Abstract
Verticillium wilt (VW) caused by Verticillium dahliae is a devastating soil-borne disease that causes severe yield losses in cotton and other major crops worldwide. Here we conducted a high-throughput screening of isolates recovered from 886 plant rhizosphere samples taken from the three main cotton-producing areas of China. Fifteen isolates distributed in different genera of bacteria that showed inhibitory activity against V. dahliae were screened out. Of these, two Pseudomonas strains, P. protegens XY2F4 and P. donghuensis 22G5, showed significant inhibitory action against V. dahliae. Additional comparative genomic analyses and phenotypical assays confirmed that P. protegens XY2F4 and P. donghuensis 22G5 were the strains most efficient at protecting cotton plants against VW due to specific biological control products they produced. Importantly, we identified a significant efficacy of the natural tropolone compound 7-hydroxytropolone (7-HT) against VW. By phenotypical assay using the wild-type 22G5 and its mutant strain in 7-HT production, we revealed that the 7-HT produced by P. donghuensis is the major substance protecting cotton against VW. This study reveals that Pseudomonas specifically has gene clusters that allow the production of effective antipathogenic metabolites that can now be used as new agents in the biocontrol of VW.
Keywords: Rhizosphere, Pseudomonas, Verticillium wilt, Biocontrol, 7-hydroxytropolone
Key points
Pseudomonas spp. isolated from high-throughput screening showed the most influential activities to multiple strains of V. dahlia.
P. protegens XY2F4 and P. donghuensis 22G5 showed that Pseudomonas spp. have developed specific mechanisms against V. dahlia.
7-hydroxytropolone produced by P. donghuensis is the major ingredient to protect cotton against verticillium wilt.
Introduction
Cotton verticillium wilt (VW) is a singularly destructive fungal disease caused by Verticillium dahliae, which is regarded as “the cancer of cotton”. The V. dahliae fungus invades the vascular system through the roots and soon causes systemic infection, leading to a series of symptoms including leaf chlorosis, necrosis or wilting, leaf or boll abscission, and even plant death. VW-related damage results in reduced cotton yield and lower fiber quality in agricultural production (Wang et al. 2016). Currently, around 50% of the cotton planting area in China (2.5 million hectares) is VW-infected, leading to direct economic losses of about 250–310 million USD annually (Wang et al. 2016). Disease management mainly includes crop rotation to non-host plants, fungicide fumigation and breeding of resistant cultivars (Klosterman et al. 2009). Crop rotation is a preventative, but not curative disease management strategy since V. dahliae can survive for extremely long periods of time in the soil as microsclerotia even in the absence of a suitable host (Termorshuizen 1995). Disease control of VW using fungicide fumigation is effective, but expensive and environmentally unfriendly (Jordan 1972). Breeding of cultivars with broad-spectrum resistance is considered to be one of the most practicable and effective approaches. However, it is difficult to apply biotechnology to breed VW-resistant cotton due to the lack of resistance markers in cotton germplasm, only a few commercial upland cotton cultivars have developed with moderate levels of VW resistance (Zhang et al. 2012a).
Rhizobacteria have great potential to improve sustainable agricultural practices due to their influence on growth, yield, nutrient uptake, and biotic/abiotic tolerance of crops. Beneficial rhizobacteria are able to colonize the rhizosphere (the root surface or intercellular spaces of plants), which impacts the plant by delivering biocontrol and other beneficial factors (Lugtenberg et al. 2001). To date, multiple isolates from genera of Enterobacter (Li et al. 2012a), Bacillus (Li et al. 2012b; Zhang et al. 2018), Serratia plymuthica (Vleesschauwer 2007), Streptomyces (Xue et al. 2013) and Pseudomonas (Erdogan and Benlioglu 2010) have documented biocontrol activities against V. dahliae in in planta assays. Thus, beneficial rhizobacteria with inhibitory action against V. dahliae are promising biocontrol agents for the management of VW in cultivated cotton (Tjamos et al. 2000). However, the specific mechanisms underlying the biocontrol of VW have yet to be determined. In this research we employed a high-throughput screening for inhibitory isolates and comparative genomic analysis to uncover the mode of action of two new Pseudomonas strains with significant V. dahliae inhibitory capacity. This study charts a path toward the development of probiotics and active ingredients for biocontrol agents (BCAs) to ameliorate cotton VW disease.
Materials and methods
Plant culture
Upland cotton (Gossypium hirsutum) cultivars Texas Marker-1 (TM-1) and Junmian1 were grown in soil consisting of 25% vermiculite and 75% artificial soil at 28 °C with a 16 h/8 h light/dark cycle in growth chambers. One-week old seedlings were used in the planta assays.
V. dahliae culture
Highly virulent strains of V. dahliae, including V07DF2, V08DF2, V15QY1, and V991 were gifts from Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences. The highly toxic and defoliant wild type pathogenic V. dahliae strain V991 was used in planta assay (Sun et al. 2013; Zhang et al. 2012b). V. dahlia strains were cultured on Czapek agar plates at 28 °C for 4–5 days after which 5 mL liquid Czapek was dispensed into Petri plates to collect the conidia. The conidia suspension was then transferred to 100 mL liquid Czapek and cultured for 7 days until cell density reached OD600 = 2, contains ~ 3 × 106 conidia/mL. Finally, the conidia were filtered through a 500-micron sieve for use in inoculation assays. The V. dahliae strain stock was composed of the conidia suspensions with 20% glycerol.
Bacteria isolation and culture
Bacterial isolates were recovered from 886 plant rhizosphere samples taken from the three main cotton-producing areas of China (the Yangtze river basin, the Yellow River basin and Xinjiang) (Additional file 1: Table S1). The samples were placed in separate, labeled 50 mL tubes filled with enough ddH2O to ensure that they were completely submerged, and then tubes were shaken approximately 4–5 times to mix. 800 µL of the mixture was aliquoted for gradient dilution (10−1, 10−2, 10−3, 10−4 and 10−5). 1:103 or 1:104 was considered a suitable dilution ratio and 100 µL solution was plated on LB media and inoculated overnight at 30 °C. Plates were stored at 4 °C for 3 days in order to enhance the formation of fluorescent pigments in bacterial colonies.
High-throughput screening for bacterial isolates with inhibitory action against V. dahliae
A plate assay was performed to screen isolates for inhibitory action against V. dahliae. In a 10-mL tube, 6 mL top agar (0.8% w/v) was cooled to less than 50 °C and gently mixed with 60 µL V. dahliae V991 culture stock (OD600 = 2), resulting in an initial density of 0.02/mL at OD600 in the top agar. This solution was then quickly poured on top of a Czapek agar plate, gently shaken by hand in a radial/rocking manner, and allowed to solidify. Afterwards, 5 µL overnight culture of candidate bacterial isolates was inoculated on top of the agar plate. 16 candidate isolates were able to be tested per plate via high-throughput screening. Plates were sealed with parafilm and cultured at 28 °C. Any inhibitory action by the bacterial isolates against V. dahliae was revealed by the appearance of a zone of inhibition on the agar plate. The size of the zone of inhibition was recorded at 72 h post inoculation and inhibitory action was qualitatively determined. For those isolates that produced a visible zone of inhibition, additional confirmational assays were performed using other highly virulent strains of V. dahliae, including V07DF2, V08DF2 and V15QY1.
16S rRNA identification and species designation
16S rDNA was amplified using the primer pair 27F and 1492R (Additional file 1: Table S2). Sequencing results were identified by using BLAST to search the NCBI 16S rRNA database. Species was designated based on the best hit for each species in BLAST and confirmed by genome-based taxonomy by Type Strain Genome Server (https://tygs.dsmz.de) (Meierkolthoff and Goker 2019).
Genomic sequencing and de-novo assembly
The genomic DNA of Pseudomonas strains were extracted using the DNeasy UltraClean Microbial Kit (Cat. no. 12224–50, QIAGEN). Sequencing library was generated using NEB Next Ultra II DNA Library Prep Kit for Illumina (Cat. no. E7645, NEB) following manufacturer’s recommendations and index codes were added to each sample. Briefly, genomic DNA sample was fragmented by sonication to a size of 350 bp. Then DNA fragments were end-polished, A-tailed, and ligated with the full-length adapter for Illumina sequencing, followed by further PCR amplification. After PCR products were purified by AMPure XP system (Beckman Coulter, USA), DNA concentration was measured by Qubit®3.0 Flurometer (Cat. no. Q33216, Invitrogen, USA), libraries were analyzed for size distribution by NGS 3 K assay (PerkinElmer, USA) and quantified by real-time PCR (3 nM) and sequenced by Illumina PE150 (Illumina, USA). Genomes were assembled de-novo using SPAdes software (Bankevich et al. 2012).
RAST annotation
Annotation of genomes was performed using the RAST Server (Rapid Annotation using Subsystem Technology) (https://rast.nmpdr.org/rast.cgi) (Overbeek et al. 2014).
Phylogenetic tree
The full-length sequences of 10 Pseudomonas housekeeping genes (acsA, aroE, dnaE, guaA, gyrB, mutL, ppsA, pyrC, recA, and rpoB) (Loper et al. 2012) were extracted using RAST (Rapid Annotation using Subsystem Technology). Protein alignment and phylogenetic tree generation were accomplished using MEGA software (version 6.06) (Tamura et al. 2007).
Core genome and unique gene analysis
Core genome and unique gene analysis was completed via BLASTP using an amino acid identity cut-off of 70% and an e-value of 1e−5.
Analysis of metabolites
A search for potential biocontrol genes/gene clusters was performed based on rough prediction of antibiotics and secondary metabolite gene clusters using antiSMASH (Medema et al. 2011) or BLAST with documented metabolite gene clusters as a reference.
Gene mutation
Construction of an in-frame deletion mutant of orf12 () from the 7-HT gene cluster in Pseudomonas donghuensis 22G5 was performed as has been described previously. 500-bp upstream and downstream sequences of orf12 were amplified separately. The upstream and downstream fragments were then concatenated by overlap extension PCR and cloned into pEX18Gm plasmids (Jiang et al. 2016) to generate a gene replacement vector for orf12 (pEX18Gm-orf12), which was then introduced into E. coli S17-1 λpir competent cells (Jiang et al. 2016). E. coli S17-1 λpir single clone carrying the pEX18Gm-orf12 plasmid was co-cultured with P. donghuensis 22G5 at 28 °C for 24 h for conjugation. Single P. donghuensis 22G5 transformants were selected on LB agar plates using 25 μg/mL chloramphenicol (P. donghuensis 22G5 could grow on the chloramphenicol-containing LB agar plate but E. coli S17-1 λpir could not) and 50 μg/mL gentamicin. Single transformants were then incubated without antibiotic overnight at 28 °C in 200 μL of liquid LB medium in a 96-well plate to complete the second step of allelic exchange. Serially diluted cultures were incubated at 28 °C on LB agar plates with 5% sucrose. Validity of the final orf12 mutants from 22G5 was verified via PCR (Wang et al. 2015) and sequencing. All primers used for vector construction and PCR verification are listed in Additional file 1: Table S2.
7-hydroxytropolone purification
7-hydroxytropolone (7-HT) purification was performed as described (Jiang et al. 2016). P. donghuensis 22G5 single clone was cultured in 3 mL MKB media overnight, which was then inoculated at a 1:100 ratio to 50 mL MKB media and cultured for 48 h. 1/100 volume of hydrochloric acid (37.5%) was added to the supernatant of P. donghuensis 22G5 culture to adjust the pH to 2, which was then extracted using 25 mL ethyl acetate for twice, NaCl (1:10 w/v, ~ 5 g) was added to reduce the formation of ethyl acetate-water emulsion. The organic phase which containing 7-HT was rotary-evaporated to dryness under vacuum. The residue was dissolved in 10 mL ethanol and purified using a Sephadex LH20 column (Cat. no. 17-0090-02, GE) eluted by ethanol. Fractions positive in CAS assay (Schwyn and Neilands 1987) were collected and dried by rotary evaporation, and were dissolved in 1 mL DMSO and stored at − 20 °C.
Planta in-vivo assay
V. dahliae strains were cultured in Czapek liquid at 28 °C for 7 days to OD600 = 2 (~ 3 × 106 CFU/mL). Pseudomonas protegens XY2F4 and P. donghuensis 22G5 were cultured in LB liquid at 28 °C for 24 h to OD600 = 2 (~ 1 × 108 CFU/mL). One seedling for each pot (5 cm × 5 cm square pot) was planted. One-week-old cotton seedlings in the experimental (XY2F4- or 22G5-protected) group was treated with a soil drench using a mixture of 50 mL V. dahliae conidia (OD600 = 0.2, equals approximately 3 × 105 colony forming unit, CFU/mL) and 50 mL 1:10 diluted XY2F4 or 22G5 overnight culture (OD600 = 0.2, equals approximately 1 × 107 CFU/mL). The control group was treated with a mixture of 50 mL V. dahliae conidia (OD600 = 0.2, equals approximately 3 × 105 CFU/mL) and 50 mL LB culture media. Plants were monitored after inoculation for disease phenotype over time and DI (Disease Index) was calculated 30 days post-infection. The disease grade was classified as 0 (no wilting), 1 (0–25% defoliated leaves), 2 (25–50% defoliated leaves), 3 (50–75% defoliated leaves), or 4 (75–100% defoliated leaves) (Zhang et al. 2017). DI (Disease Index) is calculated as follows:
dc: disease severity rating; nc: number of plants in each category of disease severity; nt: total number of plants assessed (Zhao et al. 2014). 15–20 cotton seedlings were used for each treatment. At least four independent biological replicates for each treatment were performed.
Statistical analyses
All statistical analysis was performed with the software Graphpad Prism (GraphPad Software, La Jolla, CA, USA). The Student’s t-test, Mann–Whitney U-test was used whenever appropriate. The p-values of less than 0.05 were considered statistically significant.
Public acession of culture
P. protegens XY2F4 and P. donghuensis 22G5 were deposited in China General Microbiological Culture Collection Center (CGMCC), the CGMCC No.18017 for P. protegens XY2F4 and CGMCC No.18084 for P. donghuensis 22G5.
NCBI accession
P. protegens XY2F4 (PIZE00000000), P. donghuensis 22G5 (RWIB00000000), Pseudomonas putida 25E1 (WSSD00000000), Pseudomonas lini 25D11 (RSFR00000000).
Results
High-throughput screening for isolates with inhibitory action against V. dahliae
Bacterial isolates recovered from 886 plant rhizosphere samples taken from the three main cotton-producing areas of China (the Yangtze river basin, the Yellow River basin and Xinjiang) (Additional file 1: Table S1) were cultured in order to determine their effect against V. dahliae. On average, 10 individually isolated bacterial colonies (i.e. single colonies with different color, morphologies) were tested per sample. In total, 8,736 bacterial isolates were tested in a high-throughput zone of inhibition assay (Additional file 1: Fig. S1). Isolates were considered to have shown inhibitory action against V. dahliae if a clear inhibition zone was observed upon co-incubation with conidia of V. dahliae. Of the total samples tested, 15 isolates from various bacterial genera were identified as conferring inhibitory effect on V. dahliae (Fig. 1a). Two strains (XY2F4 and 22G5) with significant, broad spectrum inhibitory action against multiple highly virulent V. dahliae strains (Fig. 1b) were selected for further genotyping. Another two strains (25E1 and 25D11) with low inhibitory capacity were randomly selected to serve as negative controls (Fig. 1b). According to identification via 16S ribosomal DNA similarity and genome-based taxonomy by Type Strain Genome Server (https://tygs.dsmz.de) (Meierkolthoff and Goker 2019), Pseudomonas protegens XY2F4, P. donghuensis 22G5, P. putida 25E1 and P. lini 25D11, were designated for study.
Fig. 1.
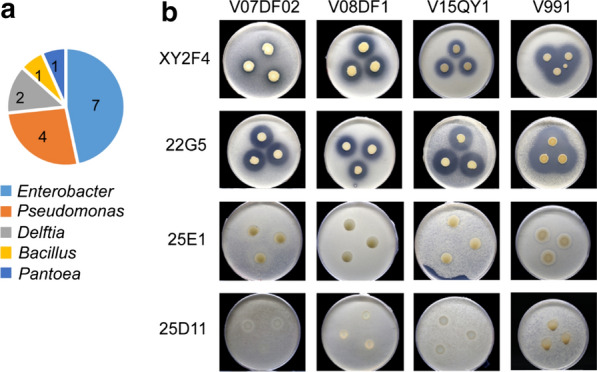
High-throughput screening of isolates with inhibitory action against V. dahliae. a Species distribution of isolates with inhibitory action against V. dahliae. Species were designated according to similarity to 16S rDNA. b Zone of inhibition assay of strains XY2F4 and 22G5 co-inoculated with conidia of various V. dahliae strains
Comparative genomic analysis of Pseudomonas spp. exhibiting inhibitory action against V. dahliae
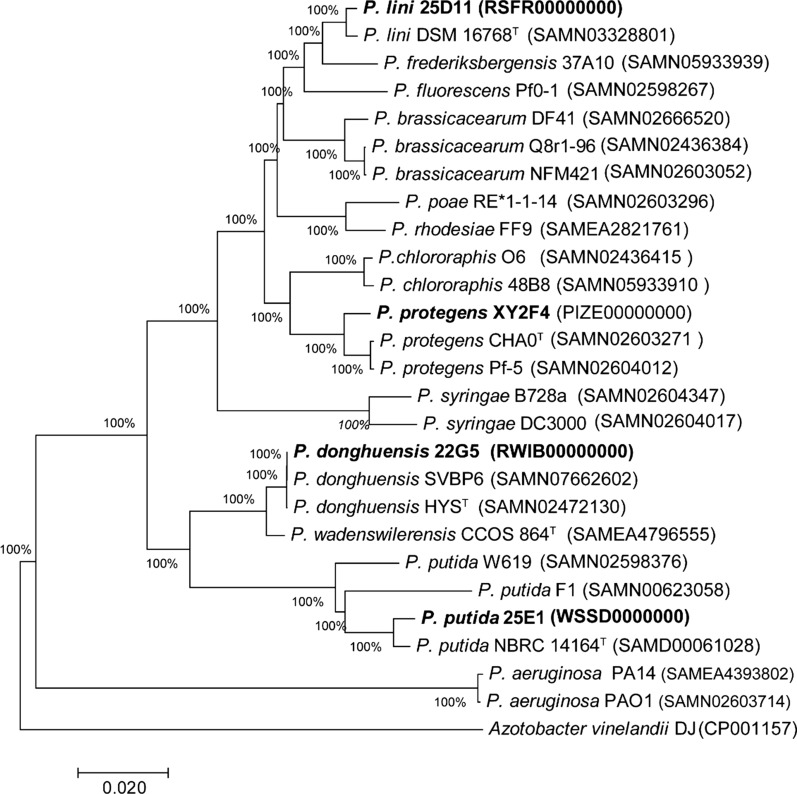
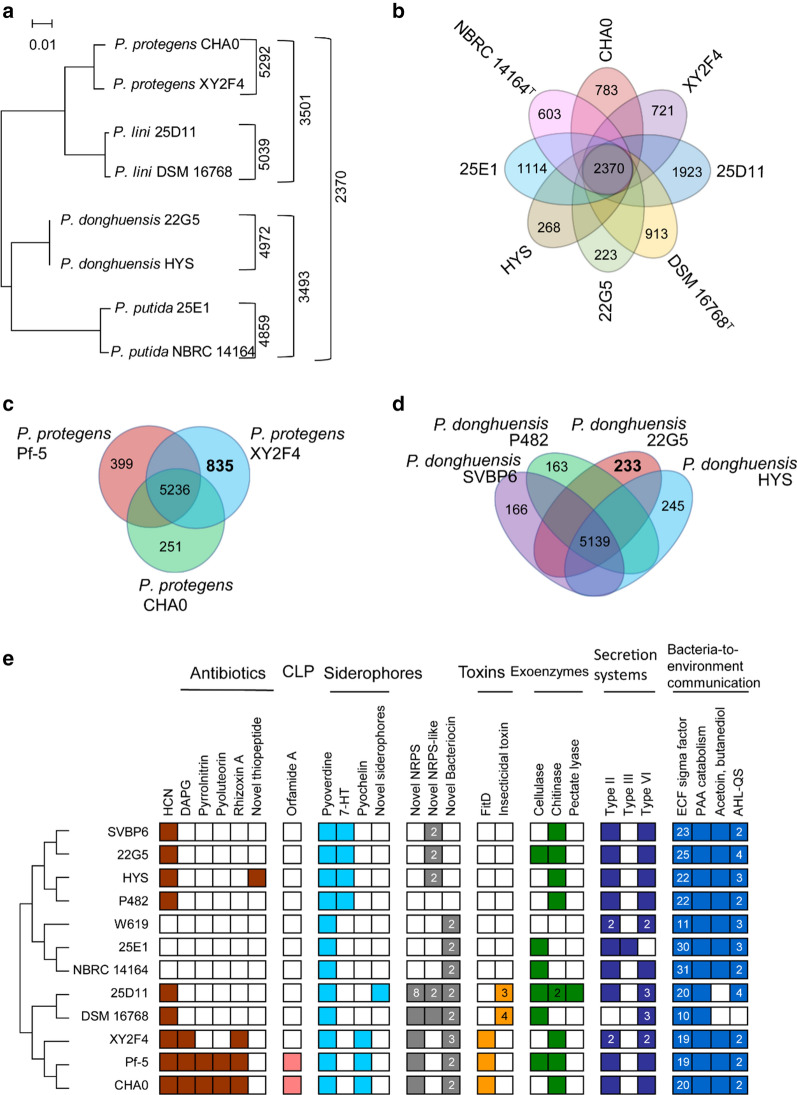
Whole genome sequencing and de novo assembly of the four Pseudomonas strains was performed (Additional file 1: Table S3). A phylogenetic tree depicting the evolutionary relationships among these four Pseudomonas strains and other well documented strains indicated that Pseudomonas isolates from different ecological environments were highly diverse (Fig. 2). From the results, P. protegens XY2F4 and P. lini 25D11 are clustered in the large group of Pseudomonas fluorescens “complex” (Garridosanz et al. 2016) according to the taxonomy analysis (Fig. 2), which have been taxonomically assigned to more than fifty different species, many of which have been described as plant growth-promoting rhizobacteria (PGPR) (Garridosanz et al. 2016). P. donghuensis 22G5 and P. putida 25E1 are close-related and clustered in the P. putida group (Fig. 2). Furthermore, we compared the genes specific to each strain and genes shared among strains. Four type strains, including P. protegens CHA0 (Shaukat and Siddiqui 2003), P. donghuensis HYS (Chen et al. 2018), P. putida NBRC 14164 T (Ohji et al. 2014), and P. lini DSM 16768 T (Kaminski et al. 2018), as reference strain for P. protegens XY2F4, P. donghuensis 22G5, P. putida 25E1 and P. lini 25D11, respectively, are incorporated together in the analysis. These eight genomes shared a core genome of 2370 genes (Fig. 3a), and with the number of genes unique to each strain ranged from 223 to 1923 (Fig. 3b). We further compared P. protegens XY2F4 and P. donghuensis 22G5 genomes respectively with more published strains in the same species, including two well-documented P. protegens strains (Paulsen et al. 2005; Shaukat and Siddiqui 2003) and three published P. donghuensis strains to date (Chen et al. 2018; Muzio et al. 2020; Ossowicki et al. 2017). Results indicated that the P. protegens XY2F4 genome had 835 specific genes compared with P. protegens CHA0 and Pf-5 (Fig. 3c), mainly including genes for widespread colonization island, general secretion pathway, orphan regulatory proteins, and so on (Additional file 1: Table S4). The P. donghuensis 22G5 genome had 233 unique genes compared with three published P. donghuensis strains (Fig. 3d), mainly including genes in iron siderophore sensor & receptor system, n-phenylalkanoic acid degradation, fatty acid metabolism cluster, and so on (Additional file 1: Table S5).
Fig. 2.
Phylogenetic tree depicting the evolutionary relationships of Pseudomonas spp. Genomes of previously reported strains were downloaded from The Pseudomonas Genome Database (https://www.pseudomonas.com/). The annotation was performed using RAST (Overbeek et al. 2014). The phylogenetic tree was generated with MEGA software using concatenated sequences of 10 housekeeping genes (acsA, aroE, dnaE, guaA, gyrB, mutL, ppsA, pyrC, recA, and rpoB) (Loper et al. 2012). Strains in this study are indicated with a star (*). Type strains or representative strains of multiple documented Pseudomonas species were compiled to build the tree. Azotobacter vinelandii DJ was used as the outgroup
Fig. 3.
Comparative genome analysis of Pseudomonas strains. a, b Number of shared genes and unique genes in genomes and of P. protegens XY2F4, P. donghuensis 22G5, P. putida 25E1 and P. lini 25D11 and their type strains. c, d Number of unique genes in P. protegens XY2F4XY2F4 and P. donghuensis 22G5 when compared with published strains from the same species (with NCBI accessions). e Presence of selected gene clusters for biosynthesis of antibiotics, cyclic lipopeptides (CLP), siderophores, insecticidal toxins, exoenzymes, secretion systems, and chemicals for bacteria-to-environment communication. Colored box/blank box indicates the presence/absence of selected gene clusters, respectively. Numbers within boxes represent the predicted number of gene clusters encoded for the corresponding products
To identify the genomic features accounting for inhibitory action against V. dahliae, gene clusters related to biocontrol traits and environmental interactions were compared. The biosynthesis of antibiotics, cyclic lipopeptides (CLP), siderophores, insecticidal toxins, exoenzymes, secretion systems, and chemicals for environmental communication and acyl-homoserine lactone (AHLs) mediated quorum-sensing was discovered. For comparison, multiple well-documented strains in the same species taxon were included in the analysis. Results indicated that genes related to the siderophore pyoverdine, type II secretion system (T2SS), type VI secretion system (T6SS), extracytoplasmic function (ECF) sigma factors for detecting environmental cues (Kwak et al. 2018), phenylacetic acid (PAA) catabolism, acetoin, butanediol catabolism for bacteria-plant communication (Mhlongo et al. 2018), and AHLs-mediated quorum-sensing were commonly distributed in the strains we analyzed (Fig. 3e). In addition to these, specific functional gene clusters were identified in the V. dahliae-inhibitory strains. 2,4-DAPG and a nonribosomal peptide synthetase (NRPS)-type gene cluster responsible for rhizoxin A, pyochelin, and FitD insecticidal toxin were specifically distributed in the P. protegens strains XY2F4 and Pf-5. However, XY2F4 had lost the gene clusters for biosynthesis of pyrrolnitrin, pyoluteorin and orfamide A, in compare with CHA0 and Pf-5 (Fig. 3e). A siderophore-type gene cluster involved in the biosynthesis of 7-hydroxytropolone (7-HT) was specifically identified in the P. donghuensis species. Gene organization in this 7-HT biosynthesis gene cluster was the same as has been previously reported in P. donghuensis HYS (Chen et al. 2018), a first reported Pseudomonas strain containing a 7-HT gene cluster. In addition, P. donghuensis 22G5 produces cellulase (Fig. 3e), which was found to be absent in P. donghuensis HYS and may function by degrading the cell walls of pathogens or by triggering plant defenses. In summary, the Pseudomonas strains screened out via V. dahliae inhibition assay have developed specific genomic characteristics that could produce certain secondary metabolites that confer inhibitory to V. dahliae.
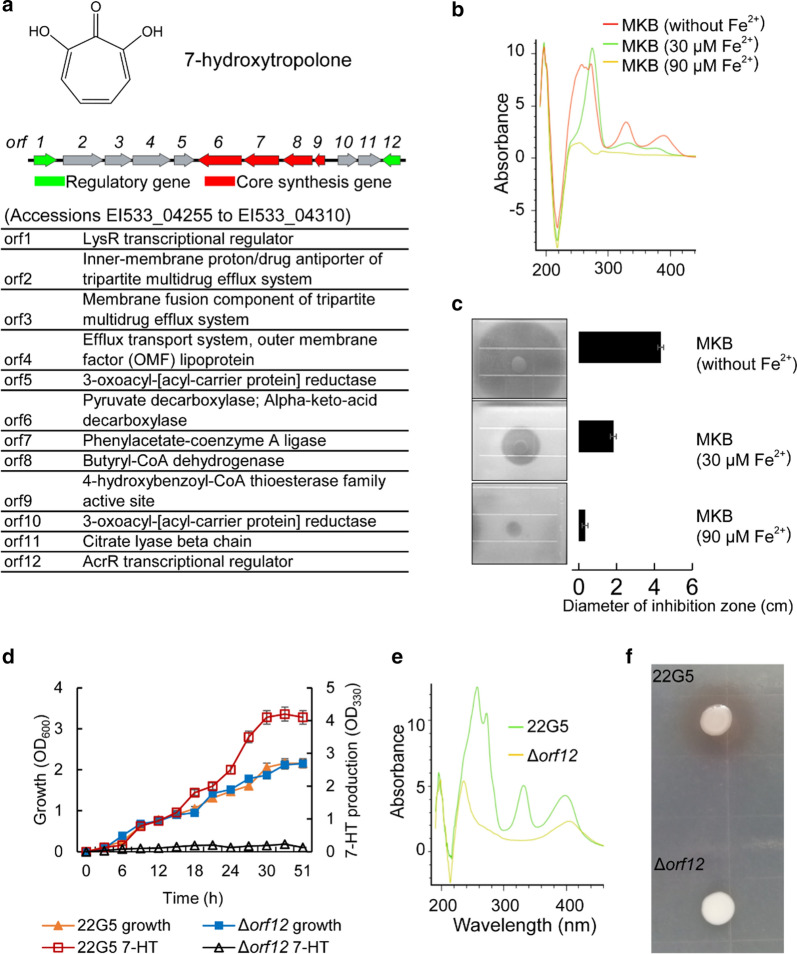
7-HT is a major factor in P. donghuensis 22G5 accounting for inhibitory action against V. dahliae
P. protegens XY2F4 has a gene cluster for biosynthesis of a biocontrol ingredient 2,4-DAPG (Fig. 3e), which is well documented in its type strain P. protegens CHA0 and Pf-5. P. donghuensis 22G5 has a gene cluster for biosynthesis of specific siderophore metabolite 7-HT, a metabolite common to P. donghuensis species which was previously reported to have fungal antagonism activity (Chen et al. 2018; Muzio et al. 2020; Ossowicki et al. 2017). We inferred that 7-HT with broad spectrum might account for their inhibitory action against V. dahliae. The same 7-HT biosynthesis cluster as described is composed of 12 ORFs (NCBI accessions EI533_04255 through EI533_04310). ORF6 through ORF9 (NCBI accessions EI533_04270 through EI533_04285) encode the core biosynthesis genes while ORF1 (NCBI accession EI533_04310) and ORF12 (NCBI accession EI533_04255) are regulatory genes (Fig. 4a). Supernatant derived from 22G5 cultures showed characteristic absorption peaks at 330 nm and 392 nm, which was identical to 7-HT (Jiang et al. 2016). As a siderophore, 7-HT’s biosynthesis was regulated by iron concentration in the culture media. Consistently, we found that the yield of 7-HT in 22G5 cultures declined dramatically as iron (FeSO4) concentration increased in the MKB media (Fig. 4b), suggesting the existence of a regulatory feedback mechanism for biosynthesis of 7-HT based on growth conditions of high iron concentration. The 7-HT was then extracted and purified from supernatant retrieved from 22G5 cultures and applied to a plate assay using V. dahliae. As expected, the inhibition zone shrunk upon reduction of 7-HT in the MKB medium (Fig. 4c). Next we generated a gene mutation of ORF12 via homologous recombination (Additional file 1: Fig. S2). As a result, the ORF12-mutated strain (Δorf12) did not show much differences in growth rate compared with wild type 22G5 (Fig. 4d), however, lost its ability to produce 7-HT (Fig. 4e), and phenotypically, lost the ability to inhibit V. dahliae (Fig. 4f). With these tests, we demonstrated that 7-HT originating from P. donghuensis 22G5 is the major factor accounting for inhibitory effects against V. dahliae.
Fig. 4.
7-HT biosynthesis and inhibition of V. dahliae. a Gene cluster for 7-HT (Jiang et al. 2016). b, c Absorption spectra of 7-HT production and phenotypes in zone of inhibition assay using 7-HT extraction from the supernatant of 22G5 culture in MKB media with or without FeSO4. d Growth trace of wild type 22G5 (22G5) and orf12 mutant (Δorf12) (displayed by OD600, left y-axis) and its corresponding 7-HT production (displayed by OD330, right y-axis). e, f Absorption spectra of 7-HT production and phenotypes in zone of inhibition assay in wild type 22G5 (22G5) and orf12 mutant (Δorf12)
In planta assays showed Pseudomonas strains XY2F4 and 22G5 significantly improve resistance to cotton verticillium wilt
To evaluate whether the strains screened via plate assay also showed potential for biocontrol in host–pathogen interaction, planta in vivo assays were conducted using P. protegens XY2F4 and P. donghuensis 22G5. Plant growth promotion or biosafety tests for XY2F4 and 22G5 were performed first using three different concentration of culture (1 × 107, 1 × 106 and 1 × 105 CFU/mL) by soil drench. Results indicated that pre-treatment with P. protegens XY2F4 cultures (1 × 107 CFU/mL) increased seedling biomass (Additional file 1: Fig. S3, P < 0.05). In contrast, P. donghuensis 22G5 cultures had no significant effect on plant biomass compared with the LB media control group (CK), indicating that it poses low biosafety risk (i.e. non-pathogenic) to cotton plants. Planta in vivo assays were also conducted using cotton seedlings co-inoculated with V. dahliae and the Pseudomonas strains with V. dahlia inhibitory activity in plate assay. Combinations of V. dahliae conidia and either P. protegens XY2F4 or P. donghuensis 22G5 were tested. It was observed 30 days after inoculation that cotton seedlings co-inoculated with either XY2F4 or 22G5 strains and V. dahliae conidia exhibited a less degree of leaf chlorosis, necrosis and wilting than seedlings inoculated with V. dahliae V991 alone (Fig. 5a, b). The disease index of XY2F4- and 22G5-protected cotton seedlings of upland cotton cultivar TM-1 was significant reduced that summarized from 4 independent biological replicates, compared to the un-protected group (Fig. 5c, d). Thus, we confirmed that the Pseudomonas species isolated via high-throughput screening significantly protect cotton plants against VW infection.
Fig. 5.
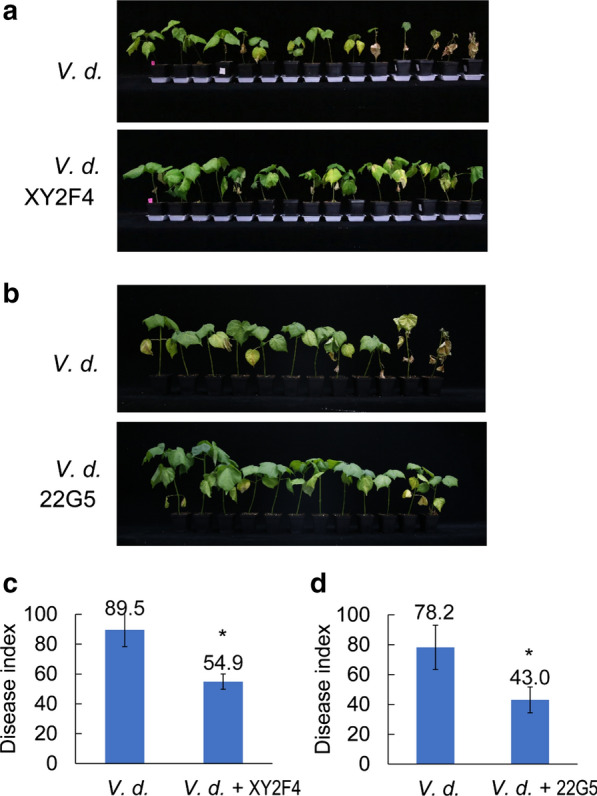
Pseudomonas strains XY2F4 and 22G5 significantly improve cotton verticillium wilt resistance. a, b Phenotypes of G. hirsutum acc. Texas Marker-1 (TM-1) treated with a mixture of V. dahliae conidia (V. d.) and culture of XY2F4 (a) or 22G5 (b), and control group treated with V. dahliae conidia only. Plants were monitored after inoculation for disease phenotype over time, and DI (Disease Index) was calculated as has been described previously (Zhao et al. 2014) 30 days post-infection. See materials and methods for details. c, d DI (Disease Index) summarized from four independent replicates of planta in-vivo assays as described in panel a, b. Mann–Whitney Non-parametric Test. P < 0.05
Discussion
Pseudomonas spp. are important operational taxonomic units in the rhizosphere with antagonistic action toward V. dahliae
Rhizosphere microbiomes are major determinants of plant health and productivity, and they have the potential to improve sustainable agricultural practices by influencing growth, yield, nutrient uptake, and biotic/abiotic tolerance (Veach et al. 2019). Plant growth–promoting rhizobacteria (PGPR) are naturally occurring soil bacteria that aggressively colonize plant rhizospheres and benefit plants by providing growth promotion or protection against pests and pathogens (Goswami et al. 2016). Both soil conditions and plant host species are commonly recognized as important determinants of the soil microbial composition (Berg et al. 2005; Rodriguez et al. 2019). Arabidopsis genotypes with a manipulated systemic expression of SA signaling have been shown to have an increased population density of Pseudomonas spp. (Doornbos et al. 2011). The microbiome structures of tomato varieties either resistant or susceptible to the soil-borne pathogen Ralstonia solanacearum differ (Kwak et al. 2018), indicating that some biochemical or molecular attributes of specific plants may constitute a host-induced filter for the microbiome in the plant–soil environment. Field research has also revealed that monoculture, but not crop rotation, leads to an enrichment of bacteria producing 2,4-DAPG, a well-known antibiotic originating from P. protegens that suppresses soil-borne pathogens (Weller et al. 2007). Until now, multiple strains from fluorescent Pseudomonas spp. and Bacillus spp. were reported to have biocontrol potential against V. dahliae (Erdogan and Benlioglu 2010; Lan et al. 2017; Li et al. 2012b; Mercadoblanco et al. 2004; Sherzad and Canming 2020; Zhang et al. 2018). However, without a parallel comparison of various hosts, rotation methods, soil types, and races of V. dahliae, the effectiveness of biocontrol among these strains has been difficult to determine. Our data provide profiles of antagonistic OTUs (operational taxonomic units) from 8736 isolates recovered from the three main cotton-producing areas of China, and we have shown via in planta assays that certain Pseudomonas spp. are important antagonistic OTUs that effectively suppress V. dahliae.
Genetic and metabolic diversity in Pseudomonas spp. reveals distinct mode of action for biocontrol of Verticillium wilt
The genus Pseudomonas belonging to the gamma subclass of proteobacteria is a group of bacteria with remarkable metabolic, genetic, and ecological diversity. It currently contains more than 100 designated species that are present in all major natural environments associated with plants and animals (Peix et al. 2018). For example, P. aeruginosa is an opportunistic pathogen in humans. In plants, Pseudomonas spp. provide promising models for plant–microbe interactions owing to the species’ metabolic, ecological, and genetic diversity (Sitaraman 2015). In the past 30 years, reference strains for different species of plant-associated Pseudomonas have been extensively documented for their many beneficial metabolites and ecological functions (David et al. 2018). However, a deep understanding of the modes of action underlying their biocontrol effects against V. dahliae is still lacking. Similar to the type strain of P. protegens (CHA0 and Pf-5 as type strains), P. protegens XY2F4 characteristically and conservatively has gene clusters for the biosynthesis of the natural phenolic antimicrobial compounds 2,4-DAPG (Nowak-Thompson et al. 1994) and rhizoxin A (Loper et al. 2008). 2,4-DAPG was commonly active against various soil-borne bacterial and fungal pathogens that cause plant diseases (Troppens et al. 2013; Yang and Cao 2012) and also showed toxicity to plant-parasitic and bacterial-feeding nematodes (Meyer et al. 2009). Moreover, 2,4-DAPG was found to mediate induced systemic resistance (ISR) by triggering the JA/ET-mediated defense system in Arabidopsis (Chae et al. 2020). P. donghuensis HYS and P. donghuensis 22G5 conservatively produce a novel siderophore, 7-HT. 7-HT was first reported as a siderophore in P. donghuensis HYS in 2016 (Jiang et al. 2016), and the gene cluster responsible for its production has been well established (Chen et al. 2018; Krzyżanowska et al. 2016). Recently, 7-HT was found to be the main metabolite responsible for the fungal antagonism of P. donghuensis SVBP6 by testing its antagonism activities against Macrophomina phaseolina, Fusarium graminearum, Fusarium semitectum and Bacillus subtilis (Muzio et al. 2020). The antagonism results were obtained from the growth inhibition of phytopathogenic fungi when they were co-cultured/co-inoculated with P. donghuensis SVBP6 or its supernatant. Iron is a necessary element in virtually all living organisms and is utilized to catalyze a wide variety of indispensable enzymatic reactions (Soares and Weiss 2015). As a siderophore, 7-HT was expected to function as an iron scavenger when interacting with phytopathogenic fungi. Our study enriched our knowledge by showing that 7-HT had broad-spectrum activity against phytopathogenic fungi, including V. dahliae. It was reported that 7-HT is virulent toward C. elegans (Gui et al. 2020); however, a possible mode of action has not been studied. In conclusion, the genetic and metabolic diversity of Pseudomonas spp. provides distinct modes of action that are dependent on plant–microbe interactions, allowing the biocontrol of cotton VW.
Discovery of antagonistic Pseudomonas strains in rhizosphere provides promising material for development of biocontrol agents
Active management of the microbiome of agriculturally important plants promises to optimize plant reliability, the use of resources, and the environmental impact related to food production by enhancing plant growth, nutrient use efficiency, abiotic stress tolerance, and disease resistance (Busby et al. 2017). Bacterial isolates displaying inhibitory action in in vitro plate assays may not demonstrate biocontrol action in the greenhouse in in planta assays or field trials, because some strains may not be able to colonize the rhizosphere (the plant root surface or intercellular spaces of plants) in order to deliver their effectors (Deketelaere et al. 2017). Rotation methods, soil types, host species, and the variety of V. dahliae all also affect rhizosphere fitness and the efficiency of biocontrol agents (BCAs). To date, multiple strains distributed across four different species of Pseudomonas have been successfully developed and registered as biopesticides that are commercially available to growers for the biocontrol of many plant diseases caused by plant pathogenic fungi and bacteria, including P. chlororaphis 63–28, P. aureofaciens Tx-1, P. fluorescens A506, and P. syringae ESC-10 (Fravel 2005). Pseudomonas has additional promising BCAs to offer for agricultural disease management. Our data identified two more effective Pseudomonas strains with broad biocontrol action against various strains of V. dahliae. In addition, our P. donghuensis 22G5 showed a higher amount of 7-HT production (the OD330 of 7-HT produced by 22G5 reached 4 to 5 when the OD600 of the culture reached 2) (Fig. 4d) compared with that reported for P. donghuensis SVBP6 (Muzio et al. 2020) (the OD330 of 7-HT produced by SVBP6 reached 1 when the OD600 of the culture reached 2). Moreover, our data from the in planta assay first showed the effectiveness of P. donghuensis against V. dahliae, and this further supported its applications in the development of BCAs for VW manipulation in cotton farming. However, several perspectives are remained to be addressed: (1) the suggested application frequency and method (e.g., seed coat or soil drench) of Pseudomonas strains used as BCAs (Angelopoulou et al. 2014); (2) the concerted effects of multiple Pseudomonas strains applied as a mixture; (3) the ecological influence of BCA Pseudomonas strains on the distribution of other OTUs in the rhizosphere (Angelopoulou et al. 2014).
Supplementary information
Additional file 1: Figure S1. Schematic diagram showing work flow for high-throughput screening. Figure S2. Schematic diagram illustrating the use of the pEX18Gm suicide plasmids to generate a gene deletion in P. donghuensis 22G5. Picture was modified from the reference (Wang et al. 2015). Refer to Table S2 for detailed primer sequences. Figure S3. Plant growth promoting and biosafety tests of P. protegens XY2F4 and P. donghuensis 22G5. 7-day old cotton seedlings with treatments of different concentrations of Pseudomonas strains (XY2F4 and 22G5) and the LB control (CK), biomass were recorded three weeks after inoculation. 1:10, 1:10 dilution of overnight culture, ~1 × 107 CFU/mL; 1:100, ~1 × 106 CFU/mL; 1:1000, ~1 × 105 CFU/mL. * P < 0.05. Student’s t test. Table S1. Informations of plant rhizosphere samples. Table S2. Primer used in this study. Table S3. Genome summary of Pseudomonas strains. Table S4. RAST Annotation of unique genes in XY2F4. Table S5. RAST Annotation of unique genes in 22G5.
Acknowledgements
We would like to thank the Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences for providing the V. dahliae strains. We would also like to thank Dr. Gaojie Hong and Dr. Shengchun Xu for collecting a portion of the plant rhizosphere samples.
Authors’ contributions
XG and XT conceptualized this project and designed the experiments. XT, HZ and ML performed the experiments and generated the data. All authors read and approved the final manuscript.
Funding
This work was financially supported in part by grants from the National Key Research and Development Program (2016YFD0100605), the China Postdoctoral Science Foundation (2018M632477, 2019M652094), the National Science Foundation of China (NSFC, 31900395, 31600989), and the JCIC-MCP project.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
The authors confirm that the work described has not been published before, that it is not under consideration for publication elsewhere, that its publication has been approved by all co-authors. The authors agree to publication in the Journal of AMB Express.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13568-020-01132-1.
References
- Angelopoulou DJ, Naska EJ, Paplomatas EJ, Tjamos SE. Biological control agents (BCAs) of verticillium wilt: influence of application rates and delivery method on plant protection, triggering of host defence mechanisms and rhizosphere populations of BCAs. Plant Pathol. 2014;63(5):1062–1069. doi: 10.1111/ppa.12198. [DOI] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Zachow C, Lottmann J, Götz M, Costa R, Smalla K. Impact of plant species and site on rhizosphere-associated fungi antagonistic to verticillium dahliae kleb. Appl Environ Microbiol. 2005;71(8):4203–4213. doi: 10.1128/aem.71.8.4203-4213.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby PE, Soman C, Wagner MR, Friesen ML, Kremer JM, Bennett AE, Morsy MR, Eisen JA, Leach JE, Dangl JL. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017;15(3):e2001793. doi: 10.1371/journal.pbio.2001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae DH, Kim DR, Cheong MS, Lee YB, Kwak YS. Investigating the induced systemic resistance mechanism of 2,4-Diacetylphloroglucinol (DAPG) using DAPG hydrolase-transgenic Arabidopsis. Plant Pathol J. 2020;36(3):255–266. doi: 10.5423/ppj.oa.02.2020.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Wang P, Xie Z. A complex mechanism involving LysR and TetR/AcrR that regulates iron scavenger biosynthesis in Pseudomonas donghuensis HYS. J Bacteriol. 2018;200(13):e00087–e118. doi: 10.1128/JB.00087-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David BV, Chandrasehar G, Selvam PN. Pseudomonas fluorescens: a plant-growth-promoting rhizobacterium (PGPR) with potential role in biocontrol of pests of crops. In: Prasad R, Gill SS, Tuteja N, editors. Crop improvement through microbial biotechnology. Amsterdam: Elsevier; 2018. pp. 221–243. [Google Scholar]
- Deketelaere S, Tyvaert L, França SC, Höfte M. Desirable traits of a good biocontrol agent against verticillium wilt. Front Microbiol. 2017;8:1186. doi: 10.3389/fmicb.2017.01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doornbos RF, Geraats BP, Kuramae EE, Van Loon LC, Bakker PA. Effects of jasmonic acid, ethylene, and salicylic acid signaling on the rhizosphere bacterial community of Arabidopsis thaliana. Mol Plant Microbe Interact MPMI. 2011;24(4):395–407. doi: 10.1094/mpmi-05-10-0115. [DOI] [PubMed] [Google Scholar]
- Erdogan O, Benlioglu K. Biological control of verticillium wilt on cotton by the use of fluorescent Pseudomonas spp. under field conditions. Biol Control. 2010;53(1):39–45. doi: 10.1016/j.biocontrol.2009.11.011. [DOI] [Google Scholar]
- Fravel DR. Commercialization and implementation of biocontrol. Annu Rev Phytopathol. 2005;43:337–359. doi: 10.1146/annurev.phyto.43.032904.092924. [DOI] [PubMed] [Google Scholar]
- Garridosanz D, Meierkolthoff JP, Goker M, Martin M, Rivilla R, Redondonieto M. Genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS ONE. 2016;11(2):e0150183. doi: 10.1371/journal.pone.0150183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami D, Thakker JN, Dhandhukia P. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): A review. Cogent Food Agric. 2016;2(1):1127500. [Google Scholar]
- Gui Z, You J, Xie G, Qin Y, Wu T, Xie Z. Pseudomonas donghuensis HYS 7-hydroxytropolone contributes to pathogenicity toward Caenorhabditis elegans and is influenced by pantothenic acid. Biochem Biophys Res Commun. 2020 doi: 10.1016/j.bbrc.2020.08.067. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Chen M, Yu X, Xie Z. 7-Hydroxytropolone produced and utilized as an iron-scavenger by Pseudomonas donghuensis. Biometals. 2016;29(5):817–826. doi: 10.1007/s10534-016-9954-0. [DOI] [PubMed] [Google Scholar]
- Jordan VWL. Evaluation of fungicides for the control of verticillium wilt (V. dahliae) of strawberry. Ann Appl Biol. 1972;70(2):163–168. doi: 10.1111/j.1744-7348.1972.tb04700.x. [DOI] [Google Scholar]
- Kaminski MA, Furmanczyk EM, Sobczak A, Dziembowski A, Lipinski L. Pseudomonas silesiensis sp. nov. strain A3T isolated from a biological pesticide sewage treatment plant and analysis of the complete genome sequence. Syst Appl Microbiol. 2018;41(1):13–22. doi: 10.1016/j.syapm.2017.09.002. [DOI] [PubMed] [Google Scholar]
- Klosterman SJ, Atallah ZK, Vallad GE, Subbarao KV. Diversity, pathogenicity, and management of verticillium species. Annu Rev Phytopathol. 2009;47(1):39–62. doi: 10.1146/annurev-phyto-080508-081748. [DOI] [PubMed] [Google Scholar]
- Krzyżanowska DM, Ossowicki A, Rajewska M, Maciąg T, Jabłońska M, Obuchowski M, Heeb S, Jafra S. When genome-based approach meets the "Old but Good": Revealing genes involved in the antibacterial activity of Pseudomonas sp. P482 against soft rot pathogens. Front Microbiol. 2016;7:782–782. doi: 10.3389/fmicb.2016.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak MJ, Kong HG, Choi K, Kwon SK, Song JY, Lee J, Lee PA, Choi SY, Seo M, Lee HJ. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol. 2018;36(11):1100–1109. doi: 10.1038/nbt.4232. [DOI] [PubMed] [Google Scholar]
- Lan X, Zhang J, Zong Z, Ma Q, Wang Y. Evaluation of the biocontrol potential of Purpureocillium lilacinum QLP12 against verticillium dahliae in eggplant. Biomed Res Int. 2017;2017:4101357. doi: 10.1155/2017/4101357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Shi L, Han Q, Hu HL, Zhao MW, Tang CM, Li SP. Biocontrol of verticillium wilt and colonization of cotton plants by an endophytic bacterial isolate. J Appl Microbiol. 2012;113(3):641–651. doi: 10.1111/j.1365-2672.2012.05371.x. [DOI] [PubMed] [Google Scholar]
- Li S, Zhang N, Zhang Z, Luo J, Shen B, Zhang R, Shen Q. Antagonist Bacillus subtilis HJ5 controls verticillium wilt of cotton by root colonization and biofilm formation. Biol Fertil Soils. 2012;49(3):295–303. doi: 10.1007/s00374-012-0718-x. [DOI] [Google Scholar]
- Loper JE, Hassan KA, Mavrodi DV, Davis EW, Lim CK, Shaffer BT, Elbourne LDH, Stockwell VO, Hartney SL, Breakwell K. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8(7):1–27. doi: 10.1371/journal.pgen.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper JE, Henkels MD, Shaffer BT, Valeriote FA, Gross H. Isolation and identification of rhizoxin analogs from Pseudomonas fluorescens Pf-5 by using a genomic mining strategy. Appl Environ Microbiol. 2008;74(10):3085–3093. doi: 10.1128/AEM.02848-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg BJ, Dekkers L, Bloemberg GV. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu Rev Phytopathol. 2001;39(1):461–490. doi: 10.1146/annurev.phyto.39.1.461. [DOI] [PubMed] [Google Scholar]
- Medema MH, Blin K, Cimermancic P, De Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39(2):339–346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierkolthoff JP, Goker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10(1):2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercadoblanco J, Rodriguezjurado D, Hervas AB, Jimenezdiaz RM. Suppression of verticillium wilt in olive planting stocks by root-associated fluorescent Pseudomonas spp. Biol Control. 2004;30(2):474–486. doi: 10.1016/j.biocontrol.2004.02.002. [DOI] [Google Scholar]
- Meyer SLF, Halbrendt JM, Carta LK, Skantar AM, Liu T, Abdelnabby HME, Vinyard BT. Toxicity of 2,4-diacetylphloroglucinol (DAPG) to plant-parasitic and bacterial-feeding nematodes. J Nematol. 2009;41(4):274–280. [PMC free article] [PubMed] [Google Scholar]
- Mhlongo MI, Piater LA, Madala NE, Labuschagne N, Dubery IA. The Chemistry of plant-microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front Plant Sci. 2018;9:112. doi: 10.3389/fpls.2018.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio FM, Agaras BC, Masi M, Tuzi A, Evidente A, Valverde C. 7-hydroxytropolone is the main metabolite responsible for the fungal antagonism of Pseudomonas donghuensis strain SVBP6. Environ Microbiol. 2020;22(7):2550–2563. doi: 10.1111/1462-2920.14925. [DOI] [PubMed] [Google Scholar]
- Nowak-thompson B, Gould S, Kraus J, Loper J. Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf-5. Can J Microbiol. 1994;40:1064–1066. doi: 10.1139/m94-168. [DOI] [Google Scholar]
- Ohji S, Yamazoe A, Hosoyama A, Tsuchikane K, Ezaki T, Fujita N. The complete genome sequence of Pseudomonas putida NBRC 14164T confirms high intraspecies variation. Genome Announcements. 2014 doi: 10.1128/genomeA.00029-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowicki A, Jafra S, Garbeva P. The antimicrobial volatile power of the rhizospheric isolate Pseudomonas donghuensis P482. PLoS ONE. 2017;12(3):e0174362. doi: 10.1371/journal.pone.0174362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M. The SEED and the rapid annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42(D1):D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GSA, Mavrodi DV, Deboy RT, Seshadri R, Ren Q, Madupu R. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol. 2005;23(7):873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peix A, Ramírez-Bahena MH, Velázquez E. The current status on the taxonomy of Pseudomonas revisited: an update. Infect Genet Evol. 2018;57:106–116. doi: 10.1016/j.meegid.2017.10.026. [DOI] [PubMed] [Google Scholar]
- Rodriguez PA, Rothballer M, Chowdhury SP, Nussbaumer T, Gutjahr C, Falter-Braun P. Systems biology of plant-microbiome interactions. Mol Plant. 2019;12(6):804–821. doi: 10.1016/j.molp.2019.05.006. [DOI] [PubMed] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160(1):47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Shaukat SS, Siddiqui IA. Impact of biocontrol agents Pseudomonas fluorescens CHA0 and its genetically modified derivatives on the diversity of culturable fungi in the rhizosphere of mungbean. J Appl Microbiol. 2003;95(5):1039–1048. doi: 10.1046/j.1365-2672.2003.02074.x. [DOI] [PubMed] [Google Scholar]
- Sherzad Z, Canming T. A new strain of Bacillus velezensis as a bioagent against verticillium dahliae in cotton: isolation and molecular identification. Egypt J Biol Pest Control. 2020;30(1):118. doi: 10.1186/s41938-020-00308-y. [DOI] [Google Scholar]
- Sitaraman R. Pseudomonas spp. as models for plant–microbe interactions. Front Plant Sci. 2015 doi: 10.3389/fpls.2015.00787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares MP, Weiss G. The Iron age of host–microbe interactions. EMBO Rep. 2015;16(11):1482–1500. doi: 10.15252/embr.201540558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Jiang H, Zhu X, Wang W, He X, Shi Y, Yuan Y, Du X, Cai Y. Analysis of sea-island cotton and upland cotton in response to verticillium dahliae infection by RNA sequencing. BMC Genomics. 2013;14(1):852–852. doi: 10.1186/1471-2164-14-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley JT, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA)software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Termorshuizen AJ. The recovery rate of microsclerotia of verticillium dahliae with different detection methods. Phytoparasitica. 1995;23:55–56. [Google Scholar]
- Tjamos E, Tsitsiyannis DI, Tjamos S (2000) Selection and evaluation of rhizosphere bacteria as biocontrol agents against verticillium dahliae. Advances in Verticillium Research and Disease Management, pp 244–248
- Troppens DM, Dmitriev RI, Papkovsky DB, O'Gara F, Morrissey JP. Genome-wide investigation of cellular targets and mode of action of the antifungal bacterial metabolite 2,4-diacetylphloroglucinol in Saccharomyces cerevisiae. FEMS Yeast Res. 2013;13(3):322–334. doi: 10.1111/1567-1364.12037. [DOI] [PubMed] [Google Scholar]
- Veach AM, Morris R, Yip DZ, Yang ZK, Engle NL, Cregger MA, Tschaplinski TJ, Schadt CW. Rhizosphere microbiomes diverge among Populus trichocarpa plant-host genotypes and chemotypes, but it depends on soil origin. Microbiome. 2019;7(1):76. doi: 10.1186/s40168-019-0668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleesschauwer D. Using Serratia plymuthica to control fungal pathogens of plants. CAB Rev. 2007 doi: 10.1079/pavsnnr20072046. [DOI] [Google Scholar]
- Wang P, Yu Z, Li B, Cai X, Zeng Z, Chen X, Wang X. Development of an efficient conjugation-based genetic manipulation system forPseudoalteromonas. Microb Cell Fact. 2015 doi: 10.1186/s12934-015-0194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liang C, Wu S, Zhang X, Tang J, Jian G, Jiao G, Li F, Chu C. Significant improvement of cotton verticillium wilt resistance by manipulating the expression of gastrodia antifungal proteins. Mol Plant. 2016;9(10):1436–1439. doi: 10.1016/j.molp.2016.06.013. [DOI] [PubMed] [Google Scholar]
- Weller DM, Landa BB, Mavrodi OV, Schroeder KL, La Fuente LD, Bankhead SB, Molar RA, Bonsall RF, Mavrodi DV, Thomashow LS. Role of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant Biol. 2007;9(1):4–20. doi: 10.1055/s-2006-924473. [DOI] [PubMed] [Google Scholar]
- Xue L, Xue Q, Chen Q, Lin C, Shen G, Zhao J. Isolation and evaluation of rhizosphere actinomycetes with potential application for biocontrol of verticillium wilt of cotton. Crop Protect. 2013;43:231–240. doi: 10.1016/j.cropro.2012.10.002. [DOI] [Google Scholar]
- Yang F, Cao Y. Biosynthesis of phloroglucinol compounds in microorganisms–review. Appl Microbiol Biotechnol. 2012;93(2):487–495. doi: 10.1007/s00253-011-3712-6. [DOI] [PubMed] [Google Scholar]
- Zhang J, Sanogo S, Flynn R, Baral JB, Bajaj S, Hughs SE, Percy RG. Germplasm evaluation and transfer of verticillium wilt resistance from Pima (Gossypium barbadense) to Upland cotton (G. hirsutum) Euphytica. 2012;187(2):147–160. doi: 10.1007/s10681-011-0549-0. [DOI] [Google Scholar]
- Zhang W, Jian G, Jiang T, Wang S, Qi F, Xu S. Cotton gene expression profiles in resistant Gossypium hirsutum cv. Zhongzhimian KV1 responding to Verticillium dahliae strain V991 infection. Mol Biol Rep. 2012;39(10):9765–9774. doi: 10.1007/s11033-012-1842-2. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ni H, Du X, Wang S, Ma XW, Nurnberger T, Guo HS, Hua C. The verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytol. 2017;215(1):368–381. doi: 10.1111/nph.14537. [DOI] [PubMed] [Google Scholar]
- Zhang F, Li X-L, Zhu S-J, Ojaghian MR, Zhang J-Z. Biocontrol potential of Paenibacillus polymyxa against verticillium dahliae infecting cotton plants. Biol Control. 2018;127:70–77. doi: 10.1016/j.biocontrol.2018.08.021. [DOI] [Google Scholar]
- Zhao Y, Wang H, Chen W, Li Y. Genetic structure, linkage disequilibrium and association mapping of verticillium wilt resistance in elite cotton (Gossypium hirsutum L.) germplasm population. PLoS ONE. 2014;9(1):e86308. doi: 10.1371/journal.pone.0086308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Schematic diagram showing work flow for high-throughput screening. Figure S2. Schematic diagram illustrating the use of the pEX18Gm suicide plasmids to generate a gene deletion in P. donghuensis 22G5. Picture was modified from the reference (Wang et al. 2015). Refer to Table S2 for detailed primer sequences. Figure S3. Plant growth promoting and biosafety tests of P. protegens XY2F4 and P. donghuensis 22G5. 7-day old cotton seedlings with treatments of different concentrations of Pseudomonas strains (XY2F4 and 22G5) and the LB control (CK), biomass were recorded three weeks after inoculation. 1:10, 1:10 dilution of overnight culture, ~1 × 107 CFU/mL; 1:100, ~1 × 106 CFU/mL; 1:1000, ~1 × 105 CFU/mL. * P < 0.05. Student’s t test. Table S1. Informations of plant rhizosphere samples. Table S2. Primer used in this study. Table S3. Genome summary of Pseudomonas strains. Table S4. RAST Annotation of unique genes in XY2F4. Table S5. RAST Annotation of unique genes in 22G5.
Data Availability Statement
All data generated or analysed during this study are included in this published article.