Abstract
Microbial rhodopsin is a large family of membrane proteins having seven transmembrane helices (TM1-7) with an all-trans retinal (ATR) chromophore that is covalently bound to Lys in the TM7. The Trp residue in the middle of TM3, which is homologous to W86 of bacteriorhodopsin (BR), is highly conserved among microbial rhodopsins with various light-driven functions. However, the significance of this Trp for the ion transport function of microbial rhodopsins has long remained unknown. Here, we replaced the W163 (BR W86 counterpart) of a channelrhodopsin (ChR), C1C2/ChRWR, which is a chimera between ChR1 and 2, with a smaller aromatic residue, Phe to verify its role in the ion transport. Under whole-cell patch clamp recordings from the ND7/23 cells that were transfected with the DNA plasmid coding human codon optimized C1C2/ChRWR (hWR) or its W163F mutant (hWR-W163F), the photocurrents were evoked by a pulsatile light at 475 nm. The ion-transporting activity of hWR was strongly altered by the W163F mutation in 3 points: (1) the H+ leak at positive membrane potential (Vm) and its light-adaptation, (2) the attenuation of cation channel activity and (3) the manifestation of outward H+ pump activity. All of these results strongly suggest that W163 has a role in stabilizing the structure involved in the gating-on and -off of the cation channel, the role of “gate keeper”. We can attribute the attenuation of cation channel activity to the incomplete gating-on and the H+ leak to the incomplete gating-off.
Keywords: microbial rhodopsin, retinal-binding pocket, cation channel, H+ channel, H+ pump
Significance
Since the first report of bacteriorhodopsin in 1971, the family of microbial rhodopsins with various light-driven functions has been expanding. The tryptophan homologous to Trp 86 of bacteriorhodopsin is highly conserved among microbial rhodopsins and forms a retinal binding pocket with other amino acids. However, the precise role of this tryptophan in the ion transport function has remained unknown for over 40 years. Nagasaka et al. revealed for the first time that this tryptophan has a role of “gate keeper” of ion transport in a channelrhodopsin. That is, it regulates the structure involved in the gating-on and -off of the cation channel.
Introduction
Microbial rhodopsin is a large family of membrane proteins with various light-driven functions, such as ion pumps, ion channels, signal transducers and enzymes [1–3]. Among them, bacteriorhodopsin (BR) was first found in 1971 from Halobacterium salinarum [4] and has 7 transmembrane helices (TM1-7) with an all-trans retinal (ATR) chromophore that is covalently bound to Lys in the TM7. Upon light absorption, the ATR photo-isomerizes to the 13-cis form with the conformational changes of the protein moiety leading to the activated state [5,6]. Meanwhile, the subsequent retinal re-isomerization returns the molecule to the basal state. During this photocycle, one H+ is transported unidirectionally through the membrane, even against its electrochemical potential gradient. Thus, the light energy is converted to the electrochemical potential gradient of the ion to be utilized by the cell for such purposes as ATP synthesis by the F-type H+-ATPase in the bacterial membrane. Similarly, the Na+ pump rhodopsins such as KR2 from Krokinobacter eikastus transport a Na+ from inside to outside during a photocycle [7], the Cl– pump rhodopsins such as NpHR/phR from Natronomonas pharaonis transport a Cl– from outside to inside [8,9] and the inward H+ pumps such as PoXeR from Parvularcula oceani transport a H+ from outside to inside [10].
On the other hand, in channelrhodopsins, the ATR photoisomerization triggers the opening of a channel permeable to either cations or anions, while the re-isomerization to ATR closes it. Therefore, a certain number of ions are transported across the membrane along the electrochemical potential gradient during the photocycle, which consists of a basal closed state (C), an activated open state (O) and several intermediate states. For example, channelrhodopsin-1 (ChR1) and -2 (ChR2) were the first channelrhodopsins found from Chlamydomonas reinhardtii [11,12] which are permeable to cations such as H+, Na+, K+ and Ca2+, whereas anion channel rhodopsins from Guillardia theta are permeable to anions such as Cl– [13]. The resultant changes in the membrane potential and/or the intracellular ion concentration mediate the light-dependent response of the cell [14,15]. The artificial expression of ChR2 in an animal neuron enabled the light-dependent depolarization of its membrane potential to trigger action potential firings [16,17], being followed by optogenetics to regulate the membrane potentials and other signaling mechanisms of various cells by light using microbial rhodopsins and other light-sensitive proteins [18–20].
Despite the differences in ion transport functions, the channelrhodopsins are structurally similar to the pump rhodopsins with homologous amino acid sequences. Among them two Trp, one in the middle of TM3 (BR W86 counterpart) and another in the middle of TM6 (BR W182 counterpart), are well conserved amino acids among microbial rhodopsins [3,21,22]. In BR, one of them, W182 is suggested to interact with the retinal to mediate coupling between ATR photoisomerization and the H+ transport [23]. This Trp is also suggested to be involved in the cation channel gating of channelrhodopsins because the photocurrent activity of C1C2/ChRWR, which is a chimera between ChR1 and 2, was completely abolished by the mutation of W262 (BR W182 counterpart) by Ala [24]. However, the significance of another Trp in TM3 in the ion transport function of microbial rhodopsins has remained unknown for over 40 years. Crystallographic structural analysis of C1C2/ChRWR revealed that this Trp (W163) is one of the aromatic amino acid residues forming the retinal-binding pocket around the ATR [25] like W86 of BR[26], W127 of NpHR/phR [27], W113 of KR2 [28], W124 of ChR2 [29] and W98 of GtACR1 [30,31]. In the present study, we replaced W163 of C1C2/ChRWR with the smaller aromatic residue, Phe to verify its role in the ion transport. This mutation strongly attenuated the passive transport of cations with the manifestation of outward H+ pump activity. It is suggested that W163 plays a role in the “gate keeper” that regulates the structure involved in channel gating at the optimal position.
Material and methods
Cell culture and molecular biology
C1C2/ChRWR is a chimeric protein that consists of the TM1-5 of ChR1 and the TM6-7 of ChR2. Human codon optimized C1C2/ChRWR (hWR) gene was inserted into pVenus-N1, as described previously [32]. Point mutations were introduced using a KOD-Plus-Mutagenesis Kit (TOYOBO, Osaka, Japan). All constructs were verified by sequencing.
The electrophysiological assays of hWR were carried out using ND7/23 cells, lined hybrid cells derived from neonatal rat dorsal root ganglion neurons fused with the mouse neuroblastoma [33]. The ND7/23 cells were grown in Dulbecco’s modified Eagle’s medium (FUJIFILM Wako Pure Chemical Co., Osaka, Japan) supplemented with 5% fetal bovine serum under a 5% CO2 atmosphere at 37°C. Twenty-four hours before the transfection, the cells were dissociated and re-plated onto collagen-coated coverslips (4912-010, AGC Techono Glass Co., Yoshida-Shizuoka, Japan), and the medium was replaced by Opti-MEM medium (Thermo Fisher Scientific Inc., Waltham, MA) containing 15% KnockOutTM Serum Replacement (Thermo Fisher Scientific Inc.), 50 ng/mL nerve growth factor-7S (Sigma-Aldrich, St. Louis, MO), and 1 mM N6,2'-O-dibutyryladenosine-3',5'-cyclic monophosphate sodium salt (Nacalai tesque, Kyoto, Japan), as previously described [34]. The expression plasmids were transiently transfected in ND7/23 cells using LipofectamineTM 2000 transfection reagent (Thermo Fisher Scientific Inc.) according to the manufacturer’s instructions. Electrophysiological recordings were then conducted at 1–2 weeks after the transfection. The transfected cells were identified by the presence of Venus fluorescence.
Imaging
Venus fluorescence in the ND7/23 cells expressing naïve hWR or hWR-W163F were imaged under a confocal laser scanning microscopy (FV1000, Olympus, Tokyo, Japan). Images were taken at 512×512 pixels (4.0323 pixels/μm) using an Ar laser at 488 nm through a water immersion objective lens (XLUMPLFLN, Olympus, 20×, NA 1.00). All images for figures were processed with ImageJ software (ImageJ 1.52p; NIH, USA).
Electrophysiology
All experiments were carried out at room temperature (20–22°C). Currents were recorded using an EPC-8 amplifier (HEKA Electronic, Lambrecht, Germany) under a whole-cell patch clamp configuration. The data were filtered at 1 kHz, sampled at 50 kHz (Digidata1440 A/D, Molecular Devices Co., Sunnyvale, CA) and stored in a computer (pClamp10.3, Molecular Devices). The pipette resistance was adjusted to 4–8 MΩ (6.5±0.2, n=28) with a series resistance of 6–22 MΩ (13.4±0.8, n=28) and a cell capacitance of 29–100 pF (58±3, n=28) with the standard extracellular/intracellular solution.
The standard internal pipette solutions for the whole-cell voltage clamp recordings from the ND7/23 cells contained (in mM) 121.2 NaOH, 90.9 glutamate, 5 Na2EGTA, 49.2 HEPES, 2.53 MgCl2, 2.5 MgATP, 0.0025 ATR (pH 7.4 adjusted with HCl). The standard extracellular Tyrode’s solution contained (in mM): 138 NaCl, 3 KCl, 2.5 CaCl2, 1 MgCl2, 10 HEPES, 4 NaOH, and 11 glucose (pH 7.4 adjusted with HCl). The liquid junction potential (LJP) was experimentally measured and determined to be 6.1 mV. All voltage readings were corrected for by this value.
The low Na+, low Cl– external solution was made by replacing these ions with N-methyl-d-glucamine (NMG) and HEPES; 73 NMG, 219 HEPES, 2.5 CaCl2, 1 MgCl2 and 11 glucose (mM, pH 7.4 adjusted with NMG). The low Na+, low Cl– pipette solution contained (in mM) 131.2 NMG, 90.9 glutamate, 5 EGTA, 50.1 HEPES, 2.53 MgSO4, 2.5 MgATP, 0.0025 ATR (pH 7.4 adjusted with H2SO4). All voltage readings were corrected for using the LJP, which was directly measured to be –0.3 mV.
Upon turning-off the light pulse, the hWR photocurrent generally returns to the baseline with complex kinetics such as a short lag phase and a small slow phase. However, the transition phase between 90 and 10% of the maximum approximately followed a single exponential function for naïve hWR [35]. Therefore, using software (Clampfit 10.7, Molecular Devices), the turning-off kinetics of each photocurrent was evaluated by fitting to the transition phase between 90 and 10% of the amplitude at the end of light pulse to a single-exponential function. Indeed, no obvious deviation was observed between the raw data and the fitted curve. However, samples were not included in the statistics if their transients deviated from the single exponential function.
Optics
To investigate the photocurrent amplitude and kinetics, two 100 ms pulse illuminations at 475±28 (nm, >90% of the maximum) with a 200 ms dark gap were given at 0.1 Hz using a SpectraX light engine (Lumencor Inc., Beaverton, OR) controlled by computer software (pCLAMP 10.3, Molecular Devices). The power of the light was directly measured under a microscope using a visible light-sensing thermopile (MIR-101Q, SSC Co., Ltd., Kuwana City, Japan) and was 1.21 mW·mm–2. The action spectrum was studied at a holding potential of 80 mV at wavelengths (nm, >90% of the maximum) with equivalent power density (mW·mm–2): 390±18 (0.23), 438±24 (0.25), 475±28 (0.28), 513±17 (0.30), 549±15 (0.32), 575±25 (0.30) and 632±22 (0.30). Each action was estimated by the maximal amplitude of the photocurrent scaled by the light power density under the assumption of a linear relationship. Then, action maxima were estimated by fitting to a Gaussian distribution for the light frequency.
Statistical analysis
Every measured current was expressed as an effective value after being divided by the whole-cell capacitance, which is proportional to the surface area of the cell. The current-voltage (I-V) relationship was experimentally fitted with a cubic polynomial function to estimate the reversal/zero-current potential (V0), the currents at –80 and 80 mV by the extrapolation. We calculated the effective inward conductance (gin) by the slope between V0 –60 and V0 –40 mV, and the effective outward conductance (gout) by the slope between V0 +40 and V0 +60 mV. The rectification index (RI) was defined as RI=gout/gin [35]. That is, the I-V relationship was inwardly rectified when RI<1, but outwardly rectified when RI>1.
All data in the text and figures were expressed as mean±standard error of the mean (SEM) with the number of samples (n) and were assessed for statistical significance using the Wilcoxon signed rank test for paired data or the Mann-Whitney U-test for unpaired data unless otherwise noted. It was judged as statistically insignificant when p>0.05.
Results
Photocurrent of Trp136 mutant
C1C2/ChRWR (hereafter abbreviated as hWR) consists of 1–245 amino acids (TM1-5) from ChR1 and 207–315 amino acids (TM6-7) from ChR2 [32]. The alignment of TM3 showed that W163 of ChR1 (and that of hWR) is well conserved in ChR2 and other microbial rhodopsins of various ion transport activities (Fig. 1A). Both the naïve hWR and its variant (hWR-W163F), in which W163 is replaced by the smaller aromatic amino acid, Phe, were tagged with a green fluorescent protein derivative, Venus, at their C-terminal ends and expressed in ND7/23 cells (Fig. 1B). The distribution of Venus fluorescence in an hWR-W163F-expressing ND7/23 cell was quite similar to that of naïve hWR, in which the fluorescence was preferentially localized to the plasma membrane. In either case, the membrane expression was stable even more than 2 weeks after transfection.
Figure 1.
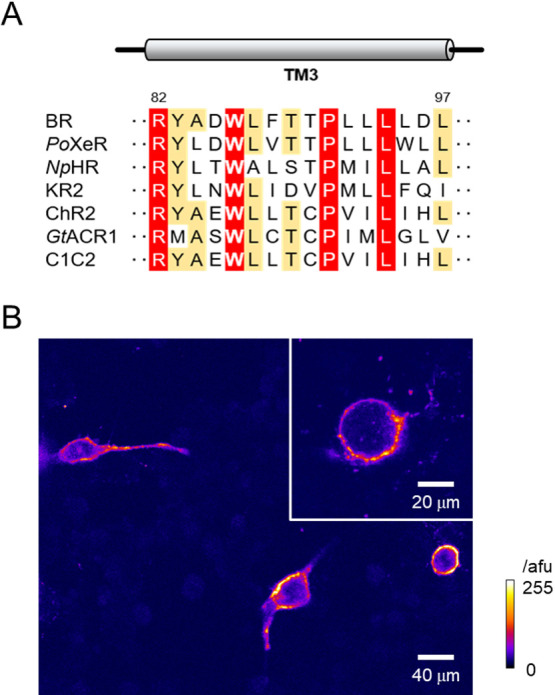
A highly conserved helix-3 tryptophan and its mutation in C1C2/ChRWR. (A) Amino acid sequence alignment of the third transmembrane domain (TM3) from microbial rhodopsins with various ion transport activities. The highly conserved and moderately conserved residues are highlighted in red and orange, respectively. The residue numbers of BR are shown on top of the alignment. (B) A typical confocal image (a single slice) of transfected C1C2/ChRWR (hWR) with W163F mutation (hWR-W163F) in ND7/23 cells (13 days after transfection). Inset, a zoom image of another sample. Note the membrane-delimited expression of Venus fluorescence the intensity of which was scaled by the arbitrary fluorescent unit (afu) and expressed in pseudocolor ratings.
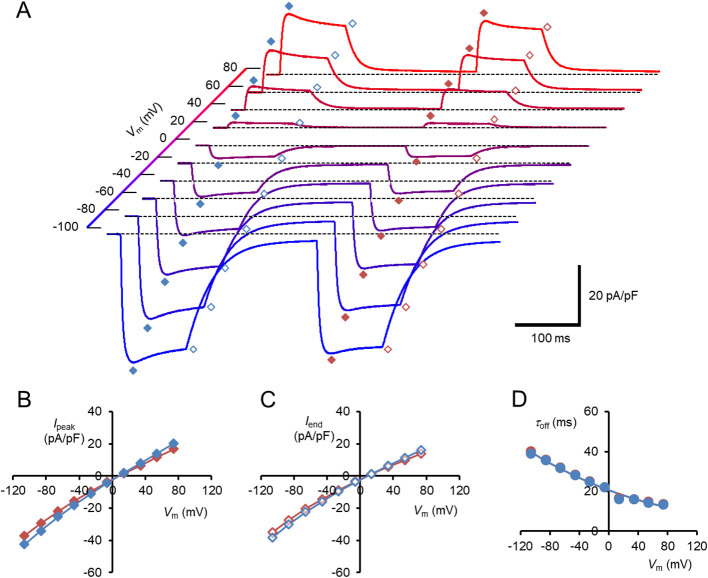
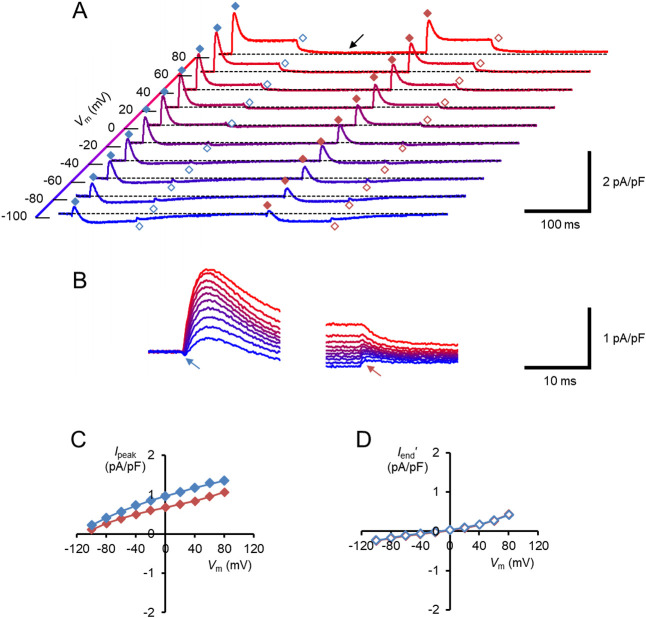
Photocurrents were evoked by two 100-ms light pulses of blue light successively illuminated with a 200 ms dark gap under a whole-cell voltage clamp of an ND7/23 cell expressing either naïve hWR or hWR-W163F. In Figure 2A, each trace shows a series of photocurrents of naïve hWR at each membrane potential (Vm) from –106.1 to 73.9 mV (holding potential, from –100 to 80 mV) at 20 mV step. At every membrane potential, the photocurrent was activated with a time constant of 2–3 ms, peaked at 30–40 ms and partially inactivated with a time constant of about 40 ms, as previously described [32,36]. For the first photocurrent, we measured the peak (Ipeak) and the average currents of the pulse-end 10 ms (Iend) when the inactivation proceeded to over 90% of its maximum, and plotted as a function of Vm (blue symbols, Fig. 2B and 2C). In summary, Ipeak and Iend reversed their signs at 9.5±0.9 mV and 9.8±0.8 mV, respectively (n=12). The turning-off kinetics of each photocurrent was approximated by a single exponential function, the time constant of which is a function of Vm (blue symbols, Fig. 2D). Similar results were obtained from the second photocurrents (red symbols).
Figure 2.
The membrane current response of naïve hWR in a standard extracellular/intracellular milieu. (A) Each sample trace was a current response to the double light pulses at the membrane potential (Vm) indicated on the depth axis. The symbols indicate the Ipeak/Iend of the first responses (blue filled/open diamonds) and the Ipeak/Iend of the second responses (red filled/open diamonds), respectively. (B) Current-voltage (I-V) relations of the peak response (Ipeak); the first responses (blue symbols) and the second responses (red symbols). (C) I-V relations of the response at pulse end (Iend); the first responses (blue symbols) and the second responses (red symbols). (D) The off-current time constant (τoff) as a function of Vm; the first responses (blue symbols) and the second responses (red symbols behind blue ones).
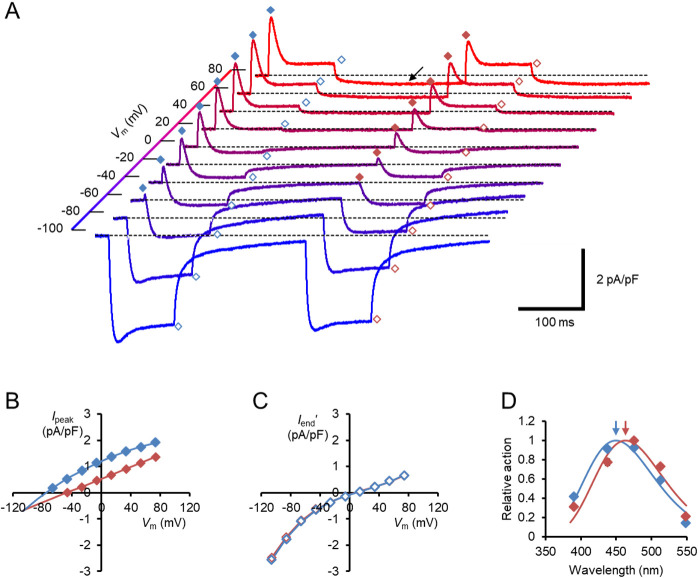
On the other hand, the hWR-W163F photocurrent was quite different. For the first photocurrent, a transient outward current with a peak time of 2–5 ms was observed at every Vm from –66.1 to 73.9 mV, but not at –86.1 and –106.1 mV (Fig. 3A). After exposure to the 100-ms blue light pulse, the holding current became smaller than that before the light-on at positive Vm although not at negative Vm. On the other hand, the second 100-ms blue light evoked a transient outward current and a steady outward/inward current, but with negligible change of the holding current. Therefore, the holding current before light contains the leak current which is sensitive to blue light. The leak current was attenuated maximally during a 100-ms light exposure since the attenuation by the second 100-ms light pulse was negligible. As shown in Figure 3B, the Ipeak was positive at every Vm from –66.1 to 73.9 mV for the first photocurrent and from –46.1 to 73.9 mV for the second one. The photocurrent amplitude at the end of the light pulse (Iend') was measured in reference to the holding current 200 ms after the light pulse and plotted as a function of Vm (Fig. 3C). For the first photocurrents, Iend' reversed its sign at 5.2±1.5 mV (n=11), whereas Ipeak was extrapolated to be 0 at –86.6±2.9 mV (n=11). The turning-off kinetics of each photocurrent was approximated by a bi-exponential function except between Vm=–26.1 and 13.9 mV. As shown in Figure 3D, the wavelength sensitivity was evaluated at 73.9 mV (holding potential, 80 mV) for the Ipeak and compared with that of naïve hWR. The estimated action maxima were 449±1 nm (n=6) for the naïve hWR whereas it was 463±2 nm (n=7) for the hWR-W163F with a significant red shift by 14 nm (p=0.001166, Mann-Whitney U-test).
Figure 3.
The membrane current response of hWR-W163F in a standard extracellular/intracellular milieu. (A) Each sample trace was a current response to the double light pulses at each Vm. The symbols indicate the Ipeak/Iend of the first responses (blue filled/open diamonds) and the Ipeak/Iend of the second responses (red filled/open diamonds), respectively. Note the holding-current shift to the negative direction after the first light pulse (arrow). (B) I-V relations of the Ipeak; the first responses (blue symbols) and the second responses (red symbols). (C) The Iend was corrected for the change in the holding current (Iend') and expressed as a function of Vm; the first responses (blue symbols) and the second responses (red symbols). (D) Comparison of the action spectrum of the average light-pulse response at 73.9 mV with SEM bars; naïve hWR (blue, n=6) vs hWR-W163F (red, n=7). Each arrow indicates the action maxima (nm): 449 (blue) and 463 (red) for naïve hWR vs hWR-W163F, respectively. Each line was drawn according to a Gaussian distribution for the light frequency.
H+ transport activity
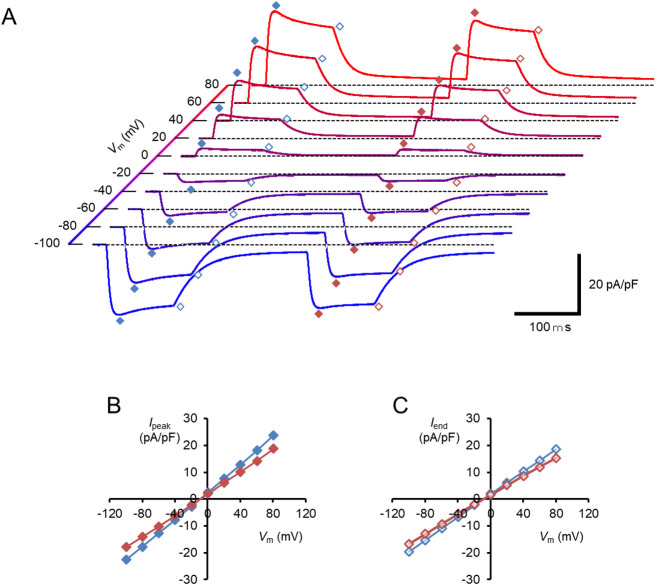
The above difference in the reversal/zero-current potential strongly suggests that the Ipeak of the hWR-W163F is generated by the pump activity, which drives electron inwardly across the membrane irrespective of the electrochemical gradient, whereas Iend' is generated by the channel activity. To verify this, the photocurrents were measured under reducing all ions (Na+ and Cl–) except Ca2+, Mg2+ and H+ by replacing them with N-methyl-d-glucamine ion (NMG+) and HEPES+/–, both of which are large in molecular size and generally unable to penetrate small-sized channels, in the intracellular and extracellular milieu (low Na+/Cl– milieu, pH 7.4). As a control, the photocurrent of naïve hWR was investigated under the identical condition (Fig. 4A). Although a significantly large photocurrent remained at every Vm, it was different from that recorded under the standard inside/outside solutions containing Na+; the negative shift of the I-V relationship and the slight outward rectification or reduced inward rectification for both the Ipeak and Iend (Fig. 4B and 4C). Indeed, the Ipeak and Iend reversed their sign at –8.6±1.2 mV and –8.1±1.3 mV (n=9), respectively with a significant difference from the reversal potentials under the standard inside/outside conditions containing Na+ (p=6.8E–6, Mann-Whitney U-test). For the Ipeak the RI was 1.08±0.03 (>1 in 7 of 9 experiments) under a low Na+/Cl– condition with significant difference from the value under the standard condition (0.70±0.04, <1 in all 12 experiments) (p=6.8E–6, Mann-Whitney U-test). For the Iend the RI was 0.97±0.02 (>1 in 2 of 9 experiments) under low Na+/Cl– condition with significant difference from the value under the standard condition (0.60±0.04, <1 in all 12 experiments) (p=6.8E–6, Mann-Whitney U-test).
Figure 4.
The membrane current response of naïve hWR in a low Na+/Cl– milieu. (A) Each sample trace was the current response to the double light pulses at each Vm. The symbols indicate the Ipeak/Iend of the first responses (blue filled/open diamonds) and the Ipeak/Iend of the second responses (red filled/open diamonds), respectively. (B) I-V relations of the Ipeak; the first responses (blue symbols) and the second responses (red symbols). (C) I-V relations of the Iend; the first responses (blue symbols) and the second responses (red symbols).
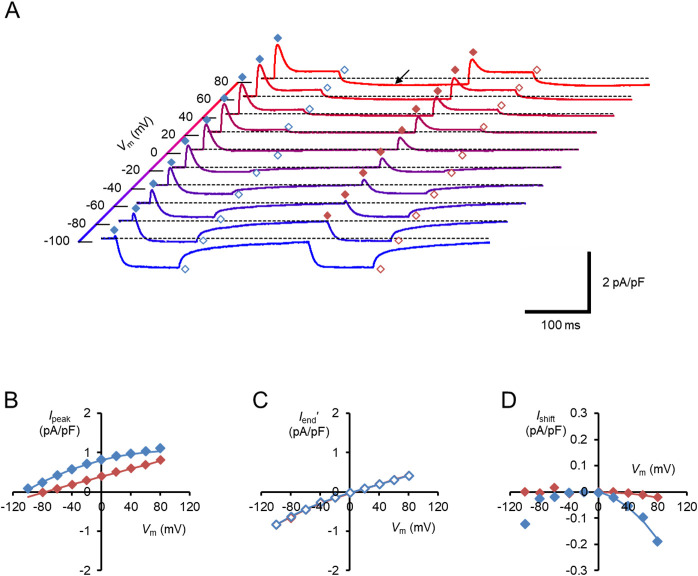
When the hWR-W163F photocurrent was measured under the above low Na+/Cl– milieu, Ipeak was always positive at every Vm from –99.7 to 80.3 mV while Iend' was relatively attenuated (Fig. 5A–C). Indeed, the extrapolated zero-current potential was significantly negative-shifted to –119.9±2.7 mV (n=11) for Ipeak vs. 1.2±2.0 mV (n=11) for Iend' (p=0.0009766, Wilcoxon signed rank test).
Figure 5.
The membrane current response of hWR-W163F in a low Na+/Cl– milieu. (A) Each sample trace was a current response to the double light pulses at each Vm. The symbols indicate the Ipeak/Iend of the first responses (blue filled/open diamonds) and the Ipeak/Iend of the second responses (red filled/open diamonds), respectively. Note the holding-current shift to the negative direction after the first light pulse (arrow). (B) I-V relations of the Ipeak; the first responses (blue symbols) and the second responses (red symbols). (C) I-V relations of the Iend'; the first responses (blue symbols) and the second responses (red symbols). (D) The change in the holding current at 200 ms after light-off (Ishift) as a function of Vm; the first responses (blue symbols) and the second responses (red symbols).
The light-dependent negative shift of the holding current (Ishift) was still observed under the low Na+/Cl– milieu. It was evaluated at 200 ms after light-off and plotted against Vm (Fig. 5D), and was negatively related to Vm at Vm>0, but was negligible at Vm<0. Since H+ is the main permeable ion in this condition, the negative Vm dependence could be attributed to the presence of a steady leak of H+ in the darkness, which is attenuated by blue light. Indeed, as shown in Figure 5A and 5B, the additional Ishift was negligible at any positive Vm by the second light pulse after the 200 ms dark gap. Therefore, this leak attenuation could be attributed to the light-adaptation of the molecule. The light-adapted state slowly recovered to the dark-adapted state since the H+ leak recovered in the darkness with a time constant of 1–2 s. One of the events associated with the light-adaptation of ChR2 is the deprotonation of E90 in the middle of TM2 [37,38], of which counterpart is E129 in the hWR-W163F. To determine whether this residue is involved in the H+ leak, we measured the photocurrents of double mutant hWR-E129A-W163F under the low Na+/Cl– milieu (Fig. 6A). At hyperpolarized Vm, the outward currents stood up with a sag waveform (blue arrow) upon the light-on and the inward currents were accompanied by a hump waveform (red arrow) upon the light-off (Fig. 6B). The hWR-E129A-W163F photocurrent consisted of an early peak and late plateau components similar to those of the hWR-W163F, but with negligible Ishift at any positive Vm. As shown in Figure 6A and 6C, Ipeak was always positive at every Vm between –99.7 and 80.3 mV with an extrapolated zero-current potential of –127.6±11.0 mV (n=9). On the other hand, as shown in Figure 6A and 6D, Iend' reversed its sign at –8.7±2.3 mV (n=9).
Figure 6.
The membrane current response of hWR-E129A-W163F in a low Na+/Cl– milieu. (A) Each sample trace was a current response to the double light pulses at each Vm. The symbols indicate the Ipeak/Iend of the first responses (blue filled/open diamonds) and the Ipeak/Iend of the second responses (red filled/open diamonds), respectively. Note the absence of the holding-current shift (arrow). (B) The same responses were magnified to show the early response to light-on -off (overlay of responses at every Vm). Note the sag waveforn (blue arrow) upon light-on and the hump waveform (red arrow) upon light-off. (C) I-V relations of the Ipeak; the first responses (blue symbols) and the second responses (red symbols). (D) I-V relations of the Iend'; the first responses (blue symbols) and the second responses (red symbols).
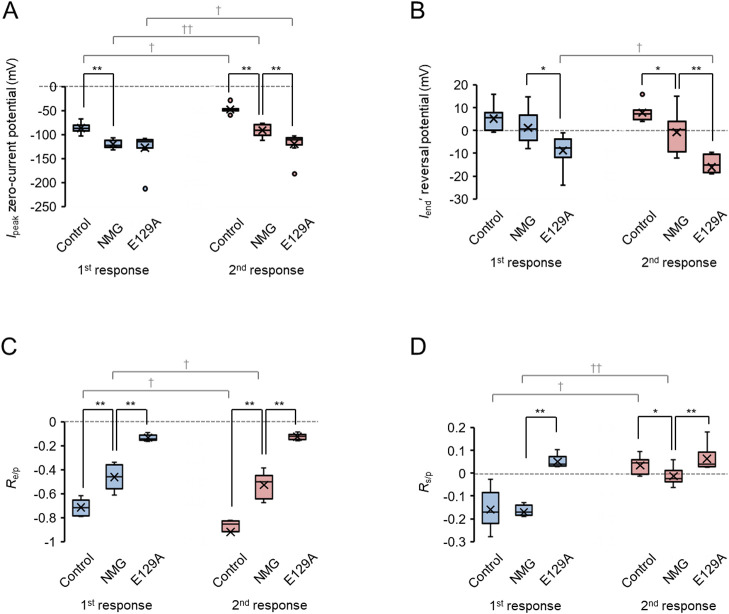
In summary, the extrapolated zero-current potential of Ipeak was significantly shifted to the negative by the Na+/Cl– reduction at the first light responses (Fig. 7A). That at the second light response was usually less negative than the first because of the inactivation of Ipeak. However, a further negative shift was induced by the additional E129A mutation. Although the reversal potential of Iend' was insignificantly changed by the Na+/Cl– reduction at the first response, that at the second response was significantly shifted to the negative. The additional E129A mutation further shifted it to the negative at both the first and second responses (Fig. 7B). Therefore, the Ipeak is assumed to be generated by the outward transport of H+. On the other hand, the Iend' is primarily dependent on the channel activity permeable to cations such as Na+ and H+ as the Cl– equilibrium potential is far more negative from 0 mV. The E129A mutation should thus selectively retard the channel activity. This hypothesis was also tested by comparing the amplitude ratio of Iend'(–80)/Ipeak(80), which is termed Re/p (Fig. 7C). For the first response of hWR-W163F, Re/p was –0.71±0.02 (n=11) under standard internal/external milieu, but was significantly increased by the Na+/Cl– reduction. It was further increased with the additional E129A mutation. Although similar traits were observed for the second response, Re/p became smaller than that of the first response because of the inactivation of the Ipeak. To verify the ionic mechanisms underlying the Ishift, it was measured at 200 ms after light-off and normalized by the Ipeak. The extrapolated value at Vm=80 mV of this ratio was termed as Rs/p and evaluated statistically (Fig. 7D). For the first response of hWR-W163F, Rs/p was –0.16±0.02 (n=11) and –0.17±0.02 (n=9) under standard internal/external milieu and by the Na+/Cl– reduction, respectively. It significantly increased to around 0 or even positive for the second response. The additional E129A mutation almost completely eliminated Ishift since Rs/p was usually positive after either the first or second response.
Figure 7.
Summary of the ion dependency and E129A mutation of hWR-W163F. (A) Comparison of the extrapolated Ipeak zero-current potential among hWR-W163F in a standard milieu (Control), in a low Na+/Cl– milieu (NMG) and that of hWR-E129A-W163F in a low Na+/Cl– milieu (E129A). Box and whisker graphs for the first responses (blue symbols) and the second responses (red symbols). (B) Similar to A, but comparison of the Iend' reversal potential. (C) Similar to A, but comparison of Re/p, the amplitude ratio of Iend'(–80)/Ipeak(80). (D) Similar to A, but comparison of Rs/p, the amplitude ratio of Ishift(80)/Ipeak(80). * and **, p<0.05 and p<0.005, Mann-Whitney U-test. † and ††, p<0.05 and p<0.005, Wilcoxon signed rank test.
Discussion
The crystal structures of ChR2 and hWR suggest that Trp in the middle of TM3 (W124 and W163, respectively) is one of the amino acid residues forming the retinal-binding pocket, which is the most common structural motif among microbial rhodopsins [25,29]. Both this Trp (BR W86 counterpart) and another Trp in the middle of TM6 (BR W182 counterpart) are highly conserved so as to sandwich ATR and are the possible determinants of ATR isomerization [3]. However, the precise roles of W86 and its counterpart have not been revealed previously. In the present study, for the first time, we characterized its role using site-directed mutagenesis of hWR by replacing W163 with another aromatic amino acid, Phe. This W163F mutation appeared not to interfere with the correct folding of the molecule for two reasons. Firstly, the hWR-W163F was well expressed in the membrane of an ND7/23 cell in a manner similar to the naïve hWR. Secondly, robust photocurrent responses were generated in the hWR-W163F-expressing cell, although red-shifted in the action spectrum. However, the ion-transporting activity of hWR was strongly altered by the W163F mutation in 3 points: (1) the H+ leak at positive Vm and its light-adaptation, (2) the attenuation of cation channel activity and (3) the manifestation of outward H+ pump activity. All of these observations strongly suggest that W163 should play a role in stabilizing the structure involved in the gating-on and -off of the cation channel, the role of “gate keeper”. We could attribute the attenuation of the cation channel activity to the incomplete gating-on and the H+ leak to the incomplete gating-off.
Upon retinal photoisomerization from the all-trans to the 13-cis form, the gating-on of a cation channel is assumed to be triggered by the deprotonation of retinal-Schiff-base (RSBH+). In the case of BR the H+ transfer from RSBH+ to D85 in TM3 is known to be the earliest process of H+ transport [39] (Fig. 8A). This H+ was then released in the extracellular water cavity via the H+-releasing complex near it. Meanwhile, the deprotonated RSB is reprotonated from the primary H+ donor D96, which is protonated at the basal state. As deprotonated D96 accepts H+ from the intracellular water cavity and the reprotonation of RSB is followed by the 13-cis to all-trans re-isomerization, one H+ is transported from an intracellular to extracellular direction as a net during a photocycle [3,40]. At the basal state D85 forms a hydrogen bond network with RSBH+ and a water molecule between them. On the other hand, in the case of ChR2, the carboxyl group at D253 in TM7 (BR D212 counterpart) can accept a H+ primarily from the RSBH+ during a photocycle as shown by the FTIR studies [41]. However, the protonation of E123 in TM3 (BR D85 counterpart) also accompanied by RSBH+ deprotonation [42]. Probably, the H+ is transferred from RSBH+ to either D253 or E123 with different probability; the higher probability to D253 and the lower probability to E123. Indeed, the ChR2 crystallography showed that E123 and D253 lie close to the RSBH+ to form the central gate with hydrogen-bonds [29]. However, the replacement of E123 with other amino acids (Gln, Ala, Thr) only partially attenuated the photocurrent with acceleration of photocycle [43–45], whereas the replacement of D253 with Thr almost abolished the channel activity [41]. It is assumed that the H+ acceptance by D253 and the disruption of hydrogen bond between E123 and RSB trigger the conformational change to open the channel gate and stabilize the open state by rearrangements of the hydrogen-bond network, although both E123 and D253 are potential H+ acceptors having flexible interaction with RSBH+ [46].
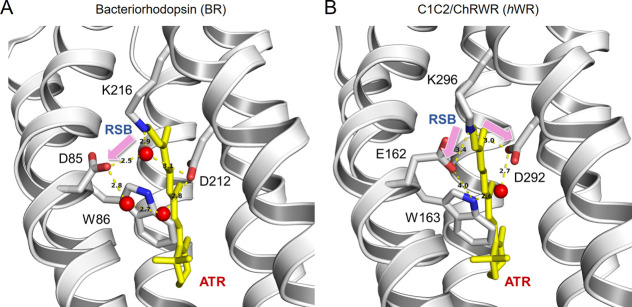
Figure 8.
H+ transfer from the retinal-Schiff-base (RSBH+) at the gating-on of ion transport. (A) In the case of BR, the primary H+ acceptor is D85. Modified from [39]. (B) In the case of hWR, E162 (BR D85 counterpart) and D292 (BR D212 counterpart), both of which are assumed to form a hydrogen-bond network with the indole-N group of W163, are candidates of the primary H+ acceptors. Modified from [25].
Among the carboxylic groups near the RSB+ E162 in TM3 (BR D85 counterpart) and D292 in TM7 (BR D212 counterpart) are also candidates of the primary H+ acceptors for hWR (Fig. 8B). Indeed, these carboxyl groups are located close to the RSBH+ in the crystallographic structure of hWR: a distance of 3.4 and 3.0 Å, respectively [25]. The FTIR spectral change partially originated from D195 protonation but the protonation of E162 or D292 is possibly also involved in [47]. The MD simulation of the closed state implies that the carboxyl groups of E162 and D292 may compete for the RSB-derived H+ [48]. Indeed, either E162A or D292A mutation significantly attenuated the hWR photocurrent. The indole-N group of W163 possibly forms a hydrogen-bond network with E162 and D292 like its counterpart of ChR2, W124, which does so with both E123 and D253 through water molecules [29]. Therefore, the W163F mutation probably changed the central gate structure consisting of RSBH+, E162 and D292 through rearrangement of hydrogen-bond networks around it. As a result, the chance of E162 to accept the RSB-derived H+ may be potentiated [48]. We found a significant outward H+ leak at the dark state of hWR-W163F. This could be attributed to a change in the hydrogen-bond network in the central gate, enabling to transfer an H+ from the intracellular to the extracellular water cavities, probably via E129 (ChR2 E90 counterpart), D292 and water molecules. The fact that this H+ leak was attenuated by the light-adaptation or by the E129A mutation suggests that the protonated E129 may be critical for this H+ leak [24,37,49]. The protonated E129 should also be involved in the passive transport of H+ as Re/p was attenuated by the E129A mutation. It is assumed that the retinal isomerization from all-trans, C=N-anti to 13-cis, C=N-syn is accompanied by the light-adaptation like ChR2 [38], although the 13-cis, C=N-syn isomer could be less stable than that of ChR2 [50]. Therefore, the possibility that the retinal isomerization might break the hydrogen bond network involved in the H+ leak in the light-adapted closed-channel state also has to be taken into consideration.
The W163F mutation strongly attenuated the passive transport of cations with the manifestation of outward H+ pump activity. Therefore, the channel and the pump activities are independent, although hWR may behave as a leaky H+ pump like ChR2 [51]. The complex photocurrent of hWR-W163F kinetics such as the sag and hump waveforms as well as the bi-exponential off time course implies that the photocycle branches in two ways by chance after ATR photoisomerization; one, in the direction of channel activity and another, in the direction of pump activity although more studies are needed to establish the photocycle model. One hypothesis is that the pump activity may be initiated by the H+ transfer from RSBH+ to E162. Since E162 is one of negatively charged residues aligned to form the extracellular water cavity [25], the H+ should be transferred in the extracellular direction. The deprotonation/reprotonation of E129 may be involved in this process because the light-dependent inactivation of the H+ pump activity was attenuated by the E129A mutation. Subsequently, the RSB would be reprotonated by the H+ transfer from somewhere in the intracellular region, although the counterpart of BR H+ donor D96 is H173, which forms a hydrogen bond with E122 in TM2 in the hWR [25]. Probably, the opening of the intracellular water cavity is accompanied by the breakdown of hydrogen bonds between E122 and H173, enabling the H+ transfer from E122 and water molecules. Consequently, the vectorial movement of a single H+ from an intracellular to an extracellular direction might be established during a photocycle. On the other hand, the H+ transfer to D292 may initiate the channel activity, probably through enlarging the pore diameter and helping cations to pass through it [25]. The W163F mutation probably affected the interaction between RSBH+ and D292, thereby decreasing the probability of RSBH+-D292 H+ transfer.
In conclusion, the present study revealed for the first time the contribution of W163 to the ion transport. Not only forming the hydrophobic retinal-binding pocket with other amino acids, based on the 14-nm red shift of the action spectrum by the W163F mutation, it also has strong π-π interaction with ATR. This Trp may have a role in locating the structure involved in channel/pump gating at the optimal position and transducing the signal of retinal photoisomerization to the protein moiety by regulating the H+ transfer from RSBH+. The above hypothesis should be verified in the future through the mutations of amino acids involved in H+ transfer, FTIR spectroscopy, structure biological analyses and computational simulation in combination.
Acknowledgments
We thank R. Kamii, M. Patti, T. Ishizuka, K. Stehfest and P. Hegemann for the preliminary research, H. Kohsaka and A. Nose for the use of the confocal microscope, and B. Bell for the language assistance. This work was supported by a Grant-in-Aid for Scientific Research (17H03007 to KI and 18H03986 to HK) of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan: http://www.jsps.go.jp/english/e-grants/grants01.html, a grant from PRESTO (JPMJPR1888 to TN) and from CREST (JPMJCR1753 to HK), Japan Science and Technology Agency, a Grant-in-Aid for JSPS Fellows of Japan Society for the Promotion of Science (JSPS) (16J09672 to SH, 19J12337 to NK) and Tohoku University Division for Interdisciplinary Advanced Research and Education (DIARE) to NK as well as Research Foundation for Opto-Science and Technology to HY.
Conflicts of Interest
The authors declare that there is no conflict of interest.
Author Contributions
HY contributed to conceiving, designing and performing the experiments. YN, SH and NK contributed to experiments and data analysis. TN contributed reagents, materials, and analysis tools. HY, HK and KI contributed to data interpretation and manuscript writing. All authors approved the final version of the manuscript.
References
- [1].Sharma A. K., Spudich J. L. & Doolittle W. F.. Microbial rhodopsins: functional versatility and genetic mobility. Trends Microbiol. 14, 463–469 (2006). DOI: 10.1016/j.tim.2006.09.006 [DOI] [PubMed] [Google Scholar]
- [2].Spudich J. L., Sineshchekov O. A. & Govorunova E. G.. Mechanism divergence in microbial rhodopsins. Biochim. Biophys. Acta 1837, 546–552 (2014). DOI: 10.1016/j.bbabio.2013.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ernst O. P., Lodowski D. T., Elstner M., Hegemann P., Brown L. S. & Kandori H.. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 (2014). DOI: 10.1021/cr4003769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Oesterhelt D. & Stoeckenius W.. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 233, 149–152 (1971). DOI: 10.1038/newbio233149a0 [DOI] [PubMed] [Google Scholar]
- [5].Lanyi J. K. Mechanism of ion transport across membranes. Bacteriorhodopsin as a prototype for proton pumps. J. Biol. Chem. 272, 31209–31212 (1997). DOI: 10.1074/jbc.272.50.31209 [DOI] [PubMed] [Google Scholar]
- [6].Neutze R., Pebay-Peyroula E., Edman K., Royant A., Navarro J. & Landau E. M.. Bacteriorhodopsin: a high-resolution structural view of vectorial proton transport. Biochim. Biophys. Acta 1565, 144–167 (2002). DOI: 10.1016/S0005-2736(02)00566-7 [DOI] [PubMed] [Google Scholar]
- [7].Inoue K., Ono H., Abe-Yoshizumi R., Yoshizawa S., Ito H., Kogure K., et al. . A light-driven sodium ion pump in marine bacteria. Nat. Commun. 4, 1678 (2013). DOI: 10.1038/ncomms2689 [DOI] [PubMed] [Google Scholar]
- [8].Matsuno-Yagi A. & Mukohata Y.. Two possible roles of bacteriorhodopsin; a comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem. Biophys. Res. Commun. 78, 237–243 (1977). DOI: 10.1016/0006-291X(77)91245-1 [DOI] [PubMed] [Google Scholar]
- [9].Schobert B. & Lanyi J. K.. Halorhodopsin is a light-driven chloride pump. J. Biol. Chem. 257, 10306–10313 (1982). [PubMed] [Google Scholar]
- [10].Inoue K., Ito S., Kato Y., Nomura Y., Shibata M., Uchihashi T., et al. . A natural light-driven inward proton pump. Nat. Commun. 7, 13415 (2016). DOI: 10.1038/ncomms13415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nagel G., Ollig D., Fuhrmann M., Kateriya S., Musti A. M., Bamberg E., et al. . Channelrhodopsin-1: a light-gated proton channel in green algae. Science 296, 2395–2398 (2002). DOI: 10.1126/science.1072068 [DOI] [PubMed] [Google Scholar]
- [12].Nagel G., Szellas T., Huhn W., Kateriya S., Adeishvili N., Berthold P., et al. . Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 100, 13940–13945 (2003). DOI: 10.1073/pnas.1936192100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Govorunova E. G., Sineshchekov O. A., Janz R., Liu X. & Spudich J. L.. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science 349, 647–650 (2015). DOI: 10.1126/science.aaa7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sineshchekov O. A., Jung K. H. & Spudich J. L.. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 99, 8689–8694 (2012). DOI: 10.1073/pnas.122243399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hegemann P. Algal sensory photoreceptors. Annu. Rev. Plant Biol. 59, 167–189 (2008). DOI: 10.1146/annurev.arplant.59.032607.092847 [DOI] [PubMed] [Google Scholar]
- [16].Boyden E. S., Zhang F., Bamberg E., Nagel G. & Deisseroth K.. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005). DOI: 10.1038/nn1525 [DOI] [PubMed] [Google Scholar]
- [17].Ishizuka T., Kakuda M., Araki R. & Yawo H.. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci. Res. 54, 85–94 (2006). DOI: 10.1016/j.neures.2005.10.009 [DOI] [PubMed] [Google Scholar]
- [18].Yawo H., Asano T., Sakai S. & Ishizuka T.. Optogenetic manipulation of neural and non-neural functions. Dev. Growth Differ. 55, 474–490 (2013). DOI: 10.1111/dgd.12053 [DOI] [PubMed] [Google Scholar]
- [19].Deisseroth K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18, 1213–1225 (2015). DOI: 10.1038/nn.4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Deisseroth K. & Hegemann P.. The form and function of channelrhodopsin. Science 357, eaan5544 (2017). DOI: 10.1126/science.aan5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Khorana H. G., Gerber G. E., Herlihy W. C., Gray C. P., Anderegg R. J., Nihei K., et al. . Amino acid sequence of bacteriorhodopsin. Proc. Natl. Acad. Sci. USA 76, 5046–5050 (1979). DOI: 10.1073/pnas.76.10.5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Adamian L., Ouyang Z., Tseng Y. Y. & Liang J.. Evolutionary patterns of retinal-binding pockets of type I rhodopsins and their functions. Photochem. Photobiol. 82, 1426–1435 (2006). DOI: 10.1562/2006-02-14-RA-802 [DOI] [PubMed] [Google Scholar]
- [23].Weidlich O., Schalt B., Friedman N., Sheves M., Lanyi J. K., Brown L. S., et al. . Steric interaction between the 9-methyl group of the retinal and tryptophan 182 controls 13-cis to all-trans reisomerization and proton uptake in the bacteriorhodopsin photocycle. Biochemistry 35, 10807–10814 (1996). DOI: 10.1021/bi960780h [DOI] [PubMed] [Google Scholar]
- [24].Takemoto M., Kato H. E., Koyama M., Ito J., Kamiya M., Hayashi S., et al. . Molecular dynamics of channelrhodopsin at the early stages of channel opening. PLoS. One 10, e0131094 (2015). DOI: 10.1371/journal.pone.0131094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kato H. E., Zhang F., Yizhar O., Ramakrishnan C., Nishizawa T., Hirata K., et al. . Crystal structure of the channelrhodopsin light-gated cation channel. Nature 482, 369–374 (2012). DOI: 10.1038/nature10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Luecke H., Schobert B., Richter H. T., Cartailler J. P. & Lanyi J. K.. Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 291, 899–911 (1999). DOI: 10.1006/jmbi.1999.3027 [DOI] [PubMed] [Google Scholar]
- [27].Kouyama T., Kanada S., Takeguchi Y., Narusawa A., Murakami M. & Ihara K.. Crystal structure of the light-driven chloride pump halorhodopsin from Natronomonas pharaonis. J. Mol. Biol. 396, 564–579 (2010). DOI: 10.1016/j.jmb.2009.11.061 [DOI] [PubMed] [Google Scholar]
- [28].Kato H. E., Inoue K., Abe-Yoshizumi R., Kato Y., Ono H., Konno M., et al. . Structural basis for Na+ transport mechanism by a light-driven Na+ pump. Nature 521, 48–53 (2015). DOI: 10.1038/nature14322 [DOI] [PubMed] [Google Scholar]
- [29].Volkov O., Kovalev K., Polovinkin V., Borshchevskiy V., Bamann C., Astashkin R., et al. . Structural insights into ion conduction by channelrhodopsin 2. Science 358, eaan8862 (2017). DOI: 10.1126/science.aan8862 [DOI] [PubMed] [Google Scholar]
- [30].Kim Y. S., Kato H. E., Yamashita K., Ito S., Inoue K., Ramakrishnan C., et al. . Crystal structure of the natural anion-conducting channelrhodopsin GtACR1. Nature 561, 343–348 (2018). DOI: 10.1038/s41586-018-0511-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li H., Huang C. Y., Govorunova E. G., Schafer C. T., Sineshchekov O. A., Wang M., et al. . Crystal structure of a natural light-gated anion channelrhodopsin. Elife 8, e41741 (2019). DOI: 10.7554/eLife.41741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang H., Sugiyama Y., Hikima T., Sugano E., Tomita H., Takahashi T., et al. . Molecular determinants differentiating photocurrent properties of two channelrhodopsins from Chlamydomonas. J. Biol. Chem. 284, 5685–5696 (2009). DOI: 10.1074/jbc.M807632200 [DOI] [PubMed] [Google Scholar]
- [33].Wood J. N., Bevan S. J., Coote P. R., Dunn P. M., Harmar A., Hogan P., et al. . Novel cell lines display properties of nociceptive sensory neurons. Proc. Biol Sci. USA 241, 187–194 (1990). DOI: 10.1098/rspb.1990.0084 [DOI] [PubMed] [Google Scholar]
- [34].Zhang Q., Hsia S. C. & Martin-Caraballo M.. Regulation of T-type Ca2+ channel expression by interleukin-6 in sensory-like ND7/23 cells post-herpes simplex virus (HSV-1) infection. J. Neurochem. 151, 238–254 (2019). DOI: 10.1111/jnc.14697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tanimoto S., Sugiyama Y., Takahashi T., Ishizuka T. & Yawo H.. Involvement of glutamate 97 in ion influx through photo-activated channelrhodopsin-2. Neurosci. Res. 75, 13–22 (2013). DOI: 10.1016/j.neures.2012.05.008 [DOI] [PubMed] [Google Scholar]
- [36].Lin J. Y., Lin M. Z., Steinbach P. & Tsien R. Y.. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys. J. 96, 1803–1814 (2009). DOI: 10.1016/j.bpj.2008.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Eisenhauer K., Kuhne J., Ritter E., Berndt A., Wolf S., Freier E., et al. . In channelrhodopsin-2 Glu-90 is crucial for ion selectivity and is deprotonated during the photocycle. J. Biol. Chem. 287, 6904–6911 (2012). DOI: 10.1074/jbc.M111.327700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kuhne J., Vierock J., Tennigkeit S. A., Dreier M. A., Wietek J., Petersen D., et al. . Unifying photocycle model for light adaptation and temporal evolution of cation conductance in channelrhodopsin-2. Proc. Natl. Acad. Sci. USA 116, 9380–9389 (2019). DOI: 10.1073/pnas.1818707116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Souvignier G. & Gerwert K.. Proton uptake mechanism of bacteriorhodopsin as determined by time-resolved stroboscopic-FTIR-spectroscopy. Biophys. J. 63, 1393–1405 (1992). DOI: 10.1016/S0006-3495(92)81722-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kandori H. Role of internal water molecules in bacteriorhodopsin. Biochim. Biophys. Acta 1460, 177–191 (2000). DOI: 10.1016/S0005-2728(00)00138-9 [DOI] [PubMed] [Google Scholar]
- [41].Lórenz-Fonfría V. A., Resler T., Krause N., Nack M., Gossing M., Fischer von Mollard G., et al. . Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc. Natl. Acad. Sci. USA 110, E1273–1281 (2013). DOI: 10.1073/pnas.1219502110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kuhne J., Eisenhauer K., Ritter E., Hegemann P., Gerwert K. & Bartl F.. Early formation of the ion-conducting pore in channelrhodopsin-2. Angew. Chem. Int. Ed. Engl. 54, 4953–4957 (2015). DOI: 10.1002/anie.201410180 [DOI] [PubMed] [Google Scholar]
- [43].Gunaydin L. A., Yizhar O., Berndt A., Sohal V. S., Deisseroth K. & Hegemann P.. Ultrafast optogenetic control. Nat. Neurosci. 13, 387–392 (2010). DOI: 10.1038/nn.2495 [DOI] [PubMed] [Google Scholar]
- [44].Watanabe H. C., Welke K., Schneider F., Tsunoda S., Zhang F., Deisseroth K., et al. . Structural model of channelrhodopsin. J. Biol. Chem. 287, 7456–7466 (2012). DOI: 10.1074/jbc.M111.320309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lórenz-Fonfría V. A., Schultz B. J., Resler T., Schlesinger R., Bamann C., Bamberg E., et al. . Pre-gating conformational changes in the ChETA variant of channelrhodopsin-2 monitored by nanosecond IR spectroscopy. J. Am. Chem. Soc. 137, 1850–1861 (2015). DOI: 10.1021/ja5108595 [DOI] [PubMed] [Google Scholar]
- [46].Guo Y., Beyle F. E., Bold B. M., Watanabe H. C., Koslowski A., Thiel W., et al. . Active site structure and absorption spectrum of channelrhodopsin-2 wild-type and C128T mutant. Chem. Sci. 7, 3879–3891 (2016). DOI: 10.1039/c6sc00468g [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Inaguma A., Tsukamoto H., Kato H. E., Kimura T., Ishizuka T., Oishi S., et al. . Chimeras of channelrhodopsin-1 and -2 from Chlamydomonas reinhardtii exhibit distinctive light-induced structural changes from channelrhodopsin-2. J. Biol. Chem. 290, 11623–11634 (2015). DOI: 10.1074/jbc.M115.642256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].VanGordon M. R., Gyawali G., Rick S. W. & Rempe S. B.. Atomistic study of intramolecular interactions in the closed-state channelrhodopsin chimera, C1C2. Biophys. J. 112, 943–952 (2017). DOI: 10.1016/j.bpj.2017.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ruffert K., Himmel B., Lall D., Bamann C., Bamberg E., Betz H., et al. . Glutamate residue 90 in the predicted transmembrane domain 2 is crucial for cation flux through channelrhodopsin 2. Biochem. Biophys. Res. Commun. 410, 737–743 (2011). DOI: 10.1016/j.bbrc.2011.06.024 [DOI] [PubMed] [Google Scholar]
- [50].Hontani Y., Marazzi M., Stehfest K., Mathes T., van Stokkum I. H. M., Elstner M., et al. . Reaction dynamics of the chimeric channelrhodopsin C1C2. Sci. Rep. 7, 7217 (2017). DOI: 10.1038/s41598-017-07363-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Feldbauer K., Zimmermann D., Pintschovius V., Spitz J., Bamann C. & Bamberg E.. Channelrhodopsin-2 is a leaky proton pump. Proc. Natl. Acad. Sci. USA 106, 12317–12322 (2009). DOI: 10.1073/pnas.0905852106 [DOI] [PMC free article] [PubMed] [Google Scholar]