Abstract
Malus sylvestris (Mill.) is the only indigenous wild apple species in Central Europe. Agriculture, forestry, and urbanization increasingly endanger Malus sylvestris natural habitats. In addition, the risks of cross‐hybridization associated with increase in the cultivation of the domesticated apple Malus × domestica (Borkh.) threaten the genetic integrity of M. sylvestris. The present study investigated the number of hybrids, genetic diversity, and genetic structure of 292 putative M. sylvestris that originate from five different natural M. sylvestris populations in Saxony, Germany. All samples were genetically analyzed using nine nuclear microsatellite markers (ncSSR) and four maternally inherited chloroplast markers (cpDNA) along with 56 apple cultivars commonly cultivated in Saxony. Eighty‐seven percent of the wild apple accessions were identified as pure M. sylvestris. The cpDNA analysis showed six private haplotypes for M. sylvestris, whereas three haplotypes were present in M. sylvestris and M. × domestica. The analysis of molecular variance (AMOVA) resulted in a moderate (ncSSR) and great (cpDNA) variation among pure M. sylvestris and M. × domestica individuals indicating a low gene flow between both species. The genetic diversity within the pure M. sylvestris populations was high with a weak genetic structure between the M. sylvestris populations indicating an unrestricted genetic exchange between these M. sylvestris populations. The clear distinguishing of M. sylvestris and M. ×domestica confirms our expectation of the existence of pure M. sylvestris accessions in this area and supports the argument for the implementation of preservation measures to protect the M. sylvestris populations in Saxony.
Keywords: chloroplast DNA, European wild apple, haplotypes, hybridization, microsatellites, species delimitation
The present study investigated the number of hybrids, genetic diversity, and genetic structure of 292 putative M. sylvestris that originate from five different natural populations in Saxony, Germany. Genetic analysis were performed with nuclear and chloroplast markers. We detected a high genetic diversity within the pure M. sylvestris populations but with a weak genetic structure between the M. sylvestris populations indicating an unrestricted genetic exchange between these M. sylvestris populations.

1. INTRODUCTION
The genus Malus Mill. comprises 25–47 species, depending on the taxonomic classification (Robinson et al., 2001). While several Malus species are indigenous to Asia, Malus sylvestris (Mill.) is the only indigenous wild apple species in Central Europe. The spatial distribution of this European crabapple ranges from South Scandinavia to the Iberian Peninsula and from the Volga to the British Isles (Robinson et al., 2001). Despite this vast distribution area, M. sylvestris is very rarely seen and in fact endangered in most European countries (Bitz et al., 2019; Coart et al., 2006; Höltken et al., 2014; Larsen et al., 2006; Reim et al., 2012, 2013; Wagner et al., 2014). Its economical insignificance and the present‐day intensive land usage by forestry and agriculture are the main reasons why wild apple trees have been displaced into areas with more or less unfavorable growing conditions. However, M. sylvestris trees have, except for their light requirements, low demands on environmental conditions. They are therefore able to survive in such niche areas. M. sylvestris populations are often very small and spatially distributed, which may lead to a reproductive isolation of individual populations (Reim et al., 2017; Ruhsam et al., 2019; Wagner et al., 2014). Most Malus species are self‐incompatible and need compatible pollination partners to maintain the natural regeneration of the populations (Hanke et al., 2020). Insects such as bees or bumblebees mainly distribute apple pollen, and most pollination events occur in a short distance (50–100 m) around the pollinator. However, if sufficient pollination partners are missing, the distance of pollination can remarkably increase (Reim et al., 2017). Since no reproductive barriers prevent the hybridization between different species within the genus Malus, the probability of admixture with domesticated apples (Malus × domestica Borkh.) increases particularly in small populations (Korban, 1986; Larsen et al., 2006; Reim et al., 2017). Such hybridization events endanger the genetic integrity of M. sylvestris by replacing pure M. sylvestris trees with hybrids (Allendorf et al., 2001). Thus, the conservation of genetic resources must extend to include the preservation of pure M. sylvestris trees.
Knowledge of population genetic diversity and genetic structures is crucial in the identification of pure genotypes, which is integral in studies for the implementation of sustainable conservation strategies (Castiglione et al., 2010). However, in closely related species such as in Malus a clear distinguishing of species remains a challenge. Delimitation of M. sylvestris and M. ×domestica is particularly difficult because of their close relationship (Duan et al., 2017).
Several morphological characteristics were described as useful for the classification of M. sylvestris like hairiness of leaf lower surface, hairiness of the flower stem; fruit size, and cover color (Wagner, 1996). However, the main disadvantage of morphological traits is their high level of variation depending on various environmental factors. For example, the intensity of hairiness, which functions as protection against evaporation, can vary during the growing season. Another example is the intensity of over color, which depends on exposure to sun light. Therefore, genetic markers are an indispensable tool for taxonomical studies of species. In conservation genetic studies, microsatellite markers or simple sequence repeats (SSRs) are still the markers of choice (Bitz et al., 2019; Coelho et al., 2018; Kaczmarczyk, 2019; Lea et al., 2018) and are useful tools for a putative identification of “hybrids” within potential wild apple populations (Koopman et al., 2007; Larsen et al., 2006). For closely related species, such as the species within the genus Malus, noncoding DNA regions of the chloroplast genome could also be an additional useful genetic tool to study the relationship among species (Fan et al., 2019; Khadivi‐Khub et al., 2014; Tang et al., 2014; Volk et al., 2015). Because of their slow rate of molecular evolution and the conserved structure of their genome, chloroplast markers were developed for several important plant species (Heinze, 2007). In Malus, several intergenic spacer and introns regions in the chloroplast genome were identified and from these regions, primers successfully generated and applied for phylogenetic studies (Coart et al., 2006; Fan et al., 2019; Khadivi‐Khub et al., 2014; Tang et al., 2014; Volk et al., 2015; Xu et al., 2019).
In our study, M. sylvestris accessions from five natural populations in Saxony (Germany) were genetically investigated using nine nuclear microsatellites (ncSSR) and four chloroplast DNA (cpDNA) markers. All putative M. sylvestris individuals were analyzed with ncSSR markers in order to identify pure accessions. Pure M. sylvestris accessions and 56 M. × domestica genotypes were further analyzed with four cpDNA markers to ascertain haplotype variation within and between these species. We further estimated the genetic diversity within the M. sylvestris populations and the genetic structure among these populations in the study area. The results of this study are the basis for the implementation of conservation measures.
2. MATERIALS AND METHODS
2.1. Study site, sampling, and DNA isolation
The study was carried out in the federal state of Saxony, in Southeast Germany. Saxony comprises an area of 18,413 square kilometers, and hills and mountains mainly compose the landscape. Two hundred and ninety‐two potential M. sylvestris trees sampled from five different areas within Saxony (Figure 1) formed the basis of this study. The mapped trees were mainly found in sparse forests, along forest edges, stonewalls, or other open landscape structures. The tree density within the populations was low in most cases with partly large distances between the single trees. The trees of some population such as in the East Ore Mountains (OEG) were distributed over several square meters. Compact populations with more than 50 individuals in a close spatial network only existed in the Leipziger Auwald (LEI) and Bahretal (BAR). Seven percent of the sampled trees were located in a nature reserve. The remaining 93% were located in unprotected areas. The selection of trees was based on several morphological characters described by Wagner (1996) including fruit diameter <3.5 cm, no fruit over color, and absence of hairiness in the leaf lower surface, for example. In addition, 56 apple cultivars (M. × domestica Borkh.) often cultivated in Germany were included as control genotypes. The location of the captured M. sylvestris trees and the names of the 56 cultivars are available in Dryad at https://doi.org/10.5061/dryad.g79cnp5n2. Leaf material of these 56 apple cultivars was provided by the Fruit Genebank of the Institute of Breeding Research on Fruit Crops of the Julius Kühn‐Institut (JKI) at Dresden‐Pillnitz.
Figure 1.
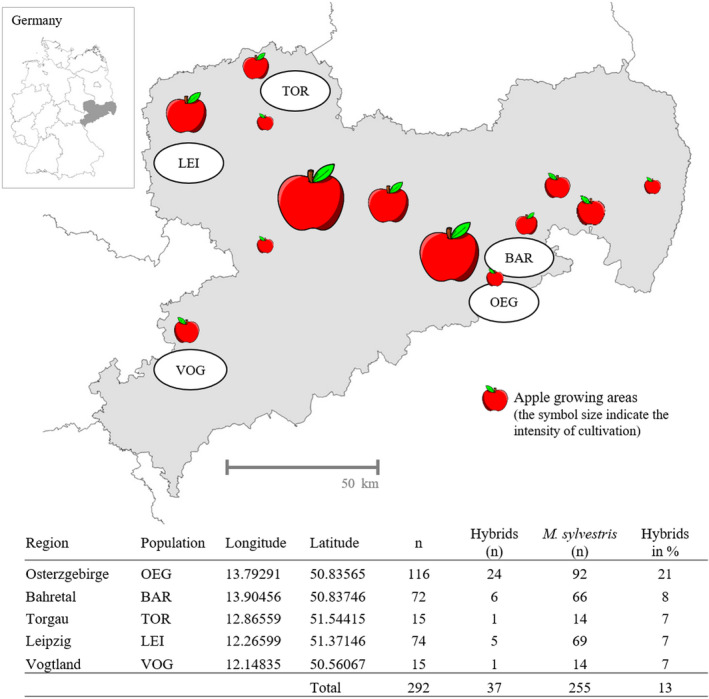
Origin, number of sampled M. sylvestris trees and number of hybrids in five natural populations in Saxony, Germany
Fresh leaf materials were collected in 2‐ml tubes and dried using silica gel according to a modified protocol of (Chase & Hills, 1991). Leaf materials were stored at room temperature until DNA isolation. LGC Genomics (Berlin, Germany) performed DNA isolation and quantification using the semi‐automated high‐throughput Genespin™ platform with the company proprietary Kleargene™ extraction chemistry.
2.2. SSR and cpDNA analysis
The genetic analysis was performed at the state‐owned enterprise Sachsenforst in Pirna. Nine ncSSR primer pairs (CH01F02, CH01F03b, CH01H01, CH01H10, CH02C09, CH04C07, GD12, GD147, Hi02C07) developed for M. × domestica (Liebhard et al., 2002) were combined in three multiplexes with three primers per multiplex (Table S1). Detailed information about the molecular characteristics of these markers is available at http://hidras.unimi.it/HiDRAS‐SSRdb/pages/index.php.
Four cpDNA primer pairs located at the matK_dup (NCBI GenBank: AF309231), rps16 intron (NCBI GenBank: JQ391664) region and the intergenic spacer region rps16_trnQ (NCBI GenBank: AB604694) and rpl2_trnH (NCBI GenBank: JQ392141), respectively, were developed based on Malus sequence data obtained from the NCBI database (www.ncbi.nlm.nih.gov; Table 1).
Table 1.
Chloroplast DNA (cpDNA) and nuclear SSR (ncSSR) markers used in this study. Table 1: Chloroplast DNA (cpDNA) and nuclear SSR (ncSSR) markers used in this study
| Primer sequence forward | Primer sequence reverse | Fragment size (bp) | |
|---|---|---|---|
| Chloroplast region | |||
| rps16_trnQ | CAGTAAGTACCATTCGCTTTTTATC | TTTCGACCAGTCTTCCGTTT | 304–312 |
| rpl2_trnH | CACTTAACACAAAAGCAGAAAAAGA | GGATCAAGGCAGTGGATTGT | 147–170 |
| matK_dup | GGTTATGCGATCGTAGAAATGG | TTCCTTCCCTATACACGACTCT | 158–178 |
| Rps16_Intron | GACAAAAAGGGTTAGAGACCACTC | CTCGTACGGCTCGAGAAAAT | 245–249 |
| ncSSR marker | |||
| CH01f02 | ACCACATTAGAGCAGTTGAGG | CTGGTTTGTTTTCCTCCAGC | 153–227 |
| CH01f03b | GAGAAGCAAATGCAAAACCC | CTCCCCGGCTCCTATTCTAC | 139–183 |
| CH01h01 | GAAAGACTTGCAGTGGGAGC | GGAGTGGGTTTGAGAAGGTT | 114–134 |
| CH02c09 | TTATGTACCAACTTTGCTAACCTC | AGAAGCAGCAGAGGAGGATG | 233–257 |
| CH01h10 | TGCAAAGATAGGTAGATATATGCCA | AGGAGGGATTGTTTGTGCAC | 094–114 |
| CH04c07 | GGCCTTCCATGTCTCAGAAG | CCTCATGCCCTCCACTAACA | 098–135 |
| GD12 | TTGAGGTGTTTCTCCCATTGGA | CTAACGAAGCCGCCATTTCTTT | 150–200 |
| GD147 | TCCCGCCATTTCTCTGC | GTTTAAACCGCTGCTGCTGAAC | 135–155 |
| Hi02c07 | AGAGCTACGGGGATCCAAAT | GTTTAAGCATCCCGATTGAAAGG | 108–149 |
Multiplex PCR was performed using the Type‐It kit (Qiagen, Germany) according to the manufacturer's protocol with three primer pairs per PCR in a total volume of 10 µl. PCR fragments were analyzed on a CEQ 8000XL Genetic Analyzer System (Beckman Coulter, Germany), for which forward primers were labeled with BMN‐5, BMN‐6, and DY751 (Biomers, Germany).
2.3. Hybridization and population structure analysis
First, samples were grouped into pure M. sylvestris, M. × domestica or hybrids using the model‐based clustering method with STRUCTURE software version 2.3.4. (Pritchard et al., 2000). In order to improve the accuracy of the inference, the analysis was performed with prior information on the population (POPINFO model) in which each individual was defined either as "M. sylvestris" or "M. × domestica." To accurately classify the individuals, 5 runs with K = 2 was performed with the program. The parameters were 50,000 burn‐in periods and 50,000 Markov Chain Monte Carlo repetitions using the admixture model with correlated allele models. The probability of membership to the M. × domestica genepool for the 56 reference M. ×domestica cultivars was > 90% indicating a low introgression from M. sylvestris. Conversely, a probability > 90% of membership to the M. sylvestris genepool was chosen as threshold for the assignment of pure M. sylvestris accessions. All M. sylvestris individuals with a probability of lower than 90% that the assigned population is right were classified as hybrid. Secondly, the number of natural M. sylvestris populations (K) was estimated with STRUCTURE. For this, we run the program again including all pure M. sylvestris accessions with K varying from 2 to 10 with 5 runs for each K value without POPINFO model. The remaining parameters were described as above. STRUCTURE HARVESTER (Earl & Vonholdt, 2012) was used for detecting the most likely value for K based on Evanno's ΔK method (Evanno et al., 2005). To examine the genetic structure of M. sylvestris, a principal coordinate analysis (PCoA) was performed based on the distance matrix data set of the 255 pure M. sylvestris accessions and the 56 reference apple cultivars in GeneAlex ver. 6.5 with 1,000 random permutations.
2.4. Diversity analysis
The mean number of alleles by locus (Na), effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He), and number of private alleles (PA) were calculated for each nuclear SSR loci within the pure M. sylvestris individuals and the apple cultivars using the software GENALEX ver. 6.5 (Peakall & Smouse, 2006, 2012). Allelic richness (Ar) was calculated with the software ADZE (Szpiech et al., 2008) using the rarefaction method to correct differences in population sizes (Kalinowki, 2005). Because of the small sample size of the TOR and VOG populations, the Ar was weighted to fourteen individuals.
The allele frequencies of the chloroplast DNA markers were compared among the M. sylvestris individuals and the apple cultivars.
An "analysis of molecular variance" (AMOVA) was performed based on nuclear and chloroplast marker data. The molecular variance (ɸ PT, an analogue of F st) and the migration rate (Nm) were calculated among the pure M. sylvestris genotypes and the apple cultivars as well as among the different M. sylvestris populations using GENALEX ver. 6.5.
The correspondence (rxy) between geographic and genetic distance was performed by Mantel test with statistical testing by 9,999 permutations using the software GENALEX ver. 6.5 (Mantel, 1967).
3. RESULTS
3.1. Genetic differentiation between M. sylvestris and M. × domestica
Of 292 putative M. sylvestris, 255 (87%) were assigned as pure M. sylvestris species. The remaining 37 individuals were assigned as hybrids or feral M. ×domestica (Figure 2).
Figure 2.
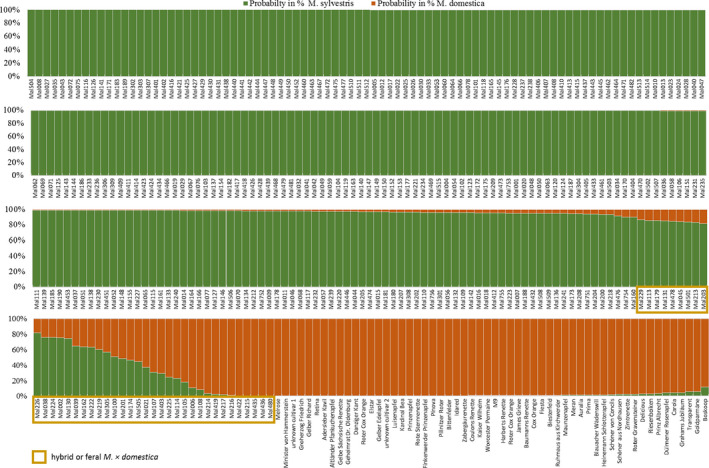
Barplot indicating the genetic affiliation of each individual to either M. sylvestris or M. ×domestica
The four cpDNA markers showed allelic patterns with 12 different alleles with two to four alleles per chloroplast region. Of the 12 different alleles, seven were present in both M. sylvestris and M × domestica samples while five alleles were species specific (Figure S1).
The combination of these different alleles resulted in 15 different haplotypes (Table S1) (Figure 3). In M. ×domestica, the most frequent haplotype was H11, which was observed for 70.0% of genotypes. The most frequent haplotype within the pure M. sylvestris accessions was H2 (57.3%). Twelve haplotypes were not observed for both species: H1, H2, H3, H5, H8, and H13 were not observed in the investigated apple cultivars whereas H4, H6, H9, H10, H14, and H15 were not found in the analyzed M. sylvestris genotypes. Only three haplotypes (H7, H711, and H12) were present in both M. sylvestris and M. × domestica.
Figure 3.
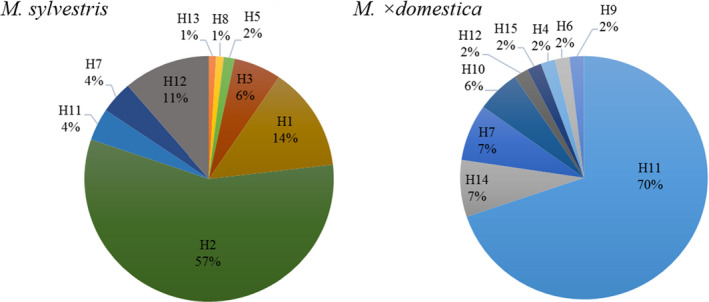
Frequency of the chloroplast 15 haplotype for M. sylvestris and M. × domestica
The analysis of molecular variance (AMOVA) showed significant results for the differentiation between M. sylvestris and M. ×domestica (Figure 4). Based on the ncSSR, a moderate variation of 20% among the species was calculated (ɸ PT = 0.20 p = .001). The effective gene flow was about one migrant per generation (Nm = 1.01). Based on cpDNA data, a great variation of 56% among pure M. sylvestris and the apple cultivars (ɸ PT = 0.56 p = .001) was detected. The effective gene flow was much less than one successful migrant per generation (Nm = 0.39).
Figure 4.
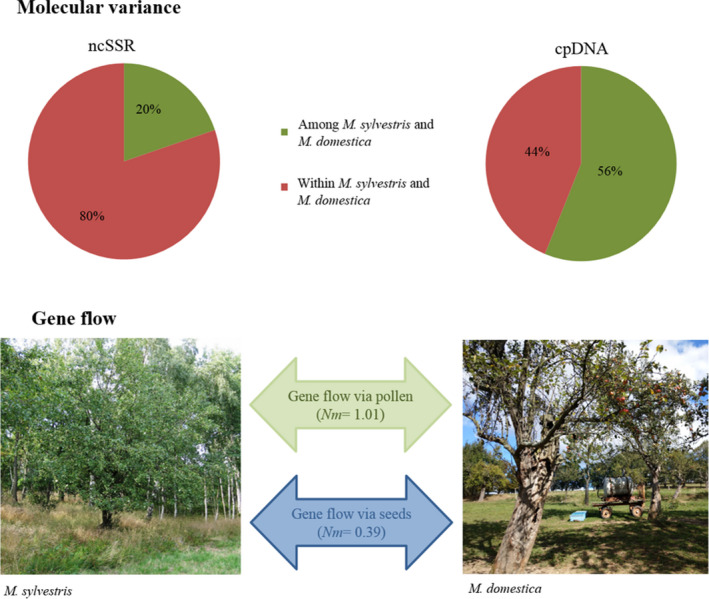
Molecular variance and estimated gene flow based on ncSSR and cpDNA data between wild apples and apple cultivars
3.2. High genetic diversity and weak genetic structure between the M. sylvestris populations
All ncSSR markers showed reproducible results with one or two amplified fragments per genotype. High genetic diversity was observed for the pure M. sylvestris genotypes with an average number of alleles of Na = 10, an average effective number of alleles of Ne = 4, a mean expected heterozygosity of He = 0.73, and a mean allelic richness of Ar = 5.03 (Table 2). These diversity parameters were only slightly lower for M. sylvestris than for the M. ×domestica reference cultivars, indicating a still high level of genetic diversity in the populations investigated. The highest level of genetic diversity was found in the both southeastern Population OEG and BAR with He = 0.78. Despite the large number of individuals, the LEI population showed the lowest genetic diversity with He = 0.68. Only one private allele with a frequency > 0.05 was found in the TOR population, and all other populations had no private alleles.
Table 2.
Number of alleles, number of effective alleles, allelic richness, observed and expected heterozygosity in five different M. sylvestris populations and the M. × domestica reference cultivars
| Population | N | Na | Ne | Ho | H e | PA | Ar |
|---|---|---|---|---|---|---|---|
| BAR | 66 | 12 | 5 | 0.74 | 0.78 | 0.00 | 5.45 |
| LEI | 68 | 12 | 4 | 0.65 | 0.68 | 0.00 | 4.48 |
| OEG | 89 | 14 | 5 | 0.76 | 0.78 | 0.00 | 5.69 |
| TOR | 14 | 8 | 4 | 0.71 | 0.72 | 0.00 | 5.22 |
| VOG | 14 | 6 | 4 | 0.69 | 0.69 | 0.00 | 4.33 |
| mean | 10 | 4 | 0.71 | 0.73 | 0.00 | 5.03 | |
| M. × domestica | 53 | 13 | 5 | 0.79 | 0.79 | 5.00 | 5.65 |
N: number of analyzed individuals; Na: number of different alleles; Ne: number of effective alleles (=1/(∑pi 2)); pi: relative frequency of the ith allele; PA: number of private alleles with a frequency >5%, Ar: Allelic richness, weighted to fourteen individuals.
After analysis of the STRUCTURE output with STRUCTURE HARVESTER based on the Evanno method, the most likely number of genetic groups was K = 3 (Earl & Vonholdt, 2012; Evanno et al., 2005). Therefore, the individuals of the five M. sylvestris population were grouped into three genetic clusters indicated by three different colors in Figure 5. No population showed individuals with allele frequencies specific to only one genetic cluster. Nevertheless, alleles of genetic cluster 1 (orange) were more frequent in the BAR, VOG, and TOR population, whereas alleles of cluster 2 (blue) were more frequent in OEG. Alleles of cluster 3 (green) were mainly attributed to the LEI population. These results indicate only weak population structures between the Saxon M. sylvestris populations.
Figure 5.
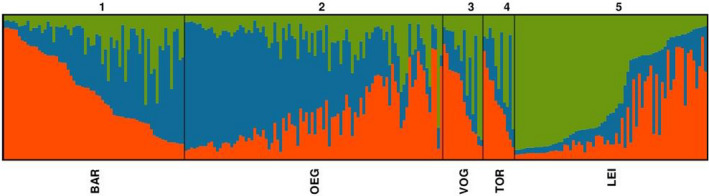
Cluster assignment (K = 3) of the examined M. sylvestris populations (N = 255, 5 population in Saxony) after model‐based clustering using STRUCTURE. Every vertical bar represents a single accession, the different colors indicate the assignment to each genetic cluster: orange= cluster 1; blue= cluster 2; green= cluster 3.
In general, PCoA supported the results of the STRUCTURE analysis and showed low genetic differences between the M. sylvestris accessions of the five population investigated in Saxony (Figure 6). However, two accessions of M. sylvestris (green symbols), previously identified by STRUCTURE as pure M. sylvestris accessions, showed a very close relationship to M. × domestica, indicating that these accessions also hybrids.
Figure 6.
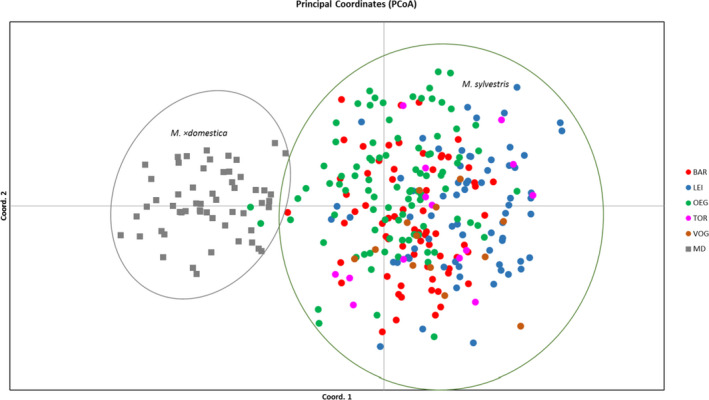
Principal coordinate analysis (PCoA) of pairwise distances between M. sylvestris accessions and the fifty‐six reference apple cultivars calculated on ncSSR markers. The M. sylvestris populations are indicated by different circle colors; red: Bahretal (BAR), blue: Leipzig (LEI), green: Osterzgebirge (OEG), pink: Torgau (TOR), brown: Vogtland (VOG), gray quadrat: M. ×domestica (MD).
Based on the cpDNA data, also weak population structures were observed (Figure 7). Of the nine different haplotypes, H2 and H1 were detected in all M. sylvestris populations with different frequencies in each population. Haplotype H3 was found in the populations LEI, VOG, and BAR, which are located in the western and eastern parts of Saxony. H13 was found in the southeastern population OEG and the northern population TOR. The both haplotypes H5 and H8 were the only private haplotypes and observed only in OEG and TOR, respectively.
Figure 7.
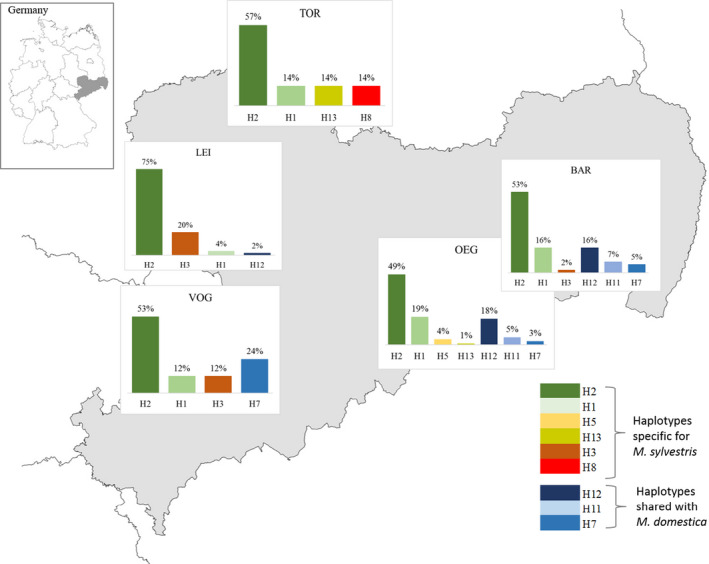
Haplotype variation in the five different pure M. sylvestris populations in Saxony
At least one of the haplotypes shared with M. × domestica (H7, H11, and H12) were also detected in the M. sylvestris populations with one exception. The population TOR showed only M. sylvestris‐specific haplotypes (H2, H1, H13, and H8). In the both southeastern populations OEG and BAR, all three M. ×domestica shared haplotypes were observed with the highest frequency for H12 (18% and 16%, respectively) (Figure 7). In the population VOG, only the M. ×domestica shared haplotype H7 was detected, but with a high frequency of 24%. The western population LEI showed only H12 as shared haplotype with a low frequency of 2%.
After AMOVA, the differences between the five M. sylvestris populations were significant, but low (Table 3). Based on ncSSR, the variation among the M. sylvestris populations was 6% and cpDNA variation was only slightly higher with 7%. The number of migrants among the M. sylvestris populations was high with Nm = 4.14 (ncSSR) and Nm = 7.07 (cpDNA) indicating random mating among each population and weak genetic structure of the populations. Mantel test resulted in a significant (p = .01) but low correlation coefficient rxy = 0.05 suggesting that the geographic distance had hardly any effect on the genetic variance in the Saxony wild apple populations.
Table 3.
Genetic variation (AMOVA) among and within M. sylvestris populations based on ncSSR and cpDNA data
| Variation | ɸPT | p | Nm | |
|---|---|---|---|---|
| Based on ncSSR data | ||||
| Among M. sylvestris populations | 6% | 0.06 | .001 | 4.14 a |
| Within M. sylvestris populations | 94% | |||
| Based on cpDNA data | ||||
| Among M. sylvestris populations | 7% | 0.07 | .001 | 7.07 b |
| Within M. sylvestris populations | 93% | |||
ɸ PT = Estimated variation among populations/(Estimated variation within populations + Estimated variation among populations).
Nm = number of migrants per generation [(1/ɸPT) − 1]/4.
Nm = number of migrants per generation [(1/ɸ PT) − 1]/2.
4. DISCUSSION
4.1. Identification of pure Malus sylvestris individuals
Knowledge of the genetic integrity of M. sylvestris and the clear identification of hybrids is one of the important prerequisites for the implementation of sustainable conservation measurements and the maintenance of pure species in natural populations. However, a clear distinguish between the European crabapple and cultivated apples is difficult because of molecular similarity between both species (Coart et al., 2006). Nevertheless, newer and more sensitive marker systems such as SNPs may be suitable for a clear differentiation of both species.
Some studies clearly demonstrated introgression of M. sylvestris DNA into the nuclear genome of M. ×domestica (Cornille et al., 2012; Harrison & Harrison, 2011). The contribution of M. sylvestris into the M. ×domestica gene pool can be very high (e.g., for the German cultivar “Landsberger Reinette” up to 48%) but depends strongly on the cultivar (Cornille et al., 2012). In our study, all 56 M. × domestica cultivars showed a low introgression from M. sylvestris (> 90% genetic information belong to the M. × domestica genepool). Based on this, all M. sylvestris samples and apple cultivars were clearly assigned in either a "M. sylvestris cluster" or "M. × domestica cluster." Putative M. sylvestris individuals with admixture genetic information of more than 10% of the M. × domestica gene pool were identified as hybrid (13%). This discrimination threshold (Q > 0.90) was very strict compared to other studies (Coart et al., 2003; Höltken et al., 2014; Schnitzler et al., 2014), and therefore, the frequency of hybrids is slightly higher in our population but corresponds well to the morphological characteristics of the sampled M. sylvestris accessions. These results indicate that a genetic differentiation of both species based on ncSSR is still possible although nuclear genome markers disclose less genetic differences in closely related species (Korotkova et al., 2014; Zheng et al., 2014).
4.2. Haplotype variation between M. sylvestris and M. × domestica
In closely related species such as apple, the investigation of phylogenetic relationships based on different cell genomes is recommended (Nikiforova et al., 2013). Chloroplast DNA is maternally inherited and hybridization events are limited to gene flow by seeds which results in a higher level of genetic differentiation among species or populations (Ennos, 1994; Korotkova et al., 2014; Shizuka et al., 2015).
In our study, 20% of the detected haplotypes were common in M. sylvestris and M. × domestica suggesting that these haplotypes are not species specific. Cross‐species shared chloroplast haplotypes were observed for several perennial species such as birch, eucalyptus and willows (Fogelqvist et al., 2015; Nevill et al., 2014; Palme et al., 2004). Within the genus Malus, shared haplotypes were also identified in studies including numerous apple individuals of different cultivars and species (Savolainen et al., 1995; Volk et al., 2015). M. × domestica is known to be an admixed species from a number of progenitor species such as M. sieversii, M. orientalis, M. sylvestris, and M. prunifolia (Cornille et al., 2013; Robinson et al., 2001; Velasco et al., 2010; Volk et al., 2015). Thus, shared haplotypes may reflect the historical introgression between different Malus species due to gene flow or retention of ancestral polymorphisms (Koopman et al., 2007; Nevill et al., 2014; Volk et al., 2015).
Nevertheless, despite the historical admixture within the genus Malus, species‐specific haplotypes exist as shown in our study in which 80% of the M. sylvestris haplotypes were absent in the 56 M. × domestica cultivars. However, that other apple cultivars not analyzed in this study could possess these haplotypes cannot be excluded. The high level of M. sylvestris‐specific haplotypes could be explained by genetic drift or selection. Descendants of M. sylvestris in our study site could have lost one or more of the identical haplotypes over time, supported by the absence or rarity of further introgression events. This assumption was confirmed by the low number of migrants per generation (Nm = 0.39; cpDNA) in our study indicating a low historical gene flow through seed dispersal between M. × domestica and M. sylvestris. Evolutionary studies showed that large fruit development in domesticated crops such as M. ×domestica correlates with a reduction in seed number/ fruit and a loss in natural seed‐dispersal mechanism (Spengler, 2019). This would also explain the remarkable haplotype variations between M. sylvestris and M. × domestica.
The results of the AMOVA also affirmed that numerous M. sylvestris individuals were hardly affected by gene flow by pollen in past times in our study area (Nm = 1.01). A similar rarity of gene flow among M. sylvestris and M. × domestica in the Rhine Valley or in Finland was observed by Schnitzler et al. (2014) and Bitz et al. (2019) and was explained by the only partial overlap in flowering time of these both species. However, the frequency of hybrids varied between the different M. sylvestris populations in our study area. In particular, the OEG population was more affected from hybridization with M. ×domestica with 21% identified hybrids, whereas in the other populations only 7%– 8% accessions originated from hybridization events. This result indicates that the frequency of introgression depends not only on the overlap in flowering time, but also on other external influences such as the occurrence of nearby cultivars or the tree density in the population (Kramer et al., 2008; Reim et al., 2017).
4.3. Gene flow between the M. sylvestris populations in Saxony
Fragmented and spatially isolated populations of rare species such as M. sylvestris are often expected to have reduced genetic diversity and strong genetic structure because of a restricted gene flow (Bacles & Jump, 2011; Kramer et al., 2008; Pierce et al., 2017). However, despite the fragmented distribution of M. sylvestris in the study area, the results of our study indicate a high genetic diversity and a weak genetic structure between the single populations. Other studies also documented that particularly long‐lived plants such as M. sylvestris can maintain connectivity even in highly fragmented populations through extensive gene flow via pollen and/or seeds (Feurtey et al., 2017; Gonzalez‐Varo et al., 2019; Lowe et al., 2016; Ozawa et al., 2013; Plue & Cousins, 2018; Schnitzler et al., 2014; Wang et al., 2014). The comparison of nuclear and chloroplast data allows the separation of the impact of pollen and seed‐mediated gene flow between populations (Ennos, 1994). The Nm‐value in our study indicated a high historical genetic exchange with a higher number of migrants by seeds (Nm = 7.07) than by pollen (Nm = 4.14.) between the M. sylvestris populations. Apple seeds are distributed by numerous wild animals such as mammals or birds, feed on the apple fruit. These animals travel over long distances and probably accounting for the large apple‐dispersal capacities (Schnitzler et al., 2014). In addition, humans dispersed wild apple fruits over long distances in recent times (Spengler, 2019). As a result, in the past genetic exchange by seeds between population occurred and even one single hybridization event retained within a lineage because the chloroplast genome is uniparental inherited (Currat et al., 2008).
Additionally, wild apple populations probably compensated their spatial isolation by higher pollen dispersal distances. Single located individuals can act as so‐called stepping stones and bridge larger distances between groups of trees (Albaladejo et al., 2012; Kramer et al., 2008). Thus, metapopulations could maintained their genetic exchange and high population genetic diversity. However, high pollen flow distances carry the risk of increased hybridization events between M. sylvestris and M. × domestica and may have negative effects on the maintenance of pure M. sylvestris individuals. Even if only less introgressed wild apples were observed in our study, the implementation of sustainable conservation strategies seems to be necessary for a long‐term preservation of this rare species.
5. CONCLUSIONS FOR CONSERVATION MEASURES
The clear genetic differentiation between M. sylvestris and M. ×domestica indicated a low gene flow between both species and the existence of pure M. sylvestris individuals in our study area. Our study has also shown that the genetic diversity of M. sylvestris is still high due to a high gene flow between the single populations. Nevertheless, introgression of M. ×domestica in wild apples takes place and endangers the preservation of the pure M. sylvestris individuals. Furthermore, most wild apples are located in unprotected areas and a reduction of the remaining population can be expected. Over time, this would lead to a loss of genetic diversity in M. sylvestris populations.
Therefore, the protection of existing M. sylvestris populations and implementation of preservations measures (e.g., replanting of young trees) are recommend. Particularly, the reintroduction of young trees and the increase of number of trees within the natural populations are recommended to reduce the risk of hybridization. An additional conservation of pure M. sylvestris accessions ex situ can be also meaningful. That can be implemented by the storage of seeds in a GenBank or the establishment of seed orchards.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTION
Stefanie Reim: Conceptualization (lead); Funding acquisition (equal); Investigation (lead); Project administration (equal); Writing‐original draft (lead). Anke Proft: Data curation (supporting); Investigation (equal); Project administration (equal); Validation (supporting). Frank Lochschmidt: Data curation (supporting); Investigation (supporting); Resources (supporting). Monika Höfer: Conceptualization (supporting); Resources (supporting); Validation (supporting).
Supporting information
Fig S1
Table S1
Table S2
ACKNOWLEDGMENTS
This work was financially supported by the German Federal Ministry of Food and Agriculture (BMEL) through the Federal Office for Agriculture and Food (BLE), grant number 2810BM018 and 2810BM025. We are grateful to Heike Wolf for her technical assistance and all further employees of the state‐owned enterprise Sachsenforst who contributed to this study. Francis Ofere Emeriewen is acknowledged for proofreading of the manuscript as a native speaker. We further thank Henryk Flachowsky for his helpful comments. Open access funding enabled and organized by Projekt DEAL.
Reim S, Lochschmidt F, Proft A, Höfer M. Genetic integrity is still maintained in natural populations of the indigenous wild apple species Malus sylvestris (Mill.) in Saxony as demonstrated with nuclear SSR and chloroplast DNA markers. Ecol Evol 2020;10:11798–11809. 10.1002/ece3.6818
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in Dryad at https://doi.org/10.5061/dryad.g79cnp5n2
REFERENCES
- Albaladejo, R. G. , Guzman, B. , Gonzalez‐Martinez, S. C. , & Aparicio, A. (2012). Extensive pollen flow but few pollen donors and high reproductive variance in an extremely fragmented landscape. PLoS One, 7(11), e49012 10.1371/journal.pone.0049012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf, F. W. , Leary, R. F. , Spruell, P. , & Wenburg, J. K. (2001). The problems with hybrids: Setting conservation guidelines. Trends in Ecology & Evolution, 16(11), 613–622. 10.1016/S0169-5347(01)02290-X [DOI] [Google Scholar]
- Bacles, C. F. E. , & Jump, A. S. (2011). Taking a tree's perspective on forest fragmentation genetics. Trends in Plant Science, 16(1), 13–18. 10.1016/j.tplants.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Bitz, L. , Heinonen, M. , Moisander, S. , Tanhuanpää, P. , & Sarvarinne, J. (2019). Differential allelic richness between Malus sylvestris l. and malus × domestica borkh. From finland as a measure of genetic loss. Sustainability, 11(24), 6949 10.3390/su11246949 [DOI] [Google Scholar]
- Castiglione, S. , Cicatelli, A. , Lupi, R. , Patrignani, G. , Fossati, T. , Brundu, G. , Sabatti, M. , van Loo, M. , & Lexer, C. (2010). Genetic structure and introgression in riparian populations of Populus alba L. Plant Biosystems, 144(3), 656–668. [Google Scholar]
- Coart, E. , Van Glabeke, S. , De Loose, M. , Larsen, A. S. , & Roldan‐Ruiz, I. (2006). Chloroplast diversity in the genus Malus: New insights into the relationship between the European wild apple (Malus sylvestris (L.) Mill.) and the domesticated apple (Malus domestica Borkh.). Molecular Ecology, 15(8), 2171–2182. 10.1111/j.1365-294X.2006.02924.x [DOI] [PubMed] [Google Scholar]
- Coart, E. , Vekemans, X. , Smulders, M. J. M. , Wagner, I. , Van Huylenbroeck, J. , Van Bockstaele, E. , & Roldan‐Ruiz, I. (2003). Genetic variation in the endangered wild apple (Malus sylvestris (L.) Mill.) in Belgium as revealed by amplified fragment length polymorphism and microsatellite markers. Molecular Ecology, 12(4), 845–857. 10.1046/j.1365-294X.2003.01778.x [DOI] [PubMed] [Google Scholar]
- Coelho, N. H. P. , Tambarussi, E. V. , Aguiar, B. I. , Roque, R. H. , Portela, R. M. , Braga, R. C. , Sanson, D. , Silva, R. A. R. , Ferraz, E. M. , Moreno, M. A. , Kageyama, P. Y. , & Gandara, F. B. (2018). Understanding genetic diversity, spatial genetic structure, and mating system through microsatellite markers for the conservation and sustainable use of Acrocomia aculeata (Jacq.) Lodd. Ex Mart. Conservation Genetics, 19(4), 879–891. 10.1007/s10592-018-1061-z [DOI] [Google Scholar]
- Cornille, A. , Gladieux, P. , & Giraud, T. (2013). Crop‐to‐wild gene flow and spatial genetic structure in the closest wild relatives of the cultivated apple. Evolutionary Applications, 6(5), 737–748. 10.1111/eva.12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornille, A. , Gladieux, P. , Smulders, M. J. M. , Roldan‐Ruiz, I. , Laurens, F. , Le Cam, B. , Nersesyan, A. , Clavel, J. , Olonova, M. , Feugey, L. , Gabrielyan, I. , Zhang, X. G. , Tenaillon, M. I. , & Giraud, T. (2012). New insight into the history of domesticated apple: Secondary contribution of the european wild apple to the genome of cultivated varieties. PLoS Genetics, 8(5), e1002703 10.1371/journal.pgen.1002703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currat, M. , Ruedi, M. , Petit, R. J. , & Excoffier, L. (2008). The hidden side of invasions: Massive introgression by local genes. Evolution, 62(8), 1908–1920. 10.1111/j.1558-5646.2008.00413.x [DOI] [PubMed] [Google Scholar]
- Duan, N. B. , Bai, Y. , Sun, H. H. , Wang, N. , Ma, Y. M. , Li, M. J. , Wang, X. , Jiao, C. , Legall, N. , Mao, L. Y. , Wan, S. B. , Wang, K. , He, T. M. , Feng, S. Q. , Zhang, Z. Y. , Mao, Z. Q. , Shen, X. , Chen, X. L. , Jiang, Y. M. , & Chen, X. S. (2017). Genome re‐sequencing reveals the history of apple and supports a two‐stage model for fruit enlargement. Nature Communications, 8 10.1038/s41467-017-00336-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl, D. A. , & Vonholdt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4(2), 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Ennos, R. A. (1994). Estimating the relative rates of pollen and seed migration among plant‐populations. Heredity, 72, 250–259. 10.1038/hdy.1994.35 [DOI] [Google Scholar]
- Evanno, G. , Regnaut, S. , & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14(8), 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Fan, L. , Qin, L. , Yan, J. , Mo, C. Y. , Rong, C. X. , Meng, Y. , & Zhang, M. R. (2019). The complete chloroplast genome sequence of Malus prattii (Rosaceae) and its phylogenetic analysis. Mitochondrial DNA B, 4(2), 2171–2172. 10.1080/23802359.2019.1623732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurtey, A. , Cornille, A. , Shykoff, J. A. , Snirc, A. , & Giraud, T. (2017). Crop‐to‐wild gene flow and its fitness consequences for a wild fruit tree: Towards a comprehensive conservation strategy of the wild apple in Europe. Evolutionary Applications, 10(2), 180–188. 10.1111/eva.12441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogelqvist, J. , Verkhozina, A. V. , Katyshev, A. I. , Pucholt, P. , Dixelius, C. , Ronnberg‐Wastljung, A. C. , Lascoux, M. , & Berlin, S. (2015). Genetic and morphological evidence for introgression between three species of willows. BMC Evolutionary Biology, 15, 193 10.1186/s12862-015-0461-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Varo, J. P. , Diaz‐Garcia, S. , Arroyo, J. M. , & Jordano, P. (2019). Seed dispersal by dispersing juvenile animals: A source of functional connectivity in fragmented landscapes. Biology Letters, 15(7), 20190264 10.1098/rsbl.2019.0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke, M.‐V. , Flachowsky, H. , Peil, A. , & Emeriewen, O. (2020). Malus × domestica apple In: Litz R.‐E.‐P.‐A.‐, F., Ignacio Hormaza J. (ed.), Biotechnology of fruit and nut crops (pp. 440–473). CAB eBooks. [Google Scholar]
- Heinze, B. (2007). A database of PCR primers for the chloroplast genomes of higher plants. Plant Methods, 3, 3–4, 10.1186/1746-4811-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltken, A. M. , Steiner, W. , & Kleinschmit, J. R. G. (2014). Species integrity and spatial genetic structures of the European crab apple (Malus sylvestris (L.) MILL.). Allgemeine Forst Und Jagdzeitung, 185(11–12), 285–296. [Google Scholar]
- Kaczmarczyk, D. (2019). Techniques based on the polymorphism of microsatellite DNA as tools for conservation of endangered populations. Applied Ecology and Environmental Research, 17(2), 1599–1615. 10.15666/aeer/1702_15991615 [DOI] [Google Scholar]
- Kalinowki, S. (2005). hp‐rare 1.0: A computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Notes, 5(1), 187–189. 10.1111/j.1471-8286.2004.00845.x [DOI] [Google Scholar]
- Khadivi‐Khub, A. , Jahangirzadeh, S. , Ahadi, E. , & Aliyoun, S. (2014). Nuclear and chloroplast DNA variability and phylogeny of Iranian apples (Malus domestica). Plant Systematics and Evolution, 300(8), 1803–1817. 10.1007/s00606-014-1007-y [DOI] [Google Scholar]
- Koopman, W. J. M. , Li, Y. H. , Coart, E. , De Weg, E. V. , Vosman, B. , Roldan‐Ruiz, I. , & Smulders, M. J. M. (2007). Linked vs. unlinked markers: Multilocus microsatellite haplotype‐sharing as a tool to estimate gene flow and introgression. Molecular Ecology, 16(2), 243–256. 10.1111/j.1365-294X.2006.03137.x [DOI] [PubMed] [Google Scholar]
- Korban, S. S. (1986). Interspecific Hybridization in Malus. HortScience, 21(1), 41–48. [Google Scholar]
- Korotkova, N. , Nauheimer, L. , Ter‐Voskanyan, H. , Allgaier, M. , & Borsch, T. (2014). Variability among the most rapidly evolving plastid genomic regions is lineage‐specific: Implications of pairwise genome comparisons in pyrus (rosaceae) and other angiosperms for marker choice. PLoS One, 9(11), e112998 10.1371/journal.pone.0112998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, A. T. , Ison, J. L. , Ashley, M. V. , & Howe, H. F. (2008). The paradox of forest fragmentation genetics. Conservation Biology, 22(4), 878–885. 10.1111/j.1523-1739.2008.00944.x [DOI] [PubMed] [Google Scholar]
- Larsen, A. S. , Asmussen, C. B. , Coart, E. , Olrik, D. C. , & Kjaer, E. D. (2006). Hybridization and genetic variation in Danish populations of European crab apple (Malus sylvestris). Tree Genetics & Genomes, 2(2), 86–97. 10.1007/s11295-005-0030-0 [DOI] [Google Scholar]
- Lea, M. V. , Syring, J. , Jennings, T. , Cronn, R. , Bruederle, L. P. , Neale, J. R. , & Tomback, D. F. (2018). Development of nuclear microsatellite loci for Pinus albicaulis Engelm. (Pinaceae), a conifer of conservation concern. PLoS One, 13(10), e0205423 10.1371/journal.pone.0205423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebhard, R. , Gianfranceschi, L. , Koller, B. , Ryder, C. D. , Tarchini, R. , Van de Weg, E. , & Gessler, C. (2002). Development and characterisation of 140 new microsatellites in apple (Malus x domestica Borkh.). Molecular Breeding, 10(4), 217–241. 10.1023/A:1020525906332 [DOI] [Google Scholar]
- Lowe, A. J. , Cavers, S. , Boshier, D. , Breed, M. F. , & Hollingsworth, P. M. (2016). The resilience of forest fragmentation genetics‐no longer a paradox‐we were just looking in the wrong place (vol 115, pg 97, 2015). Heredity, 116(3), 348 10.1038/hdy.2015.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel, N. (1967). Adaptation of Karber's method for estimating the exponential parameter from quantal data, and its relationship to birth, death, and branching processes. Biometrics, 23(4), 739–746. 10.2307/2528425 [DOI] [PubMed] [Google Scholar]
- Nevill, P. G. , Despres, T. , Bayly, M. J. , Bossinger, G. , & Ades, P. K. (2014). Shared phylogeographic patterns and widespread chloroplast haplotype sharing in Eucalyptus species with different ecological tolerances. Tree Genetics & Genomes, 10(4), 1079–1092. 10.1007/s11295-014-0744-y [DOI] [Google Scholar]
- Nikiforova, S. V. , Cavalieri, D. , Velasco, R. , & Goremykin, V. (2013). Phylogenetic analysis of 47 chloroplast genomes clarifies the contribution of wild species to the domesticated apple maternal line. Molecular Biology and Evolution, 30(8), 1751–1760. 10.1093/molbev/mst092 [DOI] [PubMed] [Google Scholar]
- Ozawa, H. , Watanabe, A. , Uchiyama, K. , Saito, Y. , & Ide, Y. (2013). Influence of long‐distance seed dispersal on the genetic diversity of seed rain in fragmented Pinus densiflora populations relative to pollen‐mediated gene flow. Journal of Heredity, 104(4), 465–475. 10.1093/jhered/est022 [DOI] [PubMed] [Google Scholar]
- Palme, A. E. , Su, Q. , Palsson, S. , & Lascoux, M. (2004). Extensive sharing of chloroplast haplotypes among European birches indicates hybridization among Betula pendula . B‐pubescens and B‐nana. Mol Ecol, 13(1), 167–178. 10.1046/j.1365-294X.2003.02034.x [DOI] [PubMed] [Google Scholar]
- Pierce, A. A. , Gutierrez, R. , Rice, A. M. , & Pfennig, K. S. (2017). Genetic variation during range expansion: effects of habitat novelty and hybridization. Proceedings of the Royal Society B: Biological Sciences, 284(1852), 20170007 10.1098/rspb.2017.0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plue, J. , & Cousins, S. A. O. (2018). Seed dispersal in both space and time is necessary for plant diversity maintenance in fragmented landscapes. Oikos, 127(6), 780–791. 10.1111/oik.04813 [DOI] [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155(2), 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim, S. , Holtken, A. , & Hofer, M. (2013). Diversity of the European indigenous wild apple (Malus sylvestris (L.) Mill.) in the East Ore Mountains (Osterzgebirge), Germany: II. Genetic characterization. Genetic Resources and Crop Evolution, 60(3), 879–892. 10.1007/s10722-012-9885-8 [DOI] [Google Scholar]
- Reim, S. , Proft, A. , Heinz, S. , & Höfer, M. (2012). Diversity of the European indigenous wild apple Malus sylvestris (L.) Mill. in the East Ore Mountains (Osterzgebirge), Germany: I. Morphological characterization. Genetic Resources and Crop Evolution, 59(6), 1101–1114. 10.1007/s10722-011-9746-x [DOI] [Google Scholar]
- Reim, S. , Proft, A. , Heinz, S. , Lochschmidt, F. , Hofer, M. , Trober, U. , & Wolf, H. (2017). Pollen movement in a Malus sylvestris population and conclusions for conservation measures. Plant Genetic Resource‐C, 15(1), 12–20. 10.1017/S1479262115000301 [DOI] [Google Scholar]
- Robinson, J. P. , Harris, S. A. , & Juniper, B. E. (2001). Taxonomy of the genus Malus Mill. (Rosaceae) with emphasis on the cultivated apple, Malus domestica Borkh. Plant Systematics and Evolution, 226(1–2), 35–58. 10.1007/s006060170072 [DOI] [Google Scholar]
- Ruhsam, M. , Jessop, W. , Cornille, A. , Renny, J. , & Worrell, R. (2019). Crop‐to‐wild introgression in the European wild apple Malus sylvestris in Northern Britain. Forestry, 92(1), 85–96. 10.1093/forestry/cpy033 [DOI] [Google Scholar]
- Savolainen, V. , Corbaz, R. , Moncousin, C. , Spichiger, R. , & Manen, J. F. (1995). Chloroplast DNA variation and parentage analysis in 55 apples. Theoretical and Applied Genetics, 90(7–8), 1138–1141. 10.1007/Bf00222934 [DOI] [PubMed] [Google Scholar]
- Schnitzler, A. , Arnold, C. , Cornille, A. , Bachmann, O. , & Schnitzler, C. (2014). Wild European apple (Malus sylvestris (l.) mill.) population dynamics: Insight from genetics and ecology in the rhine valley. Priorities for a future conservation programme. PLoS One, 9(5), e96596 10.1371/journal.pone.0096596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuka, T. , Mori, N. , Ozkan, H. , & Ohta, S. (2015). Chloroplast DNA haplotype variation within two natural populations of wild emmer wheat (Triticum turgidum ssp. dicoccoides) in southern Turkey. Biotechnology & Biotechnological Equipment, 29(3), 423–430. 10.1080/13102818.2015.1012648 [DOI] [Google Scholar]
- Spengler, R. N. (2019). Origins of the apple: The role of megafaunal mutualism in the domestication of malus and rosaceous trees. Frontiers in Plant Science, 10, 617. 10.3389/fpls.2019.00617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech, Z. A. , Jakobsson, M. , & Rosenberg, N. A. (2008). ADZE: A rarefaction approach for counting alleles private to combinations of populations. Bioinformatics, 24(21), 2498–2504. 10.1093/bioinformatics/btn478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, L. , Li, J. , Tan, S. , Li, M. X. , Ma, X. , & Zhou, Z. Q. (2014). New insights into the hybrid origin of Malus toringoides and its close relatives based on a single‐copy nuclear gene SbeI and three chloroplast fragments. Journal of Systematics and Evolution, 52(4), 477–486. 10.1111/jse.12079 [DOI] [Google Scholar]
- Velasco, R. , Zharkikh, A. , Affourtit, J. , Dhingra, A. , Cestaro, A. , Kalyanaraman, A. , Fontana, P. , Bhatnagar, S. K. , Troggio, M. , Pruss, D. , Salvi, S. , Pindo, M. , Baldi, P. , Castelletti, S. , Cavaiuolo, M. , Coppola, G. , Costa, F. , Cova, V. , Dal Ri, A. , … Viola, R. (2010). The genome of the domesticated apple (Malus x domestica Borkh.). Nature Genetics 42(10), 833 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- Volk, G. M. , Henk, A. D. , Baldo, A. , Fazio, G. , Chao, C. T. , & Richards, C. M. (2015). Chloroplast heterogeneity and historical admixture within the genus Malus. American Journal of Botany, 102(7), 1198–1208. 10.3732/ajb.1500095 [DOI] [PubMed] [Google Scholar]
- Wagner, I. (1996). Zusammenstellung morphologischer Merkmale und ihrer Ausprägung zur Unterscheidung von Wild‐ und Kulturformen des Apfels (Malus) und des Birnbaumes (Pyrus). Mitt Deut Dendr Gesells. Mitteilungen Der Deutschen Dendrologischen Gesellschaft, 82, 87–108. [Google Scholar]
- Wagner, I. , Maurer, W. D. , Lemmen, P. , Schmitt, H. P. , Wagner, M. , Binder, M. , & Patzak, P. (2014). Hybridization and genetic diversity in wild apple (Malus sylvestris (l.) Mill) from various regions in Germany and from Luxembourg. Silvae Genetica, 63(3), 81–94. 10.1515/sg-2014-0012 [DOI] [Google Scholar]
- Wang, J. , Kang, M. , & Huang, H. W. (2014). Long‐distance pollen dispersal ensures genetic connectivity of the low‐density tree species, Eurycorymbus cavaleriei, in a fragmented karst forest landscape. Conservation Genetics, 15(5), 1163–1172. 10.1007/s10592-014-0608-x [DOI] [Google Scholar]
- Xu, Y. F. , Zhao, Y. J. , Zhao, X. Q. , Chen, X. S. , & Yuan, Z. H. (2019). Characterization of complete chloroplast genome of Malus sylvestris L. Mitochondrial DNA B, 4(2), 2357–2358. 10.1080/23802359.2019.1629352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X. Y. , Cai, D. Y. , Potter, D. , Postman, J. , Liu, J. , & Teng, Y. W. (2014). Phylogeny and evolutionary histories of Pyrus L. revealed by phylogenetic trees and networks based on data from multiple DNA sequences. Molecular Phylogenetics and Evolution, 80, 54–65. 10.1016/j.ympev.2014.07.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Table S1
Table S2
Data Availability Statement
The data that support the findings of this study are openly available in Dryad at https://doi.org/10.5061/dryad.g79cnp5n2


