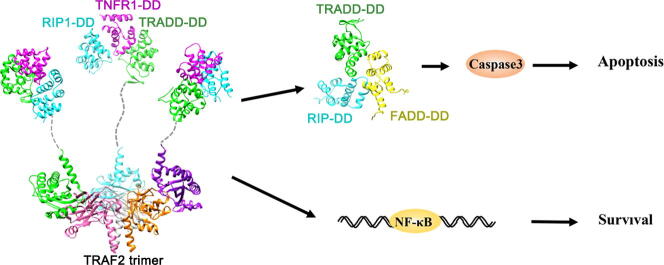
Graphical abstract
Keywords: TRADD, Adaptor protein, Signaling, Structural mechanism, Death domain
Abstract
TRADD participates in various receptor signaling pathways and plays vital roles in many biological activities, including cell survival and apoptosis, in different cellular contexts. TRADD has two distinct functional domains, a TRAF-binding domain at the N-terminus and a death domain (DD) at the C-terminus. The TRAF binding domain of TRADD folds into an α-β plait topology and is mainly responsible for binding TRAF2, while the TRADD-DD can interact with a variety of DD-containing proteins, including receptors and intracellular signaling molecules. After activation of specific receptors such as TNFR1 and DR3, TRADD can bind to the receptor through DD-DD interaction, creating a membrane-proximal platform for the recruitment of downstream molecules to propagate cellular signals. In this review, we highlight recent advances in the studies of the structural mechanism of TRADD adaptor functions for NF-κB activation and apoptosis induction. We also provide suggestions for future structure research related to TRADD-mediated signaling pathways.
1. Introduction
Signal transduction, or cell signaling, is the transmission process by which extracellular biochemical or biophysical signals are transduced through cell-surface receptors to regulate cellular functions. Studies on the mechanisms controlling cross-talk between or among signaling cascades are critical to elucidate the specificity of cell signaling. Most signaling pathways involve signaling protein complexes, which are often assembled by adaptor proteins. This class of proteins typically have two or more protein-binding domains but lack intrinsic catalytic activity for their signaling functions [1]. They rely on such domains to mediate specific protein-protein interactions (PPI). The distinctive ability of the adaptor proteins is to bind two or more proteins or domains simultaneously, leading to the formation of larger protein complexes. By linking different signaling proteins to each other, cellular signals can be propagated and amplified to induce a specific functional response to incoming extracellular signals. Subcellular localization may also contribute to the response specificity determined by the types of binding domains of the adaptor proteins.
The first example of the adaptor protein can be traced back to the discovery of the Src (or Sarcoma) homology region 2 (SH2) domain by Anthony Pawson and colleagues in the 1980s [2], [3]. SH2 domain can direct specific interactions of different proteins and control cells’ responses to external signals. Another early example is the mammalian CT10 (chicken tumor virus number 10) regulator of kinase (Crk) by Mayer and colleagues in 1988 [4], [5]. Crk contains multiple binding modules, which can connect different proteins for signal transduction. Thus, Crk may be used as a model protein to study other signaling adaptors. Adaptor proteins were also proposed as potential drug targets for therapy since they have been implicated in many human diseases, such as autoimmune disorder [5]. In the past three decades, numerous classes of adaptor proteins have been identified and characterized [5]. Among these proteins, tumor necrosis factor (TNF) receptor type 1-associated death domain protein (TRADD) is a unique adaptor molecule owing to its central role in various downstream signaling events, including cell survival and apoptosis, in different cellular contexts. Therefore, investigation of the structure-function relationship in TRADD is crucial to disclose its adaptor roles in different pathways. In this review, we focus on recent advances on the structural mechanisms of TRADD-mediated signaling for different functional responses.
2. Structure of TRADD with two functional domains
Human TRADD is a 34 kDa protein with two functional domains connected by an unstructured peptide of ~37 amino acid residues (Fig. 1A and B) [6], [7], [8]. It was first identified as a potential adaptor molecule for tumor necrosis factor receptor 1 (TNFR1) signaling from the yeast two-hybrid system in 1995 since its C-terminal domain (CTD) can specifically interact with the intracellular death domain (DD) of liganded TNFR1 [7]. TRADD also interacts with TNF receptor-associated factor 2 (TRAF2) through its the N-terminal domain (NTD) in TNFR1 signaling, consistent with its adaptor function [8], [9], [10].
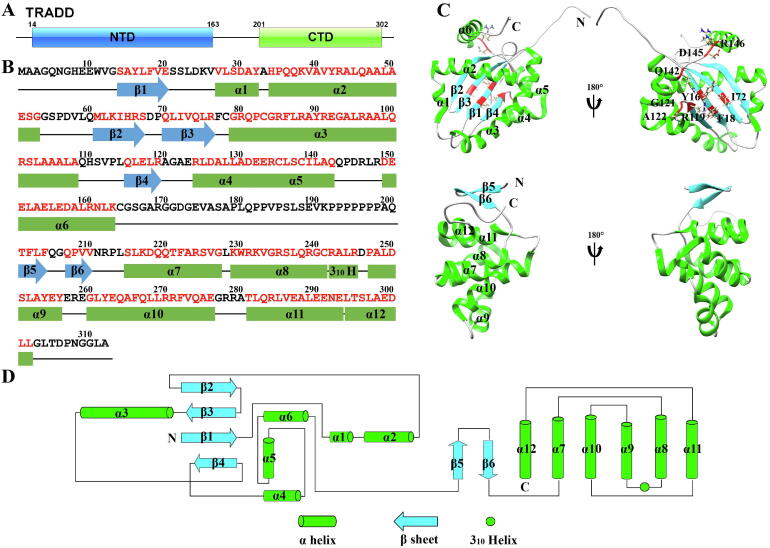
Fig. 1.
The structure of TRADD-NTD and TRADD-CTD. A, Molecular architecture of TRADD. Domain positions are indicated above the cartoon. NTD: N-terminal domain; CTD: C-terminal domain. B, Amino acid sequence of full-length TRADD. The secondary structures of TRADD are shown below the sequence. The red letters indicate the structured regions. C, Ribbon drawing of the lowest-energy structures of TRADD-NTD (PDB: 1F2H, upper) and TRADD-CTD (PDB: 5XME, lower). Key residues in ligand binding pocket for small molecule ihibitors are shown in ball-and-stick representation. D, 2D topology of full-length TRADD. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The monomeric structure of the TRADD-NTD was first determined by nuclear magnetic resonance (NMR) spectroscopy in 2000. Residues Met1-Gly13 at the N-terminus are disordered, and their function is largely unknown [9]. The topology of the TRADD-NTD is assigned to α-β plaits based on CATH (Class, Architecture, Topology, and Homologous superfamily) structure classification [11]. In the core structure of the TRADD-NTD, four antiparallel β strands form one β sheet, and six α helices pack nearly against one side of the β sheet (Fig. 1C, D). The hydrophobic center of the TRADD-NTD is formed by helix 1-3 and the β-sheet [9].
Different from the TRADD-NTD, the TRADD-CTD is composed of a novel DD, distinct from other members of the DD superfamily. The DD superfamily is one of the largest domain superfamilies known and is typically considered to consist of four subfamilies: the canonical DD, the death effector domain (DED), the caspase recruitment domain (CARD), and the pyrin domain (PYD) [6], [12], [13]. The identification of DD subfamilies is based on sequence similarity, as well as the orientation and length of the ⍺-helices. All the members of the DD superfamily contain five or six antiparallel ⍺-helices organized in a Greek key motif and hence share a common structural topology of Death Domain in CATH classification [11]. In 2017, our group solved the solution structure of the TRADD-DD by NMR spectroscopy in pure water [6]. The new structural feature of the TRADD-DD is the presence of one β-hairpin motif, which is tightly packed beside a canonical DD (Fig. 1C, D). This β structure is required for the TRADD-DD to fold into a functional domain in water solution and hence is an essential component of the TRADD-DD. The combination of β- and α-structures was not previously seen in any other DD. We, therefore, propose to place the TRADD-DD into a new subfamily, namely TDD, which will expand the DD superfamily to include five members. Hydrophobic interactions play a vital role in the folding of the extended spherical structure of the TRADD-DD, leading to the formation of four hydrophobic cores. The surface of the TRADD-DD is highly charged, suggesting a crucial role of electrostatic interactions in signaling pathways mediated by TRADD.
3. Functions of TRADD in signaling pathways
3.1. TRADD participates in TNF/TNFR1 signaling pathway
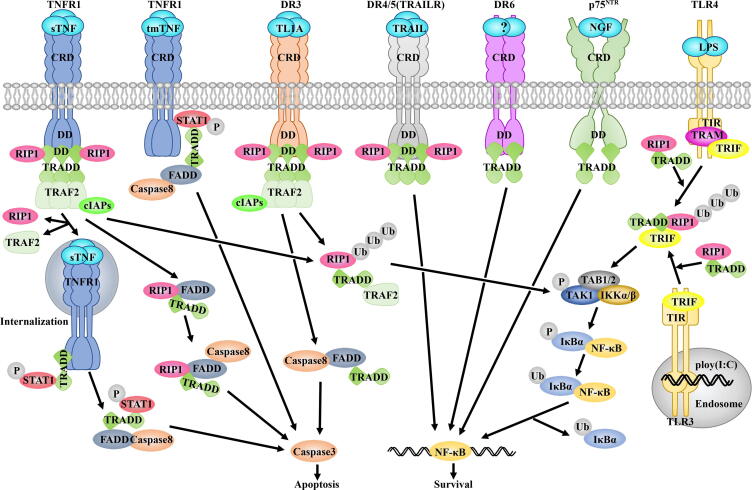
TNF, a proinflammatory cytokine, is involved in triggering apoptosis, cell proliferation, inflammation, and immune response [14], [15], [16]. Extracellular TNF can activate TNFR1, which subsequently recruits intracellular proteins to activate various signaling pathways, such as nuclear factor κB (NF-κB) and mitogen-activated protein (MAP) kinase pathways [7], [17], [18], [19]. TNFR1 belongs to the DR family, a subgroup of the tumor necrosis factor receptor superfamily (TNFRS) [20]. TNFR1 contains a cytoplasmic DD for recruiting downstream signaling proteins through an adaptor protein TRADD [14], [21], [22]. The TRADD-DD binds to the TNFR1-DD via homotypic DD interaction, and receptor interacting protein 1 (RIP1) can also associate with TNFR1-DD:TRADD-DD complex, forming a larger DD complex (Fig. 2). The TRADD-NTD interacts with the TRAF (TNF receptor-associated factor) domain of TRAF2, which further binds to the cellular inhibitor of apoptosis proteins (cIAP1 or cIAP2), thereby recruiting TRAF2/cIAPs to TNFR1 to form an active E3 ubiquitin ligase complex for RIP1 ubiquitination [23]. Ubiquitinated RIP1 can be recognized by inhibited κB kinase (IKK) and transforming growth factor-β activating kinase 1 (TAK1) complex, leading to IKK phosphorylation and activation. Subsequently, IKK phosphorylates the two serines at the N-terminus of IκBα protein, which is further recognized and ubiquitinated by E3 ubiquitin ligase and dissociates from NF-κB. Finally, the active NF-κB with exposed nuclear localization signal enters the nucleus and binds to specific sequences of DNA for pro-survival signaling [24], [25].
Fig. 2.
Cartoon models of TRADD adaptor roles in receptor signaling. TRADD participates in various DRs signaling, leading to cell survival and apoptosis. As an adaptor, the TRADD-CTD associates with the intracellular domain of the death receptor through DD-DD interactions. In TNFR1 and DR3 signaling, binding of TRADD-NTD to the TRAF domain of TRAF2 leads to the recruitment of TRAF2 and its associated molecules, including cIAPs, to the receptor for RIP1 ubiquitination and NF-κB activation. Deubiquitinated RIP1 can interact with TRADD and FADD to trigger caspase-dependent cell apoptosis. TRADD is also involved in Toll-like receptors (TLR) signaling, forming a complex with TRIF and RIP1 for NF-κB activation.
The membrane-bound complex for NF-κB activation is also known as Complex I, which is the primary signaling complex and forms within seconds after TNFR1 engagement by soluble TNF. However, when NF-κB activation is impaired or shut down, TNFR1-mediated signaling will switch from pro-survival to pro-death. RIP1 can be deubiquitinated by zinc finger protein A20 after NF-κB activation and form cytosolic Complex Ⅱa with TRADD and FADD (Fas-associated death domain protein) through DD interactions [26], [27]. The cytosolic Complex IIa, which lacks TNFR1, further activates caspase 8 for the apoptotic pathway (Fig. 2) [28], [29]. However, it is still unclear how the adaptor protein TRADD is released from Complex I for Complex IIa formation.
TNFR1 has also been shown to be internalized to cytoplasm depending on TNF receptor internalization domain (TRID) [30]. After TNFR1 internalization, STAT1 (signal transducer and activator of transcription 1) can outcompete and displace TRAF2 from the TRADD-NTD, leading to the formation of a cytosolic death-including signaling complex (DISC) (Fig. 2) [31], [32], [33], [34]. This DISC attenuates NF-κB activation and promotes apoptotic signaling. STAT1 can also bind TNFR1 at the plasma membrane in response to transmembrane TNF (tmTNF) activation [34]. TRADD is then recruited to TNFR1-bound STAT1 without interacting with TNFR1-DD for DISC formation on the membrane. It is evident that the cytosolic and the membrane-bound DISCs involve different mechanisms of complex formation for triggering apoptosis, which needs further structural investigation.
3.2. TRADD participates in the TL1A/DR3 signaling pathway
Death receptor 3 (DR3) is produced by thymus, spleen, and other tissues and organs containing lymphocytes and associated with inflammation and autoimmune diseases, such as inflammatory bowel disease and rheumatoid arthritis [35], [36], [37], [38], [39]. Similar to other members of the DR subfamily, DR3 has an intracellular DD [40]. DR3 shares the highest sequence homology to TNFR1 among all members of the tumor necrosis factor receptor superfamily (TNFRSF) [35], [41]. TNF-like ligand 1 A (TL1A), a member of the tumor necrosis factor superfamily, is an extracellular ligand of DR3 [19], [42], [43]. In the TL1A/DR3 signaling pathway, TRADD is also involved as a primary and essential adaptor molecule. Similar to TNF, soluble TL1A can self-assemble into trimers and bind to the extracellular domains (ECD) of DR3, which could result in receptors clustering and initiate recruitment of TRADD-DDs to the DR3-DD through DD-DD interactions [17], [44]. TRAF2, RIPK1, and cIAP1/2 will further bind to the complex and ubiquitinated RIP1 can trigger the activation of NF-κB signaling (Fig. 2) [44]. Although DR3 mainly promotes inflammation and cell survival, it also has the ability to induce apoptosis via the interaction between the DR3-DD and the TRADD-DD when DR3-dependent activation of NF-κB pathway is blocked. For instance, when the second mitochondria-derived activator of caspase (SMAC) mimetics are used to treat human hematopoietic progenitor cells (TF-1), TL1A/DR3 signaling will induce apoptosis because the SMAC mimetics target and inhibit TRAF2/cIAPs [42], [45]. Much like TNFR1 signaling, TRADD, FADD, and caspase 8 associate together and form Complex IIa, culminating in caspase-dependent apoptosis (Fig. 2) [44].
3.3. TRADD participates in the TRAIL/TRAILR signaling pathway
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a member of the TNF superfamily [18], [46]. As a cytokine, TRAIL is widely expressed in humans and can selectively induce tumor cell apoptosis. Three TRAIL molecules can also form a stable homo-trimeric structure and interact with TRAIL receptors (TRAILR) to initiate signaling pathways, akin to the protein-ligand binding modes involving TNF and TL1A [19], [46], [47]. There are two types of TRAILRs with cytoplasmic DDs, TRAILR1 (also known as DR4) and TRAILR2 (DR5) [48]. When TRAIL binds to the receptor, the transmembrane domain of TRAILR can drive self-assembly to form a homo-trimer [49]. TRAILR1 may also interact with TRAILR2 and form a heteroreceptor complex for TRAIL signaling [50]. Both TRAILR1 and TRAILR2 bind the adaptor molecules FADD and TRADD for apoptosis and NF-κB signaling pathways, respectively [17], [48]. TRADD and FADD could compete with each other for binding to the receptors [17]. In the NF-κB pathway, TRADD and RIP1 are detected in the TRAILR complex (Fig. 2) [51]. Recruitment of RIP1 to TRAILRs depends on TRADD. It’s possible that TRADD uses similar interfaces for DD interactions in TRAIL/TRAILR signaling pathway. The ubiquitinated RIP1 further activates IKK and resists apoptosis [51], [52]. Unlike TNFR1 and DR3 signaling, the involvement of TRAF2 and cIAPs in IKK and NF-κB activation mediated by TRAILRs depends on cell types. For example, both TRAF2 and cIAPs are involved in the activation of NF-κB signaling in Jurkat lymphoma lines. Nevertheless, TRAF2 plays little role in TRAIL-induced IKK activation in Hela cell, which could be due to the expression of other TRAF proteins with similar functions in this type of cell [53], [54].
3.4. TRADD participates in DR6-induced signaling pathways
DR6, also known as TNFRSF21, is a member of the death receptor in the TNF superfamily and is abundant in lymphoid organs. It belongs to the orphan receptor in the TNF superfamily [19], [55]. DR6 is comprised of four cysteine-rich domain (CRD) in its extracellular region, a single-pass transmembrane domain, and an intracellular segment containing a putative DD and a CARD [55]. In tumor cells, the TNF-stimulated NF-κB signaling pathway may regulate DR6 expression [56]. At present, the only reported natural ligand of DR6 is the E2 domain of amyloid precursor protein (APP). Based on the oligomeric state of APP, it was proposed that DR6 activation triggered by APP-induced dimerization could lead to axon pruning and synapse elimination [57]. This result, however, remains to be reproduced by other investigators. TRADD was shown to be involved in the DR6-mediated NF-κB signaling pathway (Fig. 2), but its extracellular activator has not been determined [58]. The interaction between DR6 and TRADD was weaker than that between DR3 and TRADD in overexpression studies. Interestingly, RIP1, FADD, and RAIDD were not detected in DR6:TRADD complex. How TRADD bridges DR6 and downstream molecules to activate NF-κB signaling is still poorly understood.
3.5. TRADD participates in p75NTR-induced signaling pathways
p75 neurotrophin receptor (p75NTR), also known as TNFRSF16, is the first receptor identified for the neurotrophins [59], [60]. It can be structurally divided into an extracellular domain, a single-pass transmembrane domain, and an intracellular domain [61]. The extracellular domain is composed of four cysteine-rich regions for extracellular signals, including all neurotrophins [62]. The transmembrane domain contains a highly conserved cysteine residue involved in the covalent stabilization of disulfide-bonded receptor dimers; it is also crucial for downstream signal propagation [63]. The intracellular domain comprises a long unstructured juxtamembrane region followed by a canonical DD [64]. Due to the lack of catalytical activity, p75NTR signaling also relies on the recruitment of intracellular molecules to the intracellular domain. In-vitro cellular and biochemical studies demonstrated that TRADD can interact with the p75NTR-DD upon nerve growth factor (NGF) stimulation to initiate NF-κB signaling pathway and antiapoptotic effects in human breast cancer cells (Fig. 2) [65]. Our NMR titration indicated that TRADD can bind to the p75NTR-DD at a region very close to the p75NTR-DD homo-dimerization site. This observation suggested that dissociation of p75NTR-DD homodimer could be necessary for TRADD recruitment, in line with TRADD recruitment by p75NTR upon NGF binding [6]. Nevertheless, the connection between TRADD and downstream molecules for p75NTR-mediated NF-κB activation is unclear. Structure determination of p75NTR-DD:TRADD-DD complex and identification of TRADD cellular interactors will provide a basis for understanding the adaptor role of TRADD in p75NTR signaling.
3.6. TRADD participates in TLR3/TLR4-induced signaling pathways
Toll-like receptors (TLRs) are pathogen-associated pattern recognition receptors on mammalian antigen-presenting cells and play a major role in innate immune response [66]. TLRs have intracellular TIR (toll-IL-1 receptor) domains responsible for stimulating downstream signaling pathways through homotypic TIR-TIR interactions [66], [67]. TLR3 and TLR4 are members of the Toll-like receptor family. TLR3 can recognize double-stranded RNA produced by some viruses such as reoviruses or its synthetic analog polyinosinic-polycytidylic acid poly(I:C) [68], while TLR4 can recognize lipopolysaccharide (LPS) from the cytoderm of Gram-negative bacteria [69]. TRADD participates in TRIF (TIR domain-containing adaptor interferon-β)-dependent signaling pathway mediated by TLR3 and TLR4 [70], [71]. TLR3 directly binds to adaptor protein TRIF, while TLR4 binds to TRIF through TRIF-related adaptor molecule (TRAM) [72]. In the NF-κB signaling pathway, the RIP homotypic interaction motif (RHIM) of TRIF-CTD can associate with both RIP1 and TRADD [73], [74], [75], [70]. During this process, RIP1 may undergo K63 polyubiquitination and facilitate activation of NF-κB signaling (Fig. 2) [76]. TRADD also exhibits cell-type specificity in the TLR3 and TLR4 signaling. In mouse embryonic fibroblasts (MEFs) [70], mouse bone marrow-derived dendritic cells (BMDCs) [70] and macrophages [77], the absence of TRADD exerts different effects on NF-κB and MAPK signaling. Currently, it is still not clear how the interaction between TRIF and TRADD could propagate the downstream signals due to the lack of their complex structure information.
3.7. TRADD-mediated apoptosis and anti-apoptosis in the nucleus
Although most studies on TRADD focus on its functions as a cytoplasmic adaptor in various receptor signaling pathways, TRADD was also found to dynamically shuttle between nucleus and cytoplasm depending on a nuclear export signal (NES) located in helix α5 (147–163) and a nuclear localization signal (NLS) in helix α7 (229–242) [78]. Nuclear TRADD plays a role in cell survival in response to DNA damage. Translocation of TRADD from the cytoplasm into the nucleus contributes to the non-homologous end-joining (NHEJ) DNA repair pathway by recruiting a complex involving NHEJ repair factors 53BP1 (p53 binding protein 1) and Ku70/80 [78]. TRADD could also be trapped in the nucleus upon inhibition of nuclear export and become associated with promyelocytic leukemia protein (PML) nuclear bodies to activate an alternative apoptotic pathway via caspase-9, which is independent of FADD and caspase-8 recruitment [79], [80].
4. Structural basis of TRADD-mediated signaling
As an adaptor protein linking binding partners together to create larger complexes, TRADD harbors two distinct functional domains and plays pivotal roles in orchestrating downstream signaling pathways in space and time. Therefore, structural insights into different adaptor complex formation will be indispensable for understanding the fundamental mechanisms of TRADD-mediated NF-κB activation and apoptosis induction. Structural characterization of TRADD complexes for its adaptor roles has been reported in only a handful of studies.
4.1. Structure of TRADD-NTD heterotypic complex
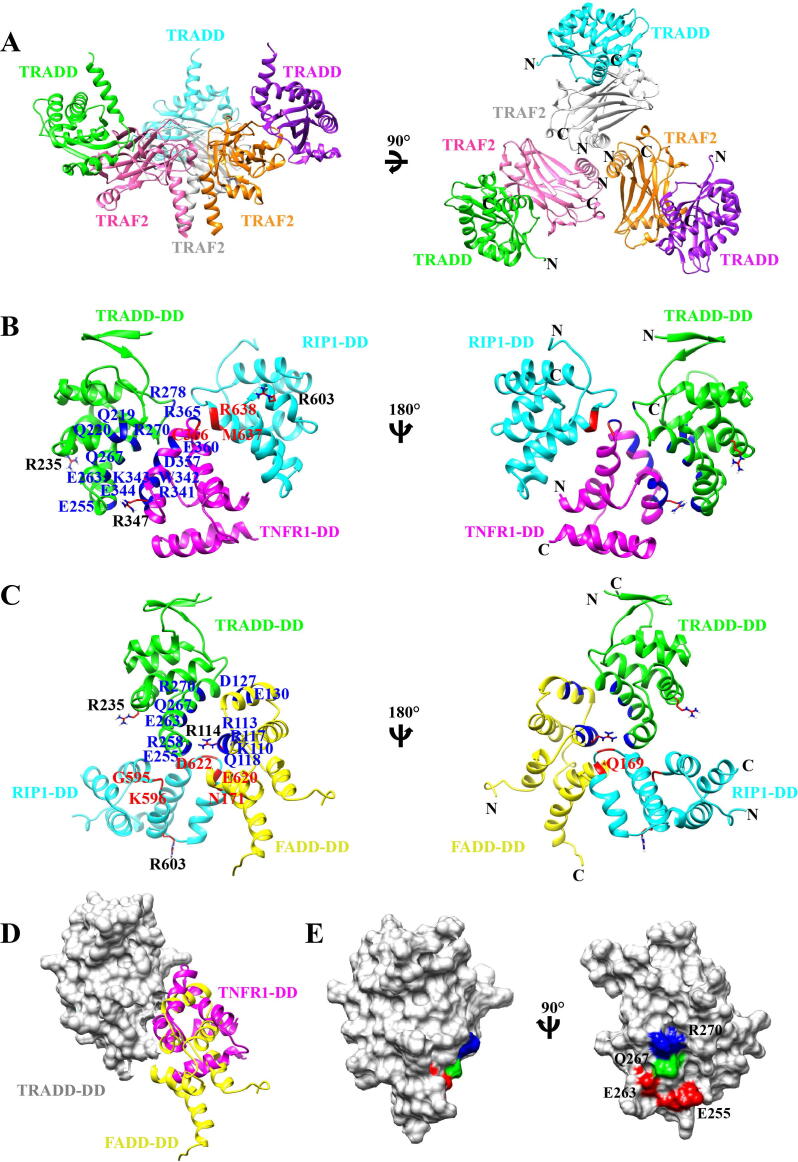
The first structure of TRADD complex with an atomic resolution was determined for TRADD-NTD bound to the TRAF domain of TRAF2 by X-ray crystallography in 2000 [10]. This complex structure, which adopts a C3 symmetry, revealed the mode of TRAF2 recruitment by TRADD (Fig. 3A). Three molecules of TRAF domain of TRAF2 form a mushroom-like trimer, sterically situating in the center of the TRADD :TRAF2 complex. Each β-sandwich subdomain of the TRAF domain binds to one TRADD-NTD. Hydrophobic interactions, hydrogen bonding and salt bridges contribute to the binding interface of the TRADD:TRAF2 complex [10]. The C-terminus of TRADD-NTD projects away from this complex, which facilitates the TRADD-DD to interact with the TNFR1-DD directly. The other TRAF family member, TRAF1, may also specifically interact with the TRADD-NTD in a similar way due to its high sequence homology to TRAF2 at the TRADD binding site. The TRAF domain of TRAF2 also contains a coiled-coil subdomain. It is constitutively associated with cIAP1 or cIAP2, leading to the recruitment of this cellular caspase inhibitor to the TNFR1 signaling Complex I for RIP1 ubiquitination. Therefore, disruption of TRAF2 association with TRADD will impair NF-κB activation and pro-survival pathway. It was found that EVER2 (Epidermodysplasia verruciformis 2) can tightly bind to the TRADD-NTD, preventing recruitment of TRAF2 to Complex I and promoting formation of TRADD:RIP:FADD complex for cell apoptosis [81]. A better understanding of the competitive binding of TRAF2 and EVER2 to TRADD requires structural determination of TRADD:EVER2 complex.
Fig. 3.
Structure models of signaling complexes involving TRADD. A, Crystal structure of trimeric TRADD-NTD:TRAF2-CTD complex showing the structural mechanism of TRAF2 recruitment by TRADD (PDB ID: 1F3V). Ribbon drawings of three TRADD-NTDs are colored in cyan, green and purple, respectively, and three TRAF2-CTDs are colored in light gray, pink, and orange, respectively. B, HADDOCK structure of heterotrimeric complex of TRADD-DD:RIP1-DD:TNFR1-DD. Ribbon drawings of TRADD-DD, TNFR1-DD, RIP1-DD are colored in green, magenta, and cyan, respectively. The free-form starting structures are NMR structure of TRADD-DD (PDB ID: 5XME), NMR structure of TNFR1-DD (PDB ID: 1ICH), and modeled structure of RIP1-DD based on the crystal structure of GlcNAcylated RIP1-DD (PDB ID:6AC5). The final docking structure with the lowest HADDOCK score and z-score was selected to represent the structure model. The GlcNAcylated sites (Arg residues) on three DDs are shown in ball-and-stick representation. Key residues involved in TRADD-DD:TNFR1-DD complex formation are highlighted and labeled in blue, and TNFR1-DD:RIP1-DD in red. C, HADDOCK structure of heterotrimeric complex of TRADD-DD:RIP1-DD:FADD-DD. NMR structure of FADD-DD (PDB ID: 1E41, yellow) was used as one of the starting structures, and the other two starting structures are the same as B. The GlcNAcylated sites (Arg residues) on three DDs are shown in ball-and-stick representation. Key residues involved in TRADD-DD:FADD-DD complex formation are highlighted and labeled in blue, and FADD-DD:RIP1-DD in red. D, Surface representation of TRADD-DD (grey) with overlapped ribbon drawings of TNFR1-DD (magenta) and FADD-DD (yellow). E, Surface representation of TRADD-DD. The positively and negatively charged residues in the overlapping interfaces, as shown in D, are colored in blue and red, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4.2. Structures of TRADD-DD heterotypic complexes
Although the TRADD-DD has been proved to interact with a few cellular interactors both in-vitro and in-vivo, no 3 dimensional (3D) complex structures between the TRADD-DD and its interactors have been reported. One of the main reasons for this difficulty is that TRADD-DD protein is particularly prone to aggregate at neutral pH in vitro. Extensive mutagenesis studies have identified a number of critical residues, including both charged and hydrophobic ones, for the formation of two signaling complexes, TRADD:RIP1:TNFR1 for NF-κB activation and TRADD:RIP1:FADD for caspase-dependent apoptosis [82], [83], [84], [85]. A structural model of the heterotrimeric complex of TRADD-DD:TNFR1-DD:RIP1-DD was previously proposed [83]. However, due to the lack of intact structures of the TRADD-DD and RIP1-DD in this model, structural mechanisms of signaling mediated by the TRADD-DD remain poorly understood.
To investigate how the TRADD-DD interacts with its binding partners, we determined the docking structure of TRADD-DD:RIP1-DD:TNFR1-DD by high ambiguity driven protein–protein docking (HADDOCK) (Fig. 3B) [86]. The experimental data acquired from all mutagenesis studies were employed as docking restraints [82], [83], [84], [85], and the lowest energy structure was selected to represent the structure model. Analysis of binding sites suggests that electrostatic interactions are dominant in the interface, while hydrophobic interactions also play an important role in the formation of the heterotrimeric complex. Similarly, we obtained the structure model of TRADD-DD:RIP1-DD:FADD-DD by HADDOCK calculation (Fig. 3C). We found that the TNFR1-DD and FADD-DD bind to the TRADD-DD at partially overlapping binding sites, where common epitopes of charged residues are consistent with previous mutagenesis studies (Fig. 3 D and E) [82], [83], [84], [85]. This comparison suggests that TRADD would have to be separated from TNFR1 and Complex I in order to form Complex II for initiating apoptotic signaling, which is in line with two different functional outputs mediated by TRADD in TNF/TNFR1 signaling. Biochemical studies indicated that the TNFR1-DD can strongly associate with the TRADD-DD with a Kd of ~25 nM [83], while the binding affinity of the FADD-DD to the TRADD-DD is unknown. Therefore, it is unclear whether the competitive binding of the FADD-DD and TNFR1-DD on the TRADD-DD is what regulates the hierarchical activation of pro-survival and pro-apoptotic pathways. The structural mechanism of TRADD shifting from Complex I to Complex II still remain largely unknown. Another adaptor protein STAT1 has been shown to interact with both TRADD and TNFR1 for pro-apoptotic signaling [34]. Therefore, structural investigation of STAT1 complexed with both TRADD and TNFR1 may shed light on the functional switching mechanism involving multiple adaptor proteins.
The TRADD-DD binding site on the TNFR1-DD also overlaps with one of TNFR1-DD homo-trimerization sites based on mutagenesis data and the model structure. In TNFR1 signaling, however, the receptor undergoes trimerization and TRADD recruitment upon extracellular TNF trimer binding. Thus, it is unlikely that the TNFR1-DD could function as a homotrimer since the self-association of the TNFR1-DD would block TRADD-DD binding and Complex I formation. A possible mechanism is that ligand-induced receptor trimerization at the N-terminal of TNFR1 could lead to TNFR1-DD equilibrium between monomer and oligomer (dimer or trimer). Strong binding of the TNFR1-DD to the TRADD-DD may enable TNFR1 receptor to recruit three TRADD adaptors locally, which would further facilitate three TRADD-NTD to associate with trimeric TRAF2 for RIP1 ubiquitination. The trimeric TNFR1-DD could represent the inactive or closed state of TNFR1. Structural determination of the homotrimeric TNFR1-DD and two heterotrimeric complexes, TRADD-DD:RIP1-DD:TNFR1-DD and TRADD-DD:RIP1-DD:FADD-DD, is necessary to uncover the detailed mechanisms underlying TRADD-dependent TNFR1 signaling.
4.3. Structure of N-GlcNAcylated TRADD-DD
The TRADD-DD is also one of the specific targets of a range of bacterial type III secretion system (T3SS) effectors, which disrupt death receptor signaling and contribute to pathogen infection and disease progression. It has been found that Non-LEE-encoded effector B (NleB) of enteropathogenic Escherichia coli (EPEC) can add an N-Acetylglucosamine (GlcNAc) group to the TRADD-DD and other DD-containing proteins, including DDs of TNFR1, FADD and RIP1 [87]. Other NleB-homologous T3SS effectors, such as Salmonella secreted effector K1 (SseK1) and SseK3 from Salmonella enterica, can also perform the same or similar modification on the TRADD-DD [88], [89]. The NleB-catalyzed N-linked GlcNAcylation site on the TRADD-DD is specifically at surface-exposed Arg235 (Fig. B and C). Our structural models of two TRADD-DD complexes indicate that bulky GlcNAc modification on the DDs of TRADD, RIP1, TNFR1, and FADD may introduce steric hindrances in the binding interfaces and thus prevent the formation of these two heterotrimeric complexes. This structural mechanism explains how bacterial pathogens antagonize host defense by blocking NF-κB activation and apoptosis induction in EPEC-infected cells.
4.4. Binding specificity of the TRADD-DD
The TRADD-DD is able to interact with a variety of DDs, while it fails to bind certain DD-containing death receptors, such as Fas (CD95). Thus, an interesting question remains about the binding specificity between the TRADD-DD and other DDs. Structural comparison among DDs has shown that most DDs exhibit similar structural fold with a conserved hydrophobic core [82]. However, lengths and orientations of helixes vary in each DD, leading to diverse surface characteristics, including surface shape and charge. These differences are believed to be critical for determining DD binding and functional specificity [90]. Since the TRADD-DD has a highly charged surface [6], electrostatic interactions may play a key role in TRADD-DD interaction with other DDs. However, none of complex structures between the TRADD-DD and its interacting DDs at the atomic level are available at present, and two TRADD-DD-containing docking structures do not provide enough resolution to analyze detailed interface network. Thus, the binding specificity of TRADD-DD still remains largely unknown.
4.5. Inhibition of TRADD signaling functions
Since the critical roles of TRADD in the direct regulation of both cell survival and apoptosis, TRADD has long been considered a promising drug target for the treatment of various human diseases, such as breast cancer and neurodegenerative diseases. Analysis of surface concavities or pockets on TRADD suggests that the TRADD-DD is less likely a good drug target, while the TRADD-NTD possesses a unique hydrophobic pocket mainly formed by sidechains of residues in the β-sheet (Fig. 1). This pocket is also involved in binding TRAF2 (Fig. 3a). Small molecule compounds, such as ICCB-19 or Apt-1 (apostatin-1), have been proved to bind the TRADD-NTD at this hydrophobic pocket, disrupting TRAF2 binding and releasing TRAF2 from TRADD [91]. Therefore, inhibition of TRADD reduces NF-κB activation in TNF-stimulated cells. Since binding of the small molecules to TRADD-NTD stabilizes Complex I, cytosolic availability of TRADD for the formation of Complex IIa is also decreased and cellular apoptosis is blocked. In autophagy pathway, the release of TRAF2/cIAP1/2 from TRADD can mediate K63-linked ubiquitination of Beclin 1, which eventually leads to activation of autophagy in cells with accumulated mutant tau and restoration of cellular homeostasis. The binding affinity of ICCB-19 or Apt-1 to the TRADD-NTD is relatively weak, and the Kd is in μM scale. Therefore, structure-based optimization of small molecule compounds targeting the TRADD-NTD is a promising strategy to develop new drugs for inhibiting cell death and restoring homeostasis. Nevertheless, we cannot rule out the possibility that cryptic drug binding sites, which are nearly invisible in the unligated structure, exist on the TRADD-DD and could be explored by using NMR techniques.
5. Summary and outlook
Structural and functional progress in the last 25 years has considerably advanced our understanding of the roles of TRADD in cellular signaling, which mainly leads to NF-κB-dependent antiapoptosis or caspases-dependent apoptosis. With two functional domains, TRADD can serve as a platform for the recruitment of signaling proteins to various receptors and the formation of different signaling complexes. The relatively long and unstructured linker between two domains of TRADD may eliminate steric hindrance and allow the stable formation of a larger complex containing both TRADD-NTD and TRADD-CTD. The receptors known to date to employ TRADD for downstream signaling include TNFR1, DR3, DR4/5, DR6, p75NTR, TLR3, and TLR4. Besides receptor signaling, TRADD can also shuttle to the nucleus and has a critical role in cell survival in response to DNA damage. The complex structure between TRADD-NTD and the TRAF domain of TRAF2 discloses the detailed structural mechanism of TRAF2 recruitment by three TRADD molecules to TNFR1 and other receptors, including DR3. Although high-resolution complex structures between TRADD-CTD and its binding partners are currently not available, heterotrimeric HADDOCK structures of TRADD-DD:RIP1-DD:TNFR1-DD and TRADD-DD:RIP1-DD:FADD-DD provide the first insight into structural mechanisms underlying TRADD functions via DD interactions. Disruption of the complexes involving the TRADD-NTD and/or the TRADD-DD by either competitive binding or structural modification (glycosylation) may lead to inhibition of TRADD-mediated signaling. This could provide clues for drug discovery targeting TRADD. Nevertheless, lack of sufficient structural and biochemical data for TRADD-mediated receptor signaling, including TRAILRs, DR6, p75NTR, and TLRs, prevents us from deciphering the whole repertoire of TRADD adaptor roles. Structural determination of complexes between TRADD, especially the TRADD-DD, and its various intracellular interactors, will be critical to bridge the structural gap between the receptors and their downstream signaling molecules.
CRediT authorship contribution statement
Zhen Li: Conceptualization, Writing - original draft. Wensu Yuan: Writing - review & editing. Zhi Lin: Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Carlos F Ibáñez for manuscript proof reading. This work was supported by the National Natural Science Foundation of China (Grant No. 21974093 to Z Lin).
References
- 1.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science (80-) 1997;278:2075–80. https://doi.org/10.1126/science.278.5346.2075. [DOI] [PubMed]
- 2.Sadowski I., Stone J.C., Pawson T. A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol Cell Biol. 1986;6(12):4396–4408. doi: 10.1128/mcb.6.12.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pawson T. Non-catalytic domains of cytoplasmic protein-tyrosine kinases: regulatory elements in signal transduction. Oncogene. 1988;3:491–495. [PubMed] [Google Scholar]
- 4.Mayer B.J., Hamaguchi M., Hanafusa H. A novel viral oncogene with structural similarity to phospholipase C. Nature. 1988;332(6161):272–275. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- 5.Flynn D.C. Adaptor proteins. Oncogene. 2001;20(44):6270–6272. doi: 10.1038/sj.onc.1204769. [DOI] [PubMed] [Google Scholar]
- 6.Zhang N., Yuan W., Fan J.-S., Lin Z. Structure of the C-terminal domain of TRADD reveals a novel fold in the death domain superfamily. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-07348-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu H., Xiong J., Goeddel D.V. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 8.Hsu H., Shu H.-B., Pan M.-G., Goeddel D.V. TRADD–TRAF2 and TRADD–FADD interactions define two distinct TNF Receptor 1 signal transduction pathways. Cell. 1996;84(2):299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 9.Tsao D.H.H., McDonagh T., Telliez J.-B., Hsu S., Malakian K., Xu G.-Y., Lin L.-L. Solution structure of N-TRADD and characterization of the interaction of N-TRADD and C-TRAF2, a key step in the TNFR1 signaling pathway. Mol Cell. 2000;5(6):1051–1057. doi: 10.1016/s1097-2765(00)80270-1. [DOI] [PubMed] [Google Scholar]
- 10.Park Y.C., Ye H., Hsia C., Segal D., Rich R.L., Liou H.-C., Myszka D.G., Wu H. A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD–TRAF2 Interaction. Cell. 2000;101(7):777–787. doi: 10.1016/s0092-8674(00)80889-2. [DOI] [PubMed] [Google Scholar]
- 11.Dawson N.L., Lewis T.E., Das S., Lees J.G., Lee D., Ashford P., Orengo C.A., Sillitoe I. CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 2017;45(D1):D289–D295. doi: 10.1093/nar/gkw1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papoff G, Trivieri N, Marsilio S, Crielesi R, Lalli C, Castellani L, et al. N-terminal and C-terminal domains of calmodulin mediate FADD and TRADD interaction. PLoS One 2015;10:e0116251. https://doi.org/10.1371/journal.pone.0116251. [DOI] [PMC free article] [PubMed]
- 13.Weber C.H., Vincenz C. The death domain superfamily: a tale of two interfaces? Trends Biochem Sci. 2001;26(8):475–481. doi: 10.1016/s0968-0004(01)01905-3. [DOI] [PubMed] [Google Scholar]
- 14.Wajant H., Pfizenmaier K., Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10(1):45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 15.Wang C., Yu X., Yan Y., Yang W., Zhang S., Xiang Y. Tumor necrosis factor-alpha: a key contributor to intervertebral disc degeneration. Acta Biochim Biophys Sin (Shanghai) 2017;49:1–13. doi: 10.1093/abbs/gmw112. [DOI] [PubMed] [Google Scholar]
- 16.Heller RA, Kronke M. Tumor necrosis factor receptor-mediated signaling pathways. J Cell Biol 1994;126:5–9. https://doi.org/10.1083/jcb.126.1.5. [DOI] [PMC free article] [PubMed]
- 17.Pobezinskaya Y.L., Liu Z. The role of TRADD in death receptor signaling. Cell Cycle. 2012;11(5):871–876. doi: 10.4161/cc.11.5.19300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodmer J.-L., Schneider P., Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27(1):19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 19.Dostert C., Grusdat M., Letellier E., Brenner D. The TNF family of ligands and receptors: communication modules in the immune system and beyond. Physiol Rev. 2019;99(1):115–160. doi: 10.1152/physrev.00045.2017. [DOI] [PubMed] [Google Scholar]
- 20.Sessler T., Healy S., Samali A., Szegezdi E. Structural determinants of DISC function: new insights into death receptor-mediated apoptosis signalling. Pharmacol Ther. 2013;140(2):186–199. doi: 10.1016/j.pharmthera.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Anderton H., Bandala-Sanchez E., Simpson D.S., Rickard J.A., Ng A.P., Di Rago L., Hall C., Vince J.E., Silke J., Liccardi G., Feltham R. RIPK1 prevents TRADD-driven, but TNFR1 independent, apoptosis during development. Cell Death Differ. 2019;26(5):877–889. doi: 10.1038/s41418-018-0166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Füllsack S., Rosenthal A., Wajant H., Siegmund D. Redundant and receptor-specific activities of TRADD, RIPK1 and FADD in death receptor signaling. Cell Death Dis. 2019;10(2) doi: 10.1038/s41419-019-1396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Condon S.M. Bivalent IAP antagonists inhibit TRAF2-bound cIAPs and limit TNF-mediated NF-kappaB signaling. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blonska M., Shambharkar P.B., Kobayashi M., Zhang D., Sakurai H., Su B. TAK1 is recruited to the tumor necrosis factor-alpha (TNF-alpha) receptor 1 complex in a receptor-interacting protein (RIP)-dependent manner and cooperates with MEKK3 leading to NF-kappaB activation. J Biol Chem. 2005;280:43056–43063. doi: 10.1074/jbc.M507807200. [DOI] [PubMed] [Google Scholar]
- 25.Jackson-Bernitsas D.G., Ichikawa H., Takada Y., Myers J.N., Lin X.L., Darnay B.G. Evidence that TNF-TNFR1-TRADD-TRAF2-RIP-TAK1-IKK pathway mediates constitutive NF-kappaB activation and proliferation in human head and neck squamous cell carcinoma. Oncogene. 2007;26:1385–1397. doi: 10.1038/sj.onc.1209945. [DOI] [PubMed] [Google Scholar]
- 26.Heyninck K., Beyaert R. A20 inhibits NF-kappaB activation by dual ubiquitin-editing functions. Trends Biochem Sci. 2005;30:1–4. doi: 10.1016/j.tibs.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Heyninck K., De Valck D., Vanden Berghe W., Van Criekinge W., Contreras R., Fiers W. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol. 1999;145:1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng M., Wang Y., Qiang L., Xu Y., Li C., Li T. Interleukin-35 inhibits TNF-alpha-induced osteoclastogenesis and promotes apoptosis via shifting the activation from TNF receptor-associated death domain (TRADD)-TRAF2 to TRADD-fas-associated death domain by JAK1/STAT1. Front Immunol. 2018;9:1417. doi: 10.3389/fimmu.2018.01417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Micheau O., Tschopp J. Induction of TNF receptor i-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114(2):181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 30.Fritsch J., Zingler P., Särchen V., Heck A.L., Schütze S. Role of ubiquitination and proteolysis in the regulation of pro- and anti-apoptotic TNF-R1 signaling. Biochim Biophys Acta (BBA) – Mol Cell Res. 2017;1864(11):2138–2146. doi: 10.1016/j.bbamcr.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 31.Kim H.S., Lee M.-S. STAT1 as a key modulator of cell death. Cell Signal. 2007;19(3):454–465. doi: 10.1016/j.cellsig.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Wu T.R., Cai S., Welte T., Chin Y.E. Stat1 as a component of tumor necrosis factor alpha receptor 1-TRADD signaling complex to inhibit NF-kappaB activation. Mol Cell Biol. 2000;20:4505–4512. doi: 10.1128/mcb.20.13.4505-4512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wesemann D.R., Qin H., Kokorina N., Benveniste E.N. TRADD interacts with STAT1-alpha and influences interferon-gamma signaling. Nat Immunol. 2004;5:199–207. doi: 10.1038/ni1025. [DOI] [PubMed] [Google Scholar]
- 34.Jiang Y., Yu M., Hu X., Han L.u., Yang K., Ba H., Zhang Z., Yin B., Yang X.-P., Li Z., Wang J. STAT1 mediates transmembrane TNF-alpha-induced formation of death-inducing signaling complex and apoptotic signaling via TNFR1. Cell Death Differ. 2017;24(4):660–671. doi: 10.1038/cdd.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chinnaiyan A.M., O'Rourke K., Yu G.-L., Lyons R.H., Garg M., Duan D.R., Xing L., Gentz R., Ni J., Dixit V.M. Signal transduction by DR3, a death domain-containing receptor related to TNFR-1 and CD95. Science. 1996;274(5289):990–992. doi: 10.1126/science.274.5289.990. [DOI] [PubMed] [Google Scholar]
- 36.Bamias G., Jia L.-G., Cominelli F. The tumor necrosis factor-like cytokine 1A/death receptor 3 cytokine system in intestinal inflammation: Curr Opin Gastroenterol. 2013;29(6):597–602. doi: 10.1097/MOG.0b013e328365d3a2. [DOI] [PubMed] [Google Scholar]
- 37.Kamada N., Hisamatsu T., Honda H., Kobayashi T., Chinen H., Takayama T. TL1A produced by lamina propria macrophages induces Th1 and Th17 immune responses in cooperation with IL-23 in patients with Crohn’s disease. Inflamm Bowel Dis. 2010;16:568–575. doi: 10.1002/ibd.21124. [DOI] [PubMed] [Google Scholar]
- 38.Cassatella M.A., da Silva G.P., Tinazzi I., Facchetti F., Scapini P., Calzetti F., Tamassia N., Wei P., Nardelli B., Roschke V., Vecchi A., Mantovani A., Bambara L.M., Edwards S.W., Carletto A. Soluble TNF-like cytokine (TL1A) production by immune complexes stimulated monocytes in rheumatoid arthritis. J Immunol. 2007;178(11):7325–7333. doi: 10.4049/jimmunol.178.11.7325. [DOI] [PubMed] [Google Scholar]
- 39.Facco M., Cabrelle A., Calabrese F., Teramo A., Cinetto F., Carraro S., Martini V., Calzetti F., Tamassia N., Cassatella M.A., Semenzato G., Agostini C. TL1A/DR3 axis involvement in the inflammatory cytokine network during pulmonary sarcoidosis. Clin Mol Allergy. 2015;13(1) doi: 10.1186/s12948-015-0022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ashkenazi A., Dixit V.M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 41.Pobezinskaya Y.L., Choksi S., Morgan M.J., Cao X., Liu Z.-G. The adaptor protein TRADD is essential for TNF-like ligand 1A/death receptor 3 signaling. J Immunol. 2011;186(9):5212–5216. doi: 10.4049/jimmunol.1002374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bittner S., Knoll G., Füllsack S., Kurz M., Wajant H., Ehrenschwender M. Soluble TL1A is sufficient for activation of death receptor 3. FEBS J. 2016;283(2):323–336. doi: 10.1111/febs.13576. [DOI] [PubMed] [Google Scholar]
- 43.Jia L.-G., Bamias G., Arseneau K.O., Burkly L.C., Wang E.C.Y., Gruszka D., Pizarro T.T., Cominelli F. A novel role for TL1A/DR3 in protection against intestinal injury and infection. JI. 2016;197(1):377–386. doi: 10.4049/jimmunol.1502466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bittner S., Ehrenschwender M. Multifaceted death receptor 3 signaling—promoting survival and triggering death. FEBS Lett. 2017;591:2543–2555. doi: 10.1002/1873-3468.12747. [DOI] [PubMed] [Google Scholar]
- 45.Bittner S., Knoll G., Ehrenschwender M. Death receptor 3 mediates necroptotic cell death. Cell Mol Life Sci. 2017;74(3):543–554. doi: 10.1007/s00018-016-2355-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim M.-H., Billiar T.R., Seol D.-W. The secretable form of trimeric TRAIL, a potent inducer of apoptosis. Biochem Biophys Res Commun. 2004;321(4):930–935. doi: 10.1016/j.bbrc.2004.07.046. [DOI] [PubMed] [Google Scholar]
- 47.Cha S.S., Kim M.S., Choi Y.H., Sung B.J., Shin N.K., Shin H.C. 2.8 A resolution crystal structure of human TRAIL, a cytokine with selective antitumor activity. Immunity. 1999;11:253–261. doi: 10.1016/s1074-7613(00)80100-4. [DOI] [PubMed] [Google Scholar]
- 48.Yuan X., Gajan A., Chu Q., Xiong H., Wu K., Wu G.S. Developing TRAIL/TRAIL death receptor-based cancer therapies. Cancer Metastasis Rev. 2018;37(4):733–748. doi: 10.1007/s10555-018-9728-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan L., Fu T.-M., Zhao W., Zhao L., Chen W., Qiu C., Liu W., Liu Z., Piai A., Fu Q., Chen S., Wu H., Chou J.J. Higher-order clustering of the transmembrane anchor of DR5 drives signaling. Cell. 2019;176(6):1477–1489.e14. doi: 10.1016/j.cell.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider P., Thome M., Burns K., Bodmer J.L., Hofmann K., Kataoka T. TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-kappaB. Immunity. 1997;7:831–836. doi: 10.1016/s1074-7613(00)80401-x. [DOI] [PubMed] [Google Scholar]
- 51.Cao X., Pobezinskaya Y.L., Morgan M.J., Liu Z.-G. The role of TRADD in TRAIL‐induced apoptosis and signaling. FASEB J. 2011;25(4):1353–1358. doi: 10.1096/fj.10-170480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J.-Y., Lee J.-Y., Kim D.-G., Koo G.-B., Yu J.-W., Kim Y.-S. TRADD is critical for resistance to TRAIL-induced cell death through NF-kappaB activation. FEBS Lett. 2011;585:2144–2150. doi: 10.1016/j.febslet.2011.05.034. [DOI] [PubMed] [Google Scholar]
- 53.Zhang L., Dittmer M.R., Blackwell K., Workman L.M., Hostager B., Habelhah H. TRAIL activates JNK and NF-kappaB through RIP1-dependent and -independent pathways. Cell Signal. 2015;27:306–314. doi: 10.1016/j.cellsig.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin Y., Devin A., Cook A., Keane M.M., Kelliher M., Lipkowitz S., Liu Z.-G. The death domain kinase RIP is essential for TRAIL (Apo2L)-induced activation of IκB kinase and c-Jun N-terminal kinase. Mol Cell Biol. 2000;20(18):6638–6645. doi: 10.1128/mcb.20.18.6638-6645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benschop R., Wei T., Na S. Tumor necrosis factor receptor superfamily member 21: TNFR-related death receptor-6, DR6. Adv Exp Med Biol. 2009;647:186–194. doi: 10.1007/978-0-387-89520-8_13. [DOI] [PubMed] [Google Scholar]
- 56.Kasof G.M., Lu J.J., Liu D., Speer B., Mongan K.N., Gomes B.C. Tumor necrosis factor-alpha induces the expression of DR6, a member of the TNF receptor family, through activation of NF-kappaB. Oncogene. 2001;20:7965–7975. doi: 10.1038/sj.onc.1204985. [DOI] [PubMed] [Google Scholar]
- 57.Xu K., Olsen O., Tzvetkova-Robev D., Tessier-Lavigne M., Nikolov D.B. The crystal structure of DR6 in complex with the amyloid precursor protein provides insight into death receptor activation. Genes Dev. 2015;29(8):785–790. doi: 10.1101/gad.257675.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu R, Du Q, Yin X, Li J, Wang T, Zhang L. Agonist antibody activates death receptor 6 downstream signaling involving TRADD recruitment. FEBS Lett 2014;588:401–7. https://doi.org/10.1016/j.febslet.2013.12.010. [DOI] [PubMed]
- 59.Herrup K., Shooter E.M. Properties of the beta nerve growth factor receptor of avian dorsal root ganglia. Proc Natl Acad Sci U S A. 1973;70:3884–3888. doi: 10.1073/pnas.70.12.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Underwood C.K., Coulson E.J. The p75 neurotrophin receptor. The Int J Biochem Cell Biol. 2008;40(9):1664–1668. doi: 10.1016/j.biocel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 61.Vilar M. Structural characterization of the p75 neurotrophin receptor: a stranger in the TNFR superfamily. Vitam Horm. 2017;104:57–87. doi: 10.1016/bs.vh.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 62.He X.-L., Garcia K.C. Structure of nerve growth factor complexed with the shared neurotrophin receptor p75. Science. 2004;304:870–875. doi: 10.1126/science.1095190. [DOI] [PubMed] [Google Scholar]
- 63.Nadezhdin K.D., García-Carpio I., Goncharuk S.A., Mineev K.S., Arseniev A.S., Vilar M. Structural basis of p75 transmembrane domain dimerization. J Biol Chem. 2016;291(23):12346–12357. doi: 10.1074/jbc.M116.723585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liepinsh E., Ilag L.L., Otting G., Ibanez C.F. NMR structure of the death domain of the p75 neurotrophin receptor. EMBO J. 1997;16:4999–5005. doi: 10.1093/emboj/16.16.4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yazidi-Belkoura I.E., Adriaenssens E., Dollé L., Descamps S., Hondermarck H. Tumor necrosis factor receptor-associated death domain protein is involved in the neurotrophin receptor-mediated antiapoptotic activity of nerve growth factor in breast cancer cells. J Biol Chem. 2003;278(19):16952–16956. doi: 10.1074/jbc.M300631200. [DOI] [PubMed] [Google Scholar]
- 66.Chen J.-Q., Szodoray P., Zeher M. Toll-like receptor pathways in autoimmune diseases. Clinic Rev Allerg Immunol. 2016;50(1):1–17. doi: 10.1007/s12016-015-8473-z. [DOI] [PubMed] [Google Scholar]
- 67.Takeda K., Akira S. Toll‐like receptors. Curr Protoc Immunol. 2015;109(1) doi: 10.1002/0471142735.2015.109.issue-1. [DOI] [PubMed] [Google Scholar]
- 68.Matsumoto M., Seya T. TLR3: Interferon induction by double-stranded RNA including poly(I:C)☆. Adv Drug Deliv Rev. 2008;60(7):805–812. doi: 10.1016/j.addr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 69.Park BS, Lee J-O. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med 2013;45:e66. https://doi.org/10.1038/emm.2013.97. [DOI] [PMC free article] [PubMed]
- 70.Ermolaeva M.A., Michallet M.-C., Papadopoulou N., Utermöhlen O., Kranidioti K., Kollias G., Tschopp J., Pasparakis M. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat Immunol. 2008;9(9):1037–1046. doi: 10.1038/ni.1638. [DOI] [PubMed] [Google Scholar]
- 71.Sharma S., Garg I., Ashraf M.Z. TLR signalling and association of TLR polymorphism with cardiovascular diseases. VascPharmacol. 2016;87:30–37. doi: 10.1016/j.vph.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 72.Funami K, Matsumoto M, Oshiumi H, Inagaki F, Seya T. Functional interfaces between TICAM-2/TRAM and TICAM-1/TRIF in TLR4 signaling. Biochem Soc Trans 2017;45:929–35. https://doi.org/10.1042/BST20160259. [DOI] [PubMed]
- 73.Ullah M.O., Ve T., Mangan M., Alaidarous M., Sweet M.J., Mansell A., Kobe B. The TLR signalling adaptor TRIF/TICAM-1 has an N-terminal helical domain with structural similarity to IFIT proteins. Acta Crystallogr D Biol Crystallogr. 2013;69(12):2420–2430. doi: 10.1107/S0907444913022385. [DOI] [PubMed] [Google Scholar]
- 74.Kawai T., Akira S. TLR signaling. Semin Immunol. 2007;19(1):24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 75.Chen N.-J., Chio I.I.C., Lin W.-J., Duncan G., Chau H., Katz D., Huang H.-L., Pike K.A., Hao Z., Su Y.-W., Yamamoto K., de Pooter R.F., Zuniga-Pflucker J.C., Wakeham A., Yeh W.-C., Mak T.W. Beyond tumor necrosis factor receptor: TRADD signaling in toll-like receptors. Proc Natl Acad Sci. 2008;105(34):12429–12434. doi: 10.1073/pnas.0806585105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.He M., Wang M., Huang Y., Peng W., Zheng Z., Xia N., Xu J., Tian D. The ORF3 protein of genotype 1 hepatitis E virus suppresses TLR3-induced NF-κB Signaling via TRADD and RIP1. Sci Rep. 2016;6(1) doi: 10.1038/srep27597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pobezinskaya Y.L., Kim Y.-S., Choksi S., Morgan M.J., Li T., Liu C., Liu Z. The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat Immunol. 2008;9(9):1047–1054. doi: 10.1038/ni.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koo G.-B., Ji J.-H., Cho H., Morgan M.J., Kim Y.-S. Nuclear TRADD prevents DNA damage-mediated death by facilitating non-homologous end-joining repair. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-03211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bender L.M., Morgan M.J., Thomas L.R., Liu Z.-G., Thorburn A. The adaptor protein TRADD activates distinct mechanisms of apoptosis from the nucleus and the cytoplasm. Cell Death Differ. 2005;12(5):473–481. doi: 10.1038/sj.cdd.4401578. [DOI] [PubMed] [Google Scholar]
- 80.Morgan M, Thorburn J, Pandolfi PP, Thorburn A. Nuclear and cytoplasmic shuttling of TRADD induces apoptosis via different mechanisms. J Cell Biol 2002;157:975–84. https://doi.org/10.1083/jcb.200204039. [DOI] [PMC free article] [PubMed]
- 81.Gaud G., Guillemot D., Jacob Y., Favre M., Vuillier F. EVER2 protein binds TRADD to promote TNF-alpha-induced apoptosis. Cell Death Dis. 2013;4 doi: 10.1038/cddis.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sandu C., Gavathiotis E., Huang T., Wegorzewska I., Werner M.H. A mechanism for death receptor discrimination by death adaptors. J Biol Chem. 2005;280(36):31974–31980. doi: 10.1074/jbc.M506938200. [DOI] [PubMed] [Google Scholar]
- 83.Park Y.-H., Jeong M.S., Jang S.B. Death domain complex of the TNFR-1, TRADD, and RIP1 proteins for death-inducing signaling. Biochem Biophys Res Commun. 2014;443(4):1155–1161. doi: 10.1016/j.bbrc.2013.12.068. [DOI] [PubMed] [Google Scholar]
- 84.Telliez J.B., Xu G.Y., Woronicz J.D., Hsu S., Wu J.L., Lin L. Mutational analysis and NMR studies of the death domain of the tumor necrosis factor receptor-1. J Mol Biol. 2000;300:1323–1333. doi: 10.1006/jmbi.2000.3899. [DOI] [PubMed] [Google Scholar]
- 85.Park Y.-H., Jeong M.S., Park H.H., Jang S.B. Formation of the death domain complex between FADD and RIP1 proteins in vitro. Biochim Biophys Acta (BBA) – Proteins Proteom. 2013;1834(1):292–300. doi: 10.1016/j.bbapap.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 86.van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A.M.J.J. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol. 2016;428(4):720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 87.Ding J., Pan X., Du L., Yao Q., Xue J., Yao H., Wang D.-C., Li S., Shao F. Structural and functional insights into host death domains inactivation by the bacterial arginine GlcNAcyltransferase effector. Mol Cell. 2019;74(5):922–935.e6. doi: 10.1016/j.molcel.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 88.Newson J.P.M., Scott N.E., Yeuk Wah Chung I., Wong Fok Lung T., Giogha C., Gan J., Wang N., Strugnell R.A., Brown N.F., Cygler M., Pearson J.S., Hartland E.L. Salmonella effectors SseK1 and SseK3 target death domain proteins in the TNF and TRAIL signaling pathways*. Mol Cell Proteom. 2019;18(6):1138–1156. doi: 10.1074/mcp.RA118.001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gunster R.A., Matthews S.A., Holden D.W., Thurston T.L.M. SseK1 and SseK3 Type III secretion system effectors inhibit NF-kappaB signaling and necroptotic cell death in salmonella-infected macrophages. Infect Immun. 2017;85 doi: 10.1128/IAI.00010-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park H.H. Structural analyses of death domains and their interactions. Apoptosis. 2011;16(3):209–220. doi: 10.1007/s10495-010-0571-z. [DOI] [PubMed] [Google Scholar]
- 91.Xu D, Zhao H, Jin M, Zhu H, Shan B, Geng J, et al. Modulating TRADD to restore cellular homeostasis and inhibit apoptosis. Nature 2020. https://doi.org/10.1038/s41586-020-2757-z. [DOI] [PubMed]