Abstract
Background:
Mechanisms that facilitate early infection and inflammation in cystic fibrosis (CF) are unclear. We previously showed that young CF children with secondhand smoke exposure (SHSe) have increased susceptibility to respiratory infections. We aimed to define the impact of SHSe and other external factors upon the fecal bacteriome in early CF.
Methods:
Twenty CF infants and children were enrolled, clinical data recorded, and hair nicotine measured as an objective surrogate of SHSe. Fecal samples were collected at clinic visits and bacteriome 16S rRNA gene sequencing performed.
Results:
SHSe was associated with increased alpha diversity and increased relative abundance of Acinetobacter and Akkermansia, along with decreased Bifidobacterium and Lactobacillus. Recent antibiotic exposure predicted bacterial population structure in children < 2 years of age and was associated with decreased Bacteroides relative abundance. Age was the strongest predictor of overall fecal bacterial composition and positively associated with Blautia and Parabacteroides. Weight for length was negatively associated with Staphylococcus relative abundance.
Conclusions:
SHSe and other external factors such as antibiotics appear to alter fecal bacterial composition in young CF children, but the strongest predictor of overall composition was age. These findings have implications for understanding the intestinal microbiome in young CF children.
INTRODUCTION
Cystic fibrosis (CF) is caused by defects in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. CFTR is widely expressed throughout the body and deficits in CFTR function result in variable disease manifestations starting in utero. Infants with CF demonstrate early pathology including progressive lung disease, recurrent sinorespiratory infections, and gastrointestinal complications.1 Emerging evidence suggests that one contributor to early CF disease is the establishment of persistent shifts in intestinal and respiratory microbial populations that can affect clinical outcomes or symptoms (dysbiosis) compared to infants without CF.2–16 Within CF there can also be a gradient of dysbiosis associated with worsened outcomes, as progressive intestinal dysbiosis distinguishes between CF infants with normal and poor linear growth.17 In particular for the fecal microbiome, decreased abundance of Bacteroides and increased abundance of Proteobacteria have been observed in several CF studies.14,17,18 However, factors that modulate early microbial composition throughout the body in CF are poorly defined.
While the early CF intestinal and respiratory microbiomes are shaped by both internal (e.g. genetic, immune cell responses) and external (e.g. diet, maternal microbiota, antibiotic usage, environmental exposures) factors,19 the specific mechanisms that facilitate the early development of the intestinal microbiome and associated inflammation in young children with CF are unclear. We previously demonstrated that young children with CF and parental-reported20 or objectively detected21 secondhand smoke exposure (SHSe) have increased susceptibility to respiratory bacterial infections. A similar role for SHSe in the regulation of intestinal bacteria in children with CF has not been determined, but likely to be present given the relationship between smoke exposure and intestinal inflammation.22 Although it is well established that SHSe should be avoided in children with CF, we have demonstrated that SHSe remains highly prevalent in young children with CF despite preventative programs.21 Therefore, continued understanding of potential alterations induced by SHSe and other external factors are needed.
The purpose of this study was to define the impact of SHSe upon the intestinal bacteriome in a pilot study of young children with CF. Secondary outcomes included the influence of age, sex, birth route and modifiable factors (antibiotic use and feeding method) upon the intestinal bacteriome. We hypothesized that SHSe shifts CF fecal bacterial populations in a differential manner compared to other modifiable factors such as diet or antibiotics. To test this hypothesis, we studied the fecal bacteriome in young children with CF and objectively measured SHSe to identify potential targets for therapeutic intervention (e.g. pre- or pro-biotics) in this vulnerable population.
METHODS
Subjects
A group of children with CF ages 3 months to 5 years were recruited during baseline health visits at the outpatient CF clinic as part of a larger study (Determining Infectious Signatures in CF, DISC) which aimed to study influence of SHSe on infection, inflammation, and clinical outcomes.21 The purpose of studying this sub-group was to specifically examine the influence of SHSe on the fecal bacteriome. The diagnosis of CF was defined by 2 disease-causing mutations or a sweat chloride test ≥ 60 mmol/L. All patients had at least one copy of the Phe508del mutation. Participants were studied as an overall group and divided into young children less than 1.3 years and those greater than 1.3 years based on microbiome differences. The study was approved by the Institutional Review Board at Nationwide Children’s Hospital (IRB12–00084). A parent or guardian of the child participant provided informed consent on their behalf.
Clinical and biochemical measures
For this study we collected clinical information using a clinical questionnaire, hair to measure nicotine concentrations, oropharyngeal swabs or sputum for respiratory bacterial growth assessment, serum lipid metabolomics, and fecal samples for bacteriome analysis. Oral antibiotic exposure in the month before fecal sample collection was recorded. Patients also underwent pulmonary function testing, and data was recorded. Lung function was measured as forced expiratory volume in 0.5 seconds (FEV0.5) and 1 second (FEV1) for infants and children, respectively. Percent predicted measurements and z scores for forced vital FEV0.5 and FEV1 were derived from reference equations.23,24 Growth was quantified as length, weight, and BMI or weight/length z scores by CF dieticians trained in anthropometric measurements using World Health Organization standardized growth charts. All information was stored in a secure REDCap database.
Secondhand smoke exposure (SHSe)
As previously described,21 hair nicotine concentrations were measured in each participant. Hair nicotine provides a long-term, stable measure of SHSe as nicotine is integrated into the growing hair shaft over multiple months.25 Hair was obtained following a standardized protocol previously described.26 Samples were processed by reverse-phase high-performance liquid chromatography with electrochemical detection as described.25 Lower cutoffs may poorly distinguish SHSe from no exposure,27 therefore a cutoff concentration of 1.0ng/mg was used to dichotomize groups into (+) and (-) SHSe.
Fecal bacteriome processing/sequencing
Fecal specimens were collected fresh during clinic visits or using a frozen diaper from the morning of the visit if an immediately fresh sample was not available. Fecal samples were aliquoted into sterile tubes containing RNAlater and immediately frozen at -80° until further processing. DNA was extracted from the fecal samples using the Zymo fecal DNA miniprep kit following the manufacturers instructions without modification, and 16S rRNA gene amplicon sequencing performed by targeted sequencing of the V4-V5 hypervariable region at the Marine Biological Laboratory as described previously.14,28
Briefly, amplification reactions were prepared as follows: 1X Platinum HiFi Taq buffer, 2 mM MgSO4, 0.2 mM dNTPs, 0.2–0.4 uM each primer, 12.5 units of Platinum HiFi Taq, and up to 25 ng of template and brought to a final volume of 100 ul with water. This reaction was split into replicate reactions (33 ul, thus reducing amplifiction bias) and a no-template negative control reaction was included for each primer set. Amplifications were performed as follows: denature for 3 min at 94°C; then 30 cycles of 30s at 94°C, 45s at 57°C, and 1 min at 72°C; with a final extension for 2 min at 72°C. Replicates were combined, the products cleaned and size-selected the products with Agencourt Ampure XP beads (0.75X sample volume), quantified using a PicoGreen assay (LifeTechnologies/Invitrogen), and finally, the products were pooled and the pool was size-selected with 1X volume of Ampure XP. The positive and negative reactions were visualized on a Perkin Elmer Caliper LabChipGX. The library pool was sequenced on an Illumina MiSeq to produce 250 nt paired-end reads, then processed as reported previously to prepare for downstream analysis.28
Fecal 16S rRNA gene amplicon quality control, taxonomy assignment, and statistical analysis
Quality control and taxonomy assignment were conducted with IM-Tornado29 and the Silva database (v128). Reads passing quality control were required to have a minimum length of 190 bases, and the reverse reads were trimmed to 200 bases. 100% similarity threshold was set for OTU binning. Alpha diversity for non-phylogenetic richness (observed OTUs), phylogenetic richness (Faith’s PD), and evenness (Pielou’s evenness), beta diversity for weighted and unweighted phylogenetic analyses (weighted and unweighted UniFrac), and gneiss were conducted in QIIME v2.2018.11, that bundles multiple external programs for these analyses (see publication).30 Gneiss31 multiple regression linear models were built initially utilizing all variables presented in Table 1. All continuous numerical variables were treated as such in the model (ex: age), and categorical variables were treated thusly (ex: SHSe). To control for overfitting, each successive model omitted the variable that explained the least variation until it would pass cross validation. 10-fold cross validation split into 10 partitions, 9 partitions were utilized to recreate the model, and the remaining partition measured the prediction accuracy (leave-one-out). The resulting within model error was required to be greater than the predicted error to prevent over-fitting. Further differential abundance analysis was tested via Wilcoxon to control for outliers. Wilcoxon test and Spearman correlation were conducted in JMP v13.0.0 with significance set at p ≤ 0.05 (Wilcoxon) or p ≤ 0.01 (Spearman) without correction for false discovery rate in this exploratory, pilot analysis.
Table 1:
Cohort Demographics
| All, n=20 | <1.3 y, n=12 | >2 y, n=8 | P value | |
|---|---|---|---|---|
| Age (years) | 1.67 ± 1.51 | 0.6 ± 0.4 | 3.3 ± 1.0 | <0.0001 |
| Sex (% Female) | 50.0% | 58.3% | 37.5% | 0.39 |
| Pancreatic Insufficiency | 95.0% | 91.7% | 100% | 0.42 |
| Genotype | ||||
| Phe508del homozygous | 55.0% | 50.0% | 62.5% | 0.61 |
| Phe508del heterozygous | 100% | 100% | 100% | n/a |
| FEV0.5/FEV1 % predicted | 93.9 ± 17.1 | 95.1 ± 20.7 | 92.0 ± 9.8 | 0.74 |
| Hx P. aeruginosa | 55% | 50.0% | 62.5% | 0.61 |
| Hx MRSA | 25% | 16.7% | 37.5% | 0.32 |
| Smoke exposure via hair nicotine | 60% | 50.0% | 75.0% | 0.29 |
| Exclusively breastfed | 25% | 16.7% | 37.5% | 0.32 |
| BMI (wt/length) z score | −0.21 ± 0.98 | −0.21 ± 1.02 | =0.22 ± 0.98 | 0.99 |
| Recent antibiotic exposure* | 55.5% | 58.3% | 50.0% | 0.73 |
Hx- history; MRSA- methicillin resistant staphylococcus aureus;
FEV–Forced expiratory volume
Within the last month
P values represent differences between the infant and child groups.
Metabolic potential of the bacterial communities was estimated utilizing the phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) 2 software package. This leverages 16S rRNA gene amplicon library data to estimate global metabolic pathway abundances on a community-wide level.32
RESULTS
Cohort characteristics
Patient demographics are listed in Table 1, with details for the whole group and separated by age (1.3 years as cutoff). Briefly, this group included children recruited between 3 months and 5 years of age, with a mean age of 1.7 years. Patients were generally pancreatic insufficient with at least one copy of the Phe508del mutation. Exclusive breastfeeding in the first year of life was reported in only 25% of patients. Objective SHSe exposure was found in 60% of the patients via hair nicotine analysis, similar to our larger published cohort.21 Infant and child groups were similar with the exception of age and non-significant increases in males, breast-feeding, bacteria present and smoke exposure in the older group. High MRSA rates are consistent with our local population.
Sequencing Reads and Rarefaction
Sequencing of 16S rRNA gene amplicons yielded 2,732,396 total paired reads. Following quality control, 1,825,744 high-quality sequences (median 94,873 sequences per sample) were considered for the analysis. To control for sequencing depth, all samples were rarefied to 74,500 sequences, resulting in the exclusion of a single sample (age 2.2 years, 6,339 sequences).
Age is the primary driver of the fecal bacteriome of young children with CF
Given that understanding of the intestinal microbiome of young children with CF is limited, the first goal was to determine which demographic and environmental variables contribute the most variability to the fecal bacteriome in this cohort of patients. Utilizing gneiss to create a linear model guided by correlation-clustering (co-occurrence) of bacterial relative abundances, about 35% of the total variation in bacterial populations could be explained by variables captured in this study. Specifically, age explained the highest amount of variation at 18% (52% of the total variation explained). Three other variables included in the model: recent antibiotic exposure, sex, and smoke exposure, each accounted for approximately 5% of the total variation.
Diversity analyses confirmed age as an important predictor of the fecal bacterial populations within these patients. Patients >2 years old had higher fecal bacterial alpha diversity by all analyses conducted (Observed OTUs [Figure 1A], Faith’s Phylogenetic Diversity and Plielou’s Evenness [not shown]) and clustered separately by beta diversity (Weighted UniFrac [Figure 1B] as well as Bray-Curtis and Unweighted UniFrac [not shown]) compared to their younger counterparts (<1.3 years old, including 3 children approximately 1 year of age).
Figure 1: Age is the primary driver of the fecal bacteriome of young children with CF.
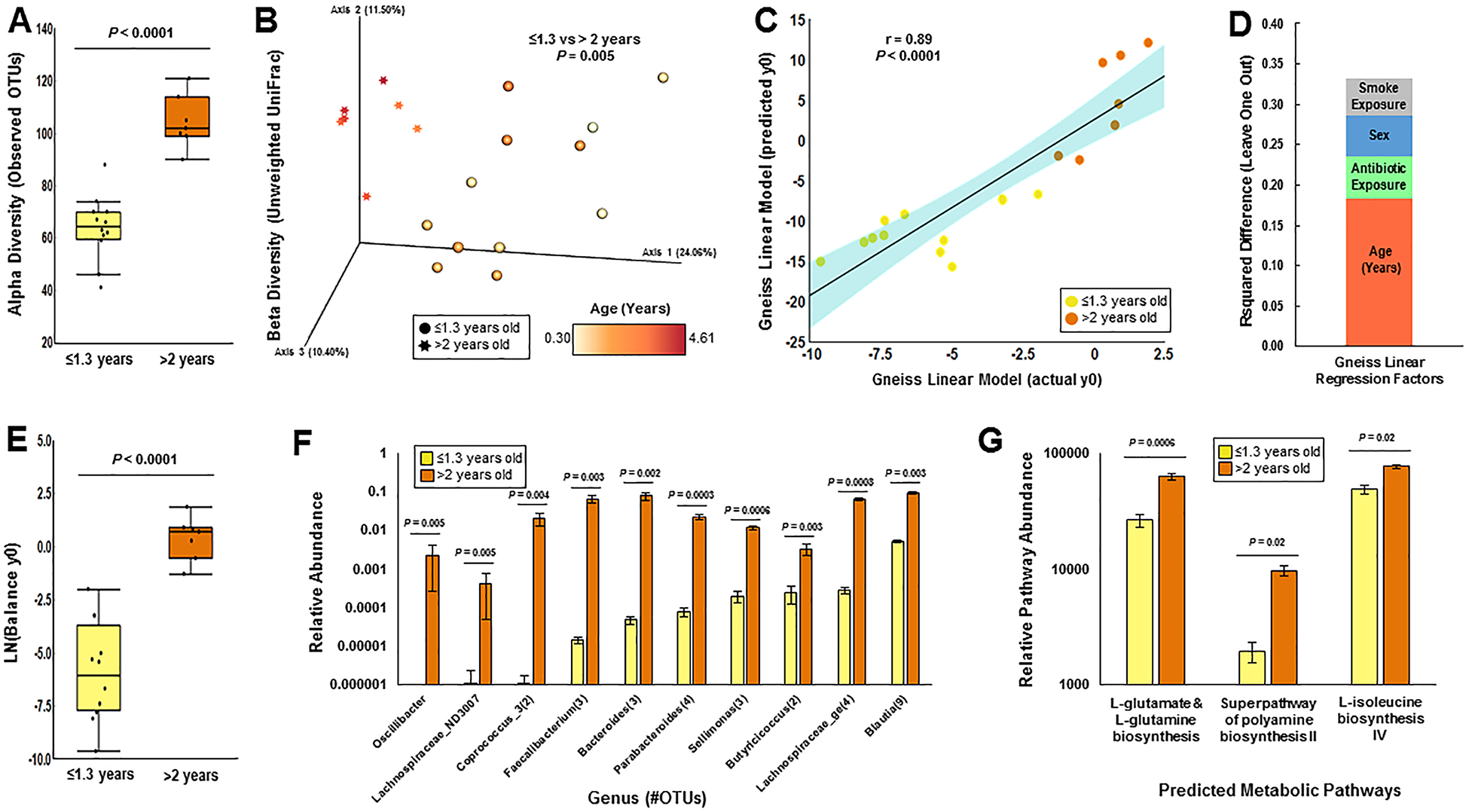
Sequencing of the bacterial 16S rRNA gene in fecal samples from children with CF revealed that children above the age of two years had (A) higher alpha diversity (as measured by observed OTUs, Kruskal-Wallis) and (B) clustered distinctly by beta diversity (as measured by unweighted UniFrac distance, PERMAVONA) compared to their younger counterparts (<1.3 years old). (C-D) Linear modeling by multiple regression using Gneiss revealed that age explained the highest amount of variation in fecal bacterial communities, while antibiotic exposure, sex, and second-hand smoke exposure explained variation to a lesser extent. (E-F) Closer inspection of the group of co-occuring bacterial taxa that best-explained variation by age (“y0”) in the linear model (E) indicated ten genera (F) that were also different by Wilcoxon test. Several bacterial metagenomic pathways predicted by PICRUSt analysis related to amino acid metabolism were higher in children with CF greater than two years old (G).
To account for the effect of age on microbial populations, a new model guided by gradient-clustering (by age of subjects) of bacterial relative abundances was constructed in gneiss. Gradient clustering by age creates balances (or groups of bacteria that change in relative proportion to one another) that diverge from youngest to oldest age, allowing the taxa that best explain age to be identified in the model. This new model utilized the same host-related variables from the original model (age, antibiotic exposure, sex, and smoke exposure; Figure 1C and 1D). The resulting balance explaining the most variation by age (y0) contained 171 total OTUs, 100 of which were relatively more abundant in patients greater than 2 years in age (Supplementary Figure 1, Figure 1E). Within this balance, the relative abundances of 32 taxa were higher by Wilcoxon test in patients above age 2 (Figure 1F). From this group of taxa the relative abundances of Blautia and Parabacteroides in particular were positively correlated to age (Spearman’s rho = 0.65 and 0.84 respectively, p values 0.004 and < 0.001). Overall, these data indicated that there was a distinct difference in the fecal bacteriomes of children with CF up to 1.3 years of age (the oldest child included under age 2) compared to their older counterparts.
We then used PICRUSt to predict metagenome functional pathways based on the changes in the above microbial populations. Three pathways were predicted to be enriched in the older age group after statistical correction for multiple hypothesis testing: 1) L-glutamate and L-glutamine biosynthesis, 2) polyamine biosynthesis, and 3) L-isoleucine biosynthesis (Figure 1G). Six pathways were predicted to be enriched in infants and children with SHSe (the top pathway being adenosine salvage), but none were significant after statistical correction for multiple hypothesis testing. There were also no significant differences in functional pathways when stratified by recent antibiotic exposure, mode of delivery, or breastfeeding.
Secondhand smoke and recent antibiotic exposure are associated with changes in the CF fecal bacteriome
Recently, we demonstrated that objective SHSe in young CF children increases the presence of bacterial pathogens in oropharyngeal cultures and decreases macrophage-mediated killing of bacteria.21 Although young children with CF often do not experience as many severe exacerbations as older patients with more severe disease, exposure to oral antibiotics (ABXe) that may also alter bacterial communities is common. To determine if SHSe and ABXe alter fecal bacteria, we examined the fecal bacteriome of the children in this study according to hair nicotine concentrations and recent antibiotic exposure (defined as exposure in the past month). As expected, ABXe was associated with decreased alpha diversity (Pielou’s Evenness only, Figure 2A), and created distinct clustering by beta diversity (Weighted UniFrac [Figure 2B] and Bray-Curtis [not shown]). While SHSe had no significant effect on diversity outcomes, it shared similarities with ABXe in the bacterial taxa affected. Children with SHSe, ABXe, or a combination had lower relative abundance of Bifidobacterium and Lactobacillus when compared to children not exposed to either ABXe or SHSe (Figure 2C and 2D).
Figure 2: Secondhand smoke and recent antibiotic exposure are associated with changes in the CF fecal bacteriome.
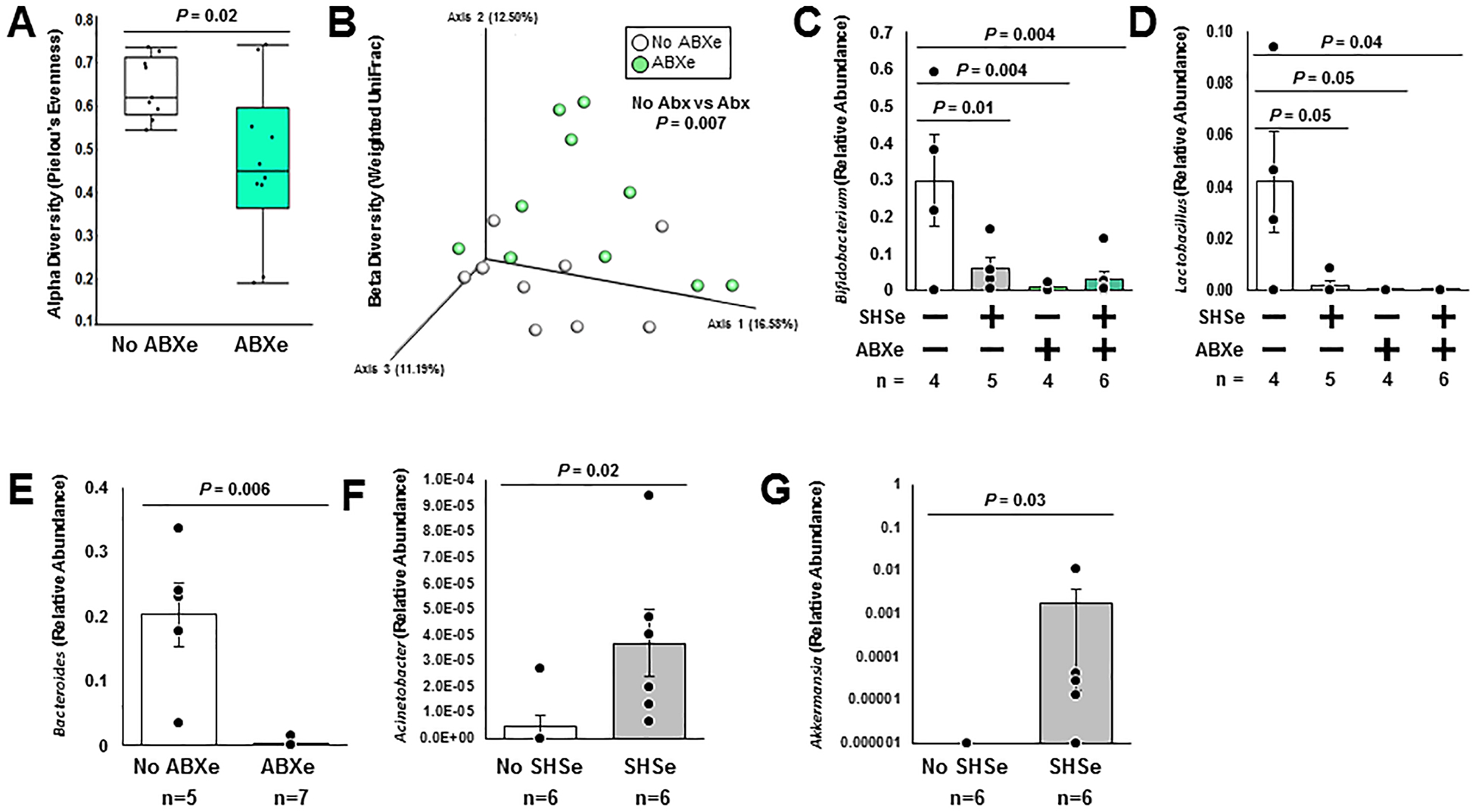
Continued analysis of the bacterial 16S rRNA gene in fecal samples from children with CF demonstrated that recent antibiotic exposure (ABXe) conferred (A) lower alpha diversity (as measured by Pielou’s Evenness, Kruskal-Wallis) and (B) distinct clustering by beta diversity (as measured by weighted UniFrac distances, PERMAVONA), whereas second-hand smoke exposure (SHSe) did not alter diversity metrics (not shown). (C-D) Both ABXe and SHSe additively reduce the relative abundance of Bifidobacterium and Lactobacillus in children less than 1.3 years old, where + indicates groups that were positive for the given exposure (Wilcoxon). (E) Children with ABXe also have a lower fecal relative abundance of Bacteroides in children less than 1.3 years old (Wilcoxon). (F-G) Children less than 1.3 years old with SHSe additionally have higher fecal relative abundances of both Acinetobacter and Akkermansia (Wilcoxon).
Given the drastic differences in bacterial composition in children less than 1.3 years old vs. those greater than 2, additional sub-analyses were conducted on the group of children less than 1.3 years old (the larger of the two age groups). Within this subset, children with ABXe had an associated lower relative abundance of the otherwise highly abundant Bacteroides (Figure 2E), and children with SHSe had higher relative abundance of both Acinetobacter (Figure 2F) and Akkermansia (Figure 2G), which had relatively low abundances. Diversity outcomes were not different from the full cohort. Combined, these results indicate that in addition to increasing the presence of airway pathogens21, SHSe and ABXe are associated with changes in the fecal bacteriome of young children with CF.
Birth mode and feeding mode influence the early CF fecal bacteriome
Next, we examined the influence of other external factors commonly known to shape the developing intestinal microbiome of young children, namely birth and feeding modes, in the subset of children less than 1.3 years old. Birth by caesarian-section was associated with higher alpha diversity than vaginal birth (Faith’s Phylogenetic Diversity, Figure 3A, and Observed OTUs). However, this difference did not extend to clustering by beta diversity. A single genus, Turicibacter, was higher in children born by caesarian-section (detected in all such individuals, albeit at a low relative abundance), and was undetected in all vaginally-born children (Figure 3B).
Figure 3: Birth mode and feeding mode influence the early CF fecal bacteriome.
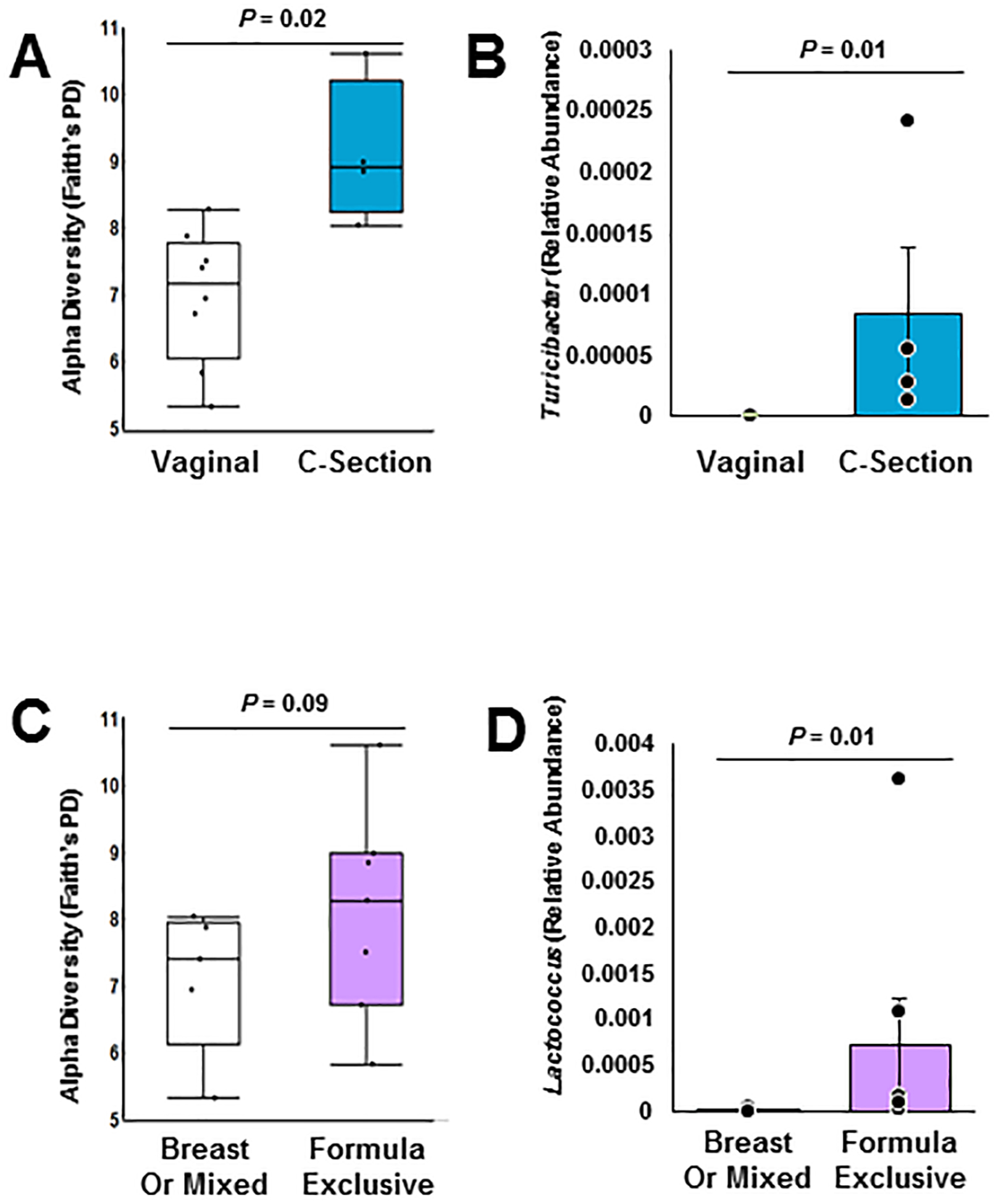
Continued analysis of the bacterial 16S rRNA gene in fecal samples from children with CF less than 1.3 years old determined that (A) birth by caesarian section (C-section) increased alpha diversity (as measured by Faith’s Phylogenetic Distance, Kruskal-Wallis), but did not alter beta diversity (data not shown). (B) Specifically, fecal relative abundance of Turicibacter was higher in children born by c-section (Wilcoxon). (C) Exclusive formula feeding also increased alpha diversity (as measured by Faith’s Phylogenetic Distance, Kruskal-Wallis), but did not alter beta diversity (data not shown). (D) Specifically, fecal relative abundance of Lactococcus were higher in exclusively formula-fed children (Wilcoxon).
Children who were exclusively formula fed vs breast fed or mixed formula and breast fed had similar alpha (Faith’s Phylogenetic Diversity p=0.09, other metrics p > 0.1, Figure 3C) and beta diversity (not shown). However, the relative abundance of the genus Lactococcus was higher in children who were exclusively formula fed (Figure 3D).
Associations of fecal bacteria with clinical outcomes and metabolites
Lastly, we determined associations between intestinal bacteria and clinical outcomes and biochemical data, beginning with growth outcomes. We found that weight for length z scores were negatively associated with relative abundances of Faecalibacterium and Staphylococcus (Spearman’s rho = −0.79 and −0.76 respectively, p values 0.002 and 0.006). There were no significant associations with weight or length scores when analyzed separately. We then analyzed associations with pulmonary outcomes. There were no significant bacterial species associations with either FEV1 or hospitalization for pulmonary exacerbation in the past 6 months. Finally, we examined associations with arachidonic acid metabolite levels from serum. We previously demonstrated that arachidonic acid metabolites were biomarkers of worsened clinical outcomes in young CF children with SHSe.21 We found that the metabolite 8-iso-PGF2α was positively associated with relative abundances of Peptoclostridium and Blautia (Spearman’s rho = 0.84 and 0.78 respectively, p values 0.006 and 0.008). There were no other significant association between intestinal bacteria and arachidonic acid metabolites.
DISCUSSION
Mechanisms that facilitate early infection and inflammation in CF remain unclear, which hinders new therapeutic development. We report fecal bacteriome findings from a small cohort of well-phenotyped infants with CF. We found that SHSe and other modifiable factors such as antibiotics were associated with changes in intestinal microbial composition in young CF children, but the strongest predictor of overall composition was age. These findings are important as we continue to differentiate controllable and non-controllable factors that shape the developing intestinal microbiome in young CF children.
SHSe has many negative impacts on lung function, nutrition, and immune function in children with CF, a summary of which can be found in a separate review.33 The impact of SHSe on different microbiomes, and particularly the intestinal microbiome, is poorly understood and has not been previously reported in CF. In contrast, a recent review of adult non-CF literature demonstrates that primary tobacco smoking alters the composition of the intestinal microbiome.34 In particular, studies have demonstrated relatively higher Clostridium and Prevotella as well as relatively lower abundance ofBifidobacterium and Lactococcus.34 There is limited data on SHSe’s influence on the intestinal microbiome in non-CF children, but maternal smoking is associated with higher abundance of Ruminococcus and Akkermansia in non-CF neonates and higher Staphylococcus in older infants (reviewed in35). In our study, SHSe was associated with higher relative abundance of opportunistic pathogens such as Acinetobacter. However, SHSe was also associated with lower relative abundance of Bifidobacterium and higher relative abundance of Akkermansia, similar to results in primary smokers and non-CF infants. Increased relative abundance of Akkermansia with smoke exposure is particularly interesting, given its intimate association with the intestinal mucus layer.36 Just as smoke exposure increases mucus secretion and thickness in the lung, smoking (in the form of nicotine exposure) can induce the same phenotype in the colon.37 The higher relative abundance of Akkermansia may be associated with changes in the colonic mucus layer in this cohort, although this requires further study. Our findings suggest that while SHSe is associated with an altered intestinal microbiome in CF, there are both similar and unique compositional changes in response to SHSe in comparison to non-CF children. It is unclear if these changes in the intestinal microbiome during SHSe translate into intestinal dysbiosis. Overall, our findings need confirmation in larger cohorts containing young children with and without CF, as well as in longitudinal cohorts to determine whether the community shifts observed may be amenable to therapeutic manipulation.
Interestingly, reductions in generally beneficial Bifidobacteria and Lactobacillus (fellow lactic acid bacteria sharing an order with Lactococcus) in young children with CF and SHSe paralleled reductions in these same genera in children with recent antibiotic exposure independent of SHSe. In this small study it was difficult to tell whether these genera are further reduced when both factors are present. This will be important to test in future studies as children with CF, who often receive antibiotics during disease exacerbations, may be at higher risk of changes in the intestinal microbiome if they are also exposed to SHSe. However, a causal relationship between SHSe, antibiotic use, and reduced Bifidobacteria and Lactobacillus has not been determined. Further, it is unclear if SHSe and antibiotics suppress specific bacterial populations through similar mechanisms or unique pathways, or how overall health changes from SHSe may influence the intestinal microbiome. Recent antibiotic use alone also greatly reduced Bacteroides and predicted overall bacterial population structure in CF children less than 1.3 years old. Reductions in Bacteroides are consistent with other studies in children with CF,5,14,38,39 including children and young adults with CF and cirrhosis.40 It is unknown whether therapeutic alteration of individual Bifidobacteria and/or Lactobacillus populations through prebiotic or probiotic formulations would have benefit in CF and non-CF children with SHSe, but this could be explored in future studies, particularly for their ability to reduce inflammation given the pro-inflammatory nature of SHSe.
Not surprisingly, we found that age was the most predictive factor of overall fecal microbial composition. This is likely reflective of a combination of factors associated with early development that have been previously described elsewhere,11,41 including dietary changes, increased environmental exposures, exposure to medications, and maturation of immune responses. These are multiple aspects that can confound microbiome analysis that we controlled for to the best of our ability in this hypothesis-generating study. Although multiple studies have demonstrated alterations between age-matched CF and non-CF fecal samples, one group compared children with CF and their healthy siblings in an attempt to control for genetic and environmental factors.38 While their cross-sectional data indicated similar species richness between siblings, longitudinal sampling revealed lower temporal stability and lower species richness in the CF fecal samples. A recent study also showed that CFTR mutations in a germ-free mouse model are sufficient to drive changes in the intestinal microbiome.42 Together, these and other studies would suggest that alterations in the CF microbiome over time are at least partially dependent on factors intrinsic to CFTR dysfunction.
Dietary factors undoubtedly drive changes in the intestinal microbiome, which may offer the best explanation for age driving differences in bacterial composition in this study. The dietary transition from primarily liquid to solid foods around the age of nine to eighteen months coincides with a large shift in the intestinal bacteriome based on the availability of new substrates entering the gastrointestinal tract,43 and is reflective of the distinction between those children above and below the age of two in this cohort. Solid foods offer increased microbially available carbohydrates, which specifically foster the growth of Bacteroides, Parabacteroides, Faecalibacterium, Butyricicoccus, Oscillibacter, Coprococcus, Blautia, and other members of Lachnospiraceae in children without CF;43,44 the majority of which are also negatively associated with the duration of exclusive breast feeding.45 All of these taxa are included in the major differential abundances explaining variability by age in the children with CF in this study.
We also found higher relative abundance of Blautia and Parabacteroides to be correlated with age. Further, the oxidative stress marker 8-iso-PGF2α also correlated with Blautia relative abundance. Blautia was previously observed to increase in relative abundance over time in both fecal and oropharyngeal samples from infants with CF12, and has been detected in fecal samples from two other CF cohorts.10,40 CF is associated with increased oxidative stress,46 and increases in Blautia with time may be reflective of the overall changes in the oxidative state of CF. In contrast to increases in Blautia in prior CF studies, the relative abundance of Parabacteroides was shown to decrease prior to the onset of P. aeruginosa colonization in a past study of CF patients.13 As over half of our CF patients had P. aeruginosa colonization and increased Parabacteroides abundance with increasing age, our findings would indicate that decreases in Parabacteroides prior to P. aeruginosa colonization are not likely to be sustained with age in young children. However, changes in Parabacteroides would need to be replicated in a larger, longitudinal cohort.
In addition to age-related changes in microbial composition, we found that relative abundances of Faecalibacterium (known butyrate-producing bacteria) and Staphylococcus (a genus with many opportunistic pathogens) were higher in individuals with lower weight/length Z scores, independent of age. Specifically, the association with Staphylococcus suggests pathobiont-associated changes in nutritional outcomes that may be reflective of a variety of factors including alterations in metabolism47 associated with high fat diets used in CF, and recurrent antibiotic use in CF patients with poor growth and associated respiratory complications. The influence of CFTR modulators upon these specific changes in the intestinal microbiome and associated clinical outcomes is a topic for future studies.
Our study results were limited by the use of a small, single-center cohort containing CF infants and young children only. Healthy controls were not available at the time of sampling, limiting a comparison of SHSe between CF and non-CF controls. Further, fecal samples were only available at a single time point during a period of clinical stability. Longitudinal comparisons within larger cohorts also containing children without CF are needed for future studies. Specific information on dietary factors that could influence intestinal bacteria was also not available, beyond measures of breast versus bottle-feeding. However, the available cohort for this pilot study was otherwise well-phenotyped including objective measures of SHSe, which are not commonly reported in other microbiome studies. Another limitation was 16S rRNA gene amplicon targeted sequencing, which does not estimate the functional potential of bacterial populations and introduces bias based on the hypervariable region amplified. Future metagenomic sequencing analyses coupled with metabolomic analyses may discover functional metabolic changes in the microbiome that our PICRUSt analyses were unable to predict. Finally, we did not correct for multiple hypothesis testing in this exploratory analysis, and confirmation of our findings in a larger cohort is warranted.
In summary, SHSe and other external factors such as antibiotics are associated with alterations in fecal bacteriome composition in young CF children, but the strongest predictor of overall composition was age. These findings have implications for the continued understanding of the developing intestinal microbiome in young CF children.
Supplementary Material
Supplementary Figure 1: Balance “y0” best-explains the differences in the CF fecal bacteriome before 1.3 years and after the age of 2. All 171 OTUs contained in gneiss multiple linear regression balance “y0” are plotted by age category, being greater than or less than 2 years old. Taxa plotted as yellow bars with white background are taxa that compose the numerator (taxa relatively higher in children >2 years old), while taxa plotted as blue bars with grey background compose the denominator (taxa relatively higher in children <1.3 years old). Taxa are collapsed within genus, with the number of OTUs within that genus indicated in parenthesis following the genus name. Error bars represent the standard error of the mean within in each bar. “*” denotes taxa that were additionally identified as different by Wilcoxon test (p<0.05).
ACKNOWLEDGEMENTS:
Thank you to Melinda Smith for assistance in sample procurement. Grant support provided by Nationwide Children’s Hospital intramural grant (BTK, RT, AM), American Academy of Pediatrics Julius B. Richmond Center New Investigator Grant (BTK), The Ohio State University Center for Clinical and Translational Science (National Center for Advancing Translational Sciences, Grant UL1TR002733), and by the Cystic Fibrosis Foundation (Cure CF Columbus Research Development Program, Grant MCCOY19RO). Support for KLR and GAO provided by NIDDK/P30‐DK117469 (DartCF; Dartmouth Cystic Fibrosis Research Center). We thank MBL for the fee-for-service sequencing, and Dr. Hilary Morrison for providing details of the methods.
Footnotes
Competing Interest Statement
There are no relevant conflicts of interest.
Data availability
Sequencing data is available in the NCBI biosample database (SUB7033953).
REFERENCES
- 1.Leung DH, Heltshe SL, Borowitz D, Gelfond D, Kloster M, Heubi JE, Stalvey M, Ramsey BW, Baby O, Nutrition Study Investigators of the Cystic Fibrosis Foundation Therapeutics Development N. Effects of Diagnosis by Newborn Screening for Cystic Fibrosis on Weight and Length in the First Year of Life. JAMA Pediatr 2017;171(6):546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frayman KB, Armstrong DS, Grimwood K, Ranganathan SC. The airway microbiota in early cystic fibrosis lung disease. Pediatr Pulmonol 2017;52(11):1384–1404. [DOI] [PubMed] [Google Scholar]
- 3.Frayman KB, Armstrong DS, Carzino R, Ferkol TW, Grimwood K, Storch GA, Teo SM, Wylie KM, Ranganathan SC. The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax 2017;72(12):1104–1112. [DOI] [PubMed] [Google Scholar]
- 4.Pittman JE, Wylie KM, Akers K, Storch GA, Hatch J, Quante J, Frayman KB, Clarke N, Davis M, Stick SM, Hall GL, Montgomery G, Ranganathan S, Davis SD, Ferkol TW, Australian Respiratory Early Surveillance Team for Cystic F. Association of Antibiotics, Airway Microbiome, and Inflammation in Infants with Cystic Fibrosis. Ann Am Thorac Soc 2017;14(10):1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Freitas MB, Moreira EAM, Tomio C, Moreno YMF, Daltoe FP, Barbosa E, Ludwig Neto N, Buccigrossi V, Guarino A. Altered intestinal microbiota composition, antibiotic therapy and intestinal inflammation in children and adolescents with cystic fibrosis. PLoS One 2018;13(6):e0198457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prevaes SM, de Winter-de Groot KM, Janssens HM, de Steenhuijsen Piters WA, Tramper-Stranders GA, Wyllie AL, Hasrat R, Tiddens HA, van Westreenen M, van der Ent CK, Sanders EA, Bogaert D. Development of the Nasopharyngeal Microbiota in Infants with Cystic Fibrosis. Am J Respir Crit Care Med 2016;193(5):504–515. [DOI] [PubMed] [Google Scholar]
- 7.Prevaes SM, de Steenhuijsen Piters WA, de Winter-de Groot KM, Janssens HM, Tramper-Stranders GA, Chu ML, Tiddens HA, van Westreenen M, van der Ent CK, Sanders EA, Bogaert D. Concordance between upper and lower airway microbiota in infants with cystic fibrosis. Eur Respir J 2017;49(3). [DOI] [PubMed] [Google Scholar]
- 8.Laguna TA, Wagner BD, Williams CB, Stevens MJ, Robertson CE, Welchlin CW, Moen CE, Zemanick ET, Harris JK. Airway Microbiota in Bronchoalveolar Lavage Fluid from Clinically Well Infants with Cystic Fibrosis. PLoS One 2016;11(12):e0167649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mika M, Korten I, Qi W, Regamey N, Frey U, Casaulta C, Latzin P, Hilty M, group Ss. The nasal microbiota in infants with cystic fibrosis in the first year of life: a prospective cohort study. Lancet Respir Med 2016;4(8):627–635. [DOI] [PubMed] [Google Scholar]
- 10.Manor O, Levy R, Pope CE, Hayden HS, Brittnacher MJ, Carr R, Radey MC, Hager KR, Heltshe SL, Ramsey BW, Miller SI, Hoffman LR, Borenstein E. Metagenomic evidence for taxonomic dysbiosis and functional imbalance in the gastrointestinal tracts of children with cystic fibrosis. Sci Rep 2016;6:22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madan JC. Neonatal Gastrointestinal and Respiratory Microbiome in Cystic Fibrosis: Potential Interactions and Implications for Systemic Health. Clin Ther 2016;38(4):740–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madan JC, Koestler DC, Stanton BA, Davidson L, Moulton LA, Housman ML, Moore JH, Guill MF, Morrison HG, Sogin ML, Hampton TH, Karagas MR, Palumbo PE, Foster JA, Hibberd PL, O’Toole GA. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. MBio 2012;3(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoen AG, Li J, Moulton LA, O’Toole GA, Housman ML, Koestler DC, Guill MF, Moore JH, Hibberd PL, Morrison HG, Sogin ML, Karagas MR, Madan JC. Associations between Gut Microbial Colonization in Early Life and Respiratory Outcomes in Cystic Fibrosis. J Pediatr 2015;167(1):138–147 e131–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antosca KM, Chernikova DA, Price CE, Ruoff KL, Li K, Guill MF, Sontag NR, Morrison HG, Hao S, Drumm ML, MacKenzie TA, Dorman DB, Feenan LM, Williams MA, Dessaint J, Yuan IH, Aldrich BJ, Moulton LA, Ting L, Martinez-Del Campo A, Stewart EJ, Karagas MR, O’Toole GA, Madan JC. Altered Stool Microbiota of Infants with Cystic Fibrosis Shows a Reduction in Genera Associated with Immune Programming from Birth. J Bacteriol 2019;201(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muhlebach MS, Zorn BT, Esther CR, Hatch JE, Murray CP, Turkovic L, Ranganathan SC, Boucher RC, Stick SM, Wolfgang MC. Initial acquisition and succession of the cystic fibrosis lung microbiome is associated with disease progression in infants and preschool children. PLoS Pathog 2018;14(1):e1006798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zemanick ET, Wagner BD, Robertson CE, Ahrens RC, Chmiel JF, Clancy JP, Gibson RL, Harris WT, Kurland G, Laguna TA, McColley SA, McCoy K, Retsch-Bogart G, Sobush KT, Zeitlin PL, Stevens MJ, Accurso FJ, Sagel SD, Harris JK. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur Respir J 2017;50(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayden HS, Eng A, Pope CE, Brittnacher MJ, Vo AT, Weiss EJ, Hager KR, Martin BD, Leung DH, Heltshe SL, Borenstein E, Miller SI, Hoffman LR. Fecal dysbiosis in infants with cystic fibrosis is associated with early linear growth failure. Nat Med 2020;26(2):215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enaud R, Hooks KB, Barre A, Barnetche T, Hubert C, Massot M, Bazin T, Clouzeau H, Bui S, Fayon M, Berger P, Lehours P, Bebear C, Nikolski M, Lamireau T, Delhaes L, Schaeverbeke T. Intestinal Inflammation in Children with Cystic Fibrosis Is Associated with Crohn’s-Like Microbiota Disturbances. J Clin Med 2019;8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stiemsma LT, Michels KB. The Role of the Microbiome in the Developmental Origins of Health and Disease. Pediatrics 2018;141(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopp BT, Sarzynski L, Khalfoun S, Hayes D Jr., Thompson R, Nicholson L, Long F, Castile R, Groner J. Detrimental effects of secondhand smoke exposure on infants with cystic fibrosis. Pediatr Pulmonol 2015;50(1):25–34. [DOI] [PubMed] [Google Scholar]
- 21.Kopp BT, Thompson R, Kim J, Konstan R, Diaz A, Smith B, Shrestha C, Rogers LK, Hayes D Jr., Tumin D, Woodley FW, Ramilo O, Sanders DB, Groner JA, Mejias A. Secondhand smoke alters arachidonic acid metabolism and inflammation in infants and children with cystic fibrosis. Thorax 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berkowitz L, Schultz BM, Salazar GA, Pardo-Roa C, Sebastian VP, Alvarez-Lobos MM, Bueno SM. Impact of Cigarette Smoking on the Gastrointestinal Tract Inflammation: Opposing Effects in Crohn’s Disease and Ulcerative Colitis. Front Immunol 2018;9:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones M, Castile R, Davis S, Kisling J, Filbrun D, Flucke R, Goldstein A, Emsley C, Ambrosius W, Tepper RS. Forced expiratory flows and volumes in infants. Normative data and lung growth. Am J Respir Crit Care Med 2000;161(2 Pt 1):353–359. [DOI] [PubMed] [Google Scholar]
- 24.Castile R, Filbrun D, Flucke R, Franklin W, McCoy K. Adult-type pulmonary function tests in infants without respiratory disease. Pediatr Pulmonol 2000;30(3):215–227. [DOI] [PubMed] [Google Scholar]
- 25.Al-Delaimy WK. Hair as a biomarker for exposure to tobacco smoke. Tob Control 2002;11(3):176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groner JA, Huang H, Joshi MS, Eastman N, Nicholson L, Bauer JA. Secondhand Smoke Exposure and Preclinical Markers of Cardiovascular Risk in Toddlers. J Pediatr 2017;189:155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S, Apelberg BJ, Avila-Tang E, Hepp L, Yun D, Samet JM, Breysse PN. Utility and cutoff value of hair nicotine as a biomarker of long-term tobacco smoke exposure, compared to salivary cotinine. International journal of environmental research and public health 2014;11(8):8368–8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newton RJ, McLellan SL, Dila DK, Vineis JH, Morrison HG, Eren AM, Sogin ML. Sewage reflects the microbiomes of human populations. MBio 2015;6(2):e02574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeraldo P, Kalari K, Chen X, Bhavsar J, Mangalam A, White B, Nelson H, Kocher JP, Chia N. IM-TORNADO: a tool for comparison of 16S reads from paired-end libraries. PLoS One 2014;9(12):e114804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodriguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vazquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37(8):852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morton JT, Sanders J, Quinn RA, McDonald D, Gonzalez A, Vazquez-Baeza Y, Navas-Molina JA, Song SJ, Metcalf JL, Hyde ER, Lladser M, Dorrestein PC, Knight R. Balance Trees Reveal Microbial Niche Differentiation. mSystems 2017;2(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31(9):814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopp BT, Ortega-Garcia JA, Sadreameli SC, Wellmerling J, Cormet-Boyaka E, Thompson R, McGrath-Morrow S, Groner JA. The Impact of Secondhand Smoke Exposure on Children with Cystic Fibrosis: A Review. International journal of environmental research and public health 2016;13(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savin Z, Kivity S, Yonath H, Yehuda S. Smoking and the intestinal microbiome. Archives of microbiology 2018;200(5):677–684. [DOI] [PubMed] [Google Scholar]
- 35.McLean C, Jun S, Kozyrskyj A. Impact of maternal smoking on the infant gut microbiota and its association with child overweight: a scoping review. World journal of pediatrics : WJP 2019;15(4):341–349. [DOI] [PubMed] [Google Scholar]
- 36.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 2013;110(22):9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finnie IA, Campbell BJ, Taylor BA, Milton JD, Sadek SK, Yu LG, Rhodes JM. Stimulation of colonic mucin synthesis by corticosteroids and nicotine. Clin Sci (Lond) 1996;91(3):359–364. [DOI] [PubMed] [Google Scholar]
- 38.Duytschaever G, Huys G, Bekaert M, Boulanger L, De Boeck K, Vandamme P. Cross-sectional and longitudinal comparisons of the predominant fecal microbiota compositions of a group of pediatric patients with cystic fibrosis and their healthy siblings. Appl Environ Microbiol 2011;77(22):8015–8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruzzese E, Callegari ML, Raia V, Viscovo S, Scotto R, Ferrari S, Morelli L, Buccigrossi V, Lo Vecchio A, Ruberto E, Guarino A. Disrupted intestinal microbiota and intestinal inflammation in children with cystic fibrosis and its restoration with Lactobacillus GG: a randomised clinical trial. PLoS One 2014;9(2):e87796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flass T, Tong S, Frank DN, Wagner BD, Robertson CE, Kotter CV, Sokol RJ, Zemanick E, Accurso F, Hoffenberg EJ, Narkewicz MR. Intestinal lesions are associated with altered intestinal microbiome and are more frequent in children and young adults with cystic fibrosis and cirrhosis. PLoS One 2015;10(2):e0116967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang YJ, LiPuma JJ. The Microbiome in Cystic Fibrosis. Clinics in chest medicine 2016;37(1):59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meeker SM, Mears KS, Sangwan N, Brittnacher MJ, Weiss EJ, Treuting PM, Tolley N, Pope CE, Hager KR, Vo AT, Paik J, Frevert CW, Hayden HS, Hoffman LR, Miller SI, Hajjar AM. CFTR dysregulation drives active selection of the gut microbiome. PLoS Pathog 2020;16(1):e1008251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laursen MF, Bahl MI, Michaelsen KF, Licht TR. First Foods and Gut Microbes. Front Microbiol 2017;8:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez I, Kim J, Duffy PR, Schlegel VL, Walter J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One 2010;5(11):e15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laursen MF, Andersen LB, Michaelsen KF, Molgaard C, Trolle E, Bahl MI, Licht TR. Infant Gut Microbiota Development Is Driven by Transition to Family Foods Independent of Maternal Obesity. mSphere 2016;1(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen AC, Burr L, McGuckin MA. Oxidative and endoplasmic reticulum stress in respiratory disease. Clin Transl Immunology 2018;7(6):e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Si X, Shang W, Zhou Z, Strappe P, Wang B, Bird A, Blanchard C. Gut Microbiome-Induced Shift of Acetate to Butyrate Positively Manages Dysbiosis in High Fat Diet. Mol Nutr Food Res 2018;62(3). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Balance “y0” best-explains the differences in the CF fecal bacteriome before 1.3 years and after the age of 2. All 171 OTUs contained in gneiss multiple linear regression balance “y0” are plotted by age category, being greater than or less than 2 years old. Taxa plotted as yellow bars with white background are taxa that compose the numerator (taxa relatively higher in children >2 years old), while taxa plotted as blue bars with grey background compose the denominator (taxa relatively higher in children <1.3 years old). Taxa are collapsed within genus, with the number of OTUs within that genus indicated in parenthesis following the genus name. Error bars represent the standard error of the mean within in each bar. “*” denotes taxa that were additionally identified as different by Wilcoxon test (p<0.05).


