Abstract
A dedicated enzyme for the formation of the central C ring in the tetracyclic ergoline of clinically important ergot alkaloids has never been found. Herein, we report a dual role catalase (EasC), unexpectedly using O2 as the oxidant, that catalyzes the oxidative cyclization of the central C ring from a 1,3-diene intermediate. Our study showcases how nature evolves the common catalase for enantioselective C–C bond construction of complex polycyclic scaffolds.
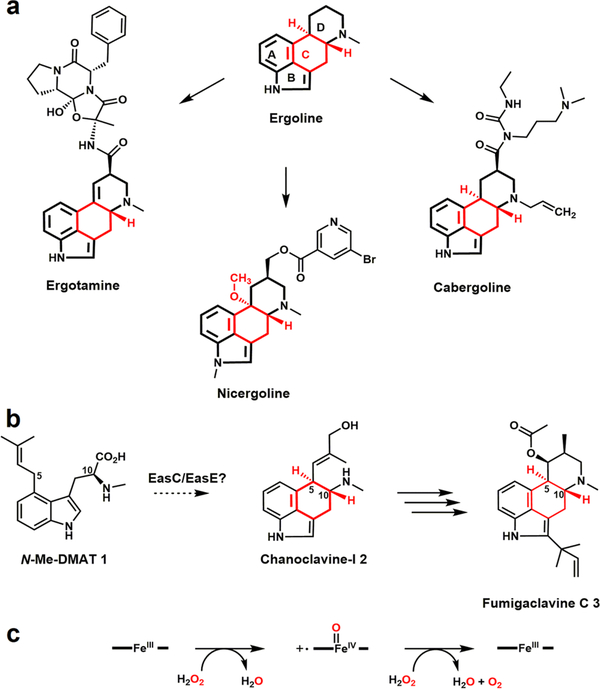
Ergot alkaloids (EAs) are among the most important pharmaceuticals and natural toxins. These bioactive compounds were initially isolated from fungi in the genus Claviceps, a group of parasitic fungi that infected rye and related crop plants.1,2 The grains infected with Claviceps and subsequently contaminated with EAs can cause ergotism (St. Anthony’s fire) in both human and livestock.3 In contrast to their toxicity to human and livestock, both natural and semisynthetic EAs (Figure 1a and Figure S1) are used for the treatment of a wide range of diseases and disorders.1 Until recently, more than a hundred EAs have been identified from different fungal genera including Aspergillus and Penicillium.4,5
Figure 1.
Chemical structures of ergot alkaloids (the central C ring is marked in red) and the catalytic cycle of a typical catalase. (a) The ergoline moiety is present in clinically used ergot alkaloids. (b) EasC and EasE are essential for the formation of the central C ring.1 (c) Catalytic cycle of a typical catalase.
EAs share a common indole-derived tetracyclic ergoline moiety that can modulate the receptors of the central nervous system, due to the structural similarities between the ergoline moiety and neurotransmitters.6 Thus, the tetracyclic-ergoline core structure is essential for the biological activity of EAs (Figure 1a). Many biosynthetic steps of ergoline formation have been established,1,2,7 yet the nature of the central C ring formation, specifically the conversion of N-methyl-dimethylallyltryptophan (N-Me-DMAT; 1) to chanoclavine-I 2 remains an enigma (Figure 1b). It is very intriguing that the regio- and enantioselective cyclization of the central C ring was enabled by creating a new C5–C10 single bond from two unactivated sp3 carbons, despite the presence of functional groups elsewhere in 1 (Figure 1b).
A biosynthetic gene cluster (BGC) of fumigaclavine C 3 has been identified in Aspergillus fumigatus (Figures 1b and S2).8 Previous studies have shown that 1 is produced by two upstream genes dmaW and easF (Figure 2d),8–10 and dmaW, easC, easE, and easF were sufficient for the production of 2.11–13 EasC was demonstrated as a functional catalase14 and proposed to catalyze the decarboxylation of 1 or its oxidative derivatives.15 By knocking out either easE or easC, 1 was found to be the only intermediate accumulated.14,16 However, the direct transformation of 1 to 2 under in vitro conditions remains elusive.7 Herein, we demonstrated that EasC, a catalase that efficiently catalyzes the decomposition of H2O2 to generate O2 in vitro, unexpectedly uses O2 as the oxidant to catalyze the oxidative cyclization of the central C ring via the construction of a new C5–C10 single bond in an enantioselective fashion.
Figure 2.
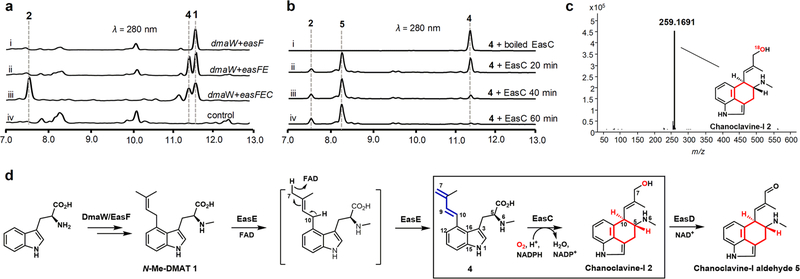
Characterization of the biosynthesis of the central C ring (in red). (a) In vivo reconstitution of dmaW, easC, easE, and easF in A. nidulans. (b) In vitro reconstitution of the activity of EasC. (c) Analysis of 18O-labeled 2 generated from EasC-reaction in 18O2 saturated buffer. LC-HRMS data: 18O-labeled chanoclavine-I 2 [M + H]+ m/z: calculated 259.1696, observed 259.1691. (d) Complete biosynthetic pathway from tryptophan to 5. See SI for reaction conditions.
First, we confirmed the production of 3 in A. fumigatus (CGMGCC 3.772),17 following 3 days of cultivation on PDA medium (Figure S3, trace ii). The production of 3 was abolished (Figure S3, trace vii) when dmaW, the prenyltransferase gene responsible for the first biosynthetic step,8 was knocked out in A. fumigatus (Figure S4). Next, 1 was produced by coexpression of dmaW and easF genes in the heterologous host Aspergillus nidulans (Figure 2a, trace i; Figure 2d). Compound 1 was purified from A. nidulans and fully characterized by MS and NMR (Table S5, Figures S5 and S26–27). Supplementation of 1 to the ΔdmaW mutant of A. fumigatus could restore the production of fumigaclavine C (Figure S3, trace vi), confirming it was an on-pathway intermediate.
Bioinformatic analysis suggested EasE is a FAD-linked oxidoreductase (Figure S6). Enzymes in this family, such as the well-characterized D-amino-acid dehydrogenase,18 catalyze electron transfers using FAD as a cofactor. We hypothesized that EasE was the immediate downstream enzyme after DmaW and EasF. To test this hypothesis, easE was coexpressed with the genes dmaW and easF in A. nidulans. Indeed, LCMS analysis of the crude extract from A. nidulans showed the emergence of a new peak (Figure 2a, trace ii). The new product was purified and characterized as compound 4 with a 1,3-diene moiety by MS and NMR (Table S5 and Figures 2d, S7, and S28–S32). Compound 4 has been previously synthesized by Floss and co-workers and demonstrated to be an intermediate for ergoline biosynthesis.19 We further confirmed 4 is an on-pathway intermediate by feeding it to the ΔdmaW strain to yield 3 (Figure S3, trace v). Compound 4 was potentially generated via base-catalyzed removal of the C10 hydrogen and capture of the C7 hydrogen as a hydride by the FAD cofactor in EasE (Figure 2d).
Whole-cell biotransformation was performed to further confirm the function of EasE. When 1 was fed to Saccharomyces cerevisiae or A. nidulans expressing easE, a 30% and 10% conversion of 1 to 4 was demonstrated, respectively (Figure S8). However, when we attempted to express EasE as a soluble protein in Escherichia coli, S. cerevisiae, or Trichoderma reesei, it was not successful, thereby precluding the direct assay of this reaction using the purified enzyme.20
EasC showed a high sequence identity with small subunit size catalases (SSCs, Figures S9 and S25), with a conserved NADPH-binding site (190H, 197S, 299W, 301L, Figure S10c) and a heme-binding site (71H, 145N, 361Y, Figure S10b). SSCs structurally bind NADPH, which is proposed to act as an electron donor for preventing and reversing the formation of Compound II.21 To evaluate its activity, EasC was purified to homogeneity as N-His6-tagged protein in E. coli BL21 DE3 (Figure S11a). Purified EasC was dark red in color and exhibited the characteristic heme absorbance at 406 nm in UV–vis spectra (Figure S12). HPLC analysis of the supernatant of the denatured protein confirmed the presence of NADPH (Figure S13). EasC was then incubated with H2O2, and the disproportionation of H2O2 by different amount of EasC was monitored by recording the absorbance of H2O2 at 240 nm. The results show that H2O2 was reduced rapidly with EasC-concentration dependencies (Figure S11b), confirming EasC is an active catalase as previously described.14
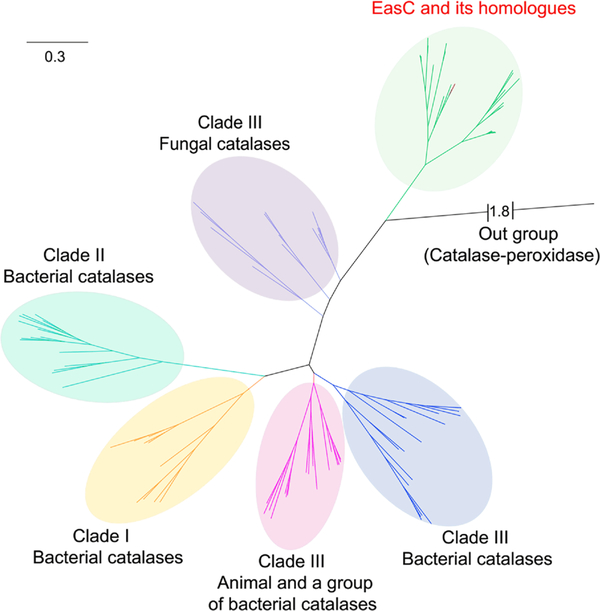
Sequence identity alignment revealed that several widely conserved amino acid residues of classic SSCs were different in EasC and its homologues (Figure S9). In particular, 112T, 114L, and 202M were observed in EasC, instead of the conserved V, G, and P, as in classic SSCs. Phylogenetic analysis of EasC and its homologues also led to a clear classification of them into a separate clade, though it remains closely related to fungal SSCs (clade III) (Figure 3). These indicate the function of EasC could be different from the classic catalases. Indeed, coexpression of easC with the genes dmaW, easF, and easE led to the detection of a new product by LCMS with m/z 257 [M + H]+ as expected for 2 (Figure 2a, trace iii, and Figure S14). Purification of the new product followed by NMR analysis confirmed the structure indeed to be 2 (Table S5, Figures 2d and S33–S38).
Figure 3.
Phylogenetic analysis of EasC and its homologues and previously categorized group I–III catalases.24 See SI for details.
To further study the exact function of EasC, we conducted in vitro reactions. In the enzymatic cycle of a heme catalase, H2O2 acts as the oxidizing substrate to generate the highly reactive heme iron-oxo species Compound I (Figure 1c). In accordance with this, a variety of catalases/peroxidases are known to use H2O2 as the oxidant to catalyze redox reactions, including the well-characterized HppE and SfmD.22,23 Thus, we proposed that EasC also uses H2O2 as the oxidant to generate Compound I to initiate the subsequent oxidative cyclization. To test this, 4 was incubated with 10 μM EasC and 2 mM NADPH, and 1 mM H2O2 in an anaerobic glovebox; however, only 20% of 4 was converted to 2 and an additional new product 5 in 4 h (Figure S15). The low in vitro peroxidase activity of EasC prompted us to consider adding O2 instead of the H2O2 into the reaction mixture. O2, the product of catalases (Figure 1c), has rarely been characterized as the oxidant for catalase/peroxidase in redox reactions.
Indeed, incubation of 4, 10 μM EasC, and 2 mM NADPH in the open air without the addition of H2O2 led to the gradual and full conversion to 2 and the additional new product 5, with a ratio of 1:3, in only 1 h (Figure 2b). In contrast, deoxygenated reaction mixtures completely inhibited the consumption of 4 by EasC (Figure S16, trace iii), indicating the reaction was O2-dependent, and the peroxidase activity of EasC in the anaerobic conditions (Figure S15) might actually have resulted from the decomposition of H2O2 and utilization of the resultant O2. To test of this possibility, EasC reactions with 4 were carried out in the open air with the presence of varying concentrations of H2O2; as shown in Figure S17, increasing the concentration of H2O2 diminished the consumption of 4 and reduced the yields of 2 and 5 with clear H2O2-concentration dependencies. This result suggests H2O2 competes with 4 and O2 for binding EasC, which results in reduced oxygenase activity.
Furthermore, we incubated 4 with EasC in the 18O2 saturated Tris-HCl buffer. The LC-HRMS indicated a 2 unit enhancement in the molecular weight of 2 (Figure 2c). In contrast, no incorporation of 18O into 2 was observed when 4 was incubated with EasC in the buffer made from H218O without 18O2 (Figure S18). When the reaction was performed without the addition of NADPH, an only trace amount of 2 and 5 was observed in a 2 h reaction (Figure S19), which suggests the reaction was also NADPH dependent, and the remaining activity was proposed due to enzyme-bound NADPH in purified EasC (Figures S10 and S13).
Considering the additional new product 5 was not detected from in vivo coexpression conditions (Figure 2a, trace iii), we opted to scale up the in vitro reaction using purified EasC. Surprisingly, 5 was determined to be the known product chanoclavine-I aldehyde 5 by MS and NMR (Table S5, Figures S20 and S39–S44), which was potentially produced via the dehydrogenation of C7 hydroxyl in 2. However, no interconversion between 2 and 5 could be observed under a variety of in vitro conditions when 2 or 5 reacted with EasC separately (Figure S21). This combined with the following results suggest that the radical species could be generated during the reaction and be responsible for the formation of the aldehyde 5: (i) in vivo coexpression only generated 2 without the detection of 5 (Figure 2a, trace iii); (ii) a dedicated NAD+-dependent dehydrogenase, EasD, was already demonstrated to convert 2 to 5 (Figure 2d);25 (iii) the radical species can catalyze the oxidation of organic substrates;26 and (iv) catalase activity is known to generate radical species, including hydroxyl radicals, under various conditions.27 Indeed, the production of 5 could be clearly observed when 2 was incubated with a mixture of H2O2, Fe2+, and ascorbic acid (Figure S22), which could generate hydroxyl radicals through the Fenton reaction.28,29
A radical addition mechanism was proposed for EasC-catalyzed cyclization by transforming 4 to 2 (Figure 4). Compound I abstracts the hydrogen from 5-carboxylic acid to generate the radical species 6. Delocalization of the radical at C5 leads to release of CO2 to yield 7, which can be stabilized via the formation of imine 8 with the adjacent secondary amine. The subsequent radical addition of C5 to C10 yields the radical 9 that can be resonance delocalized at C7 of the terminal alkene, and the final hydroxyl rebound of Compound II yields 2. Density functional theory (DFT) calculation results demonstrate that 6 undergoes barrierless release of CO2 to form the radical 7 following the EasC-catalyzed hydrogen abstraction of 4 (Figure S23). The C ring is formed by the radical addition of C5 to C10 in a low energy transition state (ΔG‡ = 6.1 kcal/mol) to form the final intermediate 9 (Figure S23). Several radial inhibitors,30,31 including DMPO (5,5-dimethyl-1-pyrroline N-oxide), 5-HTP (5-hydroxytryptophan), and L-AA (L-ascorbic acid), were added into the EasC reaction mixture, respectively, which all led to significantly reduced consumption of 4 (Figure S24), supporting the proposed radical mechanism for EasC (Figure 4).
Figure 4.
Proposed catalytic mechanism for EasC (the central C ring is marked in red).
In conclusion, we demonstrated the biosynthetic basis for the formation of the central C ring in the tetracyclic ergoline moiety, a decades-old problem since initial studies began in the 1950s.32 Our research opens the door to utilize this untapped repertoire of eukaryotic catalase-monooxygenase catalysts to generate more polycyclic indole-alkaloids, which have largely been neglected in previous studies.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Key Research and Development Program of China 2018YFA0901600, by the Strategic Priority Research Program, CAS, under grant No. XDA22050401, by the National Science Foundation of China under grant No. 31872614 and 31600270, by the Youth Innovation Promotion Association, CAS, under the grant No. Y92R011CX2, by the National Institutes of General Medical Sciences, NIH, under grant No. GM124480, and by the UCLA Chemistry-Biology Interface training program, NIH, under grant No. T32GM008496.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b10217.
Experimental details; spectroscopic and computational data; Tables S1–S6 and Figures S1–S44 (PDF)
REFERENCES
- (1).Jakubczyk D; Cheng JZ; O’Connor SE Biosynthesis of the ergot alkaloids. Nat. Prod. Rep 2014, 31, 1328. [DOI] [PubMed] [Google Scholar]
- (2).Chen J-J; Han M-Y; Gong T; Yang J-L; Zhu P Recent progress in ergot alkaloid research. RSC Adv 2017, 7, 27384. [Google Scholar]
- (3).N.J.A. de Groot A; van Dongen PWJ; Vree TB; Hekster YA; van Roosmalen J. Ergot alkaloids: Current status and review of clinical pharmacology and therapeutic use compared with other oxytocics in obstetrics and gynaecology. Drugs 1998, 56, 523. [DOI] [PubMed] [Google Scholar]
- (4).Ge HM; Yu ZG; Zhang J; Wu JH; Tan RX Bioactive alkaloids from endophytic Aspergillus fumigatus. J. Nat. Prod 2009, 72, 753. [DOI] [PubMed] [Google Scholar]
- (5).Vinokurova NG; Ozerskaia SM; Baskunov BP; Arinbasarov MU The Penicillium commune Thom and Penicillium clavigerum Demelius fungi–fumigaclavines A and B producers. Mikrobiologiia 2003, 72, 180. [PubMed] [Google Scholar]
- (6).Tudzynski P; Correia T; Keller U Biotechnology and genetics of ergot alkaloids. Appl. Microbiol. Biotechnol 2001, 57, 593. [DOI] [PubMed] [Google Scholar]
- (7).Gerhards N; Neubauer L; Tudzynski P; Li SM Biosynthetic pathways of ergot alkaloids. Toxins 2014, 6, 3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Coyle CM; Panaccione DG An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus. Appl. Environ. Microbiol 2005, 71, 3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Unsöld IA; Li S-M Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology 2005, 151, 1499. [DOI] [PubMed] [Google Scholar]
- (10).Rigbers O; Li SM Ergot alkaloid biosynthesis in Aspergillus fumigatus. Overproduction and biochemical characterization of a 4-dimethylallyltryptophan N-methyltransferase. J. Biol. Chem 2008, 283, 26859. [DOI] [PubMed] [Google Scholar]
- (11).Nielsen CA; Folly C; Hatsch A; Molt A; Schroder H; O’Connor SE; Naesby M The important ergot alkaloid intermediate chanoclavine-I produced in the yeast Saccharomyces cerevisiae by the combined action of EasC and EasE from Aspergillus japonicus. Microb. Cell Fact 2014, 13, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ryan KL; Moore CT; Panaccione DG Partial reconstruction of the ergot alkaloid pathway by heterologous gene expression in Aspergillus nidulans. Toxins 2013, 5, 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jakubczyk D; Caputi L; Hatsch A; Nielsen CA; Diefenbacher M; Klein J; Molt A; Schroder H; Cheng JZ; Naesby M; O’Connor SE Discovery and reconstitution of the cycloclavine biosynthetic pathway-enzymatic formation of a cyclopropyl group. Angew. Chem 2015, 127, 5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Goetz KE; Coyle CM; Cheng JZ; O’Connor SE; Panaccione DG Ergot cluster-encoded catalase is required for synthesis of chanoclavine-I in Aspergillus fumigatus. Curr. Genet 2011, 57, 201. [DOI] [PubMed] [Google Scholar]
- (15).Ryan KL; Akhmedov NG; Panaccione DG Identification and structural elucidation of ergotryptamine, a new ergot alkaloid produced by genetically modified Aspergillus nidulans and natural isolates of Epichloë species. J. Agric. Food Chem 2015, 63, 61. [DOI] [PubMed] [Google Scholar]
- (16).Lorenz N; Olsovska J; Sulc M; Tudzynski P Alkaloid cluster gene ccsA of the ergot fungus Claviceps purpurea encodes chanoclavine I synthase, a flavin adenine dinucleotide-containing oxidoreductase mediating the transformation of N-methyl-dimethylallyltryptophan to chanoclavine I. Appl. Environ. Microbiol 2010, 76, 1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Cole RJ; Kirksey JW; Dorner JW; Wilson DM; Johnson JC; Johnson AN; Bedell DM; Springer JP; Chexal KK Mycotoxins produced by Aspergillus fumigatus species isolated from molded silage. J. Agric. Food Chem 1977, 25, 826. [DOI] [PubMed] [Google Scholar]
- (18).Pollegioni L; Piubelli L; Sacchi S; Pilone MS; Molla G Physiological functions of D-amino acid oxidases: from yeast to humans. Cell. Mol. Life Sci 2007, 64, 1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Kozikowski AP; Chen C; Wu JP; Shibuya M; Kim CG; Floss HG Probing ergot alkaloid biosynthesis: intermediates in the formation of ring C. J. Am. Chem. Soc 1993, 115, 2482. [Google Scholar]
- (20).Walsh CT; Wencewicz TA Flavoenzymes: versatile catalysts in biosynthetic pathways. Nat. Prod. Rep 2013, 30, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kirkman HN; Galiano S; Gaetani GF The function of catalase-bound NADPH. J. Biol. Chem 1987, 262, 660. [PubMed] [Google Scholar]
- (22).Wang C; Chang W-C; Guo Y; Huang H; C Peck S; Pandelia M-E; Lin G-M; Liu H-W; Krebs C; Martin Bollinger J Evidence that the fosfomycin-producing epoxidase, HppE, is a nonheme-iron peroxidase. Science 2013, 342, 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Tang MC; Fu CY; Tang GL Characterization of SfmD as a Heme peroxidase that catalyzes the regioselective hydroxylation of 3-methyltyrosine to 3-hydroxy-5-methyltyrosine in saframycin A biosynthesis. J. Biol. Chem 2012, 287, 5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hansberg W; Salas-Lizana R; Dominguez L Fungal catalases: function, phylogenetic origin and structure. Arch. Biochem. Biophys 2012, 525, 170. [DOI] [PubMed] [Google Scholar]
- (25).Wallwey C; Matuschek M; Li SM Ergot alkaloid biosynthesis in Aspergillus fumigatus: conversion of chanoclavine-I to chanoclavine-I aldehyde catalyzed by a short-chain alcohol dehydrogenase FgaDH. Arch. Microbiol 2010, 192, 127. [DOI] [PubMed] [Google Scholar]
- (26).Huang X; Groves JT Oxygen activation and radical transformations in Heme proteins and metalloporphyrins. Chem. Rev 2018, 118, 2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Goyal MM; Basak A Hydroxyl radical generation theory: a possible explanation of unexplained actions of mammalian catalase. Int. J. Biochem. Mol. Biol 2012, 3, 282. [PMC free article] [PubMed] [Google Scholar]
- (28).Wang B; Lu J; Dubey KD; Dong G; Lai W; Shaik S How do enzymes utilize reactive OH radicals? Lessons from nonheme HppE and Fenton systems. J. Am. Chem. Soc 2016, 138, 8489. [DOI] [PubMed] [Google Scholar]
- (29).Schneider JE; Browning MM; Floyd RA Ascorbate/iron mediation of hydroxyl free radical damage to PBR322 plasmid DNA. Free Radical Biol. Med 1988, 5, 287. [DOI] [PubMed] [Google Scholar]
- (30).Keithahn C; Lerchl AJ 5-hydroxytryptophan is a more potent in vitro hydroxyl radical scavenger than melatonin or vitamin C. J. Pineal Res 2005, 38, 62. [DOI] [PubMed] [Google Scholar]
- (31).Bauer NA; Hoque E; Wolf M; Kleigrewe K; Hofmann T Detection of the formyl radical by EPR spin-trapping and mass spectrometry. Free Radical Biol. Med 2018, 116, 129. [DOI] [PubMed] [Google Scholar]
- (32).Mothes VK; Weygand F; Groger D; Grisebach H Untersuchungen zur Biosynthese der Mutterkorn-Alkaloide. Z. Naturforsch., B: J. Chem. Sci 1958, 13, 41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.