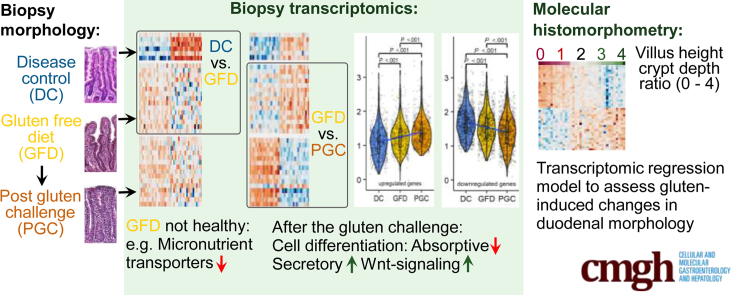
Abstract
Background & Aims
Gluten challenge studies are instrumental in understanding the pathophysiology of celiac disease. Our aims in this study were to reveal early gluten-induced transcriptomic changes in duodenal biopsies and to find tools for clinics.
Methods
Duodenal biopsies were collected from 15 celiac disease patients on a strict long-term gluten-free diet (GFD) prior to and post a gluten challenge (PGC) and from 6 healthy control individuals (DC). Biopsy RNA was subjected to genome-wide 3’ RNA-Seq. Sequencing data was used to determine the differences between the three groups and was compared to sequencing data from the public repositories. The biopsies underwent morphometric analyses.
Results
In DC vs. GFD group comparisons, 167 differentially expressed genes were identified with 117 genes downregulated and 50 genes upregulated. In PGC vs. GFD group comparisons, 417 differentially expressed genes were identified with 195 genes downregulated and 222 genes upregulated. Celiac disease patients on a GFD were not “healthy”. In particular, genes encoding proteins for transporting small molecules were expressed less. In addition to the activation of immune response genes, a gluten challenge induced hyperactive intestinal wnt-signaling and consequent immature crypt gene expression resulting in less differentiated epithelium. Biopsy gene expression in response to a gluten challenge correlated with the extent of the histological damage. Regression models using only four gene transcripts described 97.2% of the mucosal morphology and 98.0% of the inflammatory changes observed.
Conclusions
Our gluten challenge trial design provided an opportunity to study the transition from health to disease. The results show that even on a strict GFD, despite being deemed healthy, patients reveal patterns of ongoing disease. Here, a transcriptomic regression model estimating the extent of gluten-induced duodenal mucosal injury is presented.
Keywords: Gluten Challenge, RNA-Seq, Celiac Disease, Gluten-Free Diet, Morphometry
Abbreviations used in this paper: DC, disease control; GFD, gluten-free diet; GIP, gluten-specific immunogenic peptide; HLA, human leukocyte antigen; IEL, intraepithelial lymphocyte; |log2FC|, absolute log2-fold change; NGS, next-generation sequencing; PCA, principal component analysis; PGC, post–gluten challenge; RNA-Seq, RNA-sequencing; TG2, tissue transglutaminase 2; VH:CrD, villus height-to-crypt depth ratio
Graphical abstract
Summary.
Celiac disease patients, despite on a long-term gluten-free diet and consequentially healed mucosa in histomorphometric terms, still display ongoing disease at the molecular level. Gluten challenge induces hyperactive Wnt-signaling and secretory cell type signature. Molecular histomorphometry is presented as a tool to assess degree of intestinal injury in celiac disease.
Celiac disease is an autoimmune disorder in which dietary cereal gluten is an essential inducer and driving force. The overall global prevalence of celiac disease is approximately 1%.1 There are differences between countries,1,2 and at the population level, the true prevalence has been shown to be increasing over time.3 During celiac disease pathogenesis, ingested gluten induces an autoimmune-like reaction in genetically susceptible subjects carrying the DQ2 or DQ8 human leukocyte antigen (HLA) haplotypes, which results in mucosal inflammation, small intestinal villus atrophy, and crypt hyperplasia and a wide array of gastrointestinal and nongastrointestinal manifestations. The HLA genes can be considered necessary but not sufficient factors, as only a small portion of the individuals with the compatible HLA genetics will develop gluten-dependent celiac disease. Additionally, several non-HLA genes have been identified as contributing factors in celiac disease pathophysiology.4 In addition to dietary gluten and compatible HLA genetics, the auto-antigen and gluten-deamidating enzyme tissue transglutaminase 2 (TG2), is needed.5, 6, 7 It was recently hypothesized that intestinal microbiota is a factor involved in the pathogenesis and clinical presentation of the disease.8 Central to the disease pathogenesis are the abnormal intestinal T cell responses to gluten, and the ability of CD4+ T cells to recognize gluten epitopes is thought to be a driver of the disease.9,10 B cells, autoantibodies, and TG2, have also been shown to have an important function in celiac disease pathogenetic mechanisms.9,11, 12, 13, 14 Despite the well-studied role of the immune system in celiac disease development, a complete picture of the pathophysiology, especially the early steps leading to the loss of gluten tolerance, is still missing.13 Currently, a lifelong gluten-free diet (GFD) is the only accepted treatment option for patients with celiac disease.
Intestinal epithelium, as one of the components in celiac disease development, has also been the focus of research. We believe that discovering the molecular mechanisms responsible for thwarting the differentiation program of the intestinal epithelial cells could lead to a better understanding of the mechanisms of celiac disease development and the consequent deterioration of the small intestinal mucosal morphology. Transcriptomic studies using microarray15 or RNA-sequencing (RNA-Seq) technologies16, 17, 18 and proteomic19 and epigenomic20 analyses were conducted on celiac disease patient biopsies. The majority of these studies used untreated patient samples, samples from patients on a GFD, and non–celiac disease samples as a control group. Because mucosal damage is dose- and time-dependent,13,21,22 and because patients have different levels of gluten consumption at the time of their initial diagnoses, often large amount for decades, these confounding factors may lead to inconsistent results. To circumvent these shortcomings, we sought to exploit the gluten challenge study design13,21, 22, 23, 24 and perform genome-wide transcriptomic analyses of celiac disease small intestinal biopsies from patients on a strict long-term GFD and compare the results with those of biopsies taken from the same celiac disease patients after a standard amount and time on a gluten challenge. We further compared these transcriptomic results with those obtained from non–celiac disease control (DC) patient biopsies. We also aimed to correlate the gene transcript levels with the degree of gluten-induced mucosal injury to create a molecular histomorphometry tool for assessing the extent of both duodenal mucosal architectural damage and inflammation.
Results
Histomorphometry and Molecular Histomorphometry
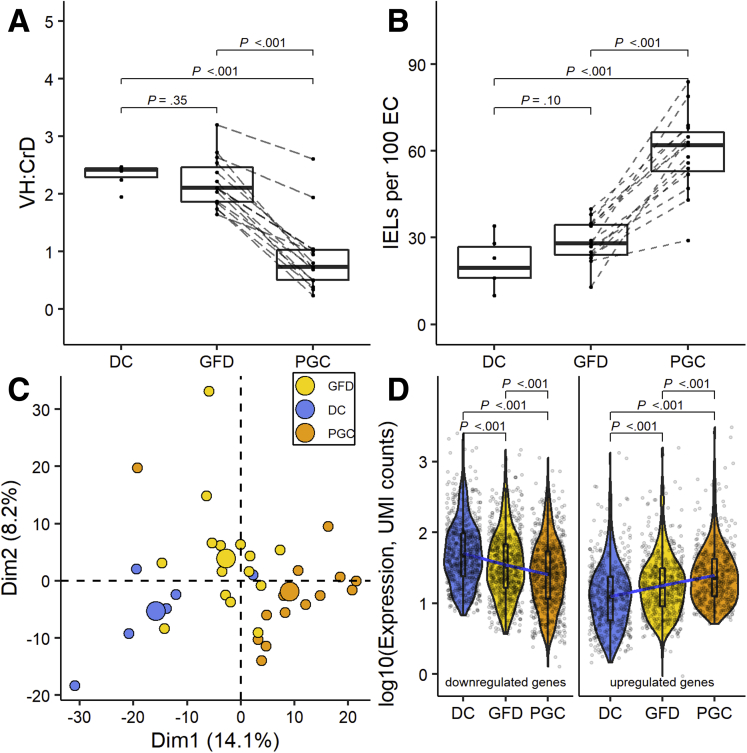
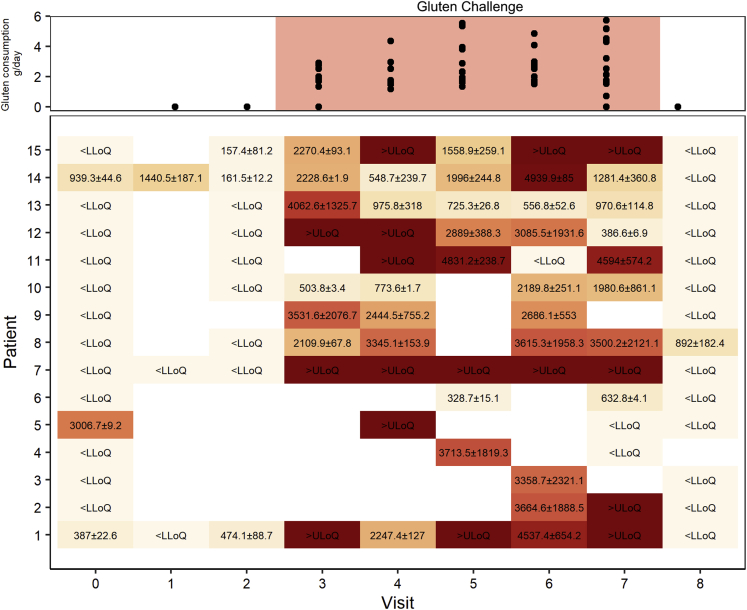
The morphology results (villus height-to-crypt depth ratio [VH:CrD]) of each study group are shown in Figure 1A, and the grade of inflammation (intraepithelial lymphocyte [IEL] density) is shown in Figure 1B. The morphometric results did not differ (P = .35) between the DC (2.33 ± 0.20) and GFD (2.19 ± 0.43) groups. Significant mucosal deterioration (P < .001) and increased inflammation (P < .001) was evident during a gluten challenge (GFD [VH:CrD: M 2.19 ± 0.43; IEL density: 28.73 ± 7.25] vs post–gluten challenge [PGC] [VH:CrD: 0.90 ± 0.63; IEL density: 59.60 ± 13.84]). Although the patients kept a strict GFD prior to the gluten challenge, the stool gluten-specific immunogenic peptide (GIP) results showed 4 of the 15 had inadvertently ingested gluten just before stool sample collection (Figure 2). The unsupervised principal component analysis (PCA) plot for the gene expression differences in all samples indicated that the combination of dimensions (Dim1 and Dim2) explains 22.3% of the variance, showing a decent separation between the groups (Figure 1C). As the GFD group was discernible from the DC and PGC groups in PCA analysis, we next analyzed how does the gene expression in GFD group differ from the DC and PGC groups. Pertaining to both significantly down- and upregulated genes in DC and PGC comparisons (false discovery rate [FDR] < .05 and absolute log2-fold change [|log2FC|] ≥ 0.5), the GFD group differs significantly from both groups and the gene expression profile in GFD group is between the DC and PGC groups (Figure 1D).
Figure 1.
Histomorphometric and molecular histomorphometric data of the DC group, celiac disease patients on a GFD, and celiac disease patients PGC. (A) VH:CrD measurements. (B) Counts of IELs per 100 enterocytes (EC). (C) PCA plot for all samples exploring biological differences between the DC (blue), GFD (yellow), and PGC (orange) groups based on the gene expression profile with bigger spheres depicting the center of a distribution. (D) Violin plots showing the log-transformed expression of differentially expressed genes detected in PGC vs DC groups comparisons (768 downregulated in the left panel and 580 upregulated genes in the right panel) in each group. Three lines (from the bottom to the top) in each violin plot show the location of the lower quartile, the median, and the upper quartile, respectively. The blue line is fitted regression model, demonstrating the linear character of mean of the log-transformed expression changes. Student’s t test was used for comparing the mean values.
Figure 2.
Gluten content (ng GIP/g stool) in patients’ stool samples taken before, during, and after gluten challenge, measured by iVYLISA GIP assay. Gluten consumption per day was assessed by number of cookies eaten, taking into account the content of gluten in them, eaten between the visits by patient divided by number of days between the visits (median 1.99 [interquartile range, 1.74 - 2.91] g/d). LLoQ, lower limit of quantification; ULoQ, upper limit of quantification.
Genes Affected by the Gluten Challenge
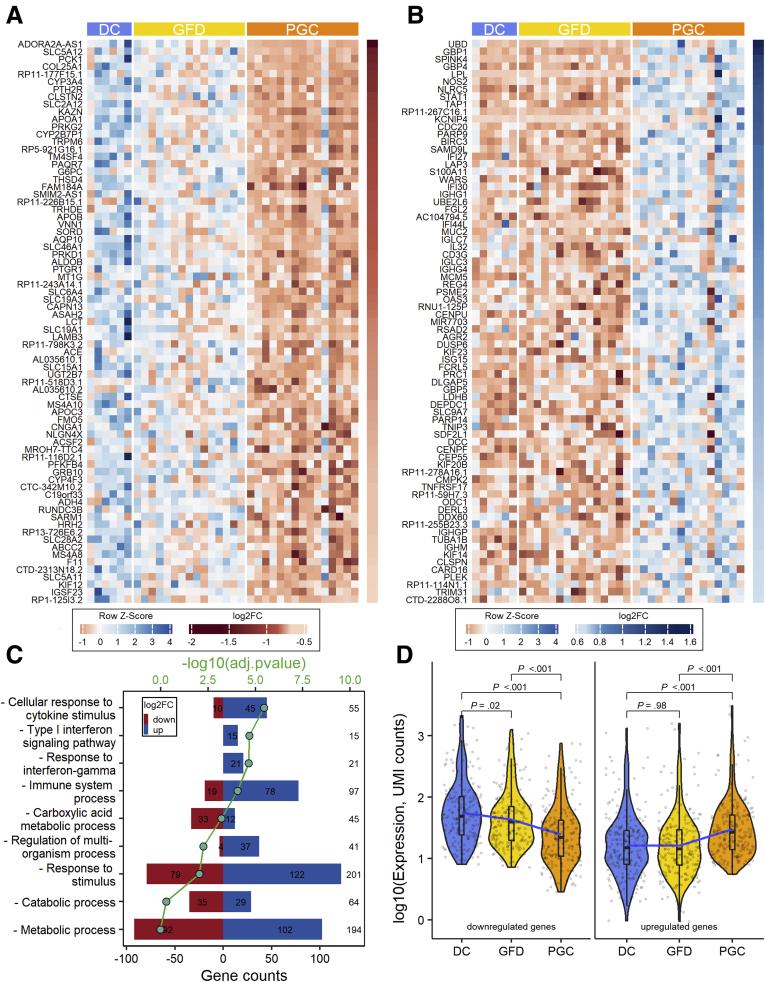
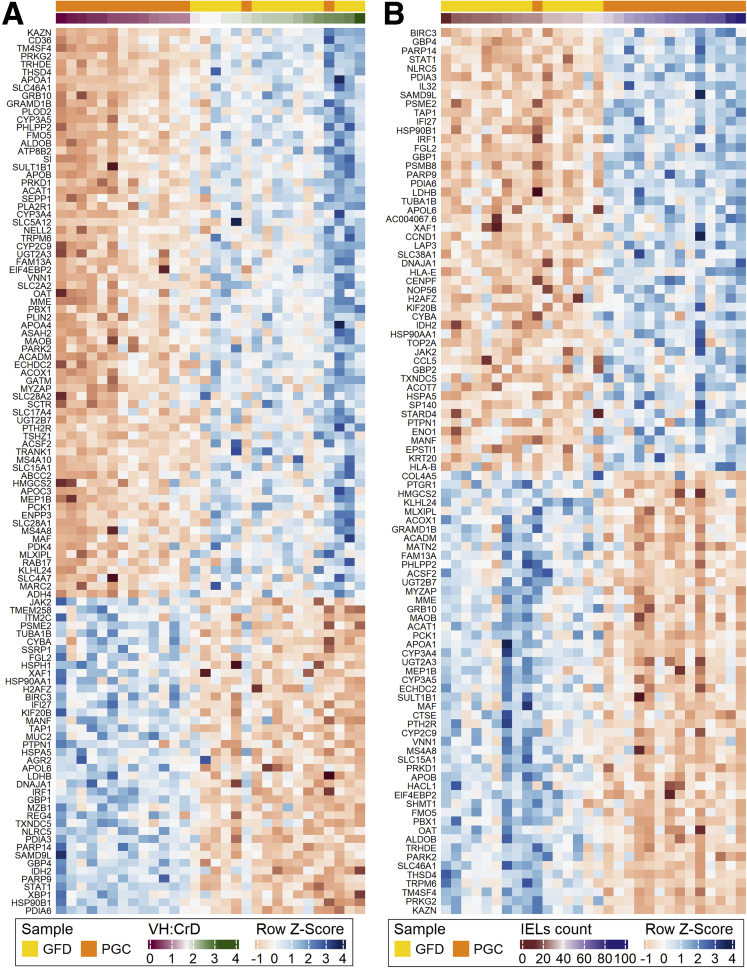
When comparing RNA expression profiles from celiac disease patients in the GFD and PGC groups, 417 differentially expressed genes were identified with 195 genes downregulated and 222 genes upregulated (FDR < .05 and |log2FC| ≥ 0.5). The top 75 differentially expressed genes, in a paired analysis of GFD vs PGC, are shown in Figures 3A and B (entire dataset in Supplementary Table 1). Gene Ontology analyses show that genes involved in cellular response to cytokines, including interferons, are overrepresented within gluten-induced gene expression profile (Figure 3C). Heatmaps organized to gene ontology terms are shown in Figure 4. Interestingly, when gluten-induced gene expression profile genes were analyzed across all the 3 groups, GFD group was positioned between the DC and PGC in downregulated genes (Figure 3D). In case of upregulated genes, however, gene expression in GFD group resemble that of in the DC group.
Figure 3.
Differential intestinal mucosal gene transcriptome after 10 weeks of the gluten challenge in celiac disease patients. (A) Heatmap of the top 75 genes that are downregulated specifically after the gluten challenge. (B) Heatmap of the top 75 genes that are upregulated after the gluten challenge. (C) Selected Gene Ontology terms enriched in identified set of genes (total 417 genes) specific for gluten-induced mucosal damage. (D) Violin plots showing the log-transformed expression of differentially expressed genes detected in PGC vs GFD comparisons (195 downregulated in the left panel and 222 upregulated genes in the right panel) in each group. Three lines (from the bottom to the top) in each violin plot show the location of the lower quartile, the median, and the upper quartile, respectively. The blue line is the fitted regression model, demonstrating the nonlinear character of mean of the log-transformed expression changes. Student’s t test was used for group mean comparisons.
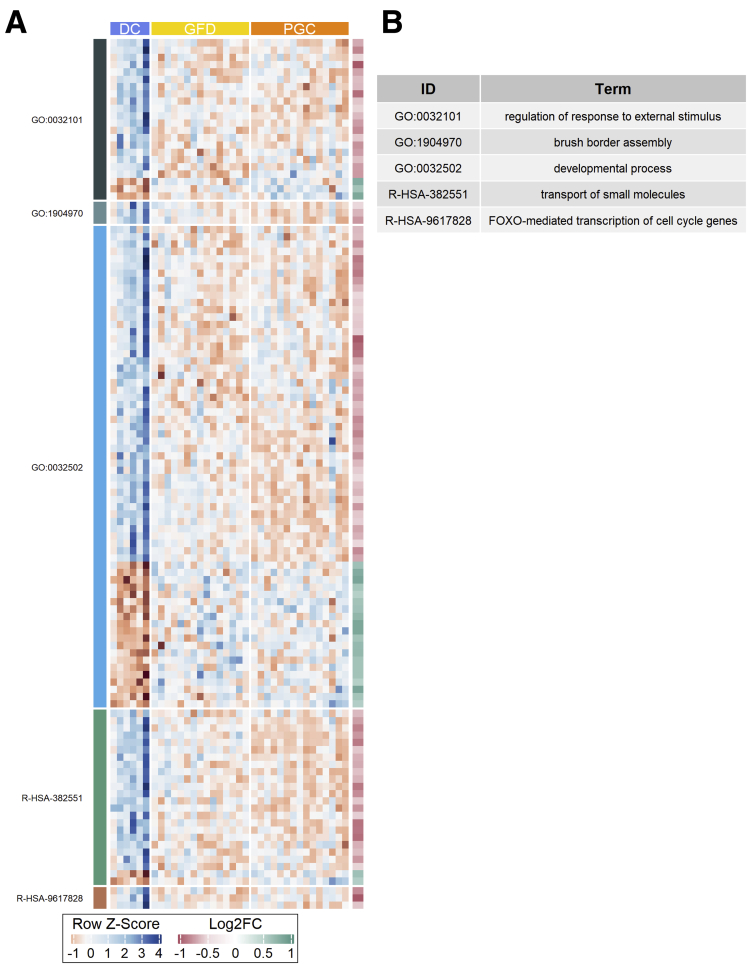
Figure 4.
Genes affected by gluten challenge shown as heatmaps organized to Gene Ontology (GO) terms.
Genes Affected by a GFD Compared With Non–Celiac Disease Control Subjects
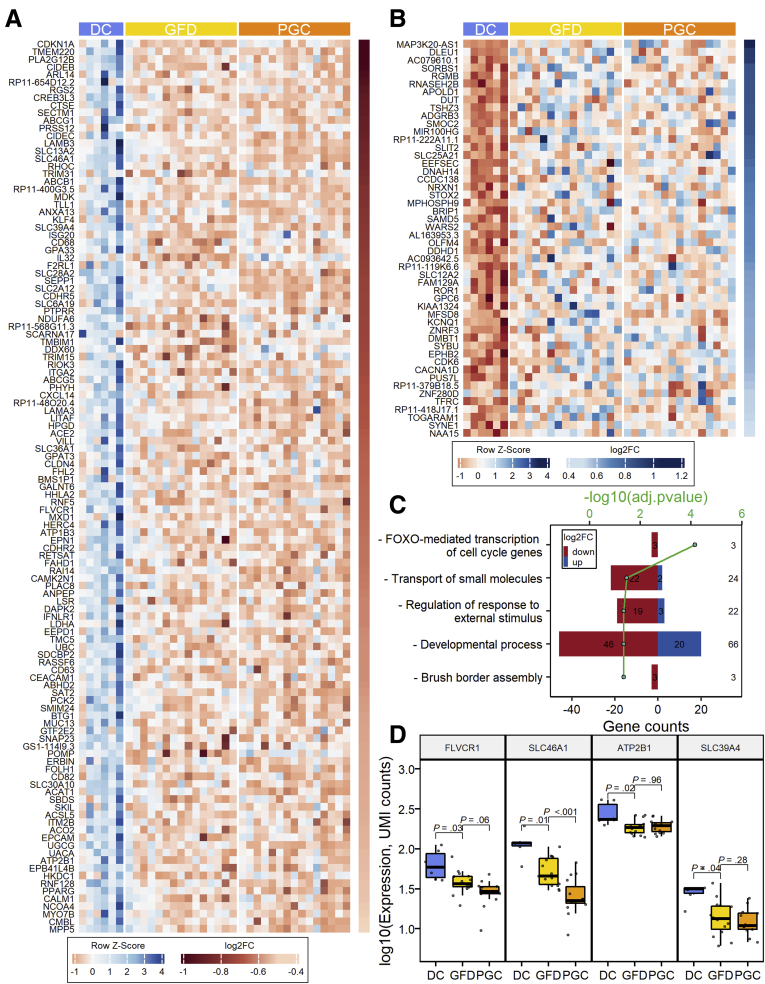
As PCA and gene expression data (Figure 1C and D), clearly discerned DC and GFD into separate groups, we analyzed the genes whose expression differ significantly already when celiac disease patients are on GFD when compared with the DC group. In total, 167 differentially expressed genes were identified (FDR < .05 and |log2FC| ≥ 0.5) with 117 genes downregulated and 50 genes upregulated in the GFD group when compared with the DC group (Figure 5A and B). Our results indicate that even though intestinal morphology in patients on a strict GFD is similar to that measured in DC patients (Figure 1A and B) based on gene transcription, the GFD group differs significantly from the DC group, showing a substantial number of differentially expressed genes. In general, the expression of these genes is not affected by the gluten challenge when comparing the GFD and PGC groups. Gene Ontology and Reactome pathway analyses reveal that patients on a GFD suffer from profoundly altered expression of genes with functions such as brush border assembly, developmental processes, transport of small molecules, and FOXO-mediated transcription of the cell cycle (Figure 5C). Heatmaps organized to gene ontology and Reactome pathway terms are shown in Figure 6. For instance, within the transporter gene group, 24 genes are differentially expressed in the GFD group vs the DC group. Of these, 22 are downregulated in the GFD group, including, for example, 4 transporter genes: FLVCR1 (heme transporter), SLC46A1 (folate and heme transporter), ATP2B1 (calcium transporter), and SLC39A4 (zinc transporter), as seen in Figure 5D. This is in accordance with the clinical data, as it has been shown that patients on a GFD quite frequently suffer from micronutrient deficiencies.25 A list of the 24 transporter genes affected by a GFD is presented in Table 1.
Figure 5.
Differential intestinal mucosal gene transcriptome between healthy control subjects and patients with celiac disease on a GFD. (A) Heatmap of 117 genes showing significantly lower expression in patients with celiac disease in both GFD and PGC groups vs the DC group. (B) Heatmap of 50 genes upregulated in both GFD and PGC groups vs the DC group. (C) Gene Ontology terms and Reactome pathways enriched in an identified set of genes (total 167) showing altered expression in the GFD group vs the DC group. (D) Changes of expression in some genes involved in the enriched “Transport of small molecules” Reactome pathway.
Figure 6.
Genes affected already in GFD shown as heatmaps organized to Gene Ontology (GO) and Reactome pathway terms.
Table 1.
List of the 24 Genes Affected Already in Gluten-Free Diet Within the Group of Transport of Small Molecules Are Grouped According to Their Specific Function
| # | ENSEMBL ID | Symbol | Function | log2-Fold change | FDR |
|---|---|---|---|---|---|
| Iron uptake and transport | |||||
| 1 | ENSG00000072274 | TFRC | delivers iron from transferrin to the cell | 0.54 | .042 |
| 2 | ENSG00000150991 | UBC | ubiquitination | –0.60 | .034 |
| 3 | ENSG00000162769 | FLVCR1 | heme transporter | –0.65 | .028 |
| 4 | ENSG00000076351 | SLC46A1 | folate transport, heme transporter in duodenal enterocytes | –0.85 | .012 |
| Lipid and lipoprotein homeostasis | |||||
| 5 | ENSG00000105699 | LSR | binds chylomicrons, LDL and VLDL in presence of free fatty acids and allows their subsequent uptake in the cells | –0.61 | .034 |
| 6 | ENSG00000160179 | ABCG1 | catalyzes the efflux of phospholipids | –0.88 | .015 |
| 7 | ENSG00000060566 | CREB3L3 | transcription factor involved in cholesterol and lipid metabolism | –0.90 | .002 |
| 8 | ENSG00000075239 | ACAT1 | catalyzes an aerobic process breaking down fatty acids into acetyl-CoA | –0.56 | .034 |
| 9 | ENSG00000187288 | CIDEC | promotes lipid transfer from the smaller to larger lipid droplets | –0.87 | .010 |
| 10 | ENSG00000198668 | CALM1 | calcium-binding protein | –0.52 | .032 |
| 11 | ENSG00000138075 | ABCG5 | mediates Mg2+- and ATP-dependent sterol transport across the cell membrane | –0.71 | .032 |
| Transport of inorganic cations/anions and amino acids/oligopeptides | |||||
| 12 | ENSG00000123643 | SLC36A1 | pH-dependent electrogenic transporter for small amino acids such as glycine, alanine and proline | –0.67 | .032 |
| 13 | ENSG00000174358 | SLC6A19 | mediates resorption of neutral amino acids across the apical membrane of renal and intestinal epithelial cells | –0.74 | .021 |
| 14 | ENSG00000198668 | CALM1 | calcium-binding protein | –0.52 | .032 |
| 15 | ENSG00000141504 | SAT2 | catalyzes the acetylation of polyamines | –0.59 | .034 |
| 16 | ENSG00000064651 | SLC12A2 | mediates sodium and chloride reabsorption | 0.64 | .032 |
| Ion homeostasis | |||||
| 17 | ENSG00000070961 | ATP2B1 | transports calcium from the cytoplasm to the extracellular space | –0.54 | .015 |
| 18 | ENSG00000069849 | ATP1B3 | maintaining the electrochemical gradients of Na and K ions across the plasma membrane | –0.64 | .015 |
| 19 | ENSG00000198668 | CALM1 | calcium-binding protein | –0.52 | .032 |
| 20 | ENSG00000204308 | RNF5 | membrane-bound ubiquitin ligase | –0.65 | .045 |
| Transport of bile salts and organic acids, metal ions, and amine compounds | |||||
| 21 | ENSG00000196660 | SLC30A10 | transports manganese | –0.56 | .042 |
| 22 | ENSG00000007216 | SLC13A2 | cotransports sodium ions and dicarboxylates such as succinate and citrate | –0.86 | .015 |
| 23 | ENSG00000174358 | SLC6A19 | mediates resorption of neutral amino acids across the apical membrane of renal and intestinal epithelial cells | –0.74 | .021 |
| 24 | ENSG00000147804 | SLC39A4 | transports zinc | –0.78 | .039 |
| Other | |||||
| 25 | ENSG00000085563 | ABCB1 | translocates drugs and phospholipids across the membrane | –0.82 | .003 |
| 26 | ENSG00000146411 | SLC2A12 | catalyzes the uptake of sugars through facilitated diffusion | –0.74 | .044 |
| 27 | ENSG00000137860 | SLC28A2 | uptake and salvage of purine nucleosides | –0.75 | .038 |
Gene ID, gene symbol, function, and numerical values for the disease control vs gluten-free diet groups as log2-fold change and adjusted P values are shown.
FDR, false discovery rate; LDL, low-density lipoprotein; VLDL, very low-density lipoprotein.
Hyperactive Intestinal Wnt Signaling Is Prominent in Celiac Disease Patients After a Gluten Challenge But Is Also Already Enhanced During a GFD
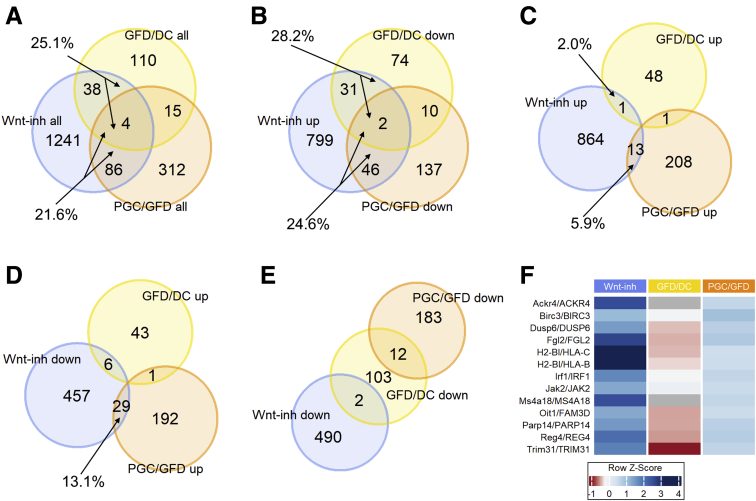
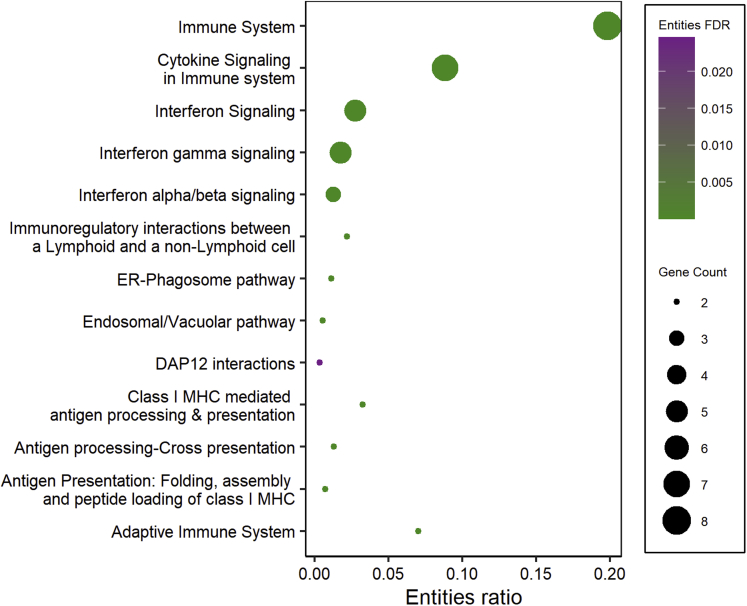
As it has been suggested that Wnt signaling is involved in celiac disease–associated villous atrophy and crypt hyperplasia.15,26 we compared the present RNA-Seq data to our mouse organoid data in which intestinal differentiation was induced in vitro with Wnt inhibitor IWP2.27 These data comparisons enable us to characterize the Wnt-mediated component of mucosal damage in celiac disease. Of all 417 differentially expressed genes after the gluten challenge, 90 (21.6%) belong to the Wnt component (Figure 7A). An approximately similar size of Wnt component is also affected when celiac patients on a GFD are compared with DC patients, as 42 genes of all 167 differentially expressed genes (25.1%) belong to the functional Wnt component in intestinal organoids (Figure 7A). To address the hypothesis that Wnt signaling is pathogenically hyperactive in celiac disease, we analyzed the directionality of the gene expression, and indeed found this to be the case. Of the 195 genes repressed by the gluten challenge, 48 (24.6%) belong to the group of genes induced by Wnt inhibition (Figure 7B). Reciprocally, only 5.9% (n = 13 of 222) of the genes upregulated by the gluten challenge are also induced in Wnt inhibition (Figure 7C). Reactome pathway analysis reveals that these 13 oppositely responding genes belong in the immune response category (Figure 8), which was specifically induced in the PGC group (Figure 7F). Interestingly, on the one hand, when comparing celiac disease patients on a GFD with DC patients, a similar kind of overactive Wnt component is also evident, as 28.2% (n = 33 of 117) of genes downregulated in the GFD group compared with the DC group belong to the Wnt inhibition induced Wnt component (Figure 7B). On the other hand, only 2% (n = 1 of 50) of genes upregulated by the gluten challenge belong to the Wnt inhibition induced Wnt component (Figure 7C). Moreover, a similar trend can be seen in genes downregulated by Wnt inhibition, as 13.1% (n = 29 of 222) of genes upregulated by a gluten challenge belong to the Wnt component repressed by Wnt inhibition (Figure 7D). Correspondingly, not a single gene of the downregulated gene group affected by the gluten challenge falls into the category of Wnt component repressed by Wnt inhibition (Figure 7E). To conclude, our data suggest that in active celiac disease, in addition to activation of immune response genes, Wnt signaling is conspicuously overactive in the small intestine, favoring immature crypt gene expression at the expense of less differentiated epithelium. Interestingly, this pathogenic “Wnt on” state, excluding the inflammatory component, seems to already be active in celiac patients on a GFD. Moreover, while different set of genes are affected in GFD vs DC and GFD vs PGC comparisons, they all seem to be, however, under the influence of Wnt signaling.
Figure 7.
Hyperactive Wnt component in celiac disease. Venn diagrams of (A) all differentially expressed genes in intestinal organoids after Wnt inhibition (blue spheres) and GFD/DC (yellow spheres) and PGC/GFD (orange spheres) comparisons. (B) Upregulated genes after Wnt inhibition and downregulated genes indicated by comparisons. (C) Upregulated genes after Wnt inhibition and upregulated genes indicated by comparisons. (D) Downregulated genes after Wnt inhibition and upregulated genes indicated by comparisons. (E) Downregulated genes after Wnt inhibition and downregulated genes indicated by comparisons. (F) The immune response category genes that are specifically induced only in the PGC group.
Figure 8.
Reactome pathway enrichment analysis of 13 oppositely responding genes (seeFigure 7F). ER, endoplasmic reticulum; MHC, major histocompatibility complex.
A Gluten Challenge Induces a Secretory Cell Type Signature and Inhibits Absorptive Enterocyte Maturation
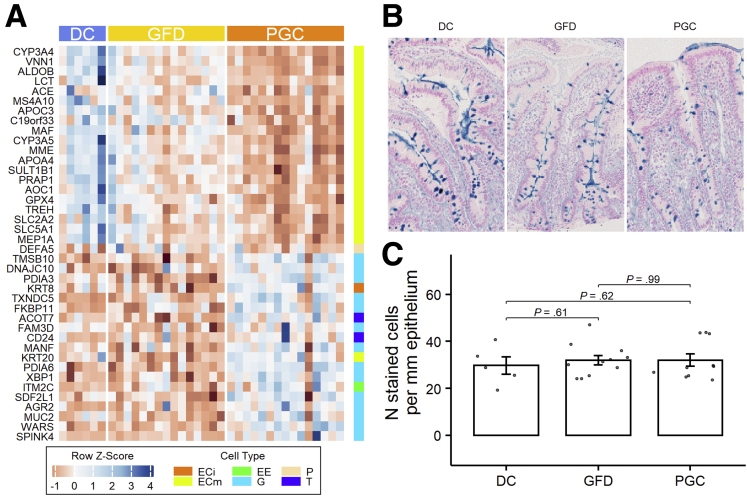
Differentiation of both absorptive and secretory epithelial cells has been shown to be affected in celiac disease.28,29 When comparing our biopsy transcriptomic data to published data on single-cell RNA-Seq studies on the small intestinal epithelium,30 we found that mature absorptive enterocyte lineage genes were almost unanimously downregulated in PGC patients. Interestingly, however, we found that genes for secretory cell signatures were upregulated during the gluten challenge (Figure 9A), and most of these genes belong to the goblet cell signature. However, the number of goblet cells per millimeter of epithelium was not significantly different between the groups (Figure 9B and C).
Figure 9.
Dietary gluten inhibits absorptive cell-specific gene expression and evokes secretory cell type gene expression. (A) Gene expression heatmap of the intestinal epithelial cell type–specific genes. (B) Alcian blue G staining in DC, GFD, and PGC patients at 40× magnification. (C) Number of G per mm of epithelium. ECi, immature enterocytes; ECm, mature enterocytes; EE, enteroendocrine cells; G, goblet cells; P, Paneth cells; T, tuft cell.
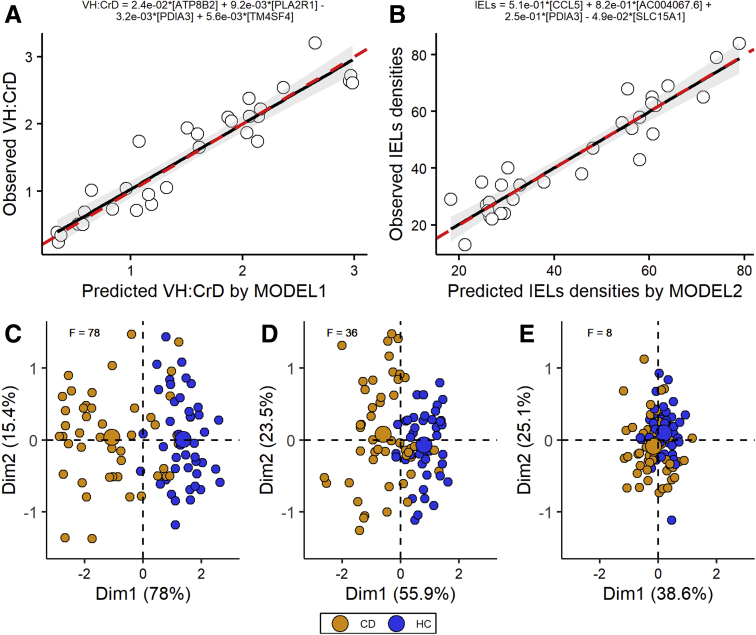
Molecular Morphometric Regression Models
Part of the differentially expressed gene expressions have high correlations with the extent of gluten-induced histological damage and inflammation and are presented as heatmaps in Figure 10. The subsequent best subset selection approach computationally allowed us to reduce the number of genes, which describes VH:CrD (model 1) and IEL density (model 2) changes to 4 genes for each model (Table 2). Created models describe 97.2% of observed VH:CrD and 98.0% of IEL number variabilities, and there is a strong correlation between the predicted and observed ratios (Figures 11A and B). Subsequently, we tested the ability of our models to discriminate between healthy controls and celiac patients on independent data. The PCA was performed on dataset GSE13490031 (Figure 11C–E). F statistics suggest that by using the expression of 4 genes predicted by our models to describe the variance in histomorphology parameters we are able to discriminate between healthy and celiac disease patients better than by using the expression of 4 randomly selected genes (F = 78 and 36 for VH:CrD and IEL density, respectively, compared with F = 8 for randomly selected genes).
Figure 10.
The gene expression correlations with the extent of (A) gluten-induced histological damage (Spearman’s rank, |ρ| > .65) and (B) inflammation (Spearman’s rank, |ρ| > .6) are presented as heatmaps.
Table 2.
Model 1 (VH:CrD) and Model 2 (CD3+ IEL Density) Coefficients
| Term | Estimate | P value |
|---|---|---|
| Model 1 (VH:CrD prediction) | ||
| ATP8B2 | 0.024 | 6.46 × 10–3 |
| PLA2R1 | 0.009 | 7.76 × 10–3 |
| PDIA3 | –0.003 | 6.92 × 10–3 |
| TM4SF4 | 0.006 | 1.01 × 10–5 |
| Model 2 (IEL density prediction) | ||
| CCL5 | 0.51 | 3.39 × 10–5 |
| AC004067.6 | 0.82 | 1.05 × 10–5 |
| PDIA3 | 0.25 | 7.68 × 10–6 |
| SLC15A1 | –0.05 | 1.37 × 10–6 |
IEL, intraepithelial lymphocyte; VH:CrD, villus height-to-crypt depth ratio.
Figure 11.
Observed vs predicted regression scatter plots (A) for model 1 predicting VH:CrD and (B) for model 2 predicting density of IELs. Red dashed line represents ideal regression case, where x = y. (C–E) For testing, a graphical representation of PCAs (Dim1 and Dim2) applied to expression of 4 genes involved in (C) model 1, (D) model 2, and (E) 4 other random genes selected from the pool of the all detected genes extracted from publicly available dataset GSE134900 is shown. CD, celiac disease; EC, epithelial cell; F, F statistics; HC, healthy control.
Discussion
To our understanding, this is the first genome-wide RNA-Seq study with paired duodenal biopsies from celiac disease patients on strict long-term GFDs before and after 10 weeks of a gluten challenge. We strived to pinpoint not only the early gluten-dependent gene transcripts but also genes that are constantly differentially expressed in celiac disease patients vs DC patients. The present results show that at the molecular level, healthy intestinal functions were not reinstated upon long-term GFD restrictions. Our novel finding is that genes, particularly those encoding proteins for transporting small molecules, were significantly less expressed in patients on a GFD compared with healthy individuals, which can contribute to micronutrient deficiency. For example, we show downregulation of 2 heme transporter expressions (FLVCR1 and SLC46A1) and upregulation of transferrin receptor (TFRC), the gene responsible for iron delivery from transferrin to the cell. The TFRC gene is also known to mediate the IgA-mediated gliadin transport during celiac disease pathogenesis.32,33 Related to folate deficiency, we found significant downregulation of the gene SLC46A1, which encodes a folate transporter. Linked to calcium and zinc deficiency, genes involved in Ca2+ homeostasis (ATP2B1 and CALM1) and the zinc transporter SLC39A4 showed decreased levels of expression in patients on a GFD compared with healthy control subjects. Our data suggest that the low expression of micronutrient transporters in the intestine explains described clinical micronutrient deficiencies (ie, iron, folate, B12, and various mineral and vitamin D deficiencies) in patients following a strict GFD.25,34, 35, 36 The low expressions might be due to inadvertent gluten ingestion and indication of disease due to early mucosal injury. Complete elimination of dietary gluten seems impossible to maintain due to issues such as contamination of gluten-free foods, resulting in the ingestion of hundreds of milligrams of gluten per day in an attempted GFD.34, 35, 36, 37 Inadvertent gluten ingestion is clinically difficult to detect, as serum TG2 or endomysial antibodies do not pick up most of the patients with persistent villous atrophy on GFDs.38 When the stool GIPs measure gluten ingestion the day or so before testing, the duodenal mucosa is reacting upon long-term and significant inadvertent gluten ingestion. Recently, quantitative histology with well-oriented biopsy sections was shown to reveal villus atrophy in the majority of patients with celiac disease compliant with well-controlled GFDs.39 The celiac disease patients in the present study did not reach a duodenal mucosal healing described as fully normal.40 Even if our 6 control patients without evidence of celiac disease had morphologically similar small intestinal mucosal findings, they clearly differed from celiac disease patients on a GFD as related to the transcriptomic findings. Furthermore, our rather small control patient group had a reason for endoscopy and, in fact, comprised DC patients, which may mean they were not fully “normal.” When the duodenal mucosa in celiac disease evidence “normalization” upon a GFD, there is still disease at a molecular and even at a clinical level. It is intriguing to speculate that transcriptomic changes such as those seen in the present study in celiac disease patients on a strict GFD could also occur in latent celiac disease (ie, patients with existing gluten-dependent disease in which the disease is not yet manifested at the mucosal level). Such patients “excluded on biopsy” for celiac disease have been shown to have micronutrient deficiencies and even bone disease with fractures, and their duodenal mucosa may deteriorate to a state typical for celiac disease only later, which includes crypt hyperplastic lesion with villous atrophy.41, 42, 43, 44
Our paired analysis indicates that genes specific for absorptive enterocytes were almost unanimously downregulated in celiac disease patients after a gluten challenge. This has also been shown in other studies where active celiac disease patients were compared with healthy control subjects or celiac patients on a GFD in a nonpaired manner.16,18 Deficient enterocyte differentiation has been suggested to be ascribed to hyperactive Wnt signaling in the disease, as Wnt signaling needs to be inhibited for enterocyte differentiation to occur.45 In fact, the number of cells expressing nuclear beta-catenin in the crypt, as a proxy for active Wnt signaling, has been shown to be increased along with expression of a few studied Wnt target genes in active celiac disease.15,26 We addressed this by comparing the RNA-Seq gene expression data from patients with our Wnt component gene expression data from intestinal organoids.27 Our data show that in active celiac disease Wnt signaling is clearly hyperactive, which is most probably causing the compromised absorptive cell differentiation. In addition, this pathogenic hyperactive Wnt signaling state (excluding the inflammatory component) seems to already be active in celiac disease patients on a GFD. In fact, contrary to compromised absorptive cell differentiation, we saw that secretory cell–type gene expression is induced after a gluten challenge. The majority of upregulated secretory cell line markers in our datasets belong to goblet cells. A reduction in the number of goblet cells has been reported in active celiac disease,46,47 but counterintuitively, some of the goblet cell markers, including SPINK448 and MUC2,49 are upregulated. Moreover, in vitro studies with organoids have shown that intestinal organoids derived from celiac disease patients are more poised for secretory cell differentiation. For example, MUC2 was expressed more in organoids derived from active celiac disease patients compared with organoids derived from non–celiac disease individuals.29 Our genome-wide in vivo data shows that secretory cell markers, mainly those of the goblet cells, are consistently upregulated in celiac disease after a gluten challenge. Contrary to absorptive cell differentiation, Notch signaling needs to be off and Wnt signaling needs to be on for secretory cell differentiation to take place.50 Because it is known that proinflammatory cytokines can induce genes involved in the immune response in intestinal enterocytes,51 one may argue that these cytokines are the primary cause for the upregulation of secretory cell markers in active celiac disease. Th1 and Th2 inflammatory cytokines are known to induce goblet cell differentiation through activation of the PI3K/Akt pathways.51,52 Celiac disease in its active phase is characterized by activation of the Th1 gene expression profile.53 Tumor necrosis factor α, interleukin-4, and interleukin-13 have been shown to induce the expression of MUC2 via nuclear factor-κB mediated by mitogen-activated protein kinase pathways in vitro.54,55 There is evidence that over-production of MUC2 in inflammatory bowel disease depletes goblet cells. In vitro studies showed that mucus hypersecretion caused severe endoplasmic reticulum stress and apoptosis of goblet cells.56 This could possibly explain our finding that in celiac disease after a gluten challenge the expression of MUC2 and other goblet cell markers are increased without any changes in goblet cell numbers. To conclude, in active celiac disease, hyperactive Wnt signaling seems to impede absorptive cell differentiation, but at the same time, together with inflammatory cytokines, it might poise secretory cell differentiation.
The basis for a celiac disease diagnosis is the characteristic gluten-triggered and dependent small intestinal mucosal lesion (villous atrophy with crypt hyperplasia), which is most often associated with inflammation seen as an increased intraepithelial T cell density. Traditional celiac disease diagnostics based on grouped classifications (so-called Marsh-Oberhuber classes) are highly subjective, and interobserver reproducibility between pathology readers is not optimal.57, 58, 59 The statement by Picarelli et al,60 in which they stressed the limits of histological interpretation due to a lack of uniformity in the use of Marsh-Oberhuber classification, is still relevant as evidenced by a recent 33-celiac center study in which one reader evaluated the mucosa to be Marsh 0 and the other graded it from Marsh 1 to 3c and vice versa—Supplementary Table 20 in Werkstetter et al.61 In the present study, we adopted our standard operating procedures and validated quantitative morphometry separately for biopsy morphology and inflammation to measure the continuum of gluten-induced mucosal injury in celiac disease.13,21,24,62 These methods were also adopted in recent clinical drug and vaccine trials.22,23,39 In the present study, we show that the expression of a selected subset of protein-coding genes correlates extremely well with the extent of morphological damage and inflammation in control patients and celiac disease patients on a GFD before and after the gluten challenge (presented as heatmaps in the order of VH:CrD and IEL density in Figure 10). It should be noted that the RNA was extracted from separate cuttings but from the same paraffin-embedded biopsy blocks that had been used for morphometry measurements, thus avoiding the patchy lesion pitfalls observed between biopsies in short gluten challenges using low and moderate amounts of gluten. Gene expression seems to determine the state of a tissue, and the changes in expression are associated with the severity of the injury. We have already, as a proof of concept, moved toward “molecular histomorphometry” using a candidate gene approach.24 Also, other attempts to build a molecular model for grouped mucosal injury classification have been presented.18,63 In our study, we modeled the behavior of continuous metric parameters (VH:CrD and the density of CD3+ IELs), which were assigned to each endoscopic biopsy, and a multiple regression analysis was applied. In conclusion, we show that by using transcriptomic data one may predict histomorphological parameters with a high accuracy. In fact, the subset selection approach computationally reduced the number of genes to 4 for both the morphology and inflammation predictions. The created models described 97.2% of observed VH:CrD and 98.0% of the IEL densities; we show a strong correlation between the predicted and observed ratios. We foresee this approach to be exploited in celiac disease diagnostics and in drug trials monitoring the efficacy of novel treatments focused on circumventing subjective biopsy readings.
We acknowledge the present study limitations. The present results are drawn from a rather small patient group. Unfortunately, we cannot provide evidence that observed low expression of micronutrient transport genes in long-term-treated celiac disease patients correlates with nutritional deficiencies, as we do not have the micronutrient or protein level data from these patients. Also, on the one hand, our designed molecular morphometry models are based on RNA-Seq data, and additional validation with reverse-transcription quantitative polymerase chain reaction is warranted in a separate clinical gluten challenge study. On the other hand, we provide genome-wide duodenal biopsy transcriptomic data from a human source in which gene expression correlated with the extent of histological injury continuum studied in DC patients and celiac disease patients on a strict GFD and after a gluten challenge. It should be noted that we addressed gluten-induced transcriptomic changes in respect to morphology and inflammation after a 10-week gluten challenge using small and moderate amount (2–4 g) of gluten and did not study, for example, newly diagnosed celiac disease patients who had ingested 10—20 g of gluten for decades. However, we were able to test our data with the published independent data (dataset GSE134900),31 suggesting that our regression model could be applicable for newly diagnosed celiac disease patients. In addition, our study design can exclude patients with the highest and most acute response to gluten with even lower and shorter gluten challenge.
To conclude, we provide transcriptomic resource data not only to determine gluten-dependent morphology and inflammation in celiac disease but also to pinpoint genes potentially important in pathogenesis, in finding genes for novel drug targets, and for biomarker and surrogate marker research. We further show that even a long and strict GFD is not sufficient to abate relatively common micronutrient deficiencies among celiac disease patients. Adherence to a strict GFD can be onerous to maintain, and because contamination of gluten-free foods is common, complete elimination of dietary gluten is difficult, if not impossible, to attain. Our results describe aforementioned adverse conditions at the molecular level and warrant the pursuit of novel adjunct treatments for celiac disease.
Materials and Methods
Patients and Biopsies
Fifteen adult HLA-DQ2 positive patients having a biopsy-proven celiac disease diagnosis were enrolled for this study.23,24 The self-reported strict GFD for at least 1 year and for 16 years on average was checked on site and the patients were found to be symptom free and they were also tested and shown to be negative for serum TG2 antibodies (GFD group). These patients were challenged, that is, consumed 2–4 g of gluten daily for 10 weeks (PGC group). The patients continued their GFD throughout the gluten challenge study, and cookies containing 1–2 g of gluten were dispensed to the patients for twice daily consumption. The ingestion of the cookies was monitored, and unused cookies were returned and counted. Diet counseling was provided if transgression in the diet was suspected. The DC group included duodenal biopsies from 6 non–celiac disease individuals undergoing endoscopies due to dyspepsia or reflux symptoms. These patients had no detectable disease and were also negative for serum TG2 autoantibodies (DC group). Before starting a gluten challenge, stool samples were collected to measure potential evidence of inadvertent gluten ingestion by using iVYLISA, a GIP assay, as described previously.34,64,65 The GIP stool testing was continued throughout the gluten challenge period (gluten stool contents are shown in Figure 2).
Upper gastrointestinal endoscopies were performed, and duodenal forceps biopsies were immersed in PaxFPE (PAXgene fixative and processed for paraffin block embedding) using a standard formalin-free paraffin-infiltration protocol. For morphology, samples were stained with hematoxylin and eosin and measured using our validated morphometry rules for VH, CrD, and VH:CrD. To measure mucosal inflammation, CD3+ IELs were counted and reported as a density of positive T cells per 100 epithelial cells, as previously described.24,62
RNA Isolation and Sequencing
Total RNA was extracted from the same duodenal PaxFPE biopsy specimens using further cuttings from where histomorphometry was to be measured and counted as previously described.24 For the extraction, an RNeasy Kit (Qiagen, Hilden, Germany) was used according to the manufacturer’s instructions. Library preparation and next-generation sequencing (NGS) was performed by Qiagen NGS Service. The library preparation was done using the QIAseq UPX 3′ Transcriptome Kit (Qiagen). A total of 10 ng of purified RNA was converted into complementary DNA NGS libraries, and a quality check was performed using TapeStation 4200 (Agilent, Santa Clara, CA) or Agilent Bioanalyzer (Agilent). Sequencing was performed on a NextSeq500 sequencing instrument according to the manufacturer’s instructions (Illumina, San Diego, CA).
RNA-Seq Data Analyses
Data quality was checked using the FastQC (Illumina). Only reads with an exact cell indices match were retained, 3′ poly-A regions were trimmed and reads with <25 bp were dropped. Reads were aligned to the human GRCh38 genome (GENCODE Release 23 genome assembly version on all regions) using splice aware aligner STAR (Spliced Transcripts Alignment to a Reference). A secondary differential expression analysis involving normalization of unique molecular identifier counts and a subsequent pairwise differential regulation analysis was done using DESeq2 package (Bioconductor, Buffalo, NY). The paired nature of samples was considered for detection of differentially expressed genes when PGC and GFD were compared. Obtained P values were adjusted for multiple testing using the Benjamini-Hochberg method.
Comparisons With Publicly Available Datasets
We took advantage of the findings of differentially expressed genes from our previous study (publicly available dataset GSE78761), obtained from mouse intestinal organoids treated with Wnt inhibitor IWP2 using Gro-Seq (global run-on sequencing) from our previous study27 (GSE78761). To detect genes directly or indirectly related to Wnt pathway, we manually checked the genes expressed differentially in GSE78761 for intersections with the differentially expressed genes obtained from RNA-Seq in the current study. We also compared our biopsy transcriptomic data to published data on single-cell RNA-seq studies on the small intestinal epithelium (GSE92332).30
Alcian Blue Staining
Tissue sections were stained with 1% (w/v) Alcian blue 8GX (Sigma-Aldrich, St Louis, MO) in 3 % (v/v) acetic acid for 30 minutes at room temperature and washed in tap water. The sections were counterstained with nuclear fast red (Sigma-Aldrich) for 3 minutes and dehydrated in an ascending series of ethanol, cleared in xylene, and mounted.
Statistics and Regression Model
A 2-tailed paired Student's t test was used to compare the differences between the PGC and GFD groups and 2-tailed unpaired Student's t test for the PGC vs DC and GFD vs DC groups comparisons. A Spearman’s rank correlation coefficient (ρ) was calculated to examine the relationships between the unique molecular identifier counts for differentially expressed genes in transcriptomes from celiac disease cohort patients (GFD and PCG groups) and the VH:CrD or density of IELs. Regression models were created by the best subset selection approach.66 Because the number of possible combinations for identified differential expressed genes was too large to iterate through all of them, the cutoff for |ρ| was set to be .65 for VH:CrD correlation and .6 for density of IELs correlation to reduce the starting gene pool to reasonable number. A total of 112 genes were used for VH:CrD and 100 genes were used for IEL density predictions. Among all possible combinations of genes from 1 to 6 fitted by a multiple linear regression, the best ones based on their cross validated prediction error, Bayesian information criterion, and adjusted R2 were selected.67 Models were evaluated by observed vs predicted regression.68 As an additional testing step, PCA was performed on independent and publicly available dataset GSE134900.31 To quantify the separation between healthy control and celiac disease groups in this independent dataset, the F statistics metric was applied for 4-dimensional data.69 All statistical testing was performed using R version 3.5.3 (R Foundation for Statistical Computing, Vienna, Austria).
Ethics Approval
The study was approved by the Ethics Committee of the Tampere University Hospital, Tampere, Finland (ETL-code R15185M). The patients gave their written consent to perform the study. The trial was registered at ClinicalTrials.gov (NCT02637141) and EudraCT (2015-003647-19).
All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
We thank patients participating for making this study possible. We also thank the expert staff for their participation in the sample collection.
Footnotes
CRediT Authorship Contributions Valeriia Dotsenko (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Investigation: Lead; Validation: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Mikko Oittinen (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Investigation: Lead; Validation: Lead; Writing – original draft: Equal; Writing – review & editing: Equal)
Juha Taavela (Data curation: Equal; Investigation: Equal; Resources: Supporting; Writing – review & editing: Supporting)
Alina Popp (Data curation: Supporting; Formal analysis: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Markku Peräaho (Investigation: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Synnöve Staff (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting)
Jani Sarin, MSc (Formal analysis: Supporting; Investigation: Supporting; Writing – review & editing: Supporting)
Francisco Leon (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Jorma Isola (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Resources: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Markku Mäki (Conceptualization: Equal; Formal analysis: Supporting; Funding acquisition: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Keijo Viiri (Conceptualization: Lead; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Lead; Project administration: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Conflicts of interest These authors disclose the following: Francisco Leon was the chief executive and medical officer of Celimmune during the conduct of the study, and currently is chief scientific officer at Provention Bio. Markku Mäki is an advisor or consultant for Janssen Research and Development, Dr. Falk Pharma, ImmunogenX, Actobio Therapeutics, Takeda, Jilab, Applied Molecular Transport, and Ukko, all outside the submitted work. Jorma Isola is the chief execute officer of Jilab Inc that produces diagnostic laboratory services for small-bowel diseases and a wide variety of cancers. Jani Sarin is an employee of Jilab Inc. The remaining authors disclose no conflicts.
Funding This work was supported by the Academy of Finland (no. 310011), Tekes (Business Finland) (no. 658/31/2015), Paediatric Research Foundation, Sigrid Jusélius Foundation, Mary och Georg C. Ehrnrooths Stiftelse, Laboratoriolääketieteen Edistämissäätiö sr, and Competitive State Research Financing of the Expert Responsibility Area of Tampere University Hospital (Grant Nos. 9V041, 9X035, and 9AA053). The funding sources played no role in the design or execution of this study or in the analysis and interpretation of the data.
Supplementary Material
Differential Intestinal Mucosal Gene Transcriptome for PGC vs GFD and PGC vs DC Group Comparisons.
References
- 1.Singh P., Arora A., Strand T.A., Leffler D.A., Catassi C., Green P.H., Kelly C.P., Ahuja V., Makharia G.K. Global prevalence of celiac disease: systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2018;16:823–836. doi: 10.1016/j.cgh.2017.06.037. [DOI] [PubMed] [Google Scholar]
- 2.Mustalahti K., Catassi C., Reunanen A., Fabiani E., Heier M., McMillan S., Murray L., Metzger M.H., Gasparin M., Bravi E., Mki M. The prevalence of celiac disease in Europe: Results of a centralized, international mass screening project. Ann Med. 2010;42:587–595. doi: 10.3109/07853890.2010.505931. [DOI] [PubMed] [Google Scholar]
- 3.Lohi S., Mustalahti K., Kaukinen K., Laurila K., Collin P., Rissanen H., Lohi O., Bravi E., Gasparin M., Reunanen A., Mäki M. Increasing prevalence of coeliac disease over time. Aliment Pharmacol Ther. 2007;26:1217–1225. doi: 10.1111/j.1365-2036.2007.03502.x. [DOI] [PubMed] [Google Scholar]
- 4.Trynka G., Hunt K.A., Bockett N.A., Romanos J., Mistry V., Szperl A., Bakker S.F., Bardella M.T., Bhaw-Rosun L., Castillejo G., De La Concha E.G., De Almeida R.C., Dias K.R.M., Van Diemen C.C., Dubois P.C.A., Duerr R.H., Edkins S., Franke L., Fransen K., Gutierrez J., Heap G.A.R., Hrdlickova B., Hunt S., Izurieta L.P., Izzo V., Joosten L.A.B., Langford C., Mazzilli M.C., Mein C.A., Midah V., Mitrovic M., Mora B., Morelli M., Nutland S., Núñez C., Onengut-Gumuscu S., Pearce K., Platteel M., Polanco I., Potter S., Ribes-Koninckx C., Ricaño-Ponce I., Rich S.S., Rybak A., Santiago J.L., Senapati S., Sood A., Szajewska H., Troncone R., Varadé J., Wallace C., Wolters V.M., Zhernakova A., Thelma B.K., Cukrowska B., Urcelay E., Bilbao J.R., Mearin M.L., Barisani D., Barrett J.C., Plagnol V., Deloukas P., Wijmenga C., Van Heel D.A. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green P.H.R., Cellier C. Medical progress: celiac disease. N Engl J Med. 2007;357:1731–1743. doi: 10.1056/NEJMra071600. [DOI] [PubMed] [Google Scholar]
- 6.Schuppan D., Junker Y., Barisani D. Celiac disease: from pathogenesis to novel therapies. Gastroenterology. 2009;137:1912–1933. doi: 10.1053/j.gastro.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Sollid L.M., Jabri B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat Rev Immunol. 2013;13:294–302. doi: 10.1038/nri3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valitutti F., Cucchiara S., Fasano A. Celiac disease and the microbiome. Nutrients. 2019;11:2403. doi: 10.3390/nu11102403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jabri B., Sollid L.M. T cells in celiac disease. J Immunol. 2017;198:3005–3014. doi: 10.4049/jimmunol.1601693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sollid L.M., Tye-Din J.A., Qiao S.W., Anderson R.P., Gianfrani C., Koning F. Update 2020: nomenclature and listing of celiac disease–relevant gluten epitopes recognized by CD4+ T cells. Immunogenetics. 2020;72:85–88. doi: 10.1007/s00251-019-01141-w. [DOI] [PubMed] [Google Scholar]
- 11.Mäki M. Oak Tree Press; Dublin, Ireland: 1994. Autoantibodies as markers of autoimmunity in coeliac disease pathogenesis. [Google Scholar]
- 12.Mäki M. The humoral immune system in coeliac disease. Baillieres Clin Gastroenterol. 1995;9:231–249. doi: 10.1016/0950-3528(95)90030-6. [DOI] [PubMed] [Google Scholar]
- 13.Garber M.E., Saldanha A., Parker J.S., Jones W.D., Kaukinen K., Laurila K., Lähdeaho M.-L., Khatri P., Khosla C., Adelman D.C., Mäki M. A B-cell gene signature correlates with the extent of gluten-induced intestinal injury in celiac disease. Cell Mol Gastroenterol Hepatol. 2017;4:1–17. doi: 10.1016/j.jcmgh.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.du Pré M.F., Blazevski J., Dewan A.E., Stamnaes J., Kanduri C., Sandve G.K., Johannesen M.K., Lindstad C.B., Hnida K., Fugger L., Melino G., Qiao S.W., Sollid L.M. B cell tolerance and antibody production to the celiac disease autoantigen transglutaminase 2. J Exp Med. 2020;217 doi: 10.1084/jem.20190860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juuti-Uusitalo K., Mäki M., Kainulainen H., Isola J., Kaukinen K. Gluten affects epithelial differentiation-associated genes in small intestinal mucosa of coeliac patients. Clin Exp Immunol. 2007;150:294–305. doi: 10.1111/j.1365-2249.2007.03500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bragde H., Jansson U., Fredrikson M., Grodzinsky E., Söderman J. Celiac disease biomarkers identified by transcriptome analysis of small intestinal biopsies. Cell Mol Life Sci. 2018;75:4385–4401. doi: 10.1007/s00018-018-2898-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonard M.M., Bai Y., Serena G., Nickerson K.P., Camhi S., Sturgeon C., Yan S., Fiorentino M.R., Katz A., Nath B., Richter J., Sleeman M., Gurer C., Fasano A. RNA sequencing of intestinal mucosa reveals novel pathways functionally linked to celiac disease pathogenesis. PLoS One. 2019;14 doi: 10.1371/journal.pone.0215132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loberman-Nachum N., Sosnovski K., Di Segni A., Efroni G., Braun T., BenShoshan M., Anafi L., Avivi C., Barshack I., Shouval D.S., Denson L.A., Amir A., Unger R., Weiss B., Haberman Y. Defining the celiac disease transcriptome using clinical pathology specimens reveals biologic pathways and supports diagnosis. Sci Rep. 2019;9:16163. doi: 10.1038/s41598-019-52733-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tutturen A.E.V.V., Dørum S., Clancy T., Reims H.M., Christophersen A., Lundin K.E.A.A., Sollid L.M., de Souza G.A., Stamnaes J. Characterization of the small intestinal lesion in celiac disease by label-free quantitative mass spectrometry. Am J Pathol. 2018;188:1563–1579. doi: 10.1016/j.ajpath.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Jimenez N., Garcia-Etxebarria K., Plaza-Izurieta L., Romero-Garmendia I., Jauregi-Miguel A., Legarda M., Ecsedi S., Castellanos-Rubio A., Cahais V., Cuenin C., Degli Esposti D., Irastorza I., Hernandez-Vargas H., Herceg Z., Bilbao J.R. The methylome of the celiac intestinal epithelium harbours genotype-independent alterations in the HLA region. Sci Rep. 2019;9:1298. doi: 10.1038/s41598-018-37746-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lähdeaho M.L., Mäki M., Laurila K., Huhtala H., Kaukinen K. Small- bowel mucosal changes and antibody responses after low- and moderate-dose gluten challenge in celiac disease. BMC Gastroenterol. 2011;11:129. doi: 10.1186/1471-230X-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lähdeaho M.-L., Kaukinen K., Laurila K., Vuotikka P., Koivurova O.-P., Kärjä-Lahdensuu T., Marcantonio A., Adelman D.C., Mäki M. Glutenase ALV003 attenuates gluten-induced mucosal injury in patients with celiac disease. Gastroenterology. 2014;146:1649–1658. doi: 10.1053/j.gastro.2014.02.031. [DOI] [PubMed] [Google Scholar]
- 23.Lähdeaho M.L., Scheinin M., Vuotikka P., Taavela J., Popp A., Laukkarinen J., Koffert J., Koivurova O.P., Pesu M., Kivelä L., Lovró Z., Keisala J., Isola J., Parnes J.R., Leon F., Mäki M. Safety and efficacy of AMG 714 in adults with coeliac disease exposed to gluten challenge: a phase 2a, randomised, double-blind, placebo-controlled study. Lancet Gastroenterol Hepatol. 2019;4:948–959. doi: 10.1016/S2468-1253(19)30264-X. [DOI] [PubMed] [Google Scholar]
- 24.Taavela J., Viiri K., Popp A., Oittinen M., Dotsenko V., Peraäho M., Staff S., Sarin J., Leon F., Mäki M., Isola J. Histological, immunohistochemical and mRNA gene expression responses in coeliac disease patients challenged with gluten using PAXgene fixed paraffin-embedded duodenal biopsies. BMC Gastroenterol. 2019;19:189. doi: 10.1186/s12876-019-1089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rondanelli M., Faliva M.A., Gasparri C., Peroni G., Naso M., Picciotto G., Riva A., Nichetti M., Infantino V., Alalwan T.A., Perna S. Micronutrients dietary supplementation advices for celiac patients on long-term gluten-free diet with good compliance: a review. Medicina (Kaunas) 2019;55:337. doi: 10.3390/medicina55070337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senger S., Sapone A., Fiorentino M.R., Mazzarella G., Lauwers G.Y., Fasano A. Celiac disease histopathology recapitulates hedgehog downregulation, consistent with wound healing processes activation. PLoS One. 2015;10 doi: 10.1371/journal.pone.0144634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oittinen M., Popp A., Kurppa K., Lindfors K., Mäki M., Kaikkonen M., Viiri K. Polycomb repressive complex 2 enacts Wnt signaling in intestinal homeostasis and contributes to the instigation of stemness in diseases entailing epithelial hyperplasia or neoplasia. Stem Cells. 2017;35:445–457. doi: 10.1002/stem.2479. [DOI] [PubMed] [Google Scholar]
- 28.Piscaglia A.C., Rutella S., Laterza L., Cesario V., Campanale M., Cazzato I.A., Ianiro G., Barbaro F., Di Maurizio L., Bonanno G., Cenci T., Cammarota G., Larocca L.M., Gasbarrini A. Circulating hematopoietic stem cells and putative intestinal stem cells in coeliac disease. J Transl Med. 2015;13:220. doi: 10.1186/s12967-015-0591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freire R., Ingano L., Serena G., Cetinbas M., Anselmo A., Sapone A., Sadreyev R.I., Fasano A., Senger S. Human gut derived-organoids provide model to study gluten response and effects of microbiota-derived molecules in celiac disease. Sci Rep. 2019;9:7029. doi: 10.1038/s41598-019-43426-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., Tirosh I., Beyaz S., Dionne D., Zhang M., Raychowdhury R., Garrett W.S., Rozenblatt-Rosen O., Shi H.N., Yilmaz O., Xavier R.J., Regev A. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abadie V., Kim S.M., Lejeune T., Palanski B.A., Ernest J.D., Tastet O., Voisine J., Discepolo V., Marietta E.V., Hawash M.B.F., Ciszewski C., Bouziat R., Panigrahi K., Horwath I., Zurenski M.A., Lawrence I., Dumaine A., Yotova V., Grenier J.-C., Murray J.A., Khosla C., Barreiro L.B., Jabri B. IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature. 2020;578:600–604. doi: 10.1038/s41586-020-2003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ménard S., Cerf-Bensussan N., Heyman M. Multiple facets of intestinal permeability and epithelial handling of dietary antigens. Mucosal Immunol. 2010;3:247–259. doi: 10.1038/mi.2010.5. [DOI] [PubMed] [Google Scholar]
- 33.Matysiak-Budnik T., Moura I.C., Arcos-Fajardo M., Lebreton C., Ménard S., Candalh C., Ben-Khalifa K., Dugave C., Tamouza H., van Niel G., Bouhnik Y., Lamarque D., Chaussade S., Malamut G., Cellier C., Cerf-Bensussan N., Monteiro R.C., Heyman M. Secretory IgA mediates retrotranscytosis of intact gliadin peptides via the transferrin receptor in celiac disease. J Exp Med. 2008;205:143–154. doi: 10.1084/jem.20071204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Syage J.A., Kelly C.P., Dickason M.A., Ramirez A.C., Leon F., Dominguez R., Sealey-Voyksner J.A. Determination of gluten consumption in celiac disease patients on a gluten-free diet. Am J Clin Nutr. 2018;107:201–207. doi: 10.1093/ajcn/nqx049. [DOI] [PubMed] [Google Scholar]
- 35.Kreutz J.M., Adriaanse M.P.M., van der Ploeg E.M.C., Vreugdenhil A.C.E. Narrative review: nutrient deficiencies in adults and children with treated and untreated celiac disease. Nutrients. 2020;12:500. doi: 10.3390/nu12020500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vici G., Belli L., Biondi M., Polzonetti V. Gluten free diet and nutrient deficiencies: a review. Clin Nutr. 2016;35:1236–1241. doi: 10.1016/j.clnu.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Melini V., Melini F. Gluten-free diet: gaps and needs for a healthier diet. Nutrients. 2019;11:170. doi: 10.3390/nu11010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silvester J.A., Kurada S., Szwajcer A., Kelly C.P., Leffler D.A., Duerksen D.R. Tests for serum transglutaminase and endomysial antibodies do not detect most patients with celiac disease and persistent villous atrophy on gluten-free diets: a meta-analysis. Gastroenterology. 2017;153:689–701. doi: 10.1053/j.gastro.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daveson A.J.M., Popp A., Taavela J., Goldstein K.E., Isola J., Truitt K.E., Mäki M., Anderson R.P., Adams A., Andrews J., Behrend C., Brown G., Chen Yi Mei S., Coates A., Daveson A.J., DiMarino A., Elliott D., Epstein R., Feyen B., Fogel R., Friedenberg K., Gearry R., Gerdis M., Goldstein M., Gupta V., Holmes R., Holtmann G., Idarraga S., James G., King T., Klein T., Kupfer S., Lebwohl B., Lowe J., Murray J., Newton E., Quinn D., Radin D., Ritter T., Stacey H., Strout C., Stubbs R., Thackwray S., Trivedi V., Tye-Din J., Weber J., Wilson S. Baseline quantitative histology in therapeutics trials reveals villus atrophy in most patients with coeliac disease who appear well controlled on gluten-free diet. GastroHep. 2020;2:22–30. [Google Scholar]
- 40.Adelman D.C., Murray J., Wu T.T., Mäki M., Green P.H., Kelly C.P. Measuring change in small intestinal histology in patients with celiac disease. Am J Gastroenterol. 2018;113:339–347. doi: 10.1038/ajg.2017.480. [DOI] [PubMed] [Google Scholar]
- 41.Popp A., Mäki M. Gluten-induced extra-intestinal manifestations in potential celiac disease—celiac trait. Nutrients. 2019;11:320. doi: 10.3390/nu11020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurppa K., Collin P., Viljamaa M., Haimila K., Saavalainen P., Partanen J., Laurila K., Huhtala H., Paasikivi K., Mäki M., Kaukinen K. Diagnosing mild enteropathy celiac disease: a randomized, controlled clinical study. Gastroenterology. 2009;136:816–823. doi: 10.1053/j.gastro.2008.11.040. [DOI] [PubMed] [Google Scholar]
- 43.De Leo L., Bramuzzo M., Ziberna F., Villanacci V., Martelossi S., Leo G Di, Zanchi C., Giudici F., Pandullo M., Riznik P., Mascio A Di, Ventura A., Not T. Diagnostic accuracy and applicability of intestinal auto-antibodies in the wide clinical spectrum of coeliac disease. EBioMedicine. 2020;51:102567. doi: 10.1016/j.ebiom.2019.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choung R.S., Khaleghi S., Cartee A.K., Marietta E.V., Larson J.J., King K.S., Savolainen O., Ross A.B., Rajkumar S.V., Camilleri M.J., Rubio-Tapia A., Murray J.A. Community-based study of celiac disease autoimmunity progression in adults. Gastroenterology. 2020;158:151–159. doi: 10.1053/j.gastro.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin X., Farin H.F., Van Es J.H., Clevers H., Langer R., Karp J.M. Niche-independent high-purity cultures of Lgr5 + intestinal stem cells and their progeny. Nat Methods. 2014;11:106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Capuano M., Iaffaldano L., Tinto N., Montanaro D., Capobianco V., Izzo V., Tucci F., Troncone G., Greco L., Sacchetti L. MicroRNA-449a overexpression, reduced NOTCH1 signals and scarce goblet cells characterize the small intestine of celiac patients. PLoS One. 2011;6 doi: 10.1371/journal.pone.0029094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ciacci C., Di Vizio D., Seth R., Insabato G., Mazzacca G., Podolsky D.K., Mahida Y.R. Selective reduction of intestinal trefoil factor in untreated coeliac disease. Clin Exp Immunol. 2002;130:526–531. doi: 10.1046/j.1365-2249.2002.02011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pietz G., De R., Hedberg M., Sjöberg V., Sandström O., Hernell O., Hammarström S., Hammarström M.-L., De R., Hedberg M., Sjöberg V., Sandström O., Hernell O., Hammarström S., Hammarström M.-L. Immunopathology of childhood celiac disease —key role of intestinal epithelial cells. PLoS One. 2017;12 doi: 10.1371/journal.pone.0185025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forsberg G., Fahlgren A., Hörstedt P., Hammarström S., Hernell O., Hammarstrôm M.L. Presence of bacteria and innate immunity of intestinal epithelium in childhood celiac disease. Am J Gastroenterol. 2004;99:894–904. doi: 10.1111/j.1572-0241.2004.04157.x. [DOI] [PubMed] [Google Scholar]
- 50.McCauley H.A., Guasch G. Three cheers for the goblet cell: maintaining homeostasis in mucosal epithelia. Trends Mol Med. 2015;21:492–503. doi: 10.1016/j.molmed.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Wang M.L., Keilbaugh S.A., Cash-Mason T., He X.C., Li N., Wu G.D. Immune-mediated signaling in intestinal goblet cells via PI3-kinase- and AKT-dependent pathways. Am J Physiol Gastrointest Liver Physiol. 2008;295:G1122–G1130. doi: 10.1152/ajpgi.90430.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim Y.S., Ho S.B. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep. 2010;12:319–330. doi: 10.1007/s11894-010-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lahdenperä A., Ludvigsson J., Fälth-Magnusson K., Högberg L., Vaarala O. The effect of gluten-free diet on Th1-Th2-Th3-associated intestinal immune responses in celiac disease. Scand J Gastroenterol. 2011;46:538–549. doi: 10.3109/00365521.2011.551888. [DOI] [PubMed] [Google Scholar]
- 54.Iwashita J., Sato Y., Sugaya H., Takahashi N., Sasaki H., Abe T. mRNA of MUC2 is stimulated by IL-4, IL-13 or TNF-α through a mitogen-activated protein kinase pathway in human colon cancer cells. Immunol Cell Biol. 2003;81:275–282. doi: 10.1046/j.1440-1711.2003.t01-1-01163.x. [DOI] [PubMed] [Google Scholar]
- 55.Ahn D., Crawley S., Hokari R., Kato S., Yang S., Li J., Kim Y. TNF-alpha activates MUC2 transcription via NF-kappaB but inhibits via JNK activation. Cell Physiol Biochem. 2005;15:29–40. doi: 10.1159/000083636. [DOI] [PubMed] [Google Scholar]
- 56.Tawiah A., Cornick S., Moreau F., Gorman H., Kumar M., Tiwari S., Chadee K. High MUC2 mucin expression and misfolding induce cellular stress, reactive oxygen production, and apoptosis in goblet cells. Am J Pathol. 2018;188:1354–1373. doi: 10.1016/j.ajpath.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 57.Corazza G.R., Villanacci V., Zambelli C., Milione M., Luinetti O., Vindigni C., Chioda C., Albarello L., Bartolini D., Donato F. Comparison of the interobserver reproducibility with different histologic criteria used in celiac disease. Clin Gastroenterol Hepatol. 2007;5:838–843. doi: 10.1016/j.cgh.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 58.Mubarak A., Nikkels P., Houwen R., Ten Kate F. Reproducibility of the histological diagnosis of celiac disease. Scand J Gastroenterol. 2011;46:1065–1073. doi: 10.3109/00365521.2011.589471. [DOI] [PubMed] [Google Scholar]
- 59.Arguelles-Grande C., Tennyson C.A., Lewis S.K., Green P.H.R., Bhagat G. Variability in small bowel histopathology reporting between different pathology practice settings: Impact on the diagnosis of coeliac disease. J Clin Pathol. 2012;65:242–247. doi: 10.1136/jclinpath-2011-200372. [DOI] [PubMed] [Google Scholar]
- 60.Picarelli A., Borghini R., Donato G., Di Tola M., Boccabella C., Isonne C., Giordano M., Di Cristofano C., Romeo F., Di Cioccio G., Marcheggiano A., Villanacci V., Tiberti A. Weaknesses of histological analysis in celiac disease diagnosis: new possible scenarios. Scand J Gastroenterol. 2014;49:1318–1324. doi: 10.3109/00365521.2014.948052. [DOI] [PubMed] [Google Scholar]
- 61.Werkstetter K.J., Korponay-Szabó I.R., Popp A., Villanacci V., Salemme M., Heilig G., Lillevang S.T., Mearin M.L., Ribes-Koninckx C., Thomas A., Troncone R., Filipiak B., Mäki M., Gyimesi J., Najafi M., Dolinšek J., Dydensborg Sander S., Auricchio R., Papadopoulou A., Vécsei A., Szitanyi P., Donat E., Nenna R., Alliet P., Penagini F., Garnier-Lengliné H., Castillejo G., Kurppa K., Shamir R., Hauer A.C., Smets F., Corujeira S., van Winckel M., Buderus S., Chong S., Husby S., Koletzko S., Socha P., Cukrowska B., Szajewska H., Wyhowski J., Brown N., Batra G., Misak Z., Seiwerth S., Dmitrieva Y., Abramov D., Vandenplas Y., Goossens A., Schaart M.W., Smit V.T.H.B.M., Kalach N., Gosset P., Kovács J.B., Nagy A., Lellei I., Kőbányai R., Khatami K., Monajemzadeh M., Dimakou K., Patereli A., Hansen T.P., Kavalar R., Bolonio M., Ramos D., Kogler H., Amann G., Kosova R., Maglio M., Janssens E., Achten R., Frűhauf P., Skálová H., Kirchner T., Petrarca L., Magliocca F.M., Martínez F., Morente V., Thanner-Lechner S., Ratschek M., Gasparetto M., Hook L., Canioni D., Wanty C., Mourin A., Laurila K., Vornane M., Friedler V.N., Morgenstern S.L., Amil Dias J., Carneiro F., João H.S., Van Biervliet S., Velde S Vande, Banoub H., Sampson S., Müller A.M., Ene A., Rafeey M., Eftekhar Sadat A.T. Accuracy in diagnosis of celiac disease without biopsies in clinical practice. Gastroenterology. 2017;153:924–935. doi: 10.1053/j.gastro.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 62.Taavela J., Koskinen O., Huhtala H., Lahdeaho M.-L., Popp A., Laurila K., Collin P., Kaukinen K., Kurppa K., Maki M. Validation of morphometric analyses of small-intestinal biopsy readouts in celiac disease. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Charlesworth R.P.G., Agnew L.L., Scott D.R., Andronicos N.M. Celiac disease gene expression data can be used to classify biopsies along the Marsh score severity scale. J Gastroenterol Hepatol. 2019;34:169–177. doi: 10.1111/jgh.14369. [DOI] [PubMed] [Google Scholar]
- 64.Comino I., Real A., Vivas S., Síglez M.Á., Caminero A., Nistal E., Casqueiro J., Rodríguez-Herrera A., Cebolla Á., Sousa C. Monitoring of gluten-free diet compliance in celiac patients by assessment of gliadin 33-mer equivalent epitopes in feces. Am J Clin Nutr. 2012;95:670–677. doi: 10.3945/ajcn.111.026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Comino I., Fernández-Bañares F., Esteve M., Ortigosa L., Castillejo G., Fambuena B., Ribes-Koninckx C., Sierra C., Rodríguez-Herrera A., Salazar J.C., Caunedo Á., Marugán-Miguelsanz J.M., Garrote J.A., Vivas S., Lo Iacono O., Nuñez A., Vaquero L., Vegas A.M., Crespo L., Fernández-Salazar L., Arranz E., Jiménez-García V.A., Antonio Montes-Cano M., Espín B., Galera A., Valverde J., Girón F.J., Bolonio M., Millán A., Cerezo F.M., Guajardo C., Alberto J.R., Rosinach M., Segura V., León F., Marinich J., Muñoz-Suano A., Romero-Gómez M., Cebolla Á., Sousa C. Fecal gluten peptides reveal limitations of serological tests and food questionnaires for monitoring gluten-free diet in celiac disease patients. Am J Gastroenterol. 2016;111:1456–1465. doi: 10.1038/ajg.2016.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.James G., Witten D., Hastie T., Tibshirani R. Springer; New York: 2013. An introduction to statistical learning. [Google Scholar]
- 67.Lumley T. Package “leaps”: regression subset selection. https://cran.r-project.org/web/packages/leaps/leaps.pdf Available at: Accessed February 16, 2020.
- 68.Piñeiro G., Perelman S., Guerschman J.P., Paruelo J.M. How to evaluate models: Observed vs predicted or predicted vs observed? Ecol Modell. 2008;216:316–322. [Google Scholar]
- 69.Goodpaster A.M., Kennedy M.A. Quantification and statistical significance analysis of group separation in NMR-based metabonomics studies. Chemom Intell Lab Syst. 2011;109:162–170. doi: 10.1016/j.chemolab.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differential Intestinal Mucosal Gene Transcriptome for PGC vs GFD and PGC vs DC Group Comparisons.