Abstract
The present study investigated the genomic constitution and antimicrobial resistance (AMR) of 238 Campylobacter from pigs and wild boars in Italy between 2012 and 2019. Campylobacter strains were genotyped using multilocus sequence typing (MLST) and whole genome MLST (wgMLST), screened for antimicrobial resistance genes, and tested for phenotypic susceptibility to six different antibiotics. C. coli was detected in 98.31% and 91.66% of pigs and wild boars, while C. jejuni was isolated in the remaining cases. MLST assigned 73 STs and 13 STs in pigs and wild boars, respectively, including 44 novel STs. The predominant ST in pigs was ST-854 (12.36%), followed by ST-9264 (6.18%). ST-1055 and ST-1417 were predominant in wild boars (30% and 13.33%, respectively). The minimum spanning tree using 1,121 global MLST profiles showed specific Italian clusters and a clear separation between pig and wild boar profiles. The wgMLST confirmed the MLST clustering and revealed a high genetic diversity within C. coli population in Italy. Minimum inhibitory concentrations (MIC) of six antibiotics revealed higher resistance in pigs to ciprofloxacin, nalidixic acid, streptomycin and tetracycline, compared to wild boar. In contrast, most strains were susceptible to gentamicin. Worrying levels of multidrug resistance (MDR) were observed mostly in pig isolates. Molecular screening of AMR mechanisms revealed the predominance of gyrA T86I substitution among fluoroquinolone- and quinolone-resistant isolates, and the 23S rRNA A2075G mutation among macrolide-resistant isolates. Other resistance determinants were observed: (i) tet(O) gene was present among tetracycline-resistant isolates; (ii) rpsL and aph(3’)-III genes conferring resistance to aminoglycosides, were identified only in streptomycin or gentamicin-resistant pig isolates; (iii) cmeA, cmeB, cmeC, cmeR genes responsible of pump efflux mechanisms, were observed in almost all the strains; (iv) OXA-61, encoding β-lactamase, was found in the half of the strains. Genotypic and phenotypic AMR profiling was fairly correlated for quinolones/fluoroquinolones. Campylobacter infection is common also in wild boar populations in Italy, suggesting that wild boars could be a reservoir of resistant and multi-resistant Campylobacter species, which may be of public health concern. The present study adds to our knowledge on the epidemiological and ecological traits of this pathogen in domesticated and wild swine.
Keywords: Campylobacter, antimicrobial resistance (AMR), multidrug resistance (MDR), multilocus sequence typing, resistance genes, wgMLST
Introduction
Campylobacter is known as the most common cause of bacterial gastrointestinal infection in Europe, with the annual number of cases exceeding those of salmonellosis and shigellosis (EFSA & ECDC, 2019). Campylobacter jejuni and Campylobacter coli are the main causative agents of campylobacteriosis, posing a threat to public health worldwide (EFSA & ECDC, 2019). Fever, bloody diarrhea, headache and abdominal pain, nausea and vomiting are the main symptoms of campylobacteriosis in humans. Generally, the infection is self-limiting after 3-5 days, but in immunocompromised individuals it can spread into the bloodstream and become potentially lethal (Whitehouse et al., 2018). In severe cases, the antibiotic treatment is required, with macrolides and fluoroquinolones being the drugs of first choice (Mourkas et al., 2019). Campylobacteriosis is a mainly food-borne disease in which foods of animal origin, such as poultry meat, beef and pork, play a primary role (Sheppard et al., 2011).
Several studies showed the possibility of wildlife or environmental sources to act as reservoirs of Campylobacter infection (Sheppard et al., 2009a; Griekspoor et al., 2013; Cody et al., 2015; Atterby et al., 2018; Marotta et al., 2019; Marotta et al., 2020). In particular, these researchers focused on agricultural settings, especially on wild birds (Sheppard et al., 2009a; Griekspoor et al., 2013; Cody et al., 2015; Atterby et al., 2018; Marotta et al., 2019) small mammals (Sippy et al., 2012) and insects (Hald et al., 2004). However, there are little data on potential spill-over between livestock and wild ungulates (Navarro-Gonzalez et al., 2014). In pig farms, campylobacteriosis often leads to a significant decrease in animal productivity and consequent economic losses (Hansson et al., 2018).
Domestic pigs and wild boars belong to the same species (Sus scrofa) making them susceptible to the same pathogens (Ruiz-Fons et al., 2006; Ruiz-Fons et al., 2008). As a result, wild boar populations infected with Campylobacter could pose a threat to the pig industry. The Eurasian wild boar is widely distributed throughout most of Europe and in the past 50 years their numbers have increased to an estimatedpopulation of over 2.2 million wild boars (Massei et al., 2015; Meier and Ryser-Degiorgis, 2018). In Italy, it is the most widespread wild ungulate with a consistent presence along the country, due to its high prolificacy, favorable climatic conditions, and to the depopulation of Apennine and Alpine areas (Apollonio et al., 2010; Stella et al., 2018). Wild boars may contract Campylobacter from avian species, due to constant contact with soil contaminated with bird droppings (Waldenström et al., 2002; Humphrey et al., 2007; Epps et al., 2013). The increasing communities of wild boars in the anthropized areas as possible reservoirs of different Campylobacter species represent a growing challenge for public and veterinary health systems (Jones et al., 2013; Miller and Sweeney, 2013). Numerous studies showed that AMR is still very common in Campylobacter strains isolated from farmed animals in many European countries (EFSA & ECDC, 2019). In particular, high level of antibiotic resistance was shown to ciprofloxacin, nalidixic acid and tetracycline (EFSA & ECDC, 2019) followed, especially in C. coli, by resistance to macrolides and aminoglycoside antibiotic classes. Moreover, an alarming trend towards multidrug resistance (MDR), particularly among C. coli, was also detected (Luangtongkum et al., 2009; Pascoe et al., 2017; Mourkas et al., 2019). In this study, we aimed to evaluate the genotypic diversity of Campylobacter in wild boar and domesticated pig populations circulating in Italy and identify AMR genes in the two species investigated in order to understand the extent to which Campylobacter species are common, indicating a potential inter-species transmission.
Material and Methods
Bacterial Strains and Species Identification
A total of 238 Campylobacter strains isolated using the bacteriological ISO method 10272-1:2017 and stored at the microbial strain collection of the National Reference Laboratory for Campylobacter (NRL, http://www.izs.it/IZS/Eccellenza/Centri_nazionali/LNR_-_Campylobacter) were included in the study. The collection comprised 178 Campylobacter pig strains isolated from carcasses and from fecal content and 60 Campylobacter wild boar strains isolated from liver, muscle and faeces, in Italy between 2012 and 2019. The strains were cultured on Columbia blood agar plates in microaerobic atmosphere at 42°C for 48 h and DNA was extracted using Maxwell instrument (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions and quantified using a Nanodrop Spectrophotometer (Nanodrop Technologies, Celbio Srl., Milan, Italy). After an initial phenotypic characterization, suspected colonies were confirmed as thermotolerant Campylobacter and identified to species level using a multiplex and a simplex PCR, as described previously (Wang et al., 2002; Marotta et al., 2019). Strains used as positive PCR controls were C. coli NCTC 11353, C. fetus ATCC 19438, C. jejuni ATCC 33291, C. upsaliensis NCTC 11541 and C. lari NCTC 11552.
Sequence Analysis and Identification of Antibiotic Resistance Genes
Total genomic DNA was used to prepare sequencing libraries using Nextera XT Library Preparation Kit (Illumina, Inc., San Diego, CA, USA). The libraries were then sequenced using Illumina NextSeq 500 sequencer. Sequence reads (150-bp, pair-end) were demultiplexed and the adapters were removed. Subsequently the reads were trimmed with Trimmomatic tool (version 0.36) and de novo assembled using SPAdes version 3.11.1 with the “careful” option selected (Bankevich et al., 2012). The sequence reads generated in this study were deposited in NCBI Sequence Read Archive (SRA) in Bioprojects PRJNA638082 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA638082) and PRJNA638084 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA638084).
C. jejuni genome assemblies, were genotyped by MLST. The assemblies were also investigated for the genomic AMR traits.
The MLST profiles were assigned using a C.jejuni/coli task template MLST 7 loci, schema available at https://pubmlst.org/Campylobacter/accessible through in Ridom SeqSphere+ v. 6.0.2. Software (RidomGmbH, Münster, Germany). Italian MLST profiles were combined with MLST data of 1,121 pig isolates from Europe, downloaded from PubMLST (http://pubmlst.org/campylobacter/) and analyzed at the time of this analysis. MLST profiles were analyzed using the goeBURST algorithm implemented in PHYLOViZ, version 2.0 (Nascimento et al., 2017). Minimum spanning trees (MST) were created using default software settings.
The wgMLST analysis was performed in Ridom SeqSphere+ v. 6.0.2. The scaffolds were analyzed using two task templates: C. jejuni/C. coli cgMLST composed of 637 gene core gene targets and C. jejuni/C. coli accessory MLST composed of 958 accessory gene targets. Scaffolds that contained less than 90% good genome targets were excluded from the analysis. UPGMA tree was constructed by pairwise analysis of identified alleles, with missing targets ignored using default settings. The tree and associated metadata were visualized using iTol v5 (Letunic and Bork, 2006).
AMR genes were identified in silico using PointFinder v. 3.1.0 and ABRicate v. 0.8 (https://github.com/tseemann/abricate/) by querying the publicly available Comprehensive Antibiotic Resistance Database (CARD) (Jia et al., 2016; Zankari et al., 2017). Prokka v1.13 (Seemann, 2014) was used to annotate the assemblies and gyrA sequences were extracted applying the query_pan_genome function in Roary v3.12.0 (Page et al., 2015). gyrA genes were aligned using Uniprot UGENE v1.18.0 (Okonechnikov et al., 2012), from which the gene variants were identified. Only mutations in the quinolone resistance-determining region (QRDR) of gyrA were assessed to be the determinants of resistance, as only these loci have been linked with phenotypic resistance to quinolones. In addition, all the strains studied were deposited in PubMLST database (http://pubmlst.org/campylobacter) and the submissions ids are: BIGSdb_20200511094837_082196_21032, BIGSdb_20200511093337_081290_49754, BIGSdb_20200508081738_149794_16751 and BIGSdb_20200508080706_045922_07760.
Antimicrobial Susceptibility
Antim icrobial susceptibility was tested by the broth microdilution method, using the Sensititre automated system (TREK Diagnostic Systems, Venice, Italy) following the manufacturer’s instructions. Briefly, colonies were subcultured on Columbia agar for 24 h and then seeded in Mueller Hinton Broth supplemented with blood (Oxoid, Basingstoke, UK). Then, they were dispensed into Eucamp2 microtiter plates (TREK Diagnostic Systems, Venice, Italy), with known scalar concentrations of the following antibiotics: ciprofloxacin (CIP) (0.12–16 μg/ml), erythromycin (ERY) (1–128 μg/ml), gentamicin (GEN) (0.12–16 μg/ml), nalidixic acid (NAL) (1–64 μg/ml), streptomycin (STR) (0.25–16 μg/ml), and tetracycline (TET) (0.5–64 μg/ml). The distribution % of MIC are reported in brackets. Following bacterial inoculation, the plates were incubated at 42°C in microaerobic atmosphere for 24 h, and then screened. The strains were classified as resistant (R), and susceptible (S) according to MIC breakpoints, by using Swin v3.3 Software (Thermo Fisher Scientific) in accordance with the epidemiological cutoff values (ECOFFs) as defined by EUCAST (European Committee on antimicrobial breakpoints) (www.eucast.org) to interpret their antimicrobial susceptibilities. C. jejuni strain NCTC 11351 was used as control. MIC breakpoints of resistance were > 0.5 μg/ml for CIP (C.jejuni and C.coli), > 4 μg/ml for STR (C.jejuni and C.coli), > 4 μg/ml for ERY (C.jejuni) and > 8 μg/ml (C.coli), > 2 μg/ml for GEN (C.jejuni and C.coli), > 16 μg/ml for NAL (C.jejuni and C.coli) and > 1 μg/ml for TET (C.jejuni) and > 2 μg/ml (C.coli). Details of the pig and wild boar isolates are summarized in Supplementary Table 1 .
Statistical Analysis
The antimicrobial resistance analysis was performed by means of a Chi-square statistic test. All values with P<0.05 were considered statistically significant (McHugh, 2013).
Results
Genus and Species Confirmation
We analyzed 178 Campylobacter strains isolated from carcasses (53.37%) and fecal content of pigs (46.62%), and 60 Campylobacter strains isolated from feces (83.33%), liver (10%), and muscle (6.67%) of wild boars ( Table 1 ). C. coli was isolated in in 98.31% of pig and 91.66% wild boar strains, while C. jejuni was isolated in 1.68% and 8.33% of pig and wild boar strains, respectively ( Table 1 ).
Table 1.
Percentages of Campylobacter coli and jejuni isolated from pigs and wild boars.
| Carcass | Feces | Muscle | Liver | |
|---|---|---|---|---|
| Pigs (n=178) | 92 (51.68%) C. coli
3 (1.68%) C. jejuni |
83 (46.62%) C. coli | – | – |
| Wild boars (n=60) | – | 46 (76.66%) C. coli
4 (6.66%) C. jejuni |
4 (6.7%) C. coli | 5 (8.33%) C. coli
1 (1.66%) C. jejuni |
MLST Analysis of C. coli and C. jejuni Isolates
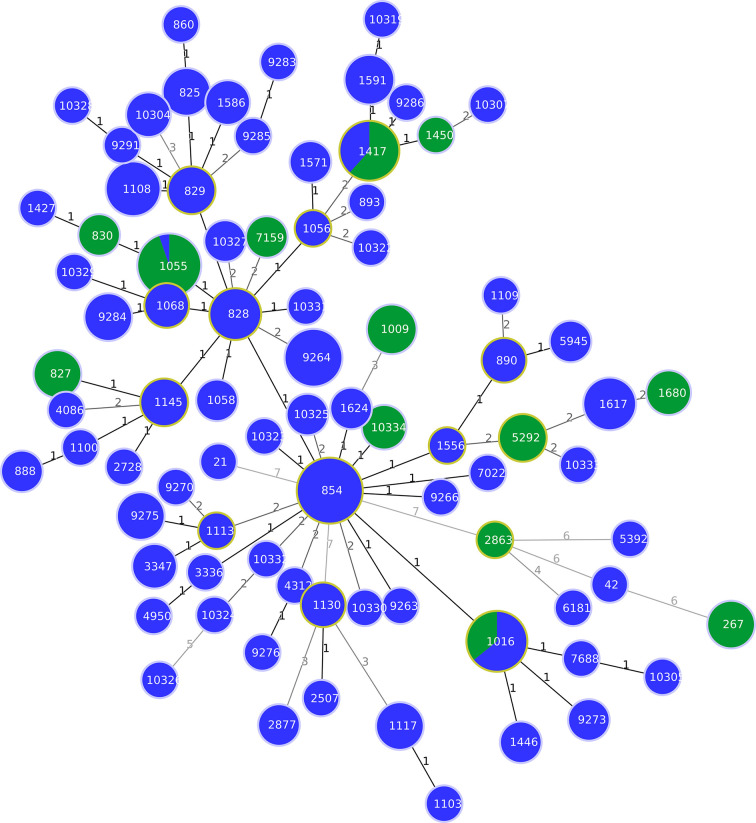
The MLST analysis showed 5 STs among the 8 C. jejuni strains studied ( Supplementary Table 1 ). One ST (ST-10326) has not been described before in the PubMLST Campylobacter database (https://pubmlst.org/campylobacter/). The ST-10326, ST-42, ST-21 were assigned to C. jejuni strains isolated from 3 pigs, while ST-267 was assigned to 4 and ST-2863 to one wild boar C. jejuni strains ( Supplementary Table 1 ). Regarding C. coli, 67 and 8 different STs were obtained from pigs and wild boars, respectively ( Supplementary Table 1 ). Fifteen STs from pigs (ST- 10304, ST-10305, ST-10307, ST-10319, ST-10323, ST-10324, ST-10325, ST-10326, ST-10327, ST-10328, ST-10329, ST-10330, ST-10331, ST-10332, and ST-10333) and one ST from wild boars (ST-10334) were identified for the first time in this study ( Supplementary Table 1 ). In particular, the novel STs contained one or more new allelic genes, and 12 novel alleles were found (aspA547, aspA548, aspA549, glnA754, gltA644, pgm1067, pgm1068, pgm1069, tkt824, tkt825, tkt826, uncA681). Fifty-five STs obtained from both hosts, belonged to the CC-828, only one ST isolated from one pig (ST-5392) belonged to CC-1150, and twenty-eight STs from pigs and wild boars did not belong to any known CC at the time of this analysis ( Supplementary Table 1 ). The ST-1055 was the most prevalent ST that grouped 18 strains isolated from wild boars (30%). The second most prevalent ST was ST-1417 assigned to 8 strains isolated from wild boars (13.3%) ( Figure 1 ). C. coli strains belonging to ST-854 were instead dominant in pigs (12.4%), followed by ST-9264 (6.18%). Out of 70 STs, 42 (60%) were obtained from pigs, and 2 STs out of the 11 STs (18.18%) isolated from wild boars, were represented by only one strain. Only three STs (ST-1016, ST-1055 and ST-1417) were shared between the two animal species ( Figure 1 ). In detail, ST-1016 was represented by 14 C. coli strains (9 from pigs and 5 from wild boars); ST-1055 was represented by 19 C. coli strains (1 from pig and 18 from wild boars) and, finally, ST-1417 was represented by 13 C. coli strains (5 from pigs and 8 from wild boars). The MLST analysis with European pig isolates found a substantial number of STs (67) circulating only on Italian territory ( Supplementary Figure 1 ). The STs most commonly shared with other European countries were: ST-854 and ST-828 shared with seven European countries (Scotland, Switzerland, Germany, UK, Portugal, Netherlands and Luxemburg), followed by ST-1016 shared with six European countries (Switzerland, Belgium, Scotland, UK, The Netherlands and Portugal). A total of 6 and 7 different STS were common with 3 and 2 other European countries, respectively, and 14 STs were shared with one other European country ( Supplementary Table 2 ). The European countries with most STs shared with Italian isolates were Scotland (13 STs), Switzerland (10 STs) and Germany (9 STs).
Figure 1.
Minimum spanning tree (MST) generated for 238 Italian strains isolated from pigs and wild boars. The tree was generated using the goeBURST algorithm in PHYLOViZ software. The distance labels correspond to the number of discriminating alleles. The blue nodes correspond to pig isolates and the green nodes to wild boar isolates.
WgMLST Analysis of C. coli
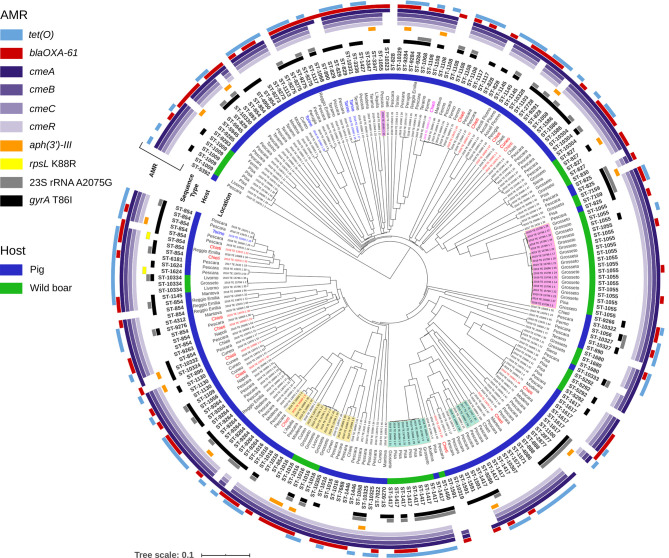
The wgMLST analysis of 213 genomes of C. coli revealed wide diversity among the strains circulating in Italy ( Figure 2 ). The maximum distance between the pair of wgMLST profiles was 583 genes. The strains isolated from domesticated pigs were scattered along most branches of the phylogenetic tree and few clusters of genetically closely related genotypes could be identified. Interestingly, even within these clusters, we did not observe clear geographic separation as they often contained strains isolated in two or more different locations. Similarly, C. coli isolates from wild boar, even though all collected in the Tuscany region, were divided into several separate lineages. The biggest cluster was found in Grosseto province and contained strains assigned to ST-1055. This sequence type was one of the three shared by both, C. coli strains from domesticated pigs and from the wild boarHowever, the isolate from the pig was distant from the wild boar ST-1055 complex by more than 400 genes demonstrating that ST determination was not sufficient to find real genetic connections between the strains. Moreover, we did not identify any clusters of closely related wgMLST profiles that contained strains from both the domesticated pig and the wild boar.
Figure 2.
Phylogenetic tree generated for 213 strains of C. coli from Italy. The UPGMA tree was constructed based on wgMLST analysis results. The presence and allelic diversity of antimicrobial resistance genes substitutions in C. coli genomes are indicated. Strains isolated from domestic pigs are marked with blue color bar and from wild boar with green bar. The isolates highlighted in yellow, green and pink, strains obtained from the two different hosts and belonging to the same MLST sequence types. The isolates in red are of Hungarian origin, those in blue of Danish origin and the only one in fuchsia of French origin. The rest of isolates in black are of Italian origin.
Antimicrobial Resistance Phenotypes
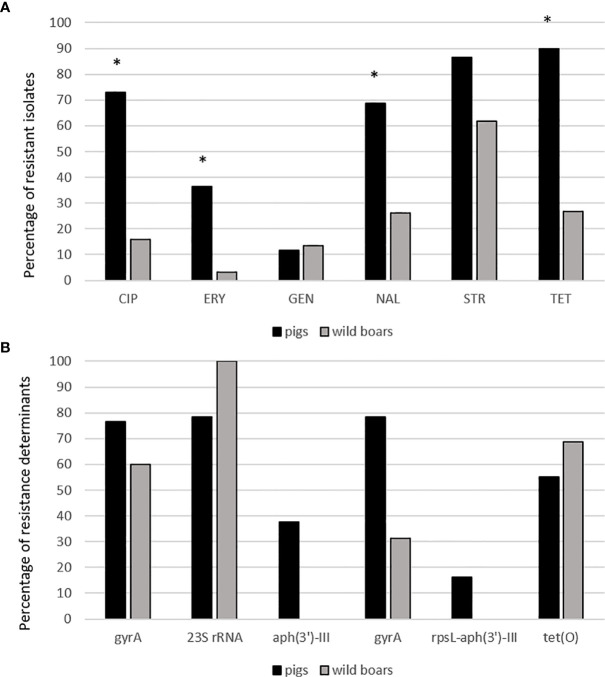
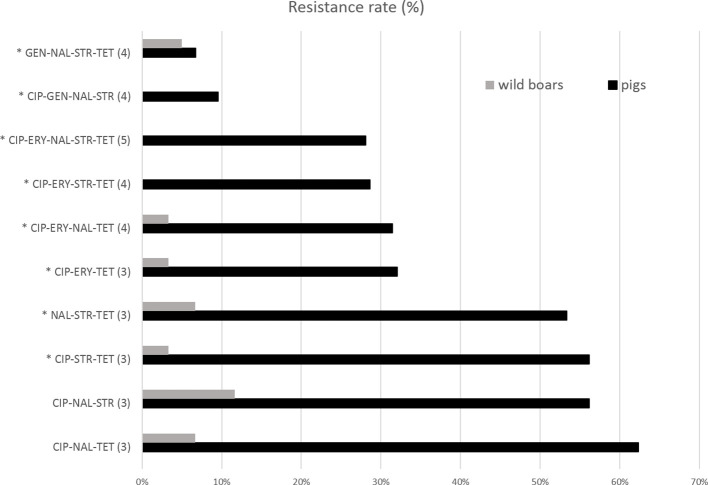
The resistance levels of pig isolates to six antibiotics were compared to genomic resistance profiles of isolates of wild boar origin in Table 2 and Figures 3 and 4 . Statistically significantly higher levels of AMR in pig isolates in respect to wild boar isolates were observed for TET (89.9% vs 26.7%), CIP (73.1% vs 16%), NAL (68.9% vs 26%) and ERY (36.5% vs 3.3%) (Chi-square test; p<0.01). The MIC test revealed that 86.5% of pig and 61.6% of wild boar isolates were resistant to STR. Lower resistance levels were observed for GEN (11.6% for pig isolates; 13.5% for wild boar isolates) ( Figure 3 ). MDR, considered as the resistance to at least three different classes of antibiotics (EFSA & ECDC, 2015), was very common ( Figure 4 ). Strains isolated from pigs were more often found to display MDR than the strains from the wild boar. The most common MDR profiles were CIP-STR-TET (56% pig isolates; 3% wild boar isolates), followed by NAL-STR-TET (53% pig isolates; 7% wild boar isolates). CIP-ERY-TET was found in the 32% and 3% of pig and wild boar isolates, respectively, while CIP-ERY-STR-TET was present only in 29% of pig isolates ( Figure 4 ).
Table 2.
Comparison of genotypic and phenotypic resistance to antibiotics in C. coli isolated from Italian pigs and wild boars.
| Antibiotic class | Antibiotics | Genes | Animals | No. of isolates with R phenotype a (n=178) | No. of isolates with R genotype b | Concordance rate d |
|---|---|---|---|---|---|---|
| Aminoglycosides | Gentamicin (GEN) | aph(3’)-III | Pig | n=24 | n= 9 | 37.5 |
| Wild boar | n=7 | n=0 | 0 | |||
| Streptomycin (STR) | rpsL- aph(3’)-III | Pig | n=154 | n=2; n=23 | 1.3–15 | |
| Wild boar | n=37 | n=0 | 0 | |||
| Beta-lactams c | – | OXA-61 | Pig | – | n=89 | – |
| Wild boar | – | n=27 | – | |||
|
Fluoroquinolones/
Quinolones |
Ciprofloxacin (CIP)/ Nalidixic acid (NAL) |
gyrA | Pig | n=129; n=121 | n=99; n=95 | 76.7–78.5 |
| Wild boar | n=10; n=16 | n=6; n=5 | 60–31.2 | |||
| Macrolides | Erythromycin (ERY) | 23S rRNA | Pig | n=65 | n=51 | 78.5 |
| Wild boar | n=2 | n=2 | 100 | |||
| Tetracyclines | Tetracycline (TET) | Tet(O) | Pig | n=160 | n=88 | 55 |
| Wild boar | n=16 | n=11 | 68.7 | |||
| Multidrug CmeABC efflux system and cmeR | cmeA,cmeB,cmeC, cmeR | Pig | – | n=153; n=132; n=130; n=129 | – | |
| Wild boar | – | n=60; n=58; n=57; n=57 | – | |||
Number of isolates expressing the resistance phenotype for the corresponding antibiotic;
Number of isolates expressing the resistance phenotype for the corresponding antibiotic, that have the indicated gene;
Antibiotic class does not tested for resistance phenotype;
Concordance rate (%).
Figure 3.
(A) Antibiotic resistance pattern between pig and wild boar isolates. CIP, ciprofloxacin, ERY, erythromycin, GEN, gentamicin, NAL, nalidixic acid, STR, streptomycin, TET, tetracycline. *statistically significant vs. wild boar isolates (χ2-test, p<0.01). (B) Percentages of resistance determinants between pig and wild boar antibiotic resistant isolates.
Figure 4.
Percentages of resistant isolates to tested antibiotics among Campylobacter pig and wild boar isolates. CIP, ciprofloxacin, ERY, erythromycin, GEN, gentamicin, NAL, nalidixic acid, STR, streptomycin, TET, tetracycline. The antibiotic numbers are reported in parenthesis. *= MDR profiles.
Detection of Resistance Genes, Mutations, and Levels of Concordance
The genome assemblies of all Campylobacter were investigated for the genomic AMR genes, 23S rRNA and gyrA-associated point mutations and RpsL substitutions. The analysis revealed the presence of 7 AMR genes including: tet(O), cmeA, cmeB, cmeC, cmeR, OXA-61, aph(3’)-III. The resistance genes for the corresponding antibiotic were observed in most but not in all resistant isolates. Regarding resistance to aminoglycosides, resistance traits associated with GEN and STR (aph(3’)-III) resistance were exclusively found in 9 and 23 pig C. coli resistant strains, respectively. RpsL substitution at amino acid 88, involved in STR resistance, was found in only two pig C. coli isolates. The concordance rate between the two types of resistances was of 37.5% and 16.3% ( Table 2 ). Although we did not test resistance to beta-lactams antibiotics class phenotypically, we detected the OXA-61 gene in the half of the pig and wild boar isolates. Tet(O) gene, conferring resistance to TET, was detected in 88 pig and 11 wild boar isolates resistant to TET. The concordance rate resulted, respectively, of 55% and 68.7%. The ERY resistant strains were screened for the presence of mutations in 23S rRNA gene. The A2075G mutation was identified in 51 and 2 isolates from pig and wild boar resistant isolates, showing a concordance rate of 78.5% and 100%, respectively. The cmeA, cmeB, cmeC, and cmeR genes, associated with efflux pump function, were present in almost all the strains. Finally, isolates resistant to fluoroquinolones and quinolones were screened for mutations in the gyrA gene. T86I mutation was detected in 99 and 6 pig and wild boar isolates with CIP resistance phenotype, showing a concordance rate of 76.7% and 60%, respectively, and in 95 and 5 pig and wild boar isolates with NAL resistance phenotype, showing a concordance rate of 78.5% and 31.2% ( Table 2 ).
Discussion
Here we presented a cross-sectional study on Campylobacter from Italian fattening pigs and wild boars using a multiplex approach that included antimicrobial susceptibility test, MLST, wgMLST, and genetic determination of AMR. The analyzed strains were representative of the Italian pigs and wild boars for the period 2012–2019. A high genomic diversity was observed among C. coli isolates in the Italian pig and wild boar populations, with 67 and 11 different STs within 175 and 55 analyzed isolates, respectively. These data are in line with other recent studies (Egger et al., 2012). In this study, MLST revealed the existence of the dominant C. coli CC-828 containing 76% of pig and wild boar isolates while the CC-1150 was detected only in one pig isolate. In addition, we observed that C. coli strains from pig and wild boar constituted two separate populations. Interestingly, only 3.7% (3/81) of STs were shared between pig and wild boar isolates. However, wgMLST analysis showed that pig isolates belonging to these three STs were genetically distant from the wild boar strains, demonstrating that ST determination was not sufficient to find real genetic connections between the strains of the two animals. In general, we did not identify any clusters of closely related wgMLST profiles that contained strains from both hosts suggesting that no exchange of Campylobacter spp. occurred between pigs and the wild boars, possibly due to the segregation of traditional pig farming and wild boar population. Interestingly, we noted that three pig strains (ST-829), isolated from pigs born in Denmark, had related wgMLST profiles, although were fattened in 2 different farms located in Pescara and Torino. Similarly, we showed several clusters in pigs with strictly related wgMLST profiles belonging to fattening farms located in different Italian regions. It is likely that fattening farms in Italy and in Europe may share the same feeder pig supplier, which would explainthe genomic relatedness observed in the distant farms. Comparison of our dataset with the strains obtained from Campylobacter MLST database revealed that C. coli population in Italian pigs and wild boars was different from other European countries. The C. coli strains featured with ST circulating only in Italy amounted for 82.7% (67/81) of the entire Italian collection, suggesting a geographical difference between the Italian and European populations. Furthermore, twenty STs were novel, likely representing geographically restricted clones, as reported also by other authors (Stone et al., 2013). Although the lack of WGS data hampered the verification of the genomic relatedness, it was surprising to observe a numerous STs shared between Italy and Scotland, indicating a possible internationally spread driven by the pig industry. However, a limitation of the study was the underrepresentation of Campylobacter isolates from wild boars in the PubMLST. As suggested in many studies we likely found several host-associated alleles that are present in Campylobacter (French et al., 2005; Miller et al., 2006; Litrup et al., 2007).
In this study, we revealed a clear separation between pig and wild boar Campylobacter, as shown by the presence of only three shared STs out of 83. It was also previously suggested that host preference or niche adaptation for certain STs play a role in acquisition and maintenance of specific clones in different host species (Schouls et al., 2003). Although our study did not allow us to draw conclusions on host association, it is likely that wild boars harbour Campylobacter STs that are rarely, if ever, transmitted to domestic pigs, possibly due to rare contact between the two hosts. Although wild boars are an environmentally destructive invasive species acting as a reservoir for zoonotic pathogens, our findings suggest that they might not be the primary source of infection of Campylobacter for traditional bio-secured domestic pig farms in Italy.
Despite the ban on the application of antibiotics as growth promoters in animal farms in the EU, C. jejuni and coli isolated from humans and animal sources show high levels of resistance to the most important antimicrobials used to treat campylobacteriosis (Castanon, 2007; EFSA & ECDC, 2019). As well as fluoroquinolones and tetracyclines, C. coli strains show a higher resistance to macrolide erythromycin and to aminoglycoside streptomycin, compared to C. jejuni (EFSA & ECDC, 2019). This is worrying because the use of fluoroquinolones, known to be the first-choice treatment for campylobacteriosis, has been recently shifted to erythromycin, against which Campylobacter resistance seemed to develop more slowly, in respect to fluoroquinolones-resistance (Lapierre et al., 2016). Campylobacter resistance mechanisms against the principal antibiotic classes are well known. Fluoroquinolone resistance is rapidly developed in Campylobacter strains because it requires only a single point mutation in gyrA gene (Luangtongkum et al., 2009). On the contrary, erythromycin resistance is due to specifics mutation in 23S rRNA and also depends on an rRNA methylation enzyme (erm B) (Wang et al., 2014). Tetracycline resistance is associated with the presence of tet(O) gene, encoding for a ribosomal protection protein (Sougakoff et al., 1987), while aminoglycosides resistance is due to several genes including rpsL and aph(3’)-III (Iovine, 2013; Zhao et al., 2016). Campylobacter is also known as a bacterium naturally resistant against Beta-lactams, (owning the ubiquitous gene OXA-61)used in combination with beta-lactamase inhibitors, when fluoroquinolones and macrolides are inefficacious (Griggs et al., 2009). Furthermore, among C. coli, which usually harbor AMR genes, a worrying trend towards MDR have been displayed. For all these reasons, Campylobacter has been categorized as a high priority pathogen on the list of bacteria for which new antimicrobials are urgently needed (WHO, 2017). In the present study, high levels of resistance to streptomycin, ciprofloxacin and tetracycline were detected in C. coli isolated from pigs, with resistance to streptomycin frequently found also among C. coli isolated from wild boars. Although the erythromycin resistance levels were lower, the existence of 36% of pig strains resistant to this antibiotic, which is the first-choice drug in the treatment of campylobacteriosis, is alarming. These resistance rates are in line with those reported by other European studies (García-Fernández et al., 2018; Di Donato et al., 2020).
In our study, we found a good correlation between phenotypic resistance to erythromycin, tetracycline, fluoroquinolones and quinolones and the presence of one or more resistance genes or nucleotide polymorphisms expected to confer resistance to the respective antimicrobials. For erythromycin, we found a correlation of 100% and 78.5% between the two types of resistances in pigs and wild boars, respectively. It is possible, that determinants of erythromycin resistance that were not analyzed in our study, such as mutations in L4 and L22 or in the regulatory region of CmeABC efflux pump, could be responsible for enhanced resistance in absence of mutations in 23S rRNA genes (Bolinger and Kathariou, 2017). For tetracyclin, the correlation varied between 68.7% and 55% of the presence of putative resistance genes and observed resistance phenotype respectively in pigs and wild boars. For fluoroquinolones and quinolones, the concordance rate varied between 77% and 45%, in pigs and wild boars. Discrepancies were found for rpsL mutation and the observed phenotype and for aminoglycosides, which could be explained with the existence of the efflux pump mechanisms or other unknown resistance mechanisms. These results suggest that, on one hand, the incidence of AMR in C. coli isolated from wild boars could be still considered low, showing that pigs, animals reared for food production, are much more exposed to antimicrobials. On the other hand, the results obtained show us the hazardous spread of AMR genes through the environment. A reassuring finding from our study was that C. coli isolated from wild boars have MDR profiles lower than 10%, in respect to MDR profiles of pigs, which were 5 times higher.
In conclusion, a rational and moderate use of antimicrobials, combined with a continuous monitoring of AMR bacteria spread in the environment, should be guaranteed to fight the increase in antibiotic resistance rates, extremely dangerous for human and animal health.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
LDM and AA carried out the experiment. FM and AJ wrote the manuscript with support from GG. EDG, RN, and GG helped supervise the project. FM and EDG conceived the original idea. GG supervised the project. GDD, AJ, and FP analyzed the data and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Italian Ministry of Health, grant No IZS AM 07/17 RC and partially by grants of the University of Pisa (PRA_2018_56).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.592512/full#supplementary-material
Minimum spanning tree (MST) generated for 1121 European and Italian strains isolated from pigs and wild boars. The tree was generated using the goeBURST algorithm in PHYLOViZ software. The distance labels correspond to the number of discriminating alleles. The blue nodes correspond to Italian isolates and the red nodes to European isolates.
List of pig and wild boar isolates with MLST and AMR genes profiles, phenotypic resistance and mutations.
References
- (OIE) W.O.f.A.H. Classical Swine Fever. Available at: https://www.oie.int/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/Disease_cards/CLASSICAL_SWINE_FEVER.pdf (Accessed on 08/05/2020).
- Apollonio M., Andersen R., Putman R. (2010). European ungulates and their management in the 21st century (UK: Cambridge University Press; ), 201–222. [Google Scholar]
- Atterby C., Mourkas E., Méric G., Pascoe B., Wang H., Waldenström J., et al. (2018). The potential of isolation source to predict colonization in avian hosts: a case study in Campylobacter jejuni strains from three bird species. Front. Microbiol. 9, 591. 10.3389/fmicb.2018.00591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 (5), 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolinger H., Kathariou S. (2017). The Current State of Macrolide Resistance in Campylobacter spp.: Trends and Impacts of Resistance Mechanisms. Appl. Environ. Microbiol. 83 (12), e00416–e00417. 10.1128/AEM.00416-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanon J. I. R. (2007). History of the use of antibiotic as growth promoters in European poultry feeds. Poult. Sci. 86, 2466–2471. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Campylobacter (Campylobacteriosis). Available at: https://www.cdc.gov/campylobacter/index.html (Accessed on 08/05/2020).
- Cody A. J., McCarthy N. D., Bray J. E., Wimalarathna H. M. L., Colles F. M., Jansen van Rensburg M. J., et al. (2015). Wild bird-associated Campylobacter jejuni isolates are a consistent source of human disease, in Oxfordshire, United Kingdom. Environ. Microbiol. Rep. 7, 782–788. 10.1111/1758-2229.12314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Donato G., Marotta F., Nuvoloni R., Zilli K., Neri D., Di Sabatino D., et al. (2020). Prevalence, Population Diversity and Antimicrobial Resistance of Campylobactercoli Isolated in Italian Swine at Slaughterhouse. Microorganisms 8 (2), 222. 10.3390/microorganisms8020222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA & ECDC (2019). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 17, 1–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger R., Korczak B. M., Niederer L., Overesch G., Kuhnert P. (2012). Genotypes and antibiotic resistance of Campylobacter coli in fattening pigs. Vet. Microbiol. 155 (2-4), 272–278. 10.1016/j.vetmic.2011.08.012 [DOI] [PubMed] [Google Scholar]
- Epps S. V., Harvey R. B., Hume M. E., Phillips T. D., Anderson R. C., Nisbet D. J. (2013). Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. Int. J. Environ. Res. Public Health 10, 6292–6304. 10.3390/ijerph10126292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority (EFSA) Aerts M., Battisti A., Hendriksen R., Kempf I., Teale C., et al. (2019). Technical specifications on harmonised monitoring of antimicrobial resistance in zoonotic and indicator bacteria from food-producing animals and food. EFSA J. 17 (6), 5709. 10.2903/j.efsa.2019.5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French N., Barrigas M., Brown P., Ribiero P., Williams N., Leatherbarrow H., et al. (2005). Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7, 1116–1126. 10.1111/j.1462-2920.2005.00782.x [DOI] [PubMed] [Google Scholar]
- García-Fernández A., Dionisi A. M., Arena S., Iglesias-Torrens Y., Carattoli A., Luzzi I. (2018). Human Campylobacteriosis in Italy: Emergence of Multi-Drug Resistance to Ciprofloxacin, Tetracycline, and Erythromycin. Front. Microbiol. 9, 1906. 10.3389/fmicb.2018.01906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griekspoor P., Colles F. M., McCarthy N. D., Hansbro P. M., Ashhurst-Smith C., Olsen B., et al. (2013). Marked host specificity and lack of phylogeographic population structure of Campylobacter jejuni in wild birds. Mol. Ecol. 22, 1463–1472. 10.1111/mec.12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs D. J., Peake L., Johnson M. M., Ghori S., Mott A., Piddock L. J. V. (2009). Lactamase-mediated ß-lactam resistance in Campylobacter species: prevalence of Cj0299 (bla. OXA-61) and evidence for a novel β-lactamase in C. jejuni. Antimicrob. Agents Chemother. 53, 3357–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hald B., Skovgard H., Bang D., Pedersen K., Dybdahl J., Jespersen J., et al. (2004). Flies and Campylobacter infection of broiler flocks. Emerg. Infect. Dis. 10, 1490–1492. 10.3201/eid1008.040129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson I., Sandberg M., Habib I., Lowman R., Olsson Engvall E. (2018). Knowledge gaps in control of Campylobacter for prevention of campylobacteriosis. Transbound Emerg. Dis. 65 (1), 30–48. 10.1111/tbed.12870 [DOI] [PubMed] [Google Scholar]
- Humphrey T., O’Brien S., Madsen M. (2007). Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 117, 237–257. 10.1016/j.ijfoodmicro.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Iovine N. M. (2013). Resistance mechanisms in Campylobacter jejuni. Virulence 4, 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B., Raphenya A. R., Alcock B., Waglechner N., Guo P., Tsang K. K., et al. (2016). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. 10.1093/nar/gkw1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. A., Grace D., Kock R., Alonso S., Rushton J., Said M. Y., et al. (2013). Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. U. S. A. 110, 8399–8404. 10.1073/pnas.1208059110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre L., Arias M. L., Fernández H. (2016). Antimicrobial Resistance in Campylobacter spp. In: Campylobacter spp. and Related Organisms in Poultry. Eds. Fonseca B., Fernandez H., Rossi D. (Cham: Springer; ). 10.1007/978-3-319-29907-5_10 [DOI] [Google Scholar]
- Letunic I., Bork P. (2006). Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23 (1), 127–128. [DOI] [PubMed] [Google Scholar]
- Litrup E., Torpdahl M., Nielsen E. M. (2007). Multilocus sequence typing performed on Campylobacter coli isolates from humans, broilers, pigs and cattle originating in Denmark. J. Appl. Microbiol. 103 (1), 210–218. 10.1111/j.1365-2672.2006.03214.x [DOI] [PubMed] [Google Scholar]
- Luangtongkum T., Jeon B., Han J., Plummer P., Logue C. M., Zhang Q. (2009). Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4, 189–200. 10.2217/17460913.4.2.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta F., Garofolo G., di Marcantonio L., Di Serafino G., Neri D., Romantini R., et al. (2019). Antimicrobial resistance genotypes and phenotypes of Campylobacter jejuni isolated in Italy from humans, birds from wild and urban habitats, and poultry [published correction appears in PLoS One. 2019 Nov 7;14(11):e0225231]. PLoS One 14 (10), e0223804. 10.1371/journal.pone.0223804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta F., Janowicz A., Di Marcantonio L., Ercole C., Di Donato G., Garofolo G., et al. (2020). Molecular Characterization and Antimicrobial Susceptibility of C. jejuni Isolates from Italian Wild Bird Populations. Pathogens 9 (4):304. 10.3390/pathogens9040304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massei G., Kindberg J., Licoppe A., Gačić D., Šprem N., Kamler J., et al. (2015). Wild boar populations up, numbers of hunters down? A review of trends and implications for Europe. Pest Manage. Sci. 71, 492–500. 10.1002/ps.3965 [DOI] [PubMed] [Google Scholar]
- McHugh M. L. (2013). The Chi-square test of independence: Lessonsin biostatistics. Biochem. Med. 23, 143–149. 10.11613/BM.2013.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier R. K. (2015). Investigations of Health and Abundance of Free-ranging Wild Boar (Sus Scrofa) in Switzerland in a European Context (Vetsuisse-Fakultät Bern; ). [Google Scholar]
- Meier R., Ryser-Degiorgis M. (2018). Wild boar and infectious diseases: evaluation of the current risk to human and domestic animal health in Switzerland: A review. Schweiz Arch. Tierheilkd. 160 (7–8), 443–460. 10.17236/sat00168 [DOI] [PubMed] [Google Scholar]
- Miller R. S., Sweeney S. J. (2013). Mycobacterium bovis (bovine tuberculosis) infection in North American wildlife: current status and opportunities for mitigation of risks of further infection in wildlife populations. Epidemiol. Infect. 141, 1357–1370. 10.1017/s0950268813000976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. G., Englen M. D., Kathariou S., Wesley I. V., Wang G., Pittenger-Alley L., et al. (2006). Identification of host-associated alleles bymultilocus sequence typing of Campylobacter coli strainsfrom food animals. Microbiology 152, 245–255. 10.1099/mic.0.28348-0 [DOI] [PubMed] [Google Scholar]
- Mourkas E., Florez-Cuadrado D., Pascoe B., Calland J. K., Bayliss S. C., Mageiros L., et al. (2019). Gene pool transmission of multidrug resistance among Campylobacter from livestock, sewage and human disease. Environ. Microbiol. 21 (12), 4597–4613. 10.1111/1462-2920.14760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento M., Sousa A., Ramirez M., Francisco A. P., Carriço J. A., Vaz C. (2017). PHYLOViZ 2.0: Providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics 33, 128–129. 10.1093/bioinformatics/btw582 [DOI] [PubMed] [Google Scholar]
- Navarro-Gonzalez N., Ugarte-Ruiz M., Porrero M. C., Zamora L., Mentaberre G., Serrano E., et al. (2014). Campylobacter shared between free-ranging cattle and sympatric wild ungulates in a natural environment (NE Spain). Ecohealth 11 (3), 333–342. 10.1007/s10393-014-0921-3 [DOI] [PubMed] [Google Scholar]
- Okonechnikov K., Golosova O., Fursov M. (2012). The UGENE team. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167. 10.1093/bioinformatics/bts091 [DOI] [PubMed] [Google Scholar]
- Page A. J., Cummins C. A., Hunt M., Wong V. K., Reuter S., Holden M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31 (22), 3691–3693. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascoe B., Méric G., Yahara K., Wimalarathna H., Murray S., Hitchings M. D., et al. (2017). Local genes for local bacteria: evidence of allopatry in the genomes of transatlantic Campylobacter populations. Mol. Ecol. 26, 4497–4508. 10.1111/mec.14176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Fons F., Vicente J., Vidal D., Höfle U., Villanúa D., Gauss C., et al. (2006). Seroprevalence of six reproductive pathogens in European wild boar (Sus scrofa) from Spain: the effect on wild boar female reproductive performance. Theriogenology 65, 731–743. 10.1016/j.theriogenology.2005.07.001 [DOI] [PubMed] [Google Scholar]
- Ruiz-Fons F., Segalés J., Gortázar C. (2008). A review of viral diseases of the European wild boar: effects of population dynamics and reservoir rôle. Vet. J. (Lond. Engl. 1997) 176, 158–169. 10.1016/j.tvjl.2007.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouls L. M., Reulen S., Duim B., Wagenaar J. A., Willems R. J., Dingle K. E., et al. (2003). Comparative genotyping of Campylobacter jejuni byamplified fragment length polymorphism, multilocussequence typing, and short repeat sequencing: strain diver-sity, host range, and recombination. J. Clin. Microbiol. 41, 15–26. 10.1128/JCM.41.1.15-26.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinform 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sheppard S. K., Dallas J. F., MacRae M., McCarthy N. D., Sproston E. L., Gormley F. J., et al. (2009. a). Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int. J. Food Microbiol. 134, 96–103. 10.1016/j.ijfoodmicro.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard S. K., Colles F. M., McCarthy N. D., Strachan N. J. C., Ogden I. D., Forbes K. J., et al. (2011). Niche segregation and genetic structure of Campylobacter jejuni populations from wild and agricultural host species. Mol. Ecol. 20, 3484–3490. 10.1111/j.1365-294X.2011.05179.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippy R., Sandoval-Green C. M. J., Sahin O., Plummer P., Fairbanks W. S., Zhang Q., et al. (2012). Occurrence and molecular analysis of Campylobacter in wildlife on livestock farms. Vet. Microbiol. 157, 369–375. 10.1016/j.vetmic.2011.12.026 [DOI] [PubMed] [Google Scholar]
- Sougakoff W., Papadopoulou B., Nordmann P., Courvalin P. (1987). Nucleotide sequence and distribution of gene tetO encoding tetracycline resistance in Campylobacter coli. FEMS Microbiol. Lett. 44, 153–159. [Google Scholar]
- Stella S., Tirloni E., Castelli E., Colombo F., Bernardi C. (2018). Microbiological Evaluation of Carcasses of Wild Boar Hunted in a Hill Area of Northern Italy. J. Food Prot. 81 (9), 1519–1525. 10.4315/0362-028X.JFP-18-077 [DOI] [PubMed] [Google Scholar]
- Stone D., Davis M., Baker K., Besser T., Roopnarine R. (2013). Sharma R. MLST genotypes and antibiotic resistance of Campylobacter spp isolated from poultry in Grenada. BioMed. Res. Int. 794643. 10.1155/2013/794643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldenström J., Broman T., Carlsson I., Hasselquist D., Achterberg R. P., Wagenaar J. A., et al. (2002). Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl. Environ. Microbiol. 68, 5911–5917. 10.1128/aem.68.12.5911-5917.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Clark C. G., Taylor T. M., Pucknell C., Barton C., Woodward D. L., et al. (2002). Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbiol. 40 (12), 4744–4747. 10.1128/jcm.40.12.4744-4747.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang M., Deng F., Shen Z., Wu C., Zhang J. (2014). Emergence of multidrug-resistant Campylobacterspecies isolates with a horizontally acquired rRNA methylase. Antimicrob. Agents Chemother. 58, 5405–5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse C. A., Zhao S., Tate H. (2018). Antimicrobial Resistance in Campylobacter Species: Mechanisms and Genomic Epidemiology. Adv. Appl. Microbiol. 103, 1–47. 10.1016/bs.aambs.2018.01.001 [DOI] [PubMed] [Google Scholar]
- WHO media centre (2017). WHO publishes list of bacteria for which new antibiotics are urgently needed. Available at: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/ [cited 2017 May 6]. [Google Scholar]
- Zankari E., Allesøe R., Joensen K. G., Cavaco L., Lund O., Aarestrup F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. 10.1093/jac/dkx217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Tyson G. H., Chen Y., Li C., Mukherjee S., Young S., et al. (2016). Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl. Environ. Microbiol. 82, 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Minimum spanning tree (MST) generated for 1121 European and Italian strains isolated from pigs and wild boars. The tree was generated using the goeBURST algorithm in PHYLOViZ software. The distance labels correspond to the number of discriminating alleles. The blue nodes correspond to Italian isolates and the red nodes to European isolates.
List of pig and wild boar isolates with MLST and AMR genes profiles, phenotypic resistance and mutations.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.