Abstract
Human embryonic stem cells (hESCs) can be progressively differentiated into definitive endoderm (DE), hepatic progenitors, and hepatocytes, and thus provide an excellent model system for the mechanistic study of hepatocyte differentiation, which is currently poorly understood. Here, we found that insulin enhanced hepatocyte differentiation from hESC-derived DE. Insulin activated the PI3K/AKT pathway, but not the mitogen-activated protein kinase pathway in the DE cells, and inhibition of the PI3K/AKT pathways by inhibitors markedly inhibited hepatocyte differentiation. In addition, insulin-like growth factor 1 (IGF1) and IGF2 also activated the PI3K/AKT pathway in DE cells and their expression was robustly upregulated during hepatocyte differentiation from DE. Furthermore, inhibition of IGF receptor 1 (IGF1R) by a small molecule inhibitor PPP or knockdown of the IGF1R by shRNA attenuated hepatocyte differentiation. Moreover, simultaneous knockdown of the IGF1R and the insulin receptor with shRNAs markedly reduced the activation of AKT and substantially impaired hepatocyte differentiation. The PI3K pathway specifically enhanced the expression of HNF1 and HNF4 to regulate hepatocyte differentiation from DE. Although inhibition of the PI3K pathway was previously shown to be required for the induction of DE from hESCs, our study revealed a positive role of the PI3K pathway in hepatocyte differentiation after the DE stage, and has advanced our understanding of hepatocyte cell fate determination.
Keywords: Insulin-like growth factor, Embryonic stem cells, Hepatocyte differentiation, Signal transduction
INTRODUCTION
Liver and other respiratory, gastrointestinal organs are derived from definitive endoderm (DE). Developmental studies in various models have revealed that the fibroblast growth factors (FGFs) secreted from cardiac mesoderm and bone morphogenic proteins (BMPs) secreted from septum transversum mesenchyme induce hepatic and suppress pancreatic specification in the foregut endoderm [1-4]. The newly specified hepatic progenitor cells, which express α-fetoprotein (AFP), undergo proliferation and differentiation to become bipotent hepatoblasts and subsequently give rise to hepatocytes (albumin+) and cholangiocytes (cytokeratin 7+). Fully mature hepatocytes, which express various cytochrome P450 enzymes, tryptophan oxygenase (TDO), and asialoglycoprotein receptor 1 (ASGPR1), appear only after the postnatal stage. Other growth factors, such as oncostatin M, hepatocyte growth factor (HGF), and glucocorticoids have been shown to regulate hepatocyte differentiation at the later stage [5-10]. Although development studies have revealed important clues concerning liver organogenesis, major gaps in our knowledge about the molecular mechanisms that regulate hepatocyte differentiation remain. For example, how these growth factors dictate cell fate is poorly understood and whether additional growth factors play roles in hepatocyte fate determination remains to be determined.
Pluripotent human embryonic stem cells (hESCs) have been actively investigated as a potential alternative cell source for hepatocytes. We and others have established protocols to differentiate hESCs to hepatocytes with less than desirable hepatic functions [4,11]. The lack of understanding of the molecular mechanisms regulating hepatocyte differentiation hinders us in achieving the full potential that hESCs provide. Previous study has revealed that insulin enhances hepatocyte differentiation from ESC-derived embryoid bodies [12]. Whether insulin-like growth factors (IGFs) act similar to insulin on hepatocyte differentiation is unknown. We aimed to identify growth factor-mediated signaling pathways that regulate hepatocyte differentiation from DE in this study. We found that the PI3K pathway, activated by insulin and/or IGFs, plays an important role in hepatocyte fate determination from DE. Understanding the molecular mechanisms regulating hepatocyte differentiation will significantly facilitate the development of stem cell-based therapy to treat liver disease.
MATERIALS AND METHODS
ESC Culture and Hepatocyte Differentiation
The hESC line, H9, was purchased from WiCell Research Institute (WiCell, Madison, WI, http://wicell.org) and cultured either on mouse embryonic fibroblasts (MEFs) or on hESC-qualified matrigel (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com)-coated plates using MEF conditioned ESC culture medium, as instructed by the provider. Hepatocyte differentiation was induced in three stages as previously described with minor modification at the DE stage [13]. Briefly, DE was induced from hESCs in RPMI medium (Gibco, Grand Island, NY, http://www.invitrogen.com) with 100 ng/ml activin A (R&D Systems Inc, Minneapolis, MN, http://www.rndsystems.com) and 0.5% bovine serum albumin (BSA) (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) for 2 days and then BSA was replaced with B27 (Gibco) for the next 6 days. Cells were lifted using trypsin and plated into collagen (Invitrogen, Carlsbad, CA, http://www.invitrogen.com)-coated culture plates and grown for 8 days in IMDM media (Gibco) with 20% fetal bovine serum (HyClone, Logan, UT, http://www.hyclone.com), 2 mM l-glutamine (Gibco), 0.3 mM 1-thioglycerol (Sigma), 0.5% DMSO (Sigma), 100 nM dexamethasone (Sigma), 0.126 U/ml human insulin, 20 ng/ml FGF4 (R&D systems), 20 ng/ml HGF (R&D systems), 10 ng/ml BMP2 (R&D systems), and 10 ng/ml BMP4 (R&D systems). At the third stage, cells were grown in HBM basal medium (Lonza, Walkersville, MD, www.lonza.com) with Hepatocyte Culture Medium HCM SingleQuots kit (Lonza), 0.5% DMSO, 100 nM dexamethasone, 20 ng/ml FGF4, 10 ng/ml HGF, and 50 ng/ml oncostatin M (R&D systems). This protocol was referred to as the P1 protocol. The P2 protocol differs from the P1 protocol only at stage 2 by the removal of FGF4, BMP2, BMP4, and HGF (R&D Systems Inc.) from stage 2 medium.
Human Primary Hepatocytes
Human primary hepatocytes (PH) were freshly isolate and directly plated in culture plates by the Liver Tissue Cell Distribution System of NIH (University of Pittsburg). Growing cells were delivered to us within 48 hours and further cultured for 1–2 days in HCM medium (Lonza) upon receiving the cells.
Quantitative RT-PCR
Total RNA isolation, cDNA generation, and quantitative real-time PCR were carried out as previously described [14]. Primers and probes for tested genes are listed in Supporting Information Table S1.
Immunoblot Analysis and Immunofluorescence Staining
Cell lysates were collected using RIPA buffer with proteinase and phosphatase inhibitor cocktail and 5 mM EDTA (Thermo Scientific, Waltham, MA, http://www.thermoscientific.com). Total proteins of 30–50 μg were used for immunoblot analysis as previously described [15]. Immunofluorescence staining was carried out as previously described [16]. Antibody against albumin ALB was purchased from Bethyl Laboratories Inc (Bethyl Laboratories Inc, Montgomery, TX, http://www.bethyl.com). Antibodies for insulin receptor (IR), phosphor-IR/IGF1R, IGF1R, phosphor-AKT, phosphor-ERK, phosphor-p38, phosphor-JNK, phosphor-FOXO1, and phosphor-FOXO3a were purchased from Cell Signaling Technology (Cell Signaling Technology, Danvers, MA, http://www.cellsignal.com). Antibody against β-actin was purchased from Sigma-Aldrich. HRP-conjugated secondary antibodies against rabbit or mouse (Cell Signaling Technology) were diluted 1:2,500 before use.
ELISA Assay for ALB
ALB in culture media at day 24 of hepatocyte differentiation from hESCs was analyzed using an ALB ELISA assay kit (Bethyl Laboratories Inc) according to the manufacturer’s instructions.
Periodic Acid Schiff Staining
Cells were first fixed in 4% paraformaldehyde for 20 minutes and stained for glycogen using the periodic acid Schiff stain kit (PolySciences Inc, Warrington, PA, http://www.polysciences.com) according to the manufacturer’s instructions.
Small Molecule Inhibitors
LY294002 (Millipore, Billerica, MA, http://www.millipore.com), AKT inhibitor IV (Millipore), and PPP (Santa Cruz Biotechnology Inc, Dallas, TX, http://www.scbt.com) were used at the concentration of 10, 0.2, and 0.2 μM, respectively.
Viral Vectors and Transduction
Lentiviral vector, pLKO.1 (Addgene, Cambridge, MA http://www.addgene.org) was used as a scrambled control vector. To construct shRNA viral vectors targeting IR or IGF1R, DNA oligos targeting IR(5′CCGGGAAGTGAGTTATCGGCGATATCT CGAGATATCGCCGATAACTCAC TTCTTTTTGCTAGCG-3′ and 5′-AATTCGCTAGCAAAAAGAAGTGAGTTATCGG CGATATCTCGAGATATCGCCGATAACTCACTTC-3′) or IGF1R (5′-CCGGCGGC AACCTGAGTTACTACATCTCGAGATGTAG TAACTCAGGTTGCCGTTTTTGCT AGCG-3′ and 5′-AATTCG CTAGCAAAAACGGCAACCTGAGTTACTACATCT CGAGAT GATGTAACTCAGGTTGCCG-3′) were annealed and then cloned into pLKO.1 between the AgeI and EcoRI sites. To generate viruses, 2.5 × 107 HEK293T cells were transfected with 25 μg of shRNA vectors, 25 μg of pCMV-dR8.91, and 5 μg of pMDG-VSV-G (gift from Kohn DB, University of California Los Angeles) by TransIT-293 transfection reagent (Mirus Bio LLC, Madison, WI, https://www.mirusbio.com). Media were changed to UltraCULTURE (Lonza) the next day. Supernatants were filtered through 0.22 μm MILLEX-GS filters (Millipore) 48 hours later, centrifuged with Centricon plus-70 (Millipore) for 35 minutes at 3,500 rpm, and stored at −80°C. Cells were transduced by viruses in the presence of 8 μg/ml polybrene at both days 6 and 8 during hepatocyte differentiation from hESCs.
Statistics
Four to five samples for each condition were analyzed and were presented as average ± SD. Probability (p) was calculated using the unpaired Student’s t test.
RESULTS
Insulin Enhances Hepatocyte Differentiation from DE
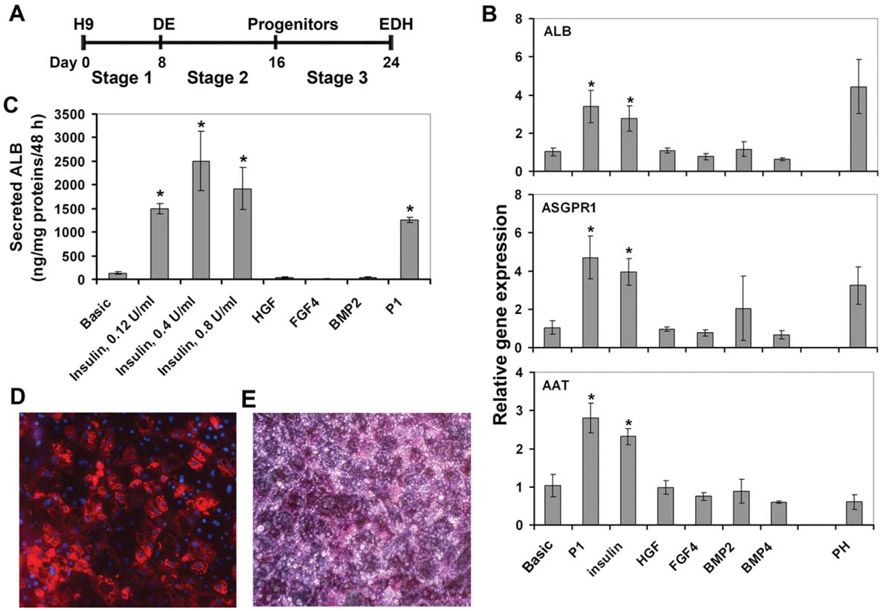
We used our previously published protocol, the P1 protocol, as a reference to derive hepatocytes from hESCs [13]. H9 cells were progressively differentiated to DE using activin A with 0.5% FBS or B27 at stage 1, and then to hepatocyte progenitors using a combination of insulin, HGF, FGF4, BMP2, and BMP4 at stage 2, and finally to ESC-derived hepatocytes (EDH) using a combination of insulin, EGF, FGF4, HGF, OSM, hydrocortisone, and dexamethasone at stage 3 (Fig. 1A).
Figure 1.
Insulin enhanced hepatocyte differentiation. (A): Schematic representation of the differentiation protocol. (B): Quantitative RT-PCR analysis of hepatic gene expression in EDHs cultured with indicated growth factors at stage 2. *, p<.005 versus Basic. (C): Secreted ALB levels in the culture media were assessed by ELISA. *, p<.005 versus Basic. (D): Immunofluorescent staining of EDHs derived using the P2 protocol for ALB. Magnification: 200. (E): Periodic acid Schiff stain of EDHs derived using the P2 protocol. Magnification: 200. Abbreviations: DE, definitive endoderm; EDH, embryonic stem cell-derived hepatocyte.
Since hepatic lineage commitment occurred at stage 2 in the presence of five growth factors, we tested the effects of these growth factors on hepatocyte differentiation by the addition of one growth factor at a time in stage 2 at the same concentration as it was used in the P1 protocol without altering the other stages. The stage 2 basic medium, which contains 20% FBS without additional growth factors, served as a control (Basic). All cells were analyzed for hepatic gene expression and function at day 24 of the differentiation. We found that the addition of insulin but not HGF, or FGF4, or BMP2, or BMP4 alone to the basic medium at stage 2 was sufficient to increase the expression of the hepatocyte markers including ALB, α-antitrypsin (AAT), and ASGPR1 to levels comparable to those in cells grown in the P1 conditions and the human PH (Fig. 1B). Moreover, insulin at three doses tested substantially increased ALB secretion compared to the basic medium, suggesting increased hepatic function in insulin-treated cells (Fig. 1C). The dose of 0.126 U/ml was used in the P1 protocol and all other experiments. In contrast, HGF, FGF4, and BMP2 alone failed to enhance ALB secretion (Fig. 1C). We thus omitted FGF4, BMP2, BMP4, and HGF but kept insulin at stage 2 medium in the following sections and refer to this modified protocol as the P2 protocol. The majority of the EDH obtained using the P2 protocol exhibited characteristic hepatocyte morphology, ALB positive staining (Fig. 1D), and glycogen storage (Fig. 1E), which is another function of hepatocytes.
Expression Profiles of Transcription Factors and Hepatic Marker Genes During Hepatocyte Differentiation from hESCs
To understand how hepatocyte differentiation was induced from hESCs at the molecular level, we examined the expression profiles of 10 transcription factors known to play roles in hepatocyte differentiation and six hepatic markers during the differentiation process using quantitative RT-PCR analysis.
Human hematopoietically expressed homeobox protein (HHEX), GATA6, forkhead box protein A2 (FOXA2), and T-box protein (TBX3) were all robustly induced in stage 1, suggesting the formation of a foregut endoderm competent for hepatocyte induction (Fig. 2A, 2B). The expression of HHEX and GATA6 rapidly declined, whereas the expression of FOXA2 and TBX3 persisted throughout the remaining differentiation process with moderate reduction.
Figure 2.
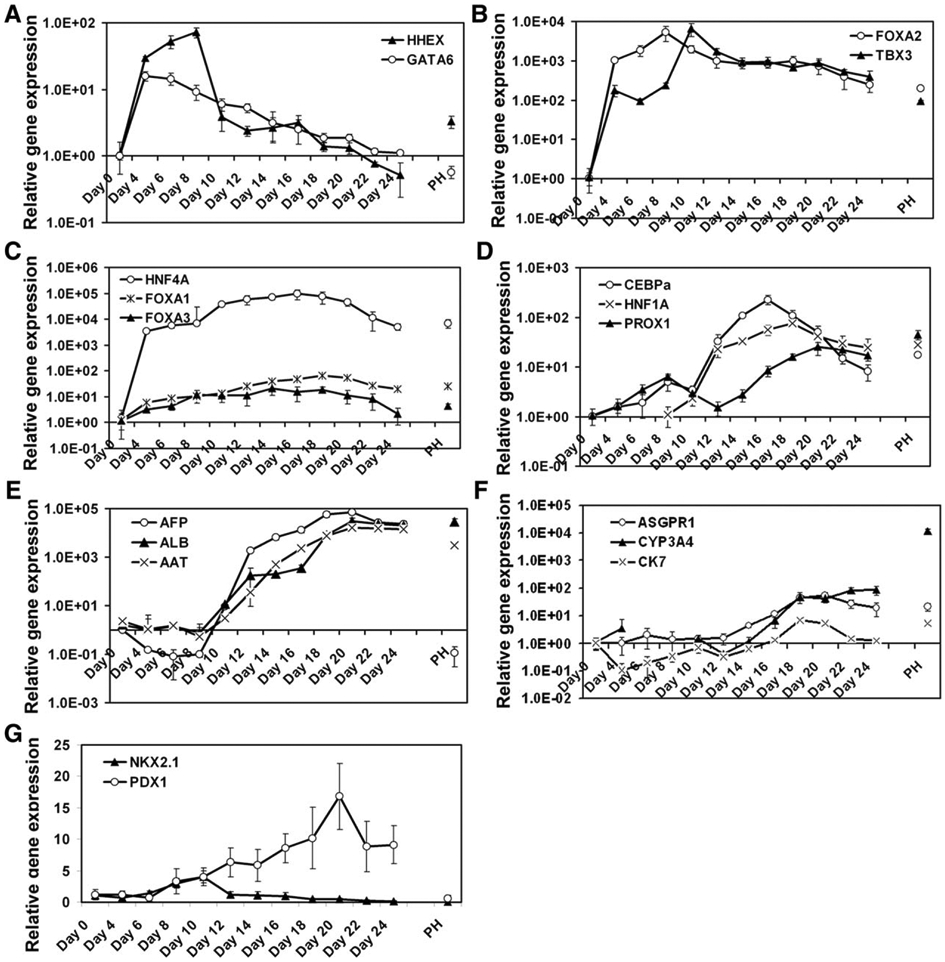
Expression profiles of transcription factors and hepatic genes during hepatocyte differentiation. Human embryonic stem cells were differentiated into embryonic stem cell-derived hepatocytes using the P2 protocol. The expression of the indicated genes was analyzed by quantitative RT-PCR at the indicated time points. The missing points for HNF1A and CYP3A4 indicated their levels not detectable. Abbreviations: AFP, α-fetoprotein; FOXA2, forkhead box protein A2.
HNF4 expression was markedly increased in stages 1 and 2 followed by a moderate reduction in stage 3 but still maintained at a level comparable to that in PHs (Fig. 2C). FOXA1 and FOXA3 levels gradually increased until day 18 of the differentiation (Fig. 2C).
HNF1A and CCAAT-enhancer-binding protein α were only moderately induced at the DE stage, but robustly induced at stage 2 and early-stage 3, followed by a reduction (Fig. 2D). The expression of prospero-related homeobox 1 increased moderately during DE induction, decreased during the first half of stage 2, then increased again thereafter (Fig. 2D).
The expression of three hepatocyte marker genes, AFP, ALB, and AAT, was not induced at the DE stage but was progressively and substantially induced at stage 2, and peaked at stage 3 at levels comparable to those in PH (Fig. 2E). The absence of AFP and the presence of FOXA2, HHEX, and GATA6 in stage 1 cells support that these cells were true DE but not visceral endoderm (Fig. 2E). However, the presence of high levels of AFP in EDHs suggests that these cells were not fully mature when compared with PH, which expressed very low amounts of AFP (Fig. 2E). The expression of ASGPR1 and cytochrome CYP3A4 increased progressively from late stage 2 to stage 3 during the differentiation (Fig. 2F). The P2 protocol did not significantly induce differentiation of cholangiocyte, lung, and pancreas lineages as demonstrated by a minor increase of CK7, NKX2.1, and PDX1 during the differentiation (Fig. 2F, 2G). The expression profiles of these tested genes evident progressive differentiation toward hepatocytes under the P2 condition, which recapitulated many aspects of liver development in vivo.
Insulin Enhances Hepatocyte Differentiation Via Activation of the PI3K/AKT Pathway
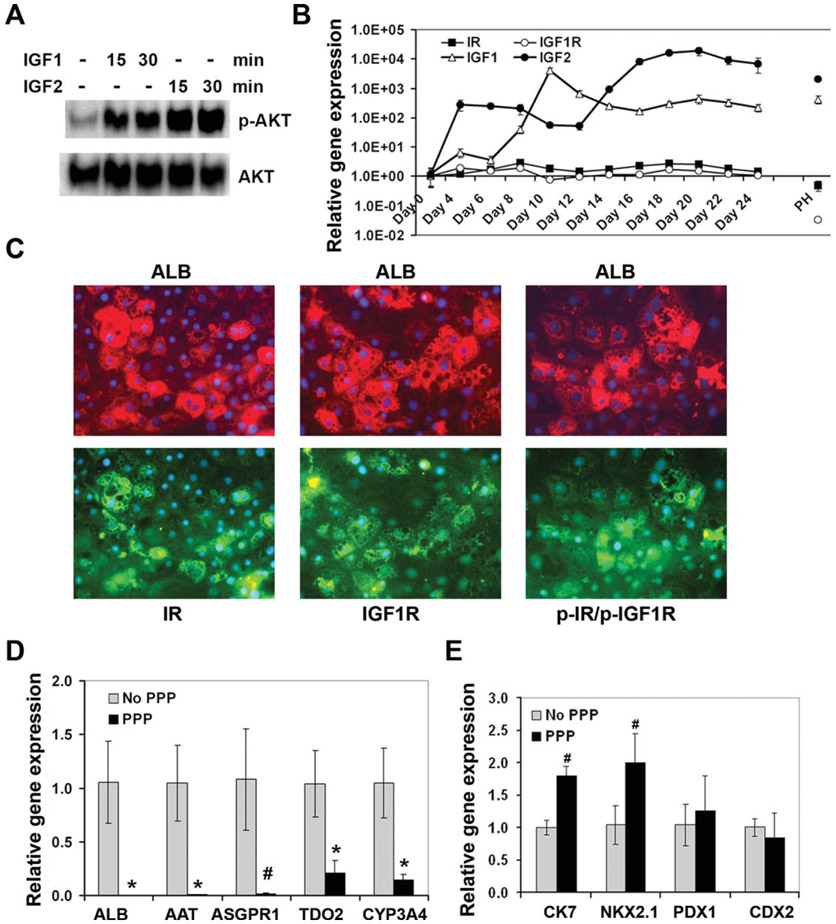
Insulin acts by binding to its receptor IR and triggers the auto-phosphorylation of IR on tyrosine residues. The phosphorylated IR recruits insulin receptor substrate-1 (IRS-1), 2 (IRS-2), Shc, and growth factor receptor-bound protein 2 (Grb2) to the receptor to further activate the signaling pathways of PI3K/AKT and mitogen-activated protein kinase (MAPK) including ERK, p38, and JNK [17]. To understand how insulin stimulates hepatocyte differentiation from DE, we investigated the signaling pathways induced by insulin in DE. For comparison, the signaling events activated by FGF and HGF were also assessed.
DE cells were serum-deprived for 6–7 hours and subsequently stimulated for 15 minutes with insulin, FGF4, or HGF individually or in combination. Cells not exposed to these growth factors served as a control. As expected, insulin induced the phosphorylation of its receptor IR in these cells (Fig. 3A). Insulin did not induce the phosphorylation of the MAPKs including ERK, p38, and JNK (Fig. 3A), but did induce the phosphorylation of AKT and its downstream targets, FoxO3a and FoxO1 in these DE cells (Fig. 3A). FGF4 and HGF induced the phosphorylation of ERK, p38, and JNK, but only weakly induced the phosphorylation of AKT, FOXO3a, and FOXO1 (Fig. 3A, 3B). The combination of insulin and FGF4 showed no significant additive effect on activation of AKT, ERK, p38, and JNK compared to the individual growth factor alone (Fig. 3A). Therefore, our data suggest that insulin primarily activates the PI3K/AKT pathway but not MAPKs in DE, whereas FGF4 and HGF are strong inducer for the MAPK pathway and weak inducers for the PI3K/AKT pathway in DE.
Figure 3.
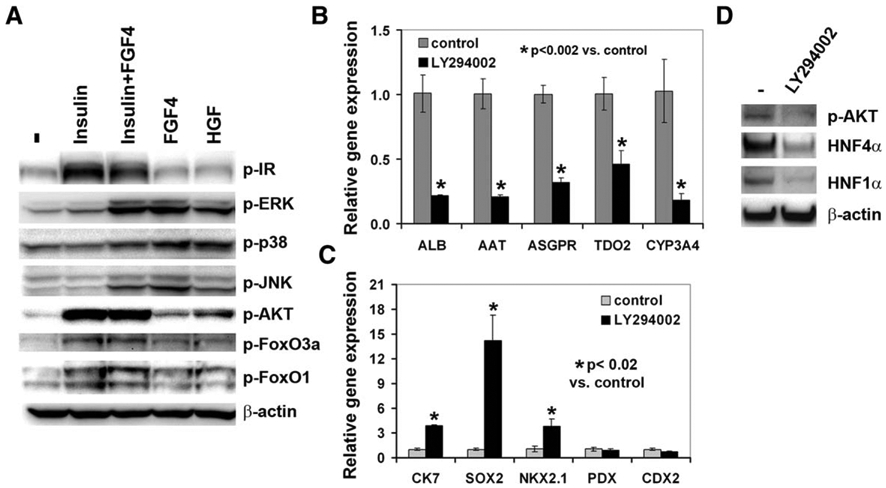
The PI3K/AKT signaling pathway was required for hepatocyte differentiation. (A): Immunoblot analysis of the indicated proteins in definitive endoderm (DE) cells treated with the indicated growth factors for 15 minutes. (B, C): Differentiated cells were treated with or without LY294002 (10 μM) from day 8 to 24. The expression of the indicated genes was analyzed by qRT-PCR. *, p<.002 versus control in (B); *, p<.02 versus control in (C). (D): Immunoblot analysis of the phosphorylated AKT, HNF4α, and HNF1α in the DE cells treated with or without LY294002 (10 μM) for 24 hours. Abbreviations: HGF, hepatocyte growth factor.
Since insulin activates the PI3K/AKT pathway, we next investigated whether blocking the PI3K/AKT pathway impairs hepatocyte differentiation. A PI3K inhibitor, LY294002 at 10 μM, was added to the media from day 8 to 24 during hepatocyte differentiation. The expression of hepatic markers, ALB, ASGPR1, AAT, TDO2, and CYP3A4, was markedly reduced in LY294002-treated cells compared to the control cells not exposed to LY294002 (Fig. 3B). We found that LY294002 enhanced the expression of cholangiocyte marker CK7 and lung lineage markers, SOX2 and NKX2.1, but had no or minimal effect on the expression of the pancreatic marker PDX1 and intestinal marker CDX2 (Fig. 3C). Therefore, the inhibition of hepatocyte differentiation by LY294002 was specific. Similar data were obtained using an AKT inhibitor, AKT IV (data not shown). To understand how LY294002 inhibited hepatocyte differentiation, we assessed the level of hepatic transcription factors, HNF4α and HNF1α in stage 2 cells treated with or without LY294002 for 24 hours. LY294002 markedly reduced the protein levels of HNF4α and HNF1α (Fig. 3D). We confirmed that LY294002 was able to inhibit the activation of AKT in these cells (Fig. 3D).
The PI3K/AKT Pathway Promotes Cell Survival
We next investigated whether the PI3K/AKT pathway regulates proliferation or survival of the DE or hepatic progenitor cells. Cells were treated with or without 10 μM LY294002 from day 8 to 10 or from day 8 to 16 during the differentiation. No significant difference was observed on the expression of a proliferation marker Ki67 in LY294002-treated and non-treated cells. In contrast, the RNA contents, which correlate with the number of cells, were partially reduced in LY294002-treated cultures at the concentration used (Fig. 4A, 4B). These data suggest a role of the PI3K/AKT pathway in preventing cell death rather than promoting proliferation in hepatic progenitor cells.
Figure 4.
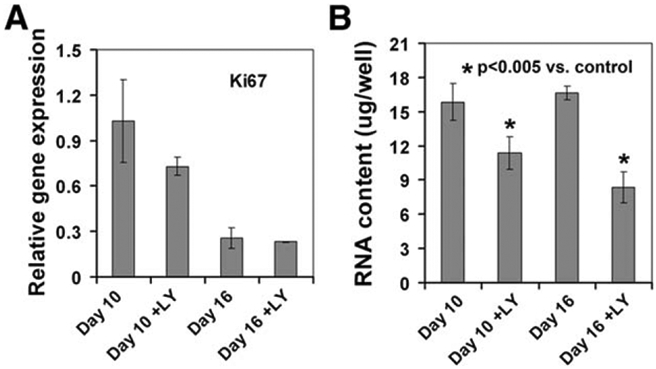
Effects of the PI3K/AKT pathway on proliferation and survival. Differentiated cells were treated with or without LY294002 (10 μM) from day 8 to 10 or from day 8 to 16. The expression of Ki67 was analyzed by quantitative RT-PCR (A) and the RNA contents in these cultures were measured (B). *, p<.005 versus control.
Role of the IGF Signaling Pathway in Hepatocyte Differentiation
IGF1 and IGF2 share considerable similarity with insulin. Both IGFs bind to IGF1 receptor (IGF1R) and trigger auto-phosphorylation of the receptor. Similar to IR, phosphorylated IGF1R can recruit IRS-1 and 2, Shc, and Grb2 to the receptor and subsequently activate the PI3K and MAPK pathway [18,19]. IGF2 also binds to the IGF2 receptor (IGF2R), which negatively regulates IGF2 signaling. We found that both IGF1 and IGF2 activated AKT but not MAPKs in the DE cells (Fig. 5A and data not shown). The expression of both IGF1 and IGF2 increased robustly very early during hepatocyte differentiation, and reached its peak expression at day 10 or day 20 of the differentiation, respectively (Fig. 5B). Furthermore, although the mRNA levels of IGF1R and IR were not markedly altered during hepatocyte differentiation (Fig. 5B), higher protein levels of IR, IGF1R, or the phosphorylated IR/phosphorylated IGF1R (p-IR/p-IGF1R) were present in the EDH with higher levels of ALB (Fig. 5C). Therefore, the stronger activation of insulin and IGF signaling pathway correlated with more differentiated hepatocytes.
Figure 5.
Role of the IGF signaling pathway on hepatocyte differentiation. (A): Immunoblot analysis of phosphorylated AKT and total AKT in definitive endoderm cells treated without or with IGF1 or IGF2 for 15 or 30 minutes. (B): The expression of IGF1, IGF2, IGF1R, and IR during hepatocyte differentiation from human embryonic stem cells assessed by quantitative RT-PCR (qRT-PCR). (C): Immunostaining of embryonic stem cell-derived hepatocytes for ALB and IR, IGF1R or phosphor-IR/phosphor-IGF1R. The same cells are shown in an upper panel and the panel under-neath. Magnification: 200. (D, E): qRT-PCR analysis of the indicated genes in the cells at day 24 of differentiation with or without exposing to PPP (0.2 μM) from day 8 to 16. *, p<.005; #, p<.01 versus no PPP. Abbreviations: IGF, insulin-like growth factor; IR, insulin receptor.
We next investigated whether inhibition of the IGF1 receptor impairs hepatocyte differentiation. An IGF1R inhibitor, PPP at 0.2 μM, was added to the media at stage 2 only during hepatocyte differentiation. Analysis of the expression of hepatic markers at the end of stage 3 revealed that the levels of ALB, ASGPR1, AAT, TDO2, and CYP3A4 were markedly decreased in PPP-treated cells compared to the control cells not exposed to PPP (Fig. 5D). Similar to the PI3K inhibitor LY294002, PPP also moderately increased the expression of the cholangiocyte marker CK7 and lung marker NKX2.1 but not the pancreatic marker PDX1 and intestine marker CDX2 (Fig. 5E).
Knockdown of the IGF1 Receptor and/or IR by shRNAs Inhibited Hepatocyte Differentiation
To directly prove that insulin and IGF signaling pathways play a critical role in hepatocyte differentiation from DE, we knocked down the expression of the β subunit of the IR or IGF1R individually or in combination in the DE cells using lentiviral vectors carrying shRNAs against these receptors. The shRNA against IR (shIR) knocked down the mRNA level of the IR but not the IGF1R, and the protein levels of both the IR and IGF1R in these cells (Fig. 6A, 6B). The shRNA against IGF1R (shIGF1R) only knocked down the expression of IGF1R but not the IR at both the mRNA and protein levels (Fig. 6A, 6B). The shIR and/or shIGF1R had no significant effect on the expression of IGF1 (Fig. 6A). However, shIR significantly reduced the level of IGF2 (Fig. 6A). Furthermore, the shIR markedly reduced the protein levels of phosphor-AKT, HNF4α, and HNF1α (Fig. 6B). To a lesser extent, shIGF1R also reduced the levels of these proteins (Fig. 6B). Either shIR or shIGF1R was sufficient to eliminate the expression of ALB at both the protein and mRNA levels (Fig. 6B, 6C), suggesting impaired hepatocyte differentiation in the IR and/or IGF1R null cells. In addition, the shIR markedly reduced the mRNA levels of ASGPR1, AAT, and TDO2 (Fig. 6C). The IGF1R knockdown cells also showed significantly reduced mRNA levels of ASGPR1 but not AAT and TDO2 (Fig. 6C). Simultaneously using shIR and shIGF1R resulted in reduced hepatic expression similar to shIR (Fig. 6A, 6C, 6D). This is because the shIR vector knocked down both IR and IGF1R at the protein levels (Fig. 6B). Therefore, our data suggest that both insulin and IGF signaling pathways play an important role in hepatocyte differentiation from DE. Similar to the inhibition of the PI3K or IGF1R by small molecule inhibitors, the knockdown of IGF1R and/or IR enhanced the expression of cholangiocyte marker CK7 and lung lineage marker NKX2.1 (Fig. 6D), supporting specific regulation of hepatocyte differentiation by insulin and the IGF signaling pathway.
Figure 6.
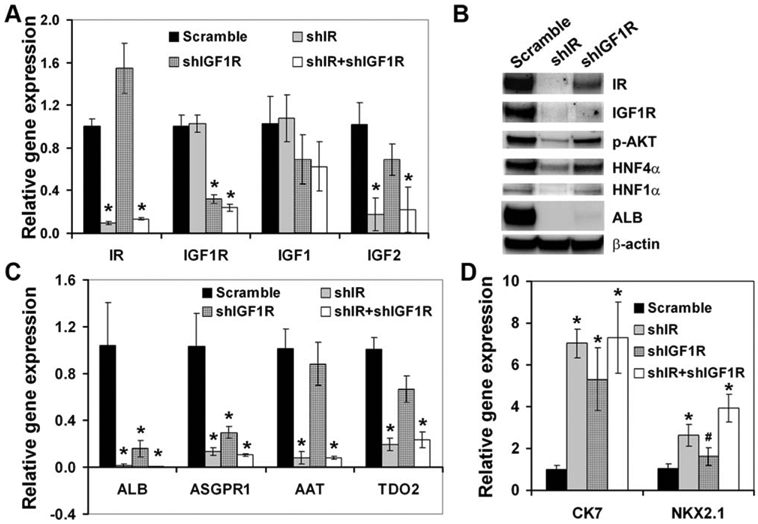
Knockdown of IR or IGF1R impaired hepatocyte differentiation. (A, C, D): qRT-PCR analysis of the indicated genes in the cells transduced with scramble or shIR, and/or shIGF1R vector. *, p<.005; #, p<.05 versus scramble. (B): Immunoblot analysis of the indicated proteins in the cells transduced with scramble or shIR, or shIGF1R vector. Abbreviations: IGF1R, insulin-like growth factor receptor 1; IR, insulin receptor.
DISCUSSION
Hepatocyte fate determination has been investigated primarily through developmental studies, which revealed that multiple growth factors, such as FGFs and BMPs, are involved in this process [1-4]. However, genetic ablation of any of these growth factors alone failed to induce significant defects in liver organogenesis. The complex and dynamic nature of hepatocyte fate determination makes it difficult to study signals that regulate this process in animals. We took advantage of the in vitro system of hepatocyte differentiation from hESCs to study growth factor-mediated signaling pathways which regulate hepatocyte differentiation from DE. We found that the addition of insulin to DE cells enhanced hepatocyte differentiation, even in the absence of several other exogenous factors thought to be critical in the process, such as FGFs and BMPs. We found that the expression of both IGF1 and IGF2 was induced robustly at the DE stage and that their levels were either maintained or further increased in later stages of hepatocyte differentiation from hESCs. Upregulation of IGF2 during hepatocyte differentiation from hESCs has also been reported by DeLaForest et al. [20]. Ablation of the IGF1R and/or IR by shRNAs markedly impaired hepatocyte differentiation. Our data demonstrated an important role of insulin and IGF in hepatocyte differentiation from DE. Although the foregut endoderm may not access insulin during early development, it may access IGFs produced by mesoderm or developing heart, which start to express IGF2 as earlier as E8 in mouse embryos [21,22].
Suppression rather than activation of the PI3K signaling pathway has been shown to be required for DE induction from ESCs [23,24]. Calmont et al. showed that the PI3K pathway is active in the foregut endoderm at the time hepatic genes are activated during embryonic development (7–8S stages). However, they found that inhibition of the MAPK but not the PI3K pathway impaired ALB expression in mouse embryo explants, while inhibition of the PI3K pathway inhibited the growth of the explants [25]. Since the embryo explants were only cultured for 48 hours in their experiment, it remains to be determined whether blockage of PI3K for longer time or at different developmental stages would affect hepatocyte differentiation in vivo. We found that inhibition of the PI3K pathway in ESC-derived DE reduced cell survival rather than proliferation. Interestingly, inhibition of the PI3K pathway by small molecule inhibitors resulted in impaired hepatocyte differentiation. Furthermore, inhibition of the insulin and IGF signaling pathways by knocking down their receptors markedly reduced the activation of the PI3K pathway and diminished hepatocyte differentiation. The impaired hepatocyte differentiation was not likely due to the reduced survival under these conditions, because the same conditions increased the expression of the cholangiocyte and lung lineage markers. Therefore, we provide compelling evidence that the PI3K signaling pathway regulates hepatocyte differentiation after the DE stage.
The importance of HNF1α and HNF4α in hepatocyte differentiation is highlighted by a number of studies. Both HNF1α and HNF4α directly promote the transcription of many hepatic specific genes [26]. Knockdown of HNF4A in hESCs inhibits hepatocyte differentiation [20]. Moreover, ectopic expression of a combination of HNF4A with a FOXA gene or a combination of HNF1A, GATA4, and FOXA and inactivation of p19(Arf) in mouse fibroblasts can directly induce hepatocyte differentiation [27,28]. We found that FOXA and GATA6 were already substantially upregulated at the DE stage and thus likely played important roles in DE rather than hepatocyte specification. The expression of HNF4α was induced initially at stage 1, but further increased at stage 2. HNF1α were primarily induced at stage 2 in cells that were exposed to exogenous insulin and endogenous IGFs. The expression of HNF1α and HNF4α was specifically downregulated by the inhibition of the PI3K or knockdown of IR and/or IGF1R, while insulin upregulated the expression of these transcription factors. Therefore, an increase of HNF1α and HNF4α induced by the PI3K pathway may be responsible for the enhanced hepatocyte differentiation through this pathway.
Although the addition of FGF4, BMP2, BMP4, and HGF to the stage 2 basic medium failed to further enhance hepatocyte differentiation, our data do not suggest that these growth factors play no role in hepatocyte differentiation. The stage 2 basic medium contains 20% FBS and may provide sufficient amounts of these growth factors. Low concentrations of FGF2 have been reported to induce hepatocyte differentiation from DE, but high concentrations of FGF2 promote pancreatic and intestinal cell fates [29]. Our previous study showed that HGF, EGF, bFGF, and aFGF counteract the effect of insulin on hepatocyte differentiation from embryoid bodies spontaneously differentiated from hESCs [12]. We showed here that both FGF4 and HGF primarily induced the activation of MAPK in DE cells. Apparently, high levels of MAPK activation may impede hepatocyte differentiation.
CONCLUSION
We revealed a nonoverlapping critical role of insulin and IGFs in hepatocyte differentiation from hESCs and demonstrated that the mechanism by which insulin and IGFs act is through activation of the PI3K pathway followed by0 upregulation of HNF1α and HNF4α. Identification of the role of the PI3K pathway in hepatocyte differentiation has significantly advanced on our understanding of hepatocyte cell fate determination.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karen Paper and Catherine Nacey for packaging lentivirus for us. This work is supported in part by an Academic Federation Innovative Developmental Award from University of California Davis (to P.Z.) and a grant from the California Institute for Regenerative Medicine (Grant TR2–01857 to M.A.Z.). Y.J. is currently affiliated with the Center for Regenerative Medicine, Massachusetts General Hospital, Harvard Stem Cell Institute, Harvard Medical School, Boston, MA.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
REFERENCES
- 1.Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science 2008;322:1490–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol 2009;25:221–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell 2010;18:175–189. [DOI] [PubMed] [Google Scholar]
- 4.Behbahan IS, Duan Y, Lam A et al. New approaches in the differentiation of human embryonic stem cells and induced pluripotent stem cells toward hepatocytes. Stem Cell Rev 2011;7:748–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamiya A, Kinoshita T, Miyajima A. Oncostatin M and hepatocyte growth factor induce hepatic maturation via distinct signaling pathways. FEBS Lett 2001;492:90–94. [DOI] [PubMed] [Google Scholar]
- 6.Clotman F, Lemaigre FP. Control of hepatic differentiation by activin/TGFbeta signaling. Cell Cycle 2006;5:168–171. [DOI] [PubMed] [Google Scholar]
- 7.Lorent K, Yeo SY, Oda T et al. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development 2004;131:5753–5766. [DOI] [PubMed] [Google Scholar]
- 8.McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development 2002;129:1075–1082. [DOI] [PubMed] [Google Scholar]
- 9.Kamiya A, Kinoshita T, Ito Y et al. Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J 1999;18:2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt C, Bladt F, Goedecke S et al. Scatter factor/hepatocyte growth factor is essential for liver development. Nature 1995;373:699–702. [DOI] [PubMed] [Google Scholar]
- 11.Snykers S, De Kock J, Rogiers V et al. In vitro differentiation of embryonic and adult stem cells into hepatocytes: State of the art. Stem Cells 2009;27:577–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirahashi H, Wu J, Yamamoto N et al. Differentiation of human and mouse embryonic stem cells along a hepatocyte lineage. Cell Transplant 2004;13:197–211. [DOI] [PubMed] [Google Scholar]
- 13.Duan Y, Ma X, Zou W et al. Differentiation and characterization of metabolically functioning hepatocytes from human embryonic stem cells. Stem Cells 2010;28:674–686. [DOI] [PubMed] [Google Scholar]
- 14.Zhou P, Lessa N, Estrada DC et al. Decellularized liver matrix as a carrier for the transplantation of human fetal and primary hepatocytes in mice. Liver Transpl 2011;17:418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P, Gross S, Liu JH et al. Flavokawain B, the hepatotoxic constituent from kava root, induces GSH-sensitive oxidative stress through modulation of IKK/NF-kappaB and MAPK signaling pathways. FASEB J 2010;24:4722–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou P, Hohm S, Olusanya Y et al. Human progenitor cells with high aldehyde dehydrogenase activity efficiently engraft into damaged liver in a novel model. Hepatology 2009;49:1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: Insights into insulin action. Nat Rev Mol Cell Biol 2006;7:85–96. [DOI] [PubMed] [Google Scholar]
- 18.Pollak M The insulin and insulin-like growth factor receptor family in neoplasia: An update. Nat Rev Cancer 2012;12:159–169. [DOI] [PubMed] [Google Scholar]
- 19.Harris LK, Westwood M. Biology and significance of signalling pathways activated by IGF-II. Growth Factors 2012;30:1–12. [DOI] [PubMed] [Google Scholar]
- 20.DeLaForest A, Nagaoka M, Si-Tayeb K et al. HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development 2011;138:4143–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JE, Pintar J, Efstratiadis A. Pattern of the insulin-like growth factor II gene expression during early mouse embryogenesis. Development 1990;110:151–159. [DOI] [PubMed] [Google Scholar]
- 22.Li P, Cavallero S, Gu Y et al. IGF signaling directs ventricular cardio-myocyte proliferation during embryonic heart development. Development 2011;138:1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLean AB, D’Amour KA, Jones KL et al. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells 2007;25:29–38. [DOI] [PubMed] [Google Scholar]
- 24.Touboul T, Hannan NR, Corbineau S et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 2010;51:1754–1765. [DOI] [PubMed] [Google Scholar]
- 25.Calmont A, Wandzioch E, Tremblay KD et al. An FGF response pathway that mediates hepatic gene induction in embryonic endoderm cells. Dev Cell 2006;11:339–348. [DOI] [PubMed] [Google Scholar]
- 26.Odom DT, Zizlsperger N, Gordon DB et al. Control of pancreas and liver gene expression by HNF transcription factors. Science 2004;303: 1378–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 2011;475:390–393. [DOI] [PubMed] [Google Scholar]
- 28.Huang P, He Z, Ji S et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 2011;475:386–389. [DOI] [PubMed] [Google Scholar]
- 29.Ameri J, Stahlberg A, Pedersen J et al. FGF2 specifies hESC-derived definitive endoderm into foregut/midgut cell lineages in a concentration-dependent manner. Stem Cells 2010;28:45–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.