SUMMARY
Many mitochondrial diseases are caused by mutations in mitochondrial DNA (mtDNA). Patients’ cells contain a mixture of mutant and nonmutant mtDNA (a phenomenon called heteroplasmy). The proportion of mutant mtDNA varies across patients and among tissues within a patient. We simultaneously assayed single-cell heteroplasmy and cell state in thousands of blood cells obtained from three unrelated patients who had A3243G-associated mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes. We observed a broad range of heteroplasmy across all cell types but also found markedly reduced heteroplasmy in T cells, a finding consistent with purifying selection within this lineage. We observed this pattern in six additional patients who had heteroplasmic A3243G without strokelike episodes. (Funded by the Marriott Foundation and others.)
SOME OF THE MOST CHALLENGING MITOCHONDRIAL DISORDERS ARISE from mutations in mitochondrial DNA (mtDNA), a high-copy-number genome that is maternally inherited. These disorders manifest with marked clinical heterogeneity, in part because tissues generally contain a mixture of both nonmutant and mutant mtDNA — a phenomenon called heteroplasmy. Heteroplasmy varies dramatically across family members, tissues, and time and is hypothesized to be shaped by a combination of random drift and selection. The molecular mechanisms governing the dynamics of mtDNA segregation remain unclear, but heritability studies involving humans1 and model systems2 indicate the presence of tissue-specific genetic influences.
The mitochondrial A3243G mutation, which disrupts the mitochondrial leucyl-tRNA gene, MT-TL1, is the most common heteroplasmic pathogenic mtDNA mutation3,4 and is classically associated with mitochondrial encephalomyopathy with lactic acidosis and strokelike episodes (MELAS).5,6 The phenotypic spectrum of heteroplasmic A3243G mutations is broad and can be associated with a variety of symptoms, including diabetes, deafness, gastrointestinal dysmotility, epilepsy, and strokelike episodes.7 Severity can range from comparatively mild disease in persons with maternally inherited diabetes and deafness to devastating multisystemic disease in persons with MELAS.1 A3243G heteroplasmy is known to vary widely among siblings and across tissues,1,8-10 thereby complicating clinical management and genetic counseling. At present, no therapies have been approved by the Food and Drug Administration (FDA), although emerging reproductive technologies to prevent transmission of the mutation are now in trials.11
Investigating the segregation of pathogenic mtDNA mutations such as A3243G by means of bulk analysis of heteroplasmy in human tissues is challenging, because tissues consist of many cell types, with distinct developmental origins and proportions. Single-cell analysis of heteroplasmy in defined cell types has been limited to at most a few dozen cells, primarily in the germline.12-15
Here, we report the application of a single-cell genomics technology,16,17 mtDNA single-cell ATAC (assay for transposase-accessible chromatin) sequencing, to determine mtDNA heteroplasmy and cell type simultaneously in many thousands of peripheral-blood mononuclear cells (PBMCs) that were obtained from three unrelated patients with MELAS. PBMCs consist of multiple cell types originating from a common stem and progenitor pool; we sought to use mtDNA single-cell ATAC sequencing to assess the segregation dynamics of pathogenic mutations in each blood-cell lineage.
CASE REPORTS
Patient 21 was a 35-year-old man with MELAS that was characterized by strokelike episodes, failure to thrive, and steatohepatitis. In this patient, clinical molecular testing identified the A3243G mutation and heteroplasmy was not quantified (Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org).
Patient 9 was a 29-year-old man with MELAS that was characterized by sensorineural hearing loss, migraine, epilepsy, ptosis, and strokelike episodes. This patient had A3243G heteroplasmy of 39% in whole blood, as assessed with clinical long-range polymerase chain reaction (PCR) and next-generation sequencing.
Patient 30 was a 60-year-old man with MELAS and associated sensorineural hearing loss, ptosis, strokelike episodes, diabetes mellitus, skeletal myopathy with ragged red fibers, and cardiomyopathy. This patient had 77% A3243G heteroplasmy in skeletal muscle, as assessed with clinical long-range PCR and next-generation sequencing.
METHODS
SINGLE-CELL ACCESSIBLE CHROMATIN AND MITOCHONDRIAL GENOTYPING
This study was approved by the institutional review board of Massachusetts General Hospital. We obtained samples of venous blood at clinical baseline and purified PBMCs from the patients. We stained cells for viability and applied antihuman CD45 antibodies before fixation and performed fluorescence-activated cell sorting (FACS) to exclude dead and nonleukocyte cells (CD45−). The mtDNA single-cell ATAC sequencing libraries were generated by a 10× Chromium Controller and a modified Chromium Single Cell ATAC Library and Gel Bead Kit protocol, which was followed by paired-end sequencing with the use of an Illumina NextSeq 500 platform (2× 72-bp reads).
DATA ANALYSIS
We demultiplexed and aligned raw sequencing reads to the hg19 reference genome using Cell-Ranger-ATAC software, version 1.0 (10× Genomics). We identified cells as bar codes that met the following criteria: at least 1000 unique fragments mapping to the nuclear genome, at least 40% of nuclear fragments overlapping a previously established chromatin accessibility peak set in the hematopoietic system,18 and a mean mtDNA coverage of at least 20×. From the output of the CellRanger-ATAC call, we quantified mtDNA using the mgatk software package, version 0.5.0.17
We computationally identified cell types on the basis of chromatin accessibility. In brief, we reprocessed cells from a healthy person19 to define axes of variation using latent semantic indexing and uniform manifold approximation and projection (UMAP). Next, we projected patient-derived cells onto this reduced-dimension space using the latent semantic indexing and UMAP loadings, as previously described.20 We used k nearest neighbors (where k = 20) to generate 12 data-driven clusters by means of Louvain community detection, which we mapped onto five major expected cell types in PBMCs (monocytes, dendritic cells, T cells, B cells, and natural killer [NK] cells). The clustering was robust to the choice of k (see the Data Analysis section in the Supplementary Appendix). We classified all cell types in the samples from patients by latent sematic indexing projection and minimum distance to cluster medoids. For visualization, we produced two-dimensional representations of patients’ PBMC data by projecting the 25 latent semantic indexing dimensions onto the pretrained UMAP model, as previously reported.20
For heteroplasmy analyses, we excluded all the cells with less than 20× coverage at position m.3243. Outliers with m.3243 coverage of more than 1.5 times the upper boundary of the interquartile range were also excluded to avoid the inclusion of artifactual sequencing multiplets. We calculated the fraction of total read fragments aligning to the mitochondrial genome in each cell as a proxy for the mtDNA copy number.
To compare the distribution of heteroplasmy in T cells with that of all PBMCs within an individual patient, we used the Kolmogorov–Smirnov two-sample test statistic, D, which is defined as the maximum difference between cumulative distributions at any given point; the D-statistic is expected to approach 0 for identical distributions and to be as high as 1 when two distributions are distinct. We evaluated significance analytically or empirically using permutation testing. We used the R base and stats software package, version 3.5.1, and base software, version 3.5.1, to perform these computations. Data analyses and visualization were also conducted with the use of R software.
BULK HETEROPLASMY ANALYSIS
We stained PBMCs with antibodies against antihuman CD45 and antihuman CD56 and used FACS to purify T-cell and T cell-depleted PBMC populations from which DNA was extracted. Small amplicons centered on m.3243 were generated by means of PCR and sequenced on an Illumina MiSeq platform. We aligned reads using the Burrows–Wheeler alignment tool21 and analyzed them with Samtools.22 We additionally purified T cells using magnetic-bead negative selection kits. DNA from purified T cells and total PBMCs was extracted and used for the generation of m.3243 region PCR amplicons for Sanger sequencing. Full details are provided in the Methods section in the Supplementary Appendix.
RESULTS
CHROMATIN ACCESSIBILITY AND HETEROPLASMY IN SINGLE CELLS
Using mtDNA single-cell ATAC sequencing, we generated high-quality sequencing Iibraries to simultaneously evaluate cell type and heteroplasmy in thousands of individual cells per patient. From Patient 21, we sequenced 7176 cells (median, 8146 nuclear fragments per cell) that passed quality control; from Patient 9, we sequenced 6003 cells (median, 6672 nuclear fragments per cell) that passed quality control; and from Patient 30, we sequenced 6007 cells (median, 6507 nuclear fragments per cell) that passed quality control.
Using accessible chromatin signatures derived from nuclear genomic reads, we defined cell states using a latent semantic indexing projection of each patient’s data set onto a single-cell reference map of healthy-donor PBMCs that had been generated by a similar single-cell ATAC sequencing protocol.18 The clusters generated by each analysis were remarkably similar and had accessible chromatin profiles that were characteristic of canonical PBMC cell types (Fig. 1). The overall distributions of the PBMC types identified by this protocol in our patients were similar to those in previously reported healthy-donor PBMC data sets.23 Furthermore, all the patients had normal representation of blood-cell types on clinical complete blood counts (Table S2). Together, these results indicated no major perturbation in lineage frequencies in these patients.
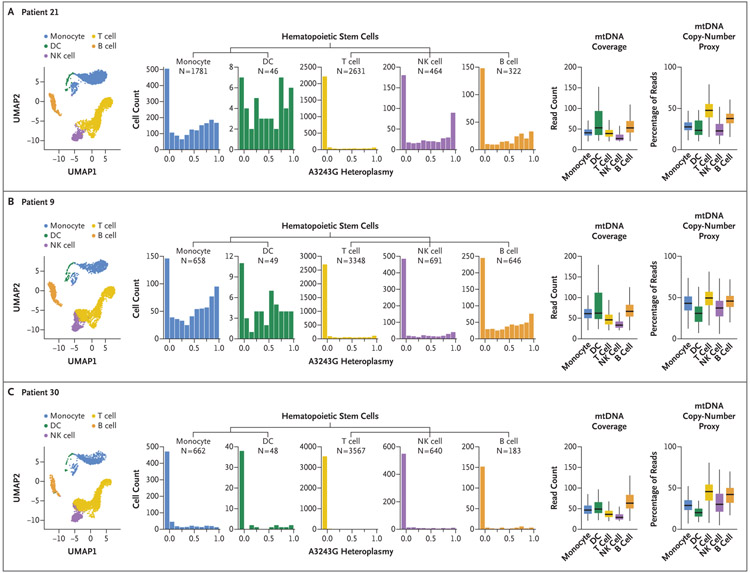
Figure 1. T-Cell-Specific Reduction in A3243G Heteroplasmy in Peripheral-Blood Mononuclear Cells (PBMCs) from Patients with MELAS.
The uniform manifold approximation and projection (UMAP) of mitochondrial DNA (mtDNA) single-cell ATAC sequencing data from Patients 21, 9, and 30 show the distribution of indicated major PBMC cell types (left-most graphs) in a single sample of blood obtained from each of these patients with MELAS (mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes). Histograms show the A3243G heteroplasmy fraction according to indicated cell types for each of the three patients, with the cell number (N) per population shown (center graphs). Box plots are shown for per-cell mtDNA coverage at m.3243 (second graphs from the right) and for a proxy of mtDNA copy number (i.e., the percentage of per-cell reads aligning to mtDNA) (right-most graphs). Our analyses excluded cells with a coverage at m.3243 of less than 20× or of more than 1.5 times the upper boundary of the interquartile range. In the right-most graphs, the horizontal line in the box indicates the median, the edges of the box the interquartile range, and the bars 1.5 times the interquartile range. DC denotes dendritic cell, and NK natural killer.
CELL TYPE-SPECIFIC HETEROPLASMY
We next examined heteroplasmy across PBMC cell types, restricting our analyses to those cells with at least 20× coverage at position m.3243. All the cell types showed a broad spectrum of heteroplasmy, ranging from no A3243G alleles detected to exclusively A3243G mutations detected within each lineage, even in patients with low (<10%) bulk heteroplasmy (Fig. 1). This observation held true even when we limited the analysis to cells with 100× coverage at m.3243 in Patient 21, in which case we still observed cells in which exclusively nonmutant or exclusively mutant alleles were detected (Fig. S1).
Levels of heteroplasmy tended to be lower within lymphocytes (B cells, T cells, and NK cells) than within myeloid (monocyte or dendritic cell) lineages, with T cells consistently showing the lowest levels (Fig. 1 and Fig. S2). For each patient, we compared the distribution of heteroplasmy for the T cells with that of all lineages combined using a two-sample Kolmogorov–Smirnov D-statistic. For Patient 21, the D-statistic comparing T cells with total PBMCs was 0.52; for Patient 9, it was 0.38; and for Patient 30, it was 0.20 (Dα = 0.03, for α = 0.05, in each patient). In all three patients, the observed D-statistics exceeded those obtained by random permutation (Fig. S3). These analyses indicated that within an individual patient, the distribution of A3243G heteroplasmy in T cells was not identical to the distribution of heteroplasmy in PBMCs. This pattern of reduced T-cell heteroplasmy was observed in CD4+ and CD8+ T cells (Fig. S4).
We validated and extended this result of reduced heteroplasmy in the T-cell lineage with traditional bulk heteroplasmy analysis involving two of these three patients and several additional patients (Table 1). In these validation studies, we purified T cells using either of two methods (FACS or bead-based negative selection). Using these orthogonal methods, we validated our findings of reduced T-cell heteroplasmy in two of the three patients discussed above (Patients 9 and 30) for whom additional blood samples were available. We then used these bulk methods to compare heteroplasmy in T cells with that in total PBMCs in six additional patients who had heteroplasmic A3243G disease but not strokelike episodes. (The patients’ clinical testing and presentations are summarized in Table S1.) In all six additionaI patients, T-cell populations showed lower heteroplasmy than all PBMCs combined or T-cell-depleted PBMCs (Table 1). Hence, our observations of reduced heteroplasmy appeared to be robust to assay with different methods.
Table 1.
Validation of Reduced A3243G Heteroplasmy in T Cells by Bulk Sequencing.*
| Patient No. | Age | Sex | Bulk Heteroplasmy Measurements | ||
|---|---|---|---|---|---|
| Total PBMCs |
T-Cell–Depleted PBMCs |
T Cells | |||
| yr | percent | ||||
| 9 | 29 | Male | 28.8 | 9.9 | |
| 30 | 60 | Male | 9.6 | 9.5 | 0.7 |
| 31 | 47 | Female | 4.9 | 1.0 | |
| 33 | 65 | Female | 6.2 | 5.7 | 2.7 |
| 36 | 53 | Female | 16.3 | 5.9 | |
| 37 | 19 | Female | 42.1 | 24.8 | |
| 38 | 33 | Male | 46.1 | 32.2 | |
| 40 | 35 | Male | 7.9 | 3.2 | |
We measured, in a single sample obtained from each patient, the percent of A3243G heteroplasmy for total peripheral-blood mononuclear cells (PBMCs), flow-sorted T-cell–depleted PBMCs, and T cells purified by negative selection as measured by next-generation sequencing of a polymerase-chain-reaction amplicon encompassing the m.3243 position. Owing to insufficient sample availability, bulk sequencing was not performed for Patient 21 (Table S1).
We investigated whether differences in the mtDNA copy number might account for the observed T-cell-specific depletion of the heteroplasmic mutation. T-cell activation induces mitochondrial biogenesis,24,25 and in worms, the regulation of mtDNA copy number is associated with mtDNA surveillance.26 Although a proxy for the mtDNA copy number varied according to cell type (Fig. 1), it did not show a relationship to heteroplasmy within any cell type (Figs. S5 and S6).
DISCUSSION
The dynamics of heteroplasmy is one of the most clinically challenging and scientifically fascinating aspects of mtDNA disease. Bulk heteroplasmy measurements across tissue types and kindreds have not provided explanations of the origin, transmission, variability, and pathogenic mechanisms of pathologic mtDNA heteroplasmy. However, blood heteroplasmy has long shown several peculiarities, including lower bulk heteroplasmy than in other tissues,1,8-10,27 a weaker direct association with disease severity than is observed with urine sediment (another clinically tested biospecimen),1,8,27 and a tendency to decrease with age.8,9,28-30 At present, the mechanisms governing these complex dynamics are not known, but previous studies have predicted the existence of genetic factors that influence tissue-specific heteroplasmy.1,2,31
Single-cell analysis of heteroplasmy holds promise for the elucidation of mechanisms regulating mtDNA heteroplasmic dynamics, but studies involving patients have, to date, largely been restricted to the study of one cell type at a time (typically oocytes) at a limited scale. In previous studies, heteroplasmy was examined in 82 oocytes15 and 8 pancreatic beta cells32 in single patients with A3243G heteroplasmy. Studies of T8993G heteroplasmy have involved restriction enzyme–based analysis in cells from single donors, including 87 oocytes,12 2 blastomeres,13 and 30 lymphocytes.14
Emerging single-cell technologies now enable the study of heteroplasmy at massive scale and high throughput,17 which has allowed us to investigate A3243G heteroplasmy and cell type simultaneously in thousands of individual cells representing multiple lineages arising from a common blood stem or progenitor pool in three unrelated patients. By investigating single-cell heteroplasmy on this scale, we made an unexpected observation regarding A3243G heteroplasmy across somatic lineages. In each patient and cell type we assessed, we were able to observe individual cells spanning a broad range of heteroplasmy. This distribution tended to be left-shifted in lymphocytes, most dramatically in T cells. In our study, in all three patients who underwent assessment by mtDNA single-cell ATAC sequencing (Fig. 1), as well as all six additional patients who underwent assessment by bulk heteroplasmy analysis (Table 1), we observed lower levels of heteroplasmy in T cells than in PBMCs in aggregate. This observation is not consistent with purely random segregation of the A3243G mutation.
The most parsimonious explanation for our observations is that they reflect the action of purifying selection against the pathogenic mtDNA allele within the T-cell lineage. Whether this selection occurs at the level of the cell, the organelle, or the molecule is yet to be determined. We found that normal relative and total numbers of T cells were maintained in patients. Given that the common lymphoid progenitor is the cell that gives rise to T-cell, B-cell, and NK-cell lineages, selection against higher heteroplasmy in T cells would be expected at this developmental stage or later. The A3243G mutation is known to cause a deficiency in the activity of complex I of the electron transport chain,33,34 and multiple previous studies in mouse models have shown that complete knockouts of nuclear-encoded mitochondrial proteins in the whole organism,35,36 at specific developmental phases,37 or selectively in T cells38 can impair T-cell development, homeostasis, or immune function. It is therefore conceivable that a cell-intrinsic or noncell autonomous T-cell-specific process in the bone marrow, the thymus, or the peripheral circulation may select against high heteroplasmy, with features enriched in T-cell biology being good candidates. Developmentally, A3243G-related mitochondrial dysfunction might, for example, present an insurmountable barrier in positive thymic selection or serve as a trigger for elimination during negative selection. Alternatively, immune mechanisms may be in place that actively surveil protein products of mutant mtDNA molecules and eliminate such cells in the T-cell lineage. For example, mutations in the mt-Nd1 gene have been shown to produce a peptide that is recognized by cytotoxic T cells in mice.39 Our observation may also represent a compensatory mechanism to guard against inflammatory responses activated by T cells with dysfunctional mitochondria.40 Further work will be necessary to determine the mechanism underlying purifying selection and whether it can be generalized to other mtDNA mutations. Moreover, future studies will be needed to investigate the effects of purifying selection on the immune system more broadly and its relationship to age-related decreases in A3243G.8,9,28-30
The understanding of dynamics of heteroplasmy within blood lineages has important clinical implications. First, future studies will be needed to determine whether the differences in the dynamics of heteroplasmy that we observed across immune cell types affect their function. This is an important area of inquiry given the long-standing observation that decompensations in mitochondrial disease are often triggered by infections. Second, with regard to the diagnosis and monitoring of heteroplasmic disease, and given the observed heterogeneity of heteroplasmy across cell types, we hypothesize that the analysis of heteroplasmy in defined blood lineages has greater diagnostic and prognostic value than the analysis of heteroplasmy in whole blood. Ultimately, a new diagnostic assay using single-cell profiling of peripheral-blood cells41 may prove to be useful and even cost-effective as the costs of sequencing decrease. Finally, given that we currently have no FDA-approved treatments for mitochondrial disease, understanding the determinants of reduced T-cell heteroplasmy may inspire new therapeutic approaches that exploit endogenous mechanisms that surveil and purify pathogenic mtDNA alleles.
Supplementary Material
Acknowledgments
Supported by the Marriott Foundation (to Dr. Mootha), the MacCurtain Family, the Tosteson Fund for Medical Discovery, the Massachusetts General Hospital Department of Neurology (to Dr. Walker), the New York Stem Cell Foundation (to Dr. Sankaran), the National Institute of Diabetes and Digestive and Kidney Diseases and National Institutes of Health (to Dr. Sankaran), the Klarman Cell Observatory (to Dr. Regev), the National Human Genome Research Institute Center of Excellence in Genome Science (to Dr. Regev), and the National Center for Advancing Translational Sciences (National Institutes of Health Award, UL 1TR002541). Dr. Sankaran is a New York Stem Cell Foundation Robertson Investigator, and Drs. Regev and Mootha are Investigators of the Howard Hughes Medical Institute.
We thank Amanda Allred, Erica Kelly, and Olga Goldberger for technical assistance and the staff of the Harvard Catalyst–Harvard Clinical and Translational Science Center for consulting services.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
Contributor Information
Melissa A. Walker, Department of Molecular Biology, Neurology, Massachusetts General Hospital, Howard Hughes Medical Institute, Harvard Medical School, Boston; Broad Institute of MIT (Massachusetts Institute of Technology) and Harvard, Cambridge, Massachusetts
Caleb A. Lareau, Division of Hematology–Oncology, Boston Children’s Hospital; Department of Pediatric Oncology, Dana–Farber Cancer Institute, Harvard Medical School; Broad Institute of MIT (Massachusetts Institute of Technology) and Harvard, Cambridge, Massachusetts
Leif S. Ludwig, Division of Hematology–Oncology, Boston Children’s Hospital; Department of Pediatric Oncology, Dana–Farber Cancer Institute, Harvard Medical School, Klarman Cell Observatory, Cambridge, Massachusetts
Amel Karaa, Genetics Unit, Department of Pediatrics, Harvard Medical School, Boston
Vijay G. Sankaran, Division of Hematology–Oncology, Boston Children’s Hospital; Department of Pediatric Oncology, Dana–Farber Cancer Institute, Harvard Medical School; Broad Institute of MIT (Massachusetts Institute of Technology) and Harvard, Harvard Stem Cell Institute, Cambridge, Massachusetts
Aviv Regev, Massachusetts General Hospital, Howard Hughes Medical Institute, Klarman Cell Observatory; Department of Biology and Koch Institute of Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, Massachusetts
Vamsi K. Mootha, Department of Molecular Biology, Medicine, Massachusetts General Hospital, Howard Hughes Medical Institute; Department of Systems Biology, Harvard Medical School; Broad Institute of MIT (Massachusetts Institute of Technology) and Harvard, Cambridge, Massachusetts
REFERENCES
- 1.Pickett SJ, Grady JP, Ng YS, et al. Phenotypic heterogeneity in m.3243A>G mitochondrial disease: the role of nuclear factors. Ann Clin Transl Neurol 2018;5: 333–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet 1997;16:93–5. [DOI] [PubMed] [Google Scholar]
- 3.Manwaring N, Jones MM, Wang JJ, et al. Population prevalence of the MELAS A3243G mutation. Mitochondrion 2007;7:230–3. [DOI] [PubMed] [Google Scholar]
- 4.Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet 2008;83:254–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goto Y, Nonaka I, Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 1990; 348:651–3. [DOI] [PubMed] [Google Scholar]
- 6.Hirano M, Ricci E, Koenigsberger MR, et al. MELAS: an original case and clinical criteria for diagnosis. Neuromuscul Disord 1992;2:125–35. [DOI] [PubMed] [Google Scholar]
- 7.Kaufmann P, Engelstad K, Wei Y, et al. Protean phenotypic features of the A3243G mitochondrial DNA mutation. Arch Neurol 2009;66:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grady JP, Pickett SJ, Ng YS, et al. mtDNA Heteroplasmy level and copy number indicate disease burden in m.3243A>G mitochondrial disease. EMBO Mol Med 2018;10(6):e8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Laat P, Koene S, van den Heuvel LPWJ, Rodenburg RJT, Janssen MCH, Smeitink JAM. Clinical features and heteroplasmy in blood, urine and saliva in 34 Dutch families carrying the m.3243A > G mutation. J Inherit Metab Dis 2012;35:1059–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeda K, Kawai H, Sanada M, et al. Clinical phenotype and segregation of mitochondrial 3243A>G mutation in 2 pairs of monozygotic twins. JAMA Neurol 2016; 73:990–3. [DOI] [PubMed] [Google Scholar]
- 11.Hyslop LA, Blakeley P, Craven L, et al. Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease. Nature 2016;534:383–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blok RB, Gook DA, Thorburn DR, Dahl HHM. Skewed segregation of the mtDNA nt 8993 (T→) mutation in human oocytes. Am J Hum Genet 1997;60:1495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steffann J, Frydman N, Gigarel N, et al. Analysis of mtDNA variant segregation during early human embryonic development: a tool for successful NARP preimplantation diagnosis. J Med Genet 2006; 43:244–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gigarel N, Ray PF, Burlet P, et al. Single cell quantification of the 8993T>G NARP mitochondrial DNA mutation by fluorescent PCR. Mol Genet Metab 2005; 84:289–92. [DOI] [PubMed] [Google Scholar]
- 15.Brown DT, Samuels DC, Michael EM, Turnbull DM, Chinnery PF. Random genetic drift determines the level of mutant mtDNA in human primary oocytes. Am J Hum Genet 2001;68:533–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludwig LS, Lareau CA, Ulirsch JC, et al. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell 2019;176(6):1325–1339.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lareau CA, Ludwig LS, Muus C, et al. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat Biotechnol (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ulirsch JC, Lareau CA, Bao EL, et al. Interrogation of human hematopoiesis at single-cell and single-variant resolution. Nat Genet 2019;51:683–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satpathy AT, Granja JM, Yost KE, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol 2019;37:925–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granja JM, Klemm S, McGinnis LM, et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat Biotechnol 2019; 37:1458–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludwig LS, Lareau CA, Bao EL, et al. Transcriptional states and chromatin accessibility underlying human erythropoiesis. Cell Rep 2019;27(11):3228–3240.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ron-Harel N, Santos D, Ghergurovich JM, et al. Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab 2016;24:104–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filograna R, Koolmeister C, Upadhyay M, et al. Modulation of mtDNA copy number ameliorates the pathological consequences of a heteroplasmic mtDNA mutation in the mouse. Sci Adv 2019;5(4):eaav9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haroon S, Li A, Weinert JL, et al. Multiple molecular mechanisms rescue mtDNA disease in C. elegans. Cell Rep 2018;22:3115–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fayssoil A, Laforêt P, Bougouin W, et al. Prediction of long-term prognosis by heteroplasmy levels of the m.3243A>G mutation in patients with the mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes syndrome. Eur J Neurol 2017;24:255–61. [DOI] [PubMed] [Google Scholar]
- 28.Rahman S, Poulton J, Marchington D, Suomalainen A. Decrease of 3243 →G mtDNA mutation from blood in MELAS syndrome: a longitudinal study. Am J Hum Genet 2001;68:238–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pyle A, Taylor RW, Durham SE, et al. Depletion of mitochondrial DNA in leucocytes harbouring the 3243A->G mtDNA mutation. J Med Genet 2007;44:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehrazin M, Shanske S, Kaufmann P, et al. Longitudinal changes of mtDNA A3243G mutation load and level of functioning in MELAS. Am J Med Genet A 2009;149A:584–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jokinen R, Marttinen P, Sandell HK, et al. Gimap3 regulates tissue-specific mitochondrial DNA segregation. PLoS Genet 2010;6(10):e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynn S, Borthwick GM, Charnley RM, Walker M, Turnbull DM. Heteroplasmic ratio of the A3243G mitochondrial DNA mutation in single pancreatic beta cells. Diabetologia 2003;46:296–9. [DOI] [PubMed] [Google Scholar]
- 33.Nishizawa M, Tanaka K, Shinozawa K, et al. A mitochondrial encephalomyopathy with cardiomyopathy: a case revealing a defect of complex I in the respiratory chain. J Neurol Sci 1987;78:189–201. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka M, Nishikimi M, Suzuki H, et al. Deficiency of subunits of complex I or IV in mitochondrial myopathies: immunochemical and immunohistochemical study. J Inherit Metab Dis 1987;10:284–8. [DOI] [PubMed] [Google Scholar]
- 35.Cabon L, Bertaux A, Brunelle-Navas MN, et al. AIF loss deregulates hematopoiesis and reveals different adaptive metabolic responses in bone marrow cells and thymocytes. Cell Death Differ 2018;25:983–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramstead AG, Wallace JA, Lee SH, et al. Mitochondrial pyruvate carrier 1 promotes peripheral T cell homeostasis through metabolic regulation of thymic development. Cell Rep 2020;30(9):2889–2899.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simula L, Pacella I, Colamatteo A, et al. Drp1 controls effective T cell immune-surveillance by regulating T cell migration, proliferation, and cMyc-dependent metabolic reprogramming. Cell Rep 2018; 25(11):3059–3073.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarasenko TN, Pacheco SE, Koenig MK, et al. Cytochrome c oxidase activity is a metabolic checkpoint that regulates cell fate decisions during T cell activation and differentiation. Cell Metab 2017; 25(6):1254–1268.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loveland B, Wang CR, Yonekawa H, Hermel E, Lindahl KF. Maternally transmitted histocompatibility antigen of mice: a hydrophobic peptide of a mitochondrially encoded protein. Cell 1990;60:971–80. [DOI] [PubMed] [Google Scholar]
- 40.Desdín-Micó G, Soto-Heredero G, Aranda JF, et al. T cells with dysfunctional mitochondria induce multimorbidity and premature senescence. Science 2020;368:1371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Regev A, Teichmann SA, Lander ES, et al. The human cell atlas. Elife 2017;6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.