Abstract
Objective
We investigated the proportions of and reclassified BRCA1/2 variants of unknown significance (VUS) in Korean patients with epithelial ovarian, tubal, and primary peritoneal cancers.
Methods
Data from 805 patients who underwent genetic testing for BRCA1/2 from January 1, 2006 to August 31, 2018 were included. The VUS in BRCA1/2 were reclassified using the 2015 American College of Medical Genetics and Genomics and the Association for Molecular Pathology standards and guidelines.
Results
A BRCA1 pathogenic variant was found in 17.0% (137/805) of the patients, and BRCA1 VUS were found in 15.9% (128/805) of the patients. Further, 8.7% (69/805) of the patients possessed a BRCA2 pathogenic variant and 18.4% (148/805) of the patients possessed BRCA2 VUS. Fifty-three specific BRCA1 VUS were found and 20 were further reclassified as benign (n=11), likely benign (n=5), likely pathogenic (n=3), and pathogenic (n=1). The remaining 33 remained classified as VUS. For BRCA2, 55 specific VUS were detected; among these, 14 were reclassified as benign or likely benign, and 2 were reclassified as likely pathogenic. Among the 805 patients, 195 were found to have only VUS and no pathogenic variants (PV), and 41.5% (81/195) were reclassified as benign or likely benign, and 10.3% (20/195) as pathogenic or likely pathogenic variants.
Conclusions
Approximately 33.3% (36/108) of the specific BRCA1/2 variants analyzed in this study that were initially classified as VUS over a 13-year period were reclassified. Among these, 5.6% (6/108) were reclassified as pathogenic or likely pathogenic variants.
Keywords: Genes, BRCA1; Genes, BRCA2; Ovarian Neoplasms; Genetic Testing
INTRODUCTION
Ovarian cancer is often deadly and is ranked second in terms of mortality and third in terms of incidence among gynecological malignancies worldwide [1]. Particularly in Korea, where the prevalence of ovarian cancer is increasing, it is already the second most common gynecological malignancy [2]. Ovarian carcinoma is the most common hereditary gynecological cancer, and BRCA1 and BRCA2 are associated with most hereditary ovarian cancers. An inherited disposition is found in 10% of ovarian cancers [3]. The average cumulative risk for developing ovarian cancer for BRCA1-mutation carriers by the age of 70 years is 39% (95% confidence interval=18%–54%), and the corresponding estimate for BRCA2 is 11% (2.4%–19%) [4].
Accurate risk assessment for potential ovarian cancer cases is vital for cancer prevention, and a suitable management program for the proband in each family is important. Thus, detection of BRCA1/2 mutation is an indispensable part of ovarian cancer management. Therefore, some study groups, such as the National Comprehensive Cancer Network, recommend that patients with ovarian cancer in their family should have a genetic risk evaluation and BRCA1/2 testing [5].
Genetic testing categorizes ovarian cancer into three main types: benign, pathologic, or uncertain clinical significance; each type has various clinical implications. BRCA1/2 variants which have not been established or ruled out as a disease risk are referred to as “variants of unknown significance” (VUS) [6]. The field of genetic testing is rapidly expanding, and interpretation of results is a greater challenge now that a larger number of genetic VUS have been isolated [7]. In, South Korea, which is a unitary country, approximately 3.9%–24.6% of patients tested for BRCA1/2 possessed one or more VUS, primarily in breast cancer patients [8,9,10,11,12]. Although BRCA1 and BRCA2 gene testing plays a pivotal role in selecting treatment options for patients with hereditary gynecological cancer, VUS therapeutic decision-making faces constraints. Moreover, the clinical significance of each VUS has not yet been reported.
Although a substantial number of studies have been conducted for VUS re-classification and multifarious tools, such as in silico tests, have been utilized to confirm the pathogenicity of VUS, standardized methods have not yet been established. In this study, we aimed to elucidate BRCA1/2 VUS in gynecological cancer patients and reclassify them using the 2015 American College of Medical Genetics and Genomics and the Association for Molecular Pathology standards and guidelines (ACMG/AMP 2015 guidelines).
MATERIALS and METHODS
1. Subjects
Medical records, including genetic testing of patients with gynecological cancer, were retrospectively reviewed between January 2006 and August 2018. A total of 870 patients were tested, and their BRCA1 or BRCA2 status was recorded.
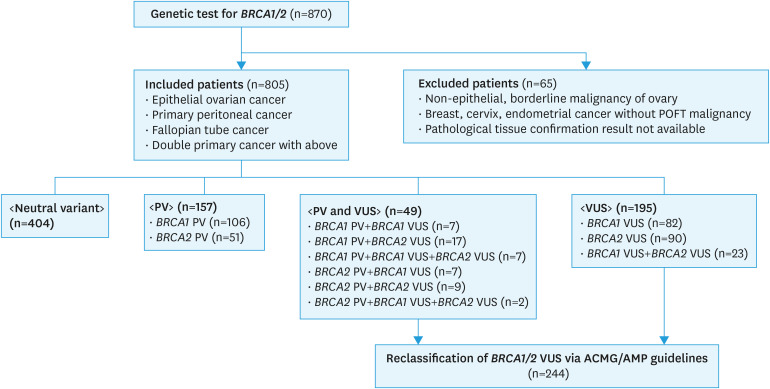
All patients with epithelial ovarian cancer, fallopian tube cancer, and primary peritoneal carcinoma were included, while patients with borderline malignancy of ovarian tumor, non-epithelial ovarian cancer, and breast cancer without gynecological malignancy were excluded (Fig. 1). However, a familial mutational test for the member proband with gynecological cancer was not incorporated into this analysis.
Fig. 1. Flow chart for reclassification of BRCA1/2 VUS via the ACMG/AMP 2015 guidelines.
ACMG/AMP, American College of Medical Genetics and Genomics and the Association for Molecular Pathology; POFT, peritoneal, ovarian, and fallopian tube; PV, pathogenic variants; VUS, variants of unknown significance.
This study adhered to the ethical tenets of the Declaration of Helsinki and was approved by the Institutional Review Board (IRB) of the National Cancer Center in Goyang, Korea (IRB number: 2018-0259). The need for informed consent was waived because of the low risk designated by the IRB.
2. BRCA1/2 sequencing
Germline or somatic mutations of BRCA1/2 were tested by Sanger sequencing or by next generation sequencing (NGS). Briefly, genomic DNA was extracted from the peripheral blood of participants using a Chemagic DNA Blood 200 Kit (Chemagen, Baesweiler, Germany) or a QIAamp DNA Blood Mini kit (Qiagen, Valencia, CA, USA). Amplified PCR products were sequenced on an ABI 3500xl analyzer (Applied Biosystems, Foster City, CA, USA), using the BigDye Terminator v3.1 Cycle Sequencing Kit. Sequences were analyzed using Sequencer v5.0 software. NGS was done on the Illumina MiSeqDX (Illumina Inc., San Diego, CA, USA) generating 2×150 bp reads. Alignment was conducted with BWA-MEM (version 0.7.10), duplicated reads were marked with Picard (version 1.138) and local alignment, base quality recalibration, and variant calling were performed with the Genome Analysis Tool kit (GATK, version 3.5), SAMtools (version 0.1.19), FreeBayes (v0.9.21-26-gbfd9832) and Scalpel (version 0.5.3). Variant annotation was conducted with a Variant Effect Predictor and dbNSFP. All genetic variants were determined according to the Breast Cancer Information Core database and both BIC database and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) database were used from the 2014 onwards. All variants were described according to HUGO-approved systematic nomenclature.
3. Reclassification of VUS
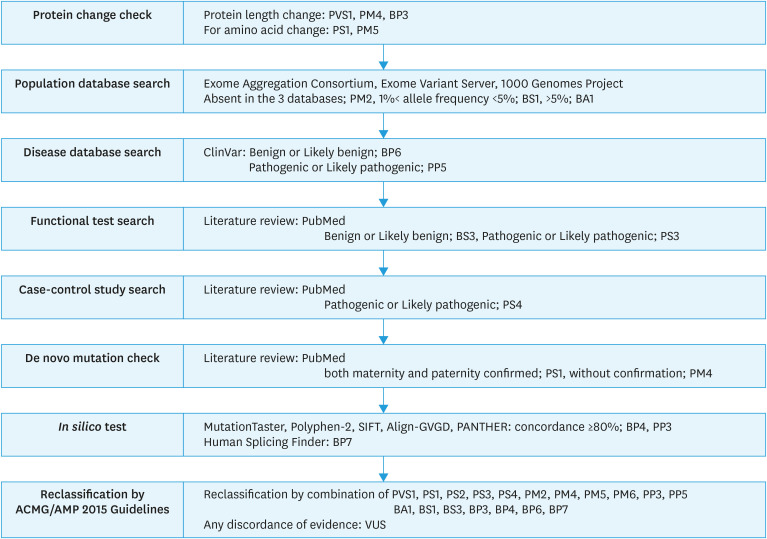
According to the ACMG/AMP 2015 guidelines, variant pathogenicity was reviewed (Fig. 2). The detailed methods used in this study were described in Supplementary Tables 1 and 2.
Fig. 2. Flow chart for describing patient distribution in the present study.
BA, benign stand-alone evidence; BP, benign supporting evidence; BS, benign strong evidence; PM, pathogenic moderate evidence; PP, pathogenic supporting evidence; PS, pathogenic strong evidence; PV, pathogenic variants evidence; PVS, pathogenic very strong evidence; VUS, variants of unknown significance.
The variant pathogenicity in the absence of controls (or at an extremely low frequency, if recessive) was determined using the Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium was assigned pathogenic moderate (PM) evidence 2. Databases were used for benign strong evidence 1 (BS1) when the allele frequency was greater than 1% and less than 5%.
For pathogenic strong evidence 4 (PS4), we conducted a literature survey and looked up information from well-established previous case-control studies. If association analysis showed OR >5 and its confidential interval >1, according to the ACMG/AMP guidelines, we regarded the results as PS4 (Supplementary Table 1).
For benign supporting (BP) evidence 6 or pathogenic supporting (PP) evidence 5, we used the ClinVar database as a source. The database uses a five-tier system, “pathogenic,” “likely pathogenic,” “uncertain significance,” “conflicting,” “likely benign,” and “benign.” We decided to classify “pathogenic” and “likely pathogenic” as evidence of PP5, and “likely benign” and “benign” as evidence of BP6.
BP4 and PP3 required multiple lines of computational evidence to support any effect on the gene or gene product; thus, we used MutationTaster, Polyphen-2, SIFT, Align-GVGD, and PANTHER. If more than 4 out of 5 of the results matched, we regarded the results as BP4 or PP3. Human Splicing Finder was used for splicing variants of BP7. We conducted a literature survey for information from previous functional studies on results regarding PP3 and BP4.
Finally, we reclassified the patients with reclassified VUS. If they have multiple VUS, we assigned the status which is more pathogenic. The detailed methods used in this study were described in Supplementary Tables 1 and 2.
RESULTS
A total of 870 patients were tested for BRCA1/BRCA2 variants, regardless of the type of cancer. A total of 805 patients were included in this study, which was conducted between January 1, 2006 and August 31, 2018. Among the 805 patients, the Sanger method was used for 647 patients. In addition, next-generation sequencing (NGS) was used for 190 patients: 137 germline, 54 somatic, and 1 both germline and somatic NGS. Thirty-two patients were tested utilizing the Sanger method and NGS simultaneously. Five among these were tested via germline NGS and 27 via somatic NGS (Supplementary Table 3).
Among the 805 patients, 751 patients exhibited epithelial ovarian cancer, 12 primary peritoneal cancer, and 42 fallopian tube cancer. Patient characteristics, Histologic subtype, and surgical stages are depicted in Supplementary Tables 4, 7, 8, 9.
Total 97 BRCA1 or BRCA2 specific pathogenic variants (59 BRCA1 PV and 38 BRCA2 PV) were detected and 108 BRCA1 or BRCA2 specific VUS (53 BRCA1 VUS and 55 BRCA2 VUS) from the included 805 patients after removing duplicates (Supplementary Table 5). Among the 805 patients, 50.2% (404/805) exhibited only neutral variants that were neither pathogenic nor VUS. 25.6% (206/805) of patients were found to possess a pathogenic variant, 30.3% (244/805) VUS, and 6.1% (49/805) both PV and VUS simultaneously. In the BRCA1 gene, 17.0% (137/805) of patients exhibited PV, 15.9% (128/805) VUS, and 1.7% (14/805) both variants. Further, in the BRCA2 gene, 8.6% (69/805) of patients exhibited PV, 18.4% (148/805) VUS, and 1.4% (11/805) both variants. No patients were found to have BRCA1 PV, BRCA1 VUS, BRCA2 PV, and BRCA2 VUS simultaneously (Fig. 1).
1. Reclassification of specific BRCA1 and BRCA2 “VUS”
In total, of the specific BRCA1 or BRCA2 VUS that were reclassified, 5.6% (6/108) were reclassified as pathogenic/likely pathogenic and 27.8% (30/108) were reclassified as benign/likely benign. 66.7% (72/108) of the specific BRCA1/2 VUS were not reclassified.
The BRCA1 VUS consisted of 25 missense, 10 synonymous, 17 intronic, and 1 in-frame deletion variants. The frequencies of the specific BRCA1 VUS ranged from 0.12% (1/805) to 2.9% (23/805). Using the ACMG/AMP 2015 guidelines, 11 specific VUS were reclassified as benign, 5 as likely benign, 3 as likely pathogenic (c.5215G>C, c.5363G>T, and c.5017_5019delCAC), and 1 as pathogenic (c.5339T>C). Thirty-three specific VUS were not reclassified (Table 1, Supplementary Fig. 1).
Table 1. Reclassification of variants of unknown significance in BRCA1 via the ACMG/AMP 2015 guidelines.
| Exon | HGVSc (NM_07294.3) | HGVSp | No. of variants (%) | Allele frequency | ClinVar | Functional study/case control study | Type of evidence | Re-classification | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 1000G (EAS) | ExAC (East Asian) | ESP | ||||||||
| 5 | c.154C>T | p.Leu52Phe | 12 (1.49) | OV | 0.001752 | - | CIP | - | PP3 | VUS |
| 6 | c.220C>A | p.Gln74Lys | 1 (0.12) | - | - | - | VUS | - | PM2 | VUS |
| 6 | c.288C>A | p.Asp96Glu | 1 (0.12) | - | - | - | - | - | PM2, PP3 | VUS |
| 7 | c.427G>A | p.Glu143Lys | 1 (0.12) | OV | 0.000578 | - | B | - | BP6, BP4 | LB |
| 10 | c.626C>T | p.Pro209Leu | 1 (0.12) | 0.001 | - | - | VUS | - | BP4 | VUS |
| 11 | c.756T>C | p.Arg252= | 1 (0.12) | - | - | - | LB | - | PM2, BP6 | VUS |
| 11 | c.795T>C | p.Ser265= | 4 (0.49) | 0.001 | - | - | B | - | BP6 | VUS |
| 11 | c.811G>A | p.Val271Met | 1 (0.12) | 0.001 | 0.001387 | - | CIP | Not deleterious [13] | BS3, BP4 | LB |
| 11 | c.824G>A | p.Gly275Asp | 2 (0.24) | OV | - | - | CIP | - | PM2 | VUS |
| 11 | c.1251T>C | p.Asn417= | 3 (0.37) | - | - | - | LB | - | PM2, BP6, BP4 | VUS |
| 11 | c.1676G>C | p.Gly559Ala | 1 (0.12) | - | - | - | - | - | PM2 | VUS |
| 11 | c.1930T>A | p.Cys644Ser | 1 (0.12) | - | - | - | VUS | - | PM2 | VUS |
| 11 | c.2082C>T | p.Ser694= | 23 (2.85) | - | 0.3783 | - | B | - | BA1, BP4, BP6 | B |
| 11 | c.2171C>T | p.Pro724Leu | 1 (0.12) | - | - | - | VUS | - | PM2 | VUS |
| 11 | c.2311T>C | p.Leu771= | 21 (2.60) | 0.371 | 0.3785 | - | B | - | BA1, BP4, BP6 | B |
| 11 | c.2481A>C | p.Glu827Asp | 1 (0.12) | OV | - | - | CIP | - | PM2 | VUS |
| 11 | c.2566T>C | p.Tyr856His | 20 (2.48) | 0.014 | 0.0208 | 0.00008 | B | Not deleterious [13] | BS1, BS3, BP6 | B |
| 11 | c.2726A>T | p.Asn909Ile | 3 (0.37) | OV | 0.0006935 | - | CIP | - | - | VUS |
| 11 | c.3524C>T | p.Ala1175Val | 1 (0.12) | - | - | - | VUS | - | PM2, PP3 | VUS |
| 11 | c.3747C>T | p.Thr1249= | 1 (0.12) | - | - | - | LB | - | PM2, BP6, BP4 | VUS |
| 11 | c.3975G>A | p.Arg1325= | 1 (0.12) | - | - | - | LB | - | PM2, BP6, BP4 | VUS |
| 12 | c.4158C>T | p.Ser1386= | 1 (0.12) | - | - | - | - | - | PM2 | VUS |
| 13 | c.4308T>C | p.Ser1436= | 18 (2.36) | 0.371 | 0.3777 | 0.27956 | B | - | BA1, BP4, BP6 | B |
| 14 | c.4422T>C | p.Ala1474= | 1 (0.12) | - | - | - | LB | - | PM2, BP6 | VUS |
| 16 | c.4729T>C | p.Ser1577Pro | 3 (0.37) | OV | 0.0002313 | - | CIP | - | BP4 | VUS |
| 16 | c.4883T>C | p.Met1628Thr | 20 (2.48) | 0.012 | 0.006355 | - | B | Not deleterious [14] | BS1, BS3, BP4, BP6 | B |
| 16 | c.4985T>C | p.Phe1662Ser | 1 (0.12) | OV | 0.0001156 | - | B | Not deleterious [15] | BS3, BP4, BP6 | LB |
| 17 | c.5017_5019delCAC | p.His1673del | 3 (0.37) | - | - | - | CIP | - | PS4 [16], PM4 | LP |
| 17 | c.5068A>C | p.Lys1690Gln | 1 (0.12) | OV | 0.000579 | - | VUS | - | - | VUS |
| 19 | c.5165C>A | p.Ser1722Tyr | 1 (0.12) | OV | - | - | VUS | - | - | VUS |
| 20 | c.5215G>C | p.Asp1739His | 1 (0.12) | OV | - | - | VUS | Deleterious [15] | PS3, PM5, PP3 | LP |
| 20 | c.5254G>A | p.Ala1752Thr | 1 (0.12) | OV | - | - | VUS | - | PM5 | VUS |
| 22 | c.5339T>C | p.Leu1780Pro | 10 (1.24) | OV | - | - | CIP | Deleterious [15] | PP3, PS3, PS4 [6] | P |
| 22 | c.5363G>T | p.Gly1788Val | 1 (0.12) | OV | 0 | - | P | Deleterious [15] | PP5, PM5, PS3 | LP |
| 22 | c.5372T>A | p.Val1791Glu | 1 (0.12) | - | - | - | - | - | PM2 | VUS |
| 24 | c.5516T>C | p.Leu1839Ser | 1 (0.12) | OV | - | - | VUS | - | PP3 | VUS |
| IVS1 | c.-19-115T>C | - | 1 (0.12) | 0.371 | - | - | B | - | BA1, BP6 | B |
| IVS2 | c.81-9C>G | - | 1 (0.12) | - | - | - | VUS | - | PM2, PP3 | VUS |
| IVS5 | c.212+11T>C | - | 1 (0.12) | - | - | - | - | - | PM2, BP7 | VUS |
| IVS6 | c.213-161A>G | - | 4 (0.49) | 0.371 | - | - | B | - | BA1, BP6, BP7 | B |
| IVS7 | c.442-18C>A | - | 1 (0.12) | - | - | - | - | - | PM2, BP7 | VUS |
| IVS8 | c.547+14delG | - | 2 (0.24) | - | - | - | CIP | Not deleterious [17] | PM2, BS3, BP7 | VUS |
| IVS8 | c.547+30A>G | - | 5 (0.62) | - | - | - | - | - | PM2, BP7 | VUS |
| IVS8 | c.547+146A>T | - | 2 (0.24) | 0.371 | - | - | B | - | BA1, BP6, BP7 | B |
| IVS9 | c.548-58delT | - | 2 (0.24) | 0.371 | - | 0.2784 | B | - | BA1, BP6, BP7 | B |
| IVS9 | c.548-64delT | - | 2 (0.24) | - | - | - | - | - | PM2, BP7 | VUS |
| IVS9 | c.594-34T>C | - | 2 (0.24) | 0.003 | 0.002439 | 0.0001 | LB | - | BP6, BP7 | LB |
| IVS11 | c.4097-141A>C | - | 2 (0.24) | 0.371 | - | - | B | - | BA1, BP6, BP7 | B |
| IVS11 | c.4097-38C>G | - | 1 (0.12) | - | - | - | - | - | PM2, BP7 | VUS |
| IVS14 | c.4484+14A>G | - | 1 (0.12) | 0.01 | 0.009722 | - | B | - | BP4, BP6, BP7 | LB |
| IVS18 | c.5152+66G>A | - | 7 (0.86) | 0.371 | - | - | B | - | BA1, BP6, BP7 | B |
| IVS22 | c.5406+1G>T | - | 1 (0.12) | - | - | - | - | - | PM2, BP7 | VUS |
| IVS23 | c.5467+10_5467+13dupCTGC | - | 1 (0.12) | - | - | - | - | - | PM2, BP7 | VUS |
1000G (EAS), minor allele frequency from East-Asian population in the 1000 Genome Phase 3 database; ACMG/AMP, American College of Medical Genetics and Genomics and the Association for Molecular Pathology; B, benign; BA, benign stand-alone evidence; BP, benign supporting evidence; BS, benign strong evidence; c, coding sequence; CIP, conflicting interpretations of pathogenicity; ESP, Exome Sequencing Project; ExAC, Exome Aggregation Consortium; HGVS, Human Genome Variation Society; IVS, intervening sequence; LB, likely benign; LP, likely pathogenic; OV, observed variant(s) without frequency or population information; P, pathogenic; p, protein sequence; PM, pathogenic moderate evidence; PP, pathogenic supporting evidence; PS, pathogenic strong evidence; PVS, pathogenic very strong evidence; VUS, variants of unknown significance.
The 55 specific BRCA2 VUS consisted of 39 missense, 7 synonymous, 8 intronic, and one in-frame deletion variants. The frequencies of these specific variants ranged from 0.12% (1/805) to 3.4% (27/805). Six specific VUS were reclassified as benign, 8 as likely benign, 2 as likely pathogenic (c.9154C>T, c.8488-1G>A). Thirty-nine specific VUS were not reclassified. An overview of these results is found in (Table 2, Supplementary Fig. 1).
Table 2. Reclassification of variants of unknown significance in BRCA2 via the ACMG/AMP 2015 guidelines.
| Exon | HGVSc (NM_000059) | HGVSp | No. of variants (%) | Allele frequency | ClinVar | Functional study/case control study | Type of evidence | Re-classification | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 1000G (EAS) | ExAC (East Asian) | ESP | ||||||||
| 2 | c.53G>A | p.Arg18His | 4 (0.49) | 0.002 | 0.0004688 | - | B | No deleterious [18] | BS3, BP6 | LB |
| 3 | c.78A>G | p.Pro26= | 1 (0.12) | - | - | - | LB | - | BP6, PM2 | VUS |
| 7 | c.623T>G | p.Val208Gly | 1 (0.12) | OV | 0.0008096 | - | VUS | - | - | VUS |
| 8 | c.673A>G | p.Thr225Ala | 1 (0.12) | OV | - | - | VUS | - | PM2, BP4 | VUS |
| 9 | c.734G>A | p.Arg245Lys | 5 (0.62) | - | - | - | - | - | BP4 | VUS |
| 10 | c.943T>A | p.Cys315Ser | 1 (0.12) | 0.008 | 0.005101 | - | CIP | - | BP4 | VUS |
| 10 | c.964A>C | p.Lys322Gln | 2 (0.24) | 0.003 | 0.0008124 | - | CIP | - | - | VUS |
| 10 | c.1738A>G | p.Ile580Val | 1 (0.12) | - | - | - | - | - | PM2 | VUS |
| 10 | c.1744A>C | p.Thr582Pro | 5 (0.62) | 0.003 | 0.003134 | - | B | Not deleterious [18] | BS3, BP6 | LB |
| 10 | c.1796C>T | p.Ser599Phe | 4 (0.49) | - | - | - | VUS | - | PM2 | VUS |
| 11 | c.2127G>C | p.Leu709= | 4 (0.49) | OV | 0.001156 | - | B | - | BP6 | VUS |
| 11 | c.2186T>C | p.Ile729Thr | 1 (0.12) | 0.002 | 0.0001156 | - | VUS | - | BP4 | VUS |
| 11 | c.2229T>C | p.His743= | 7 (0.87) | 0.095 | 0.1002 | 0.0313 | B | - | BA1, BP6 | B |
| 11 | c.2350A>G | p.Met784Val | 14 (1.74) | 0.018 | 0.004393 | - | B | - | BS1, BP4, BP6 | LB |
| 11 | c.2490C>T | p.Asn830= | 5 (0.62) | - | 0.0003468 | 0.0001 | LB | - | BP6 | VUS |
| 11 | c.2550A>G | p.Gln850= | 3 (0.37) | 0.003 | 0.002083 | - | CIP | - | - | VUS |
| 11 | c.2585A>G | p.Lys862Arg | 1 (0.12) | - | - | - | VUS | - | PM2 | VUS |
| 11 | c.2971A>G | p.Asn991Asp | 1 (0.12) | 0.097 | 0.1002 | 0.0373 | B | Not deleterious [18] | BA1, BS3, BP6 | B |
| 11 | c.3007C>G | p.His1003Asp | 1 (0.12) | - | - | - | VUS | - | PM2, BP4 | VUS |
| 11 | c.3109C>A | p.Gln1037Lys | 1 (0.12) | - | - | - | VUS | - | PM2 | VUS |
| 11 | c.3220A>T | p.Ser1074Cys | 1 (0.12) | 0.001 | 0.0001157 | - | VUS | - | - | VUS |
| 11 | c.3484G>A | p.Ala1162Thr | 1 (0.12) | - | - | - | VUS | - | PM2, BP4 | VUS |
| 11 | c.3568C>T | p.Arg1190Trp | 1 (0.12) | OV | 0 | - | B | Not deleterious [18] | BS3, BP6 | LB |
| 11 | c.4427A>G | p.Asp1476Gly | 1 (0.12) | - | - | - | VUS | - | PM2 | VUS |
| 11 | c.4649A>G | p.Glu1550Gly | 1 (0.12) | 0 | - | VUS | - | BP4 | VUS | |
| 11 | c.4854T>A | p.Asp1618Glu | 3 (0.37) | OV | 0.0001161 | - | VUS | - | BP4 | VUS |
| 11 | c.5436A>C | p.Glu1812Asp | 1 (0.12) | - | - | - | - | - | PM2, BP4 | VUS |
| 11 | c.5590G>A | p.Asp1864Asn | 1 (0.12) | - | - | - | VUS | - | PM2, BP4 | VUS |
| 11 | c.5764G>C | p.Ala1922Pro | 1 (0.12) | OV | - | - | VUS | - | BP4 | VUS |
| 11 | c.5785A>G | p.Ile1929Val | 1 (0.12) | 0.006 | 0.009827 | - | B | Not deleterious [18] | BS1, BS3, BP4, BP6 | B |
| 11 | c.5882G>A | p.Ser1961Asn | 1 (0.12) | OV | 0.00006057 | - | CIP | - | BP4 | VUS |
| 11 | c.5969A>C | p.Asp1990Ala | 1 (0.12) | 0.001 | 0.000578 | - | VUS | - | PP3 | VUS |
| 11 | c.6131G>T | p.Gly2044Val | 1 (0.12) | 0.003 | 0.0005782 | - | CIP | - | - | VUS |
| 11 | c.6325G>A | p.Val2109Ile | 6 (0.74) | 0.002 | 0.003835 | - | CIP | - | BP4 | VUS |
| 11 | c.6448A>C | p.Lys2150Gln | 1 (0.12) | - | - | - | VUS | - | PM2, BP4 | VUS |
| 14 | c.7052C>G | p.Ala2351Gly | 4 (0.49) | 0.004 | 0.001735 | - | CIP | - | - | VUS |
| 14 | c.7188G>A | p.Leu2396= | 3 (0.37) | - | - | - | LB | - | BP6, PM2 | VUS |
| 15 | c.7469T>C | p.Ile2490Thr | 1 (0.12) | 0.003 | - | 0.0016 | B | - | BP6, BP4 | LB |
| 18 | c.8187G>T | p.Lys2729Asn | 27 (3.35) | 0.012 | 0.009711 | - | B | No deleterious [18] | BS1, BS3, BP6 | B |
| 23 | c.8976A>G | p.Pro2992= | 1 (0.12) | - | - | - | - | - | PM2 | VUS |
| 24 | c.9110_9112delAAC | p.Gln3037del | 1 (0.12) | OV | - | - | VUS | - | PM4, PP3 | VUS |
| 24 | c.9154C>T | p.Arg3052Trp | 2 (0.24) | OV | 0.00006066 | - | P | Deleterious [18] | PS3, PP5, PP3 | LP |
| 24 | c.9232G>T | p.Val3078Phe | 1 (0.12) | - | - | - | - | - | PM2 | VUS |
| 26 | c.9617A>G | p.Gln3206Arg | 1 (0.12) | - | - | - | - | - | PM2 | VUS |
| 27 | c.10131A>C | p.Glu3377Asp | 1 (0.12) | - | - | - | VUS | - | PM2, BP4 | VUS |
| 27 | c.10150C>G | p.Arg3384Gly | 1 (0.12) | - | - | - | VUS | - | PM2, BP4 | VUS |
| 27 | c.10234A>G | p.Ile3412Val | 6 (0.74) | 0.021 | - | - | B | - | BS1, BP4, BP6 | LB |
| IVS1 | c.-26G>A | - | 1 (0.12) | 0.20927 | 0.3805 | 0.209 | B | - | BA1, BP6, BP7 | B |
| IVS5 | c.476-24A>G | - | 1 (0.12) | - | - | - | - | - | PM2, BP7 | VUS |
| IVS10 | c.1909+12delT | - | 1 (0.12) | OV | - | - | CIP | - | BP7 | VUS |
| IVS12 | c.6938-25_6938-19delT | - | 1 (0.12) | - | - | - | - | - | PM2, PP3 | VUS |
| IVS17 | c.7806-14T>C | - | 17(2.11) | 0.534 | 0.5544 | - | B | - | BA1, BP6, BP7 | B |
| IVS17 | c.7976+24G>A | - | 1 (0.12) | 0.001 | 0.0009315 | - | LB | - | BP6, BP7 | LB |
| IVS17 | c.7976+45G>C | - | 8 (0.99) | 0.001 | 0.01271 | - | B/LB | - | BS1, BP6, BP7 | LB |
| IVS20 | c.8488-1G>A | - | 3 (0.37) | - | - | - | P/LP | Deleterious [19] | PS3, PM2, PP5 | LP |
1000G (EAS), minor allele frequency from East-Asian population in the 1000 Genome Phase 3 database; ACMG/AMP, American College of Medical Genetics and Genomics and the Association for Molecular Pathology; B, benign; BA, benign stand-alone evidence; BP, benign supporting evidence; BS, benign strong evidence; c, coding sequence; CIP, conflicting interpretations of pathogenicity; ESP, Exome Sequencing Project; ExAC, Exome Aggregation Consortium; HGVS, Human Genome Variation Society; IVS, intervening sequence; LB, likely benign; LP, likely pathogenic; OV, observed variant(s) without frequency or population information; P, pathogenic; p, protein sequence; PM, pathogenic moderate evidence; PP, pathogenic supporting evidence; PS, pathogenic strong evidence; PVS, pathogenic very strong evidence; VUS, variants of unknown significance.
2. Reclassified “patients” due to the reclassification of BRCA1 or BRCA2 VUS
Excluding patients with both PV and VUS, there were 195 patients with only VUS. 82 were with BRCA1 VUS only, 90 were with BRCA2 VUS only and 23 were with BRCA1 and BRCA2 simultaneously (Fig. 1). Among the 195 patients, the status for 25.1% (49/195) changed to benign, that for 16.4% (32/195) changed to likely benign, that for 5.1% (10/195) changed to likely pathogenic, and that for 5.1% (10/195) changed to pathogenic. Each reclassification rate for BRCA1 VUS, BRCA2 VUS, and BRCA1 plus BRCA2 subset was reported in Table 3.
Table 3. Percentage of reclassified patients in each subset.
| Reclassification | Patients with BRCA1 VUS (n=82) | Patients with BRCA2 VUS (n=90) | Patients with BRCA1 VUS+BRCA2 VUS (n=23) | Total (n=195) |
|---|---|---|---|---|
| Benign | 26 (31.7) | 20 (22.2) | 3 (13.0) | 49 (25.1) |
| Likely benign | 3 (3.7) | 24 (26.7) | 5 (21.7) | 32 (16.4) |
| VUS | 39 (47.6) | 42 (46.6) | 13 (56.5) | 94 (48.2) |
| Likely pathogenic | 5 (6.1) | 4 (4.4) | 1 (4.3) | 10 (5.1) |
| Pathogenic | 9 (11.0) | 0 (0.0) | 1 (4.3) | 10 (5.1) |
VUS, variants of unknown.
DISCUSSION
In the current study, 108 specific BRCA1/2 VUS were detected. There were 195 people with only VUS, and the 51.8% (101/195) patients were reclassified. The status of 41.5% (81/195) patients was changed to benign or likely benign, and that of 10.3% (20/195) to pathogenic or likely pathogenic.
Several studies have shown that about 10%–15% of the cases tested for BRCA exhibited a pathogenic variant [20]. Further, previous studies have reported a range of prevalence for BRCA VUS (3.9%–22.5%) [7,8,9,10,11,21,22]. However, these results are primarily from breast cancer studies, as there are very few studies that have solely focused on gynecological oncology. Moreover, in Korea, genetic studies of patients with breast cancer have been conducted on a large scale through research conducted by the Korean Hereditary Breast Cancer (Korean Hereditary Breast Cancer) Study [23,24], which has reported a pathogenic variant prevalence of 16.5% (418/2,526). However, in the case of gynecological cancers, there is no such large-scale study. Several studies have reported that the frequency of BRCA1/2 germline mutations among patients with epithelial ovarian cancer ranges from 31.9% (37/116) [25] to 33% (13/40) [26]. However, in each of those studies, only a small fraction of ovarian cancer patients was subject to genetic testing (116 out of 711 and 40 out of 337, respectively). Thus, selection biases might have occurred due to lack of these genetic tests. In the present study, we detected a BRCA1 PV prevalence of 17.0% (137/805) and a BRCA2 PV prevalence of 8.6% (69/805). In addition, this study found BRCA1 VUS and BRCA2 VUS prevalence of 15.9% (128/805), and 18.4% (148/805), which was slightly lower than the previously reported incidence.
The ACMG/AMP 2015 guidelines were created to standardize the classification of genetic variants, and they proposed multiple criteria which can be combined to assign pathogenicity status. However, from their disclaimer, adherence to these standards and guidelines is voluntary and does not necessarily assure a successful medical outcome [12]. They suggested 16 criteria as evidence of pathogenicity, and 12 criteria as evidence for the benign effect, even with different degrees of evidence available. Thus, the determination of pathogenicity based on these criteria could vary depending on the researcher. For instance, for in silico tests, which were classified as criteria for PP3 or BP4, the use of “multiple lines of computational evidence” could result in arbitrary classification. The current study used tools such as “Mutation taster,” “Align-GVGD,” “PANTHER,” “Polyphen,” and “SIFT,” and 4 out of 5 matches were required for certainty. However, in a previous study [9], “nsSNPAnalyzer” was used in addition to the tools used in the current study and considered up to 80% as evidence of BP4 or PP3.
In another study [10], Human Splicing Finder and a criteria of 5 concordances out of 6 was used for the criteria for PP5 “Reputable source recently reported variants as pathogenic, but the evidence was not available to the laboratory to perform an independent evaluation,” or BP6 “Reputable source recently reported a variant as benign, but the evidence was not available to the laboratory to perform an independent evaluation.” A previous study [9] utilized 6 population databases and used information with 50% agreement between databases. However, the present study and a previous study [10] only used the ClinVar database.
In the case of BRCA1/2, differences regarding BP1 (missense variant in a gene for which primarily truncating variants are known to cause disease) could lead to inaccurate results for all missense variants [24]. A previous study [10] and the present study do not utilized this criterion because several single amino acid changes caused by single nucleotide variations could cause a pathogenic result. Conversely, another study [9] used BP1 as benign supporting evidence.
There were 4 reclassified BRCA1 variants (c.5215G>C [p.Asp1739His], c.5363G>T [p.Gly1788Val], c.5339T>C [p.Leu1780Pro], and c.5017_5019delCAC [p.His1673del]). Only a few studies have reported VUS found in patients with malignant disease rather than in a healthy cohort, regardless of organ, and reclassified the BRCA1/2 VUS. The rate at which specific VUS were reclassified as pathogenic or likely pathogenic variants was 5.6% (6/108) and that specific VUS reclassified as benign or likely benign variants was 27.8% (30/108). These rates were comparable to those observed in previous studies (Table 4) [21].
Table 4. Reclassification frequency of BRCA1 and BRCA2 VUS.
| Study | Type of cancer | Total No. of patients | No. of initial VUS type | No. of reviewed VUS type | Reclassified class | Reclassification method | Country | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B (%) | LB (%) | VUS (%) | LP (%) | P (%) | |||||||
| Mighton et al. [21] | Breast, colon | 6,090 | 453 | 154 | 26.6 | 14.9 | 56.5 | 1.3 | 0.6 | Nationwide database | Canada |
| So et al. [10] | Breast, ovary | 423 | 48 | 48 | 6.3 | 20.8 | 68.8 | 4.2 | 0.0 | ACMG/AMP 2015 guideline | Korea |
| Lee et al. [11] | Breast | 2,115 | 286 | 83 | 0.0 | 67.4 | 27.7 | 3.5 | 1.2 | Multifactorial probability-based model | Korea |
| ACMG/AMP 2015 guideline | |||||||||||
| Park et al. [7] | HBOC | 745 | 86 | 86 | 10.4 | 41.9 | 40.7 | 1.7 | 5.8 | ACMG/AMP 2015 guideline | Korea |
| Present study | EOC, FTC, PPC | 805 | 108 | 108 | 15.7 | 12.0 | 66.7 | 4.6 | 1.0 | ACMG/AMP 2015 guideline | Korea |
ACMG/AMP, American College of Medical Genetics and Genomics and the Association for Molecular Pathology; B, benign; EOC, epithelial ovarian cancer; FTC, fallopian tube cancer; HBOC, hereditary breast and ovarian cancer syndrome; LB, likely benign; LP, likely pathogenic; P, pathogenic; PPC, primary peritoneal cancer; VUS, variants of unknown significance.
c.5215G>C is a missense change at an amino acid residue where a different missense change that was determined to be pathogenic has been seen before (c.5216A>T [p.Asp1739Val]). This could be used for evidence of PM5 pathogenicity using ACMG/AMP 2015 criteria. c.5017_5019delCAC (p.His1673del) is an in-frame deletion and causes protein length change. Its allele frequency could not be found in the 1000 Genomes Project, Exome Aggregation Consortium, and Exome Variant Server databases; however, 0.37% (3/805) of patients had this variant in the present study.
Further, 2 BRCA2 VUS were reclassified as likely pathogenic (c.9154C>T [p.Arg3052Trp], c.9110_9112delAAC [p.Gln3037del] and c.8488-1G>A). In the case of c.8488-1G>A, several functional studies have reported it as deleterious [19,27], and its allele frequency could not be found in the 1000 Genomes Project, Exome Aggregation Consortium, and Exome Variant Server databases. Discordant with other reports, in silico testing using the Human Splicing Finder, detected no significant splicing motif alteration and showed that this mutation potentially had no impact on splicing.
In the present study, 17 synonymous variants (10 in BRCA1 and 7 in BRCA2) were detected, and 4 of these variants were reclassified as benign while the others remained VUS. The degeneracy of the genetic code means that many mutations in coding sequences, particularly at the third base codon, do not affect the protein sequence and are, therefore, considered silent. However, there is increasing evidence that they could still have effects on transcription, splicing, mRNA transport or translation, any of which could alter the phenotype [28]. For example, c.9117G>C (p.Pro3089=) is a known pathogenic variant. However, since it is a simple synonymous variant, it could easily be misinterpreted as a benign variant. Synonymous variants should, therefore, not be judged as benign without definite evidence.
Current evidence suggests that several candidate pathogenic/likely pathogenic variants could not be classified as such due to lack of evidence to fulfill the ACMG/AMP guidelines. For example, the BRCA1 variant c.81-9C>G in the present study showed activation of the intron cryptic acceptor site and potential alteration of splicing (PP3) as the result of in silico testing by Human Splicing Finder. Moreover, the frequency of this variant could not be found in the 1000 Genomes Project, Exome Sequencing Project, or the Exome Aggregation Consortium; therefore, we assigned it to PM2 [29]. c.81-9C>G is an intronic variant which had the effect of creation and use of a novel acceptor site for frameshift mutations. Therefore, a co-segregation study, pedigree analysis, and functional study are required to determine pathogenicity.
Literally, the variants which have not been established or ruled out as a disease risk are referred to as VUS. Under the present circumstances, VUS is treated the same as benign. For example, there may be patients who have a strong family history and consistently present novel variants from the family. However, pathogenicity cannot be easily determined unless the variant is a frame-shift or nonsense mutation. If the pathogenicity of the variant has not been demonstrated without uncertainty, we cannot use poly (ADP-ribose) polymerase-1 inhibitor, cannot perform preventive procedure such as risk-reducing salpingo-oophorectomy. The best way to determine the pathogenicity of a new variant is to conduct a functional study. However, because the number of variants is infinite in theory, it is currently unreasonable to conduct a functional study on all variants. Therefore, by consistently conducting the same attempts as in this study, we selected VUS as a candidate for a pathogenic variant. After that, if the functional study of the VUSs is done, it would be a good challenge to effectively prevent and treat hereditary gynecological cancer.
The limitations of the present study are that this was a study performed in a single institution and may have selection bias due to the population undergoing genetic testing at this institution. The female relatives of the ovarian cancer patients tend to visit the same hospital; therefore, the subjects in the present data could be closely related each other, and the frequency of a certain variant could be exaggerated. In addition, we used a retrospective chart review for data collection, and data for the pedigree analysis or co-segregation test was partly insufficient.
In conclusion, the clinical significance of VUS in BRCA1/2 is changing over time, and in this study, we reclassified 5.6% (6/108) of specific VUS identified over a 13-year period as pathogenic or likely pathogenic. Among the 195 patients with VUS, the BRCA1/2 variants of 13.8% (27/195) were reclassified as benign, those of 27.7% (54/194) to likely benign, and those of 10.3% (20/195) to pathogenic or likely pathogenic. Therefore, continued monitoring of patients with mutations, and further investigation into the pathogenicity of VUS is required in the future.
Footnotes
Funding: This work was supported by the National Cancer Center Grant (NCC-1911274-2).
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
- Conceptualization: H.H.I., R.J.S., S.H., K.S.Y., L.M.C.
- Data curation: H.H.I.
- Formal analysis: H.H.I., L.M.C.
- Investigation: H.H.I.
- Methodology: H.H.I., L.M.C.
- Project administration: L.M.C.
- Resources: H.H.I., L.M.C.
- Supervision: L.M.C.
- Validation: R.J.S., S.H., K.S.Y.
- Visualization: H.H.I.
- Writing - original draft: H.H.I., L.M.C.
- Writing - review & editing: R.J.S., S.H., K.S.Y.
SUPPLEMENTARY MATERIALS
Criteria for reclassification of variants of unknown significance in present study (for pathogenic evidence)
Criteria for reclassification of variants of unknown significance in present study (for benign evidence)
The number of implemented genetic testing methods
Patients characteristics and composition of the study cohort
All of the detected variants among 805 patients
Percentage of the reclassified specific “VUS”
Frequency of variants by histologic subtype
Frequency of variants by stage
Frequency of variants by age at diagnosis
The number of reclassified specific BRCA1 and BRCA2 variants of uncertain significance.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Lim MC, Won YJ, Ko MJ, Kim M, Shim SH, Suh DH, et al. Incidence of cervical, endometrial, and ovarian cancer in Korea during 1999–2015. J Gynecol Oncol. 2019;30:e38. doi: 10.3802/jgo.2019.30.e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramus SJ, Gayther SA. The contribution of BRCA1 and BRCA2 to ovarian cancer. Mol Oncol. 2009;3:138–150. doi: 10.1016/j.molonc.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Network NC. Ovarian cancer: including fallopian tube cancer and primary peritoneal cancer. Version 1.2019. Fort Washington, PA: National Comprehensive Cancer Network; 2018. [cited 2020 Feb 10]. Available from: https://www.nccn.org/professionals/physician_gls/pdf/ovarian.pdf. [Google Scholar]
- 6.Ryu JM, Kang G, Nam SJ, Kim SW, Yu J, Lee SK, et al. Suggestion of BRCA1 c.5339T>C (p.L1780P) variant confer from ‘unknown significance’ to ‘Likely pathogenic’ based on clinical evidence in Korea. Breast. 2017;33:109–116. doi: 10.1016/j.breast.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Park JS, Nam EJ, Park HS, Han JW, Lee JY, Kim J, et al. Identification of a novel BRCA1 pathogenic mutation in Korean patients following reclassification of BRCA1 and BRCA2 variants according to the ACMG standards and guidelines using relevant ethnic controls. Cancer Res Treat. 2017;49:1012–1021. doi: 10.4143/crt.2016.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi MC, Jang JH, Jung SG, Park H, Joo WD, Song SH, et al. Unclassified variants of BRCA1 and BRCA2 in Korean patients with ovarian cancer. Int J Gynecol Cancer. 2018;28:308–315. doi: 10.1097/IGC.0000000000001161. [DOI] [PubMed] [Google Scholar]
- 9.Park KS, Cho EY, Nam SJ, Ki CS, Kim JW. Comparative analysis of BRCA1 and BRCA2 variants of uncertain significance in patients with breast cancer: a multifactorial probability-based model versus ACMG standards and guidelines for interpreting sequence variants. Genet Med. 2016;18:1250–1257. doi: 10.1038/gim.2016.39. [DOI] [PubMed] [Google Scholar]
- 10.So MK, Jeong TD, Lim W, Moon BI, Paik NS, Kim SC, et al. Reinterpretation of BRCA1 and BRCA2 variants of uncertain significance in patients with hereditary breast/ovarian cancer using the ACMG/AMP 2015 guidelines. Breast Cancer. 2019;26:510–519. doi: 10.1007/s12282-019-00951-w. [DOI] [PubMed] [Google Scholar]
- 11.Lee JS, Oh S, Park SK, Lee MH, Lee JW, Kim SW, et al. Reclassification of BRCA1 and BRCA2 variants of uncertain significance: a multifactorial analysis of multicentre prospective cohort. J Med Genet. 2018;55:794–802. doi: 10.1136/jmedgenet-2018-105565. [DOI] [PubMed] [Google Scholar]
- 12.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouwman P, van der Gulden H, van der Heijden I, Drost R, Klijn CN, Prasetyanti P, et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 2013;3:1142–1155. doi: 10.1158/2159-8290.CD-13-0094. [DOI] [PubMed] [Google Scholar]
- 14.Phelan CM, Đapić V, Tice B, Favis R, Kwan E, Barany F, et al. Classification of BRCA1 missense variants of unknown clinical significance. J Med Genet. 2005;42:138–146. doi: 10.1136/jmg.2004.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuntini R, Cortesi L, Calistri D, Pippucci T, Martelli PL, Casadio R, et al. BRCA1 p.His1673del is a pathogenic mutation associated with a predominant ovarian cancer phenotype. Oncotarget. 2017;8:22640–22648. doi: 10.18632/oncotarget.15151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steffensen AY, Dandanell M, Jønson L, Ejlertsen B, Gerdes AM, Nielsen FC, et al. Functional characterization of BRCA1 gene variants by mini-gene splicing assay. Eur J Hum Genet. 2014;22:1362–1368. doi: 10.1038/ejhg.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guidugli L, Carreira A, Caputo SM, Ehlen A, Galli A, Monteiro AN, et al. Functional assays for analysis of variants of uncertain significance in BRCA2 . Hum Mutat. 2014;35:151–164. doi: 10.1002/humu.22478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acedo A, Sanz DJ, Durán M, Infante M, Pérez-Cabornero L, Miner C, et al. Comprehensive splicing functional analysis of DNA variants of the BRCA2 gene by hybrid minigenes. Breast Cancer Res. 2012;14:R87. doi: 10.1186/bcr3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vallée MP, Francy TC, Judkins MK, Babikyan D, Lesueur F, Gammon A, et al. Classification of missense substitutions in the BRCA genes: a database dedicated to Ex-UVs. Hum Mutat. 2012;33:22–28. doi: 10.1002/humu.21629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mighton C, Charames GS, Wang M, Zakoor KR, Wong A, Shickh S, et al. Variant classification changes over time in BRCA1 and BRCA2 . Genet Med. 2019;21:2248–2254. doi: 10.1038/s41436-019-0493-2. [DOI] [PubMed] [Google Scholar]
- 22.Lerner-Ellis J, Wang M, White S, Lebo MS Canadian Open Genetics Repository Group. Canadian Open Genetics Repository (COGR): a unified clinical genomics database as a community resource for standardising and sharing genetic interpretations. J Med Genet. 2015;52:438–445. doi: 10.1136/jmedgenet-2014-102933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han SA, Kim SW, Kang E, Park SK, Ahn SH, Lee MH, et al. The prevalence of BRCA mutations among familial breast cancer patients in Korea: results of the Korean Hereditary Breast Cancer study. Fam Cancer. 2013;12:75–81. doi: 10.1007/s10689-012-9578-7. [DOI] [PubMed] [Google Scholar]
- 24.Son BH, Ahn SH, Kim SW, Kang E, Park SK, Lee MH, et al. Prevalence of BRCA1 and BRCA2 mutations in non-familial breast cancer patients with high risks in Korea: the Korean Hereditary Breast Cancer (KOHBRA) Study. Breast Cancer Res Treat. 2012;133:1143–1152. doi: 10.1007/s10549-012-2001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eoh KJ, Park HS, Park JS, Lee ST, Han J, Lee JY, et al. Comparison of clinical outcomes of BRCA1/2 pathologic mutation, variants of unknown significance, or wild type epithelial ovarian cancer patients. Cancer Res Treat. 2017;49:408–415. doi: 10.4143/crt.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim MC, Kang S, Seo SS, Kong SY, Lee BY, Lee SK, et al. BRCA1 and BRCA2 germline mutations in Korean ovarian cancer patients. J Cancer Res Clin Oncol. 2009;135:1593–1599. doi: 10.1007/s00432-009-0607-3. [DOI] [PubMed] [Google Scholar]
- 27.Acedo A, Hernández-Moro C, Curiel-García Á, Díez-Gómez B, Velasco EA. Functional classification of BRCA2 DNA variants by splicing assays in a large minigene with 9 exons. Hum Mutat. 2015;36:210–221. doi: 10.1002/humu.22725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goymer PJ. Synonymous mutations break their silence. Nat Rev Genet. 2007;8:92. [Google Scholar]
- 29.Joosse SA, van Beers EH, Tielen IH, Horlings H, Peterse JL, Hoogerbrugge N, et al. Prediction of BRCA1-association in hereditary non-BRCA1/2 breast carcinomas with array-CGH. Breast Cancer Res Treat. 2009;116:479–489. doi: 10.1007/s10549-008-0117-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Criteria for reclassification of variants of unknown significance in present study (for pathogenic evidence)
Criteria for reclassification of variants of unknown significance in present study (for benign evidence)
The number of implemented genetic testing methods
Patients characteristics and composition of the study cohort
All of the detected variants among 805 patients
Percentage of the reclassified specific “VUS”
Frequency of variants by histologic subtype
Frequency of variants by stage
Frequency of variants by age at diagnosis
The number of reclassified specific BRCA1 and BRCA2 variants of uncertain significance.