Abstract
Purpose
The molecular mechanism underlying the tumorigenesis and progression of lung adenocarcinoma (LUAD) in nonsmoking patients remains unclear. This study was conducted to select crucial therapeutic and prognostic biomarkers for nonsmoking patients with LUAD.
Methods
Microarray datasets from the Gene Expression Omnibus (GSE32863 and GSE75037) were analyzed for differentially expressed genes (DEGs). Gene Ontology (GO) enrichment analysis of DEGs was performed, and protein-protein interaction network was then constructed using the Search Tool for the Retrieval of Interacting Genes and Cytoscape. Hub genes were then identified by the rank of degree. Overall survival (OS) analyses of hub genes were performed among nonsmoking patients with LUAD in Kaplan-Meier plotter. The Cancer Genome Atlas (TCGA) and The Human Protein Atlas (THPA) databases were applied to verify hub genes. In addition, we performed Gene Set Enrichment Analysis (GSEA) of hub genes.
Results
We identified 1283 DEGs, including 743 downregulated and 540 upregulated genes. GO enrichment analyses showed that DEGs were significantly enriched in collagen-containing extracellular matrix and extracellular matrix organization. Moreover, 19 hub genes were identified, and 12 hub genes were closely associated with OS. Although no obvious difference was detected in ITGB1, the downregulation of UBB and upregulation of RAC1 were observed in LUAD tissues of nonsmoking patients. Immunohistochemistry in THPA database confirmed that UBB and ITGB1 were downregulated, while RAC1 was upregulated in LUAD. GSEA suggested that ribosome, B cell receptor signaling pathway, and cell cycle were associated with UBB, RAC1, and ITGB1 expression, respectively.
Conclusions
Our study provides insights into the underlying molecular mechanisms of the carcinogenesis and progression of LUAD in nonsmoking patients and demonstrated UBB, RAC1, and ITGB1 as therapeutic and prognostic indicators for nonsmoking LUAD. This is the first study to report the crucial role of UBB in nonsmoking LUAD.
1. Introduction
Lung cancer (LC) is the most prevalent cancer type, with approximately 228,820 cancer cases and 135,720 death cases in 2020 worldwide, thus causing considerable socioeconomic burdens [1]. Patients with non-small-cell lung cancer (NSCLC) constitute approximately 85% of the total LC cases. Lung adenocarcinoma (LUAD) is the most common histological type of NSCLC [2]. It is widely known that tobacco smoking is a crucial risk factor for LC; however, approximately 20% of LUAD cases occur among nonsmoking patients [3]. Patients with smoking-related LUAD have some altered genes such as KRAS and STK11. Moreover, these known cancer-related genes may alter simultaneously, leading to a larger tumor mutational burden (TMB) [4, 5]. However, the molecular mechanism underlying the carcinogenesis and progression of nonsmoking-related LUAD remains unclear. In addition, nonsmoking patients are easily ignored as they are not exposed to smoking [6], leading to a high rate of missed diagnosis at early stages. Hence, it is extremely necessary to identify key indicators in the carcinogenesis and development of nonsmoking-related LUAD.
In the past two decades, gene chip technologies and bioinformatics analyses have made great progress in screening genetic alterations at the genome level [7]. These technologies are adopted to find differentially expressed genes (DEGs) that play crucial roles in the occurrence and adverse progression of nonsmoking-related LUAD. However, false-positive rates of independent microarray studies probably weaken the reliability of outcomes [8]. Thus, we selected two microarray datasets on the same platform from the Gene Expression Omnibus (GEO) to acquire DEGs between LUAD tissues of nonsmoking patients and matched adjacent lung tissue samples. Gene Ontology (GO) enrichment analyses of DEGs were then performed, and a protein-protein interaction (PPI) network was constructed for a better understanding of the molecular mechanism underlying tumorigenesis and invasion of nonsmoking-related LUAD. Hub genes were then identified from the PPI network, which are candidates for therapeutic and prognostic biomarkers for nonsmoking-related LUAD. Subsequently, the overall survival (OS) analysis of hub genes was performed. Finally, we validated the findings using The Cancer Genome Atlas (TCGA), The Human Protein Atlas (THPA) database, and Gene Set Enrichment Analysis (GSEA).
2. Materials
2.1. Microarray Data
GEO (http://www.ncbi.nlm.nih.gov/geo) [9] is a public functional genomics data repository of high-throughput gene expression data, chip, and microarray. All microarray datasets were selected only if they met the following criteria: (1) the topic was on nonsmoking-related LUAD and matched adjacent lung tissues; (2) the platform was GPL6884 Illumina HumanWG-6 v3.0 expression BeadChip; (3) the organism was Homo sapiens; (4) the size of matched adjacent lung tissue sample was more than 3; and (5) the last update time was in 2019. In this study, two gene expression profiles (GSE32863 [10] and GSE75037 [11]) from GEO were selected, which met these criteria. GSE32863 included 29 nonsmoking LUAD samples and 30 matched adjacent lung samples, whereas GSE75037 contained 30 nonsmoking LUAD samples and 30 matched adjacent lung tissue samples.
2.2. Identification of Differentially Expressed Genes
The DEGs between nonsmoking LUAD and adjacent lung tissues were obtained using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r) [12]. As an interactive web tool, GEO2R allows users to compare at least two datasets in one GEO series for selecting DEGs across experimental conditions. Both adjusted P values (adj.P) and Benjamini-Hochberg's false discovery rates were adopted to balance the finding of a statistically significant gene and false-positive limitation. Probe sets with no corresponding gene symbols were deleted, whereas genes with at least one probe set were averaged. The cutoff criteria were set as adj.P < 0.05 and ∣logFC(fold change) | <1.
2.3. Gene Ontology Enrichment Analysis of Differentially Expressed Genes
GO is an important bioinformatics tool for annotating genes and analyzing the biological process of genes [13]. GO enrichment analyses consisted of 3 terms: biological processes (BP), cellular component (CC), and molecular function (MF). All these were performed using the clusterProfiler and GOplot packages in the R software to analyze the functions and signaling pathways of DEGs [14]. P value of < 0.05 was regarded statistically significant.
2.4. Protein-Protein Interaction Network and Significant Module Construction
The PPI network of DEGs was constructed in the Search Tool for the Retrieval of Interacting Genes (http://string-db.org, version 11.0) [15], and the interaction with a combined score of >0.90 was considered statistically significant. The analysis of the function of PPI provided insights into the mechanism of the development of the disease. Cytoscape (version 3.7.2), a bioinformatics software, was adopted to construct visual networks of molecular interactions [16]. The plug-in Molecular Complex Detection (MCODE) (version 1.4.2) of Cytoscape, a crucial application, was adopted to find closely correlated modules from the PPI network [17]. Genes in significant modules were graphically shown through MCODE plug-in. The selection criteria were set as follows: MCODE score > 5, node score cutoff = 0.2, degree cutoff = 2, k − score = 2, and max depth = 100.
2.5. Hub Gene Selection and Analysis
The cytoHubba plug-in of Cytoscape was used to calculate the degree rank of hub genes, and hub genes with degrees greater than 20 were selected. The Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) is a crucial database to understand high-level functions and biological systems from large-scale molecular datasets generated by high-throughput experimental technology [18]. To explore their biological function, the BP and KEGG pathway of these hub genes were analyzed and visualized through the ClueGO plug-in [19]. Subsequently, a heat map of hub genes was created and visualized in TCGA database (https://portal.gdc.cancer.gov/). Then, the Kaplan-Meier plotter was used to perform the survival analysis of these hub genes for understanding their prognostic roles (http://kmplot.com/analysis/) [20]. Moreover, TCGA database is an external and relatively authoritative database, which was used for the verification of the difference in the expression levels of hub genes between LUAD and normal lung tissues.
2.6. Gene Set Enrichment Analysis
AS a computing approach, GSEA can identify whether previously defined gene sets were statistically significant and concordantly different between the two biological states [21]. Nonsmoking-related LUAD samples were categorized into two groups (high and low expression) by the median expression levels of hub genes. The timing of their expression on many gene sets was then explored to find related KEGG pathways using the molecular signatures database (MSigDB) (c2.cp.kegg.all.v7.1.symbols.gmt) [22]. The number of permutations was set as 1000 times in every analysis. ∣Normalized enrichment score | >1, NOM P value < 0.05, and FDR q value < 0.25 were considered statistically significant.
3. Results
3.1. Identification of Differentially Expressed Genes in Nonsmoking Lung Adenocarcinoma
The volcano plots illustrated the selection process for DEGs in GSE32863 (Figure 1(a)) and GSE75037 (Figure 1(b)). After normalization of microarray outcomes, we identified DEGs in nonsmoking LUAD and adjacent lung tissues. In addition, the Venn diagram in Figure 1(c) shows that the overlap between both datasets included 1283 DEGs, including 743 downregulated and 540 upregulated genes.
Figure 1.
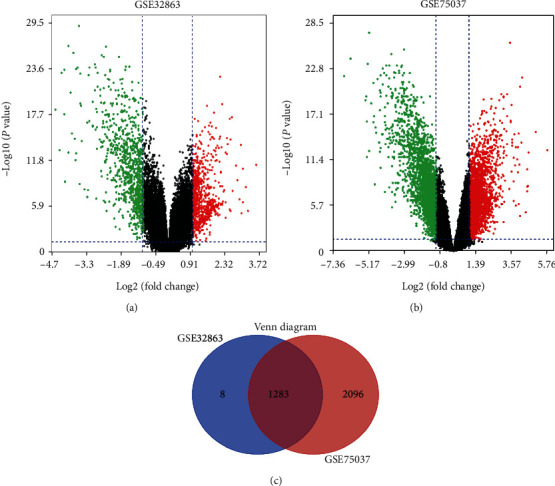
Volcano plot, Venn diagram, and function enrichment analysis of DEGs. The selection process for DEGs with adj.P < 0.01 and ∣logFC | <1 in GSE32863 (a) and GSE75037 (b). Upregulated genes are marked in red, and downregulated genes are marked in green. The two datasets display an overlap of 576 DEGs (c). adj.P: adjusted P values; DEGs: differentially expressed genes; FC: fold change.
3.2. Gene Ontology Enrichment Analysis of Differentially Expressed Genes
GO enrichment analyses of downregulated DEGs revealed that collagen-containing extracellular matrix (ECM) was considerably enriched in BP, extracellular structure organization in CC, and ECM structural constituent in MF (Figure 2(a)). Moreover, GO enrichment analyses of upregulated DEGs suggested that the ECM organization was primarily enriched in BP, apical plasma membrane in CC, and cell adhesion module binding in MF (Figure 2(b)).
Figure 2.
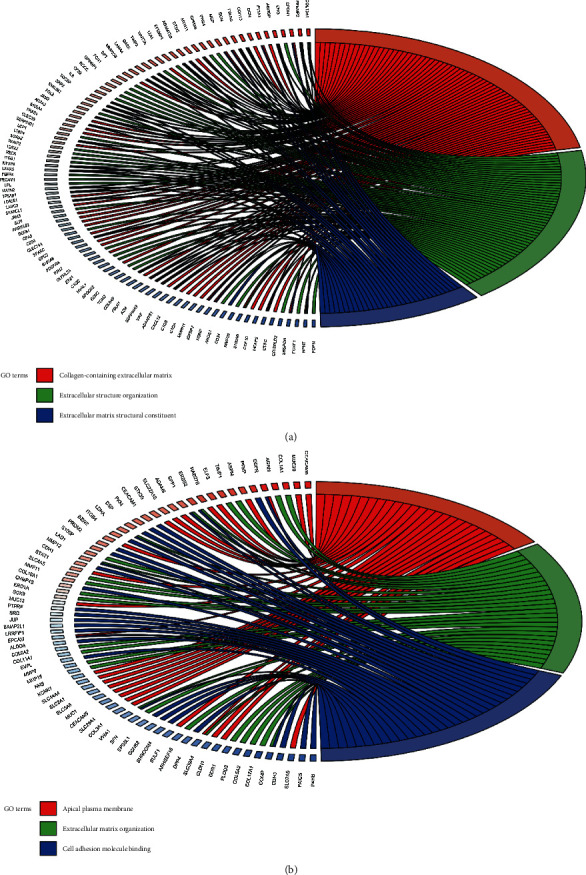
GO enrichment analysis of the DEGs using the clusterProfiler package in the R software. GO enrichment analyses of (a) downregulated DEGs and (b) upregulated DEGs are performed. DEGs: differentially expressed genes; GO: Gene Ontology.
3.3. Protein-Protein Interaction Network and Significant Module Construction
The PPI network of these DEGs is illustrated in Figure 3(a), which consisted of 607 nodes and 1841 edges. The most significant module was then detected through MCODE plug-in. Moreover, this module consisted of 48 nodes and 247 edges, as shown in Figure 3(b), wherein upregulated genes are marked in red and downregulated genes in blue.
Figure 3.
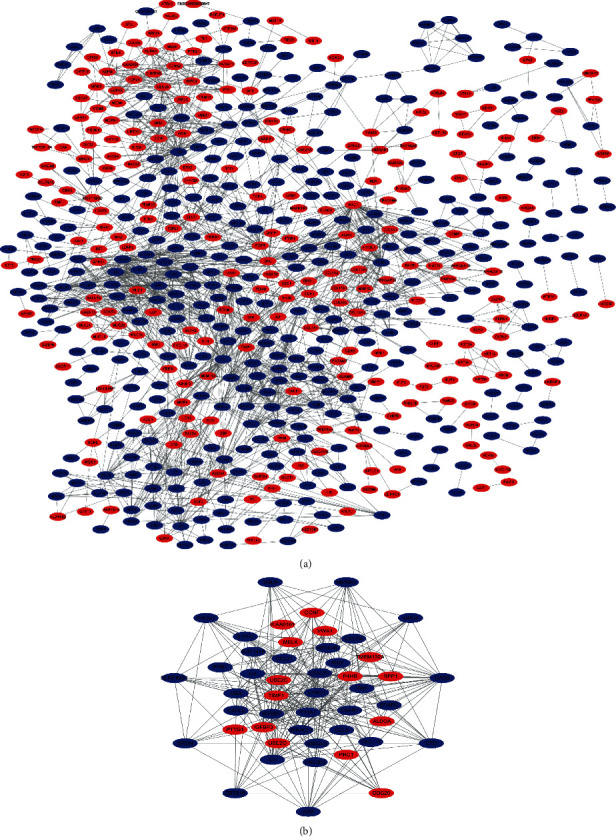
PPI network and the most significant module of DEGs. The PPI network of DEGs is constructed using Cytoscape (a). The most significant module is obtained from the PPI network with 48 nodes and 257 edges (b). Upregulated genes are marked in red and downregulated genes in blue. DEGs: differentially expressed genes; PPI: protein-protein interaction network.
3.4. Hub Gene Selection and Analysis
A total of 19 DEGs with degrees ≥ 20 were selected as hub genes. Table 1 shows the gene symbol, degree, full name, and function of these hub genes. BP (Figure 4(a)) and KEGG analysis (Figure 4(b)) of these hub genes are clearly shown in figures. Moreover, a heat map demonstrated the upregulation or downregulation of 19 hub genes in nonsmoking LUAD samples using TCGA dataset (Figure 4(c)). In addition, Figure 5 reveals that UBB, RAC1, ITGB1, CDC20, EGFR, UBE2C, TIMP1, P4H8, and MMP9 are negatively correlated with OS in nonsmoking-related LUAD, whereas CXCL12, GAS6, and FPR1 are positively correlated with OS. Table S1 displays the hazard ratio (HR), 95% confidence interval (CI), and log-rank P value of hub genes.
Table 1.
Functional roles of 19 hub genes with degrees > 20.
| No. | Gene symbol | Degree | Full name | Function |
|---|---|---|---|---|
| 1 | UBB | 51 | Ubiquitin B | UBB is lowly expressed in some cancers, such as nonsmoking LUAD and endometrial carcinoma. |
| 2 | RAC1 | 50 | Rac Family Small GTPase 1 | RAC1 serves a key role in the EMT, cell proliferation, and worse invasion in LUAD. |
| 3 | ITGB1 | 42 | Integrin subunit beta 1 | ITGB1 encodes the beta subunit of integrins and participates in the carcinogenesis and progression of LUAD. |
| 4 | SRC | 40 | SRC protooncogene, nonreceptor tyrosine kinase | SRC participates in the cell proliferation, migration, and invasion of LUAD. |
| 5 | C3 | 40 | Complement C3 | C3 is obviously altered in serum among patients with LUAD and C3 may be a candidate diagnostic biomarker. |
| 6 | IL6 | 33 | Interleukin 6 | IL6 promotes Kras driven of the carcinogenesis of LUAD and induces the resistance of gefitinib in EGFR-mutant LC. |
| 7 | CDC20 | 33 | Cell division cycle 20 | CDC20 is associated with protein ubiquitination and modification and promotes the progression of NSCLC. |
| 8 | EGFR | 32 | Epidermal growth factor receptor | EGFR mutation is the driven alteration of nonsmoking LUAD and participates in the cell proliferation in LUAD. |
| 9 | UBE2C | 29 | Ubiquitin-conjugating enzyme E2 C | UBE2C contributes to the EMT, cell proliferation, and malignant invasion of LUAD. |
| 10 | TIMP1 | 28 | TIMP metallopeptidase inhibitor 1 | TIMP1 participates in the progression and inhibits the apoptosis of tumor cells in LUAD. |
| 11 | GNG11 | 26 | G protein subunit gamma 11 | Low expression of GNG11 is correlated with poorer prognosis among woman patients with nonsmoking LC. |
| 12 | CXCL12 | 26 | C-X-C motif chemokine ligand 12 | CXCL12/CXCR4 plays an important role in the propagation of NSCLC. |
| 13 | GAS6 | 25 | Growth arrest specific 6 | AXL/GAS6 axis contributes to cell migration in NSCLC, which is a candidate prognostic biomarker in NSCLC. |
| 14 | P4HB | 25 | Prolyl 4-hydroxylase subunit beta | P4HB participates in the invasion and metastasis of gastric cancer. |
| 15 | CXCR4 | 24 | C-X-C motif chemokine receptor 4 | CXCR4 serves a procarcinogenic role by interacting with CXCL12 in NSCLC. |
| 16 | FPR1 | 22 | Formyl peptide receptor 1 | FPR1 induces the translocation of NF-κB to promote the progression by upregulating IL6 and IL8 in cervical cancer. |
| 17 | ADRB2 | 22 | Adrenoceptor beta 2 | ADRB2 is overexpressed in LUAD, which promotes the adverse progression in LUAD. |
| 18 | LYZ | 21 | Lysozyme | Hypermethylated LYZ is observed in gastric cancer. |
| 19 | MMP9 | 21 | Matrix metallopeptidase 9 | MMP9 regulates the cell proliferation and metastasis of LUAD. |
Abbreviations: LC: lung cancer; NSCLC: non-small-cell lung cancer; LUAD: lung adenocarcinoma; TSA: trichostatin A; EMT: epithelial-mesenchymal transition; NF-κB: nuclear factor kappa B.
Figure 4.
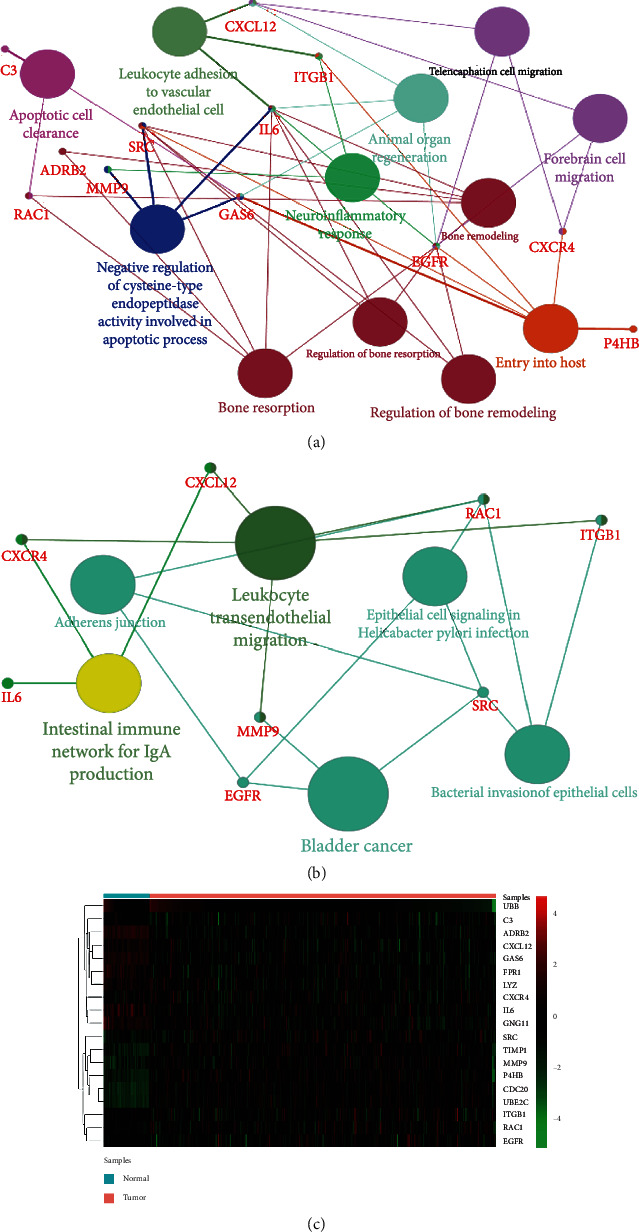
The BP, KEGG pathway analysis, and heat map of hub genes. The BP analysis of hub genes is visualized using the ClueGO plug-in (a). KEGG pathway analysis of hub genes is visualized using the ClueGO plug-in (b). The color depth of nodes refers to the corrected P value of ontologies, and the size of nodes refers to the number of genes that participate in the ontologies. P < 0.01 is considered statistically significant. Heat map of hub genes is created on the basis of data from TCGA and visualized using pheatmap package (c). Upregulated genes are marked in red and downregulated genes in green. BP: biological process; KEGG: Kyoto Encyclopedia of Genes and Genomes pathway; TCGA: The Cancer Genome Atlas.
Figure 5.
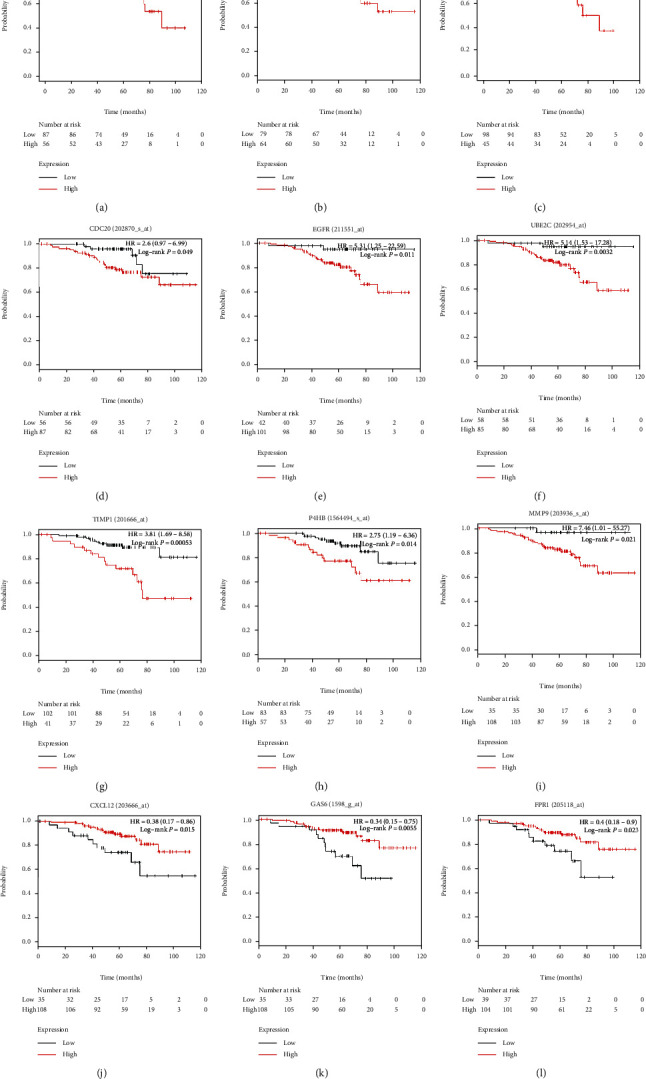
OS analyses of hub genes are performed in a Kaplan-Meier plotter online platform. UBB, RAC1, ITGB1, CDC20, EGFR, UBE2C, TIMP1, P4H8, and MMP9 are negatively associated with OS in nonsmoking patients with LUAD (a–h), whereas CXCL12, GAS6, and FPR1 are positively correlated with OS (i–k). P < 0.05 is considered statistically significant. LUAD: lung adenocarcinoma; OS: overall survival.
UBB, RAC1, and ITGB1 had the highest degrees among these hub genes, indicating their pivotal roles in the occurrence and progression of nonsmoking-related LUAD. The overexpression of UBB, RAC1, and ITGB1 had worse OS (P = 0.00044, P = 0.0021, and P = 0.00044, respectively), suggesting their potential prognostic implications. Moreover, according to data from TCGA, we found that UBB was significantly downregulated (Figure 6(a)), and RAC1 was upregulated (Figure 6(b)) in nonsmoking-related LUAD. Moreover, ITGB1 was not significantly different in nonsmoking LUAD and normal lung tissues (Figure 6(c)); however, further studies on this are warranted. Immunohistochemistry (IHC) in THPA database verified that UBB (Figure 6(d)) and ITGB1 (Figure 6(e)) were downregulated, and RAC1 (Figure 6(f)) was upregulated in LUAD.
Figure 6.
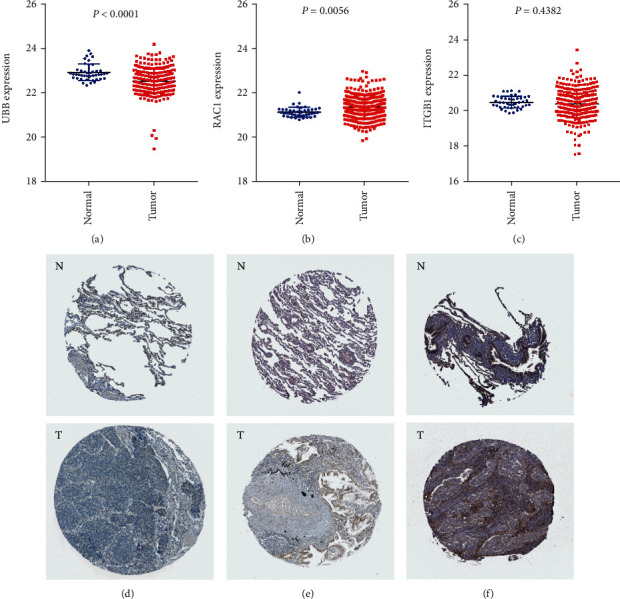
The expression of UBB, RAC1, and ITGB1 in normal lung and LUAD tissues using TCGA and THPA databases. The expression of (a) UBB, (b) RAC1, and (c) ITGB1 in normal lung and nonsmoking LUAD samples is shown using TCGA database. The results of immunohistochemistry of (d) UBB, (e) RAC1, and (e) ITGB1 in normal lung and LUAD tissues using THPA database are displayed. LUAD: lung adenocarcinoma; TCGA: The Cancer Genome Atlas; THPA: The Human Protein Atlas.
3.5. Gene Set Enrichment Analysis
GSEA showed that ribosome was associated with UBB expression (Figure 7(a)). Furthermore, it suggested that ECM receptor interaction, B cell receptor signaling pathway, T cell receptor signaling pathway, toll-like receptor signaling pathway, and focal adhesion were correlated with RAC1 expression (Figures 7(b)–7(f)). In addition, cell cycle, spliceosome, DNA replication, RNA degradation, mismatch repair, and pyrimidine metabolism were associated with ITGB1 expression (Figures 7(g)–7(m)). The detailed outcomes of the analysis were shown in Table 2.
Figure 7.
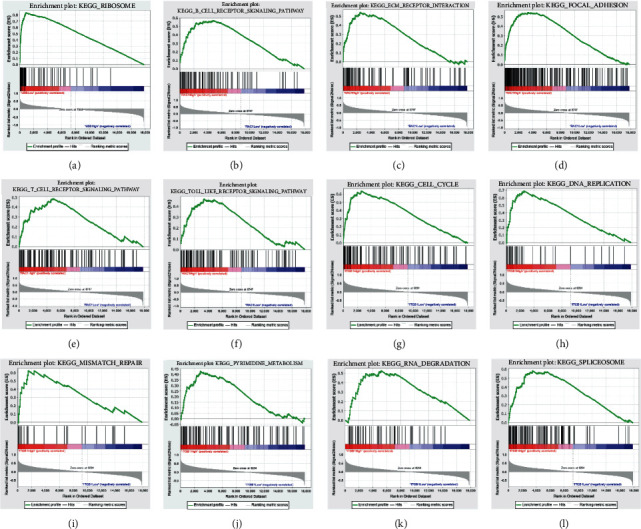
Enrichment plots by GSEA. Relative pathways associated with the expression of (a) UBB, (b–f) RACI, and (g–l) ITGB1 are displayed. GSEA: Gene Set Enrichment Analysis; KEGG: Kyoto Encyclopedia of Genes and Genomes pathway.
Table 2.
KEGG pathways associated with the expression of UBB, RAC1, and ITGB1 using GSEA.
| Gene | Name | ES | NES | NOM P value | FDR q value |
|---|---|---|---|---|---|
| UBB | KEGG_ribosome | 0.84 | 2.00 | ≤0.001 | 0.012 |
|
| |||||
| RAC1 | KEGG_ECM_receptor_interaction | 0.54 | 1.74 | 0.032 | 0.100 |
| KEGG_B_cell_receptor_signaling_pathway | 0.57 | 1.79 | 0.006 | 0.111 | |
| KEGG_T_cell_receptor_signaling_pathway | 0.49 | 1.64 | 0.026 | 0.132 | |
| KEGG_toll_like_receptor_signaling_pathway | 0.47 | 1.64 | 0.019 | 0.133 | |
| KEGG_focal_adhesion | 0.54 | 1.81 | 0.010 | 0.188 | |
|
| |||||
| ITGB1 | KEGG_cell_cycle | 0.64 | 2.09 | ≤0.001 | 0.005 |
| KEGG_spliceosome | 0.59 | 1.97 | ≤0.001 | 0.022 | |
| KEGG_DNA_replication | 0.59 | 1.82 | 0.018 | 0.056 | |
| KEGG_RNA_degradation | 0.53 | 1.83 | ≤0.001 | 0.066 | |
| KEGG_mismatch_repair | 0.62 | 1.68 | 0.032 | 0.204 | |
| KEGG_pyrimidine_metabolism | 0.43 | 1.65 | 0.010 | 0.232 | |
Abbreviations: GSEA: Gene Set Enrichment Analysis; NES: normalized enrichment score; NOM: nominal; FDR: false discovery rate.
4. Discussion
LC commonly has a high mortality rate and results in great socioeconomic pressure for patients, families, and countries. Certainly, smoking contributes to the occurrence and development of LC. However, one in five LUAD cases occurs among patients who do not smoke [3]. The alterations of some genes, including EGFR, ERBB2, ALK, ROS1, and RET, are evidently associated with the occurrence and progression of LUAD [23]. However, the underlying molecular mechanism of nonsmoking-related LUAD remains unclear. Therefore, it is essential to identify crucial biomarkers for understanding the molecular mechanism of nonsmoking-related LUAD. Microarray technology is available for finding new biomarkers, which will be the basis of future studies on the potential mechanism of nonsmoking-related LUAD.
We analyzed two microarray datasets to obtain DEGs between nonsmoking LUAD and matched adjacent lung tissues. A total of 1283 DEGs were selected, containing 743 downregulated and 540 upregulated genes. Function enrichment analyses manifested that DEGs were mainly enriched in collagen-containing ECM, ECM organization, and apical plasma membrane. Moreover, 19 hub genes with degrees greater than 20 were selected from the PPI network: UBB, RAC1, ITGB1, SRC, C3, IL6, CDC20, EGFR, UBE2C, TIMP1, GNG11, CXCL12, GAS6, P4HB, CXCR4, FPR1, ADRB2, LYZ, and MMP9. BP enrichment analysis of these hub genes suggested that apoptotic cell clearance, negative regulation of cysteine-type endopeptidase activity involved in apoptotic process, and leukocyte adhesion to vascular endothelial cell were primarily enriched, and the most enriched KEGG pathway was leukocyte transendothelial migration, bladder cancer, and intestinal immune network for IgA production. Among 19 hub genes, 12 hub genes were closely associated with poorer OS in nonsmoking patients with LUAD. From the results of GO enrichment analysis, KEGG pathway analysis, and survival analysis and degree rank, UBB, RAC1, and ITGB1 were believed to be the core genes in the occurrence and development of nonsmoking-related LAUD at the molecular level. Besides, IHC outcomes in THPA database confirmed that UBB and ITGB1 were downregulated, and RAC1 was upregulated in nonsmoking-related LUAD.
This study was the first one to report the key role of UBB in nonsmoking-related LUAD. Ubiquitin B (UBB) is a crucial member of gene families encoding ubiquitin. Ubiquitin is involved in several cellular processes, and aberrant events in ubiquitin-mediated processes promote the carcinogenesis and progression of NSCLC [24]. UBB is strongly suppressed in some cancers, including endometrial carcinoma and ovarian cancer [25]. Tang et al. revealed that ubiquitin was highly expressed in NSCLC tissues; however, increased ubiquitin was attributed to the increased transcripts of ubiquitin C (UBC) rather than UBB. Moreover, they showed that no significant difference was observed in the UBB mRNA level between NSCLC and normal lung tissues (P = 0.167) [26]. This finding contradicted our results, and there can be two probable reasons for this difference. First, Tang et al. compared NSCLC and normal lung tissues, whereas we compared nonsmoking-related LUAD and normal lung tissues, thus suggesting higher accuracy of our study. Second, this study compared nonsmoking-related LUAD and matched adjacent lung tissues with similar features instead of unmatched normal lung tissues, revealing higher reliability of our study. However, future studies on the expression level of UBB are warranted to confirm findings. Additionally, according to the survival analysis, UBB may play a crucial prognostic role in nonsmoking-related LUAD.
As one member of the RAS superfamily of small GTP-binding proteins, Rac Family Small GTPase 1, RAC1, can interact with effector proteins, and downstream kinases were activated to regulate multiple cellular processes [27]. In addition, RAC1 improved nuclear factor kappa B activity to regulate cell proliferation and migration in NSCLC [28]. RAC1 is upregulated in various cancers, such as LUAD, breast cancer, and kidney cancer [29], and the overexpression of RAC1 is frequently reported to be associated with worse prognosis [30]. One study suggested that high expression of RAC1 plays a key role in the epithelial-mesenchymal transition and malignant progression in LUAD [31]. In addition, KIF18B promotes cell proliferation and invasion through activating RAC1 and mediating the AKT/mTOR signaling pathway in LUAD, indicating the crucial role of RAC1 in the cell proliferation and adverse progression of LUAD [32]. Similarly, Li et al. showed that intracellular mature interleukin 37 can inhibit tumor metastasis through inhibiting RAC1 activation [33], suggesting a crucial role of RAC1 in the tumor metastasis. In addition, RAC1 may predict the prognosis of nonsmoking patients with LUAD in light of our survival analysis.
Integrin subunit beta 1 (ITGB1) encodes the beta subunit of integrins, which is a heterodimeric cell surface receptor and participates in the carcinogenesis, migration, and invasion of LUAD [34]. ITGB1 is abnormally expressed in several cancers, including LUAD and breast cancer [35]. MicroRNA- (miR-) 134 inhibits the migration and metastasis by targeting ITGB1 in NSCLC [36]. Similarly, high expression of miR-493–5p is correlated with better survival of NSCLC via targeting ITGB1 [37], indicating the crucial role of ITGB1 in the carcinogenesis and worse prognosis of NSCLC. Zheng et al. observed that ITGB1 is a predictive biomarker of NSCLC after matching clinical factors (odds ratio (OR) = 1.31, 95% CI: 1.10–1.55) [38]. Our survival analysis also confirmed the prognostic value of ITGB1. Furthermore, GSEA revealed that ribosome was correlated with UBB expression; ECM receptor interaction, B cell receptor signaling pathway, and T cell receptor signaling pathway with RAC1 expression; and cell cycle, spliceosome, and DNA replication with ITGB1 expression. From these findings, we concluded that UBB, RAC1, and ITGB1 were therapeutic and prognostic biomarkers of nonsmoking-related LUAD.
Undeniably, this study was not the first one to select pivotal genes and pathways in nonsmoking-related LUAD using bioinformatics methods. In fact, three similar analyses were previously published [39–41]. However, previous studies had several differences and/or disadvantages compared with this study. First, two analyses [40, 41] compared NSCLC/LC in nonsmoking females and normal lung tissues rather than nonsmoking LUAD, suggesting lower accuracy of the study. Second, LUAD/NSCLC/LC in nonsmoking females and normal lung tissues were compared in the previous studies [39–41], whereas male patients with LUAD were excluded. However, this study has several advantages compared with previous studies: (1) this study included both male and female patients with nonsmoking LUAD, which may have potential benefits for nonsmoking male patients with LUAD; (2) this study compared nonsmoking-related LUAD and matched adjacent lung tissues with similar features, thus having higher reliability; (3) this study revealed some novel findings regarding UBB, UBE2C, GAS6, and P4HB; and (4) some advanced analyses such as GSEA were also performed in this study.
This study also has some limitations. (1) Only two datasets were included. Several similar microarray datasets exist, but they did not meet the selection criteria of our study. Therefore, to decrease the bias, these similar datasets had to be excluded from this study. (2) The included datasets did not reveal detailed information about survival time; therefore, survival analysis of hub genes had to be performed using the Kaplan-Meier plotter. (3) These findings were not verified by performing experiments, which was warranted in future studies.
5. Conclusion
Our study was conducted to select DEGs that may correlate with the carcinogenesis and malignant invasion of nonsmoking-related LUAD. A total of 19 hub genes were selected from the PPI network, and 12 hub genes correlated with the prognosis of nonsmoking-related LUAD. Furthermore, UBB, RAC1, and ITGB1 were potential therapeutic and prognostic indicators of nonsmoking-related LUAD. Moreover, this study is the first to report the key role of UBB in nonsmoking-related LUAD. This study provided evidence for future genomic-based individualized treatments of LUAD in nonsmoking patients. However, future studies are warranted to explore further the biological relationships among these DEGs in nonsmoking-related LUAD.
Acknowledgments
The authors gratefully acknowledge the contributions from the GEO, TCGA, and THPA database. This study is supported by the National Natural Science Foundation of China (grant No. 81672972) and Major Program of Provincial and Ministerial Co-construction, Ministry of Health Science Foundation (grant No. WKJ-ZJ-1701), without the involvement of commercial entities.
Abbreviations
- LC:
Lung cancer
- NSCLC:
Non-small-cell lung cancer
- LUAD:
Lung adenocarcinoma
- TMB:
Tumor mutation burden
- DEGs:
Differentially expressed genes
- GEO:
Gene Expression Omnibus
- TCGA:
The Cancer Genome Atlas
- THPA:
The Human Protein Atlas
- GO:
Gene Ontology
- KEGG:
Kyoto Encyclopedia of Genes and Genomes pathway
- PPI:
Protein-protein interaction network
- adj.P:
Adjusted P values
- FC:
Fold change
- STRING:
Search Tool for the Retrieval of Interacting Genes
- MCODE:
Molecular Complex Detection
- KM:
Kaplan-Meier plotter
- BP:
Biological processes
- CC:
Cellular component
- MF:
Molecular function
- OS:
Overall survival
- HR:
Hazard ratio
- CI:
Confidence interval
- GSEA:
Gene Set Enrichment Analysis
- miRNA:
MicroRNA
- OR:
Odds ratio.
Data Availability
The datasets downloaded and analyzed during the current study are available on the GEO database: GSE32863, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32863;GSE75037, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75037.
Disclosure
The funder had no role in the design or performance of the study; the collection, management, analysis, and interpretation of the data; the preparation, review, and approval of the manuscript; or the decision to submit the manuscript for publication.
Conflicts of Interest
The authors declare that they have no competing interests.
Authors' Contributions
Huan Deng had full access to all of the data in the manuscript and takes responsibility for the integrity of the data and the accuracy of the data analysis. Concept and design was done by all authors. Acquisition, analysis, and interpretation of data were done by all authors. Huan Deng and Ming Chen contributed to the drafting of the manuscript. Critical revision of the manuscript for important intellectual content was done by all authors. Huan Deng, Yichao Huang, and Li Wang contributed to the statistical analysis. Supervision was done by Ming Chen and Yichao Huang. Revision was done by Huan Deng, Yichao Huang, and Ming Chen. Huan Deng and Yichao Huang are co-first authors of this study, and they contribute to this study equally.
Supplementary Materials
Table S1: overall survival analysis of hub genes using K-M plotter.
References
- 1.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2020. CA: A Cancer Journal for Clinicians. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z., Fillmore C. M., Hammerman P. S., Kim C. F., Wong K. K. Non-small-cell lung cancers: a heterogeneous set of diseases. Nature Reviews Cancer. 2014;14(8):535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu M., Lipka M. B., Bhateja P., Fu P., Dowlati A. A detailed smoking history and determination of MYC status predict response to checkpoint inhibitors in advanced non-small cell lung cancer. Translational Lung Cancer Research. 2020;9(1):55–60. doi: 10.21037/tlcr.2020.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H., Deng Y. M., Chen Z. C., et al. Clinical significance of tumor mutation burden and DNA damage repair in advanced stage non-small cell lung cancer patients. European Review for Medical and Pharmacological Sciences. 2020;24(14):776–7672. doi: 10.26355/eurrev_202007_22267. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pros E., Saigi M., Alameda D., et al. Genome-wide profiling of non-smoking-related lung cancer cells reveals common RB1 rearrangements associated with histopathologic transformation in EGFR-mutant tumors. Annals of Oncology. 2020;31(2):274–282. doi: 10.1016/j.annonc.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Dawany N. B., Dampier W. N., Tozeren A. Large-scale integration of microarray data reveals genes and pathways common to multiple cancer types. International Journal of Cancer. 2011;128(12):2881–2891. doi: 10.1002/ijc.25854. [DOI] [PubMed] [Google Scholar]
- 8.Manoli T., Gretz N., Gröne H. J., Kenzelmann M., Eils R., Brors B. Group testing for pathway analysis improves comparability of different microarray datasets. Bioinformatics. 2006;22(20):2500–2506. doi: 10.1093/bioinformatics/btl424. [DOI] [PubMed] [Google Scholar]
- 9.Barrett T., Wilhite S. E., Ledoux P., et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Research. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selamat S. A., Chung B. S., Girard L., et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Research. 2012;22(7):1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girard L., Rodriguez-Canales J., Behrens C., et al. An expression signature as an aid to the histologic classification of non-small cell lung cancer. Clinical Cancer Research. 2016;22(19):4880–4889. doi: 10.1158/1078-0432.CCR-15-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis S., Meltzer P. S. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 13.Ashburner M., Ball C. A., Blake J. A., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu G., Wang L. G., Han Y., He Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franceschini A., Szklarczyk D., Frankild S., et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Research. 2013;41(Database issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reimand J., Isserlin R., Voisin V., et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nature Protocols. 2019;14(2):482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bandettini W. P., Kellman P., Mancini C., et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. Journal of Cardiovascular Magnetic Resonance. 2012;14(1):p. 83. doi: 10.1186/1532-429X-14-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mlecnik B., Galon J., Bindea G. Automated exploration of gene ontology term and pathway networks with ClueGO-REST. Bioinformatics. 2019;35(19):3864–3866. doi: 10.1093/bioinformatics/btz163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Győrffy B., Surowiak P., Budczies J., Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12, article e82241) doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A., Tamayo P., Mootha V. K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., Mesirov J. P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dagogo-Jack I., Robinson H., Mino-Kenudson M., et al. Expediting comprehensive molecular analysis to optimize initial treatment of lung Cancer patients with minimal smoking history. Journal of Thoracic Oncology. 2019;14(5):835–843. doi: 10.1016/j.jtho.2018.12.032. [DOI] [PubMed] [Google Scholar]
- 24.Zhang K., Yang L., Wang J., et al. Ubiquitin-specific protease 22 is critical to in vivo angiogenesis, growth and metastasis of non-small cell lung cancer. Cell Communication and Signaling. 2019;17(1):p. 167. doi: 10.1186/s12964-019-0480-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haakonsen D. L., Rape M. Ubiquitin levels: the next target against gynecological cancers? The Journal of Clinical Investigation. 2017;127(12):4228–4230. doi: 10.1172/JCI98262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y., Geng Y., Luo J., et al. Downregulation of ubiquitin inhibits the proliferation and radioresistance of non-small cell lung cancer cells in vitro and in vivo. Scientific Reports. 2015;5(1, article 9476) doi: 10.1038/srep09476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan X., Guan X., Dong C., Jiao Z. Rho GTPases and related signaling complexes in cell migration and invasion. Experimental Cell Research. 2020;388(1, article 111824) doi: 10.1016/j.yexcr.2020.111824. [DOI] [PubMed] [Google Scholar]
- 28.Gastonguay A., Berg T., Hauser A. D., Schuld N., Lorimer E., Williams C. L. The role of Rac1 in the regulation of NF-κB activity, cell proliferation, and cell migration in non-small cell lung carcinoma. Cancer Biology & Therapy. 2012;13(8):647–656. doi: 10.4161/cbt.20082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De P., Aske J. C., Dey N. RAC1 takes the lead in solid tumors. Cell. 2019;8(5):p. 382. doi: 10.3390/cells8050382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bid H. K., Roberts R. D., Manchanda P. K., Houghton P. J. RAC1: an emerging therapeutic option for targeting cancer angiogenesis and metastasis. Molecular Cancer Therapeutics. 2013;12(10):1925–1934. doi: 10.1158/1535-7163.MCT-13-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seiz J. R., Klinke J., Scharlibbe L., et al. Different signaling and functionality of Rac1 and Rac1b in the progression of lung adenocarcinoma. Biological Chemistry. 2020;401(4):517–531. doi: 10.1515/hsz-2019-0329. [DOI] [PubMed] [Google Scholar]
- 32.Ji Z., Pan X., Shang Y., Ni D. T., Wu F. L. KIF18B as a regulator in microtubule movement accelerates tumor progression and triggers poor outcome in lung adenocarcinoma. Tissue & Cell. 2019;61:44–50. doi: 10.1016/j.tice.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Li Y., Zhao M., Guo C., et al. Intracellular mature IL-37 suppresses tumor metastasis via inhibiting Rac1 activation. Oncogene. 2018;37(8):1095–1106. doi: 10.1038/onc.2017.405. [DOI] [PubMed] [Google Scholar]
- 34.Xia Z. J., Hu W., Wang Y. B., Zhou K., Sun G. J. Expression heterogeneity research of ITGB3 and BCL-2 in lung adenocarcinoma tissue and adenocarcinoma cell line. Asian Pacific Journal of Tropical Medicine. 2014;7(6):473–477. doi: 10.1016/S1995-7645(14)60077-8. [DOI] [PubMed] [Google Scholar]
- 35.Sun Q., Zhou C., Ma R., et al. Prognostic value of increased integrin-beta 1 expression in solid cancers: a meta-analysis. Oncotargets and Therapy. 2018;Volume 11:1787–1799. doi: 10.2147/OTT.S155279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin Q., Wei F., Zhang J., Li B. miR-134 suppresses the migration and invasion of non-small cell lung cancer by targeting ITGB1. Oncology Reports. 2017;37(2):823–830. doi: 10.3892/or.2017.5350. [DOI] [PubMed] [Google Scholar]
- 37.Liang Z., Kong R., He Z., et al. High expression of miR-493–5p positively correlates with clinical prognosis of non small cell lung cancer by targeting oncogene ITGB1. Oncotarget. 2017;8(29):47389–47399. doi: 10.18632/oncotarget.17650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng W., Jiang C., Li R. Integrin and gene network analysis reveals that ITGA5 and ITGB1 are prognostic in non-small-cell lung cancer. Oncotargets and Therapy. 2016;9:2317–2327. doi: 10.2147/OTT.S91796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H., Zhang Z., Xu K., Wei S., Li L., Wang L. Exploration of estrogen receptor-associated hub genes and potential molecular mechanisms in non-smoking females with lung adenocarcinoma using integrated bioinformatics analysis. Oncology Letters. 2019;18(5):4605–4612. doi: 10.3892/ol.2019.10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi K., Li N., Yang M., Li W. Identification of key genes and pathways in female lung cancer patients who never smoked by a bioinformatics analysis. Journal of Cancer. 2019;10(1):51–60. doi: 10.7150/jca.26908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou W., Yin M., Cui H., et al. Identification of potential therapeutic target genes and mechanisms in non-small-cell lung carcinoma in non-smoking women based on bioinformatics analysis. European Review for Medical and Pharmacological Sciences. 2015;19(18):3375–3384. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: overall survival analysis of hub genes using K-M plotter.
Data Availability Statement
The datasets downloaded and analyzed during the current study are available on the GEO database: GSE32863, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32863;GSE75037, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75037.


