Abstract
Meningitic Escherichia coli invasion of the host brain can lead to increased blood-brain barrier (BBB) permeability. Circular RNAs (circRNAs) are non-coding RNAs, highly abundant in the brain, that are widely involved in the pathological processes of central nervous system (CNS) disorders; however, whether circRNAs participate in the regulation of BBB permeability during E. coli meningitis remains unknown. Here, we identified a novel circRNA, circ_2858, that was significantly upregulated in human brain microvascular endothelial cells (hBMECs) upon meningitic E. coli infection. We also found that circ_2858 regulated BBB permeability in hBMECs by competitively binding miR-93-5p, thereby inducing the upregulation of vascular endothelial growth factor A and finally resulting in downregulation as well as altered distribution of tight junction proteins such as ZO-1, Occludin, and Claudin-5. These findings provide novel insights into the influence of circ_2858 on BBB permeability during the pathogenic process of E. coli meningitis, suggesting potential nucleic acid targets for future prevention and therapy of CNS infection induced by meningitic E. coli.
Keywords: blood-brain barrier, brain microvascular endothelial cells, circ_2858, miR-93-5p, vascular endothelial growth factor A, permeability
Graphical Abstract
Yang and colleagues found that meningitic Escherichia coli-induced circular RNA circ_2858 acts as a competing endogenous RNA for the miR-17 family to facilitate VEGFA expression in human brain microvascular endothelial cells, thus leading to tight junction protein disruption and blood-brain barrier dysfunction.
Introduction
Bacterial meningitis is currently recognized as one of the top ten causes of infection-related deaths worldwide, and 30%–50% of survivors sustain severe neurological sequelae.1 Escherichia coli is widely accepted as one of the most common gram-negative pathogens inducing bacterial meningitis, especially during the neonatal stage.2 Although great progress has been achieved in the field of antimicrobial agents, the presence of the blood-brain barrier (BBB) in the central nervous system (CNS) makes it difficult for drugs to penetrate into the brain, bringing great challenges to the clinical therapy of bacterial meningitis.
Brain microvascular endothelial cells (BMECs) are the most direct and functional structural components of the BBB. They are characterized by the presence of several tight junction (TJ) proteins, such as Claudins, Occludins, and ZOs,3 which increase the trans-endothelial electric resistance (TEER) of the BMECs monolayer and impede paracellular transportation of macromolecules.4 Decrease or redistribution of TJ proteins could lead to increased BBB permeability, an important indicator of BBB damage.5 Accumulating evidence has suggested that meningitic pathogens enhance BBB permeability by degrading TJ proteins and that infection-induced neuroinflammatory responses largely aggravate CNS dysfunction.6,7 Meanwhile, several host factors, such as transforming growth factor β1 (TGF-β1), matrix metallopeptidases (MMPs), and vascular endothelial growth factor A (VEGFA) are involved in the regulation of BBB permeability.8, 9, 10 The identification of the key molecules involved in BBB disruption induced by different CNS-infecting bacteria is of great significance; however, the underlying mechanisms by which these key factors regulate BBB permeability in response to infection remain largely unclear.
Circular RNAs (circRNAs) are a class of special RNA molecules that lack 5′ and 3′ ends and a poly(A) tail and form covalently closed loops.11 They have recently been identified as a naturally occurring family of non-coding RNAs that are stable, abundant, conserved, and specifically expressed in eukaryotic cells,12 especially in the brain tissue.13 Many studies have demonstrated the essential regulatory roles of circRNAs in multiple pathological processes.14, 15, 16, 17 For example, circ-Vav3 was shown to be associated with epithelial-mesenchymal transition, whereas circHIPK3 is involved in cell growth.18,19 Moreover, some studies have disclosed different working mechanisms of circRNAs, which can act as miRNA sponges, regulators of transcription and splicing, protein-protein interaction adapters, ribosomal RNA processing factors, etc.20, 21, 22, 23 Noticeably, our previous whole-transcriptome sequencing and transcriptome-wide bioinformatic analyses suggested a novel significantly upregulated circRNA, circ_2858, acting as a sponge for multiple members of the miR-17 family, which has been studied for its role in multiple infectious diseases.24,25 However, the specific role of the miR-17 family in meningitic E. coli-induced BBB dysfunction remains unknown, and whether circ_2858 contributes to this pathogenic process has yet to be determined.
Here, we report for the first time a novel circRNA, circ_2858, which is involved in the pathogenesis of bacterial meningitis. We demonstrated that meningitic E. coli infection could significantly increase circ_2858 expression in human BMECs (hBMECs), which substantially upregulated VEGFA by competitive sponging of multiple miRNAs of the miR-17 family, thereby disrupting the BBB. These findings substantiate the important role of host circRNAs in the context of meningitic bacterial infection, which could serve as potential nucleic acid targets for further study of bacterial meningitis and for better prevention and therapy of bacterial-induced CNS dysfunction.
Results
Differential miRNAs Profiling and Regulatory miRNA-mRNA Networks in hBMECs upon Meningitic E. coli Infection
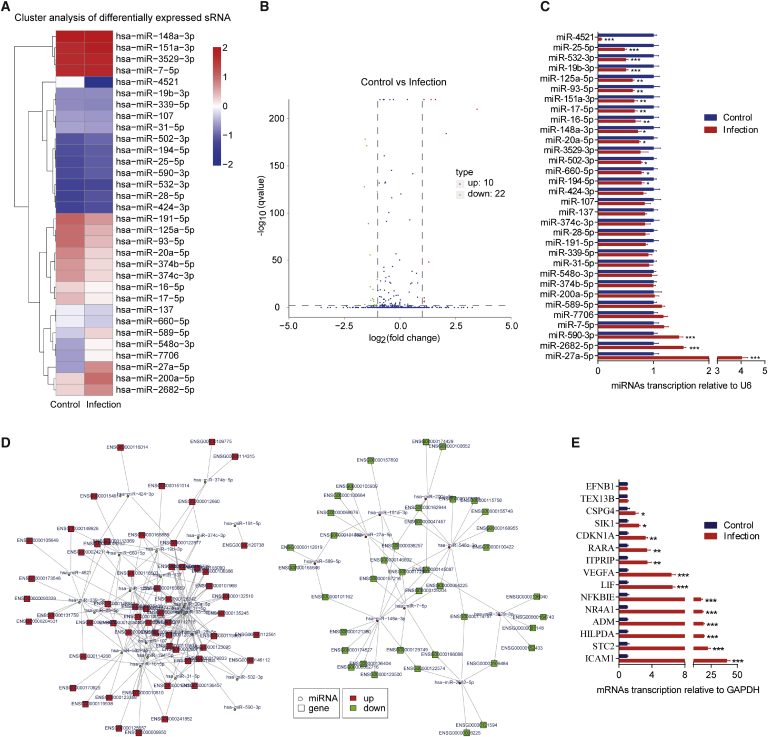
The microRNA (miRNA) sequencing was performed to identify differentially expressed miRNAs in hBMECs in response to meningitic E. coli infection. As shown in Figures 1A and 1B, a total of 32 miRNAs were found to have significant changes in their expression levels (≥2-fold increase or ≤0.5-fold decrease, p < 0.05), including 10 upregulated miRNAs and 22 downregulated miRNAs. Among them, miR-27a-5p (11.18-fold) and miR-200a-5p (4.27-fold) were the most highly upregulated, whereas miR-4521 (202.29-fold) and miR-20a-5p (3.01-fold) were the most highly downregulated (Table 1). We next employed poly(A) tailing quantitative polymerase chain reaction (qPCR) to verify differential expression of all 32 miRNAs emerging from the sequencing data. For most miRNAs, the trend of expression changes detected by qPCR was consistent with that obtained by miRNA sequencing (Figure 1C). Since we have previously reported a total of 366 significantly differentially expressed mRNAs (≥2-fold increase or ≤0.5-fold decrease, p < 0.05) in hBMECs after meningitic E. coli infection,26 we next sought to predict the potential mRNA targets of these differentially expressed miRNAs using the miRanda and RNAhybrid software. By using miRanda, 473 miRNA-mRNA pairs were matched (Table S1), among which 269 pairs showed a negative correlation (Figure 1D; Table S2). When both miRanda and RNAhybrid were applied, a total of 23 miRNA-mRNA pairs with negative correlation were obtained (Table 2). Notably, multiple downregulated miRNAs, such as miR-93-5p, miR-20a-5p, miR-17-5p, miR-125a-5p, and miR-16-5p, all targeted VEGFA, suggesting multiple miRNA-mediated regulation of VEGFA during meningitic E. coli infection. The transcription of mRNAs in these 23 negative-matched pairs was subsequently verified by qPCR, and most qPCR results were highly consistent with those from RNA sequencing except for EFNB1 and TEX13B (Figure 1E). In addition, two genes, SLA2 and POU5F1, were not well amplified by qPCR because of their low expression levels in hBMECs. Taken together, we provided a comprehensive expression profile of miRNAs in hBMECs challenged by meningitic E. coli and suggested their potential target mRNAs; in particular, we observed five significantly downregulated miRNAs that likely targeted VEGFA during infection.
Figure 1.
Differential miRNAs and the Regulatory Network of miRNAs-mRNAs in hBMECs during Meningitic E. coli Infection
(A) Heatmap indicating the differential expression of miRNAs in hBMECs with or without meningitic E. coli infection. (B) Volcano plot of the overall distribution of miRNAs. Red dots represent upregulated miRNAs, and green dots represent downregulated miRNAs (≥2-fold or ≤0.5-fold, p < 0.05). (C) qPCR verification of 32 differentially expressed miRNAs in hBMECs in response to the infection. Data are expressed as mean ± SD from three independent experiments. (D) The negative regulatory network between different miRNAs and different mRNAs predicted by miRanda. (E) qPCR verification of the miRNA targets in hBMECs upon meningitic E. coli infection. Data are expressed as mean ± SD from three independent experiments.
Table 1.
The Most Significantly Changed miRNAs in hBMECs upon Infection
| miRNAs | Read Count |
Fold Change | p Value | q Value | |
|---|---|---|---|---|---|
| Control | Infection | ||||
| hsa-miR-27a-5p | 115.270307 | 1288.638585 | 11.17962584 | 9.60E-213 | 6.33E-211 |
| hsa-miR-200a-5p | 503.7817248 | 2150.681355 | 4.269005437 | 6.40E-187 | 3.86E-185 |
| hsa-miR-151a-3p | 5827.250573 | 17805.29679 | 3.055482038 | 0 | 0 |
| hsa-miR-148a-3p | 8890.222425 | 23681.72671 | 2.663826278 | 0 | 0 |
| hsa-miR-2682-5p | 520.6095069 | 1294.152409 | 2.48578238 | 2.86E-50 | 5.92E-49 |
| hsa-miR-548o-3p | 122.9479825 | 264.6635266 | 2.152629453 | 1.94E-08 | 1.85E-07 |
| hsa-miR-589-5p | 186.8935543 | 402.0738115 | 2.151286992 | 4.57E-12 | 5.02E-11 |
| hsa-miR-7706 | 117.2686061 | 250.7338673 | 2.138056533 | 6.15E-08 | 5.72E-07 |
| hsa-miR-3529-3p | 5643.617402 | 12061.63394 | 2.137167525 | 0 | 0 |
| hsa-miR-7-5p | 5737.011592 | 12204.55805 | 2.127265346 | 0 | 0 |
| hsa-miR-191-5p | 2072.551706 | 1027.747675 | 0.495892737 | 5.95E-125 | 1.87E-123 |
| hsa-miR-16-5p | 500.4161684 | 248.1220562 | 0.495823996 | 1.66E-31 | 2.80E-30 |
| hsa-miR-502-3p | 47.11778971 | 22.78079697 | 0.483470578 | 0.00024986 | 0.0018115 |
| hsa-miR-28-5p | 34.39177954 | 16.10616856 | 0.468298395 | 0.0013104 | 0.0087965 |
| hsa-miR-25-5p | 41.96428146 | 19.29838215 | 0.459870641 | 0.00031512 | 0.0021968 |
| hsa-miR-194-5p | 44.38327513 | 20.31408647 | 0.457708186 | 0.00020004 | 0.0014649 |
| hsa-miR-590-3p | 42.0694551 | 19.15328153 | 0.455271798 | 0.00027697 | 0.0019882 |
| hsa-miR-19b-3p | 93.28901668 | 41.49877665 | 0.444852819 | 3.53E-08 | 3.33E-07 |
| hsa-miR-107 | 118.1099952 | 52.52642359 | 0.444729497 | 5.50E-10 | 5.61E-09 |
| hsa-miR-424-3p | 32.70900134 | 14.36496115 | 0.439184791 | 0.0009805 | 0.0066436 |
| hsa-miR-137 | 264.0910044 | 114.7745886 | 0.434612026 | 3.74E-21 | 5.21E-20 |
| hsa-miR-31-5p | 129.1532272 | 55.42843595 | 0.429163712 | 2.69E-11 | 2.87E-10 |
| hsa-miR-339-5p | 98.23217766 | 39.90266986 | 0.406210659 | 1.55E-09 | 1.52E-08 |
| hsa-miR-660-5p | 266.0893035 | 106.0685515 | 0.398623868 | 7.77E-24 | 1.10E-22 |
| hsa-miR-17-5p | 671.1129825 | 266.6949353 | 0.397382432 | 9.04E-58 | 1.99E-56 |
| hsa-miR-374c-3p | 1000.201295 | 376.5361028 | 0.376468223 | 3.69E-91 | 8.91E-90 |
| hsa-miR-374b-5p | 1001.463379 | 376.8263041 | 0.376285604 | 2.51E-91 | 6.28E-90 |
| hsa-miR-125a-5p | 1810.353827 | 646.7134529 | 0.357223773 | 6.38E-174 | 3.31E-172 |
| hsa-miR-93-5p | 1783.008681 | 606.3754812 | 0.340092713 | 5.92E-181 | 3.30E-179 |
| hsa-miR-532-3p | 34.39177954 | 11.60804941 | 0.337532891 | 6.38E-05 | 0.00050292 |
| hsa-miR-20a-5p | 1249.988685 | 415.8583702 | 0.332678376 | 1.94E-130 | 6.40E-129 |
| hsa-miR-4521 | 322.8830679 | 1.596106794 | 0.004943334 | 1.44E-76 | 3.38E-75 |
Table 2.
miRNA-mRNA Pairs Predicted by miRanda and RNAhybrid
| miRNA | miRNA Fold Change | mRNA | mRNA Fold Change |
|---|---|---|---|
| hsa-miR-27a-5p | 11.17962584 | SLA2 | −4.41 |
| hsa-miR-93-5p | 0.340092713 | HILPDA | 6.65 |
| hsa-miR-93-5p | 0.340092713 | VEGFA | 7.55 |
| hsa-miR-125a-5p | 0.357223773 | LIF | 4.00 |
| hsa-miR-125a-5p | 0.357223773 | STC2 | 5.18 |
| hsa-miR-125a-5p | 0.357223773 | VEGFA | 7.55 |
| hsa-miR-20a-5p | 0.332678376 | VEGFA | 7.55 |
| hsa-miR-4521 | 0.004943334 | ADM | 11.90 |
| hsa-miR-17-5p | 0.397382432 | VEGFA | 7.55 |
| hsa-miR-16-5p | 0.495823996 | VEGFA | 7.55 |
| hsa-miR-107 | 0.444729497 | SIK1 | 2.77 |
| hsa-miR-339-5p | 0.406210659 | CDKN1A | 2.64 |
| hsa-miR-339-5p | 0.406210659 | RARA | 3.50 |
| hsa-miR-339-5p | 0.406210659 | POU5F1 | 2.55 |
| hsa-miR-339-5p | 0.406210659 | NFKBIE | 2.86 |
| hsa-miR-339-5p | 0.406210659 | CSPG4 | 2.25 |
| hsa-miR-339-5p | 0.406210659 | ITPRIP | 2.16 |
| hsa-miR-339-5p | 0.406210659 | ICAM1 | 8.75 |
| hsa-miR-532-3p | 0.337532891 | LIF | 4.00 |
| hsa-miR-532-3p | 0.337532891 | EFNB1 | 2.03 |
| hsa-miR-532-3p | 0.337532891 | NR4A1 | 7.48 |
| hsa-miR-532-3p | 0.337532891 | PDLIM2 | 2.60 |
| hsa-miR-532-3p | 0.337532891 | TEX13B | 2.97 |
miR-93-5p Negatively Regulated VEGFA Expression in hBMECs
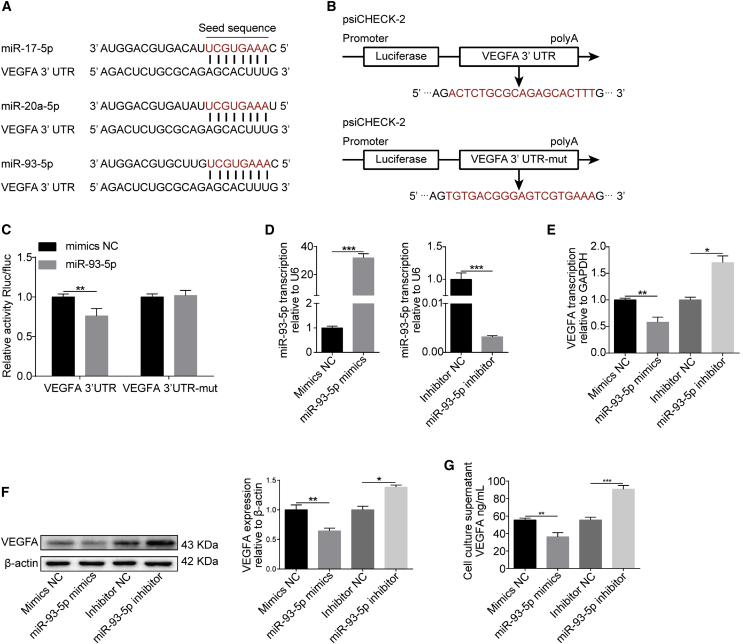
Based on the above prediction, we next sought to investigate the regulatory relationship between these miRNAs and VEGFA. Bioinformatic analysis showed that the VEGFA 3′-UTR contains conserved miRNA response elements (MREs) for miR-17-5p, miR-20a-5p, and miR-93-5p (Figure 2A), which belong to the miR-17 family. Because the seed sequences of miR-17-5p, miR-20a-5p, and miR-93-5p are identical, and miR-93-5p was more abundant than the other two miRNAs in hBMECs according to our miRNA sequencing data, we next chose miR-93-5p, which was further verified independently to be significantly downregulated upon infection (Figure S1), as the representative miRNA to investigate its role in the regulation of VEGFA expression. Dual-luciferase reporter assays were first performed to analyze the possible interaction between miR-93-5p and VEGFA. As shown in Figure 2B, both wild-type and mutant VEGFA 3′-UTR were cloned into the psiCHECK-2 luciferase reporter plasmid for the assays. Results showed that co-transfection with miR-93-5p and VEGFA 3′-UTR led to a significantly decreased luciferase activity with respect to that of the transfection with a control miRNA mimic, whereas, in contrast, luciferase activity did not significantly differ when miR-93-5p or control miRNA mimic were co-transfected with mutant VEGFA 3′-UTR (Figure 2C), indicating the direct binding of miR-93-5p to VEGFA 3′-UTR. We next used miR-93-5p mimic or inhibitor to significantly increase or decrease the miR-93-5p level, respectively (Figure 2D), and observed that transfection with the miR-93-5p mimic significantly decreased the expression of VEGFA, whereas the miR-93-5p inhibitor significantly increased VEGFA expression, both at the mRNA and protein levels (Figures 2E and 2F). Moreover, the level of secreted VEGFA in cell culture supernatant exhibited similar changes upon transfection with miR-93-5p mimic or inhibitor, compared with their respective controls (Figure 2G). Together, these results largely confirm the negative regulation of VEGFA expression by miR-93-5p.
Figure 2.
miR-93-5p Negatively Affects VEGFA Expression
(A) Putative binding sites of miR-17 family members including miR-17-5p, miR-20a-5p, and miR-93-5p in the 3′-UTR of VEGFA mRNA. (B) Generation of the VEGFA 3′-UTR wild-type and mutant constructs by cloning into the psiCHECK-2 plasmid. (C) Dual-luciferase reporter assays showing that miR-93-5p significantly reduced luciferase activity of VEGFA 3′-UTR but not the luciferase activity of the mutant VEGFA 3′-UTR. The Renilla luciferase activity was normalized to the firefly luciferase activity, and data were presented as mean ± SD from three independent experiments. (D) qPCR determining the level of miR-93-5p expression in hBMECs transfected with an miR-93-5p mimic or inhibitor. U6 was used as internal reference, and data were presented as mean ± SD from three independent experiments. (E) qPCR determining the effects of transfection with an miR-93-5p mimic or inhibitor on the expression of VEGFA. GAPDH was used as internal reference, and data were presented as mean ± SD from three independent experiments. (F) Western blot analysis of VEGFA expression in hBMECs treated with miR-93-5p mimic or inhibitor. β-actin was used as loading control, and densitometry was performed to quantify differences. (G) Levels of secretory VEGFA in culture supernatant of hBMECs transfected with miR-93-5p mimic or inhibitor, as determined by ELISA. Results were calculated as mean ± SD from three independent experiments.
The miR-93-5p/VEGFA Axis Contributes to the Regulation of BBB Permeability
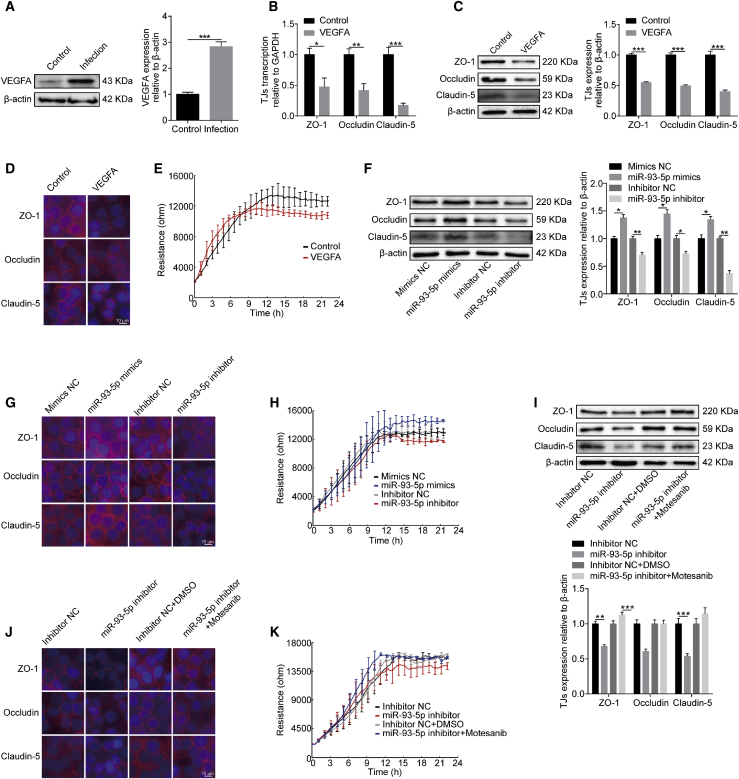
We previously reported that meningitic E. coli infection can induce VEGFA expression, which aggravates infection-dependent BBB dysfunction.9 Here, we further confirmed our previous reports using a recombinant VEGFA protein (Novoprotein, Summit, NJ, USA). As shown in Figure 3A, a significantly high level of VEGFA was detected in hBMECs challenged with meningitic E. coli. Upon treatment with 100 ng/mL of recombinant VEGFA for 24 h, we found that the transcriptional levels of genes encoding TJ proteins, including ZO-1, Occludin, and Claudin-5, were significantly decreased as compared to that of the control (Figure 3B), and the levels of the corresponding proteins also decreased significantly in response to VEGFA treatment (Figure 3C). Moreover, these TJ proteins were found by immunofluorescence analysis to be well arranged and distributed around the hBMECs. In contrast, these proteins became inconsecutive, irregularly distributed, or scattered around the cells upon VEGFA treatment, indicating a direct breakdown of TJ proteins between adjacent endothelial cells (Figure 3D). We also evaluated the possible influence of VEGFA treatment on the TEER of hBMECs through the electric cell-substrate impedance sensing (ECIS) system. The results indicate that VEGFA treatment appreciably decreased the TEER values of hBMECs when reaching the monolayer after 12 h of growth (Figure 3E). These observations further confirm that VEGFA is an important effector leading to BBB damage by downregulating TJ proteins.
Figure 3.
Effects of the miR-93-5p/VEGFA Axis on BBB Permeability
(A) Western blotting and densitometry analysis revealed a significant increase of VEGFA levels in hBMECs in response to infection. β-actin was used as loading control. (B) qPCR determining the expression of ZO-1, Occludin, and Claudin-5 in hBMECs upon VEGFA treatment. (C) Western blot determining the expression of ZO-1, Occludin, and Claudin-5 in hBMECs upon VEGFA treatment. β-actin was used as loading control, and densitometry was performed to analyze differences. (D) Immunofluorescence analysis determining the distribution of ZO-1, Occludin, and Claudin-5 in hBMECs upon VEGFA treatment. Scale bar, 10 μm. (E) TEER changes of hBMECs in response to recombinant VEGFA monitored by the ECIS system. Data were collected and are presented as mean ± SD from three replicated wells. (F) Western blot analysis of TJ protein expression in hBMECs transfected with miR-93-5p mimic or inhibitor. β-actin was used as loading control, and densitometry was performed to analyze differences. (G) Immunofluorescence analysis of TJ protein distribution in hBMECs transfected with miR-93-5p mimic or inhibitor. Scale bar, 10 μm. (H) TEER changes of monolayer hBMECs transfected with miR-93-5p mimic or inhibitor as recorded by the ECIS system. Data were collected and are presented as mean ± SD from three replicated wells. (I) The expression of TJ proteins in response to miR-93-5p inhibitor in hBMECs with or without treatment with the VEGFR inhibitor Motesanib, as determined by western blot and densitometry. β-actin was used as loading control. (J) The expression of TJ proteins in response to miR-93-5p inhibitor in hBMECs with or without treatment with the VEGFR inhibitor Motesanib, as determined by immunofluorescence analysis. Scale bar, 10 μm. (K) TEER changes of hBMECs in response to miR-93-5p inhibitor with or without treatment with the VEGFR inhibitor Motesanib, as monitored by the ECIS system. Data were collected and are presented as mean ± SD from three replicated wells at each time point.
Since VEGFA could be targeted by miR-93-5p, we also examined whether miR-93-5p worked in the regulation of BBB permeability in the hBMECs model. Transfection of the miR-93-5p mimic in hBMECs led to an increased expression of TJ proteins, including ZO-1, Occludin, and Claudin-5, whereas transfection of the miR-93-5p inhibitor in hBMECs caused decreased expression of these TJ proteins (Figure 3F). By immunofluorescence analysis, we also demonstrated that the TJ proteins between adjacent endothelial cells could be well maintained when overexpressing miR-93-5p, whereas transfection with the miR-93-5p inhibitor resulted in a significant breakdown of ZO-1, Occludin, and Claudin-5 between adjacent cells (Figure 3G). Meanwhile, ECIS analysis showed that transfection with the miR-93-5p mimic led to higher TEER values in the cells, whereas transfection with the miR-93-5p inhibitor led to a decreased resistance of the hBMEC monolayer, compared with their respective controls (Figure 3H). The effects of the miR-93-5p/VEGFA axis on BBB integrity were further analyzed using the VEGFR inhibitor Motesanib. As shown in Figure 3I, the downregulation of ZO-1, Occludin, and Claudin-5 in miR-93-5p inhibitor-transfected hBMECs was completely prevented by treatment with Motesanib. In addition, immunofluorescence and ECIS assays showed that the negative regulatory effects of the miR-93-5p inhibitor on BBB integrity were well blocked when Motesanib was applied (Figures 3J and 3K). Taken together, these results indicate that miR-93-5p functions by targeting VEGFA to regulate BBB permeability.
Characterization of a Novel circ_2858 that Would Potentially Bind miR-17 Family
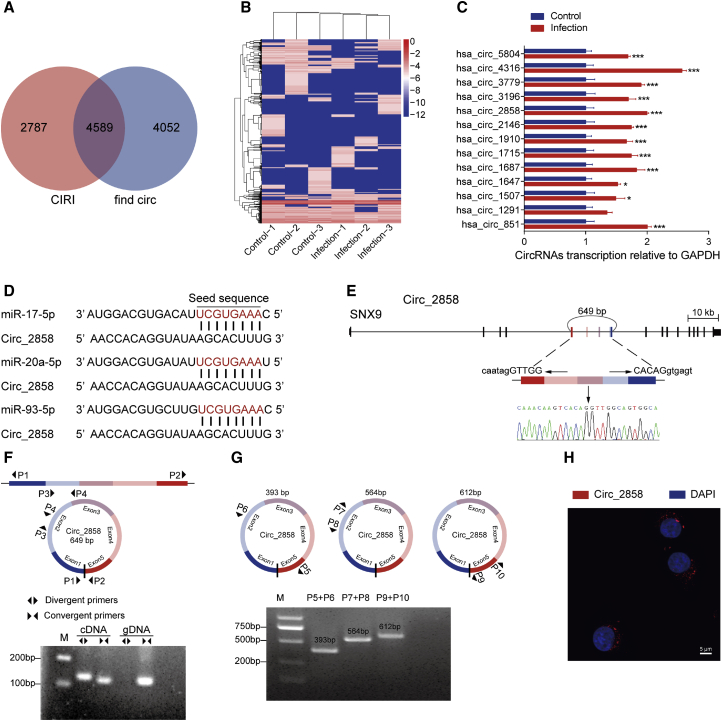
By reanalyzing our previous long non-coding RNA sequencing data,26 a total of 7,376 circRNAs were identified by CIRI, and 8,641 circRNAs were identified by find_circ, with 4,589 circRNAs overlapping between the results of these two algorithms (Figure 4A; Table S3). We built heatmaps to show the expression levels of these circRNAs in hBMECs in the presence or absence of meningitic E. coli (Figure 4B). Subsequently, 13 upregulated circRNAs were randomly selected for validation through qPCR with specific divergent primers (Table S4), and the results showed the same trends of upregulation as the sequencing data (Figure 4C). Noticeably, by competitive endogenous RNA (ceRNA) analysis, we found that one circRNA, circ_2858, may potentially act as a miRNA sponge to bind the miR-17 family members, including miR-17-5p, miR-20a-5p, and miR-93-5p herein, with completely the same seed sequence (Figure 4D). And, more importantly, the infections by another neonatal meningitis-causing E. coli strain, RS218, as well as the non-meningitic E. coli HB101, were also implemented to check the transcription of circ_2858, and we found that this circ_2858 was specifically induced by these two meningitis-causing strains, but not by the non-meningitic E. coli strain (Figure S2). circ_2858 derives from the parental gene SNX9, which was not affected by the infection (Figure S3); it consists of five exons of the SNX9 mRNA, and its length was predicted to be 649 bp (Figure 4E). By using both the divergent primers P1/P2 and the convergent primers P3/P4 (Table S5) for PCR amplification of cDNA and genomic DNA (gDNA) from hBMECs, we demonstrated that the convergent primers P3/P4 yielded a specific product of identical size from both cDNA and gDNA templates. However, the divergent primers P1/P2 only produced an amplicon from cDNA, but not from the gDNA template (Figure 4F), suggesting that circ_2858 was indeed a circRNA formed by head-to-tail fusion of these exons. We designed three different pairs of divergent primers, P5/P6, P7/P8, and P9/P10 (Figure 4G; Table S5), to verify the predicted length of circ_2858. By PCR amplification and sequencing (data not shown), we confirmed that the whole sequence of circ_2858 was the same as the predicted one (Figure 4G). Additionally, fluorescence in situ hybridization (FISH) using a specific probe indicated that circ_2858 was preferentially localized in the cytoplasm (Figure 4H). Altogether, we identified and characterized a novel cytoplasm-located circRNA, circ_2858, which is likely to act as a sponge for members of the miR-17 family.
Figure 4.
Characterization of circ_2858 in hBMECs
(A) Venn diagram demonstrating the overlap between circRNAs predicted by CIRI and find_circ. (B) Heatmaps of these 4,589 overlapping circRNAs in hBMECs based on their expression levels (RPKM, Reads Per Kilobase of exon model per Million mapped reads.). (C) qPCR verification of the 13 circRNAs upregulated in hBMECs in response to meningitic E. coli infection. Data are expressed as mean ± SD from three independent experiments. (D) Putative binding sites of miR-17 family members (miR-17-5p, miR-20a-5p, and miR-93-5p) on circ_2858. (E) The genomic loci of circ_2858 in its parental gene SNX9. The presence of circ_2858 was validated by sequencing. (F) Both divergent and convergent primer pairs were applied for amplification of cDNA as well as of genomic DNA (gDNA) to verify the formation of circ_2858. (G) Three different divergent primer pairs were additionally designed to check the length of circ_2858. (H) RNA FISH showing the subcellular location of circ_2858. Nuclei were stained with DAPI, while circ_2858 was probed in red. Scale bar, 5 μm.
circ_2858 Positively Regulates VEGFA Expression by Competitively Sponging miR-93-5p, thus Increasing BBB Permeability
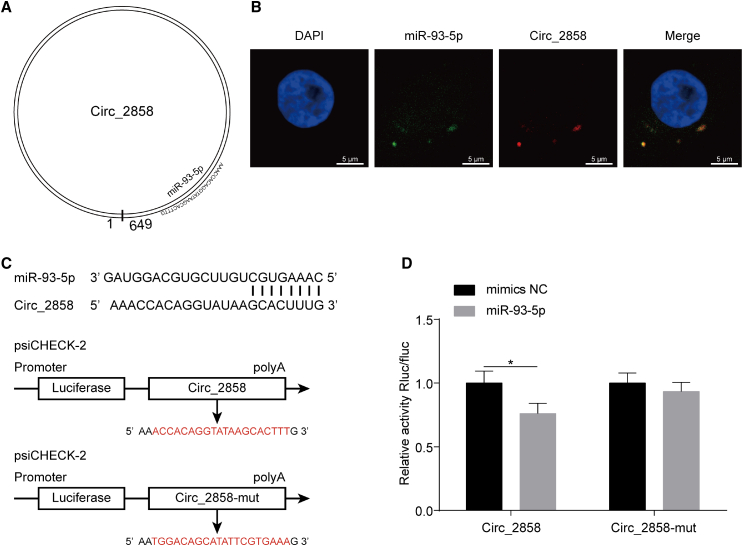
Since we demonstrated the potential binding of the miR-17 family by circ_2858 (Figure 5A, miR-93-5p as a representative member), we next investigated the possible intracellular interaction between these two molecules. By using RNA FISH assays, we observed the co-localization of miR-93-5p and circ_2858 in hBMECs (Figure 5B). Meanwhile, dual-luciferase reporter assays were performed to confirm whether there was a direct interaction between miR-93-5p and circ_2858. The potential miRNA binding sites on circ_2858 and its respective mutant sequences were cloned into psiCHECK-2 plasmid after bioinformatic analysis (Figure 5C). As shown in Figures 5D and S4, there was a significant and dose-dependent reduction in the relative luciferase activity after co-transfection of miR-93-5p and wild-type circ_2858, whereas no significant change in luciferase activity was observed when miR-93-5p was co-transfected with the mutant circ_2858 construct. These results indicate a direct physical interaction between miR-93-5p and circ_2858.
Figure 5.
circ_2858 Sponges with miR-93-5p
(A) Schematic drawing showing the putative binding sites of miR-93-5p on circ_2858. (B) Co-localization of miR-93-5p and circ_2858 in hBMECs by RNA FISH assay. Nuclei were stained with DAPI. miR-93-5p was probed with FAM (green), while circ_2858 was probed with Cy3 (red). Scale bars, 5 μm. (C) Putative miR-93-5p-binding site on circ_2858. These binding sequences and the respective mutant sequences (labeled in red) were cloned into the psiCHECK-2 vector. (D) Dual-luciferase reporter assays showing that miR-93-5p significantly reduced the luciferase activity of the wild-type circ_2858 construct but not of the mutant construct. The Renilla luciferase activity was normalized to the firefly luciferase activity, and data are presented as mean ± SD from three independent experiments.
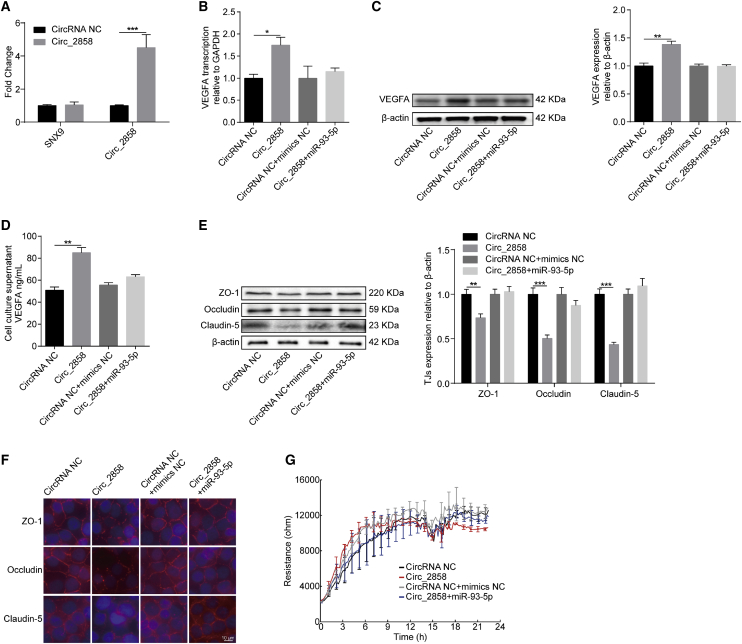
Subsequently, we aimed to examine the possible role of circ_2858 in BBB permeability. circ_2858 levels were significantly increased by transforming hBMECs with a circ_2858 overexpression construct, without affecting the transcription of its parental gene SNX9 (Figure 6A). We observed that circ_2858 overexpression significantly increased VEGFA expression at both mRNA and protein levels, whereas circ_2858-mediated upregulation of VEGFA was completely prevented by co-transfection with miR-93-5p mimic (Figures 6B and 6C). In addition, the amount of secretory VEGFA in the cell culture supernatant showed similar trends to those of VEGFA mRNA and protein levels (Figure 6D). As expected, the overexpression of circ_2858 resulted in a significant decrease in the level of TJ proteins, including ZO-1, Occludin, and Claudin-5, whereas such downregulation was prevented by co-transfection with the miR-93-5p mimic (Figure 6E). In addition, immunofluorescence and ECIS assays were performed to further examine the effect of circ_2858 on BBB integrity. As shown in Figure 6F, the levels of TJ proteins (ZO-1, Occludin, and Claudin-5) between adjacent endothelial cells were significantly decreased upon overexpression of circ_2858, whereas this damage effect was completely prevented by the miR-93-5p mimic. ECIS results also indicated that circ_2858 overexpression led to significantly lower TEER values of the cells when they reached confluence, whereas this reduced TEER could be well restored when the miR-93-5p mimic was co-transfected (Figure 6G). Therefore, these findings indicate that circ_2858 promotes the disruption of BBB integrity through the upregulation of VEGFA via competitive sponging of miR-93-5p.
Figure 6.
Involvement of the circ_2858/miR-93-5p Axis in VEGFA-Mediated BBB Damage
(A) qPCR analyzing the transcription of circ_2858, and its parental gene SNX9, in hBMECs with circ_2858 overexpression. Data are expressed as mean ± SD from three independent experiments. (B) The expression of VEGFA in circ_2858-overexpressing hBMECs co-transfected or not with miR-93-5p determined by qPCR analysis. (C) The expression of VEGFA in circ_2858-overexpressing hBMECs co-transfected or not with miR-93-5p, as determined by western blot analysis. β-actin was used as loading control. (D) ELISA determining the secreted VEGFA levels in circ_2858-overexpressing hBMECs co-transfected or not with miR-93-5p. Data were collected and calculated as mean ± SD from three independently replicated wells. (E) Expression of TJ proteins in circ_2858-overexpressing hBMECs co-transfected or not with miR-93-5p determined by western blot. β-actin was used as loading control. (F) Distribution of TJ proteins in circ_2858-overexpressing hBMECs co-transfected or not with miR-93-5p, as determined by immunofluorescence analysis. Scale bar, 10 μm. (G) ECIS system monitoring TEER changes of the circ_2858-overexpressing hBMECs co-transfected or not with miR-93-5p. Data were collected and are presented as mean ± SD from three independently replicated wells at each time point.
Discussion
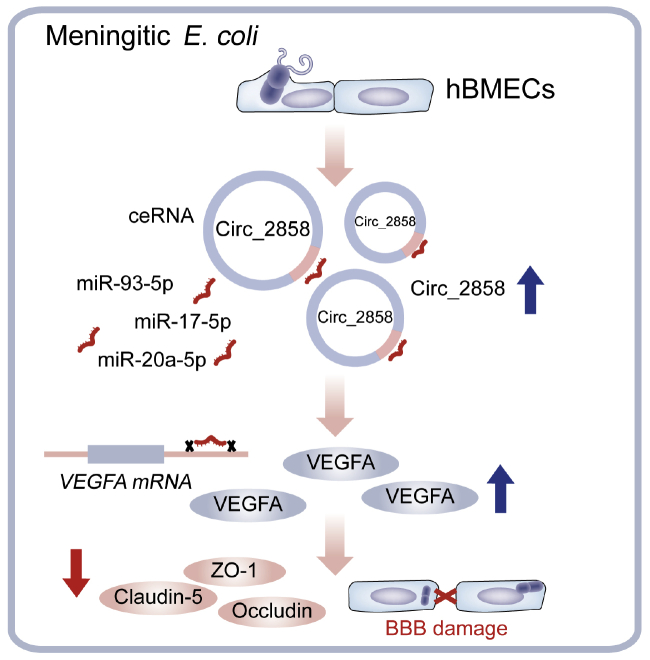
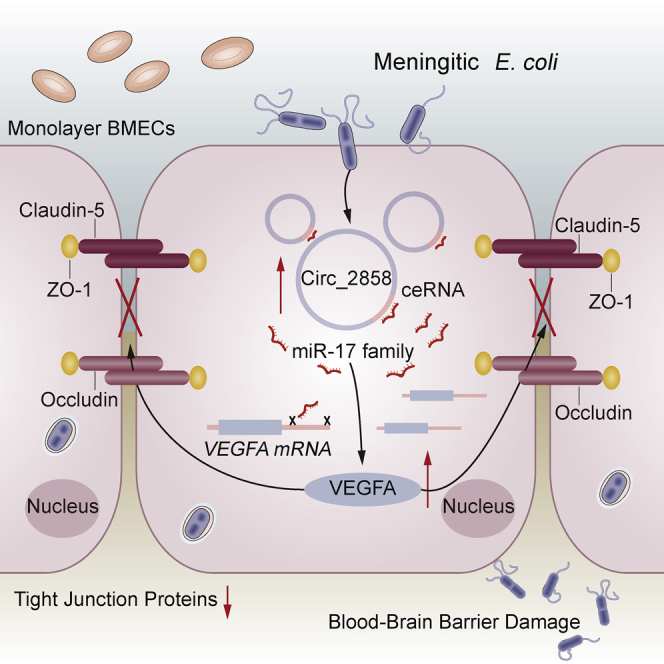
Due to their extensive involvement in different physiological and pathological processes, non-coding RNAs have attracted increasing attention in recent years. As an important type of non-coding RNAs, circRNAs are considered to have significant regulatory potential due to their high expression in brain tissue and have been experimentally shown to be involved in a variety of CNS-related diseases, such as multiple sclerosis, Alzheimer’s disease, stroke, and depression.27, 28, 29, 30 However, the roles of circRNAs in the process of host CNS invasion by pathogenic microbes have not been well documented. By combining transcriptomic and bioinformatic approaches, we have previously identified many differentially expressed circRNAs in hBMECs challenged with meningitic E. coli. Here, we provide novel insights into the function of circ_2858, showing that it is involved in meningitic E. coli-induced BBB dysfunction by sponging miR-93-5p to regulate VEGFA expression, thus leading to TJ protein breakdown (Figure 7).
Figure 7.
Schematic Presentation of the Working Mechanism of circ_2858 in Meningitic E. coli-Induced BBB Disruption
Meningitic E. coli infection of hBMECs increases the expression of a novel circRNA, circ_2858, which acts as an endogenous sponge sequestering miR-93-5p, thereby increasing VEGFA production and resulting in damage to BBB integrity.
Although the functions of circRNAs are largely unclear, accumulating evidence suggests that they are important endogenous regulators of diverse biological processes by acting as miRNA sponges.31,32 One well-known circRNA is ciRS-7, which harbors more than 70 conventional miR-7 binding sites.33 Previous studies have reported that miR-7-mediated suppression of the epidermal growth factor receptor (EGFR)-RAF1-mitogen-activated protein kinase (MAPK) pathway can be alleviated by ciRS-7 overexpression in colorectal cancer34 and that upregulation of ciRS-7 can promote hepatocellular carcinoma proliferation and invasion by sponging miR-7.35 In lung adenocarcinoma, both circRNF13 and circCRIM1 regulate invasion and metastasis by targeting miR-93-5p.36,37 Moreover, the circ-MTO1/miR-17/QKI-5 feedback loop was also reported to inhibit lung adenocarcinoma progression.38 In a recent study, circ-ITCH promoted prostate cancer growth, invasion, and metastasis by regulating the miR-17-5p/HOXB13 axis.39 Moreover, circRNAs are also involved in many CNS-associated diseases. In the mouse stroke model obtained by transient middle cerebral artery occlusion, circDLGAP4 functioned as an endogenous miR-143 sponge to increase the expression of the miR-143 target HECTD1, thus decreasing infarct area and BBB damage.40 Similarly, circHECW2 upregulates ATG5 expression by competitively sponging miR-30d, thereby promoting the transformation of BMECs into mesenchymal cells and leading to BBB dysfunction.41 In our study, 4,859 circRNAs were predicted from our previous transcriptomic data by CIRI and find_circ, and bioinformatic analysis revealed that circ_2858 had potential for miR-93-5p binding. The interaction between circ_2858 and miR-93-5p was further confirmed by FISH and dual-luciferase reporter assays. Meanwhile, we demonstrated that circ_2858, acting as a ceRNA to sponge miR-93-5p, promoted BBB permeability by inducing downregulation and redistribution of TJ proteins in hBMECs.
Since increasing BBB permeability has been commonly recognized as a critical step in the development of bacterial meningitis, it is worth wondering what the connection between miR-93-5p and BBB permeability (or TJ proteins) is upon challenge by meningitic E. coli. Previously, multiple miRNAs have been reported to regulate BBB integrity, and these miRNAs may be potential therapeutic targets for certain CNS diseases. For example, miR-34a was reported to regulate BBB permeability and mitochondrial function by targeting cytochrome c.42 Similarly, miR-29b mediates BBB dysfunction by targeting DNMT3B and MMP9 during hyperhomocysteinemia.43 Furthermore, in traumatic brain injury, miR-21 exerts a protective effect on the BBB by activating the Ang-1/Tie-2 axis in BMECs.44 In addition, HIV-1 Tat C protein modulates the expression of miR-101 to suppress vascular endothelial cadherin (VE-cadherin) in hBMECs.45 In the present study, we observed that miR-93-5p expression was significantly decreased in hBMECs in response to meningitic E. coli infection, and miR-93-5p was subsequently confirmed to be an important regulator of VEGFA expression. VEGFA can promote angiogenesis, as it regulates the stability of the vascular tone and acts as a trophic factor for vascular endothelial cells.46 VEGFA not only regulates normal growth and development of vessels but also plays important biological roles in many disease models.47 Indeed, several studies have found that VEGFA can activate many regulatory factors in endothelial cells, such as MAPK (extracellular signal-regulated kinase [ERK], p38), phospholipase, protein kinase C, phosphatidylinositol 3-kinase (PI3K)-AKT, and endothelial nitric oxide synthase (eNOS), to enhance the permeability of endothelial cells, leading to the accumulation of tissue fluid in tumors, psoriatic lesions, tissue edema, neovascular eye disease with vision loss, etc.48 We have previously shown in vivo that meningitic E. coli infection can induce VEGFA to enhance BBB permeability.9 In the present study, we confirmed this effect in vitro by stimulating hBMECs with recombinant VEGFA, further consolidating the essential role of VEGFA in regulating BBB permeability in response to meningitic E. coli infection. To our knowledge, this is the first demonstration that miR-93-5p, by targeting the expression of VEGFA, participates in meningitic E. coli-induced BBB dysfunction.
In summary, we reported meningitic E. coli-induced upregulation of circ_2858 in hBMECs, which facilitated VEGFA expression by competitively sponging miR-93-5p, thus leading to TJ protein disruption and BBB dysfunction. Our study provides new insights into the pathogenic process of meningitic E. coli infection. Further exploration of the regulatory roles of different circRNAs during meningitic E. coli infection will help deepen our understanding of CNS infectious diseases. More importantly, specific blocking of these circRNAs, such as circ_2858, might be envisioned as a potential prevention and therapeutic strategy for future treatment of CNS dysfunction.
Materials and Methods
Cell Culture and Bacterial Strain
The hBMECs used in this study were kindly provided by Prof. Kwang Sik Kim (Johns Hopkins University School of Medicine, USA). Cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 1 mM sodium pyruvate, essential amino acids, nonessential amino acids, vitamins, and penicillin and streptomycin (100 U/mL)49 and incubated in a 37°C incubator under 5% CO2 until monolayer confluence. Confluent cells were washed three times with Hank’s balanced salt solution and starved in serum-free medium (1:1 mixture of Ham’s F-12 and M-199) for 16–18 h before further treatment. The 293T cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum in a 37°C incubator under 5% CO2 until 70% confluence prior to the transfection.
Meningitic E. coli strain PCN033 used herein was originally isolated from pig cerebrospinal fluid from a diseased farm in China, 2006.50 The strain was grown aerobically at 37°C in Luria-Bertani medium overnight.
Meningitic E. coli Infection of hBMECs
Meningitic E. coli infection of hBMECs was performed following previously described methods.51 Briefly, overnight cultures of E. coli were resuspended and diluted in serum-free medium, then added to the confluent hBMEC monolayer grown in 100-mm dishes at a multiplicity of infection of 10 (approximately 108 colony-forming units [CFUs] per dish) to allow bacterial invasion at 37°C. Cells were washed three times with pre-chilled PBS and subjected to RNA extraction using TRIzol reagent (Aidlab Biotech, Beijing, China).
miRNA Library Preparation and Sequencing
Two libraries (Infection versus Control) were constructed and sequenced by Novogene (Beijing, China). RNA sample preparation was based on 3 μg RNA for each sample. First, sequencing libraries were generated by using NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs, Beverly, MA, USA) according to manufacturer’s instructions, and index codes were added to indicate sequences to each sample. Next, first strand cDNA was synthesized by using M-MuLV Reverse Transcriptase (RNase H−). PCR amplification was performed using LongAmp Taq 2 × Master Mix, SR Primer for Illumina and index (X) primer. PCR products were purified on 8% polyacrylamide gel (100 V, 80 min). DNA fragments corresponding to 140–∼160 bp (the length of small noncoding RNA plus the 3′ and 5′ adaptors) were recovered and dissolved in 8 μL elution buffer. Last, library quality was evaluated by Agilent Bioanalyzer 2100 system using DNA High Sensitivity Chips.
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq SR Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s guidelines. After cluster generation, the library preparations were sequenced on an Illumina HiSeq 2500/2000 platform and 50 bp single-end reads were generated. Differential expression analysis for the control and infection groups was performed using DEGseq (2010) R package. miRNAs with an adjusted p value of < 0.05 and an increase in their expression of ≥2-fold or a decrease to ≤0.5-fold were considered differentially expressed. The transcriptomic data have been deposited at the BioProject database of National Center for Biotechnology Information (accession number: NCBI: SRR5868085).
Reverse Transcription and Real-Time PCR
For miRNAs, cDNA synthesis was carried out using miRcute plus miRNA First-Strand cDNA kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The qPCR was performed using the miRcute plus miRNA qPCR detection kit (Tiangen, Beijing, China). Primers for the miRNA qPCR are listed in Table S6. For mRNAs, aliquots (500 ng) of the total RNAs in each sample were subjected to cDNA synthesis using the HiScript II Q RT SuperMix for qPCR gDNA wiper (Vazyme, Nanjing, China). For circRNAs, the linear-stranded RNAs were removed by RNase R treatment (Epicenter, Madison, WI, USA), and RNAs (500 ng) were reverse-transcribed into cDNAs with HiScript II Q RT SuperMix for qPCR (Vazyme, Nanjing, China). Primers for circRNAs and mRNAs qPCR are listed in Tables S4 and S7.
The qPCR amplification was performed with a qPCR thermal cycler qTOWER3 (Analytik Jena, Germany) by using MonAmp SYBR Green qPCR Mix (Monad Biotech, Wuhan, China) according to manufacturer’s recommendations. Expression levels of the mRNAs and circRNAs were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and the miRNA expression levels were normalized to U6. Each assay was performed three times independently.
Transfection
The miR-93-5p mimics and inhibitor were obtained from GenePharma (Shanghai, China). The circ_2858 overexpression plasmid was generated by cloning the whole circ_2858 sequence into pcircRNA-2.2-hsa purchased from BersinBio (Guangzhou, China). Transfection was performed using the jetPRIME reagent (Polyplus Transfection, Illkirch, France) following the manufacturer’s protocol. The medium was replaced into the complete medium after 5 h of transfection, and the cells were collected after another 48 h of incubation.
Western Blot
Cells were collected and lysed in radioimmunoprecipitation assay (RIPA) buffer (Epizyme, Shanghai, China) with protease inhibitor cocktail (MedChemexpress, Monmouth, NJ, USA) and centrifuged at 12,000 rpm for 10 min at 4°C to remove insoluble cell debris. The soluble protein concentration in the supernatants was measured using the bicinchoninic acid (BCA) protein assay kit (NCM Biotech, China) and applied to western blot analyses. Aliquots from each sample were separated by 12% SDS-PAGE and then transferred to polyvinylidene difluoride membranes. The blots were blocked with 5% bovine serum albumin in Tris-buffered saline with Tween-20 at room temperature for 1 h and then incubated overnight at 4°C with primary antibodies against ZO-1, Claudin-5, Occludin (Abcam, Cambridge, MA, USA), VEGFA, and β-actin (Proteintech, Chicago, IL, USA). The blots were subsequently washed and incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (Biodragon, Beijing, China) at 37°C for 1 h and visualized with electrochemiluminescence kit (US Everbright, Suzhou, China). Densitometry analysis was analyzed in three independent blots by using Image Lab software version 5.2.1 (Bio-Rad, Hercules, CA, USA).
Dual-Luciferase Assay
The sequences of circ_2858, circ_2858-mut, VEGFA 3′ UTR, and VEGFA 3′ UTR-mut were synthesized by Genscript (Nanjing, China) and cloned into psiCHECK-2 plasmid for dual-luciferase activity assay. Briefly, 293T cells were seeded in 24-well plates and cultured until 70% confluence before transfection. For each experimental group, 200 ng plasmids and 50 nM of miRNA mimics or negative control were used for the transfection. After 24 h of incubation, cells were collected, and both firefly luciferase activity and Renilla luciferase activity were detected by using Dual-Luciferase kit (Promega, Madison, WI, USA) following the manufacturer’s instructions. Results were calculated as the ratio of Renilla luciferase activity and the firefly luciferase activity and were recorded as mean ± standard deviation (mean ± SD) from three replicated wells.
Secretory VEGFA Detection by Enzyme-Linked Immunosorbent Assay (ELISA)
The mimics or inhibitor of miR-93-5p and the circ_2858 overexpression plasmid were transfected to hBMECs following the above procedure, and cell culture supernatant was collected after 48 h of transfection. The secretory VEGFA from supernatant was quantified using the VEGFA ELISA Kit purchased from 4A Biotech (Beijing, China), following the manufacturer’s procedures.
FISH
The commercial FISH Kit purchased from Bersinbio (Guangzhou, China) was used for the FISH assays following the manufacturer’s procedures. Briefly, hBMECs were fixed in 4% paraformaldehyde for 30 min at room temperature and washed with PBS twice. Cells were incubated with Proteinase K solution for 15 min at 37°C, washed twice with PBS for 5 min, and then mixed with 1% fixation solution for 10 min at room temperature. Fixed samples were incubated with 70% ethanol for 5 min, 85% ethanol for 5 min, and 100% ethanol for 5 min at −20°C before drying. Samples were next incubated with pre-hybridization solution for 30 min at 37°C, followed by incubating in the hybridization buffer with specific probes for circ_2858 (5′-Cy3-TGCCACTGCCAACCTGTGACTTGTTT-3′) as well as miR-93-5p (5′-FAM-CTACCTGCACGAACAGCACTTTG-3′) for 18 h at 37°C. After strict wash in SSC buffer, 1 μg/mL of 4′,6-diamidino-2-phenylindole (DAPI, Beyotime Biotechnology, China) was used for nucleus staining. Fluorescent images were acquired with an Olympus FV10 laser scanning confocal microscope (Olympus, Tokyo, Japan).
Immunofluorescence Assay
The hBMECs grown in 6-well dishes were fixed with 4% paraformaldehyde for 30 min followed by three washes in PBS. Cells were incubated with the primary rabbit ZO-1, Occludin, or Claudin-5 antibody (Abcam, Cambridge, MA, USA) overnight at 4°C and then incubated with Alexa Fluor 594 goat anti-rabbit antibody (Bioss, Woburn, MA, USA) for another 1 h. Cells were counterstained with DAPI to visualize the nucleus morphology and mounted and photographed by using BX41 fluorescence microscopy (Olympus, Tokyo, Japan).
ECIS
The miR-93-5p mimics or inhibitor and the circ_2858 overexpression plasmid were transfected into hBMECs following the above procedures. The cells were seeded on collagen-coated, gold-plated electrodes in 8-well chamber slides (8W1E+) linked to ECIS Zθ equipment (Applied BioPhysics, Troy, NY, USA), which monitored continuously to reflect any alterations of the monolayer barrier. The TEER changes of the monolayer cells were automatically recorded within ECIS system.
Statistical Analysis
Data were expressed as mean ± SD from three different independent experiments, and the significance of differences between groups was evaluated by one-way analysis of variance. ∗p <0.05 was considered significant, and ∗∗p <0.01 as well as ∗∗∗p <0.001 were all considered extremely significant. Graphs were plotted and analyzed by using GraphPad Prism version 6.0 (GraphPad Software, La Jolla, CA, USA).
Author Contributions
R.Y. performed the experiments, analyzed the data, and drafted the manuscript. C.T., H.C., and B.X. provided the technical support and helped with the statistical analysis. S.X., J.F., J.C., and B.Y. participated in the experiments. X.W. conceived and designed the experiments and revised the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was funded by grants from the National Natural Science Foundation of China, China (31772736); the National Key R&D Program of China, China (2016YFD0500406); the outstanding youth project of Natural Science Foundation of Hubei Province, China (2018CFA070); the Hubei Postdoctoral Preferential Foundation, China (590320102); and the China Postdoctoral Science Foundation, China (2020M672379).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.09.034.
Supplemental Information
References
- 1.Kim K.S. Mechanisms of microbial traversal of the blood-brain barrier. Nat. Rev. Microbiol. 2008;6:625–634. doi: 10.1038/nrmicro1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sukumaran S.K., Prasadarao N.V. Escherichia coli K1 invasion increases human brain microvascular endothelial cell monolayer permeability by disassembling vascular-endothelial cadherins at tight junctions. J. Infect. Dis. 2003;188:1295–1309. doi: 10.1086/379042. [DOI] [PubMed] [Google Scholar]
- 3.Edwards V.L., Wang L.C., Dawson V., Stein D.C., Song W. Neisseria gonorrhoeae breaches the apical junction of polarized epithelial cells for transmigration by activating EGFR. Cell. Microbiol. 2013;15:1042–1057. doi: 10.1111/cmi.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballabh P., Braun A., Nedergaard M. The blood-brain barrier: an overview: structure, regulation, and clinical implications. Neurobiol. Dis. 2004;16:1–13. doi: 10.1016/j.nbd.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 5.Candelario-Jalil E., Yang Y., Rosenberg G.A. Diverse roles of matrix metalloproteinases and tissue inhibitors of metalloproteinases in neuroinflammation and cerebral ischemia. Neuroscience. 2009;158:983–994. doi: 10.1016/j.neuroscience.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim B.J., Hancock B.M., Bermudez A., Del Cid N., Reyes E., van Sorge N.M., Lauth X., Smurthwaite C.A., Hilton B.J., Stotland A. Bacterial induction of Snail1 contributes to blood-brain barrier disruption. J. Clin. Invest. 2015;125:2473–2483. doi: 10.1172/JCI74159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Obaidi M.M.J., Desa M.N.M. Mechanisms of blood brain barrier disruption by different types of bacteria, and bacterial-host interactions facilitate the bacterial pathogen invading the brain. Cell. Mol. Neurobiol. 2018;38:1349–1368. doi: 10.1007/s10571-018-0609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMillin M.A., Frampton G.A., Seiwell A.P., Patel N.S., Jacobs A.N., DeMorrow S. TGFβ1 exacerbates blood-brain barrier permeability in a mouse model of hepatic encephalopathy via upregulation of MMP9 and downregulation of claudin-5. Lab. Invest. 2015;95:903–913. doi: 10.1038/labinvest.2015.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang R., Liu W., Miao L., Yang X., Fu J., Dou B., Cai A., Zong X., Tan C., Chen H., Wang X. Induction of VEGFA and Snail-1 by meningitic Escherichia coli mediates disruption of the blood-brain barrier. Oncotarget. 2016;7:63839–63855. doi: 10.18632/oncotarget.11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang R.C., Qu X.Y., Xiao S.Y., Li L., Xu B.J., Fu J.Y., Lv Y.J., Amjad N., Tan C., Kim K.S. Meningitic Escherichia coli-induced upregulation of PDGF-B and ICAM-1 aggravates blood-brain barrier disruption and neuroinflammatory response. J. Neuroinflammation. 2019;16:101. doi: 10.1186/s12974-019-1497-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santer L., Bär C., Thum T. Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol. Ther. 2019;27:1350–1363. doi: 10.1016/j.ymthe.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett S.P., Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsiao K.Y., Sun H.S., Tsai S.J. Circular RNA - New member of noncoding RNA with novel functions. Exp. Biol. Med. (Maywood) 2017;242:1136–1141. doi: 10.1177/1535370217708978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bei Y., Yang T., Wang L., Holvoet P., Das S., Sluijter J.P.G., Monteiro M.C., Liu Y., Zhou Q., Xiao J. Circular RNAs as potential theranostics in the cardiovascular system. Mol. Ther. Nucleic Acids. 2018;13:407–418. doi: 10.1016/j.omtn.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J., Qi X., Liu L., Hu X., Liu J., Yang J., Yang J., Lu L., Zhang Z., Ma S. Emerging epigenetic regulation of circular RNAs in human cancer. Mol. Ther. Nucleic Acids. 2019;16:589–596. doi: 10.1016/j.omtn.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang F., Li A., Qin Y., Che H., Wang Y., Lv J., Li Y., Li H., Yue E., Ding X. A novel circular RNA mediates pyroptosis of diabetic cardiomyopathy by functioning as a competing endogenous RNA. Mol. Ther. Nucleic Acids. 2019;17:636–643. doi: 10.1016/j.omtn.2019.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X., Yan Y., Lin W., Li A., Zhang H., Lei X., Dai Z., Li X., Li H., Chen W. Circular RNA Vav3 sponges gga-miR-375 to promote epithelial-mesenchymal transition. RNA Biol. 2019;16:118–132. doi: 10.1080/15476286.2018.1564462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng X., Li X., Zhang P., Wang J., Zhou Y., Chen M. Circular RNA: an emerging key player in RNA world. Brief. Bioinform. 2017;18:547–557. doi: 10.1093/bib/bbw045. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S., Zhu D., Li H., Li H., Feng C., Zhang W. Characterization of circRNA-Associated-ceRNA networks in a senescence-accelerated mouse prone 8 brain. Mol. Ther. 2017;25:2053–2061. doi: 10.1016/j.ymthe.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meganck R.M., Borchardt E.K., Castellanos Rivera R.M., Scalabrino M.L., Wilusz J.E., Marzluff W.F., Asokan A. Tissue-dependent expression and translation of circular RNAs with recombinant AAV vectors in vivo. Mol. Ther. Nucleic Acids. 2018;13:89–98. doi: 10.1016/j.omtn.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X., Abraham J.M., Cheng Y., Wang Z., Wang Z., Zhang G., Ashktorab H., Smoot D.T., Cole R.N., Boronina T.N. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol. Ther. Nucleic Acids. 2018;13:312–321. doi: 10.1016/j.omtn.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar R., Sahu S.K., Kumar M., Jana K., Gupta P., Gupta U.D., Kundu M., Basu J. MicroRNA 17-5p regulates autophagy in Mycobacterium tuberculosis-infected macrophages by targeting Mcl-1 and STAT3. Cell. Microbiol. 2016;18:679–691. doi: 10.1111/cmi.12540. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z., Yu H., Guo Q. MicroRNA-20a promotes inflammation via the nuclear factor-κB signaling pathway in pediatric pneumonia. Mol. Med. Rep. 2018;17:612–617. doi: 10.3892/mmr.2017.7899. [DOI] [PubMed] [Google Scholar]
- 26.Yang R., Huang F., Fu J., Dou B., Xu B., Miao L., Liu W., Yang X., Tan C., Chen H., Wang X. Differential transcription profiles of long non-coding RNAs in primary human brain microvascular endothelial cells in response to meningitic Escherichia coli. Sci. Rep. 2016;6:38903. doi: 10.1038/srep38903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iparraguirre L., Muñoz-Culla M., Prada-Luengo I., Castillo-Triviño T., Olascoaga J., Otaegui D. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis. Hum. Mol. Genet. 2017;26:3564–3572. doi: 10.1093/hmg/ddx243. [DOI] [PubMed] [Google Scholar]
- 28.Lukiw W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD) Front. Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta S.L., Pandi G., Vemuganti R. Circular RNA expression profiles alter significantly in mouse brain after transient focal ischemia. Stroke. 2017;48:2541–2548. doi: 10.1161/STROKEAHA.117.017469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang G., Ma Y., An T., Pan Y., Mo F., Zhao D., Liu Y., Miao J.N., Gu Y.J., Wang Y., Gao S.H. Relationships of circular RNA with diabetes and depression. Sci. Rep. 2017;7:7285. doi: 10.1038/s41598-017-07931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Z.B., Huang G.X., Fu Q., Han B., Lu J.J., Chen A.M., Zhu L. circRNA.33186 Contributes to the Pathogenesis of Osteoarthritis by Sponging miR-127-5p. Mol. Ther. 2019;27:531–541. doi: 10.1016/j.ymthe.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sang Y., Chen B., Song X., Li Y., Liang Y., Han D., Zhang N., Zhang H., Liu Y., Chen T. CircRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol. Ther. 2019;27:1638–1652. doi: 10.1016/j.ymthe.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 34.Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., Cai S., Qin H., Ma Y., Goel A. Circular RNA ciRS-7-a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu L., Gong X., Sun L., Zhou Q., Lu B., Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS ONE. 2016;11:e0158347. doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Wang L., Liu S., Mao Y., Xu J., Yang S., Shen H., Xu W., Fan W., Wang J. CircRNF13 regulates the invasion and metastasis in lung adenocarcinoma by targeting miR-93-5p. Gene. 2018;671:170–177. doi: 10.1016/j.gene.2018.04.069. [DOI] [PubMed] [Google Scholar]
- 37.Wang L., Liang Y., Mao Q., Xia W., Chen B., Shen H., Xu L., Jiang F., Dong G. Circular RNA circCRIM1 inhibits invasion and metastasis in lung adenocarcinoma through the microRNA (miR)-182/miR-93-leukemia inhibitory factor receptor pathway. Cancer Sci. 2019;110:2960–2972. doi: 10.1111/cas.14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B., Chen M., Jiang N., Shi K., Qian R. A regulatory circuit of circ-MTO1/miR-17/QKI-5 inhibits the proliferation of lung adenocarcinoma. Cancer Biol. Ther. 2019;20:1127–1135. doi: 10.1080/15384047.2019.1598762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Wang X., Wang R., Wu Z., Bai P. Circular RNA ITCH suppressed prostate cancer progression by increasing HOXB13 expression via spongy miR-17-5p. Cancer Cell Int. 2019;19:328. doi: 10.1186/s12935-019-0994-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Bai Y., Zhang Y., Han B., Yang L., Chen X., Huang R., Wu F., Chao J., Liu P., Hu G. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J. Neurosci. 2018;38:32–50. doi: 10.1523/JNEUROSCI.1348-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang L., Han B., Zhang Y., Bai Y., Chao J., Hu G., Yao H. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy. 2018;14:404–418. doi: 10.1080/15548627.2017.1414755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bukeirat M., Sarkar S.N., Hu H., Quintana D.D., Simpkins J.W., Ren X. MiR-34a regulates blood-brain barrier permeability and mitochondrial function by targeting cytochrome c. J. Cereb. Blood Flow Metab. 2016;36:387–392. doi: 10.1177/0271678X15606147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalani A., Kamat P.K., Familtseva A., Chaturvedi P., Muradashvili N., Narayanan N., Tyagi S.C., Tyagi N. Role of microRNA29b in blood-brain barrier dysfunction during hyperhomocysteinemia: an epigenetic mechanism. J. Cereb. Blood Flow Metab. 2014;34:1212–1222. doi: 10.1038/jcbfm.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge X., Han Z., Chen F., Wang H., Zhang B., Jiang R., Lei P., Zhang J. MiR-21 alleviates secondary blood-brain barrier damage after traumatic brain injury in rats. Brain Res. 2015;1603:150–157. doi: 10.1016/j.brainres.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 45.Mishra R., Singh S.K. HIV-1 Tat C modulates expression of miRNA-101 to suppress VE-cadherin in human brain microvascular endothelial cells. J. Neurosci. 2013;33:5992–6000. doi: 10.1523/JNEUROSCI.4796-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maharaj A.S., D’Amore P.A. Roles for VEGF in the adult. Microvasc. Res. 2007;74:100–113. doi: 10.1016/j.mvr.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koch S., Tugues S., Li X., Gualandi L., Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Biochem. J. 2011;437:169–183. doi: 10.1042/BJ20110301. [DOI] [PubMed] [Google Scholar]
- 48.Ferrara N., Mass R.D., Campa C., Kim R. Targeting VEGF-A to treat cancer and age-related macular degeneration. Annu. Rev. Med. 2007;58:491–504. doi: 10.1146/annurev.med.58.061705.145635. [DOI] [PubMed] [Google Scholar]
- 49.Zhu L., Pearce D., Kim K.S. Prevention of Escherichia coli K1 penetration of the blood-brain barrier by counteracting the host cell receptor and signaling molecule involved in E. coli invasion of human brain microvascular endothelial cells. Infect. Immun. 2010;78:3554–3559. doi: 10.1128/IAI.00336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu C., Zheng H., Yang M., Xu Z., Wang X., Wei L., Tang B., Liu F., Zhang Y., Ding Y. Genome analysis and in vivo virulence of porcine extraintestinal pathogenic Escherichia coli strain PCN033. BMC Genomics. 2015;16:717. doi: 10.1186/s12864-015-1890-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang R., Xu B., Yang B., Fu J., Liu L., Amjad N., Cai A., Tan C., Chen H., Wang X. Circular RNA transcriptomic analysis of primary human brain microvascular endothelial cells infected with meningitic Escherichia coli. Mol. Ther. Nucleic Acids. 2018;13:651–664. doi: 10.1016/j.omtn.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.