Abstract
The regimens containing levofloxacin (LVX) have been recommended as an alternate to standard triple therapy to treat Helicobacter pylori infections and H pylori mixed infection always lead to H pylori chronic infection. Although the molecular mechanism of LVX resistance with gyrA gene mutation has been clearly understood in H pylori, other genes involved in antibiotic resistance remain unclear. Efflux pump plays an important role in clinically relevant multidrug resistance. Furthermore, the relationship between the strains with different LVX level-resistances from individuals is also unknown.
Helicobacter pylori monoclonal strains were isolated from patients with eradication failure. E test was used to detect the minimal inhibitory concentration of LVX. One lower-level LVX-resistant clone and 2 higher-level LVX-resistant clones from the same patient were selected to sequence the complete genomes. Single-nucleotide variants (SNVs) and mutations were extracted and analyzed from gryA and resistance-nodulation-division family efflux genes.
Two clones with higher-level resistance had the mutation pattern of Asn87Lys and one lower-level LVX-resistant clone had an Asp91Asn mutation. Compared to clones with higher-level resistance, the higher genetic variations were found in genes belonging to the resistance-nodulation-division family in H pylori strains with lower-level resistance to LVX. There were significantly more SNVs of Hp0970 (hefE) and Hp1329 (hefI) in the lower-level LVX-resistant clone than those in the higher-level LVX-resistant clones (P = .044).
The mutation pattern of the Asn87Lys of the gyrA gene confers a higher resistance to LVX than that of the Asp91Asn in H pylori. Increase in the number of SNVs of the Hp0970 (hefE) and Hp1329 (hefI) genes change the resistance to LVX. Twelve mutations verified by Sanger sequencing in Hp0970 (hefE) and Hp1329 (hefI) may decrease resistant levels to LVX.
Keywords: Helicobacter pylori, higher-/lower-level resistance, levofloxacin, resistance-nodulation-division family, single-nucleotide variants
1. Introduction
Helicobacter pylori, colonized in human gastric mucosa, infects more than 50% of the world's population and is confirmed to cause many serious diseases such as chronic gastritis, peptic ulcer diseases, mucosa-associated lymphoid tissue lymphoma, and gastric cancer.[1,2] Many meta-analyses show that H pylori eradication can not only improve the cure rates of peptic ulcers but also prevents its recurrence and reduces the incidence of gastric cancer.[3,4] The standard triple therapy using proton pump inhibitors, clarithromycin (CLR), and amoxicillin or metronidazole is recommended as the 1st line of therapy for H pylori treatment.[5,6] Nevertheless, with the increasing prevalence of resistance to CLR and metronidazole, the eradication rate has dropped below 70% in numerous countries.[7–9] In recent years, triple or quadruple therapy with levofloxacin (LVX) confers obvious advantages for clinical treatment of H pylori infections.[10,11] However, the prevalence of LVX resistance in H pylori has increased because of the abuse of fluoroquinolone.
The majority of H pylori LVX-resistant clones have been identified to be linked to mutations in the fluoroquinolone resistance-determining region within the gyrA gene: Asn87 and Asp91.[12] Simultaneously, some other mutations that have also been involved in LVX resistance include mutations at positions Ala88, Ala97, and Met191 of gyrA and Phe438, Glu463, Asp481, and Arg484 of gyrB.[13–15] In addition, other mechanisms, such as the presence of efflux pumps, have been reported to explain the fluoroquinolone resistance in many gram-negative bacteria.[16,17] Resistance to fluoroquinolones in gram-negative bacteria is acquired by active export of the agents via antibiotic efflux pumps. Using Pseudomonas aeruginosa, Oh et al found that an efflux pump is involved in fluoroquinolones resistance.[18] Genome analysis suggests that Campylobacter jejuni contains at least 9 other putative efflux pumps, which influence antimicrobial resistance.[17]
Additionally, the resistance-nodulation-division (RND) family, which is one of the efflux pumps, plays an important role in clinically relevant multidrug resistance (MDR) in H pylori.[19] RND efflux pumps, including the AcrAB-TolC system, and 4 gene clusters, consisting of Hp0605-0607 (hefABC), Hp0969-0971 (hefDEF), Hp1327-1329 (hefGHI), and Hp1487-1489[20,21] have been considered as RND family candidates in H pylori. Although, the molecular mechanism of efflux pumps leading to H pylori antibiotic resistance has been studied, such as CLR,[22] studies on LVX resistance have not been previously reported.
In recent years, many studies have found that H pylori-mixed infections and heteroresistance is one of the reasons for refractory H pylori infection. Although H pylori clones from individual patients always have an antibiotic-susceptible or resistant phenotype, both of them could exist in a single population of H pylori in each patient.[23] When the colonization of H pylori in the stomach is identified, susceptible or resistant clones could simultaneously be present in the same region or in different regions of the stomach.[23] As early as 2010, mixed-infection of antibiotic susceptible and resistant H pylori clones were found in a single patient, which can lead to 34% with resistant strains being misclassified as susceptible if a biopsy of the antrum alone was used for antimicrobial susceptibility testing.[24] To date, there are few studies on the drug-resistance mechanism of H pylori-mixed infections, especially the antibiotic resistance genes of different drug resistant strains in the same patient.
In this study, we applied Sanger sequencing to detect the genotype of gyrA and NGS to analyze the genomic variations in clinically isolated H pylori clones. The LVX susceptibility test was performed via the E test. Three H pylori monoclonal strains in the same patient meeting our requirements were applied to whole-genome sequencing (WGS), including one lower-level LVX-resistant clone and 2 higher-level LVX-resistant clones.
2. Methods
2.1. Helicobacter pylori clones and growth conditions
Gastric mucosal tissues were collected from the lesser curvature of the antrum, the greater curvature of the antrum and the gastric corpus from patients with eradication failure due to H pylori at Taizhou Hospital (Taizhou, Zhejiang Province). The following principles must be followed as a patient selection: Patients failed 2nd time or more times after a standardized treatment, such as standard quadruple therapy; Patients have completed the entire course of treatment each time and the treatment time is 10 to 14 days per treatment.[25] In this study, a total of 31 patients participated in the 1st phase of the study. The collection and genomic analysis of the DNA were approved by the ethics committee of Taizhou Hospital, and written informed consent, in accordance with the declaration of Helsinki, was obtained. Isolation and culture of H pylori were performed at the laboratory of the Hangzhou Zhiyuan Medical Inspection Institute (Hangzhou, Zhejiang Province). Briefly, grinded gastric mucosa tissue was inoculated onto Columbia agar plates supplemented with 5% fresh defibrinated sheep blood. The plates were cultured at 37°C under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) for 3 to 7 days. Suspicious colonies were identified by gram staining and were confirmed to be positive by their urease, oxidase, and catalase traits. To ensure consistency of drug susceptibility results, we performed a monoclonal separation.
2.2. Monoclonal culture preservation
Multiple scattered monoclonal colonies were picked on the culture-positive plate and then inoculated on fresh blood-agar medium in sections. The medium was cultured in a microaerobic environment (5% O2, 10% CO2, and 85% N2) for 2 days. Different monoclonal moss was again transferred and inoculated on different fresh blood-agar medium. The labeled strains were expanded and cultured according to the growth of the strain until the growth of the strain reached a certain level. The strains were collected and stored in a storage tube containing brain heart infusion for future study.
2.3. Antibiotic susceptibility testing
The minimal inhibitory concentration (MIC) of LVX for H pylori was determined using the E test in this study. Briefly, the concentration of H pylori was equivalent to a 2.0 McFarland standard (8.8 × 107) by adjusting the concentration of bacteria solution with physiologic saline. The suspensions (200 μL) were spread on Columbia agar plates supplemented with 5% sheep blood. The plate was placed facing up in a microaerobic environment (5% O2, 10% CO2, and 85% N2) for 15 minutes, making sure the bacterial solution penetrates into the medium. Subsequently, LVX E test strips were attached on the plates, with the largest scale facing the edge of the flat plate and the test strip should be soaked with bacterial solution. The medium was cultured at 37°C for 3 to 5 days under microaerophilic conditions. The MIC of LVX for H pylori was determined according to the drug concentration value at the intersection of the inhibition zone and the E test strip. The E test strip has a series of 2-fold dilution gradients, with a concentration range of 0.016 to 32 μg/mL. Clones were considered resistant when the MIC value was more than 2 μg/mL.[26] To understand H pylori mixed infection, we selected a monoclonal with the different susceptibility results for the next experiment. In this study, strains with different resistance levels were found in the same patient. So, the patient with one lower-level LVX-resistant clone and 2 higher-level LVX-resistant clones was the subject of a 2nd phase of the study.
2.4. DNA extraction and Sanger sequencing of gyrA
We used the Invitrogen Purelink Genomic DNA mini kit (Life Technologies, Carlsbad, CA) to extract H pylori genomic DNA following the manufacturer's instructions. The concentration of each extract was determined by the Qubit dsDNA HS assay kit (Life Technologies). According to the reference sequence, ATCC26695 (GenBank accession no: NC_000915), the HP-gyrA forward primer (5′-ACAGCGATGTGGTCTCAGC-3′) and the HP-gyrA reverse primer (5′-CCACCAAGCATTGTCCTGC-3′) were employed to detect gyrA gene mutations at the 87 and 91 positions. Each 25 μL of reaction contained 5 μL of 5× PrimeSTAR buffer (Mg2+ plus), 2 μL of dNTP mixture (2.5 mM each), 0.5 μL each of 10 μM primer, 2 μL of genomic DNA, 0.25 μL of PrimeSTAR HS DNA polymerase (2.5 IU/μL) (Takara Bio Inc, Otsu, Japan), and 13.75 μL of sterilized water. The polymerase chain reaction (PCR) conditions were performed using 30 cycles of denaturing at 98°C for 10 seconds, annealing at 58°C for 15 seconds, and extending at 72°C for 40 seconds. The amplified PCR products were analyzed by 1.5% agarose gel electrophoresis to verify the size of 502 bp and subsequently sequenced with a Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) using an ABI 3730XL DNA analyzer. The sequences of the gyrA gene were then aligned with the reference sequence of H pylori 26695 deposited in GenBank using MEGA 6.0 software.
2.5. WGS and analysis of the complete genome
Genomic DNA libraries were prepared and WGS was performed using Illumina HiSeq 2500 sequencing (Illumina, San Diego, CA), as described by Ni et al.[27] The remaining clean reads were mapped against the reference genome with GenBank accession NC_000915 using BWA (version 0.7.12) after we trimmed the low-quality reads using Trimmomatic (version 0.30). Single-nucleotide variants (SNVs) and insertions and deletions (InDels) were named using the Genome Analysis Toolkit. A phylogenetic tree was constructed based on the identification results of the SNV to intuitionally observe the phylogenetic relationships and relative genetic distances.
2.6. Variations in efflux pump genes
To validate the role of multidrug efflux transporter genes that contribute to antibiotic resistance and to assess their association with novel mechanisms of LVX resistance, variations of the RND family were analyzed, including Hp0605-0607, Hp0969-0971, Hp1327-1329, and Hp1487-1489.
2.7. Verification experiment
We designed primers based on SNVs in Hp0970 (hefE) and Hp1329 (hefI) (Table 1). The lower-level LVX resistant and higher-level LVX resistant clones were prepared as a template for PCR amplification. To validate the SNVs, Sanger sequencing was performed with an ABI 3730XL DNA Analyzer (Applied Biosystems) using BigDye Terminator V3.1 according to the manufacturer's instructions.
Table 1.
The primers in Hp0970 and Hp1329.

2.8. Statistical analysis
All data analyses were performed using the SPSS statistical software package, version 19.0 for Windows (SPSS Inc, Chicago, IL). The relationships between variations of the MDR genes and higher/lower-resistance to LVX were determined by Chi-squared tests. Statistical significance was considered for P values of ≤.05.
3. Results
3.1. Genotype of gyrA and phenotype of H pylori that are resistant to LVX
To detect whether the patient was infected with mixed H pylori clones in this study, monoclonal strains from samples of the lesser curvature of the gastric antrum, the greater curvature of the gastric antrum, and gastric body were isolated and cultured. An LVX susceptibility test of the 3 clinically isolated monoclonal strains from the same patient indicated that 1 clone was lower-level LVX resistant and 2 clones were higher-level LVX resistant (Table 2). The gastric mucosal tissues of the same individual could isolate H pylori clones with different traits, such as negative or positive clones and resistant or sensitive clones.[28] The genotype of gyrA was verified by Sanger sequencing. All of the 3 clones had mutations in the gyrA gene. Two of them had a mutation of Asn87Lys, and 1 clone had an Asp91Asn mutation (Table 2). Both mutation sites had been reported previously.[14] The mutation pattern of Asn87Lys caused a higher resistance to LVX than that of Asp91Asn in H pylori.
Table 2.
Minimal inhibitory concentration and Sanger sequencing results.

3.2. Overview of WGS of the clinical H pylori clones
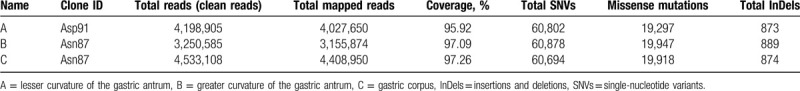
In this study, 3 H pylori genomes were successfully assembled by a Hiseq sequencer. After trimming the low-quality reads, clean reads ranged from 3.2 to 4.5 million (Table 3). Coverage depths were over 95% mapped to the reference genome. Therefore, the efficient reads were sufficient for subsequent analysis of the SNVs. The number of SNVs, InDels, and missense mutations is summarized in Table 2.
Table 3.
Overview of whole genome sequencing for the 3 samples.
To understand the phylogenetic relationships of the 3 clones from different regions of the patient, we constructed a phylogenetic tree. As a result, 2 clones with the Asn87Lys mutation were in the same branch, and the Asp91Asn mutation was in another, indicating that the patient was infected with 2 different H pylori clones.
3.3. Identification of mutations in gyrA and gyrB genes
To analyze the relationship between LVX of lower-level-resistant and higher-level-resistant clones of H pylori with different mutations of the gyrA gene from the patient, mutations in the gyrA and gyrB genes were investigated. Consistent with Sanger sequencing, WGS showed that 2 of the clones had a mutation of Asn87Lys and 1 clone had a mutation of Asp91Asn. We did not find any mutations associated with the known LVX-resistant sites in the gyrA gene, except Asn87Lys or Asp91Asn, and the gyrB gene. However, lower-level-resistant (Asp91Asn) clones have fewer missense mutations than those of higher-level (Asn87Lys)-resistant clones in both gyrA and gyrB genes, and the mutations of 2 higher-level LVX-resistant clones were exactly the same both in loci and numbers.
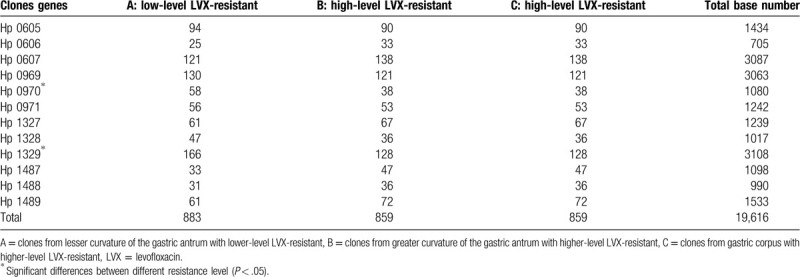
3.4. Identification of RND family efflux pump genes
To identify the mutations of the multidrug efflux transporter genes, a study of the RND family efflux pump was focused on H pylori. Prior to identification of the gene mutation in the RND family efflux pump genes, all mutations of these genes were presented after InDels mutations were filtered in the CDS region (Table 4).
Table 4.
Single-nucleotide variants of the resistance-nodulation-division family efflux pump transporter genes.
Regardless of whether the clone is higher-level LVX-resistant or lower-level LVX-resistant, SNVs were detected in all 4 clusters of the RND family, and there were numerically more SNVs in the lower-level LVX-resistant clone than those in the higher-level LVX-resistant clones, without significantly difference (P = .202). There were significantly more SNVs of Hp0970 (hefE) and Hp1329 (hefI) in the lower-level LVX-resistant clone than those in the higher-level LVX-resistant clones (P = .044). These results suggest that the SNVs of Hp0970 (hefE) and Hp1329 (hefI) may change resistant levels to LVX.
3.5. Verification of SNVs in Hp0970 (hefE) and Hp1329 (hefI)
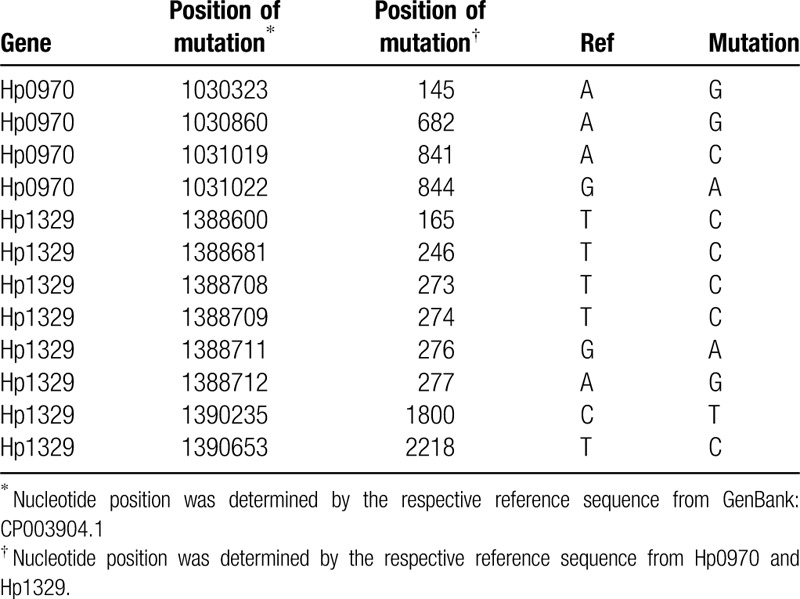
To determine whether the predicted SNVs was mutated in clinical samples, we used Sanger sequencing to verify the lower-level and higher-level LVX-resistant clone in Hp0970 and Hp1329. According to mutation type (homozygous), base depth and mutation frequency (=1), we finally selected 9 SNVs of Hp0970 and 13 SNVs of Hp1329, which were present in lower-level LVX-resistant clone, but not in higher-level LVX-resistant clone. Through PCR identification and Sanger sequencing, we identified a total of 12 mutations, including 4 in Hp1970 (Fig. 1) and 8 in Hp1329 (Fig. 2). The positions of mutation are showed in Table 5.
Figure 1.
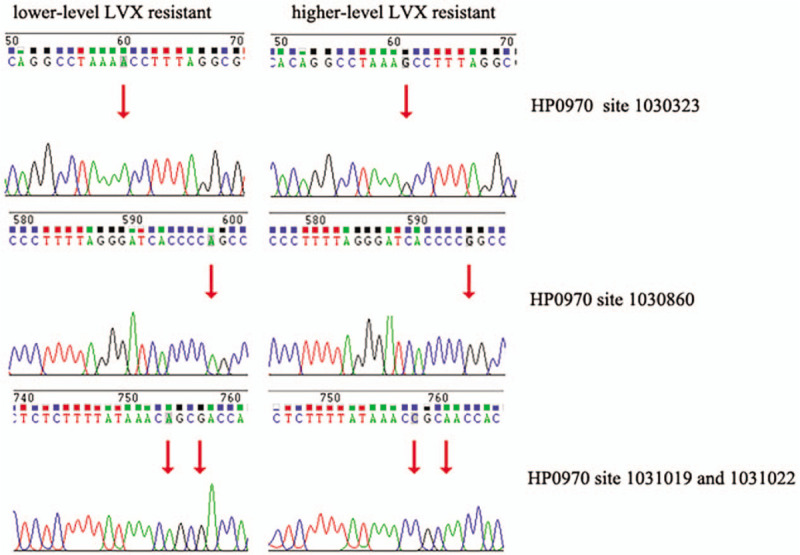
The results of Sanger sequencing for Hp0970 in lower-level and higher-level levofloxacin (LVX)-resistant clones. The mutation site is marked by an arrow.
Figure 2.
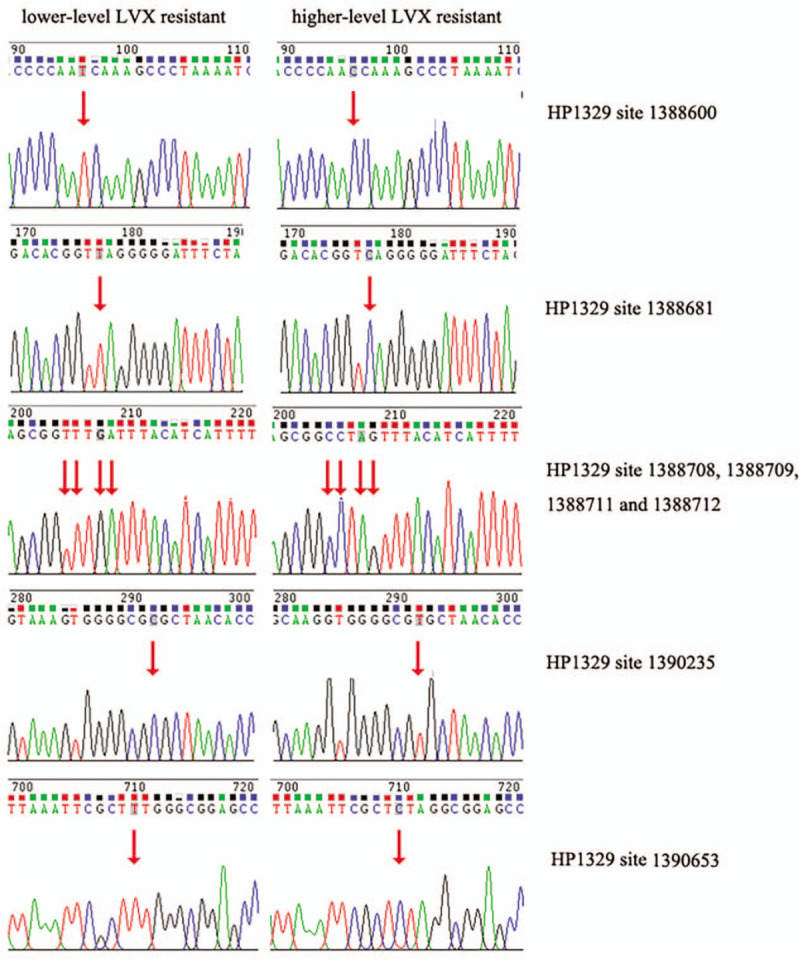
The results of Sanger sequencing for Hp1329 in lower-level and higher-level levofloxacin (LVX)-resistant clones. The mutation site is marked by an arrow.
Table 5.
The special variations were identified in Hp0970 and Hp1329.
4. Discussion
In recent years, due to a pre-existing antibiotic resistant H pylori clone or the generation of a new resistant clone derived from an original susceptible clone, treatment failure has been prevalent.[29] It seems to be difficult to eradicate H pylori infections in patients using failed eradication therapies.[30] The patient who was studied in this research failed 2nd eradication therapies, and 2 different H pylori clones of both the phenotypes and genotypes were detected in different gastric regions. As a result, antibiotic susceptibility tests should be based on multiple sampling for patients infected with H pylori who have undergone failed eradication therapies.
Regimens containing fluoroquinolone have been proposed as an alternate to the classic standard triple therapy for the treatment of H pylori infections. Although the mechanism of LVX resistance of positions 87 and 91 mutations of gyrA has been reported,[12] it is difficult to explain the different levels of the bacterial antibiotic resistance. A study in Latin America showed that the occurrence of the Asn87Ile mutation was associated with a high resistance (MIC ≥ 32 μg/mL) to LVX.[31] In agreement with a previous study,[14] we found that the Asn87Lys mutation of the gyrA gene confers a higher resistance to LVX than that of the Asp91Asn mutation. We only investigated one type of mutation at each position, and double mutation sites, such as Asn87Lys with Asp91Asn, showed a high-level resistance to LVX in Chang's study.[32]
The RND family is grouped into MDR efflux pumps in gram-negative bacteria. Many studies have been reported on the association of RND family efflux pumps with bacterial antibiotic resistance in some gram-negative bacteria, such as Bacteroides fragilis, Escherichia coli, and Burkholderia cepacia.[33–35] In previous surveys, the role of RND efflux pumps in metronidazole and CLR resistance of H pylori has been studied.[20,36] In LVX resistance, the studies only concentrated on C jejuni, Campylobacter coli, P aeruginosa, and Streptococcus pneumoniae strains.[18,17,37] The influence of these pumps on LVX resistance to H pylori should be established. Efflux pumps have been confirmed to increase the resistance levels synergistically with other resistance mechanisms. In the current study, the SNVs of 4 RND efflux pump genes in H pylori were measured by WGS at different level resistances of LVX. The SNVs were detected in all 4 clusters of the RND family genes. In 2005, Ge et al,[17] found that inactivation of efflux pumps gene could decrease the MIC and increate the susceptibilities to fluoroquinolone. In E coli isolates, the inhibition of the efflux pump gene expression decreased the MIC of fluoroquinolone.[38] Furthermore, in this study, there were significantly more SNVs of Hp0970 (hefE) and Hp1329 (hefI) in the lower-level LVX-resistant clone than those in the higher-level LVX-resistant clones. In other words, a large number of SNVs may increase the risk of gene inactivation or activation. At the same time, the increased risk of gene inactivation increases the probability of MIC reduction. Through PCR identification and Sanger sequencing, we identified 4 mutations in Hp1970 and 8 mutations in Hp1329, which were present in lower-level LVX-resistant clone, but not in higher-level LVX-resistant clone. Therefore, we speculated that the 12 mutations of Hp0970 (hefE) and Hp1329 (hefI) may decrease resistant levels to LVX.
Our research also has several limitations. Firstly, the clones we isolated in this study were all monoclonal strains, and the susceptibility results of each monoclonal in the same site are consistent, which led to a small sample size for this study. There were only 3 H pylori clones, including 1 lower-level LVX-resistant clone and 2 higher-level LVX-resistant clones in 1 patient in our study. Secondly, we only study from genomics perspective to speculate the role of efflux pump genes, especially in a different number of SNVs in RND efflux pump genes between lower-level and higher-level resistant strains. Despite certain data support for other strains, such as E coli, C jejuni, and C coli, but lacks clear biologic validation. Future studies are needed to collect more H pylori monoclonal with same drug susceptibility. We believed that these genes may be responsible for regulating the susceptibility of H pylori to fluoroquinolone.
5. Conclusion
In this study, we successfully isolated 3 monoclonal strains with the different susceptibility results (1 lower-level LVX-resistant clone and 2 higher-level LVX-resistant clones) from the same H pylori patient with eradication failure. Compared with Asp91Asn of the gyrA gene, the mutation pattern of the Asn87Lys might make a higher resistance to LVX in H pylori. SNVs from RND family efflux genes change resistance to LVX, especially Hp0970 (hefE) gene and Hp13289 (hefI) gene. Twelve mutations verified by Sanger sequencing in Hp0970 (hefE) and Hp1329 (hefI) may decrease resistant levels to LVX.
Author contributions
Conceptualization: Liping Ye.
Data curation: Xinli Mao, Yu Zhang, Jun Wang, Wei Zhu.
Formal analysis: Fei Meng, Yunhui Liu.
Funding acquisition: Liping Ye.
Investigation: Binbin Gu, Qin Huang.
Methodology: Binbin Gu.
Resources: Xinli Mao, Yu Zhang, Jun Wang, Wei Zhu, Qin Huang.
Software: Fei Meng.
Validation: Liping Ye, Fei Meng, Xinli Mao.
Visualization: Yu Zhang.
Writing – original draft: Yunhui Liu.
Writing – review & editing: Liping Ye.
Footnotes
Abbreviations: CLR = clarithromycin, hefABC = Hp0605-0607 gene clusters, hefDEF = Hp0969-0971 gene clusters, hefGHI = Hp1327-1329 gene clusters, InDels = insertions and deletions, LVX = levofloxacin, MDR = multidrug resistance, MIC = minimal inhibitory concentration, NGS = next generation sequencing, RND family = resistance-nodulation-division family, SNVs = single-nucleotide variants, WGS = whole-genome sequencing.
How to cite this article: Ye L, Meng F, Mao X, Zhang Y, Wang J, Liu Y, Zhu W, Gu B, Huang Q. Using next-generation sequencing to analyze Helicobacter pylori clones with different levofloxacin resistances from a patient with eradication failure. Medicine. 2020;99:32(e20761).
Our research has been reviewed by the Medical Ethics Committee of Taizhou Hospital of Zhejiang Province. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients for being included in the study.
This study was supported by Public Technology Application Research of Zhejiang Province Science and Technology Hall (2016C33232).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Venerito M, Goni E, Malfertheiner P. Helicobacter pylori screening: options and challenges. Expert Rev Gastroenterol Hepatol 2016;10:497–503. [DOI] [PubMed] [Google Scholar]
- [2].Dore MP, Lu H, Graham DY. Role of bismuth in improving Helicobacter pylori eradication with triple therapy. Gut 2016;65:870–8. [DOI] [PubMed] [Google Scholar]
- [3].Zullo A, Gatta L, De Francesco V, et al. High rate of Helicobacter pylori eradication with sequential therapy in elderly patients with peptic ulcer: a prospective controlled study. Aliment Pharmacol Ther 2005;21:1419–24. [DOI] [PubMed] [Google Scholar]
- [4].Misra V, Pandey R, Misra SP, et al. Helicobacter pylori and gastric cancer: Indian enigma. World J Gastroenterol 2014;20:1503–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kim N, Kim JJ, Choe YH, et al. Diagnosis and treatment guidelines for Helicobacter pylori infection in Korea. Korean J Gastroenterol 2009;54:269–78. [DOI] [PubMed] [Google Scholar]
- [6].Malfertheiner P, Megraud F, O’Morain C, et al. Current concepts in the management of Helicobacter pylori infection; the Maastricht III Consensus Report. Gut 2007;56:772–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fakheri H, Saberi FM, Bari Z. Eradication of Helicobacter pylori in Iran: a review. Middle East J Dig Dis 2018;10:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Vakil N, Megraud F. Eradication therapy for Helicobacter pylori. Rev Basic Clin Gastroenterol 2007;133:985–1001. [DOI] [PubMed] [Google Scholar]
- [9].Miftahussurur M, Yamaoka Y. Appropriate first-line regimens to combat Helicobacter pylori antibiotic resistance: an Asian perspective. Molecules 2015;20:6068–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cheng H, Hu FL, Zhang GX, et al. Levofloxacin-based triple therapy for first-line Helicobacter pylori eradication treatment: a multi-central, randomized, controlled clinical study [in Chinese]. Zhonghua Yi Xue Za Zhi 2010;90:79–82. [PubMed] [Google Scholar]
- [11].Tursi A, Picchio M, Elisei W. Efficacy and tolerability of a third-line, levofloxacin-based, 10-day sequential therapy in curing resistant Helicobacter pylori infection. J Gastrointestin Liver Dis 2012;21:133–8. [PubMed] [Google Scholar]
- [12].Lee JW, Kim N, Nam RH, et al. Mutations of Helicobacter pylori associated with fluoroquinolone resistance in Korea. Helicobacter 2011;16:301–10. [DOI] [PubMed] [Google Scholar]
- [13].Liu G, Xu X, He L, et al. Primary antibiotic resistance of Helicobacter pylori isolated from Beijing children. Helicobacter 2011;16:356–62. [DOI] [PubMed] [Google Scholar]
- [14].Rimbara E, Noguchi N, Kawai T, et al. Fluoroquinolone resistance in Helicobacter pylori: role of mutations at position 87 and 91 of GyrA on the level of resistance and identification of a resistance conferring mutation in GyrB. Helicobacter 2012;17:36–42. [DOI] [PubMed] [Google Scholar]
- [15].Teh X, Khosravi Y, Lee WC, et al. Functional and molecular surveillance of Helicobacter pylori antibiotic resistance in Kuala Lumpur. PLoS One 2014;9:e101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shen J, Yang J, Gu Q, et al. The role of AcrAB-TolC efflux pump in mediating fluoroquinolone resistance in naturally occurring Salmonella isolates from China. Foodborne Pathog Dis 2017;14:728–34. [DOI] [PubMed] [Google Scholar]
- [17].Ge B, McDermott PF, White DG, et al. Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother 2005;49:3347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Oh H, Stenhoff J, Jalal S, et al. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb Drug Resist 2003;9:323–8. [DOI] [PubMed] [Google Scholar]
- [19].Mehrabadi JF, Sirous M, Daryani NE, et al. Assessing the role of the RND efflux pump in metronidazole resistance of Helicobacter pylori by RT-PCR assay. J Infect Dev Ctries 2011;5:88–93. [DOI] [PubMed] [Google Scholar]
- [20].Iwamoto A, Tanahashi T, Okada R, et al. Whole-genome sequencing of clarithromycin resistant Helicobacter pylori characterizes unidentified variants of multidrug resistant efflux pump genes. Gut Pathog 2014;6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Van Amsterdam K, Bart A, van der Ende A. A Helicobacter pylori TolC efflux pump confers resistance to metronidazole. Antimicrob Agents Chemother 2005;49:1477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hirata K, Suzuki H, Nishizawa T, et al. Contribution of efflux pumps to clarithromycin resistance in Helicobacter pylori. J Gastroenterol Hepatol 2010;25:S75–9. [DOI] [PubMed] [Google Scholar]
- [23].Alebouyeh M, Yadegar A, Farzi N, et al. Impacts of H. pylori mixed-infection and heteroresistance on clinical outcomes. Gastroenterol Hepatol Bed Bench 2015;8: Suppl 1: S1–5. [PMC free article] [PubMed] [Google Scholar]
- [24].Kim JJ, Kim JG, Kwon DH. Mixed-infection of antibiotic susceptible and resistant Helicobacter pylori isolates in a single patient and underestimation of antimicrobial susceptibility testing. Helicobacter 2010;8:202–6. [DOI] [PubMed] [Google Scholar]
- [25].The principle and strategy for the treatment of refractory Helicobacter pylori infection [in Chinese]. Zhonghua Yi Xue Za Zhi 2017;97:721–3. [DOI] [PubMed] [Google Scholar]
- [26].Osato MS, Reddy R, Reddy SG, et al. Comparison of the Etest and the NCCLS-approved agar dilution method to detect metronidazole and clarithromycin resistant Helicobacter pylori. Int J Antimicrob Agents 2001;17:39–44. [DOI] [PubMed] [Google Scholar]
- [27].Ni M, Ding H, Liu S, et al. Application of next generation sequencing for molecular diagnosis in a large family with osteogenesis imperfecta type I. Mol Med Rep 2017;16:6846–9. [DOI] [PubMed] [Google Scholar]
- [28].Secka O, Antonio M, Berg DE, et al. Mixed infection with cagA positive and cagA negative strains of Helicobacter pylori lowers disease burden in the Gambia. PLoS One 2011;6:e27954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].De Francesco V, Zullo A, Ierardi E, et al. Phenotypic and genotypic Helicobacter pylori clarithromycin resistance and therapeutic outcome: benefits and limits. J Antimicrob Chemother 2010;65:327–32. [DOI] [PubMed] [Google Scholar]
- [30].Kobayashi I, Saika T, Muraoka H, et al. Helicobacter pylori isolated from patients who later failed H. pylori eradication triple therapy readily develop resistance to clarithromycin. J Med Microbiol 2006;55(Pt 6):737–40. [DOI] [PubMed] [Google Scholar]
- [31].Lopez-Gasca M, Pena J, Garcia-Amado MA, et al. Point mutations at gyrA and gyrB genes of levofloxacin-resistant Helicobacter pylori isolates in the esophageal mucosa from a Venezuelan population. Am J Trop Med Hyg 2018;98:1051–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chang WL, Kao CY, Wu CT, et al. Gemifloxacin can partially overcome quinolone resistance of H. pylori with gyrA mutation in Taiwan. Helicobacter 2012;17:210–5. [DOI] [PubMed] [Google Scholar]
- [33].Li XZ, Plesiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev 2015;28:337–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Opperman TJ, Kwasny SM, Kim HS, et al. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob Agents Chemother 2014;58:722–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tseng SP, Tsai WC, Liang CY, et al. The contribution of antibiotic resistance mechanisms in clinical Burkholderia cepacia complex isolates: an emphasis on efflux pump activity. PLoS One 2014;9:e104986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hirata K, Suzuki H, Nishizawa T, et al. Contribution of efflux pumps to clarithromycin resistance in Helicobacter pylori. J Gastroenterol Hepatol 2010;25: Suppl 1: S75–9. [DOI] [PubMed] [Google Scholar]
- [37].Broskey J, Coleman K, Gwynn MN, et al. Efflux and target mutations as quinolone resistance mechanisms in clinical isolates of Streptococcus pneumoniae. Antimicrob Chemother 2000;45: Suppl 1: 95–9. [DOI] [PubMed] [Google Scholar]
- [38].Shaheen BW, Boothe DM, Oyarzabal OA, et al. Evaluation of the contribution of gyrA mutation and efflux pumps to fluoroquinolone and multidrug resistance in pathogenic Escherichia coli isolates from dogs and cats. Am J Vet Res 2011;72:25–32. [DOI] [PubMed] [Google Scholar]


