Abstract
Cyclin-dependent kinase 6 (CDK6) is a potential drug target that plays an important role in the progression of different types of cancers. We performed in silico and in vitro screening of different natural compounds and found that quercetin has a high binding affinity for the CDK6 and inhibits its activity with an IC50 = 5.89 μM. Molecular docking and a 200 ns whole atom simulation of the CDK6-quercetin complex provide insights into the binding mechanism and stability of the complex. Binding parameters ascertained by fluorescence and isothermal titration calorimetry studies revealed a binding constant in the range of 107 M–1 of quercetin to the CDK6. Thermodynamic parameters associated with the formation of the CDK6–quercetin complex suggested an electrostatic interaction-driven process. The cell-based protein expression studies in the breast (MCF-7) and lung (A549) cancer cells revealed that the treatment of quercetin decreases the expression of CDK6. Quercetin also decreases the viability and colony formation potential of selected cancer cells. Moreover, quercetin induces apoptosis, by decreasing the production of reactive oxygen species and CDK6 expression. Both in silico and in vitro studies highlight the significance of quercetin for the development of anticancer leads in terms of CDK6 inhibitors.
Introduction
Cancer cells frequently show a gain of proliferation, cell cycle aberrations, loss of genomic and chromosomal stabilities, and metabolic alterations.1,2 More than 80% of the cellular pathways such as cell cycle progression, transcription, DNA repair, and metabolic events are regulated by protein kinases through phosphorylation/dephosphorylation.2−5 Out of different protein kinases, overexpression of cyclin-dependent kinases (CDKs) (EC 2.7.11.22) is associated with the majority of cancer types, and thus, identification and establishment of CDKs targeted inhibitors become an attractive approach for cancer therapy.6−10
At present, >20 members of the CDK family are reported.11 The basic structure of CDKs contains a conserved catalytic domain with an ATP-binding site, PSTAIRE-like cyclin-binding domain, and activating T-loop motif.12 Cell cycle progression is regulated by specific CDKs, cyclin proteins, and respective CDK inhibitors (CKIs). Proper coordination between all three components allows the smooth progression of the cell cycle.11−13 CDK6 and cyclin D are responsible for the G1-to-S phase transition.13 Normally, CDK6 regulates the cell cycle by phosphorylating the retinoblastoma (Rb) protein in the Rb-E2F pathway, which results in the release of the transcription factor (E2F). When E2F enters the nucleus, it initiates the synthesis of different proteins required for DNA replication (S phase).14,15 However, during cancer conditions, the upregulation of CDK6 leads to the permanent activation of the Rb-E2F pathway, which results in uncontrolled cell growth.14,16 CDK6 not only regulates the cell cycle but also controls the metabolic alterations in the cancer cells.17 It regulates the production of reactive oxygen species (ROS), by inhibiting the activity of phosphofructokinase (PFK) (EC 2.7.1.11) and other protein kinases of the glycolytic pathways.17 This reprogramming of glycolysis directs the glycolytic intermediates toward pentose phosphate (PPP) or serine pathways and preventing the cell apoptosis.3,17 These studies suggested that CDK6 is the key regulator of cell cycle, metabolism, and its overexpression is associated with cancer, diabetes, and inflammatory diseases. All these evidences favor the role of CDK6 in a variety of diseases, thus making it a potential drug target.6,18
Medicinal plants are an important source of diverse natural compounds and phytonutrients that have been used in the treatment of cancer and are known for their potential therapeutic applications to manage different diseases.19−21 Most of the chemopreventive agents include plant-derived components because they show less or no side effects.22 Plant-derived natural products have been used for several decades, including flavonoids, polyphenols, alkaloids, and sesquiterpenes. The therapeutic potential of these plant-based products has been explored in terms of their kinase targeting capabilities.23−27 Around 80% of the population still relies on plant-derived formulations to manage different medical issues.19 In recent years, there is a switch in the screening and discovery of natural products as potential kinase inhibitors.25,28−31 Plant-derived products have several important biological properties such as antioxidant, anticancerous, anti-inflammatory, antidiabetic, antimicrobial, and hepatoprotective24,25,32 and thus are used in treating cancer. One such class of compound is natural polyphenols like flavonoids, which are widely distributed in plants and are generally present in food like herbs, nuts, vegetables, fruits, plant-derived beverages like tea, coffee, and red wine.33,34 These plant-based phenolic compounds target many cancer-associated signaling pathways like PI3K (EC2.7.1.137), Akt (EC 2.7.11.1), B-Raf kinase (EC2.7.11.25), and ERK or MAPK (EC 2.7.11.24).35−39
Quercetin is a ubiquitous pentahydroxyflavone, found mainly in fruits and vegetables.40 Many studies have reported diverse activities of quercetin, viz., anti-inflammatory, antithrombotic, antiobesity, antihypercholesterolemic, antiatherosclerotic, and anticancer activities.40,41 Quercetin inhibits cancer cell proliferation by inducing apoptosis, in a way reducing the growth of different tumors by unknown mechanisms, and remains a molecule of high interest and investigation with promising biological activities.29,42,43 Quercetin is a molecule of immense importance especially in context with its anticancer potential. It shows different types of efficacies, specificities, and targets in different cancers through the diverse mode of action.43 Recently, it was reported that quercetin reverses the irregular DNA methylation patterns of different cancer cells and thus also exerts anticancer activities through epigenetic modifications.44,45 Quercetin decreases the population of tumor-initiating cells through the modulation of the death-associated factor 6 (DAXX) protein.46 Interestingly, when used in combination, quercetin increases the efficiencies of other anticancer molecules like losartan, paclitaxel, and resveratrol in different cancers.47−49 The nanoformulations of quercetin are very effective against different cancers including lung, liver, breast, and glioblastomas.48,50,51 These studies reported the anticancer properties of quercetin, but the exact mechanism of its action remains elusive, and this study is the first of its kind that investigated the potential of quercetin as an inhibitor of CDK6.
Previously, we have reported the anticancer potential of quercetin using different models,28,29 but the present study provides newer insights into the anticancer activity of quercetin by establishing CDK6 as a target. Docking and molecular dynamics (MD) simulation studies were performed to ascertain the interaction analysis and stability of the CDK6-quercetin complex. The enzyme assay of recombinant CDK6 with quercetin suggested a significant inhibition profile of quercetin. Cell-based anticancer analysis of CDK6 overexpressing cancer cell lines suggested that quercetin decreases the viability of cancer cell lines and induces apoptosis probably by decreasing the expression of CDK6.
Results and Discussion
Screening of Natural Molecules
Molecular docking, fluorescence-based binding, and enzyme inhibition studies were used to screen out the best possible inhibitor of CDK6 amongst different natural products. We screened gallic acid, ferulic acid, caffeic acid, rosmarinic acid, capsaicin, tocopherol, limonene, and quercetin for the identification of a probable CDK6 inhibitor (Table S1, Figure S1). Based on in silico binding studies (binding energy and binding residues), fluorescence binding, and enzyme assay, the best compounds that interacted with functionally important residues possess high affinity and high inhibition potential for CDK6 was selected (Table S1, Figure S1). Screening of these compounds suggested that quercetin acts as the best inhibitor of CDK6 and thus subjected to detailed analysis.
Molecular Docking
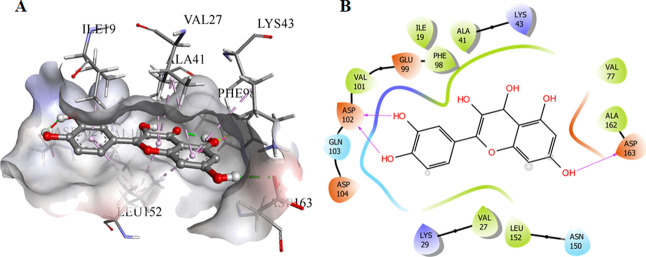
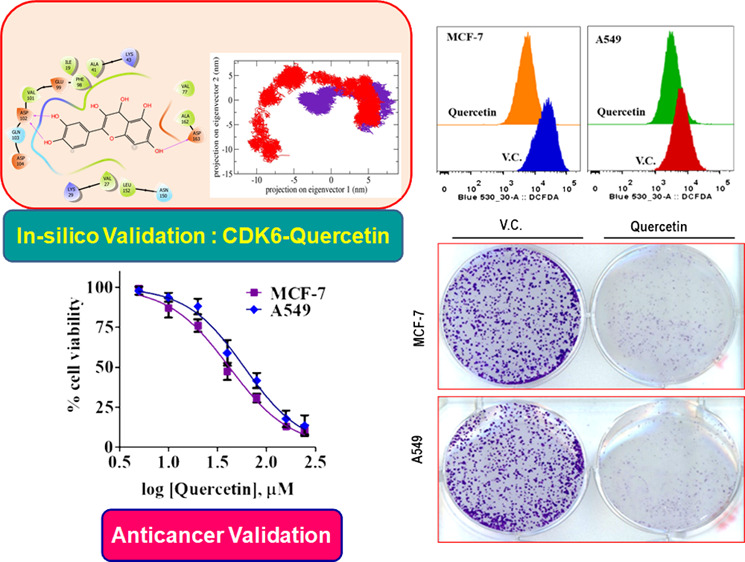
There are around 16 crystal structures of CDK6 that were present in the Protein Data Bank (PDB), but on the basis of calculated docking RMSD values of the quercetin against a variety of the deposited structures, the 3NUP was selected for the current in silico analysis because it showed relatively lower RMSD values for the generated docked conformations The validation of the current protocol using the SAnDReS algorithms showed the reliability of the current study, which can be estimated with a docking accuracy of 76.3% (Table S2). The information about the active site of CDK6 was collected,52 and the ATP-binding site was deeply analyzed, which was occupied by Lys43, Phe98, His100, Asp104, Thr107, and Asp163. The significant binding was observed for quercetin to CDK6, which can be inferred from the free energy of a binding of −5.87 kcal/mol, with three hydrogen bonds offered by hydroxyl groups and side chains of Asp102 and Asp163 (Figure 1). The 40th docked pose was selected based on combined scoring. Furthermore, the validation of the predictive performance of the semiempirical free energy scoring function implemented in the AutoDock4 was performed by assessing the Spearman’s rank correlation coefficient between the predicted free energy and experimental binding affinity. The process involved the collection of eight CDK6 inhibitors deposited in the binding DB,53 followed by the calculation of the predicted binding energies with the CDK6. Spearman’s rank correlation coefficient was calculated to be around 0.964, indicating that the association between predicted free energy of binding and experimentally IC50 values can be considered statistically significant. These observations indicated that quercetin inhibits the CDK6 activity, and thus, the docked system was selected and subjected to MD simulations.
Figure 1.
Molecular docking studies of quercetin with CDK6. (A) The docked conformation of quercetin and CDK6 in the 3D view showing the bound orientation of the inhibitor. (B) The 2D view of the docked complexes highlighting the observed hydrogen bonds between the quercetin and CDK6.
MD Simulations
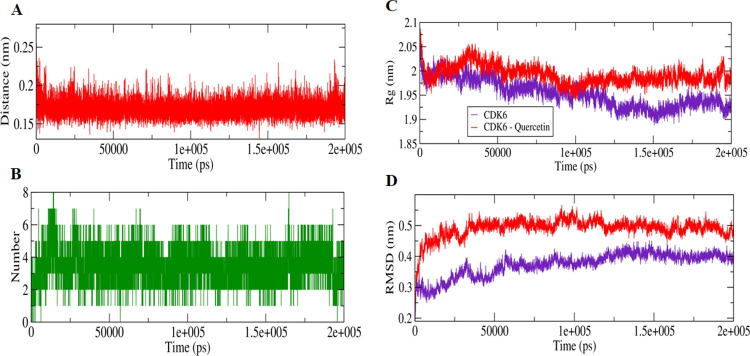
The extent of quercetin interaction with the CDK6 in the docked complex was assessed by 200 ns MD simulations, which were performed for both apo- and quercetin-bound CDK6 separately. The distance parameters were calculated between the quercetin and CDK6, which was indicative of the attainment of closely bound confirmation with the computed distance, fluctuated between 0.15 and 0.25 nm. This observation was validated by the computation of the hydrogen bonds with their magnitude reached up to 8 between the CDK6 and quercetin during MD simulations (Figure 2A,B). The stabilities of the studied apo CDK6 and docked systems were further assessed by calculating Rg, and RMSD values for both the system reached an equilibrium confirmation after 120 ns (Figure 2C,D). Both Rg and RMSD calculations suggest that the unbound apo form of CDK6 is more stable than the quercetin-bound form, highlighting the effect of the quercetin interaction on the structure of CDK6.
Figure 2.
MD simulation studies of quercetin with CDK6. (A) Plot showing changes in the computed distance between the CDK6 and quercetin. (B) Hydrogen-bond fluctuation curve highlighting the changes in the observed number. (C) The Rg plot showing the difference in the compactness between the quercetin bound and unbound CDK6. (D) The RMSD plot highlighting the changes between the stabilities in the observed systems.
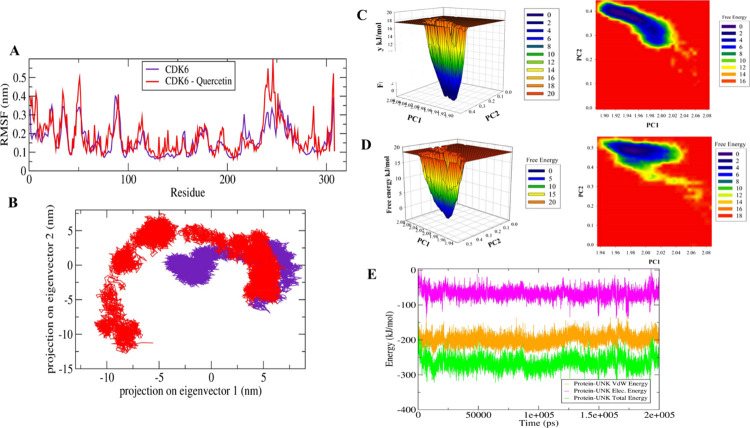
Furthermore, a significant difference in the fluctuation of active site residues was observed between the quercetin bound and apo CDK6 conformations (Figure 3A). The active site residues Lys43, Phe98, His100, Asp104, Thr107, and Asp163 from the corresponding secondary structure elements of β3, β5, L8, β6, α3, and L12 showed relatively lower motion in the unbound form of the CDK6, indicating the presence of lower relative energy as compared to the bound form. Moreover, the PCA was used for the assessment of the difference in the flexibility parameters between the quercetin bound and unbound conformations. In MD simulations, the PCA is a significant statistical technique employed for the reduction of data complexity and in understanding the variations in the atomic motion present in biomolecules.54 A set of eigenvectors and eigenvalues were projected as a result of PCA analyses with the quercetin-bound form occupies a larger conformational space as compared to the unbound apo CDK6 conformation (Figure 3B). These observations indicated the presence of higher structural stability in the unbound CDK6 form as compared to the quercetin bound form.
Figure 3.
MD simulation studies of quercetin with CDK6. (A) The graphical representation of the changes observed in the fluctuation of the constituent residues between the quercetin bound and unbound CDK6. (B) The 2D eigenvector projection plot showing the differences between the flexibility of the two studied forms. (C) The plots of the free energy landscape and contour map for the unbound form of CDK6. (D) The graphical representation of the free energy landscape of quercetin bounds form. (E) The MMPBSA-based generated curves highlighting the changes in the total, electrostatic, and van der Waals energies calculated between the CDK6 and quercetin.
Additionally, the free energy landscapes were plotted to understand the differences in the protein-folding patterns between the quercetin bound and unbound CDK6 forms. A distinct difference in the projection of free energy was observed with energetically favored and relatively stable conformation for the unbound form of CDK6 as a compared complex (Figure 3C,D). These observations indicated that the interaction of quercetin with CDK6 slightly perturbs its conformation and subsequently inhibits the enzyme activity. Besides the mentioned observations, the MMPBSA-based algorithm was used to calculate the energy difference between the CDK6 and quercetin (Figure 3E). The total free energy of binding of CDK6 to quercetin was observed between −200 and −300 kJ/mol in which electrostatic energy is a major contributor.
Fluorescence Measurements
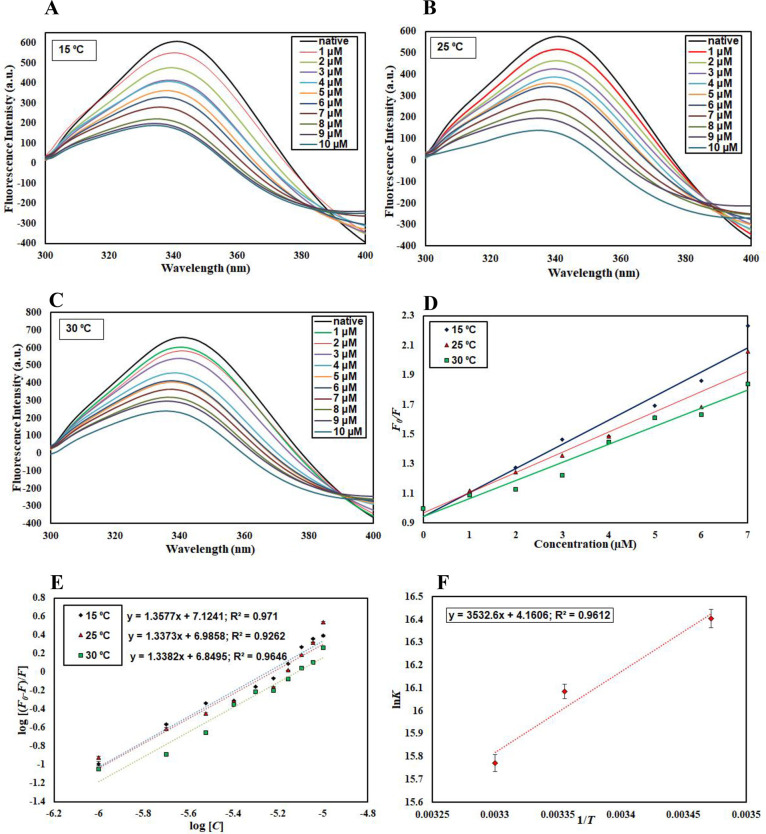
In silico observations were further complemented by in vitro assays. To get insights into the actual binding affinity of quercetin with CDK6, fluorescence measurements were carried out. A significant gradual decrease in the fluorescence intensity of CDK6 with increasing concentrations of quercetin suggested a strong binding affinity. Temperature dependency of binding parameters provides information about the type of quenching taking place for the protein–ligand interaction (static or dynamic or a combination of both), and thus, fluorescence measurements were carried out at three different temperatures.55,56 A similar pattern of decrease in fluorescence intensity with increasing concentrations of quercetin was observed at all three temperatures (15, 25, and 30 °C), indicating the formation of a stable complex between the CDK6 and quercetin (Figure 4A–C).
Figure 4.
Fluorescence binding studies of quercetin with CDK6. Steady-state fluorescence of CDK6 in the absence and presence of quercetin (1–10 μM) at (A) 15 °C, (B) 25 °C, and (C) 30 °C. (D) Stern–Volmer plots, (E) modified Stern–Volmer plots, and (F) van’t Hoff plots of the CDK6–quercitin interaction at three temperatures (15, 25, and 30 °C).
The addition of quercetin causes an alteration in the microenvironment of fluorophores, which causes fluorescence quenching. This decrease in fluorescence intensity was mathematically calculated by applying various equations to find out quenching and binding parameters. The quenching data was mathematically fitted to the Stern-Volmer and modified Stern-Volmer equations to obtain binding parameters. Figure 4D shows the Stern–Volmer plot of F0/F versus [C] in which the slope gives the value of Stern–Volmer constant Ksv at a fixed intercept after linear regression fitting. For the calculation of Ksv, only linear points were taken into consideration.
Table 1 represents the values of the obtained Ksv at three different temperatures. We observed that Ksv decrease with an increase in temperature, suggesting that static quenching governs the CDK6–quercetin complex formation. Further, equation 1 was used to validate the static mode of quenching (Kq=Ksv/ τ0 Equation 1). The value of the bimolecular quenching rate constant (Kq) was estimated by this equation. The value of Kq for the CDK6–quercetin interaction was found to be substantially higher than the maximum scatter collision quenching constant of quenchers with biopolymer (2 × 1010 M–1 s–1),57 supporting the earlier observations of the static mode of quenching to be operative for the CDK6–quercetin interaction.58
Table 1. Stern–Volmer Constants Calculated at Different Temperatures Obtained from Fluorescence Spectroscopy.
| temperature (°C) | Ksv (104 M–1) | Kq (1013 M–1 s–1) | R2 |
|---|---|---|---|
| 15 | 16.3 | 6.03 | 0.96 |
| 25 | 13.6 | 5.03 | 0.96 |
| 30 | 12.2 | 4.51 | 0.96 |
The binding constant for the CDK6–quercetin interaction was estimated by modified Stern-Volmer equation. Figure 4E shows the modified Stern–Volmer plot in which the intercept and slope give the value of binding constant (K) and the number of binding sites (n), respectively. The value of K was estimated as 1.3 × 107 M1 at 15 °C, suggesting that quercetin binds to CDK6 with a high affinity. Table 2 shows the K obtained at different temperatures, suggesting that a less stable complex is formed at higher temperatures as values of K decrease with increasing temperature. These observations are following our in silico observations, suggesting a strong binding affinity of CDK6 for quercetin.
Table 2. Thermodynamic Parameters Obtained from Fluorescence Quenching Studies Carried Out at Different Temperatures.
| temperature (°C) | K (107 M–1) | n | ΔG (kcal mol–1) | ΔS (cal mol–1 K–1) | ΔH (kcal mol–1) | TΔS (kcal mol–1) |
|---|---|---|---|---|---|---|
| 15 | 1.3 | 1.35 | –9.400 | 8.267 | –7.019 | 2.38 |
| 25 | 0.96 | 1.33 | –9.482 | 2.46 | ||
| 30 | 0.70 | 1.33 | –9.524 | 2.50 |
The major forces involved in protein–ligand interactions are hydrogen bonds, van der Waals, electrostatic, and hydrophobic interactions. The van't Hoff equation was used to find the thermodynamic parameters associated with the CDK6–quercetin interaction such as ΔH, ΔS, and ΔG, an indicative of prevailing forces for the specific protein–ligand interaction. When the values of ΔH and ΔS are negative, the dominant forces are van der Waals and hydrogen bonding, while the positive values of ΔH and ΔS suggest hydrophobic interactions.59 On the contrary, the negative ΔH and positive ΔS values indicate predominant electrostatic interactions involved. The van’t Hoff plot shows the dependence of the binding constant (K) on 1/T, the slope of which is equal to −ΔH/R, and the intercept gives an estimate of ΔS/R (Figure 4F). Table 2 provides all thermodynamic parameters obtained for CDK6–quercitin binding. The negative ΔH and positive ΔS values indicated that electrostatic interactions are predominantly involved.
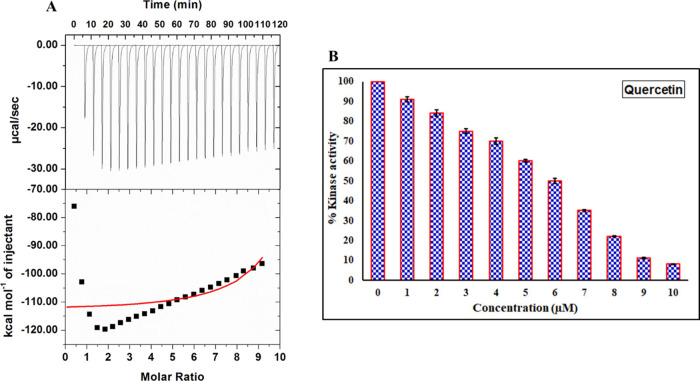
Isothermal Titration Calorimetry
To complement fluorescence binding studies, ITC is employed to evaluate ligand-binding affinity60 and associated thermodynamic parameters such as a change in entropy (ΔS), change in enthalpy (ΔH), Gibbs free energy (ΔG), number of binding sites (n), and binding affinity constant (K).56 A representative ITC profile for the CDK6–quercetin interaction is depicted in Figure 5A. Every peak of the binding isotherm represents a single injection of quercetin into the CDK6 solution filled in the sample cell (Figure 5A). The thermodynamic parameters obtained for the CDK6–quercetin interaction were: Ka = 5.88 × 105 ± 1.40 × 106 M–1, ΔH = −1.12 × 105 ± 4.57 × 102 cal/mol, and ΔS = −351 cal/mol/deg. This data was obtained for the model of one binding site. The variation in thermodynamic parameters obtained from fluorescence and ITC experiments is a common trait that is owed to the assumption made in noncalorimetric approaches that ΔH does not depend on temperature.60,61 Additionally, the ITC measured a global change in the property, whereas the fluorescence spectroscopy measured only local changes around the fluorophore (Trp214), which might be the reason for the differences in the values obtained through fluorescence spectroscopy and ITC.58,62
Figure 5.
Binding and enzyme inhibition studies of quercetin with CDK6. (A) Binding isotherm for the titration of quercetin with CDK6 at 25 °C. The CDK6 (15 μM) in the sample cell of ITC was titrated with quercetin (500 μM, taken in the syringe), and heat change was measured. (B) ATPase inhibition assay of CDK6 with increasing concentration of quercetin (0–10 μM). The activity of native CDK6 was taken as 100% for reference.
Enzyme Inhibition Assay
The kinase inhibition assay of recombinant CDK6 was performed with increasing concentrations of quercetin (Figure 5B). The enzyme inhibition assay results show a significant decrease in kinase activity of CDK6 with increasing concentration of quercetin, suggesting the inhibitory potential of quercetin. At 5.89 μM concentrations of quercetin, around 50% of the ATPase activity of CDK6 was lost as estimated by the AAT Bioquest calculator (https://www.aatbio.com/tools/ic50-calculator). These observations are in close agreement with other experiments that suggest a considerable binding between quercetin and CDK6.
Cell Viability Studies
Overexpression of CDK6 supports the growth and viability of different cancer cells. Thus, we evaluated the consequences of CDK6 inhibition through quercetin on the cell viability of MCF-7 and A549 cell lines. The reason behind the selection of breast cancer and lung cancer lines is the reported overexpression of CDK6 in these cells. Due to the prominent role of CDK6 in these cancers, the inhibitors of CDK6 are under clinical trials for breast and lung cancer patients.64,65 These studies rationalized the evaluation of the anticancer potential of quercetin on the cancer cell types where CDK6 supported their growth. Cell viability results of quercetin with selected cancer cell lines showed a decrease in the viability in a concentration-dependent manner (Figure 6A). The observed IC50 dose of quercetin for MCF-7 cells is 41.78 ± 2.21 μM and for A549 cells is 52.35 ± 2.44 μM. These results suggested that quercetin decreases the viability of breast and lung cancer cells. The cell viability results are consistent with previous reports, which suggested that inhibition of CDK6 decreases the growth of cancer cells.65,66
Figure 6.
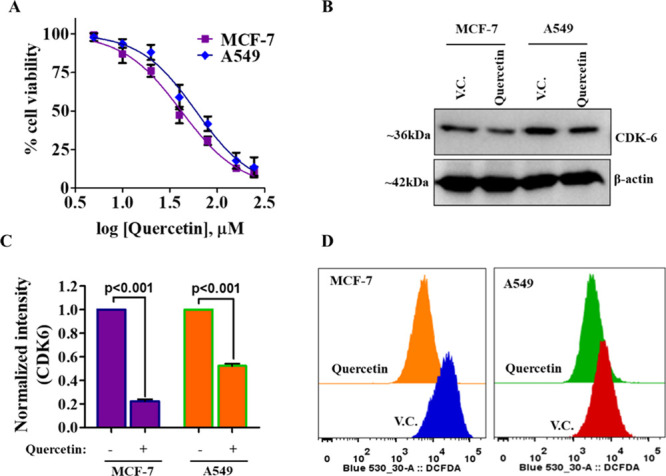
Quercetin decreases the cell viability of cancer cells, CDK6 expression, and ROS production. (A) The cell–viability curve showing inhibition potential of quercetin on the growth of MCF-7 and A549 cells. The cells were treated with an increasing dose of quercetin for 72 h, and the MTT assay was performed to measure respective cell viabilities. (Each data point shown is the mean ±SD from n = 3). (B) Protein expression studies of CDK6 in quercetin and vehicle-treated MCF-7 and A549 cells. (C) Quantification of CDK6 expression after performing normalization with loading control β-actin. (D) Representative histogram of quercetin/vehicle-treated MCF-7 and A549 cells stained with H2DCFDA for ROS measurements. A shift toward left shows a decrease in the levels of ROS.
Quercetin Decreases the Expression of CDK6
Our in silico and in vitro studies showed that quercetin has a high binding affinity for CDK6, and it also decreases its enzyme activity. Further, cell viability studies also suggested that the treatment of quercetin decreases the viability of selected cancer cells. Next, we wanted to see the effect of quercetin on the cellular expression of CDK6 and thus, check the expression of CDK6 in the quercetin-treated cells. Interestingly, we found that quercetin decreases the CDK6 expression in both MCF-7 and A549 cells (Figure 6B,C). These results complement both in silico and enzyme assay results and further suggest that quercetin may act by decreasing the expression and activity of CDK6.
Quercetin Inhibited ROS Production
Overexpression of CDK6 induces the production of ROS.17 To see the effect of CDK6 inhibition on cellular ROS, we have quantified ROS in quercetin-treated cells. For total ROS measurements, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) staining was used. Results of ROS measurements showed that the treatment of quercetin decreases the production of ROS (Figure 6D). Earlier studies suggested that quercetin behaves as an antioxidant.67 The results of our study corroborate with these studies and provide a possible explanation for the antioxidant potential of quercetin. These results suggested that inhibition of CDK6 through quercetin decreases the production of ROS and this adds up to the anticancer properties of quercetin.
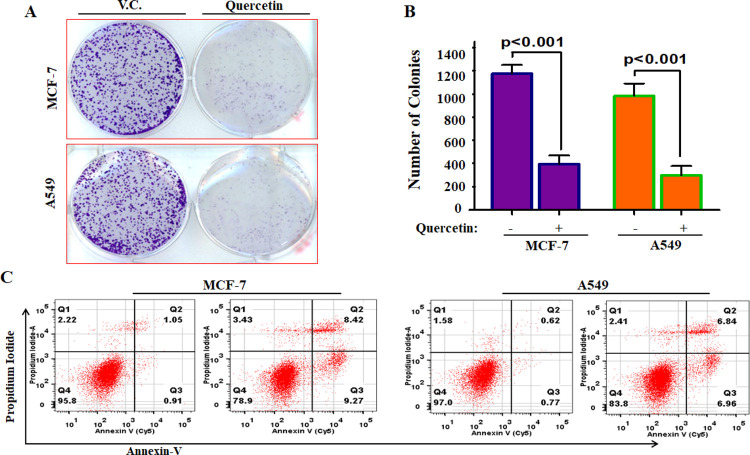
Quercetin Decreases the Colonogenic Potential and Induces Apoptosis
CDK6 overexpression acts as a survival mechanism and supported the apoptotic evasion as well as colonization of cancer cells.17,67 To see whether the quercetin-mediated inhibition of CDK6 has any effect on colonization of MCF-7/A549 cells and apoptosis induction, we performed the colony formation and apoptosis studies and found that quercetin significantly decreases the colonization of MCF-7 and A549 cells (Figure 7A,B). Interestingly, results of apoptosis studies further revealed that quercetin induces apoptosis in ∼21.10% of the MCF-7 and ∼16.22% of the A549 cells after 48 h of treatment (Figure 7C). Our results of apoptosis studies are in agreement with previous reports, which suggested that the treatment of quercetin induces apoptosis in different cancer cells.67 Previous reports showed that quercetin treatment leads to mitotic catastrophe and failure of cytokinesis.43,67 CDK6 plays an important role in cytokinesis/mitotic regulations,11 and thus, inhibition of CDK6 through quercetin explains the antimitotic or cytokinesis inhibitory activities. In our previous studies, we also reported that quercetin inhibited calcium/calmodulin-dependent protein kinase IV, pyruvate dehydrogenase kinase isozyme 3, and sphingosine kinase 1.28,29,68 In these studies, we used different cancer cell lines (as per the expression profile of selected kinase), it means that quercetin may target different molecules; but at the same time, the results of the present study argued with these reports and suggest that the activity of quercetin might be kinase-specific as per the expression of specific kinases in different cancers. These observations further provide the preclinical rationale for the optimization of quercetin analogs for the development of CDK6 inhibitors.
Figure 7.
Quercetin decreases colonization potential of cancer cells and induces apoptosis. (A) Colony formation studies of quercetin with selected cancer cells. (B) Graphical representation of the number of colonies obtained in quercetin/vehicle-treated groups. (C) Representative dot plots of Annexin-V/PI staining studies performed to quantify the induction of apoptosis in quercetin treated cells. (Statistical analysis was performed using Student’s t-test for unpaired samples compared to vehicle control).
Conclusions
High expression of CDK6 is directly associated with different types of cancers, and thus, it may be targeted for the design and development of small molecule inhibitors to address anticancer therapy. Natural molecules are the source of different bioactive molecules and are frequently implicated in the therapeutic management of cancer. This study identifies quercetin as a potent inhibitor of CDK6. Although anticancer properties of quercetin were also reported previously, the inhibition of CDK6 by quercetin provides a new avenue for cancer therapy. In addition, the structure of quercetin may be further chemically modified to develop effective preclinical leads with high selectivity and improved drug likeness.
Material and Methods
Materials
Luria–Bertani broth (Merck, Darmstadt, Germany), kanamycin, IPTG (Sigma, Saint Louis, MO, USA), Ni-NTA column (Biorad), and Ni-NTA beads (Qiagen QIA express) were purchased. The Ni-NTA column purchased from Biorad (Qiagen QIA express). Anti-Cdk6 antibody (Cat#mAb #13331) is from Cell Signaling Technology, Danvers, USA, and anti-β-Actin monoclonal antibody (Cat#A2228) is from Millipore Sigma, Darmstadt, Germany. Cancer cells were procured from National Centre for Cell Sciences (NCCS), Pune, India. Primary antibody goat anti-rabbit IgG (31635) secondary antibodies and dihydroethidium were taken from Invitrogen; Dulbecco minimal essential medium (DMEM), RPMI-1640, and fetal bovine serum (FBS) were purchased from Gibco life sciences. All reagents used were molecular biology grades.
Expression and Purification
The CDK6 (981nucleotieds) gene was successfully cloned in the pET28a+ vector and confirmed by the gene-sequencing method. CDK6 expressed in Codon+ competent cells, which were purified by our previously reported protocol.66 In brief, the recombinant cells were grown and induction was done at 18 °C by 0.5 mM IPTG (Sigma, Saint Louis, USA). The pellet obtained from this culture was dissolved in 50 mM Tris–HCl, 200 mM MgCl2 buffer, pH 8.0. The cell lysate was sonicated and subjected to 30 min centrifugation at 13,000 rpm at 4 °C. The obtained pellet was solubilized, centrifuged, and loaded on pre-equilibrated Ni-NTA column chromatography. Following the binding step, the column was washed, and protein was eluted with elution buffer (50 mM Tris buffer pH 8.0, 200 mM NaCl, and 0.1% N-lauroylsarcosine, 300 mM imidazole). The purity of the recombinant CDK6 protein was checked on 12% SDS-PAGE.
Kinase Inhibition Assay
The ATPase assay was performed to measure the inhibitory effect on purified CDK6 by gradually increasing concentrations of quercetin. Freshly prepared ATP (50 μM) was mixed with CDK6 (1 μM), and a final reaction mixture of a 100 μL volume was incubated at 25 °C for 1 h. Malachite green (200 μL) was further added to the reaction mixture to block the reaction followed by incubation of samples at room temperature for 15–20 min for the development of color. From the final reaction mixture, 100 μL was transferred to a 96-well plate in triplicates to measure spectrophotometrically at 620 nm.69
Fluorescence Measurements
To examine the binding affinity of quercetin with purified recombinant CDK6, a fluorescence-based experiment was performed.25,70 Each titration of protein with quercetin was conducted in triplicates, and the average was taken for analysis of binding parameters. To measure the binding constant (K) and the total number of binding sites (n) on the protein molecule, and different thermodynamic parameters. the quenching data of fluorescence intensity of CDK6 with an increasing concentration of quercetin was analyzed using the Stern–Volmer equation, modified Stern–Volmer, and van’t Hoff equations as described in earlier studies.62,71
Isothermal Titration Calorimetry
Isothermal titration calorimetry (ITC) experiment was carried out at 25 °C on a VP-ITC microcalorimeter from MicroCal, Inc. (GE, MicroCal, USA). The CDK6 protein was dialyzed against 25 mM Tris with 200 mM MgCl2 buffer, and the ligand was dissolved properly in the dialyzing buffer. DMSO was added to the protein solution (1% v/v) to prevent signal stability problems during the ITC experiment. The sample cell contained 15 μM CDK6, while 500 μM quercetin present in the syringe was titrated into the cell that was filled with CDK6 and analyzed for binding parameters.66 To nullify the heat of dilution of solvent, the titration of the ligand was performed in buffer and substracted from the protein–ligand dilutions before final fitting of binding data. The MicroCal Origin 8.0 was used to analyze the stoichiometry of binding (n), enthalpy change (ΔH), and an association constant (Ka).
Structural Modeling and Molecular Docking
The structural coordinates of CDK6 (PDB ID: 3NUP) were taken from the Protein Data Bank (PDB) and subsequently refined using the “PRIME” module of Schrödinger.72 The structure of quercetin was drawn in the MAESTRO (Schrödinger Release 2018-1: Maestro, Schrödinger, LLC, New York, NY, 2018), and its resulted structure was optimized using the density functional theory (DFT)-based protocols present in JAGUAR.73 Docking was performed between the optimized structures of CDK6 and quercetin using the AutoDock 4 package.74 The prediction of the bound conformation was performed on the basis of the free-energy function, which was calculated by the combined empirical force-field and Lamarckian genetic algorithm.75 The grid dimensions of 44 × 54 × 50 Å were set along the XYZ directions using the AutoGrid module with a spacing of 0.375 Å, and the parameters associated with the Lamarckian genetic algorithm were set to the maximum efficiency values. The number of individuals in the population was fixed to 250 and the maximum number of energy evaluations to a “longer” interval. As a result, 100 docked conformations were generated for the CDK6 and quercetin system, grouped according to the RMSD tolerance of 2.0 Å. The rescoring of the generated docked conformations was performed using the DrugScoreX server,76 and the docked conformation with the highest score was selected for the MD simulations.
Validation of Molecular Docking Protocol
The validation of the used docking protocol was performed using the SAnDReS algorithm,77 which carried out the re-docking using the Autodock 4.0 module and calculated the average RMSD and the docking accuracy. In the primary step, the validation process involved the creation of the SAnDReS format file from the docking output using the “Docking Hub” option, which ranked the docking conformations on the basis of the RMSD and calculated energy parameters. After the successful generation of the SAnDReS format file, the option of the “Statistical analysis of scoring functions versus RMSD” was selected, which invoked the inbuilt “strmsd.in” module that calculated the protocol performance parameters. Furthermore, the validation of the predictive performance of the protocol was performed by calculating the Spearman’s rank correlation coefficient (rs). This analysis involved the classification of the available CDK6 inhibitors in the BindingDBdatabase (https://www.bindingdb.org/bind/tabLuceneResult.jsp?thisInput=CDK6&submit=Go). Then, docking of the inhibitors was performed using similar setup parameters in the active site of the CDK6. Then, rs was calculated using the Microsoft Excel module.
MD Simulation
The MD simulations were performed on the quercetin bound and apo-form of CDK6 using GROMACS version 2018-2.78 Initially, the GROMOS96 53a6 force-field79 was used for the generation of topology parameters of the protein structure in the docked complex. The DFT method implemented in GAUSSIAN, which utilized the B3LYP6-31G(d,p) basis set, and CHELPG program80 was used for the charge correction. The topology generation stage was followed by the production of the solvated systems using the SPC/E water model,81 along with the neutralization was performed by adding a suitable number of Na+ and Cl– ions. Subsequently, the solvated neutralized systems undergo the process of energy minimization using combined the steepest descent and conjugate gradient algorithms with a convergence criterion of 0.005 kcal/mol, and position restraints were applied to the structure of the ligand.
The equilibration step was performed in the combined stages of constant volume (NVT) and constant pressure (NPT) ensemble conditions, each at a 100 ps time scale. The temperature of 300 K was maintained for the system using the Berendsen weak coupling method, and the pressure of 1 bar was maintained utilizing Parrinello–Rahman barostat in the equilibration stage. The LINCS algorithm was used for the generation of the final conformational production stage for a 200 ns timescale, and trajectories were generated, which were analyzed to understand the behavior of each complex in the explicit water environment. The changes in the protein–ligand distance, hydrogen (H)-bonds, root-mean-square deviation (RMSD), the radius of gyration (Rg), root-mean-square fluctuation (RMSF), principal component analysis (PCA), and free-energy landscape (FEL) of the complex system were analyzed. Furthermore, the molecular mechanic Poisson–Boltzmann surface area (MM-PBSA) protocols implemented in the g_mmpbsa package82 were used for the calculation of free energy of the binding protein and ligand molecules.
Cell Viability Studies
The effect of quercetin treatment on the viability of breast cancer (MCF-7) and human adenocarcinoma (A549) cells was studied using the MTT-assay.24,83 Briefly, the MCF-7 and A549 cells were seeded in 96-well culture plate at a density of 5000–5500 cells/well and treated with quercetin (0–250 μM) for 72 h. Following the treatment, cells were incubated with 20 μL of MTT (from 5 mg/mL stock solution in PBS, pH 7.4) for 4–5 h and the formazan were dissolved in 100 μL of DMSO. The optical density of the dissolved purple reaction product was recorded (570 nm) and transformed into percentage cell viability.
Total Protein Isolation and Expression Studies
The MCF-7 and A549 cells were treated with an IC50 dose of quercetin for a stipulated time and harvested using trypsinization. For total protein isolation, the collected cell line samples were lysed using RIPA buffer (Thermo Fisher Scientific, USA). The isolated protein was quantified (using BCA-protein estimation kit) and expression studies were performed using the Western blot as described.84,85
ROS Determination
2′,7′-Dichlorodihydrofluorescein diacetate (H2DCFDA) staining was used to see the effect of quercetin on ROS production.25,86 Briefly, the MCF-7 and A549 cells were treated with an IC50 dose of quercetin and vehicle control (DMSO) for 5–6 h. After the stipulated time of treatment, cells were further incubated with 10 μMH2DCFDA (Invitrogen Grand Island, NY) for 30 min. The cells were trypsinized, washed twice with PBS, and analyzed for ROS production using flow cytometry.
Colony Formation Studies
The colony formation assay of selected cancer cells with quercetin was performed.87 For the colony formation assay, nearly 1000–1500 (MCF-7/A549 cells) cells/well of 6-well cell culture plates were seeded and each type of cell line was treated with IC50 concentrations of quercetin for 8–10 days, (at 37 °C, in a 5% CO2 incubator). DMSO is taken as vehicle control. The colonies obtained in each treatment group were fixed using 100% methanol, and staining was performed using crystal violet. The number of colonies was quantified using ImageJ software (https://imagej.nih.gov/ij/) and compared with vehicle control.
Annexin-V/PI Staining
The apoptotic potential of quercetin was studied using annexin-V/PI staining as described previously.85,88 Briefly, the selected cells (MCF-7 and A549) were treated with the IC50 concentration of quercetin or vehicle control for 48 h. Following the quercetin treatment, cells were harvested and stained using annexin-V/PI (Annexin-V kit, BD-Biosciences, San Jose, USA). The stained cells were analyzed using flow cytometry.
Acknowledgments
M.Y. is thankful to the Indian Council of Medical Research, Government of India for the award of Senior Research Fellowship (F.no.45/54/2018-PHA/BMS/OL). Authors thank the Department of Science and Technology, Government of India for the FIST support (FIST program No. SR/FST/LSI-541/2012).
Glossary
List of Abbreviations
- CDKs
cyclin-dependent kinases
- CDK6
cyclin-dependent kinase 6
- ATP
adenosine triphosphate
- UBA domain
ubiquitin-associated domain
- KA
kinase-associated domain
- DMSO
dimethyl sulfoxide
- H2DCFDA
2′,7′-Dichlorodihydrofluorescein diacetate
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- ITC
isothermal titration calorimetry
- CKIs
CDK inhibitors
- PDB
Protein Data Bank
- Rb
retinoblastoma
- DMEM
Dulbecco minimal essential medium
- RPMI
Roswell Park Memorial Institute
- MD simulation
molecular dynamic simulation
- RMSD
root-mean-square deviation
- Rg
radius of gyration
- RMSF
root-mean-square fluctuation
- PCA
principal component analysis
- FEL
free-energy landscape
- MM-PBSA
molecular mechanics Poisson–Boltzmann surface area.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c03975.
(Figure SI) ATPase enzyme assay of CDK6 with in silico screened natural molecules, (Table S1) binding parameters of all the screened natural compounds with CDK6, and (Table S2) the list of parameters generated regarding the validation of the protocol using the SAnDReS algorithm (PDF)
Author Contributions
Conceptualization was done by M.Y. and M.I.H. Methodology was done by M.Y., P.K., A.S., and M.S. The software part was done by M.S. and A.C. Validation was done by M.S., A.I., Q.M.R.H., and MIH. The formal analysis was done by A.I., A.C., and M.S. Investigation was done by P.K., A.S., and M.I.H. The resource part was done by A.I. and M.I.H. Data curation was done by A.S., M.Y., P.K., and M.S. Writing of original draft preparation was done by M.Y., A.S., and P.K. Writing review and editing were done by M.S. and M.I.H. The visualization part was done by Q.M.R.H. Supervision was done by A.C. and M.I.H. Project administration was done by M.I.H. Funding acquisition was done by A.I. and M.I.H.
This work is funded by the Indian Council of Medical Research.
The authors declare no competing financial interest.
Notes
All data generated or analyzed during this study are included in this manuscript and supplementary materials attached to this article.
Supplementary Material
References
- Malumbres M.; Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- Sever R.; Brugge J. S. Signal transduction in cancer. Cold Spring Harbor Perspect. Med. 2015, 5, a006098. 10.1101/cshperspect.a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icard P.; Fournel L.; Wu Z.; Alifano M.; Lincet H. Interconnection between metabolism and cell cycle in cancer. Trends Biochem. Sci. 2019, 44, 490–501. 10.1016/j.tibs.2018.12.007. [DOI] [PubMed] [Google Scholar]
- Naz F.; Anjum F.; Islam A.; Ahmad F.; Hassan M. I. Microtubule affinity-regulating kinase 4: structure, function, and regulation. Cell Biochem. Biophys. 2013, 67, 485–499. 10.1007/s12013-013-9550-7. [DOI] [PubMed] [Google Scholar]
- Naz H.; Islam A.; Ahmad F.; Hassan M. I. Calcium/calmodulin-dependent protein kinase IV: A multifunctional enzyme and potential therapeutic target. Prog. Biophys. Mol. Biol. 2016, 121, 54–65. 10.1016/j.pbiomolbio.2015.12.016. [DOI] [PubMed] [Google Scholar]
- Noble M. E.; Endicott J. A.; Johnson L. N. Protein kinase inhibitors: insights into drug design from structure. Science 2004, 303, 1800–1805. 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- dos Santos Paparidis N. F.; Canduri F. The emerging picture of CDK11: genetic, functional and medicinal aspects. Curr. Med. Chem. 2018, 25, 880–888. 10.2174/0929867324666170815102036. [DOI] [PubMed] [Google Scholar]
- Bernhardt Levin N. M.; Oliveira Pintro V.; Boff de Avila M.; Boldrini de Mattos B.; Filgueira D. A. J. Understanding the structural basis for inhibition of cyclin-dependent kinases. New pieces in the molecular puzzle. Curr. Drug Targets 2017, 18, 1104–1111. 10.2174/1389450118666161116130155. [DOI] [PubMed] [Google Scholar]
- Volkart P. A.; Bitencourt-Ferreira G.; Souto A. A.; de Azevedo W. F. Cyclin-dependent kinase 2 in cellular senescence and cancer. A structural and functional review. Curr. Drug Targets 2019, 20, 716–726. 10.2174/1389450120666181204165344. [DOI] [PubMed] [Google Scholar]
- Bitencourt-Ferreira G.; da Silva A. D.; de Azevedo W. F. Application of Machine Learning Techniques to Predict Binding Affinity for Drug Targets. A Study of Cyclin-Dependent Kinase 2. Curr. Med. Chem. 2020, 10.2174/2213275912666191102162959. [DOI] [PubMed] [Google Scholar]
- Malumbres M.; Harlow E.; Hunt T.; Hunter T.; Lahti J. M.; Manning G.; Morgan D. O.; Tsai L.-H.; Wolgemuth D. J. Cyclin-dependent kinases: a family portrait. Nat. Cell Biol. 2009, 11, 1275–1276. 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton-Fessler S.; Parker L. L.; Geahlen R. L.; Piwnica-Worms H. Mechanisms of p34cdc2 regulation. Mol. Cell. Biol. 1993, 13, 1675–1685. 10.1128/MCB.13.3.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M. Cell cycle checkpoints and their inactivation in human cancer. Cell proliferation 2000, 33, 261–274. 10.1046/j.1365-2184.2000.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topacio B. R.; Zatulovskiy E.; Cristea S.; Xie S.; Tambo C. S.; Rubin S. M.; Sage J.; Ko̅ivomägi M.; Skotheim J. M. Cyclin D-Cdk4,6 Drives Cell-Cycle Progression via the Retinoblastoma Protein’s C-Terminal Helix. Mol. Cell 2019, 74, 758–770.e4. 10.1016/j.molcel.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinsson H. S.; Starborg M.; Erlandsson F.; Zetterberg A. Single cell analysis of G1 check points-the relationship between the restriction point and phosphorylation of pRb. Exp. Cell Res. 2005, 305, 383–391. 10.1016/j.yexcr.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Calzone L.; Gelay A.; Zinovyev A.; Radvanyi F.; Barillot E. A comprehensive modular map of molecular interactions in RB/E2F pathway. Mol. Syst. Biol. 2008, 4, 0174. 10.1038/msb.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Nicolay B. N.; Chick J. M.; Gao X.; Geng Y.; Ren H.; Gao H.; Yang G.; Williams J. A.; Suski J. M.; Keibler M. A.; Sicinska E.; Gerdemann U.; Haining W. N.; Roberts T. M.; Polyak K.; Gygi S. P.; Dyson N. J.; Sicinski P. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 2017, 546, 426–430. 10.1038/nature22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G.; Whyte D. B.; Martinez R.; Hunter T.; Sudarsanam S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Ekor M. The growing use of herbal medicines: issues relating to adverse reactions and challenges in monitoring safety. Front. Pharmacol. 2014, 4, 177. 10.3389/fphar.2013.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivoira M. A.; Rodriguez V.; Talamoni G.; de Talamoni N. T. New Perspectives in the Pharmacological Potential of Naringin in Medicine. Curr. Med. Chem. 2020, 10.2174/0929867327666200604171351. [DOI] [PubMed] [Google Scholar]
- Papakosta K.; Grafakou M.-E.; Barda C.; Kostopoulos I. V.; Tsitsilonis O.; Skaltsa H. Cytotoxicity and Anti-cancer Activity of the Genus Achillea L. Curr. Med. Chem. 2020, 10.2174/0929867327666200505092514. [DOI] [PubMed] [Google Scholar]
- Kumar V.; Gupta P.; Hassan M. I. Mechanism and implications of traditional Chinese medicine in amyotrophic lateral sclerosis therapy. J. Proteins Proteomics 2019, 1–17. 10.1007/s42485-019-00009-7. [DOI] [Google Scholar]
- Anwar S.; Mohammad T.; Shamsi A.; Queen A.; Parveen S.; Luqman S.; Hasan G. M.; Alamry K. A.; Azum N.; Asiri A. M.; Hassan M. I. Discovery of Hordenine as a Potential Inhibitor of Pyruvate Dehydrogenase Kinase 3: Implication in Lung Cancer Therapy. Biomedicines 2020, 8, 119. 10.3390/biomedicines8050119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulzar M.; Ali S.; Khan F. I.; Khan P.; Taneja P.; Hassan M. I. Binding mechanism of caffeic acid and simvastatin to the integrin linked kinase for therapeutic implications: a comparative docking and MD simulation studies. J. Biomol. Struct. Dyn. 2019, 37, 4327–4337. 10.1080/07391102.2018.1546621. [DOI] [PubMed] [Google Scholar]
- Jameel E.; Naz H.; Khan P.; Tarique M.; Kumar J.; Mumtazuddin S.; Ahamad S.; Islam A.; Ahmad F.; Hoda N.; Hassan I. Design, synthesis, and biological evaluation of pyrimidine derivatives as potential inhibitors of human calcium/calmodulin-dependent protein kinase IV. Chem. Biol. Drug Des. 2017, 89, 741–754. 10.1111/cbdd.12898. [DOI] [PubMed] [Google Scholar]
- Roy S.; Mohammad T.; Gupta P.; Dahiya R.; Parveen S.; Luqman S.; Hasan G. M.; Hassan M. I. Discovery of Harmaline as a Potent Inhibitor of Sphingosine Kinase-1: A Chemopreventive Role in Lung Cancer. ACS Omega 2020, 21550. 10.1021/acsomega.0c02165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad T.; Batra S.; Dahiya R.; Baig M. H.; Rather I. A.; Dong J.-J.; Hassan I. Identification of high-affinity inhibitors of cyclin-dependent kinase 2 towards anticancer therapy. Molecules 2019, 24, 4589. 10.3390/molecules24244589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P.; Khan S.; Fakhar Z.; Hussain A.; Rehman M. T.; AlAjmi M. F.; Islam A.; Ahmad F.; Hassan M. I. Identification of Potential Inhibitors of Calcium/Calmodulin-Dependent Protein Kinase IV from Bioactive Phytoconstituents. Oxid. Med. Cell. Longevity 2020, 2020, 2094635. 10.1155/2020/2094635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya R.; Mohammad T.; Roy S.; Anwar S.; Gupta P.; Haque A.; Khan P.; Kazim S. N.; Islam A.; Ahmad F.; Hassan M. I. Investigation of inhibitory potential of quercetin to the pyruvate dehydrogenase kinase 3: Towards implications in anticancer therapy. Int. J. Biol. Macromol. 2019, 136, 1076–1085. 10.1016/j.ijbiomac.2019.06.158. [DOI] [PubMed] [Google Scholar]
- Khan P.; Queen A.; Mohammad T.; Smita; Khan N. S.; Hafeez Z. B.; Hassan M. I.; Ali S. Identification of alpha-Mangostin as a Potential Inhibitor of Microtubule Affinity Regulating Kinase 4. J. Nat. Prod. 2019, 82, 2252–2261. 10.1021/acs.jnatprod.9b00372. [DOI] [PubMed] [Google Scholar]
- Gulzar M.; Syed S. B.; Khan F. I.; Khan P.; Ali S.; Hasan G. M.; Taneja P.; Hassan M. I. Elucidation of interaction mechanism of ellagic acid to the integrin linked kinase. Int. J. Biol. Macromol. 2019, 122, 1297–1304. 10.1016/j.ijbiomac.2018.09.089. [DOI] [PubMed] [Google Scholar]
- Han D. H.; Lee M. J.; Kim J. H. Antioxidant and apoptosis-inducing activities of ellagic acid. Anticancer Res. 2006, 26, 3601–3606. [PubMed] [Google Scholar]
- Yun D.-Y.; Kang Y.-G.; Yun B.; Kim E.-H.; Kim M.; Park J. S.; Lee J. H.; Hong Y.-S. Distinctive metabolism of flavonoid between cultivated and semiwild soybean unveiled through metabolomics approach. J. Agric. Food Chem. 2016, 64, 5773–5783. 10.1021/acs.jafc.6b01675. [DOI] [PubMed] [Google Scholar]
- Zhao S.; Park C. H.; Li X.; Kim Y. B.; Yang J.; Sung G. B.; Park N. I.; Kim S.; Park S. U. Accumulation of rutin and betulinic acid and expression of phenylpropanoid and triterpenoid biosynthetic genes in mulberry (Morus alba L.). J. Agric. Food Chem. 2015, 63, 8622–8630. 10.1021/acs.jafc.5b03221. [DOI] [PubMed] [Google Scholar]
- Pramanik K. C.; Kudugunti S. K.; Fofaria N. M.; Moridani M. Y.; Srivastava S. K. Caffeic acid phenethyl ester suppresses melanoma tumor growth by inhibiting PI3K/AKT/XIAP pathway. Carcinogenesis 2013, 34, 2061–2070. 10.1093/carcin/bgt154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen Y.; Schewe T.; Sies H. (−)-Epicatechin elevates nitric oxide in endothelial cells via inhibition of NADPH oxidase. Biochem. Biophys. Res. Commun. 2007, 359, 828–833. 10.1016/j.bbrc.2007.05.200. [DOI] [PubMed] [Google Scholar]
- Tsao S.-m.; Hsia T.-c.; Yin M.-c. Protocatechuic Acid Inhibits Lung Cancer Cells by Modulating FAK, MAPK, and NF-κ B Pathways. Nutr. Cancer 2014, 66, 1331–1341. 10.1080/01635581.2014.956259. [DOI] [PubMed] [Google Scholar]
- Lin H. H.; Chen J. H.; Chou F. P.; Wang C. J. Protocatechuic acid inhibits cancer cell metastasis involving the down-regulation of Ras/Akt/NF-κB pathway and MMP-2 production by targeting RhoB activation. Br. J. Pharmacol. 2011, 162, 237–254. 10.1111/j.1476-5381.2010.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowska H.; Kalinowska M.; Lewandowski W.; Stȩpkowski T. M.; Brzóska K. The role of natural polyphenols in cell signaling and cytoprotection against cancer development. J. Nutr.Biochem. 2016, 32, 1–19. 10.1016/j.jnutbio.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Erdman J. W. Jr.; Balentine D.; Arab L.; Beecher G.; Dwyer J. T.; Folts J.; Harnly J.; Hollman P.; Keen C. L.; Mazza G.; Messina M.; Scalbert A.; Vita J.; Williamson G.; Burrowes J. Flavonoids and heart health: proceedings of the ILSI North America flavonoids workshop, May 31–June 1, 2005, Washington, DC. J. Nutr. 2007, 137, 718S–737S. 10.1093/jn/137.3.718S. [DOI] [PubMed] [Google Scholar]
- Pratheeshkumar P.; Son Y.-O.; Divya S. P.; Wang L.; Turcios L.; Roy R. V.; Hitron J. A.; Kim D.; Dai J.; Asha P.; Zhang Z.; Shi X. Quercetin inhibits Cr (VI)-induced malignant cell transformation by targeting miR-21-PDCD4 signaling pathway. Oncotarget 2017, 8, 52118. 10.18632/oncotarget.10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Kang J.; Da W.; Ou Y. Combination with water-soluble antioxidants increases the anticancer activity of quercetin in human leukemia cells. Die Pharmazie-An International Journal of Pharmaceutical Sciences 2004, 59, 859–863. [PubMed] [Google Scholar]
- Yang F.; Song L.; Wang H.; Wang J.; Xu Z.; Xing N. Quercetin in prostate cancer: Chemotherapeutic and chemopreventive effects, mechanisms and clinical application potential (Review). Oncol. Rep. 2015, 33, 2659–2668. 10.3892/or.2015.3886. [DOI] [PubMed] [Google Scholar]
- Ghazi T.; Arumugam T.; Foolchand A.; Chuturgoon A. A. The Impact of Natural Dietary Compounds and Food-Borne Mycotoxins on DNA Methylation and Cancer. Cell 2020, 9, 2004. 10.3390/cells9092004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdeos G. C.; Blank R.; Wolffram S. Influence of quercetin on the global DNA methylation pattern in pigs. Food Funct. 2020, 7421. 10.1039/D0FO00896F. [DOI] [PubMed] [Google Scholar]
- Peiffer D. S.; Ma E.; Wyatt D.; Albain K. S.; Osipo C. DAXX-inducing phytoestrogens inhibit ER+ tumor initiating cells and delay tumor development. NPJ Breast Cancer 2020, 6, 37. 10.1038/s41523-020-00178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiailanis A. D.; Renziehausen A.; Kiriakidi S.; Vrettos E. I.; Markopoulos G. S.; Sayyad N.; Hirmiz B.; Aguilar M. I.; Del Borgo M. P.; Kolettas E.; Widdop R. E.; Mavromoustakos T.; Crook T.; Syed N.; Tzakos A. G. Enhancement of glioblastoma multiforme therapy through a novel Quercetin-Losartan hybrid. Free Radical Biol. Med. 2020, 160, 391–402. 10.1016/j.freeradbiomed.2020.08.007. [DOI] [PubMed] [Google Scholar]
- Liu M.; Fu M.; Yang X.; Jia G.; Shi X.; Ji J.; Liu X.; Zhai G. Paclitaxel and quercetin co-loaded functional mesoporous silica nanoparticles overcoming multidrug resistance in breast cancer. Colloids Surf., B 2020, 196, 111284. 10.1016/j.colsurfb.2020.111284. [DOI] [PubMed] [Google Scholar]
- Singh C. K.; Chhabra G.; Ndiaye M. A.; Siddiqui I. A.; Panackal J. E.; Mintie C. A.; Ahmad N. Quercetin-Resveratrol Combination for Prostate Cancer Management in TRAMP Mice. Cancers 2020, 12, 2141. 10.3390/cancers12082141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Huang Y.; Song H.; Canup B. S. B.; Gou S.; She Z.; Dai F.; Ke B.; Xiao B. Inhibition of growth and lung metastasis of breast cancer by tumor-homing triple-bioresponsive nanotherapeutics. J. Controlled Release 2020, 454. 10.1016/j.jconrel.2020.08.066. [DOI] [PubMed] [Google Scholar]
- Abbaszadeh S.; Rashidipour M.; Khosravi P.; Shahryarhesami S.; Ashrafi B.; Kaviani M.; Moradi Sarabi M. Biocompatibility, Cytotoxicity, Antimicrobial and Epigenetic Effects of Novel Chitosan-Based Quercetin Nanohydrogel in Human Cancer Cells. Int. J. Nanomed. 2020, Volume 15, 5963–5975. 10.2147/IJN.S263013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. S.; Borland M.; Brain C.; Chen C. H. T.; Cheng H.; Chopra R.; Chung K.; Groarke J.; He G.; Hou Y.; Kim S.; Kovats S.; Lu Y.; O’Reilly M.; Shen J.; Smith T.; Trakshel G.; Vögtle M.; Xu M.; Xu M.; Sung M. J. 4-(Pyrazol-4-yl)-pyrimidines as Selective Inhibitors of Cyclin-Dependent Kinase 4/6. J. Med. Chem. 2010, 53, 7938–7957. 10.1021/jm100571n. [DOI] [PubMed] [Google Scholar]
- Liu T.; Lin Y.; Wen X.; Jorissen R. N.; Gilson M. K. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007, 35, D198–D201. 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jairajpuri D. S.; Mohammad T.; Adhikari K.; Gupta P.; Hasan G. M.; Alajmi M. F.; Rehman M. T.; Hussain A.; Hassan M. I. Identification of Sphingosine Kinase-1 Inhibitors from Bioactive Natural Products Targeting Cancer Therapy. ACS Omega 2020, 5, 14720–14729. 10.1021/acsomega.0c01511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsi A.; Ahmed A.; Khan M. S.; Al Shahwan M.; Husain F. M.; Bano B. Understanding the binding between Rosmarinic acid and serum albumin: In vitro and in silico insight. J. Mol. Liq. 2020, 113348. 10.1016/j.molliq.2020.113348. [DOI] [Google Scholar]
- Shamsi A.; Mohammad T.; Anwar S.; Alajmi M. F.; Hussain A.; Hassan M. I.; Ahmad F.; Islam A. Probing the interaction of Rivastigmine Tartrate, an important Alzheimer’s drug, with serum albumin: Attempting treatment of Alzheimer’s disease. Int. J. Biol. Macromol. 2020, 533. 10.1016/j.ijbiomac.2020.01.134. [DOI] [PubMed] [Google Scholar]
- Shamsi A.; Ahmed A.; Khan M. S.; Husain F. M.; Amani S.; Bano B. Investigating the interaction of anticancer drug temsirolimus with human transferrin: Molecular docking and spectroscopic approach. J. Mol. Recognit. 2018, 31, e2728 10.1002/jmr.2728. [DOI] [PubMed] [Google Scholar]
- Shamsi A.; Ahmed A.; Bano B. Probing the interaction of anticancer drug temsirolimus with human serum albumin: Molecular docking and spectroscopic insight. J. Biomol. Struct. Dyn. 2018, 36, 1479–1489. 10.1080/07391102.2017.1326320. [DOI] [PubMed] [Google Scholar]
- Ross P. D.; Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry 1981, 20, 3096–3102. 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- de Azevedo J.; Walter F.; Dias R. Experimental approaches to evaluate the thermodynamics of protein-drug interactions. Curr. Drug Targets 2008, 9, 1071–1076. 10.2174/138945008786949441. [DOI] [PubMed] [Google Scholar]
- Rehman M. T.; Shamsi H.; Khan A. U. Insight into the binding mechanism of imipenem to human serum albumin by spectroscopic and computational approaches. Mol. Pharmaceutics 2014, 11, 1785–1797. 10.1021/mp500116c. [DOI] [PubMed] [Google Scholar]
- Shamsi A.; Al Shahwan M.; Ahamad S.; Hassan M. I.; Ahmad F.; Islam A. Spectroscopic, calorimetric and molecular docking insight into the interaction of Alzheimer’s drug donepezil with human transferrin: Implications of Alzheimer’s drug. J. Biomol. Struct. Dyn. 2020, 38, 1094–1102. 10.1080/07391102.2019.1595728. [DOI] [PubMed] [Google Scholar]
- Wolff A. C. CDK4 and CDK6 Inhibition in Breast Cancer - A New Standard. N. Engl. J. Med. 2016, 375, 1993–1994. 10.1056/NEJMe1611926. [DOI] [PubMed] [Google Scholar]
- Pernas S.; Tolaney S. M.; Winer E. P.; Goel S. CDK4/6 inhibition in breast cancer: current practice and future directions. Ther. Adv. Med. Oncol. 2018, 10, 1758835918786451. 10.1177/1758835918786451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousuf M.; Shamsi A.; Khan P.; Shahbaaz M.; AlAjmi M. F.; Hussain A.; Hassan G. M.; Islam A.; Rizwanul Haque Q. M.; Hassan M. I. Ellagic Acid Controls Cell Proliferation and Induces Apoptosis in Breast Cancer Cells via Inhibition of Cyclin-Dependent Kinase 6. Int. J. Mol. Sci. 2020, 21, 3526. 10.3390/ijms21103526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimaszewska-Wiśniewska A.; Hal̷as-Wiśniewska M.; Izdebska M.; Gagat M.; Grzanka A.; Grzanka D. Antiproliferative and antimetastatic action of quercetin on A549 non-small cell lung cancer cells through its effect on the cytoskeleton. Acta Histochem 2017, 119, 99–112. 10.1016/j.acthis.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Gupta P.; Mohammad T.; Dahiya R.; Roy S.; Noman O. M. A.; Alajmi M. F.; Hussain A.; Hassan M. I. Evaluation of binding and inhibition mechanism of dietary phytochemicals with sphingosine kinase 1: Towards targeted anticancer therapy. Sci. Rep. 2019, 9, 18727. 10.1038/s41598-019-55199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voura M.; Khan P.; Thysiadis S.; Katsamakas S.; Queen A.; Hasan G. M.; Ali S.; Sarli V.; Hassan M. I. Probing the Inhibition of Microtubule Affinity Regulating Kinase 4 by N-Substituted Acridones. Sci. Rep. 2019, 9, 1676. 10.1038/s41598-018-38217-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P.; Mohammad T.; Khan P.; Alajmi M. F.; Hussain A.; Rehman M. T.; Hassan M. I. Evaluation of ellagic acid as an inhibitor of sphingosine kinase 1: A targeted approach towards anticancer therapy. Biomed. Pharmacother. 2019, 118, 109245. 10.1016/j.biopha.2019.109245. [DOI] [PubMed] [Google Scholar]
- Shamsi A.; Anwar S.; Mohammad T.; Alajmi M. F.; Hussain A.; Rehman M.; Hasan G. M.; Islam A.; Hassan M. MARK4 Inhibited by AChE Inhibitors, Donepezil and Rivastigmine Tartrate: Insights into Alzheimer’s Disease Therapy. Biomolecules 2020, 10, 789. 10.3390/biom10050789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. P.; Pincus D. L.; Rapp C. S.; Day T. J. F.; Honig B.; Shaw D. E.; Friesner R. A. A hierarchical approach to all-atom protein loop prediction. Proteins: Struct., Funct., Bioinf. 2004, 55, 351–367. 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- Bochevarov A. D.; Harder E.; Hughes T. F.; Greenwood J. R.; Braden D. A.; Philipp D. M.; Rinaldo D.; Halls M. D.; Zhang J.; Friesner R. A. Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013, 113, 2110–2142. 10.1002/qua.24481. [DOI] [Google Scholar]
- Bitencourt-Ferreira G.; Pintro V. O.; de Azevedo W. F. Jr. Docking with AutoDock4. Methods Mol. Biol. 2019, 2053, 125–148. 10.1007/978-1-4939-9752-7_9. [DOI] [PubMed] [Google Scholar]
- Morris G. M.; Huey R.; Lindstrom W.; Sanner M. F.; Belew R. K.; Goodsell D. S.; Olson A. J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudert G.; Klebe G. DSX: a knowledge-based scoring function for the assessment of protein-ligand complexes. J. Chem. Inf. Model. 2011, 51, 2731–2745. 10.1021/ci200274q. [DOI] [PubMed] [Google Scholar]
- Bitencourt-Ferreira G.; de Azevedo W. F. Jr. SAnDReS: A Computational Tool for Docking. Methods Mol. Biol. 2019, 2053, 51–65. 10.1007/978-1-4939-9752-7_4. [DOI] [PubMed] [Google Scholar]
- Pronk S.; Páll S.; Schulz R.; Larsson P.; Bjelkmar P.; Apostolov R.; Shirts M. R.; Smith J. C.; Kasson P. M.; van der Spoel D.; Hess B.; Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostenbrink C.; Villa A.; Mark A. E.; van Gunsteren W. F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004, 25, 1656–1676. 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Mennucci B.; Petersson G. A.; Nakatsuji H.; Caricato M.; Li X.; Hratchian H. P.; Izmaylov A. F.; Bloino J.; Zheng G.; Sonnenberg J. L.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Montgomery J. A. Jr.; Peralta J. E.; Ogliaro F.; Bearpark M. J.; Heyd J.; Brothers E. N.; Kudin K. N.; Staroverov V. N.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A. P.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Rega N.; Millam N. J.; Klene M.; Knox J. E.; Cross J. B.; Bakken V.; Adamo C.; Jaramillo J.; Gomperts R.; Stratmann R. E.; Yazyev O.; Austin A. J.; Cammi R.; Pomelli C.; Ochterski J. W.; Martin R. L.; Morokuma K.; Zakrzewski V. G.; Voth G. A.; Salvador P.; Dannenberg J. J.; Dapprich S.; Daniels A. D.; Farkas Ö.; Foresman J. B.; Ortiz J. V.; Cioslowski J.; Fox D. J.. Gaussian 09, Gaussian, Inc.: Wallingford, CT, USA, 2009.
- Zielkiewicz J. Structural properties of water: comparison of the SPC, SPCE, TIP4P, and TIP5P models of water. J. Chem. Phys. 2005, 123, 104501. 10.1063/1.2018637. [DOI] [PubMed] [Google Scholar]
- Kumari R.; Kumar R. g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Shamsi F.; Hasan P.; Queen A.; Hussain A.; Khan P.; Zeya B.; King H. M.; Rana S.; Garrison J.; Alajmi M. F.; Rizvi M. M. A.; Zahid M.; Imtaiyaz Hassan M.; Abid M. Synthesis and SAR studies of novel 1,2,4-oxadiazole-sulfonamide based compounds as potential anticancer agents for colorectal cancer therapy. Bioorg. Chem. 2020, 98, 103754. 10.1016/j.bioorg.2020.103754. [DOI] [PubMed] [Google Scholar]
- Khan P.; Idrees D.; Moxley M. A.; Corbett J. A.; Ahmad F.; von Figura G.; Sly W. S.; Waheed A.; Hassan M. I. Luminol-based chemiluminescent signals: clinical and non-clinical application and future uses. Appl. Biochem. Biotechnol. 2014, 173, 333–355. 10.1007/s12010-014-0850-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar S.; Shamsi A.; Shahbaaz M.; Queen A.; Khan P.; Hasan G. M.; Islam A.; Alajmi M. F.; Hussain A.; Ahmad F.; Hassan M. I. Rosmarinic Acid Exhibits Anticancer Effects via MARK4 Inhibition. Sci. Rep. 2020, 10, 10300. 10.1038/s41598-020-65648-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parveen I.; Khan P.; Ali S.; Hassan M. I.; Ahmed N. Synthesis, molecular docking and inhibition studies of novel 3-N-aryl substituted-2-heteroarylchromones targeting microtubule affinity regulating kinase 4 inhibitors. Eur. J. Med. Chem. 2018, 159, 166–177. 10.1016/j.ejmech.2018.09.030. [DOI] [PubMed] [Google Scholar]
- Khan N. S.; Khan P.; Inam A.; Ahmad K.; Yousuf M.; Islam A.; Ali S.; Azam A.; Husain M.; Hassan M. I. Discovery of 4-(2-(dimethylamino) ethoxy) benzohydrazide derivatives as prospective microtubule affinity regulating kinase 4 inhibitors. RSC Adv. 2020, 10, 20129–20137. 10.1039/D0RA00453G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen A.; Khan P.; Idrees D.; Azam A.; Hassan M. I. Biological evaluation of p-toluene sulphonylhydrazone as carbonic anhydrase IX inhibitors: An approach to fight hypoxia-induced tumors. Int. J. Biol. Macromol. 2018, 106, 840–850. 10.1016/j.ijbiomac.2017.08.082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.