilities & resources () D11 - International collaboration & harmonization () |
O1 - Insufficient validation () O2 - Complexity of interpretation () O3 - Lack of standardization () O4 - Lack of expertise () O5 - Difficulty of coordination () O6 - Resistance to change () O7 - High level of required investment () O8 - Lack of organizational support () |
| Performance uncertainties (generic obstacle) | D1 - Superior scientific understanding () D2 - New applications () D4 - Scientific and technological advances () D6 - Stakeholder commitment & investment () D9 - Enabling laws & regulations () D10 - Accessibility of capabilities & resources () D11 - International collaboration & harmonization () |
O1 - Insufficient validation () O2 - Complexity of interpretation () O3 - Lack of standardization () O4 - Lack of expertise () O5 - Difficulty of coordination () O6 - Resistance to change () O7 - High level of required investment () O8 - Lack of organizational support () O9 - Uncertain economic benefits () O11 - Concerns about litigation () |
| Skill development (generic driver) | D1 - Superior scientific understanding () D2 - New applications () D3 - Reduced cost & increased efficiency () D4 - Scientific and technological advances () D6 - Stakeholder commitment & investment () D9 - Enabling laws & regulations () D10 - Accessibility of capabilities & resources () D11 - International collaboration & harmonization () |
O1 - Insufficient validation () O2 - Complexity of interpretation () O3 - Lack of standardization () O4 - Lack of expertise () O6 - Resistance to change () O7 - High level of required investment () O8 - Lack of organizational support () |
| Organizational learning (generic driver) | D1 - Superior scientific understanding () D2 - New applications () D3 - Reduced cost & increased efficiency () D4 - Scientific and technological advances () D6 - Stakeholder commitment & investment () D9 - Enabling laws & regulations () D10 - Accessibility of capabilities & resources () D11 - International collaboration & harmonization () |
O1 - Insufficient validation () O2 - Complexity of interpretation () O3 - Lack of standardization () O4 - Lack of expertise () O6 - Resistance to change () O7 - High level of required investment () O8 - Lack of organizational support () O11 - Concerns about litigation () |
Note: () indicates that the empirically derived driver of or obstacle to adoption of toxicogenomics tools is likely to contribute positively to or increase the generic driver or obstacle from Attewell’s framework. () indicates that the empirically derived driver of or obstacle to adoption of toxicogenomics tools is likely to contribute negatively to or decrease the generic driver or obstacle from Attewell’s framework. For the sources of drivers and obstacles, please see Tables 2 and 3 respectively.
Empirically derived drivers mapped onto Attewell’s generic drivers and obstacles almost as richly but less symmetrically (Table 7). Several empirically derived drivers with very high salience, i.e., Superior scientific understanding (D1), New applications (D2) and Scientific and technological advances (D4) contribute positively to generic obstacles and negatively to generic drivers, just like empirically derived obstacles. The more advanced, sophisticated, and novel that toxicogenomics becomes, the more knowledge barriers and performance uncertainties are increased and the more skill development and organizational learning are required and difficult. Most other empirical drivers map to generic drivers and obstacles in the opposite way, i.e., decreasing knowledge barriers and performance uncertainties and/or contributing positively to skill development and organizational learning. For example, consider Reduced cost & increased efficiency (D3). As toxicogenomics tools become cheaper and more efficient, more organizations will be able to afford to acquire, experiment with, and engage in learning-by-doing with them, which contributes positively to skill development and organizational learning.
Recommendations for policy and practice.
We argue that because toxicogenomics innovations are complex ones that require new expertise, know-how, and learning by doing, two generic obstacles to adoption, knowledge barriers and performance uncertainties, are to be expected. These obstacles can however be overcome by two generic drivers: individual skill development and organizational learning. When potential adopters lament performance uncertainties and the knowledge barriers to which they give rise, we recommend that toxicogenomics’ proponents focus more on understanding adopter needs and working toward skill development and organizational learning in the adopting system. They could also attend to and foster the emergence of mediating institutions to bridge the divide between innovators and potential adopters, which might include specialized consulting organizations and contract laboratories with the requisite know-how, bioinformatics platforms, and databases that reduce knowledge asymmetries and performance uncertainties associated with toxicogenomics innovations, and accessible training programs to speed skill development.
Indeed, numerous entities already appear to be playing the facilitating role of mediating institutions. For example, publicly available omics databases, such as TG-GATEs (Igarashi et al. 2015) and, more broadly, NCBI’s Gene Expression Omnibus (GEO), create opportunities for potential users to experiment with omics data. Similarly, online bioinformatics platforms, such as EcoToxXplorer.ca, developed for the EcoToxChip project (Basu et al. 2019), offer tutorials that support skill development and allow potential users to generate their own bioinformatics data. In reducing knowledge barriers and mitigating performance uncertainties, these and similar initiatives facilitate the adoption of toxicogenomics and other NAMs in chemical risk assessment.
Conclusion
We observed that an innovation-centric perspective appears to have dominated discourse about the adoption of toxicogenomics in chemical risk assessment during the period 1998–2017, with proponents extolling the tools’ putative superior and novel functionality but overlooking the tools’ and data sets’ understandability, ease of use, and fit with users’ routines. We recommend that more attention be placed on ensuring the simplicity and compatibility of toxicogenomics tools and data, as well as creating opportunities for potential adopters to experiment with them directly (trialability) and vicariously (observability). We also conclude that the innovation-centric perspective would be usefully balanced with an adopter-centric one that highlights the importance of skill development and organizational learning in the adopting system.
The asymmetric focus on honing the innovations themselves rather than engaging with, understanding and intervening in the adopting system might reflect toxicogenomics’ origins in “basic” science, i.e., biology, which tends to be more abstracted from and less embedded in the context of its potential use than “applied” sciences. Even further embedded in a specific context of use are “regulatory” sciences such as toxicology and ecotoxicology, which are accountable to normative demands of diverse stakeholders and epistemic demands of the scientific community (Jasanoff 1994, 2011). Regulatory knowledge is not produced simply as a result of curiosity but, rather, for application in regulatory decision making involving multiple stakeholders (Balbus and Environmental Defense 2005; Boverhof and Zacharewski 2006; Buesen et al. 2017) who are relevant to evaluating the merits or shortcomings of toxicogenomics innovations. We therefore propose that proponents of toxicogenomics would benefit greatly from a heightened sensitivity to the workings of the system into which they hope their innovations will be adopted.
Supplementary Material
Acknowledgments
This study informs the GE3LS (Genomics and its Ethical, Environmental, Economic, Legal, and Social aspects) component of the project “EcoToxChip: A toxicogenomics tool for chemical prioritization and environmental management” funded by Genome Canada under grant no. 10501. The authors thank the financial sponsors of this project (Genome Canada; Génome Québec; Genome Prairie; the Government of Canada; Environment and Climate Change Canada; Ministère de l’Économie, de la Science et de l’Innovation du Québec; the University of Saskatchewan; and McGill University) and acknowledge funding from the McGill Sustainable Systems Initiative (MSSI). The authors also thank T. Onookome-Okome for her research assistance.
References
- Andersen ME, Krewski D. 2010. The vision of toxicity testing in the 21st century: moving from discussion to action. Toxicol Sci 117(1):17–24, PMID: 20573784, 10.1093/toxsci/kfq188. [DOI] [PubMed] [Google Scholar]
- Ankley G, Daston GP, Degitz SJ, Denslow ND, Hoke RA, Kennedy SW, et al. 2006. Toxicogenomics in regulatory ecotoxicology. Environ Sci Technol 40(13):4055–4065, PMID: 16856717, 10.1021/es0630184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold C. 2015. ToxCast™ wants you: recommendations for engaging the broader scientific community. Environ Health Perspect 123(1):A20, PMID: 25561607, 10.1289/ehp.123-A20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attewell P. 1992. Technology diffusion and organizational learning: the case of business computing. Organization Sci 3(1):1–19, 10.1287/orsc.3.1.1. [DOI] [Google Scholar]
- Bahamonde PA, Feswick A, Isaacs MA, Munkittrick KR, Martyniuk CJ. 2016. Defining the role of omics in assessing ecosystem health: perspectives from the Canadian environmental monitoring program: omics for ecosystem health. Environ Toxicol Chem 35(1):20–35, PMID: 26771350, 10.1002/etc.3218. [DOI] [PubMed] [Google Scholar]
- Balbus JM, Environmental Defense. 2005. Toxicogenomics: Harnessing the Power of New Technology. New York, NY: Environmental Defense. [Google Scholar]
- Balbus JM. 2005. Ushering in the new toxicology: toxicogenomics and the public interest. Environ Health Perspect 113(7):818–822, PMID: 16002368, 10.1289/ehp.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu N, Crump D, Head J, Hickey G, Hogan N, Maguire S, et al. 2019. EcoToxChip: a next-generation toxicogenomics tool for chemical prioritization and environmental management. Environ Toxicol Chem 38(2):279–288, PMID: 30693572, 10.1002/etc.4309. [DOI] [PubMed] [Google Scholar]
- Bergeson LL. 2008. Challenges in applying toxicogenomic data in federal regulatory settings. In: Genomics and Environmental Regulation: Science, Ethics, and Law. Sharp RR, Marchant, GE, and Grodsky, JA, eds. Baltimore, MD: Johns Hopkins University Press, 67–80. [Google Scholar]
- Bhaskarabhatla A. 2016. The moderating role of submarket dynamics on the product customization–firm survival relationship. Organization Sci 27(4):1049–1064, 10.1287/orsc.2016.1074. [DOI] [Google Scholar]
- Birnbaum LS. 2013. 15 years out: reinventing ICCVAM. Environ Health Perspect 121(2):a40, PMID: 23380598, 10.1289/ehp.1206292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boverhof DR, Zacharewski TR. 2006. Toxicogenomics in risk assessment: applications and needs. Toxicol Sci 89(2):352–360, PMID: 16221963, 10.1093/toxsci/kfj018. [DOI] [PubMed] [Google Scholar]
- Brockmeier EK, Hodges G, Hutchinson TH, Butler E, Hecker M, Tollefsen KE, et al. 2017. The role of omics in the application of adverse outcome pathways for chemical risk assessment. Toxicol Sci 158(2):252–262, PMID: 28525648, 10.1093/toxsci/kfx097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buesen R, Chorley BN, da Silva Lima B, Daston G, Deferme L, Ebbels T, et al. 2017. Applying ’omics technologies in chemicals risk assessment: report of an ECETOC workshop. Regul Toxicol Pharmacol 91:S3–S13, PMID: 28958911, 10.1016/j.yrtph.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zhang M, Borlak J, Tong W. 2012. A decade of toxicogenomic research and its contribution to toxicological science. Toxicol Sci 130(2):217–228, PMID: 22790972, 10.1093/toxsci/kfs223. [DOI] [PubMed] [Google Scholar]
- Clark JP. 2003. 15: How to peer review a qualitative manuscript. In: Peer Review in Health Sciences. Godlee F and Jefferson, T, eds. London, UK: BMJ Books, 219–235. [Google Scholar]
- Cohen WM, Levinthal DA. 1990. Absorptive capacity: a new perspective on learning and innovation. Adm Sci Q 35(1):128–152, 10.2307/2393553. [DOI] [Google Scholar]
- Compagni A, Mele V, Ravasi D. 2015. How early implementations influence later adoptions of innovation: social positioning and skill reproduction in the diffusion of robotic surgery. Acad Manag J 58(1):242–278, 10.5465/amj.2011.1184. [DOI] [Google Scholar]
- Cote I, Andersen ME, Ankley GT, Barone S, Birnbaum LS, Boekelheide K, et al. 2016. The Next Generation of Risk Assessment multi-year study—highlights of findings, applications to risk assessment, and future directions. Environ Health Perspect 124(11):1671–1682, PMID: 27091369, 10.1289/EHP233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creswell JW. 2013. Qualitative Inquiry and Research Design: Choosing Among Five Approaches. Los Angeles, CA: SAGE Publications. [Google Scholar]
- Cusumano MA, Kahl SJ, Suarez FF. 2015. Services, industry evolution, and the competitive strategies of product firms. Strat Mgmt J 36(4):559–575, 10.1002/smj.2235. [DOI] [Google Scholar]
- De Abrew KN, Shan YK, Wang X, Krailler JM, Kainkaryam RM, Lester CC, et al. 2019. Use of connectivity mapping to support read across: a deeper dive using data from 186 chemicals, 19 cell lines and 2 case studies. Toxicology 423:84–94, PMID: 31125584, 10.1016/j.tox.2019.05.008. [DOI] [PubMed] [Google Scholar]
- Dunn RT, Kolaja KL. 2003. Gene expression profile databases in toxicity testing. In: An Introduction to Toxicogenomics. Burczynski ME, ed. Boca Raton, FL: CRC Press, 213–224. [Google Scholar]
- ECETOC (European Centre for Ecotoxicology and Toxicology of Chemicals). 2007. Workshop on the Application of Omic Technologies in Toxicology and Ecotoxicology: Case Studies and Risk Assessment, 6-7 December 2007, Malaga. Brussels, Belgium: ECETOC. [Google Scholar]
- ECHA (European Chemicals Agency). 2016a. New Approach Methodologies in Regulatory Science, 19–20 April 2016. Proceedings of a scientific workshop. Helsinki, Finland: ECHA. [Google Scholar]
- ECHA. 2016b. Topical Scientific Workshop on New Approach Methodologies in Regulatory Science, 19–20 April 2016. Background document. Helsinki, Finland: ECHA. [Google Scholar]
- ECVAM (European Centre for the Validation of Alternative Methods), ICCVAM (Interagency Coordinating Committee on the Validation of Alternative Methods). 2003. Workshop on the Validation Principles for Toxicogenomics-Based Test Systems: An overview. Ispra, Italy: ECVAM. [Google Scholar]
- Farmahin R, Williams A, Kuo B, Chepelev NL, Thomas RS, Barton-Maclaren TS, et al. 2017. Recommended approaches in the application of toxicogenomics to derive points of departure for chemical risk assessment. Arch Toxicol 91(5):2045–2065, PMID: 27928627, 10.1007/s00204-016-1886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fent K, Sumpter JP. 2011. Progress and promises in toxicogenomics in aquatic toxicology: is technical innovation driving scientific innovation? Aquat Toxicol 105(suppl 3–4):25–39, PMID: 22099342, 10.1016/j.aquatox.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Fielden MR, Zacharewski TR. 2001. Challenges and limitations of gene expression profiling in mechanistic and predictive toxicology. Toxicol Sci 60(1):6–10, PMID: 11222867, 10.1093/toxsci/60.1.6. [DOI] [PubMed] [Google Scholar]
- Freeman K. 2004. Toxicogenomics data: the road to acceptance. Environ Health Perspect 112(12):A678–A685, PMID: 15345379, 10.1289/ehp.112-a678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frueh FW. 2006. Impact of microarray data quality on genomic data submissions to the FDA. Nat Biotechnol 24(9):1105–1107, PMID: 16964222, 10.1038/nbt0906-1105. [DOI] [PubMed] [Google Scholar]
- Gant TW, Sauer UG, Zhang S-D, Chorley BN, Hackermüller J, Perdichizzi S, et al. 2017. A generic Transcriptomics Reporting Framework (TRF) for ’omics data processing and analysis. Regul Toxicol Pharmacol 91:S36–S45, 10.1016/j.yrtph.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gant TW. 2016. Analysing Data: Towards Developing a Framework for Transcriptomics and Other Big Data Analysis for Regulatory Application. Helsinki, Finland: ECHA. [Google Scholar]
- Garud R, Rappa MA. 1994. A socio-cognitive model of technology evolution: the case of cochlear implants. Organization Sci 5(3):344–362, 10.1287/orsc.5.3.344. [DOI] [Google Scholar]
- Gershon D. 2002. Toxicogenomics gains impetus. Nature 415(6869):4–5, PMID: 11796961, 10.1038/nj6869-04a. [DOI] [PubMed] [Google Scholar]
- Goodsaid FM, Amur S, Aubrecht J, Burczynski ME, Carl K, Catalano J, et al. 2010. Voluntary exploratory data submissions to the US FDA and the EMA: experience and impact. Nat Rev Drug Discov 9(6):435–445, PMID: 20514070, 10.1038/nrd3116. [DOI] [PubMed] [Google Scholar]
- Government of Canada. 2016. Integrating New Approach Methodologies within the CMP: Identifying Priorities for Risk Assessment, Existing Substances Risk Assessment Program. Ottawa, ON: Government of Canada. [Google Scholar]
- Grodsky JA. 2007. Genomics and toxic torts: dismantling the risk-injury divide. Stanford Law Rev 59(6):1671–1734, PMID: 17593590, https://ssrn.com/abstract=1000971. [PubMed] [Google Scholar]
- Grondin CJ, Davis AP, Wiegers TC, Wiegers JA, Mattingly CJ. 2018. Accessing an expanded exposure science module at the comparative toxicogenomics database. Environ Health Perspect 126(1):014501, PMID: 29351546, 10.1289/EHP2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill J, Shah I, Setzer RW, Haggard D, Auerbach S, Judson R, et al. 2019. Considerations for strategic use of high-throughput transcriptomics chemical screening data in regulatory decisions. Curr Opin Toxicol 15:64–75, PMID: 31501805, 10.1016/j.cotox.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T. 2009. Toxicology for the twenty-first century. Nature 460(7252):208–212, PMID: 19587762, 10.1038/460208a. [DOI] [PubMed] [Google Scholar]
- Hartung T. 2011. From alternative methods to a new toxicology. Eur J Pharm Biopharm 77(3):338–349, PMID: 21195172, 10.1016/j.ejpb.2010.12.027. [DOI] [PubMed] [Google Scholar]
- Hattis D. 2009. High-throughput testing—the NRC vision, the challenge of modeling dynamic changes in biological systems, and the reality of low-throughput environmental health decision making. Risk Anal 29(4):483–484, PMID: 19076322, 10.1111/j.1539-6924.2008.01167.x. [DOI] [PubMed] [Google Scholar]
- Iannaccone PM. 2001. Toxicogenomics: “the call of the wild chip.” Environ Health Perspect 109(1):A8–A11, PMID: 11277097, 10.1289/ehp.109-a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICCVAM (Interagency Coordinating Committee on the Validation of Alternative Methods). 2018. A Strategic Roadmap for Establishing New Approaches to Evaluate the Safety of Chemicals and Medical Products in the United States. Research Triangle Park, NC: NTP, NICEATM. [Google Scholar]
- Igarashi Y, Nakatsu N, Yamashita T, Ono A, Ohno Y, Urushidani T, et al. 2015. Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res 43(Database issue):D921–D927, PMID: 25313160, 10.1093/nar/gku955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCS (International Programme on Chemical Safety). 2003. Workshop Report: Toxicogenomics and the Risk Assessment of Chemicals for the Protection of Human Health. Geneva, Switzerland: WHO. [Google Scholar]
- Jasanoff S. 1994. The Fifth Branch: Science Advisers as Policymakers. Cambridge, MA: Harvard University Press. [Google Scholar]
- Jasanoff S. 2011. Constitutional moments in governing science and technology. Sci Eng Ethics 17(4):621–638, PMID: 21879357, 10.1007/s11948-011-9302-2. [DOI] [PubMed] [Google Scholar]
- Kavlock RJ, Bahadori T, Barton-Maclaren TS, Gwinn MR, Rasenberg M, Thomas RS. 2018. Accelerating the pace of chemical risk assessment. Chem Res Toxicol 31(5):287–290, PMID: 29600706, 10.1021/acs.chemrestox.7b00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JA, Kolaja KL. 2002. Toxicogenomics: an opportunity to optimise drug development and safety evaluation. Expert Opin Drug Saf 1(3):275–286, PMID: 12904143, 10.1517/14740338.1.3.275. [DOI] [PubMed] [Google Scholar]
- Krewski D, Andersen ME, Mantus E, Zeise L. 2009. Toxicity testing in the 21st century: implications for human health risk assessment. Risk Anal 29(4):474–479, PMID: 19144067, 10.1111/j.1539-6924.2008.01150.x. [DOI] [PubMed] [Google Scholar]
- LaLone CA, Ankley GT, Belanger SE, Embry MR, Hodges G, Knapen D, et al. 2017. Advancing the adverse outcome pathway framework—an international horizon scanning approach. Environ Toxicol Chem 36(6):1411–1421, PMID: 28543973, 10.1002/etc.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane L, Pray L. 2002. Better living through toxicogenomics? Scientist. 4 March 2002. https://www.the-scientist.com/technology-profile/better-living-through-toxicogenomics-53604 [accessed 22 September 2018].
- Leung KM. 2018. Joining the dots between omics and environmental management. Integr Environ Assess Manag 14(2):169–173, PMID: 29171160, 10.1002/ieam.2007. [DOI] [PubMed] [Google Scholar]
- Li A, Lu X, Natoli T, Bittker J, Sipes NS, Subramanian A, et al. 2019. The Carcinogenome Project: in vitro gene expression profiling of chemical perturbations to predict long-term carcinogenicity. Environ Health Perspect 127(4):47002, PMID: 30964323, 10.1289/EHP3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lühe A, Suter L, Ruepp S, Singer T, Weiser T, Albertini S. 2005. Toxicogenomics in the pharmaceutical industry: hollow promises or real benefit? Mutat Res 575(1–2):102–115, PMID: 15924886, 10.1016/j.mrfmmm.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Malloy T, Zaunbrecher V, Beryt E, Judson R, Tice R, Allard P, et al. 2017. Advancing alternatives analysis: the role of predictive toxicology in selecting safer chemical products and processes. Integr Environ Assess Manag 13(5):915–925, PMID: 28247928, 10.1002/ieam.1923. [DOI] [PubMed] [Google Scholar]
- Malterud K. 2001. Qualitative research: standards, challenges, and guidelines. Lancet 358(9280):483–488, 10.1016/S0140-6736(01)05627-6. [DOI] [PubMed] [Google Scholar]
- Mav D, Shah RR, Howard BE, Auerbach SS, Bushel PR, Collins JB, et al. 2018. A hybrid gene selection approach to create the S1500+ targeted gene sets for use in high-throughput transcriptomics. PLoS One 13(2):e0191105, PMID: 29462216, 10.1371/journal.pone.0191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NASEM (National Academies of Sciences, Engineering, and Medicine). 2017. Using 21st Century Science to Improve Risk-Related Evaluations. Washington, DC: NASEM. [PubMed] [Google Scholar]
- Nature Publishing Group. 2006. Making the most of microarrays. Nat Biotechnol 24(9):1039, PMID: 16964193, 10.1038/nbt0906-1039. [DOI] [PubMed] [Google Scholar]
- NRC (National Research Council). 2005. Toxicogenomic Technologies and Risk Assessment of Environmental Carcinogens: A Workshop Summary. Washington, DC: National Academies Press. [PubMed] [Google Scholar]
- NRC. 2007. Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: National Academies Press. [Google Scholar]
- NRC. 2012. Exposure Science in the 21st Century: A Vision and a Strategy. Washington, DC: National Academies Press. [PubMed] [Google Scholar]
- NTP (National Toxicology Program). 2004. A National Toxicology Program for the 21st Century – A Roadmap for the Future. Research Triangle Park, NC: NTP. [Google Scholar]
- O’Brien BC, Harris IB, Beckman TJ, Reed DA, Cook DA. 2014. Standards for reporting qualitative research: a synthesis of recommendations. Acad Med 89(9):1245–1251, PMID: 24979285, 10.1097/ACM.0000000000000388. [DOI] [PubMed] [Google Scholar]
- OECD (Organization for Economic Cooperation and Development). 2005. Report of the OECD/IPCS Workshop on Toxicogenomics. Paris, France: OECD. [Google Scholar]
- OECD. 2010. Report of The Focus Session on Current And Forthcoming Approaches for Chemical Safety and Animal Welfare. Paris, France: OECD. [Google Scholar]
- Olden K, Guthrie J, Newton S. 2001. A bold new direction for environmental health research. Am J Public Health 91(12):1964–1967, PMID: 11726375, 10.2105/ajph.91.12.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olden K, Wilson S. 2000. Environmental health and genomics: visions and implications. Nat Rev Genet 1(2):149–153, PMID: 11253655, 10.1038/35038586. [DOI] [PubMed] [Google Scholar]
- Orphanides G. 2003. Toxicogenomics: challenges and opportunities. Toxicol Lett 140–141:145–148, PMID: 12676460, 10.1016/S0378-4274(02)00500-3. [DOI] [PubMed] [Google Scholar]
- Pennie W, Pettit SD, Lord PG. 2004. Toxicogenomics in risk assessment: an overview of an HESI collaborative research program. Environ Health Perspect 112(4):417–419, PMID: 15033589, 10.1289/ehp.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit S, Des Etages SA, Mylecraine L, Snyder R, Fostel J, Dunn RT, et al. 2010. Current and future applications of toxicogenomics: results summary of a survey from the HESI Genomics State of Science Subcommittee. Environ Health Perspect 118(7):992–997, PMID: 20100679, 10.1289/ehp.0901501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EM. 1962. Diffusion of Innovations. New York, NY: Free Press of Glencoe. [Google Scholar]
- Rogers EM. 2003. Diffusion of Innovations. New York, NY: Free Press. [Google Scholar]
- Sauer UG, Deferme L, Gribaldo L, Hackermüller J, Tralau T, van Ravenzwaay B, et al. 2017. The challenge of the application of ’omics technologies in chemicals risk assessment: background and outlook. Regul Toxicol Pharmacol 91:S14–S26, 10.1016/j.yrtph.2017.09.020. [DOI] [PubMed] [Google Scholar]
- Schmidt CW. 2002. Toxicogenomics: an emerging discipline. Environ Health Perspect 110(12):A750–A755, PMID: 12460812, 10.1289/ehp.110-a750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SETAC (Society of Environmental Toxicology and Chemistry). 2019. Technical Issue Paper: Omics: Complete Systems, Complete Analyses. Pensacola, FL: SETAC. [Google Scholar]
- SOT (Society of Toxicology). 2015. Alternative Toxicity Test Methods: Reducing, Refining, and Replacing Animal Use for Safety Testing. Reston, VA: SOT; https://www.toxicology.org/pubs/docs/pr/ToxTopics/TT3_InVitro_SOT.pdf [accessed 22 September 2018]. [Google Scholar]
- Soufan O, Ewald J, Viau C, Crump D, Hecker M, Basu N, et al. 2019. T1000: a reduced gene set prioritized for toxicogenomic studies. PeerJ 7:e7975, PMID: 31681519, 10.7717/peerj.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. 2017. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171(6):1437–1452.e17, PMID: 29195078, 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF, Paton NW, Lilley KS, Binz P-A, Julian RK, Jones AR, et al. 2007. The minimum information about a proteomics experiment (MIAPE). Nat Biotechnol 25(8):887–893, 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- Thomas RS, Bahadori T, Buckley TJ, Cowden J, Deisenroth C, Dionisio KL, et al. 2019. The next generation blueprint of computational toxicology at the U.S. Environmental Protection Agency. Toxicol Sci 169(2):317–332, PMID: 30835285, 10.1093/toxsci/kfz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, Bucher JR. 2013. Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect 121(7):756–765, PMID: 23603828, 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A, Flemming K, McInnes E, Oliver S, Craig J. 2012. Enhancing transparency in reporting the synthesis of qualitative research: ENTREQ. BMC Med Res Methodol 12:181, PMID: 23185978, 10.1186/1471-2288-12-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A, Sainsbury P, Craig J. 2007. Consolidated criteria for reporting qualitative research (COREQ): a 32-item checklist for interviews and focus groups. Int J Qual Health Care 19(6):349–357, PMID: 17872937, 10.1093/intqhc/mzm042. [DOI] [PubMed] [Google Scholar]
- Tralau T, Oelgeschläger M, Gürtler R, Heinemeyer G, Herzler M, Höfer T, et al. 2015. Regulatory toxicology in the twenty-first century: challenges, perspectives and possible solutions. Arch Toxicol 89(6):823–850, PMID: 25820917, 10.1007/s00204-015-1510-0. [DOI] [PubMed] [Google Scholar]
- Trosko JE, Upham BL. 2010. A paradigm shift is required for the risk assessment of potential human health after exposure to low level chemical exposures: a response to the toxicity testing in the 21st century report. Int J Toxicol 29(4):344–357, PMID: 20634539, 10.1177/1091581810371384. [DOI] [PubMed] [Google Scholar]
- Tsuji JS, Garry MR. 2009. Advances in toxicity testing herald improvements and challenges for risk assessment. Risk Anal 29(4):490–491, PMID: 19076320, 10.1111/j.1539-6924.2008.01170.x. [DOI] [PubMed] [Google Scholar]
- Vachon J, Campagna C, Rodriguez MJ, Sirard M-A, Levallois P. 2017. Barriers to the use of toxicogenomics data in human health risk assessment: a survey of Canadian risk assessors. Regul Toxicol Pharmacol 85:119–123, PMID: 28137640, 10.1016/j.yrtph.2017.01.008. [DOI] [PubMed] [Google Scholar]
- van Leeuwen K. 2007. Strategic Testing in REACH – A Testing Paradigm Shift. The McKim Conferences. Knife River, MN: The International QSAR Foundation. [Google Scholar]
- Viant MR, Ebbels TMD, Beger RD, Ekman DR, Epps DJT, Kamp H, et al. 2019. Use cases, best practice and reporting standards for metabolomics in regulatory toxicology. Nat Commun 10(1):, 10.1038/s41467-019-10900-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, Garcia-Reyero N, Escalon BL, Jensen KM, Cavallin JE, Makynen EA, et al. 2012. Ecotoxicogenomics to support ecological risk assessment: a case study with bisphenol A in fish. Environ Sci Technol 46(1):51–59, PMID: 21786754, 10.1021/es201150a. [DOI] [PubMed] [Google Scholar]
- Wakefield J. 2003. Toxicogenomics: roadblocks and new directions. Environ Health Perspect 111(6):A334, PMID: 12760837, 10.1289/ehp.111-a334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks MF, Roth N, Aicher L, Faust M, Papadaki P, Marchis A, et al. 2015. White paper on the promotion of an integrated risk assessment concept in European regulatory frameworks for chemicals. Sci Total Environ 521–522:211–218, 10.1016/j.scitotenv.2015.03.065. [DOI] [PubMed] [Google Scholar]
- Willett CE, Antczak P, Burgoon L, Falciani F, Gutsell S, Hodges G, et al. 2014. Using Adverse Outcome Pathways for Regulatory Applications. 9th World Congress on Alternatives to Animal Use in the Life Sciences, Prague, Czech Republic. https://htpconsortium.files.wordpress.com/2014/07/willett_wc9_aop_reguseposter_ii_1-781_smaller.pdf [accessed 22 September 2018].
- Wittwehr C, Aladjov H, Ankley G, Byrne HJ, de Knecht J, Heinzle E, et al. 2017. How adverse outcome pathways can aid the development and use of computational prediction models for regulatory toxicology. Toxicol Sci 155(2):326–336, PMID: 27994170, 10.1093/toxsci/kfw207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaunbrecher V, Beryt E, Parodi D, Telesca D, Doherty J, Malloy T, et al. 2017. Has toxicity testing moved into the 21st century? A survey and analysis of perceptions in the field of toxicology. Environ Health Perspect 125(8):087024, PMID: 28934728, 10.1289/EHP1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger E. 1999. Federal interagency activities toward validation and regulatory acceptance of alternative tests. In: Toxicity Assessment Alternatives: Methods, Issues, Opportunities. Totowa, NJ: Humana Press, 247–256. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


