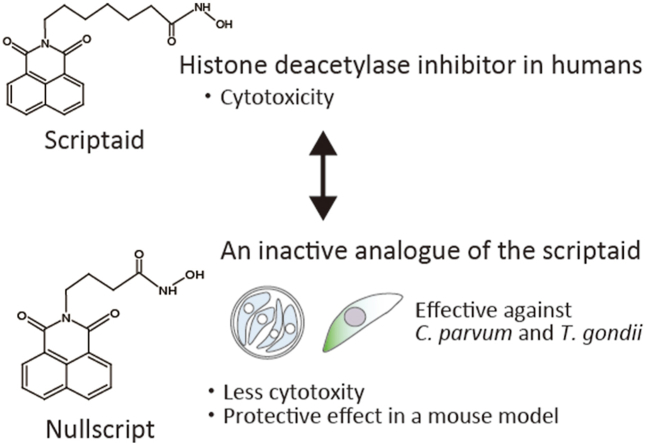
Abstract
Cryptosporidium and Toxoplasma are parasites that have caused problems worldwide. Cryptosporidium causes severe watery diarrhoea and may be fatal in immunocompromised patients and in infants. Nitazoxanide is the only agent currently approved by the FDA, but its efficacy is limited. Toxoplasmosis is also a problem in the immunocompromised, as currently available treatment options have limited efficacy and patient tolerance can be poor. In the present investigation, we screened libraries of epigenetic compounds to identify those that inhibited C. parvum growth. Nullscript was identified as a compound with an inhibitory effect on C. parvum and T. gondii growth, and was less toxic to host cells. Nullscript was also able to significantly decrease oocyst excretion in C. parvum-infected SCID mice.
Keywords: Cryptosporidium, HDAC inhibitor, Nullscript, Toxoplasma
Graphical abstract
Highlights
-
•
A library of epigenetic compounds was used to screen for compounds effective against Cryptosporiium parvum.
-
•
Nullscript has been shown to be effective in inhibiting the growth of C. parvum and T. gondii.
-
•
Nullscript significantly decreased oocyst excretion in C. parvum-infected SCID mice.
1. Introduction
Cryptosporidium parvum and Toxoplasma gondii are pathogenic protozoan parasites belonging to the Apicomplexa; they cause cryptosporidiosis and toxoplasmosis, respectively.
C. parvum causes severe watery diarrhoea. Its oocysts are resistant to chlorine; therefore, waterborne outbreaks of cryptosporidiosis often occur. In the absence of effective therapeutic agents and vaccines, infections in immunocompromised patients are sometimes fatal (Bohne et al., 1994). Nitazoxanide (NTZ) is an FDA-approved drug for the treatment of cryptosporidiosis; however, its effectiveness is limited in immunocompromised patients (Amadi et al., 2009).
Toxoplasmosis is caused by ingestion of meat from infected animals that contains tissue cysts (bradyzoites) or by ingestion of oocysts in food or water. Toxoplasma proliferates in the host as a tachyzoite causing acute disease, but then converts to a bradyzoite and remains in the tissue. Bradyzoites usually do not cause symptoms; however, they may re-activate (i.e., convert to tachyzoites) in immunocompromised individuals and cause morbidity (Hill and Dubey, 2002). Pyrimethamine and sulfadiazine are the standard drug treatment for toxoplasmosis; however, these agents cannot remove T. gondii bradyzoites (Luft and Remington, 1988).
Accordingly, it is important to search for new, effective agents against these parasites.
Gene expression is controlled by modifications to DNA or histones in addition to the gene sequence. Histone deacetylase (HDAC) removes acetylated sites on histones by hydrolysis, and inhibiting transcription. Histone acetylases (HAT), on the other hand, add acetyl groups to histones, thereby weakening the association between histones and DNA, making it easier for transcription factors to bind to DNA and promoting gene expression. It has also been reported that non-histone proteins undergo acetylation modification by histone acetylases (HATs)/histone deacetylases (HDACs), which alters the stability of the protein, thereby regulating its bioactivity. In humans, the histone deacetylase inhibitor (HDACi) SAHA was the first approved anticancer drug in the United States (Duvic and Vu, 2007). In protozoan parasites, apicidin was first reported to be effective against Plasmodium malaria parasites (Darkin-Rattray et al., 1996) and various HDAC inhibitors have since been reported to be effective against protozoan parasites (e.g. Campo, 2017; Carrillo et al., 2015; Chua et al., 2017; Engel et al., 2015; Guo et al., 2018; Sereno et al., 2005; Strobl et al., 2007; Vergnes et al., 2005). However, it should be noted that as HDAC inhibitors can also target human HDACs, any potential HDAC inhibitor developed for parasitic diseases for humans would need to address the question of selectivity/off target activity.
Here, we used an epigenetic library to screen compounds with growth inhibitory effects on C. parvum. This screening led to the identification of nullscript, which was not toxic to host cells and showed growth-inhibitory effects on Cryptosporidium and Toxoplasma. Nullscript also showed a protective effect against Cryptosporidium in a mouse model.
2. Material and methods
2.1. Compounds
An epigenetics library (containing various compounds with epigenetic activity and different structures and mechanisms of action) (ENZO; CB-BML-28360100) was provided by the Cancer Research Institute of Kanazawa University (http://ganken.cri.kanazawa-u.ac.jp/). This library contains 43 compounds. Of these, 42 commercially available compounds were used in the experiments. The compounds were dissolved in DMSO at a concentration of 10 mM and stored at −80 °C. Pyrimethamine (Wako, Osaka, Japan) and nitazoxanide (Sigma-Aldrich, St. Louis, USA) served as positive controls for T. gondii and C. parvum, respectively. Apicidin (CaymanChemical, USA) was used as a positive control as an HDAC inhibitor of C. parvum.
2.2. Culture of Toxoplasma gondii and Cryptosporidium parvum
As host cells for T. gondii, the human foreskin fibroblast (HFF) cell line (ATCC: SCRC-1041) were maintained in DMEM (1.0 g/l Glucose) with L-Gln and sodium pyruvate (Nacalai Tesque, Tokyo, Japan) with 10% foetal bovine serum (FBS) and Penicillin (100 U/ml) -Streptomycin (100 μg/ml) Solution (Nacalai). T. gondii RH-2F (ATCC: 50839), expressing β-galactosidase was maintained as previously described (Sugi et al., 2014). C. parvum oocysts, strain HNJ-1 (Masuda et al., 1991; Satoh et al., 2005), were kindly provided by Dr. M. Matsubayashi (Osaka Prefecture University). Oocysts were maintained by passage in experimentally infected SCID mice (CB17/Icr-Prkdcscid/CrlCrlj) (Charles River Laboratories, Shizuoka, Japan) and were purified from faeces by using discontinuous sucrose and caesium chloride gradients as described previously (Arrowood and Donaldson, 1996). C. parvum oocysts less than 3 months since harvest were used in all experiments. This animal experiments were approved by the Ethical Committee of the Obihiro University of Agriculture and Veterinary Medicine (No. 29-72) and the Committee on the Animal Experiments of the Kyoto Prefectural University of Medicine (M2020-243).
As host cells for C. parvum, The HCT-8 cell line derived from human ileocecal adenocarcinoma (ATCC # CCL-225) were maintained in RPMI 1640 with L-Gln and (Nacalai Tesque, Tokyo, Japan) with 1 mM sodium pyruvate, 15 mM HEPES, 10% foetal bovine serum (FBS) and Penicillin (100 U/ml) -Streptomycin (100 μg/ml) - Amphotericin B (0.25 μg/ml) Solution (Nacalai).
2.3. In initial epigenetic library compound screening for C. parvum
An initial epigenetic library compound screening for C. parvum was performed as follows: HCT-8 cells were seeded in 96-well plates at a density of 2 × 104 cells/well and allowed to grow overnight. Cryptosporidium oocysts were excited to sporozoites immediately before infecting cells. Briefly, oocysts were suspended in 1 ml of 0.1% sodium hypochlorite with PBS, incubated for 5 min in 4 °C, centrifuged at 3000 rpm for 10 min, and washed three times with PBS. Subsequently, the oocysts were suspended in excystation solution (consisting of 0.75% sodium taurocholate and 0.25% trypsin diluted in phosphate buffer (PB); pH 8.0) and incubated for 1 h at 37 °C. Subsequently, the oocyst/sporozoite suspensions were washed once in PBS, then filtered to remove the oocyst wall. HCT-8 cells were incubated with C. parvum sporozoites (6 × 104 sporozoites/well) suspended with 5 μM library compound or 0.1% DMSO in RPMI medium. Parasite-infected cells were cultured for 48 h. The cells were fixed with ice-cold 100% methanol for 10 min, and then washed with PBS three times. The cells were then blocked with 1% BSA in PBS for 30 min and then stained with Sporo-Glo (an anti-Cryptosporidium polyclonal antibody) (Waterborne Environmental, Inc., VA, USA) for 1 h. Images were acquired by using a Keyence BZ-X810 (KEYENCE, Osaka, Japan). All Cryptosporidium in the well were counted. Nine compounds that inhibited C. parvum growth by more than 70% compared with DMSO control, were defined as “candidate compounds” and the following experiments were conducted.
2.4. Analyses of C. parvum growth-inhibitory effects of hit compounds identified in primary screening
HCT-8 cells (2 × 104 cells/well) were plated in 96-well plates and cultured for 24 h, and subsequently infected with 5 × 104 C. parvum oocysts in the presence of 5 μM of 9 compounds hit in the initial screening, scriptaid or DMSO control. Experiments were also conducted using nullscript at concentrations of 0 (DMSO Control), 0.5, 2, 5, 10 or 20 μM to determine the IC50 of nullscript. Three hours after inoculation, the oocysts that did not invade were washed with RPMI-1640 and incubated for additional 45 h with RPMI-1640 with each concentration of the compound added.
In addition, to investigate the effect of nullscript after C. parvum invaded the cells, the following experiments were performed. The cells were infected with 5 × 104 C. parvum oocysts in RPMI-1640 medium for 3 h. The cells were washed with new RPMI-1640 medium in order to wash out the oocysts that did not invade and then new RPMI-1640 medium containing nullscript or nitazoxanide (0 or 5 μM) was added and the cells were incubated for further 45 h. That is, nullscript was added after C. parvum invaded the host cells, and the growth inhibitory effect of nullscript on C. parvum after invasion of the host cells was examined. Sporo-Glo was used for C. parvum staining as described above. Images of parasites were acquired by the IN Cell Analyzer 2200 (GE Healthcare, Chicago, IL, USA) (42 field images were acquired per well using a 20× objective lens.), and the number of Sporo-Glo positive C. parvum and cell nuclei visible was calculated by IN Cell Developer Toolbox software (GE Healthcare).
2.5. The quantitative determination of cell viability
Cytotoxicity to HCT-8 cells induced by candidate compounds was measured. Briefly, HCT-8 cells (5 × 103 cells/well) were plated in 96-well plates and cultured for 24 h, and subsequently treated with 0–100 μM candidate compounds for 48 h. Then, add 10 μl of Cell Count Reagent SF (nacalai) to each well and carry out a color reaction in a CO2 incubator for 1–4 h, and then cell viability was measured according to the manufacturer's instructions.
Cytotoxicity to HFF cells induced by nullscript was measured as previously described (Bando et al., 2018). The cell viability was monitored by measuring lactate dehydrogenase (LDH) leakage into the culture medium by using CytoTox96 Non-Radio Cytotoxicity Assay kit (Promega). HFF cells (1 × 104 cells/well) were plated in 96-well plates and cultured for 24 h, and subsequently treated with 100 μM of nullscript for 48 h. Culture supernatant was collected and centrifuged at 3000 rpm for 5 min, and then the activity of LDH in Cell free supernatant was measured according to the manufacturer's instructions. Cell-free medium was used for negative control. Culture supernatant of TritonX-100 (0.1%) treated cells to kill the all cells was used for positive control.
2.6. Growth assay and estimating EC50 of T. gondii
HFF cells (1 × 104 cells/well) were plated in 96-well plates and cultured for 24 h, and subsequently infected with 5 × 103 T. gondii RH-2F strain in the presence or absence of nullscript (0, 2, 5, 12.5, 25, 50, 65 or 100 μM) or pyrimethamine (5 μM) for 48 h. The number of T. gondii were measured by Beta-Glo® Assay System (Promega). To measure the number of T. gondii, all infected cells were lysed and the activity of beta-galactosidase in the cell lysates was measured according to the manufacturer's instructions. The percentages of the activities in nullscript or pyrimethamine treated cells over those in unstimulated cells were shown as “Relative T. gondii numbers” in figures.
2.7. Measurement of acetylated histone in extracellular parasites treated with nullscript
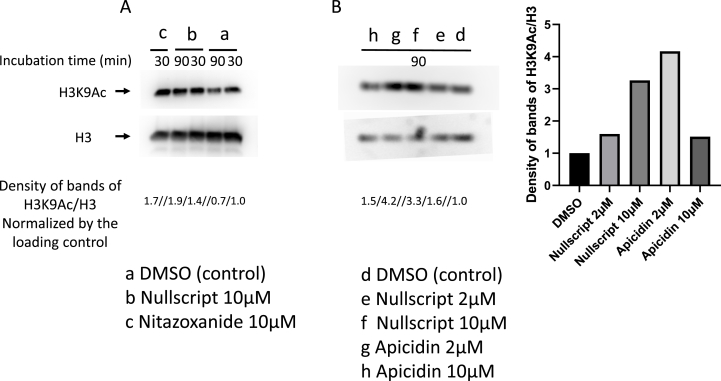
C. parvum oocysts were encysted and 8 × 106 C. parvum sporozoites were incubated in 1% DMEM, 2 or 10 μM of nullscript, nitazoxanide and apicidin with DMEM for 30 or 90 min at 37 °C. Apicidin was used as a control for HDACi effective against C. parvum. Parasites were centrifuged at 3000 rpm for 10 min at 4 °C, resuspended in 20 μl of SDS lysis buffer (50 mM Tris-HCl (pH 8.0), 1% SDS and 10 mM EDTA) and then, 20 μl of dilution buffer (50 mM Tris-HCl (pH 8.0), 167 mM 5 M NaCl and 1.1% Triton X-100) was added and incubated for 5min on ice. 40 μl of SDS-sample buffer was then added to the samples and boiled for 5min. SDS-PAGE was carried out with 20 μl of each sample using 16.5% polyacrylamide gel. In Western blotting analysis, acetylated histone H3K9 was detected by a mouse monoclonal antibody (MABI 0305); an anti-histone H3 mouse monoclonal antibody (MABI 0301) was used as a loading control. The band density obtained by Western blotting was quantified using Image J software 1.53c. Furthermore, the density of the acetylated histone H3K9Ac bands was normalized by the density of the histone H3 loading control bands.
2.8. Statistical analysis
Except for the result of Fig. 1, each point represents mean (from three technical replicates) of one individual experiment. Therefore, three points in all graphs represent three means derived from three independent experiments (three biological replicates). The experimental points and n values represent an average of each three biological replicates (three independent experiments). Statistical analyses were performed using Prism7 (GraphPad). The statistical significance of differences in mean values was analyzed by using an unpaired two-tailed Student's or Welch's t-test or Tukey's multiple comparisons test. P values < 0.05 were considered to be statistically significant.
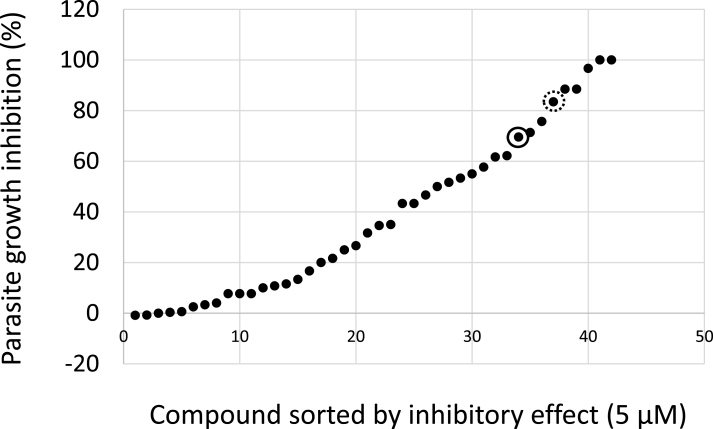
Fig. 1.
Screening for anti-Cryptosporidium activity. C. parvum HNJ-1 strain-infected HCT-8 cells were incubated with 5 μM epigenetic library compounds for 48 h. A circle indicates a nullscript. A dashed circle indicates a positive control, apicidin. This is a single experiment and single technical replicate.
2.9. Animal experiments
Five-week-old female SCID mice (Charles River Laboratories, Shizuoka, Japan) were used in this study. For the in vivo infection analysis, 1.0 × 105 C. parvum oocysts were inoculated orally (PO) with nullscript (10 mg/kg) in 50% DMSO or with 50% DMSO in PBS (solvent control) (Day 0). Five mice per a group were used in this experiment. Thereafter, until Day 5, 100 μl of nullscript (10 mg/kg) or DMSO was administered PO once daily. From Day 6 to Day 9, the compounds were administered in the diet. Faecal samples were collected daily from Day 1 to Day 9. The number of oocysts in the faeces was counted by using the sugar flotation method and the total number of oocysts per gram (OPG) was determined.
The animal experiments described in this study were approved by the Ethical Committee of The Obihiro University of Agriculture and Veterinary Medicine (No. 29-72) and were performed at the Obihiro University of Agriculture and Veterinary Medicine.
3. Results
3.1. Screening of the epigenetics library for anti-Cryptosporidium compounds
A total of 42 compounds were screened for their ability to inhibit the growth of Cryptosporidium at a concentration of 5 μM (Fig. 1). The Sporo-glo fluorescence in wells to which only DMSO was added were considered as 0% inhibition. The epigenetic libraries used and the results of the initial screening were described separately (Supplementary table). Apicidin has been reported to be effective against Cryptosporidium, with a minimum concentration of 45 μM at which all C. parvum growth was inhibited (Darkin-Rattray et al., 1996). It was also among the candidate compounds obtained. Nine compounds showed a growth inhibition rate of >70% (Fig. 1), which was the cut-off selected to define a hit compound for further analyses. Table 1 shows C. parvum growth inhibition values at concentrations of 5 μM and host cell IC50 values with HCT-8 cells. Most of the compounds with >50% inhibition are HDAC inhibitors (including one SIRT2 inhibition). Some compounds (Apicidin, TrichostatinA, M-344, Oxamflatin and SAHA) had low host cell IC50 values, indicating that the growth of C. parvum was inhibited by cell death. SAHA and Oxamflatin also showed a 100% parasite inhibitory effect in the first screen (Table 1 (a)), but show no inhibitory effect in a subsequent detailed analysis. This was thought to be due to variations in the effects of compounds on host cells caused by experimental errors due to the very low host cell IC50 values. For CI -994, there was an effect on growth inhibition of the parasite in primary screening, but when the experiment was repeated, there was no effect on growth inhibition of C. parvum. Therefore, among the compound libraries used in this study, nullscript had the highest host cell IC50 value and had comparable growth-inhibiting effects on C. parvum compared to other compounds. BML -266 and NSC -3852 were also suggested to have certain inhibitory effects on C. parvum. Nullscript is an analog of scriptaid (Fig. 2A) (Su et al., 2000), it does not have the same HDAC inhibitory activity as scriptaid and is therefore used as a control for that compound but scriptaid did not inhibit the growth of C. parvum. For these reasons, we focused on nullscript, which has been shown to be non-toxic to host cells, and conducted the following experiments.
Table 1.
Effects of hit compounds and scriptaid on Cryptosporidium and HCT-8 cells.
| ID | Compound | Parasite inhibition (%) (primary screening) (a) | Parasite inhibition (%) (b) | Host cell IC50 (μM) (c) | Effects in mammalian cells |
|---|---|---|---|---|---|
| A6 | Apicidin | 83 | 83.5 ± 1.4 | 1.5 ± 0.1 | HDAC inhibitor |
| A1 | TrichostatinA | 76 | 75.7 ± 0.8 | 1.6 ± 0.1 | HDAC inhibitor |
| B8 | M-344 | 71 | 71.3 ± 1.7 | 2.3 ± 0.2 | HDAC inhibitor, which shows selectivity for HDAC6 |
| A9 | Nullscript | 70 | 67.6 ± 10.4 | 87.1 ± 2.5 | Analog of scriptaid |
| C10 | BML-266 | 97 | 64.7 ± 3.9 | 51.2 ± 0.6 | Sirtuin Type 2 (SIRT2) Inhibitor |
| D4 | NSC-3852 | 88 | 64.4 ± 0.4 | 36.7 ± 1.0 | HDAC inhibitor |
| C10 | Oxamflatin | 100 | 49.8 ± 4.0 | 2.5 ± 0.1 | HDAC inhibitor |
| A12 | SAHA | 100 | −0.8 ± 5.8 | 2.5 ± 0.3 | HDAC inhibitor |
| D3 |
CI-994 |
88 |
−15.7 ± 8.4 |
24.6 ± 2.4 |
Class I HDAC inhibitor |
| A8 | Scriptaid | 10 | 4.0 ± 5.7 | 11.9 ± 1.3 | HDAC inhibitor |
(a) Results of the primary screening in Fig. 1. (b) Parasite growth inhibition values with C. parvum at concentrations of 5 μM. (c) Host cell IC50 values with HCT-8 cells.
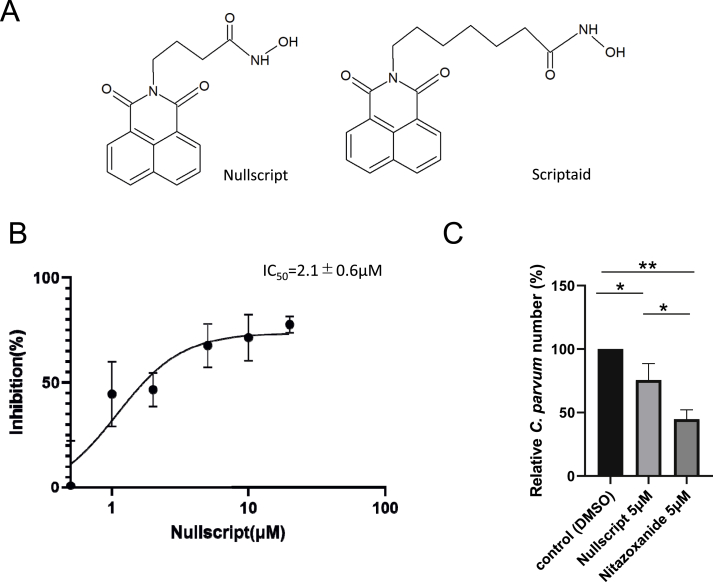
Fig. 2.
Dose-response curves and growth inhibitory effects of nullscript on C. parvum. (A) The structures of nullscript and scriptaid. (B) Dose-response curves and IC50 values of nullscript for C. parvum. (C) This is the relative C. parvum number when nullscript is added after invasion. C. parvum oocysts were incubated with 5 μM of nullscript or nitazoxanide (positive control) 45 h after a 3 h infection window. **p < 0.01; *p < 0.05; (Tukey's multiple comparisons test).
3.2. Dose-response curves and growth inhibitory effects of nullscript on C. parvum
We investigated the IC50 value of nullscript on C. parvum. Nullscript showed a dose-dependent growth inhibitory effect on C. parvum. The IC50 value of nullscript was 2.1 ± 0.6 μM and the Selectivity Index (SI) was 41 (Fig. 2B).
To investigate the step at which nullscript interferes with the life cycle of Cryptosporidium, after C. parvum invaded HCT-8, 5 μM nullscript (2.4 × IC50) or nitazoxanide (1.4 × IC50) was added to investigate the antiproliferative effect. That is, nullscript was added after C. parvum invaded the host cells, and the growth inhibitory effect of nullscript on C. parvum after invasion of the host cells was examined. IC50 of nitazoxanide was reported to be 3.7 μM (Bessoff et al., 2013) and 5 μM (1.4 × IC50) was used as the positive control in this study. As a result, the relative number of C. parvum was 75.7 ± 12.9% (24.3% inhibition) and 44.9 ± 7.3% (55.1% inhibition), respectively when nullscript or nitazoxanide was added compared with the DMSO control (Fig. 2C). In comparison to the above, in the experiment in Fig. 2B in which nullscript was added at the same time as C. parvum inoculation, the growth inhibitory effect of 5 μM nullscript was 67.5%. In other words, nullscript was found to be effective against C. parvum even at during entry into host cells. Thus, a nullscript has a certain growth inhibitory effect. However, it may be more effective when C. parvum is outside the cell.
3.3. Effects of nullscript on Toxoplasma and its host cell, HFF
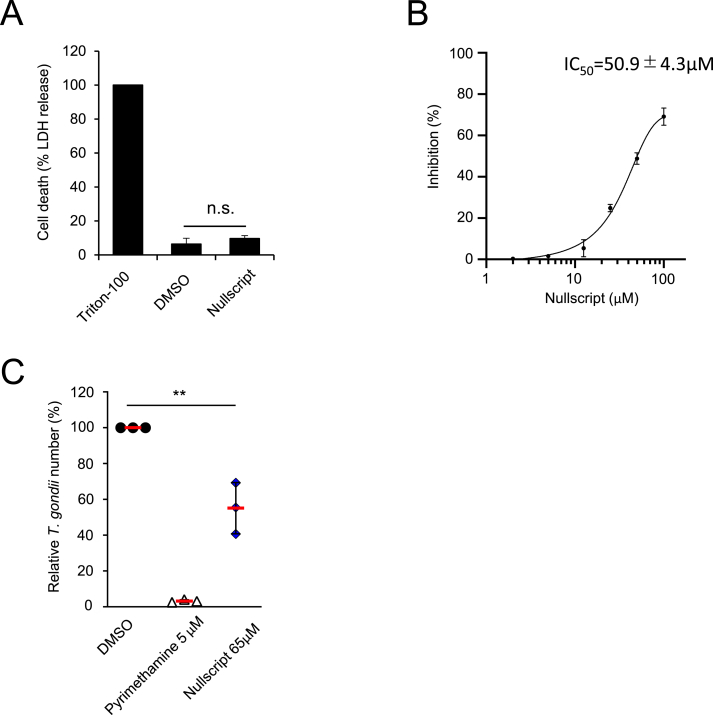
Because nullscript was found to inhibit the growth of C. parvum in cells, we examined its effects on Toxoplasma gondii, a member of the same Apicomplexa phylum, and its host cell, HFF. The results showed that 100 μM nullscript was not toxic to HFF (Fig. 3A). The IC50 value for nullscript against in vitro cultured T. gondii was 50.9 ± 4.3 μM (Fig. 3B). The above results were confirmed again in Fig. 3C. 5 μM of pyrimethamine was used as a positive control at the concentration that completely inhibited the growth of T. gondii.
Fig. 3.
Effects of Nullscript on Toxoplasma and its host cell, HFF. (A) Cell viability was measured by the LDH assay. HFF cells were cultured in the presence or absence of nullscript (100 μM) for 48 h. The release of LDH was measured. (B) Dose-response curves and IC50 values of nullscript for Toxoplasma. HFF cells were untreated or treated with indicated concentration of nullscript, and then infected with T. gondii. The parasite number after 48 h post infection was measured by Beta-Glo assay. (C) HFF cells were untreated or treated with presence or absence of nullscript (65 μM) or pyrimethamine (positive control), and then infected with T. gondii. The parasite number after 48 h post infection was measured by Beta-Glo assay. Indicated values represent means ± standard deviations (SD) (three biological replicates per group from three independent experiments). **p < 0.01; N.S., not significant; (Student's t-test).
3.4. Nullscript was suggested to inhibit histone deacetylation in C. parvum
To investigate whether nullscript inhibits HDACs of Cryptosporidium, we quantified the amount of acetylated histones by western blotting. When C. parvum sporozoites were incubated with nullscript for 90 min, acetylated histone H3K9 levels were unchanged, whereas in the positive control group, the acetylated histone H3K9 level was reduced (Fig. 4A). Acetylation of histone H3K9 was increased in the 30 min incubation with nitazoxanide, probably because C. parvum was killed early by nitazoxanide. A similar experiment was then performed using apicidin reported as HDACi of C. parvum (Darkin-Rattray et al., 1996). As a result, nullscript inhibited the deacetylation of histone H3K9 of C. parvum (Fig. 4B). This inhibitory effect of nullscript on deacetylation was dose-dependent, but not apicidin. The reason is unknown, but it is possible that C. parvum was killed early by the treatment with high concentrations of apicidin and that the amount of acetylation was again reduced by prolonged incubation.
Fig. 4.
Nullscript was suggested to inhibit histone deacetylation in C. parvum. (A) C. parvum sporozoites were incubated in 1% DMSO solvent control, 10 μM nullscript, or 10 μM nitazoxanide in DMSO for 30 or 90 min at 37 °C. (B) C. parvum sporozoites were incubated in 1% DMSO solvent control, 2-10 μM of nullscript or apicidin in DMSO for 90 min at 37 °C. The density of the acetylated histone H3K9Ac bands was normalized by the density of the histone H3 loading control bands.
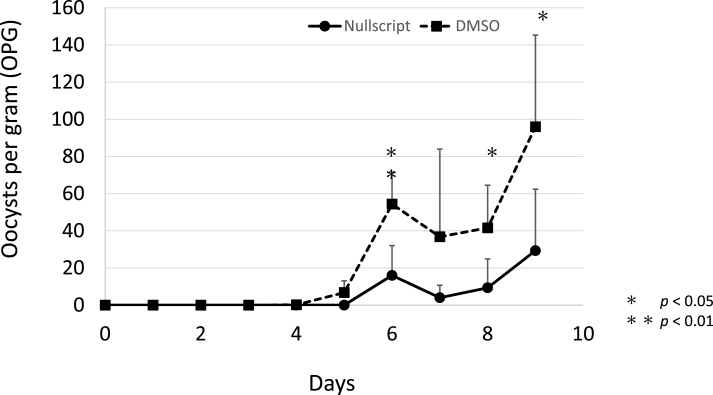
3.5. Nullscript decreases Cryptosporidium oocyst excretion in mice
To determine whether nullscript reduces C. parvum oocyst excretion, we conducted experiments in mice. C. parvum was orally administered with nullscript or with DMSO in PBS. From Day 6 to Day 9, the compounds were administered in the diet. In the control group, oocysts were detected 5 days after inoculation. In the nullscript group, oocysts were excreted 1 day later than in the control group. The excretion of oocysts was significantly lower in the nullscript group than in the control group on Days 6, 8, and 9 after inoculation (Fig. 5).
Fig. 5.
Nullscript delays and reduces oocyst shedding in mice. Effect of nullscript on the production of C. parvum oocysts in mice. C. parvum oocysts were inoculated orally with nullscript (10 mg/kg) or DMSO in PBS (solvent control). Thereafter, until Day 5, 100 μl of nullscript (10 mg/kg) or DMSO was administered orally once daily. From Day 6 to Day 9, the compounds were administered in the diet. The number of oocysts in the faeces was counted by using the sugar flotation method, and the total number of oocysts per gram (OPG) was determined. Asterisks indicate levels of statistical significance as evaluated by Welch's t-test.
4. Discussion
Histone deacetylases (HDACs) are very important in the regulation of transcription and maintenance of cells in organisms. Three HDACs have been identified in C. parvum (Rider and Zhu, 2009). The HDACi vorinostat (SAHA) was reported to inhibit the deacetylation activity of CpHDAC3 in C. parvum (Guo et al., 2018). On the other hand, five nicotinamide adenine dinucleotide independent HDACs have been found in T. gondii (Sullivan and Hakimi, 2006). The HDAC inhibitor FR 235222 has been shown to inhibit TgHDAC3 (Maubon et al., 2010). The effects of HDAC inhibitors have also been studied in various protozoan parasites (Campo, 2017; Carrillo et al., 2015; Chua et al., 2017; Engel et al., 2015; Guo et al., 2018; Sereno et al., 2005; Strobl et al., 2007; Vergnes et al., 2005). However, HDAC inhibitors approved for use in humans may have side effects that limit their use even if they are effective against parasites. In fact, in the present study, some of the drugs that were effective against Cryptosporidium were toxic to the host cells. Most of the compounds that appeared effective in the initial screen were highly cytotoxic. Therefore, it is considered that the growth of C. parvum was apparently suppressed by the low cell number (Table 1). Nullscript cytotoxicity was examined in HCT-8 and HFF cells, the host cells of each parasite, and showed less cytotoxicity in either cell type (Table 1 and Fig. 3A). This finding is consistent with nullscript being used as a negative control for scriptaid, an HDAC inhibitor in host cells (Su et al., 2000; Tong et al., 2006). Thus, nullscript was the least cytotoxic compound to host cells among the compounds that exhibited high C. parvum inhibitory effects in the primary screening. Yet, in our study, it specifically inhibited Cryptosporidium and Toxoplasma. The IC50 value for C. parvum of nullscript was 2.1 ± 0.6 μM and SI = 41 (Fig. 2B). The IC50 value for T. gondii of nullscript was 50.9 ± 4.3 μM and SI could not be calculated because the concentration of 100 μM was not toxic to the host cells. Nitazoxanide, an FDA-approved drug, has an IC50 against C. parvum of ~3.7 μM (Bessoff et al., 2013), suggesting that nullscript may have an equivalent inhibitory effect. In contrast to previous studies (Guo et al., 2018), SAHA did not inhibit the growth of C. parvum in our experiments. The reason for this is unknown, but it may be due to differences in compound manufacturing. Nullscript showed a certain growth inhibitory effect when added after C. parvum had invaded the cells (Fig. 2C). The addition of nullscript after C. parvum cell invasion was less effective in inhibiting the growth of C. parvum, suggesting that nullscript is effective both at the time of entry and at the time of growth, and that the effect of the nullscript is reduced after C. parvum cell invasion.
Then, to know how nullscript inhibits growth of C. parvum, we showed that nullscript inhibits the deacetylation of C. parvum H3K9 (Fig. 4). In human cells, scriptaid inhibits lysine acetylation of the N-terminal tail domains of histone H3 and H4, but nullscript do not (Gaur et al., 2016). Therefore, it is considered that the reason why nullscript has an inhibitory effect on histone deacetylation of C. parvum is the difference in the structure of HDAC due to the difference in the amino acid sequence. As we were unable to obtain active recombinant CpHDAC, further analysis is required.
In the treatment of cryptosporidiosis, nitazoxanide is the only agent with limited activity currently approved by the FDA. For nitazoxanide, a dose of 100 or 200 mg/kg was ineffective in reducing the parasite in C. parvum-infected SCID mice (Theodos et al., 1998). In the present study, nullscript showed a significant reduction in oocyst excretion at 10 mg/kg. For the above reasons, our findings thus suggest that nullscript is an effective lead compound against Cryptosporidium.
Declaration of competing interest
The authors have no affiliation with any organization with a direct or indirect financial interest in the subject matter discussed in the manuscript.
Acknowledgments
We thank the Cancer Research Institute of Kanazawa University (Ishikawa, Japan) for providing the chemical library. We also thank Takeshi Yaoi (Department of Pathology and Applied Neurobiology, Graduate School of Medical Science, Kyoto Prefectural University of Medicine) for valuable suggestions regarding cell analysis using the IN Cell Analyzer 2200. This study was funded by grants-in-aid for Scientific Research (B) and (C), Scientific Research on Innovative Areas (3805) and Young Scientists (B) from the Ministry of Education, Culture, Science, Sports, and Technology (MEXT) of Japan; and by the Livestock Promotional Subsidy from the Japan Racing Association.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2020.10.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Amadi B., Mwiya M., Sianongo S., Payne L., Watuka A., Katubulushi M., Kelly P. High dose prolonged treatment with nitazoxanide is not effective for cryptosporidiosis in HIV positive Zambian children: a randomised controlled trial. BMC Infect. Dis. 2009;9:195. doi: 10.1186/1471-2334-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowood M.J., Donaldson K. Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. J. Eukaryot. Microbiol. 1996;43:89S. doi: 10.1111/j.1550-7408.1996.tb05015.x. [DOI] [PubMed] [Google Scholar]
- Bando H., Lee Y., Sakaguchi N., Pradipta A., Ma J.S., Tanaka S., Cai Y., Liu J., Shen J., Nishikawa Y., Sasai M., Yamamoto M. Inducible nitric oxide synthase is a key host factor for Toxoplasma GRA15-dependent disruption of the gamma interferon-induced antiparasitic human response. mBio. 2018;9 doi: 10.1128/mBio.01738-18. e01738-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessoff K., Sateriale A., Lee K.K., Huston C.D. Drug repurposing screen reveals FDA-approved inhibitors of human HMG-CoA reductase and isoprenoid synthesis that block Cryptosporidium parvum growth. Antimicrob. Agents Chemother. 2013;57:1804–1814. doi: 10.1128/AAC.02460-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohne W., Heesemann J., Gross U. Reduced replication of Toxoplasma gondii is necessary for induction of bradyzoite-specific antigens: a possible role for nitric oxide in triggering stage conversion. Infect. Immun. 1994;62:1761–1767. doi: 10.1128/iai.62.5.1761-1767.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo V.A. Comparative effects of histone deacetylases inhibitors and resveratrol on Trypanosoma cruzi replication, differentiation, infectivity and gene expression. Int. J. Parasitol. Drugs. Drug. Resist. 2017;7:23–33. doi: 10.1016/j.ijpddr.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo A.K., Guiguemde W.A., Guy R.K. Evaluation of histone deacetylase inhibitors (HDACi) as therapeutic leads for human African trypanosomiasis (HAT) Bioorg. Med. Chem. 2015;23:5151–5155. doi: 10.1016/j.bmc.2014.12.066. [DOI] [PubMed] [Google Scholar]
- Chua M.J., Arnold M.S., Xu W., Lancelot J., Lamotte S., Späth G.F., Prina E., Pierce R.J., Fairlie D.P., Skinner-Adams T.S., Andrews K.T. Effect of clinically approved HDAC inhibitors on Plasmodium, Leishmania and Schistosoma parasite growth. Int. J. Parasitol. Drugs. Drug. Resist. 2017;7:42–50. doi: 10.1016/j.ijpddr.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darkin-Rattray S.J., Gurnett A.M., Myers R.W., Dulski P.M., Crumley T.M., Allocco J.J., Cannova C., Meinke P.T., Colletti S.L., Bednarek M.A., Singh S.B., Goetz M.A., Dombrowski A.W., Polishook J.D., Schmatz D.M. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13143–13147. doi: 10.1073/pnas.93.23.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvic M., Vu J. Vorinostat: a new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expet Opin. Invest. Drugs. 2007;16:1111–1120. doi: 10.1517/13543784.16.7.1111. [DOI] [PubMed] [Google Scholar]
- Engel J.A., Jones A.J., Avery V.M., Sumanadasa S.D., Ng S.S., Fairlie D.P., Skinner-Adams T., Andrews K.T. Profiling the anti-protozoal activity of anti-cancer HDAC inhibitors against Plasmodium and Trypanosoma parasites. Int. J. Parasitol. Drugs. Drug. Resist. 2015;5:117–126. doi: 10.1016/j.ijpddr.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur V., Connor T., Sanigorski A., Martin S.D., Bruce C.R., Henstridge D.C., Bond S.T., McEwen K.A., Kerr-Bayles L., Ashton T.D., Fleming C., Wu M., Pike, Winer L.S., Chen D., Hudson G.M., Schwabe J.W.R., Baar K., Febbraio M.A., Gregorevic P., Pfeffer F.M., Walder K.R., Hargreaves M., McGee S.L. Disruption of the class IIa HDAC corepressor complex increases energy expenditure and lipid oxidation. Cell Rep. 2016;16:2802–2810. doi: 10.1016/j.celrep.2016.08.005. [DOI] [PubMed] [Google Scholar]
- Guo F., Zhang H., McNair N.N., Mead J.R., Zhu G. The existing drug vorinostat as a new lead against cryptosporidiosis by targeting the parasite histone deacetylases. J. Infect. Dis. 2018;217:1110–1117. doi: 10.1093/infdis/jix689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D., Dubey J.P. Toxoplasma gondii: transmission, diagnosis and prevention. Clin. Microbiol. Infect. 2002;8:634–640. doi: 10.1046/j.1469-0691.2002.00485.x. [DOI] [PubMed] [Google Scholar]
- Luft B.J., Remington J.S. AIDS commentary. Toxoplasmic encephalitis. J. Infect. Dis. 1988;157:1–6. doi: 10.1093/infdis/157.1.1. [DOI] [PubMed] [Google Scholar]
- Masuda G., Maeda Y., Ohtomo H., Kimata I., Uni S., Iseki M., Takada S. Cryptosporidium diarrhea developing in two Japanese adults, one in AIDS and the other in a normal host. Kansenshogaku Zasshi. 1991;65:1614–1619. doi: 10.11150/kansenshogakuzasshi1970.65.1614. (in Japanese) [DOI] [PubMed] [Google Scholar]
- Maubon D., Bougdour A., Wong Y.S., Brenier-Pinchart M.P., Curt A., Hakimi M.A., Pelloux H. Activity of the histone deacetylase inhibitor FR235222 on Toxoplasma gondii: inhibition of stage conversion of the parasite cyst form and study of new derivative compounds. Antimicrob. Agents Chemother. 2010;54:4843–4850. doi: 10.1128/AAC.00462-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider S.D., Jr., Zhu G. An apicomplexan ankyrin-repeat histone deacetylase with relatives in photosynthetic eukaryotes. Int. J. Parasitol. 2009;39:747–754. doi: 10.1016/j.ijpara.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M., Kimata I., Iseki M., Nakai Y. Gene analysis of Cryptosporidium parvum HNJ-1 strain isolated in Japan. Parasitol. Res. 2005;97:452–457. doi: 10.1007/s00436-005-1474-8. [DOI] [PubMed] [Google Scholar]
- Sereno D., Alegre A.M., Silvestre R., Vergnes B., Ouaissi A. In vitro antileishmanial activity of nicotinamide. Antimicrob. Agents Chemother. 2005;49:808–812. doi: 10.1128/AAC.49.2.808-812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl J.S., Cassell M., Mitchell S.M., Reilly C.M., Lindsay D.S. Scriptaid and suberoylanilide hydroxamic acid are histone deacetylase inhibitors with potent anti-Toxoplasma gondii activity in vitro. J. Parasitol. 2007;93:694–700. doi: 10.1645/GE-1043R.1. [DOI] [PubMed] [Google Scholar]
- Su G.H., Sohn T.A., Ryu B., Kern S.E. A novel histone deacetylase inhibitor identified by high-throughput transcriptional screening of a compound library. Canc. Res. 2000;60:3137–3142. [PubMed] [Google Scholar]
- Sugi T., Masatani T., Murakoshi F., Kawazu S., Kato K. Microplate assay for screening Toxoplasma gondii bradyzoite differentiation with DUAL luciferase assay. Anal. Biochem. 2014;464:9–11. doi: 10.1016/j.ab.2014.06.018. [DOI] [PubMed] [Google Scholar]
- Sullivan W.J., Jr., Hakimi M.A. Histone mediated gene activation in Toxoplasma gondii. Mol. Biochem. Parasitol. 2006;148:109–116. doi: 10.1016/j.molbiopara.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Theodos C.M., Griffiths J.K., D’Onfro J., Fairfield A., Tzipori S. Efficacy of nitazoxanide against Cryptosporidium parvum in cell culture and in animal models. Antimicrob. Agents Chemother. 1998;42:1959–1965. doi: 10.1128/aac.42.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong M., Ding Y., Tai H.H. Histone deacetylase inhibitors and transforming growth factor-beta induce 15-hydroxyprostaglandin dehydrogenase expression in human lung adenocarcinoma cells. Biochem. Pharmacol. 2006;72:701–709. doi: 10.1016/j.bcp.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Vergnes B., Vanhille L., Ouaissi A., Sereno D. Stage-specific antileishmanial activity of an inhibitor of SIR2 histone deacetylase. Acta Trop. 2005;94:107–115. doi: 10.1016/j.actatropica.2005.03.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.