Abstract
Maintaining genomic integrity in mammalian early embryos, which are deficient in DNA damage repair, is critical for normal preimplantation and subsequent development. Abnormalities in DNA damage repair in preimplantation embryos can cause not only developmental arrest, but also diseases such as congenital disorders and cancers. Histone H4 lysine 20 monomethylation (H4K20me1) is involved in DNA damage repair and regulation of gene expression. However, little is known about the role of H4K20me1 during mouse preimplantation development. In this study, we revealed that H4K20me1 mediated by SETD8 is involved in maintaining genomic integrity. H4K20me1 was present throughout preimplantation development. In addition, reduction in the level of H4K20me1 by inhibition of SETD8 activity or a dominant-negative mutant of histone H4 resulted in developmental arrest at the S/G2 phase and excessive accumulation of DNA double-strand breaks. Together, our results suggest that H4K20me1, a type of epigenetic modification, is associated with the maintenance of genomic integrity and is essential for preimplantation development. A better understanding of the mechanisms involved in maintaining genome integrity during preimplantation development could contribute to advances in reproductive medicine and technology.
Keywords: Genomic integrity, Histone H4 lysine 20 monomethylation (H4K20me1), Preimplantation embryo
The cellular genome is constantly sustaining damage due to DNA replication errors, mutagens, or ionizing radiation. Damaged cells arrest cell-cycle progression by activating cell-cycle checkpoints, and then repair their DNA during the arrest. Abnormalities in DNA damage repair mechanisms have serious effects, including developmental failures and cancers [1]. Loss of maternally derived BCAS2 (breast carcinoma amplified sequence 2), which is involved in DNA damage repair, causes increased expression of the p53 effector p21, a major cell-cycle checkpoint regulator, resulting in arrest of embryonic development [2]. In addition, phosphorylation of H2AX (γH2AX), a marker of DNA double-strand breaks (DSBs), occurs in embryos damaged by oxidative stress or that lack a DNA damage repair pathway [2, 3]. Recent studies suggested that repair of DSBs is insufficient in oocytes and early embryos, especially in primates [4]. Therefore, DNA damage repair mechanisms in preimplantation embryos remain controversial, and an understanding of these pathways is crucial for advancement of reproductive medicine and technology.
Epigenetic alterations such as histone modifications and localization of linker histones contribute to DNA damage repair [5,6,7]. Histone H4K20 methylation is conserved from yeast to human; H4K20 can be monomethylated by SETD8/PR-SET7 and di- and trimethylated by SUV4-20H1 and SUV4-20H2 [8,9,10,11]. The expression of SETD8 protein and the level of H4K20 monomethylation (H4K20me1) have strong correlation; they change dynamically during the cell cycle, peaking at G2/M phase [12, 13]. DSB repair mechanisms include homologous recombination (HR) and nonhomologous end-joining (NHEJ); SETD8 is involved in DNA damage repair via NHEJ throughout the cell cycle, and inhibition of SETD8 activity triggers accumulation of DNA damage [14]. In Setd8-deficient Drosophila cells, DNA damage accumulates and cell-cycle arrest occurs. The arrest is dependent on ATR (ataxia telangiectasia–mutated [ATM] and rad3-related), which is an important checkpoint regulator [15].
Mouse preimplantation development is associated with dynamic changes in the level of histone methylation, including H4K20 methylation [16,17,18]. H4K20me2 is absent at the one- and two-cell stages, and appears only after the four-cell stage [19, 20]. By contrast, H4K20me3 is detected exclusively in the maternal pronucleus and becomes almost undetectable at the two-cell stage or later [11, 19, 20]. On the other hand, H4K20me1 is detected at all stages of preimplantation development [20]. Setd8-null embryos derived from Setd8+/− intercrosses are arrested by the eight-cell stage, emphasizing the importance of SETD8 in preimplantation development; however, its function remains unknown [13]. In addition, SETD8 methylates not only a lysine (Lys) residue of H4K20 but also Lys residues of non-histone proteins such as Lys382 of p53, Lys158 and Lys163 of NUMB, Lys248 of PCNA, and Lys385 of UHRF1 [21,22,23,24]. This is the main reason why the importance of H4K20me1 during preimplantation development remains largely unclear.
In this study, we analyzed the role of H4K20me1 during mouse preimplantation development. First, we examined the transition of H4K20me1 level during preimplantation development. To investigate the effect of SETD8 and H4K20 methylation on preimplantation development, we used UNC0379, a specific inhibitor of SETD8 and performed overexpression experiments in which Lys20 on histone H4 is replaced by methionine (H4K20M) [25], and analyzed the level of γH2AX, a marker of DSBs. Finally, we clarified the function of H4K20me1 in the cell-cycle checkpoint using inhibitors of p53 and ATR. In summary, our study suggests that SETD8-mediated H4K20me1 plays a key role in maintaining genomic integrity during mouse preimplantation development.
Materials and Methods
Collection of oocytes, in vitro fertilization, and embryo culture
Six- to twelve-week-old ICR female mice (Japan SLC, Shizuoka, Japan) were superovulated by injection of 7.5 IU of equine chorionic gonadotropin (eCG; ASUKA Animal Health, Tokyo, Japan) followed 46–48 h later by 7.5 IU of human chorionic gonadotropin (hCG; ASUKA Animal Health). Cumulus-oocyte complexes (COCs) were collected from the ampullae of the excised oviducts 14 h after the hCG injection. COCs were placed in a 100-μl droplet of human tubal fluid (HTF) medium supplemented with 4 mg/ml BSA (A3311; Sigma-Aldrich, St. Louis, MO, USA) [26]. Spermatozoa were collected from the cauda epididymis of 12- to 18-week-old ICR male mice (Japan SLC) and cultured for at least 1 h in a 100-μl droplet of HTF. After preincubation, sperm suspension was added into fertilization droplets at a final concentration of 1 × 106 cells/ml. At 2 hours post-insemination (hpi), morphologically normal fertilized embryos were collected and cultured in potassium simplex optimized medium (KSOM) supplemented with amino acids [27] and 1 mg/ml BSA under paraffin oil (Nacalai Tesque, Kyoto, Japan). All incubations were performed at 37°C under 5% CO2 in air.
Inhibitor treatments
UNC0379 (Sigma-Aldrich) is a substrate-competitive inhibitor of SETD8 and selective for SETD8 over 15 other methyltransferases [28]. Embryos were cultured in KSOM containing 0, 5, 10, 20, and 40 µM UNC0379 from 6 hpi, and they were morphologically observed at 24 hpi or collected at 15 hpi to stain with H4K20me1 antibody. In order to determine the optimal concentration of UNC0379 with the least side effect, H4K20me1 levels were examined measured by immunostaining. After this, to inhibit methylation by SETD8, embryos were cultured in KSOM containing 20 µM UNC0379 from 6 hpi. In the control, the same protocol was applied without UNC0379. For inhibition of p53-dependent transactivation and apoptosis, pifithrin-α hydrobromide (Adipogen, San Diego, CA, USA) [29] was used at a concentration of 20 µM in KSOM. Embryos were cultured in the presence or absence of pifithrin-α hydrobromide from 6 hpi. For inhibition of ATR activation, VE-821 (Cayman Chemical, Ann Arbor, MI, USA) [30] was used at a concentration of 10 µM in KSOM. Embryos were cultured in the presence or absence of VE-821 from 6 hpi. The number of analyzed embryos is given in each figure legend.
In vitro transcription and microinjection of mRNA
Total RNA was isolated from mouse ovaries by using TRIzol (Invitrogen, Carlsbad, CA, USA). After treatment with RNase-free DNase I (Roche, Indianapolis, IN, USA), cDNA was synthesized by using ReverTra Ace (Toyobo, Osaka, Japan) with random primers (Invitrogen). The coding region of histone H4 were amplified by PCR with KOD Plus (Toyobo) from the total cDNA. The initial denaturation was performed at 94°C for 2 min; followed by 30 cycles of denaturation at 94°C for 15 sec, annealing at 53°C for 30 sec, and extension at 68°C for 25 sec. The primer sequences used for PCR were as follows: 5 ´- AAAGGATCCGGTGGTTCTGGACGTGGTAAGGGT -3 ´ (forward) and 5 ´- TTTTCTAGATTAACCGCCGAATCCGTA -3 ´ (reverse). FLAG tag were added to pBlueScript SK(-) vector plasmid, and then the CDS region of histone H4 were subcloned into the downstream of FLAG tag of the plasmid. Then, a point mutation from AAG (K: Lysine) to ATG (M: Methionin) at Lysine 20 of histone H4 was induced by KOD-plus-Mutagenesis Kit (Toyobo). The mRNAs were made in vitro transcription by using T7 mMESSAGE mMACHINE kit (Ambion, Foster City, CA, USA). Poly(A) tails were added, using a Poly(A) Tailing Kit (Ambion). Synthesized mRNAs were purified by RNA Mini Kit (Qiagen, Hilden, Germany), diluted in nuclease-free water, and stored at −80°C. Approximately 3–5 pl of 100 ng/μl mRNA in nuclease-free water was microinjected into the cytoplasm of zygotes.
RNA extraction and RT-qPCR
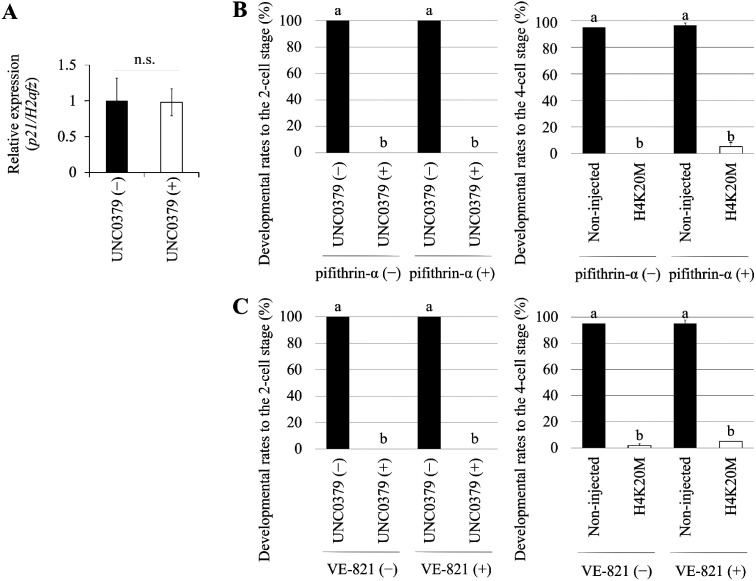
Total RNA was isolated from 50 embryos by using TRIzol (Invitrogen). cDNA was synthesized from total RNA using ReverTra Ace® qPCR RT Master Mix with gDNA Remover (Toyobo). Synthesized cDNA was mixed with specific primers and KOD SYBR qPCR Mix (Toyobo), and then amplified by RT-qPCR (reverse transcription-quantitative polymerase chain reaction). The initial denaturation was performed at 98°C for 2 min, followed by 40 cycles of denaturation at 98°C for 10 sec, annealing at 60°C for 10 sec, and extension at 68°C for 30 sec. Transcript levels were determined on a Step One Plus RT-PCR (Applied Biosystems, Foster City, CA, USA) and normalized against the internal control H2afz. Relative gene expression was calculated by the 2−ΔΔCt method [31]. The primer sequences used for qRT-PCR were as follows: H2afz, 5´ - TCCAGTGGACTGTATCTCTGTGA -3´ (forward) and 5´ - GACTCGAATGCAGAAATTTGG -3´ (reverse) ; p21, 5´ - CTTGCACTCTGGTGTCTG -3´ (forward) and 5´- CTTGGAGTGATAGAAATCTGTCA -3´ (reverse).
Immunofluorescence analysis
Embryos were fixed in 4% paraformaldehyde in PBS at room temperature (RT) for 20 min after the removal of the zona pellucidae with acid Tyrode’s solution (pH 2.5). After washing three times in PBS containing 0.3% polyvinylpyrrolidone (PVP K-30; Nacalai Tesque), fixed embryos were treated with 0.5% Triton X-100 (Sigma-Aldrich) in PBS at RT for 40 min, and were blocked in PBST containing 1.5% BSA, 0.2% sodium azide, and 0.02% Tween20 (antibody dilution buffer) at RT for 1 h. Embryos were incubated with primary antibody in antibody dilution buffer at 4°C overnight, for H4K20me1 (1:3000 dilution; ab9051, Abcam, Cambridge, UK), H4K20me3 (1:5000 dilution; ab9053, Abcam), γH2AX (1:200 dilution; AB_315794, Biolegend, San Diego, CA, USA), and FLAG (1:5000 dilution; F1804, Sigma-Aldrich) staining. Embryos were washed three times in antibody dilution buffer, and then were incubated with appropriate secondary antibody in antibody dilution buffer (1:500 dilution; Alexa Fluor 488-conjugated goat anti-mouse IgG or Alexa Fluor 594-conjugated goat anti-rabbit IgG, Invitrogen) at RT for 1 h. After stained with Hoechst 33342 (Sigma-Aldrich), embryos were mounted on slides in 50% glycerol in PBS and signals were observed using a fluorescence microscopy (BX50 or FSX100, Olympus, Tokyo, Japan). The number of analyzed embryos is given in each figure legend.
EdU staining
According to the manufacturer’s instructions, the procedures were performed using Click-iT™ Plus EdU Alexa Fluor™ 594 Imaging Kit (Invitrogen). After in vitro fertilization, embryos non-treated and treated with UNC0379 were cultured in presence of 50 µM 5-ethynyl-2'-deoxyuridine (EdU) for 2 h (7–9 hpi) and embryos overexpressing H4K20WT and H4K20M were cultured in presence of 50 µM EdU 2 h (20–22 hpi). They were fixed with 4% paraformaldehyde at RT for 20 min, and then permeabilized with 0.5% Triton X-100 in PBS at RT for 40 min. Embryos were stained with Click-iT reaction cocktail at RT for 30 min, followed by hoechst staining. Embryos were mounted on glass slides as mentioned above.
Statistical analyses
Data was analyzed by Student’s t-test when two-group compassions were made (Figs. 3E, 5A). Data was analyzed by one-way analysis of variance (ANOVA) followed by Tukey-Kramer test when multiple comparisons were made (Figs. 5B, 5C). P value < 0.05 was considered to be statistically significant.
Ethical approval for the use of animals
All experimental procedures were approved by the Animal Research Committee of Kyoto University (Permit no. 30-17) and were performed in accordance with the committee’s guidelines.
Results
SETD8 monomethylates H4K20 and is essential for preimplantation development
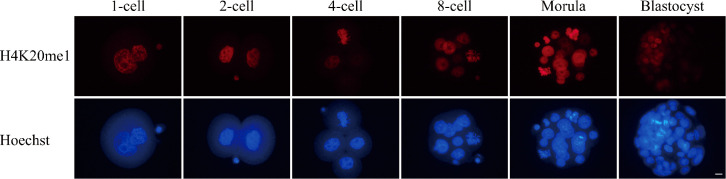
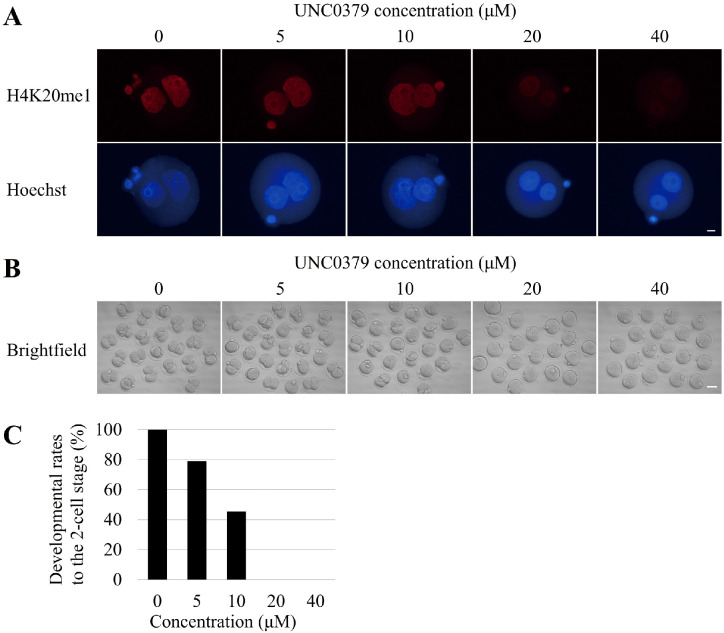
To analyze SETD8 function, we first monitored the amount of H4K20me1 during preimplantation development. Immunostaining revealed that H4K20me1 was present continuously throughout preimplantation development (Fig. 1). The level of H4K20me1 varied among blastomeres at the late stage of preimplantation development, including the morula and blastocyst stages (Fig. 1). In this experiment, we used UNC0379, a selective and substrate-competitive inhibitor of SETD8, to inhibit SETD8. To determine the optimal concentration of UNC0379 with the least side effects, we treated embryos with UNC0379 at the indicated concentrations (0, 5, 10, 20, and 40 µM) for 9 h (from 6 to 15 hpi), and then subjected them to immunofluorescence staining. Treatment of embryos with UNC0379 at 20 or 40 µM caused a significant decrease in the H4K20me1 level (Fig. 2A). Next, using the same technique, we observed the development of embryos treated with UNC0379 from 6 to 24 hpi. We found that 20 µM UNC0379 was the minimum effective concentration for inhibition of SETD8, and that embryos treated with this concentration underwent developmental arrest at the one-cell stage (Figs. 2B, 2C).
Fig. 1.
Localization of H4K20me1 through preimplantation development. Embryos were collected at the one-cell [15 h post-insemination (hpi), n = 9), two-cell (34 hpi, n = 11), four-cell (48 hpi, n = 11), eight-cell (56 hpi, n = 11), morula (72 hpi, n = 12), and blastocyst (96 hpi, n = 11) stages. Embryos were stained with anti-H4K20me1 antibody. H4K20me1 was present at all stages of preimplantation development. Scale bar, 10 μm.
Fig. 2.
Effect of SETD8 inhibition on preimplantation development. (A) Zygotes were treated with UNC0379 at the indicated concentrations (0 µM, n = 13; 5 µM, n = 17; 10 µM, n = 21; 20 µM, n = 18; and 40 µM, n = 11) for 9 h [from 6 to 15 h post-insemination (hpi)], collected at 15 hpi, and stained with anti-H4K20me1 antibody. The amount of H4K20me1 gradually decreased as the concentration of UNC0379 increased. Scale bar, 10 μm. (B) Morphology of embryos treated with UNC0379 at the concentrations (0 µM, n = 21; 5 µM, n = 24; 10 µM, n = 23; 20 µM, n = 20; and 40 µM, n = 20) for 18 h (from 6 to 24 hpi). Scale bar, 50 μm. (C) Graph shows the developmental rates of embryos treated with various concentrations of UNC0379 to the two-cell stage. Embryos treated with a low concentration (< 10 µM) of UNC0379 developed to the two-cell stage, whereas embryos treated with a high concentration (> 20 µM) of UNC0379 did not develop beyond the one-cell stage.
H4K20me1 is necessary for preimplantation development
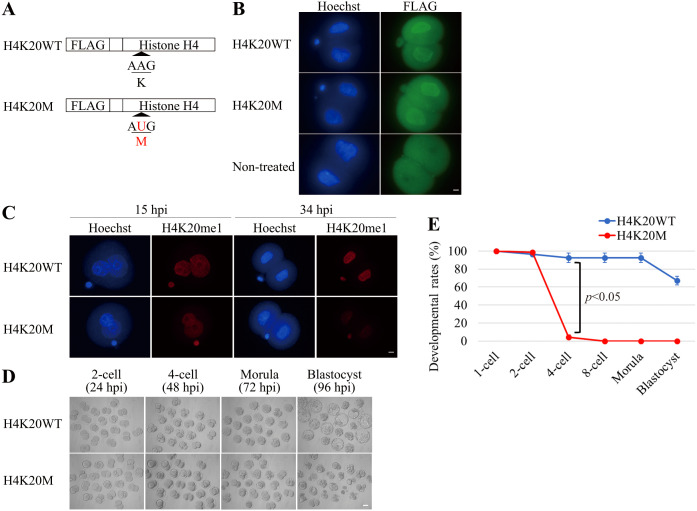
SETD8 methylates not only H4K20, but also non-histone proteins [21,22,23,24]. To determine whether developmental arrest by inhibition of SETD8 was due to a decrease in the level of H4K20me1, mRNA encoding wild-type histone H4 (H4K20WT) with a FLAG tag or a mutated histone H4 in which the 20th lysine was replaced by methionine (H4K20M), also with a FLAG tag, was overexpressed in embryos (Fig. 3A). Immunofluorescence with the anti-FLAG antibody revealed that the two mRNAs were translated to the same extent (Fig. 3B). There was no significant difference in H4K20me1 level between these embryos at 15 hpi, whereas at 34 hpi the level was much lower in embryos overexpressing H4K20M than in those overexpressing H4K20WT (Fig. 3C). Morphological observation revealed that embryos overexpressing H4K20WT developed normally to the blastocyst stage, whereas those overexpressing H4K20M were arrested at the two-cell stage (Figs. 3D, 3E).
Fig. 3.
Effects of H4K20M overexpression on H4K20me1 level and preimplantation development. (A) Schematic diagram of mRNAs microinjected into zygotes. H4K20M mRNA sequence contains a substitution in which AAG (K: Lysine) is replaced by AUG (M: Methionine) at residue 20. (B) Embryos were microinjected with mRNA encoding histone H4K20WT or H4K20M at 3 h post-insemination (hpi), and then fixed at 22 hpi to stain with FLAG antibody. The expression level of FLAG was almost the same between embryos overexpressing H4K20WT (n = 8) and H4K20M (n = 9). Scale bar, 10 μm. (C) The amount of H4K20me1 in embryos overexpressing H4K20WT (at 15 hpi, n = 12; at 34 hpi, n = 33) and H4K20M (at 15 hpi, n = 14; at 34 hpi, n = 29) were examined. The amount of H4K20me1 in embryos overexpressing H4K20M was almost the same at 15 hpi but dramatically decreased at 34 hpi, relative to embryos overexpressing H4K20WT. Scale bar, 10 μm. (D) Morphology of embryos overexpressing H4K20WT and H4K20M. Developmental abnormalities were observed in embryos overexpressing H4K20M after the two-cell stage. Scale bar, 50 μm. (E) Graph shows developmental rates of embryos overexpressing H4K20WT (n = 60) or H4K20M (n = 65). Almost all embryos overexpressing H4K20M stopped development at the two-cell stage. Data were obtained from three independent experiments and were expressed as mean ± s.e.m. Statistical analysis was performed using Student’s t-test. P value < 0.05 was considered to be statistically significant.
Reduction of the H4K20me1 level by inhibition of SETD8 or overexpression of H4K20M results in accumulation of DNA damage
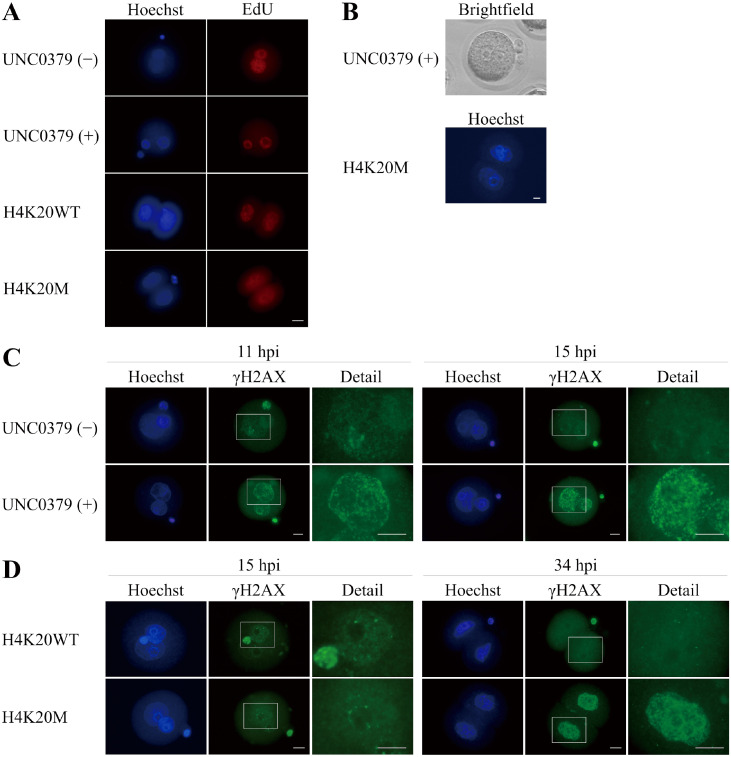
EdU uptake experiments showed that embryos treated with UNC0379 or overexpressing H4K20M were able to develop to S phase (Fig. 4A). In addition, pronuclei were observed even at 24 hpi in embryos treated with UNC0379, and nuclei were observed at 48 hpi in embryos overexpressing H4K20M, indicating that these embryos did not enter M phase (Fig. 4B). Next, we performed immunofluorescence staining for γH2AX to investigate the level of DNA damage in embryos treated with UN0379 and embryos overexpressing H4K20M. At late S phase of the one-cell stage (11 hpi), the number of γH2AX foci was larger in embryos treated with UNC0379 than in non-treated embryos. At G2 phase of the one-cell stage (15 hpi), the γH2AX foci decreased in non-treated embryos, but increased in UNC0379-treated embryos (Fig. 4C). On the other hand, there was no difference in the number of γH2AX foci between embryos overexpressing H4K20WT and those overexpressing H4K20M at G2 phase of the one-cell stage (15 hpi). The γH2AX foci was barely detectable in embryos overexpressing H4K20WT, whereas accumulation of γH2AX foci was observed in embryos overexpressing H4K20M at G2 phase of the two-cell stage (34 hpi) (Fig. 4D).
Fig. 4.
Cell-cycle progression and DNA damage in embryos lacking H4K20me1. (A) Incorporation of EdU in embryos treated with (n = 10) or without (n = 9) UNC0379 at 9 h post-insemination (hpi) and in embryos overexpressing H4K20WT (n = 16) and H4K20M (n = 16) at 22 hpi. Scale bar, 25 μm. (B) Upper photo shows the presence of two pronuclei at 24 hpi in a one-cell embryo treated with UNC0379 for 18 h (6–24 hpi, n = 20) and the lower photo shows the presence of two nuclei at 48 hpi (n = 8) in a two-cell embryo overexpressing H4K20M. Scale bar, 10 μm. (C) The number of γH2AX foci was larger in embryos treated with UNC0379 (n = 27) than in non-treated embryos (n = 24) at 11 hpi; at 15 hpi, the γH2AX foci almost disappeared in non-treated embryos (n = 27) but increased in embryos treated with UNC0379 (n = 23). Details show larger magnification of the white boxed regions. Scale bars, 16 μm. (D) There was no difference in the number of γH2AX foci at 15 hpi between embryos overexpressing H4K20WT (n = 19) and H4K20M (n = 21). However, larger number of γH2AX foci was observed at 34 hpi in embryos overexpressing H4K20M (n = 24) than in embryos overexpressing H4K20WT (n = 23). Details show larger magnification of the white boxed regions. Scale bar, 16 μm.
Developmental arrest due to a decrease in H4K20me1 level is not due to p53 or ATR activation
To determine whether developmental arrest resulting from a decrease in the H4K20me1 level depends on p53 activity, which promotes cell-cycle arrest at G1/S and S/G2 phases to enable DNA damage repair, we examined the expression of p21, a downstream gene of p53. UNC0379 caused no significant difference in the level of p21 (Fig. 5A). In addition, we investigated whether developmental arrest in response to UNC0379 treatment or H4K20M overexpression was rescued by a p53-specific inhibitor, pifithrin-α; however, the developmental arrest caused by UNC0379 or H4K20M overexpression could not be rescued by p53-inhibitor (Fig. 5B). Next, we sought to determine whether the activation of ATR, which also functions in cell-cycle arrest at S/G2 phase to allow DNA damage repair, is the cause of the developmental arrest of embryos in response to UNC0379 treatment or H4K20M overexpression. To this end, embryos were cultured in medium containing VE-821, a specific inhibitor of ATR. However, developmental arrest was not rescued by inhibitor in either case (Fig. 5C).
Fig. 5.
Reduction in H4K20me1 level and p53 and ATR activation. (A) qRT-PCR of p21 was performed using embryos treated or not treated with UNC0379. There was no significant difference between the expression levels in these embryos. Data were obtained from three independent experiments and were expressed as mean ± s.e.m. Statistical analysis was performed using Student’s t-test. P value < 0.05 was considered to be statistically significant. (B) Left graph shows developmental rates at 24 h post-insemination (hpi) to the two-cell stage in embryos treated with pifithrin-α and UNC0379 for 18 h (from 6 to 24 hpi). Right graph shows the developmental rates at 48 hpi to the four-cell stage of embryos overexpressing H4K20M and treated with pifithrin-α for 42 h (from 6 to 48 hpi). Non-injected embryos were used as negative control. The number of embryos analyzed were sixty. Data were from three independent experiments and were expressed as mean ± s.e.m. Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Tukey-Kramer test. P value < 0.05 was considered to be statistically significant, represented by different letters (a and b). (C) Left graph shows the developmental rates at 24 hpi to the two-cell stage of embryos treated with VE-821 and UNC0379 for 18 h (from 6 to 24 hpi). Right graph shows the developmental rates at 48 hpi to the four-cell stage of embryos overexpressing H4K20M and treated with VE-821 for 42 h (from 6 to 48 hpi). Non-injected embryos were used as negative control. The number of embryos analyzed were sixty. Data were from three independent experiments and were expressed as mean ± s.e.m. Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Tukey-Kramer test. P value < 0.05 was considered to be statistically significant, represented by different letters (a and b).
Discussion
In this study, we revealed that SETD8-mediated H4K20me1 is essential for preimplantation development, and that it is involved in maintenance of genomic integrity through repair of DNA damage caused by the in vitro culture environment.
Maintaining the integrity of the genome of mammalian preimplantation embryos, which can be damaged by both internal and external factors, is critical for normal preimplantation and subsequent development. Epigenetic factors contribute to the maintenance of genome stability, e.g., by preventing excessive activation of ATM by linker histone H1.2 and promoting DNA damage repair by histone-modifying enzymes G9a or SETD8 [5, 7, 14]. However, few studies have examined the relationship between epigenetics and genomic integrity during mouse preimplantation development. In this study, we showed that H4K20me1 was present at all stages of preimplantation development, and elucidated the function of H4K20me1 generated by the methyltransferase SETD8 during preimplantation development. As in the case of SETD8 inhibition, overexpression of H4K20M decreased the H4K20me1 level and resulted in developmental arrest. Furthermore, the decrease in H4K20me1 level triggered accumulation of DNA damage, especially at S/G2 phase. H4K20me1 is required for repair of DNA damage [14, 32], implying that H4K20me1 contributes to DNA damage repair during preimplantation development. Taken together, our findings reveal that H4K20me1 generated by SETD8 is essential for DNA damage repair during preimplantation development. This clearly demonstrates that epigenetics plays an important role in maintaining genomic integrity in mouse preimplantation embryos.
H4K20me1 was present consistently from the very early zygote stage to the blastocyst stage, suggesting that it contributes to preimplantation development in some way. On the other hand, contrary to a previous study using cultured cells [13], the H4K20me1 level did not change dramatically with cell-cycle progression at the one-cell stage. Unlike somatic cells, preimplantation embryos, especially at the one-cell stage, exhibit a loosening of chromatin structure and stage-specific gene regulation [33]. Therefore, one explanation for these observations might be the plastic state of zygotic chromatin, followed by an abnormal balance between the activities of the H4K20-modifying and -removing enzymes.
Treatment with the SETD8-specific inhibitor UNC0379 caused a decrease in the level of H4K20me1 and developmental arrest at the one-cell stage. The reason why SETD8-inhibited embryos were arrested at an earlier stage than Setd8-null embryos [13] is likely that the activity of maternally derived SETD8 is also blocked in SETD8-inhibited embryos, or that the effect of SETD8 inhibition varies depending on mouse strain [34]. However, SETD8 methylates not only histone H4K20 but also many non-histone proteins, including p53 and PCNA [21,22,23,24]; thus, it is difficult to determine from conventional methyltransferase inhibition experiments which SETD8-catalyzed methylation is important for preimplantation development. To analyze the function of H4K20me1 from a different point of view, we carried out dominant-negative experiments using histone lysine–methionine substituents. The H4K20me1 level decreased in embryos overexpressing H4K20M, and most of them (95% or more) were arrested at the two-cell stage. Histone lysine–methionine substituents can decrease the levels of mono-, di-, and trimethylation, but we detected no difference in H4K20me3 level between embryos overexpressing H4K20WT and H4K20M (Supplementary Fig. 1: online only). Considering the fact that H4K20me2 is not detected at all until the two-cell stage [20], developmental arrest at the two-cell stage in embryos overexpressing H4K20M is likely to be due to a decrease in the H4K20me1 level. However, it is possible that the decrease in H4K20me1 level due to H4K20M overexpression is not mediated by a dominant-negative effect, but by the direct inhibition of SETD8’s H4K20 mono-methyltransferase activity [35]. Embryos overexpressing H4K20M were arrested at the two-cell stage, not the one-cell stage, whereas embryos treated with UNC0379 were arrested at the one-cell stage. This was probably because it took a great deal of time for H4K20M mRNA to be translated sufficiently to inhibit H4K20me1 (Fig. 3C). In Drosophila cells, 80% of total histone H4K20 is in a dimethylated state, which is important for DNA damage repair through 53BP1 recruitment, and decreasing the level of both H4K20me1 and H4K20me2 causes cell-cycle arrest [15, 36]. During mouse preimplantation development, H4K20me2 is absent at the one- and two-cell stages, implying that DNA damage repair is highly dependent on H4K20me1 during development.
DSBs can form at stalled replication forks during DNA replication, and high levels of DSBs can have a serious impact on the organism’s survival [37]. Strong signals of DSBs were detected at S phase in mouse zygotes (Supplementary Fig. 2: online only), suggesting that the maintenance of genomic integrity at that time is critical for cell survival. In fact, mouse preimplantation embryos undergo developmental arrest at the two-cell stage when culture conditions are inadequate [38, 39] or transcription is inhibited [40], whereas developmental arrest at the one-cell stage occurs when DNA replication is inhibited [41]. Thus, development arrest at the one-cell stage upon treatment with a SETD8 inhibitor may be due to insufficient repair of damage during DNA replication. In this study, because a decrease in H4K20me1 level leads to cell-cycle arrest at S/G2 phase during preimplantation development (Figs. 4A, 4B), we hypothesized that the loss of H4K20me1 in in vitro fertilized embryos would result in accumulation of DSBs triggered by DNA replication errors. Consistent with our hypothesis, we observed accumulation of DSBs at the S/G2 phase in embryos treated with UNC0379 or overexpressing H4K20M (Figs. 4C, 4 D). The p53-binding protein 53BP1 promotes DNA damage repair via NHEJ, not HR, via an interaction with RIF1 (replication timing regulatory factor 1) [42]. Given that SETD8 is required to recruit 53BP1 and repair DSBs through the NHEJ pathway, rather than HR pathway [14], SETD8-mediated H4K20me1 may contribute to repair of DSBs through NHEJ-mediated repair during preimplantation development. Because NHEJ-mediated repair is activated in mouse preimplantation development [43], it is not surprising that NHEJ is important for the maintenance of genomic integrity during preimplantation development.
p53 pathway and its downstream gene, p21, contribute to activation of the G1/S and G2/M checkpoints [44]. Pifithrin-α, a p53-dependent transcription inhibitor, cannot alleviate developmental arrest in embryos treated with UNC0379 or overexpressing H4K20M [2], suggesting that the developmental arrest is probably not due to activation of p53 pathway. The observation that the p21 level did not change in embryos treated with UNC0379 supports the idea that developmental arrest is not p53-dependent. In addition, previous studies showed that DNA damage accumulates and cell-cycle arrest occurs in an ATR-dependent manner in SETD8-deficient Drosophila cells [15], whereas ATR inhibitor cannot alleviate the developmental arrest. This observation implies that the developmental arrest is not due to activation of ATR pathway although there is a possibility that the ATR inhibitor is not effective. The localization of γH2AX and H4K20me1 is almost completely identical (Supplementary Fig. 2), implying that H4K20me1 contributes significantly to DNA damage repair. In preimplantation development, however, it is possible that H4K20me1 is involved in other functions such as the regulation of gene expression, control of chromatin structural organization, and X-chromosome inactivation [13, 15, 45,46,47,48,49]. Thus, although the loss of genomic integrity may not be the only cause of the developmental arrest, we investigated the function of H4K20me1 in maintenance of genomic integrity in preimplantation development.
In summary, we revealed that H4K20me1 catalyzed by SETD8 is essential and is involved in the maintenance of genomic integrity during mouse preimplantation development. These findings emphasize the importance of correlations between epigenetic modifications and genomic integrity during preimplantation development. Furthermore, a better understanding of the mechanisms underlying maintenance of genomic integrity in mammalian preimplantation embryos will greatly advance reproductive medicine and technology.
Supplementary
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (no. 19H03136 and 16H05042 to NM) from the Japan Society for the Promotion of Science.
References
- 1.O’Driscoll M, Jeggo PA. The role of double-strand break repair - insights from human genetics. Nat Rev Genet 2006; 7: 45–54. [DOI] [PubMed] [Google Scholar]
- 2.Xu Q, Wang F, Xiang Y, Zhang X, Zhao ZA, Gao Z, Liu W, Lu X, Liu Y, Yu XJ, Wang H, Huang J, Yi Z, Gao S, Li L. Maternal BCAS2 protects genomic integrity in mouse early embryonic development. Development 2015; 142: 3943–3953. [DOI] [PubMed] [Google Scholar]
- 3.Xiao J, Liu Y, Li Z, Zhou Y, Lin H, Wu X, Chen M, Xiao W. Effects of the insemination of hydrogen peroxide-treated epididymal mouse spermatozoa on γH2AX repair and embryo development. PLoS One 2012; 7: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Liu D, He D, Suo S, Xia X, He X, Han JJ, Zheng P. Transcriptome analyses of rhesus monkey preimplantation embryos reveal a reduced capacity for DNA double-strand break repair in primate oocytes and early embryos. Genome Res 2017; 27: 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginjala V, Rodriguez-Colon L, Ganguly B, Gangidi P, Gallina P, Al-Hraishawi H, Kulkarni A, Tang J, Gheeya J, Simhadri S, Yao M, Xia B, Ganesan S. Protein-lysine methyltransferases G9a and GLP1 promote responses to DNA damage. Sci Rep 2017; 7: 16613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keum YS, Kim HG, Bode AM, Surh YJ, Dong Z. UVB-induced COX-2 expression requires histone H3 phosphorylation at Ser10 and Ser28. Oncogene 2013; 32: 444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Li Y, Tang M, Peng B, Lu X, Yang Q, Zhu Q, Hou T, Li M, Liu C, Wang L, Xu X, Zhao Y, Wang H, Yang Y, Zhu WG. Destabilization of linker histone H1.2 is essential for ATM activation and DNA damage repair. Cell Res 2018; 28: 756–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao B, Jing C, Kelly G, Walker PA, Muskett FW, Frenkiel TA, Martin SR, Sarma K, Reinberg D, Gamblin SJ, Wilson JR. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev 2005; 19: 1444–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couture JF, Collazo E, Brunzelle JS, Trievel RC. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev 2005; 19: 1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callén E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, Espejo A, Bedford MT, Nussenzweig A, Busslinger M, Jenuwein T. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 2008; 22: 2048–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eid A, Rodriguez-Terrones D, Burton A, Torres-Padilla ME. SUV4-20 activity in the preimplantation mouse embryo controls timely replication. Genes Dev 2016; 30: 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev 2002; 16: 2225–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 2009; 29: 2278–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dulev S, Tkach J, Lin S, Batada NN. SET8 methyltransferase activity during the DNA double-strand break response is required for recruitment of 53BP1. EMBO Rep 2014; 15: 1163–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Armstrong RL, Duronio RJ, MacAlpine DM. Methylation of histone H4 lysine 20 by PR-Set7 ensures the integrity of late replicating sequence domains in Drosophila. Nucleic Acids Res 2016; 44: 7204–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ancelin K, Syx L, Borensztein M, Ranisavljevic N, Vassilev I, Briseño-Roa L, Liu T, Metzger E, Servant N, Barillot E, Chen CJ, Schüle R, Heard E. Maternal LSD1/KDM1A is an essential regulator of chromatin and transcription landscapes during zygotic genome activation. eLife 2016; 5: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, Preissl S, Jermstad I, Haugen MH, Suganthan R, Bjørås M, Hansen K, Dalen KT, Fedorcsak P, Ren B, Klungland A. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016; 537: 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q, Liu W, Kou X, Zhao Y, He W, Li C, Chen B, Li Y, Wang Q, Ma J, Yin Q, Kee K, Meng A, Gao S, Xu F, Na J, Xie W. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 2016; 537: 553–557. [DOI] [PubMed] [Google Scholar]
- 19.van der Heijden GW, Dieker JW, Derijck AA, Muller S, Berden JH, Braat DD, van der Vlag J, de Boer P. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Dev 2005; 122: 1008–1022. [DOI] [PubMed] [Google Scholar]
- 20.Wongtawan T, Taylor JE, Lawson KA, Wilmut I, Pennings S. Histone H4K20me3 and HP1α are late heterochromatin markers in development, but present in undifferentiated embryonic stem cells. J Cell Sci 2011; 124: 1878–1890. [DOI] [PubMed] [Google Scholar]
- 21.Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 2007; 27: 636–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West LE, Roy S, Lachmi-Weiner K, Hayashi R, Shi X, Appella E, Kutateladze TG, Gozani O. The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression. J Biol Chem 2010; 285: 37725–37732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Gao Q, Tan S, You J, Lyu C, Zhang Y, Han M, Chen Z, Li J, Wang H, Liao L, Qin J, Li J, Wong J. SET8 prevents excessive DNA methylation by methylation-mediated degradation of UHRF1 and DNMT1. Nucleic Acids Res 2019; 47: 9053–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhami GK, Liu H, Galka M, Voss C, Wei R, Muranko K, Kaneko T, Cregan SP, Li L, Li SS. Dynamic methylation of Numb by Set8 regulates its binding to p53 and apoptosis. Mol Cell 2013; 50: 565–576. [DOI] [PubMed] [Google Scholar]
- 25.Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, Sarkaria J, Zhang Z. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 2013; 27: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minami N, Sasaki K, Aizawa A, Miyamoto M, Imai H. Analysis of gene expression in mouse 2-cell embryos using fluorescein differential display: comparison of culture environments. Biol Reprod 2001; 64: 30–35. [DOI] [PubMed] [Google Scholar]
- 27.Ho Y, Wigglesworth K, Eppig JJ, Schultz RM. Preimplantation development of mouse embryos in KSOM: augmentation by amino acids and analysis of gene expression. Mol Reprod Dev 1995; 41: 232–238. [DOI] [PubMed] [Google Scholar]
- 28.Ma A, Yu W, Li F, Bleich RM, Herold JM, Butler KV, Norris JL, Korboukh V, Tripathy A, Janzen WP, Arrowsmith CH, Frye SV, Vedadi M, Brown PJ, Jin J. Discovery of a selective, substrate-competitive inhibitor of the lysine methyltransferase SETD8. J Med Chem 2014; 57: 6822–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 1999; 285: 1733–1737. [DOI] [PubMed] [Google Scholar]
- 30.Prevo R, Fokas E, Reaper PM, Charlton PA, Pollard JR, McKenna WG, Muschel RJ, Brunner TB. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther 2012; 13: 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 32.Oda H, Hübner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell 2010; 40: 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Z, Zheng H, Huang B, Ma R, Wu J, Zhang X, He J, Xiang Y, Wang Q, Li Y, Ma J, Zhang X, Zhang K, Wang Y, Zhang MQ, Gao J, Dixon JR, Wang X, Zeng J, Xie W. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 2017; 547: 232–235. [DOI] [PubMed] [Google Scholar]
- 34.Okayasu R, Suetomi K, Yu Y, Silver A, Bedford JS, Cox R, Ullrich RL. A deficiency in DNA repair and DNA-PKcs expression in the radiosensitive BALB/c mouse. Cancer Res 2000; 60: 4342–4345. [PubMed] [Google Scholar]
- 35.Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013; 340: 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, Pesavento JJ, Starnes TW, Cryderman DE, Wallrath LL, Kelleher NL, Mizzen CA. Preferential dimethylation of histone H4 lysine 20 by Suv4-20. J Biol Chem 2008; 283: 12085–12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.So A, Le Guen T, Lopez BS, Guirouilh-Barbat J. Genomic rearrangements induced by unscheduled DNA double strand breaks in somatic mammalian cells. FEBS J 2017; 284: 2324–2344. [DOI] [PubMed] [Google Scholar]
- 38.Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 1989; 86: 679–688. [DOI] [PubMed] [Google Scholar]
- 39.Gardner DK, Lane M. Alleviation of the ‘2-cell block’ and development to the blastocyst of CF1 mouse embryos: role of amino acids, EDTA and physical parameters. Hum Reprod 1996; 11: 2703–2712. [DOI] [PubMed] [Google Scholar]
- 40.Warner CM, Versteegh LR. In vivo and in vitro effect of α-amanitin on preimplantation mouse embryo RNA polymerase. Nature 1974; 248: 678–680. [DOI] [PubMed] [Google Scholar]
- 41.Howlett SK. The effect of inhibiting DNA replication in the one-cell mouse embryo. Rouxs Arch Dev Biol 1986; 195: 499–505. [DOI] [PubMed] [Google Scholar]
- 42.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 2013; 339: 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiorenza MT, Bevilacqua A, Bevilacqua S, Mangia F. Growing dictyate oocytes, but not early preimplantation embryos, of the mouse display high levels of DNA homologous recombination by single-strand annealing and lack DNA nonhomologous end joining. Dev Biol 2001; 233: 214–224. [DOI] [PubMed] [Google Scholar]
- 44.Medema RH, Macůrek L. Checkpoint control and cancer. Oncogene 2012; 31: 2601–2613. [DOI] [PubMed] [Google Scholar]
- 45.Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2004; 2: E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakaguchi A, Steward R. Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. J Cell Biol 2007; 176: 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo YB, Ma JY, Zhang QH, Lin F, Wang ZW, Huang L, Schatten H, Sun QY. MBTD1 is associated with Pr-Set7 to stabilize H4K20me1 in mouse oocyte meiotic maturation. Cell Cycle 2013; 12: 1142–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shoaib M, Walter D, Gillespie PJ, Izard F, Fahrenkrog B, Lleres D, Lerdrup M, Johansen JV, Hansen K, Julien E, Blow JJ, Sørensen CS. Histone H4K20 methylation mediated chromatin compaction threshold ensures genome integrity by limiting DNA replication licensing. Nat Commun 2018; 9: 3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka H, Takebayashi SI, Sakamoto A, Igata T, Nakatsu Y, Saitoh N, Hino S, Nakao M. The SETD8/PR-Set7 methyltransferase functions as a barrier to prevent senescence-associated metabolic remodeling. Cell Reports 2017; 18: 2148–2161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.