Abstract
Mitochondrial metabolism plays an essential role in various biological processes of cancer cells. Herein, we established an experimental procedure for the metabolic assessment of mitochondria in cancer cells. We examined procedures for mitochondrial isolation coupled with various mitochondrial extraction buffers in three major cancer cell lines (PANC1, A549, and MDA-MB-231) and identified a potentially optimal and generalized approach. The purity of the mitochondrial fraction isolated by the selected protocol was verified using specific protein markers of cellular components, and the ultrastructure of the isolated mitochondria was also analyzed by transmission electron microscopy. The isolation procedure, involving a bead beater for cell lysis, a modified sucrose buffer, and differential centrifugation, appeared to be a suitable method for the extraction of mitochondria from cancer cells. Electron micrographs indicated an intact two-layer membrane and inner structures of mitochondria isolated by this procedure. Metabolomic and lipidomic analyses were conducted to examine the metabolic phenotypes of the mitochondria-enriched fractions and associated bulk cancer cells. A total of 44 metabolites, including malate and succinate, occurred at significantly higher levels in the mitochondrial fractions, whereas 51 metabolites, including citrate, oxaloacetate, and fumarate of the Krebs cycle and the oncometabolites glutamine and glutamate, were reduced in mitochondria compared to that in the corresponding bulk cells of PANC1. Similar patterns were observed in mitochondria and bulk cells of MDA-MB-231 and A549 cell lines. A clear difference between the lipid profiles of bulk PANC1, MDA-MB-231, and A549 and corresponding mitochondrial fractions of these cell lines was detected by principal component analysis. In conclusion, we developed an experimental procedure for a large-scale metabolic assessment for suborganelle metabolic profiling and multiple omics data integration in cancer cells with broad applications.
Introduction
The reprogramming of energy metabolism is an acknowledged hallmark of cancer.1 A large body of evidence has demonstrated that metabolic rewiring, mainly mediated by mitochondria, plays an essential role in the initiation, progression, and metastasis of cancer cells.2,3 The neoplastic cells rewire their metabolism by reducing the rate of oxidative phosphorylation and simultaneously accelerating glycolysis and glutaminolysis. These effects mainly result from the aberrant metabolism in mitochondria, among other contributing factors.4 The multifaceted interactions of mitochondria with other organelles in mitochondrial disorders are exceptionally complex and highly convoluted.5 Furthermore, mitochondrial metabolism appears to be essential in the oncogenesis of cancer cells. It is considered a promising therapeutic target for anticancer therapy.6 The search for new immunotherapeutic approaches targeting mitochondrial metabolism has gained momentum in recent years.7
Analysis of biomolecules at the cellular level is essential to capture a more precise mechanistic process of a disease, and conventional and low-throughput assays do not provide adequate information about the metabolism of cells. The application of metabolomics, as well as its subdiscipline, lipidomics, is known to be more suitable for a comprehensive examination of metabolic disturbances regardless of the origin of the disease.8 With the advancement of analytical instruments and data mining, metabolomics, and lipidomics can be employed to characterize endogenous metabolites and lipids at cellular and subcellular levels, respectively.9,10 Furthermore, the information derived from particular organelles is needed for the development of accurate diagnostic and therapeutic strategies. Nonetheless, omics studies that selectively focus on mitochondria suffer from several obstacles. First, the metabolic network of the mammalian cells is compartmentalized. Thus, bulk analysis using whole cells does not reflect the metabolic state of the mitochondria, which represents only a fraction of cellular biomaterials.11 Second, limited efforts have been directed toward the comprehensive metabolic profiling of cancer cell mitochondria. Finally, but importantly, there are a few technical methodologies for fractionation and high-throughput analytical analysis of isolated mitochondria.12,13
The objective of this study was to develop stepwise sucrose- and differential centrifugation-based procedure for the large-scale in vitro metabolic assessment of the mitochondria-enriched fraction of cancer cells. First, a comparison of nine mitochondria-enriched extraction protocols for different cancer cell lines was conducted prior to the selection of the optimal experimental procedure. Next, we investigated the similarities and differences in metabolic signatures of mitochondria and bulk cancer cells using metabolomic and lipidomic approaches.
Results
Extraction Procedure for a Mitochondria-Enriched Fraction
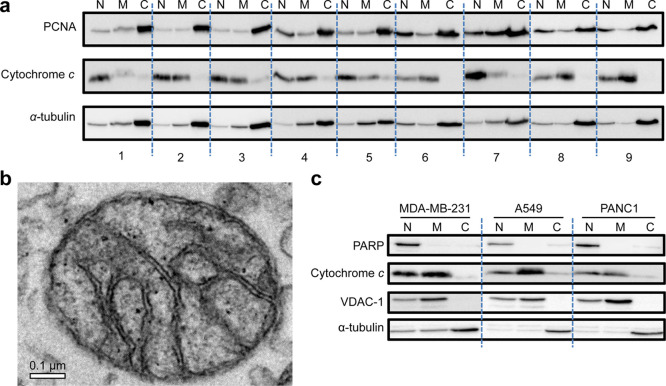
To optimize extraction, one chemical and eight physical (four different lysis buffers and two other homogenization times) homogenization procedures were first compared using the PANC1 (pancreas cells) cell line. Isolated nuclear (N), mitochondria-enriched (M), and cytosol (C) fractions from the cell line were examined. The isolation quality was checked using western blots of one representative protein marker for each fraction. Proliferating cell nuclear antigen (PCNA), a DNA clamp that regulates DNA replication and repair, was used as a representative nuclear marker.14 Cytochrome c, a component of the electron transport system located at the mitochondrial inner membrane, served as the mitochondrial marker.15 α-Tubulin, a cytoskeletal microtubule protein, was employed as the cytosolic marker.16 The sucrose-based buffer was the most appropriate buffer as it efficiently isolated the mitochondria-enriched fraction (Figure 1a). No significant difference was observed by varying the homogenization time in our experiments. Thus, we concluded that 5 s was adequate for the cell lysis step, resulting in efficient mitochondrial isolation. We further examined the mitochondrial fraction derived from the sucrose-based buffer using transmission electron microscopy (TEM). As shown in Figure 1b, we observed intact inner and outer membranes of isolated mitochondria. The data demonstrate that the buffer and applied procedure were adequate for obtaining mitochondrial fractions from PANC1 cells with subtle influences on the structure of the mitochondria.
Figure 1.
Mitochondrial isolation method development. (a) Isolated nuclear (N), mitochondria-enriched (M), and cytosol (C) fractions of the following tested methods: (1) chemical lysis method, (2) lysis buffer from the commercial kit and homogenized for 5 s, (3) lysis buffer from the commercial kit and homogenized for 10 s, (4) normal saline as lysis buffer and homogenized for 5 s, (5) normal saline as lysis buffer and homogenized for 10 s, (6) mannitol-based lysis buffer and homogenized for 5 s, (7) mannitol-based lysis buffer and homogenized for 10 s, (8) sucrose-based lysis buffer and homogenized for 5 s, and (9) sucrose-based lysis buffer and homogenized for 10 s. PCNA, nuclear marker; cytochrome c, mitochondrial marker; and α-tubulin, cytosolic marker. (b) Transmission electron micrograph of a mitochondrion isolated from PANC1 cells. (c) Western blot of isolated mitochondria from A549, MDA-MB-231, and PANC1 using the optimal protocol, showing representative markers for each fraction. PARP, nuclear marker; cytochrome c and VDAC1, mitochondrial markers; and α-tubulin, cytosolic marker.
We further validated our isolation method using two other cancer cell lines. Primarily, the method was employed to isolate mitochondria from A549 (alveolar basal epithelial cells) and MDA-MB-231 (breast cancer cells), in addition to PANC1, wherein we utilized a more comprehensive set of markers. Poly (ADP-ribose) polymerase (PARP), which recognizes single-strand DNA breaks and is involved in DNA repair in the nucleus,17 was used as the nuclear marker. Voltage-dependent anion channel 1 (VDAC1) is the most abundant protein on the mitochondrial outer membrane and thus could be used as a mitochondrial marker.18 To ensure the quality of the isolated intact mitochondrial fraction, in case the TEM imaging was not available, VDAC1 was employed together with cytochrome c as mitochondrial markers. Finally, α-tubulin was utilized as the cytosolic marker. The results showed that mitochondria-enriched fractions with little nuclear and cytosolic contamination were obtained with our optimized method in all three different cancer lines (Figure 1c).
Metabolomic Profiles of Mitochondria and Whole Cancer Cells
We employed a standard large-scale metabolomics approach to analyze endogenous metabolites and demonstrated that there is a difference in the metabolic components of mitochondria and associated bulk cancer cells. In total, 212 transitions of endogenous metabolites were monitored in PANC1, MDA-MB-231, and A549 cells. Low-quality peaks, e.g., RSD > 30% in working quality control (QC) samples, were excluded prior to the downstream statistical analysis. There were 62, 56, and 61 statistically different metabolites between the mitochondrial fraction and the bulk cancer cells in PANC1, MDA-MB-231, and A549, respectively (p value < 0.05, FDR < 0.05), as confirmed by the Student’s t-test.
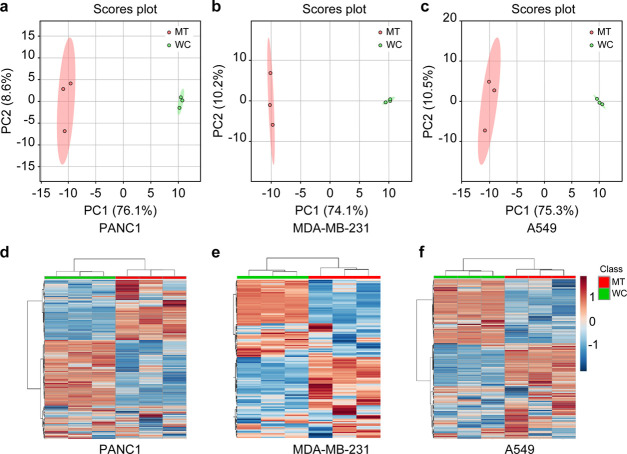
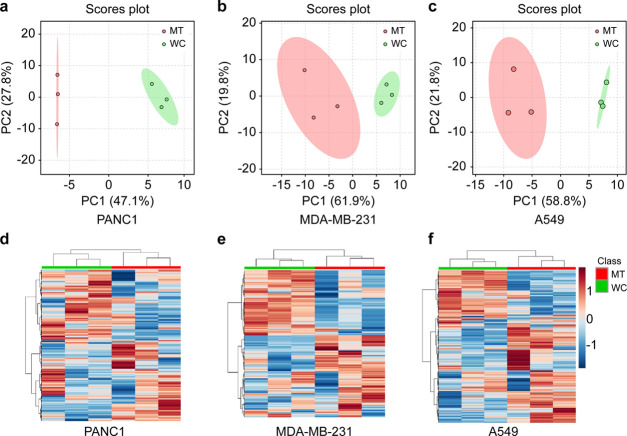
In the unsupervised principal component analysis (PCA) plots, the metabolic profiles of mitochondria were significantly different from that of the bulk cancer cells in all three tested cell lines (Figure 2a–c). In the heatmap of the mitochondria and bulk PANC1 cells (Figure 2d), 44 metabolites including malate, succinate, and important oncometabolites involved in the Krebs cycle occurring within mitochondria, were significantly higher in the mitochondrial fraction than in the bulk cells. In contrast, 51 metabolites, including citrate, oxaloacetate, and fumarate of the Krebs cycle, and oncometabolites glutamine and glutamate, were expressed at a relatively lower concentration in mitochondria than in bulk cells of PANC1. Of note, several metabolites such as lactate and fructose-1,6-biphosphate showed no apparent differences between the two comparison groups. Similar patterns could also be found in the comparative heatmap of mitochondria and bulk cells of MDA-MB-231 (Figure 2e) and A549 (Figure 2f) cell lines.
Figure 2.
Hydrophilic metabolic profiles of mitochondria and bulk cells of different cell lines. Principal component analysis (PCA) score plots of (a) PANC1, (b) MDA-MB-231, and (c) A549. Heatmaps of (d) PANC1, (e) MDA-MB-231, and (f) A549. MT (red) indicates mitochondria, and WC (green) indicates the whole cells or bulk cancer cells.
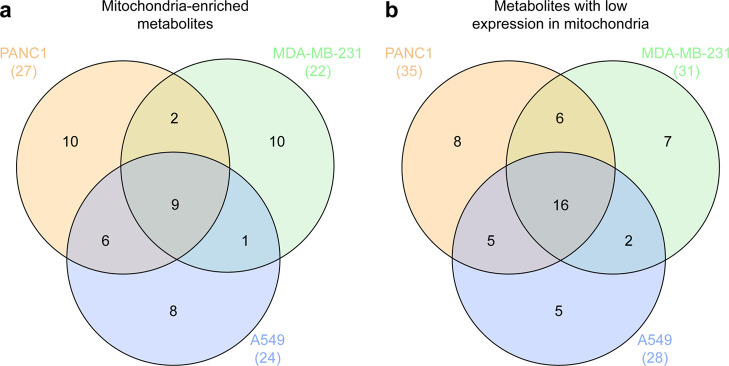
Interestingly, nine metabolites were found relatively enriched (Figure 3a) and 16 metabolites were found at relatively low concentrations (Figure 3b) in the mitochondrial fractions among the three cancer cell lines. Collectively, we demonstrated that bulk cell analysis could not represent the metabolome of the mitochondria using univariate and multivariate analyses. Notably, the metabolic components were dependent on the tissue of origin. Detailed information on significant differences in metabolite composition of the three cell lines is presented in Table S1.
Figure 3.
Venn diagrams representing the overlap of metabolite compositions among three cell lines. (a) Mitochondria-enriched metabolites. (b) Metabolites with low expression in mitochondria.
Lipidomic Profiles of Mitochondria and Whole Cancer Cells
It is well known that the lipid composition of mitochondria is significantly different from that of whole cells. For instance, cardiolipin is exclusively present in the mitochondrial membrane.19 Hence, the objective of this section was to examine relative differences in detected lipids between bulk cancer cells and associated mitochondria. Our results demonstrate that the composition of commonly detected lipid classes of bulk cancer cells may not be representative of mitochondria. In particular, the relative differences were examined using an established nontargeted lipidomics approach, as described in the Methodssection. Of note, we detected lipids from more than ten subclasses, composed of various complex species including acylcarnitine (Acar), lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), bismonoacylglycerophosphate (BMP), sphingomyelin (SM), ceramide non-hydroxyfatty acid-dihydrosphingosine (Cer-NDS), ceramide non-hydroxyfatty acid-sphingosine (Cer-NS), hexosylceramide non-hydroxyfatty acid-sphingosine (HexCer-NS), phosphatidylcholine (PC), ether-linked phosphatidylcholine (PCe), phosphatidylethanolamine (PE), ether-linked phosphatidylethanolamine (PEe), diacylglycerol (DAG), triacylglycerol (TAG), and sphingosine 18:1.
There were 35 significantly different lipids found between bulk PANC1 cells and associated mitochondrial fractions as deduced by the Student’s t-test (p value < 0.05 and FDR < 0.2). Among the 35 lipids, were 3 ACars subclass, 1 BMP, 1 CE, 1 Cer-NDS, 4 DAGs, 5 LPCs, 4 PCs, 3 PCes, 1 PE, 1 SM, and 11 TAGs. These results demonstrated a difference that spread across lipid species, although some species showed subtle divergence. Similarly, 60 lipids (2 ACars, 1 CE, 3 DAGs, 1 LPC, 37 PCs, 1 PCe, 1 PE, 1 SM, and 13 TAGs) were detected with significant differences between bulk MDA-MB-231 cells and associated mitochondria (p value < 0.05 and FDR < 0.2). Finally, 101 significantly different lipophilic molecules comprised of 3 ACars, 1 BMP, 1 CE, 1 Cer-NDS, 1 Cer-NS, 4 DAGs, 1 HexCer-NS, 5 LPCs, 3 LPCes, 8 PCs, 19 PCes, 2 PEes, 3 PSs, 15 SMs, sphingosine 18:1, 29 TAGs, and 4 TAGs were detected between bulk A549 cells and associated mitochondria (p value < 0.05 and FDR < 0.2). Detailed information on these significantly different species is included in Table S2. In the PCA plots, there was a clear difference between the lipid profiles of bulk PANC1, MDA-MB-231, and A549 cells and their corresponding mitochondrial fractions (Figure 4a–c). The clustering of the heatmaps for the three comparison groups showed a relatively straightforward contrast in the overlapping lipid compositions from the bulk cancer cells and the mitochondria (Figure 4d–f). Collectively, we demonstrated that the concentrations of detected overlapping lipids between bulk cells and mitochondria were significantly different by univariate and multivariate analyses.
Figure 4.
Lipophilic metabolic profiles of mitochondria and bulk cells. Principal component analysis (PCA) score plots of (a) PANC1, (b) MDA-MB-231, and (c) A549. Heatmaps of (d) PANC1, (e) MDA-MB-231, and (f) A549. MT (red) indicates mitochondria, and WC (green) indicates whole cells or bulk cancer cells.
Western blotting was performed to analyze protein levels of fatty acid synthase (FASN), acetyl-CoA carboxylase (ACC), tricarboxylate transport protein (SLC25A1), and ATP-citrate synthase (ACLY). The results suggest that there were, to a certain extent, different expression levels of these proteins among tested cell lines. ACLY, which is responsible for the conversion of citrate to acetyl-CoA, was noticeably different among the three tested cell lines. These data might partly explain the differences in differentially expressed lipids across cell lines (Figure S1).
Metabolic Profiles of Mitochondria in Cancer Cells
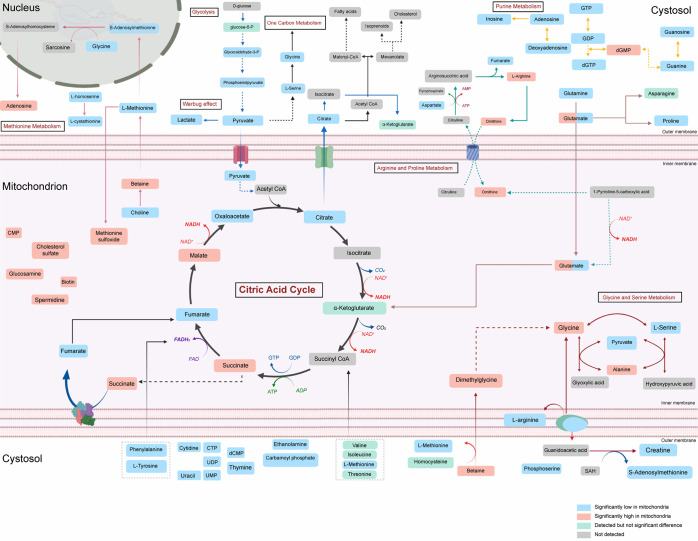
From the results of the metabolomic and lipidomic analyses described above, we constructed a metabolic landscape displaying the relative differences between mitochondria and bulk cancer cells. In our study, there was a general agreement regarding the increase or decrease of metabolites in the mitochondrial fractions isolated from the three tested cancer cell lines. However, the metabolites did not exhibit complete overlap, which is perhaps due to the unique characteristics of the metabolism of each tissue of origin (Figure 5).
Figure 5.
Hydrophilic metabolic landscape of mitochondria in cancer cells (created with BioRender.com).
Similarly, differences in lipid profiles between whole cancer cells and associated mitochondria clearly depended on the tissue of origin. In A549 cells (Figure S2), PEs, PSs, and SMs appeared to be enriched in the mitochondrial matrix. In contrast, ACars, TGs, DGs, and CEs seemed to be less concentrated within the mitochondria. Finally, PCs and LPCs appeared to have varying enrichment tendencies, wherein some species were more enriched inside mitochondria while others were not. PEs and SMs were consistently found to be enriched in mitochondria upon analysis of MDA-MB-231 cells. The decreased presence of CEs and ACars was also noticed in all three cell lines. However, DGs and TGs appeared to exhibit varying enrichment tendencies within the cell lines (Figure S3). The lipidome profile of PANC1 was quite similar to that of MDA-MB-231 (Figure S4); however, some ether lipids were either enriched or impoverished in the mitochondria compared to that in the whole cells.
Discussion
The advancement of omics technologies is beneficial for elucidating the mechanisms of human mitochondrial diseases as well as cancer.5,20 Nevertheless, efforts have been limited in developing a suitable platform to characterize the hydrophilic and lipophilic metabolite networks within cancer cells at the suborganellar level.12 A recent study suggested that high-throughput metabolic profiling is appropriate to track mitochondrial metabolism.21 Several protocols have been developed for the metabolomic and lipidomic analysis of enriched mitochondrial fractions of cultured cells, including fibroblasts,22 muscle cells,23 and liver cancer cells (HepG2).24 However, comprehensive method comparisons and an expeditious omic-suitable experimental procedure for the enrichment and partial purification of mitochondria for different human cancer cell lines have not been developed. In a recent review, Liu et al. argued that there are three fundamental obstacles (the speed of the fractionation procedure, the purity of the mitochondrial fraction, and the number of mitochondria in the biospecimens) that require a better resolution to improve the precision of suborganellar omics experiments.12 To this end, we established a procedure that facilitated the examination of nine different isolation strategies and determined the best isolation procedure for mitochondria with high purity and acceptable yield for subsequent high-throughput metabolic phenotyping experiments. The immunoblotting of specific protein markers is generally employed as an indicator of the quality and integrity of isolated mitochondria. In addition to immunoblotting, we employed TEM imaging to observe the ultrastructure of mitochondria before the metabolic phenotyping experiments. Furthermore, one of the biggest challenges in mitochondrial isolation is that the mitochondrial metabolome after isolation may not be representative of the cell as metabolites are altered during isolation. The procedure takes approximately 5 min per sample with the traditional method using a Dounce homogenizer. Therefore, it is crucial to shortening the experimental time to minimize alteration. To overcome this, we developed a method using a bead beater, successfully shortening the homogenization step to 10 s for 24 samples. The evaluation method developed in the present study simplifies the experimental process of metabolome analysis in isolated mitochondria. We also focused on the relative differences in lipid composition between mitochondrial fractions and associated whole cancer cells. We observed a significant difference in the levels of detected lipids, and these differences were dependent on the pathological conditions of the tissue of origin of the tested cells. Further studies are warranted to explore the metabolome and lipidome, as well as the dynamic conversion of metabolites in other organelles of cancer cells. In conclusion, the experimental workflow, with proper modifications, may be scalable to facilitate analysis of other suborganellar metabolic profiles, including stable-isotope tracing of intracellular metabolism of cancer cells with wide application.
The current experimental platform can also be used to isolate other cellular fractions from cultured cells, such as nuclei and Golgi apparatus. However, we noticed that the endoplasmic reticulum and the contact sites might not be thoroughly washed out, which may introduce bias to the hydrophilic and lipophilic profiles of mitochondria. Therefore, after the high-throughput metabolic profiling analysis, it is recommended to perform subsequent validation experiments using orthogonal assays. Additionally, it is recommended to examine the TEM images of the mitochondria within the cells before and after isolation to help ensure that there are no unexpected structural changes in the mitochondria.
Conclusions
In this study, different mitochondrial fractionation protocols were examined, and a practical method for mitochondrial isolation from cancer cells with high purity and integrity of isolated mitochondria was developed. The endogenous metabolites and commonly detected lipids appeared to be divergent in mitochondria and bulk cells in the three cancer cell lines tested. We expect the experimental platform will facilitate the precise characterization of the metabolic signature of suborganelles, which provides a framework for translational precision medicine.
Methods
Cell Lines
A549 (alveolar basal epithelial cells) and MDA-MB-231 (breast cancer cells) cell lines were obtained from the Korean Cell Line Bank (KCLB, Seoul, Korea), and the PANC1 (pancreas cells) cell line was kindly provided by Soon-Sun Hong of Inha University (Incheon, Korea).
Materials and Reagents
Ammonium acetate, ammonium formate, and formic acid were purchased from Sigma-Aldrich (St. Louis, MO). High-performance liquid chromatography (HPLC)-grade solvents, including acetonitrile, methanol, and water, were purchased from J.T. Baker (Phillipsburg, NJ). Liquid chromatography–mass spectrometry (LC–MS)-grade solvents, including 2-isopropanol, acetonitrile, methanol, and water, were purchased from Fisher Scientific (Hampton, NH). All the reagents used for the preparation of TEM samples were obtained from the National Instrumentation Center for Environmental Management (NICEM, Seoul, Korea). Lipid standards including lysophosphatidylcholine 12:0 (LPC 12:0), phosphatidylcholine 10:0/10:0 (PC(10:0/10:0)), lysophosphatidylethanolamine 14:0 (LPE(14:0)), phosphatidylethanolamine 10:0/10:0 (PE(10:0/10:0)), sphingomyelin 18:1/17:0 (SM(d18:1/17:0)), ceramide d18:1/17:0 (Cer(d18:1/17:0)), and triglyceride 17:0/17:0/17:0 (TAG(17:0/17:0/17:0)) were purchased from Avanti (Alabaster, AL). All other standards for the identification of metabolites were purchased from Sigma-Aldrich.
Adherent Cell Culture
All cells were cultured in Dulbecco’s modified eagle medium (DMEM, Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% antibiotic–antimycotic solution (Gibco) under standard conditions of 37 °C and 5% CO2 in a cell incubator. We optimized the seeding density and culture period for each cell line prior to the omics experiments. For extraction of metabolites and lipids, the PANC1 and A549 cells were seeded into a 15 cm dish at a density of 2 × 106 cells per plate and harvested 120 h or 96 h, respectively, after seeding. MDA-MB-231 cells were seeded at a density of 4 × 106 cells per dish and harvested 96 h after seeding.
Optimization of Mitochondrial Isolation
To isolate mitochondria from each cell line, cells were washed with cold phosphate-buffered saline (PBS) two times and homogenized by either chemical or physical methods. For chemical lysis, cells were collected with 800 μL of reagent A from the commercial kit (Mitochondria Isolation Kit for Cultured Cells, Thermo Fisher Scientific, Waltham, MA) and incubated for 2 min on ice. Cells were treated with 10 μL of reagent B from the commercial kit and incubated for 5 min on ice with intermittent vortexing every minute. For physical lysis, cells were collected in 1 mL lysis buffer and homogenized using a Precellys 24 (Bertin Technologies, Montigny-le-Bretonneux, France). For optimization of lysis buffer composition, four different lysis buffers were compared: buffer from the commercial kit, sucrose-based buffer (250 mM sucrose, 20 mM Tris, and 1 mM ethylenediaminetetraacetic acid), mannitol-based buffer (225 mM mannitol, 75 mM sucrose, and 30 mM Tris),25 and normal saline.22 Additionally, the homogenization process was performed at 4000g for 5 or 10 s for comparison. The homogenized cells were transferred to a new tube and centrifuged at 1000g for 10 min at 4 °C. The pellet containing the nuclear fraction was stored for further western blot analysis, and the supernatant was transferred to a new tube and centrifuged at 3000g for 15 min at 4 °C. The pellet containing the mitochondria-enriched fraction was washed with lysis buffer and centrifuged at 10 900g for 15 min at 4 °C. The pellet and supernatant were kept as the mitochondria-enriched fraction and cytosolic fraction, respectively.
Western Blot Analysis
The mitochondria-enriched fraction was lysed using radioimmunoprecipitation assay buffer (RIPA) buffer (Thermo Fisher Scientific) containing the Complete Protease Inhibitor Cocktail (Roche Diagnostics, Basel, Switzerland). Protein concentration was determined using the Micro BCA Protein Assay Kit (Pierce, Rockford, IL) following the manufacturer’s instructions. Next, proteins (10 μg) from each fraction were separated using an 8 or 13.5% SDS-polyacrylamide gel by electrophoresis and subsequently transferred onto poly(vinylidene difluoride) membrane (Merck Millipore, Burlington, MA). The membrane was blocked with 5% nonfat dry milk in Tween 20–Tris-buffered saline (TBST, Bio-Rad, Hercules, CA) for 1 h at room temperature (20–25 °C), followed by overnight incubation at 4 °C with the specific primary antibody, anti-PARP, -α-tubulin (#9542, #3873, Cell Signaling Technologies, Danvers, MA), -VDAC1, -cytochrome c (ab14734, ab13575, Abcam, Cambridge, U.K.), and -PCNA (sc-56, Santa Cruz Biotechnology, Dallas, TX). All antibodies were diluted at a ratio of 1:1000. After washing three times with TBST, membranes were incubated with the appropriate secondary antibody for 1 h at room temperature. Membranes probed with specific antibodies were incubated with SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific) and visualized using an ImageQuant LAS 4000 (GE Healthcare, Chicago, IL).
Visualization of Mitochondria Using TEM
The isolated mitochondria-enriched fraction from 1 × 107 cells was fixed in 1% glutaraldehyde with 0.05 M sodium cacodylate buffer for 2 h and washed three times with 0.05 M sodium cacodylate buffer. The pellet was postfixed in 1% OsO4, and en-bloc-stained with 0.5% uranyl acetate at 4 °C overnight. The sample was dehydrated by sequentially increasing ethanol concentration (50, 70, 80, 90, and 100%) and incubated in a transition solvent, propylene oxide. Next, the sample was embedded in Spurr’s resin and polymerized at 70 °C overnight. The resin-embedded specimen was sectioned into 70 nm thick sections using an ultramicrotome (EM UC7, Leica, Wetzlar, Germany) and observed with an energy-filtering transmission electron microscope (LIBRA 120, Carl Zeiss, Oberkochen, Germany).
Metabolite Extraction for Large-Scale Targeted Metabolomics
For each experiment, the mitochondrial pellet was resuspended in 500 μL of MeOH (−80 °C) containing the internal standards LPC(12:0), PC(10:0/10:0), LPE(14:0), PE(10:0/10:0), SM(d18:1/17:0), and succinic acid (d4) and then vortexed vigorously. Samples were snap-frozen in liquid nitrogen followed by a 10 min incubation at 37 °C.26 This cycle was performed twice. After sonication on ice for 30 s and vortexing for 20 s, 300 μL was transferred to another 2 mL tube. Methyl tert-butyl ether (MTBE; 1 mL at −20 °C) containing TAG (17:0/17:0/17:0) as the internal standard was added to each sample, followed by vortexing for 20 s. After shaking at 1500 rpm for 10 min at 4 °C, 250 μL pure water (HPLC grade) was added to the sample and then mixed by vortexing for 20 s. Samples were centrifuged at 16 000g for 2 min at 4 °C. The upper lipid extract layer (1 mL) was transferred to another tube and evaporated under constant nitrogen flow at room temperature until completely dry. Lipid extracts were stored at −80 °C until analysis. The lower layer, which contained polar metabolites, was also collected, transferred to a new tube, and evaporated under constant nitrogen flow at room temperature until completely dry. Metabolite extracts were stored at −80 °C until further analysis. For quality assessment purposes, QC samples were made by mixing 15 μL of extracts from each sample. Three biological samples, i.e., three different passages of mitochondrial extracts and whole cells, were analyzed per cell line.
Large-Scale Endogenous Metabolite Analysis
Triple quadrupole LC–MS (Agilent 6400, Agilent Technologies, Santa Clara, CA) using electrospray ionization (ESI) with the positive/negative ion-switching mode was employed for metabolomics analysis.27 The parental and product transitions of tracked metabolites were adopted from a previous study.27 In addition, parental and product transitions were curated using available literature, and commercially available standards were used for mass spectrometry optimization using an Agilent Optimizer (Agilent Technologies). This platform showed comparable or better performance compared to an established in vitro gas chromatography–mass spectrometry (GC-MS) untargeted metabolomic analysis described in our previous study.28 The chromatographic separation was achieved using an Amide XBridge HPLC (100 mm × 4.6 mm; 3.5 μm) plus a guard column (Waters, Milford, MA). The column temperature was held at 30 °C during the analysis. Mobile phase A contained 95% water:5% acetonitrile (v/v) with 20 mM ammonium hydroxide and 20 mM ammonium acetate, and mobile phase B contained only acetonitrile. A flow rate of 0.4 mL/min was employed. Gradient elution was as follows: 0–3 min, linear gradient from 85 to 30% B; 3–12 min, linear gradient from 30 to 2% B; 12–15 min, hold at 2% B; 15–16 min, linear gradient from 2 to 85% B; and 16–23 min, keep at 85% B. A sample volume of 5 μL was injected for each run. The mass spectrometer was operated in MRM transitions mode with a polarity switching time of 30 ms. The dwell time was 3 ms in each transition, which was adequate for the relative comparison of the integrated peak areas between groups. ESI sources were set as follows: capillary voltage of 3.5 kV, nozzle voltage of 500 V, gas temperature at 325 °C, drying gas at 5 L/min, nebulizer gas at 45 psi, sheath gas temperature of 350 °C, and sheath gas flow at 11 L/min. Agilent MassHunter Workstation Software was used to acquire the data.
All samples were randomly allocated in the sampling chamber for the analysis. We ran eight QC samples before the real samples for each analysis. There were five other QC samples analyzed in the beginning, during the course, and at the end of the course of each cell line. These working QC samples were used for the peak filtering process. The internal standard was used to assess the quality of the data acquisition procedure and was removed before statistical analysis.
Untargeted Cellular Lipid Profiling
Essentially, the lipid profiling followed a well-validated protocol with proven reproducibility across different mass spectrometers, including an Agilent 6530 Q-ToF mass spectrometer, with a slight modification in the chromatographic separation protocol.29 In short, the samples were suspended in methanol/toluene (9:1). After 1 min of vortexing, a small amount of the lipid suspension from whole-cell and mitochondrial samples were collected in a single microcentrifuge tube to prepare QC samples. Samples were then centrifuged at 16 000g for 2 min, and the supernatant was collected for analysis. Data acquisition was performed on the Agilent 1290 Infinity II UPLC system (Agilent Technologies) coupled with an Agilent 6530 Q-ToF MS (Agilent Technologies). An Acquity UPLC CSH C18 column (100 mm × 2.1 mm; 1.7 μm) coupled to an Acquity UPLC CSH C18 VanGuard Pre-column (5 mm × 2.1 mm; 1.7 μm) (Waters) was employed for lipid separation. Gradient condition by mobile phase (A) 60:40 (v/v) of acetonitrile/water with ammonium formate (10 mM) and formic acid (0.1%) and (B) 90:10 (v/v) of isopropanol/acetonitrile with ammonium formate (10 mM) and formic acid (0.1%) was conducted as following mobile phase (B) percentage: 0–2 min, 15 to 30%; 2–2.5 min, 30 to 48%; 2.5–11 min, 48 to 82%; 11–11.5 min, 82 to 99%; 11.5–12 min, maintained at 99%; 12–13 min, 99 to 15%; and 13–16 min, maintained at 15%. The column system was maintained at 65 °C. A constant flow rate of 0.6 mL/min was employed. The autosampler was kept at 4 °C and 5 μL of sample was injected for each analytical run. Auto MS/MS data-dependent acquisition on ESI positive mode was performed on peak acquisition. Specific parameters were set as follows: capillary voltage, 3.5 kV; fragmentor voltage, 135 V; collision energy, 20 eV; nozzle voltage, 1000 V; gas temperature, 325 °C; drying gas, 8 L/min; nebulizer gas, 35 psi; sheath gas temperature, 350 °C; sheath gas flow, 11 L/min; MS resolution, 4 GHz; MS1 range (m/z), 300–1200; MS1 acquisition speed, 2 spectra/s; MS/MS range (m/z), 50–1200; MS/MS acquisition speed, 4 spectra/s; precursor ions per cycle, 4; isolation width, narrow (1.3 m/z); precursor ion threshold, 500 count (absolute) or 0.01% (relevant); and exclusion criteria, disable. Eight QC samples were run before the actual samples to stabilize the column. To support the lipid annotation, targeted MS/MS analysis was also performed at the end of the batch. Internal standards and QC samples were used to assess the quality of the overall experimental procedure and were removed before statistical analysis.
Mass Spectrometry Data Alignment, Processing, and Normalization
MSConvert software was used to convert the raw data into mzML files, then XCMS-MRM online (https://xcmsonline-mrm.scripps.edu/) was used for data processing. The parameters of this processing were set as 8 s for “average peak width” and 4 for “scans per second”; other parameters were set as default. Data imputation with average value was employed for missing data. The peak areas of metabolites that had the same nominal mass transitions were manually integrated using the Vendor’s software. Next, we removed the metabolites with a peak intensity lower than the limit of detection (LOD) generated from XCMS-MRM in both mitochondria and whole-cell samples.
Raw lipidomics data files (.d format) were converted to.abf format files by AbfConverter 4.0.0 (Reifycs), imported into MS-DIAL version 3.7.0, and processed as previously described.28 Detailed parameters can be found in Table S3.
Due to significant technical variance between mitochondria and whole-cell samples, both large-scale targeted metabolomics and untargeted lipidomics data were further normalized using quantile normalization to allow cross-sample comparison.30 Given the data matrix X = (xst, 1 ≤ s ≤ p, 1 ≤ t ≤ N) of dimension p and sample size n, let Xord = (x(s)t, 1 ≤ s ≤ p, 1 ≤ t ≤ N) be the sorted data matrix where p values are ordered within a sample. Define the averaged distribution function F and the sth (1 ≤ s ≤ p) cross-sample empirical distribution Gs by
where x ®(s) = N–1 ∑t=1Nx(s)t. Next, the normalized data matrix is returned by
Peaks with a relative standard deviation of 30% or above that of working QC samples were filtered out. Moreover, normalized data were log-transformed and scaled by Pareto scaling before downstream statistical analysis.
Lipid Annotation Protocol
For lipid data processing, multiplatform driven data processing based on MS-DIAL31 and LipiDex/Mzmine32,33 was conducted and combined to reinforce the quality and accuracy of the lipid identification. More reliable identification of lipids was expected as the two platforms rely on different libraries and mechanisms by which data are processed. Regarding the quantitative criteria of the annotation using MS-DIAL, the m/z score of approximately 950 or above and MS/MS reverse product score of roughly 700 or above (with a good match of the MS/MS) were the two essential scores for the identification. It is worth mentioning that other additional factors, such as the appearance of sodium adducts and matching of experimental retention time with reference retention times (within 0.5 min), were also taken into account during the annotation. Reinspection was performed on MS-DIAL for the lipids with mismatched identification between two platforms. Importantly, if the difference in aligned retention time was over 0.15 min, it was considered to be a low-quality annotated peak and removed. When applicable, the annotation with detailed information of acyl chains (carbon number and the degree of unsaturation) was reported. We basically followed the nomenclature given in MS-DIAL version 3.7.0 to report the examined lipids.
Data Exploration and Visualization
For data exploration, we applied PCA and showed the two-dimensional (2D) score plot to observe sample distribution along with the first two principal components. This visualization technique is frequently used to illustrate the dispersal of biological features across groups. The first two principal components were computed as described below, and others were sequentially defined similarly
where Sn is a sample covariance matrix based on n sample vectors. A heatmap was also generated to observe the contrast regarding the relative differences of all features between comparative groups. Both data visualization techniques were conducted using MetaboAnalyst 4.0.34
Statistical Analysis
Continuous data were summarized by the mean and standard deviation (SD). Multiple two-tailed t-tests were conducted unless otherwise stated. p values were adjusted using the Benjamini–Hochberg procedure to control the false discovery rate (FDR). Assume there are r hypotheses {Hk:1 ≤ k ≤ r} to test and individual p values are denoted by p1,..., pr and p(1) ≤ p(2) ≤ ... ≤ p(r) are their ordered statistics. The Benjamini–Hochberg procedure rejects the null hypothesis {H(k):1 ≤ k ≤ s} that satisfies p(k) ≤ kα/r for all 1 ≤ k ≤ s and p(s+1) > (s + 1)α/r where α is a prespecified significance level. If p(k) ≤ kα/r for all 1 ≤ k ≤ r (i.e., the case s = r), the hypothesis is rejected. A p value of 0.05 and an FDR of 0.05 (hydrophilic metabolites)35 or 0.2 (lipophilic metabolites)36 were set as statistical significance levels.
Acknowledgments
This work was supported by the Bio-Synergy Research Project of the Ministry of Science, ICT and Future Planning through the National Research Foundation (NRF-2012M3A9C4048796) and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1D1A1A02046560). This work was also supported by the BK21 Plus Program in 2020.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c03612.
Metabolite composition of mitochondria as compared to that of bulk cancer cells; differentially expressed lipids among the three indicated cell lines; MS-DIAL lipid data alignment and processing; expression levels of FASH, ACC, SCL25A1, and ACLY in the three indicated cancer cell lines; lipid compositions of mitochondria and bulk A549 cells; lipid compositions of mitochondria and bulk MDA-MB-231 cells; and lipid compositions of mitochondria and bulk PANC1 cells (PDF)
Author Contributions
⊥ N.P.L. and J.E.M. contributed equally to this work.
The authors declare the following competing financial interest(s): N.P.L., J.E.M., N.H.A., S.J.K., S.P., J.L., and S.W.K. hold the Korean patent related to the isolation technique of mitochondria from cancer cells. Patent registration ID: 10-2019-0125693. The authors declare no other potential conflicts of interest.
Supplementary Material
References
- Hanahan D.; Weinberg R. A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Vyas S.; Zaganjor E.; Haigis M. C. Mitochondria and cancer. Cell 2016, 166, 555–566. 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neagu M.; Constantin C.; Popescu I. D.; Zipeto D.; Tzanakakis G.; Nikitovic D.; Fenga C.; Stratakis C. A.; Spandidos D. A.; Tsatsakis A. M. Inflammation and Metabolism in Cancer Cell-Mitochondria Key Player. Front. Oncol. 2019, 9, 348. 10.3389/fonc.2019.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaghello L.; Longo V.. Chapter One - Metabolic Alterations at the Crossroad of Aging and Oncogenesis. In International Review of Cell and Molecular Biology; Galluzzi L., Ed.; Academic Press: New York, 2017; Vol. 332, pp 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman J.; Rahman S. Mitochondrial medicine in the omics era. Lancet 2018, 391, 2560–2574. 10.1016/S0140-6736(18)30727-X. [DOI] [PubMed] [Google Scholar]
- Porporato P. E.; Filigheddu N.; Pedro J. M. B.-S.; Kroemer G.; Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. 2018, 28, 265–280. 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustylnikov S.; Costabile F.; Beghi S.; Facciabene A. Targeting mitochondria in cancer: current concepts and immunotherapy approaches. Transl. Res. 2018, 202, 35–51. 10.1016/j.trsl.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterhuizen K.; van der Westhuizen F. H.; Louw R. Metabolomics of mitochondrial disease. Mitochondrion 2017, 35, 97–110. 10.1016/j.mito.2017.05.012. [DOI] [PubMed] [Google Scholar]
- Brügger B. Lipidomics: analysis of the lipid composition of cells and subcellular organelles by electrospray ionization mass spectrometry. Annu. Rev. Biochem. 2014, 83, 79–98. 10.1146/annurev-biochem-060713-035324. [DOI] [PubMed] [Google Scholar]
- Nagrath D.; Caneba C.; Karedath T.; Bellance N. Metabolomics for mitochondrial and cancer studies. Biochim. Biophys. Acta, Bioenerg. 2011, 1807, 650–663. 10.1016/j.bbabio.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Matuszczyk J.-C.; Teleki A.; Pfizenmaier J.; Takors R. Compartment-specific metabolomics for CHO reveals that ATP pools in mitochondria are much lower than in cytosol. Biotechnol. J. 2015, 10, 1639–1650. 10.1002/biot.201500060. [DOI] [PubMed] [Google Scholar]
- Liu X.; Xu G. Recent advances in using mass spectrometry for mitochondrial metabolomics and lipidomics - a review. Anal. Chim. Acta 2018, 1037, 3–12. 10.1016/j.aca.2017.11.080. [DOI] [PubMed] [Google Scholar]
- Chen W. W.; Freinkman E.; Wang T.; Birsoy K.; Sabatini D. M. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell 2016, 166, 1324–1337.e11. 10.1016/j.cell.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan G.-L.; Pfander B.; Jentsch S. PCNA, the maestro of the replication fork. Cell 2007, 129, 665–679. 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Ow Y.-L. P.; Green D. R.; Hao Z.; Mak T. W. Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol. 2008, 9, 532–542. 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- Downing K. H.; Nogales E. Tubulin and microtubule structure. Curr. Opin. Cell Biol. 1998, 10, 16–22. 10.1016/S0955-0674(98)80082-3. [DOI] [PubMed] [Google Scholar]
- Kraus W. L.; Lis J. T. PARP goes transcription. Cell 2003, 113, 677–683. 10.1016/S0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- Tomasello F.; Messina A.; Lartigue L.; Schembri L.; Medina C.; Reina S.; Thoraval D.; Crouzet M.; Ichas F.; De Pinto V.; De Giorgi F. Outer membrane VDAC1 controls permeability transition of the inner mitochondrial membrane in cellulo during stress-induced apoptosis. Cell Res. 2009, 19, 1363–1376. 10.1038/cr.2009.98. [DOI] [PubMed] [Google Scholar]
- Paradies G.; Paradies V.; Ruggiero F. M.; Petrosillo G. Role of cardiolipin in mitochondrial function and dynamics in health and disease: molecular and pharmacological aspects. Cells 2019, 8, 728. 10.3390/cells8070728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil S.; Kuman M. M.; Palvai S.; Sengupta P.; Basu S. Impairing powerhouse in colon cancer cells by hydrazide–hydrazone-based small molecule. ACS Omega 2018, 3, 1470–1481. 10.1021/acsomega.7b01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roede J. R.; Park Y.; Li S.; Strobel F. H.; Jones D. P. Detailed mitochondrial phenotyping by high resolution metabolomics. PLoS One 2012, 7, e33020 10.1371/journal.pone.0033020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrat-Durebex C.; Bocca C.; Chupin S.; Kouassi Nzoughet J.; Simard G.; Lenaers G.; Reynier P.; Blasco H. Metabolomics and lipidomics profiling of a combined mitochondrial plus endoplasmic reticulum fraction of human fibroblasts: A robust tool for clinical studies. J. Proteome Res. 2018, 17, 745–750. 10.1021/acs.jproteome.7b00637. [DOI] [PubMed] [Google Scholar]
- Shaham O.; Slate N. G.; Goldberger O.; Xu Q.; Ramanathan A.; Souza A. L.; Clish C. B.; Sims K. B.; Mootha V. K. A plasma signature of human mitochondrial disease revealed through metabolic profiling of spent media from cultured muscle cells. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 1571. 10.1073/pnas.0906039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler L.; Li J.; Häring H.-U.; Weigert C.; Lehmann R.; Xu G.; Hoene M. Purity matters: a workflow for the valid high-resolution lipid profiling of mitochondria from cell culture samples. Sci. Rep. 2016, 6, 21107 10.1038/srep21107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieckowski M. R.; Giorgi C.; Lebiedzinska M.; Duszynski J.; Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 2009, 4, 1582–1590. 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Gao Y.; Sun J.; Fan S.; Yao X.; Ran X.; Zheng C.; Huang M.; Bi H. Optimization of lipid extraction and analytical protocols for UHPLC-ESI-HRMS-based lipidomic analysis of adherent mammalian cancer cells. Anal. Bioanal. Chem. 2017, 409, 5349–5358. 10.1007/s00216-017-0483-7. [DOI] [PubMed] [Google Scholar]
- Yuan M.; Breitkopf S. B.; Yang X.; Asara J. M. A positive/negative ion–switching, targeted mass spectrometry–based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012, 7, 872–881. 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S. J.; Kim J. Y.; Long N. P.; Min J. E.; Kim H. M.; Yoon J. H.; Anh N. H.; Park M. C.; Kwon S. W.; Lee S. K. Comprehensive multi-omics analysis reveals aberrant metabolism of Epstein–Barr-virus-associated gastric carcinoma. Cells 2019, 8, 1220. 10.3390/cells8101220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajka T.; Smilowitz J. T.; Fiehn O. Validating quantitative untargeted lipidomics across nine liquid chromatography–high-resolution mass spectrometry platforms. Anal. Chem. 2017, 89, 12360–12368. 10.1021/acs.analchem.7b03404. [DOI] [PubMed] [Google Scholar]
- Bolstad B. M.; Irizarry R. A.; Åstrand M.; Speed T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Tsugawa H.; Cajka T.; Kind T.; Ma Y.; Higgins B.; Ikeda K.; Kanazawa M.; VanderGheynst J.; Fiehn O.; Arita M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins P. D.; Russell J. D.; Coon J. J. LipiDex: an integrated software package for high-confidence lipid identification. Cell Syst. 2018, 6, 621–625.e5. 10.1016/j.cels.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskal T.; Castillo S.; Villar-Briones A.; Orešič M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 2010, 11, 395 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.; Wishart D. S.; Xia J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinf. 2019, 68, e86 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- Kang Y. P.; Lee S. B.; Lee J.-M.; Kim H. M.; Hong J. Y.; Lee W. J.; Choi C. W.; Shin H. K.; Kim D.-J.; Koh E. S.; Park C.-S.; Kwon S. W.; Park S.-W. Metabolic profiling regarding pathogenesis of idiopathic pulmonary fibrosis. J. Proteome Res. 2016, 15, 1717–1724. 10.1021/acs.jproteome.6b00156. [DOI] [PubMed] [Google Scholar]
- Yang R.; Zhang Y.; Qian W.; Peng L.; Lin L.; Xu J.; Xie T.; Ji J.; Zhan X.; Shan J. Surfactant lipidomics of alveolar lavage fluid in mice based on ultra-high-performance liquid chromatography coupled to hybrid quadrupole-exactive orbitrap mass spectrometry. Metabolites 2019, 9, 80. 10.3390/metabo9040080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.