Abstract
In December 2019, a new coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) led to the outbreak of a pulmonary disease called COVID-19, which killed thousands of people worldwide. Therefore, the necessity to find out the potential therapeutic pharmaceuticals is imperious. This study investigates the inhibitory effect of SARS-CoV-2 3-chymotrypsin-like protease (3CLpro) using caffeine and caffeine-containing pharmaceuticals (3CPs) based on molecular dynamics simulations and free energy calculations by means of molecular mechanics-Poisson–Boltzmann surface area (MMPBSA) and molecular mechanics-generalized-Born surface area (MMGBSA). Of these 3CPs, seven drugs approved by the US-Food and Drug Administration have shown a good binding affinity to the catalytic residues of 3CLpro of His41 and Cys145: caffeine, theophylline, dyphylline, pentoxifylline, linagliptin, bromotheophylline and istradefylline. Their binding affinity score ranged from –4.9 to –8.6 kcal/mol. The molecular dynamic simulation in an aqueous solution of docked complexes demonstrated that the 3CPs conformations bound to the active sites of 3CLpro during 200 ns molecular dynamics simulations. The free energy of binding also confirms the stability of the 3CPs–3CLpro complexes. To our knowledge, this in silico study shows for the first time very inexpensive drugs available in large quantities that can be potential inhibitors against 3CLpro. In particular, the repurposing of linagliptin, and caffeine are recommended for COVID-19 treatment after in vitro, in vivo and clinical trial validation.
Communicated by Ramaswamy H. Sarma
Keywords: Coronavirus SARS-CoV-2, COVID-19, 3-chymotrypsin-like protease, caffeine-containing pharmaceuticals, molecular dynamics
1. Introduction
The cause of the first infection of COVID-19 is not yet clear. However, some researchers have proposed that that the reason might be attributed to some seafood and animal sold in Wuhan in China; then the infection dramatically increased via human-to-human transmission (Heymann & Shindo, 2020; Lai et al., 2020; Sabino-Silva et al., 2020). On 11 March 2000, the outbreak-pandemic of COVID-19 was declared by the World Health Organization. On 30 May, the confirmed cases worldwide went up to 5,819,962 with 362,786 confirmed deaths (Herten-Crabb & Davies, 2020; World Health Organization, 2020). Coronavirus mainly targets the upper respiratory tract system in humans. It was recently found that the new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus can cause the severe acute respiratory syndrome, which is known as COVID-19 (Chang, Lin, et al., 2020; Rothan & Byrareddy, 2020; Velavan & Meyer, 2020).
Coronaviruses are positive-stranded RNA viruses, with the largest known viral RNA genomes (27–31 kb). SARS-CoV-2 belongs to class b of the genus Betacoronavirus. The RNA genomes of SARS-CoV-2 were found to be identical to about 82% of the SARS-CoV genomes (Enayatkhani et al., in press; Lu et al., 2020; Wu, Liu et al., 2020; Zhou et al., 2020; Zhu et al., 2020). The 229E gene encodes two polyproteins involved in releasing an essential functional polypeptide for viral replication and transcription. These polypeptides released by extensive proteolytic processing achieved 3-chymotrypsin-like protease of SARS-CoV-2 (3CLpro), as it cleaves at least 11 sites on these polyproteins. Thus, the inhibition of 3CLpro can interrupt the replication and transcription of SARS-CoV-2 (Anand et al., 2003; Shereen et al., 2020; Sk et al., in press).
Recently, several studies have investigated the inhibitory effect of some natural products, synthetic pharmaceuticals for 3CLpro using the molecular docking approach. Of these tested substances were chloroquine phosphate, Remdesivir, Indinavir, Lopinavir, Carfilzomib, Eravacycline, Elbasvir, Valrubicin, Darunavir, Favipiravir isoflavone, myricitrin, α-ketoamides and methyl rosmarinate (Ahmad et al., in press; Beck et al., 2020; Chang, Tung et al., 2020; Das et al., in press; Duan et al., 2020; Ul Qamar et al., 2020; Wahedi et al., in press; Wang, 2020; Wu, Liu et al., 2020). Remdesivir was approved by US-Food and Drug Administration (US-FDA) on 1 May 2020 for emergency use to treat COVID-19 (US-Food & drug Administration, 2020).
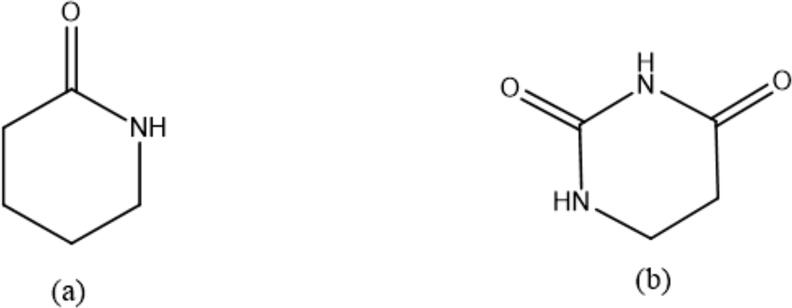
On the other hand, in some countries, some plants used in traditional medicine are allegedly used for treating COVID-19. Among these assumptions, what was widespread among Sudanese on social media was the description of hot sugar-free tea for COVID-19. This claim may find a scientific basis to support it. Zhang et al. (in press) highlighted the importance of the pyridone ring in synthetic materials, which is active against SARS-CoV-2. Accordingly, the pyridone-containing pharmaceuticals were studied. The pyridone ring played a key role to form hydrogen bonds with the active residues of 3CLpro (Elzupir, 2020). 4-Pyrimid-dione ring has an extra carbonyl group as an advantage over pyridone (Scheme 1), which can be found in the natural and cheap material caffeine (Nair, in press). This study demonstrates the inhibitory effect of 3CLpro by means of approved caffeine and caffeine-containing pharmaceuticals (3CPs) using the molecular docking approach.
Scheme 1.
Chemical structures of (a) pyridone and (b) 4-pyrimid-dione
2. Materials and methods
2.1. Caffeine and caffeine-containing pharmaceuticals
The DrugBank database search engine was used to find 3CPs. The research results showed that eleven drugs from this family were approved by the US-FDA. One of them, Xaninthol, was canceled post-marketing and is therefore not included in this study. The investigated drugs are aminophylline, bromotheophylline, caffeine, dimenhydrinate, dyphylline, istradefylline, pentoxifylline, linagliptin, oxtriphylline and theophylline.
2.2. Buildup and energy minimization of the molecular structures
The three-dimensional structures of the 3CPs were generated and minimized using the UCSF Chimera software (v 1.10.2.), utilizing the available canonical SMILES at the PubChem database site. The 3CLpro of the SARS-CoV-2 crystal structure was obtained from the Protein Data Bank database (PDB ID: 6Y2E).
2.3. Molecular docking
Molecular docking experiments were performed using the AutoDock Vina tool plugin UCSF Chimera. The default values for the parameters were adopted with a grid box (–16.5 × −24.0 × 17) Å, centered at (35, 65, 65) Å. Water was added as a solvent to represent the real environment. The total solvent accessible surface area was 14,358.5 Å2. The predicted affinity score values were explored using the View Dock tool. The binding sites and image processing were achieved by the UCSF Chimera (Edgar et al., 2011; Pettersen et al., 2004; Trott & Olson, 2010).
2.4. MD simulation
The docked complexes were separated by UCSF Chimera to ligands and receptor and saved as PDB using AMBER large-structure serial numbering, after adding hydrogen to the ligands. Then the topology files and parameters of the ligands and the receptor were made utilizing antechamber and leap of Amber Tools 20 (Case et al., 2006; Da Silva & Vranken, 2012). The missing hydrogen atoms of 3CLpro were added utilizing the Leap. The systems were solvated using TIP3P water molecules (Jorgensen et al., 1983), and were neutralized using sodium ions. Afterward, molecular dynamics (MD) simulations of the selected pharmaceuticals were performed by means of the Nanoscale Molecular Dynamics (NAMD) Simulation 2.6 program (Nelson et al. 1996; Phillips et al. 2005), using the Amber force field of ff14SB (Maier et al., 2015), GAFF2 (Wang et al., 2004) to assign 3CLpro structure and inhibitors, respectively. Each system tested was minimized for 1 ps at 273.15 K by the NVE ensemble. Then the temperature was gradually increased to 310 K using the NVT ensemble in a protocol consisting of 10,000 minimization steps, followed by 200 ns of MD simulations control at 310 K and a time step of 2 fs using the NVT ensemble. For electrostatics calculation, periodic boundary conditions besides the particle mesh Ewald process were applied (de Leeuw et al., 1980; Essmann et al., 1995). In order to obtain the root-mean-square deviation (RMSD) and the root-mean-square fluctuation (RMSF) for each system, the trajectory was analyzed with the VMD 1.8 program (Humphrey et al., 1996).
2.5. The binding free energies
The binding free energies of the complexes was calculated by means of molecular mechanics-Poisson–Boltzmann surface area (MMPBSA) and molecular mechanics-generalized-Born surface area (MMGBSA) utilizing MMPBSA.py module of Amber Tools 20 (Miller et al., 2012). The MD simulation output over 200 ns provided several structures that were sampled after equilibrium at about each 7 ns. The snapshots were obtained using CPPTRAJ (Roe & Cheatham, 2013). The quasi-harmonic entropy approximation was used to evaluate the conformational changes (Numata et al., 2007). The binding free energy of the interaction between inhibitors and 3CLpro system can be obtained through the following equations:
where the ΔH, –TΔS, Evdw, Eele, ΔGsol, Epb/gb and Enp represent enthalpy change, the entropic contribution, van der Waals interaction energy, electrostatic interaction energy, the polar solvation energy and the non-polar solvation energy, respectively.
3. Results and discussion
3.1. Molecular docking
The inhibitory effect of CLpro was investigated based on van der Walls interactions and hydrogen-bonds between the chosen drugs and the catalytic residues of Cys145 and His41, the important residues of Ser1 and Glu166 for maintaining the enzyme on the correct conformation (Zhang et al., in press). The results of 3CPs, including structure, binding affinity and RMSD, are presented in Table 1. The lower RMSD values indicate the favorability of these drugs as good ligands to binding with the 3CLpro. Of the ten drugs tested, aminophylline, oxtriphylline and dimenhydrinate were not docked with the targeted enzyme 3CLpro.
Table 1.
The binding affinity of caffeine and caffeine-containing pharmaceuticals with 3-chymotrypsin-like protease (CLpro).
| Pharmaceutical name | Binding percentage a | Score ± SD b (kcal/mol) | RMSD | Hydrogen bond | Vander Val (distance) |
|---|---|---|---|---|---|
| Remdesivir | 11 | –6.7 ± 0.00 | 29.278–32.562 | – | Cys145 (4 side contacts, distance range 3.572–3.852 Å). |
| Glu166 (6 side contacts, distance range: 3.518–3.991 Å). | |||||
| Linagliptin | 56 | –8.6 ± 0.55 | 0.000–0.000 | – | His41 (3.453 Å/3.498 Å/3.834 Å). |
| Glu166 (3.562 Å/3.902 Å/3.987 Å). | |||||
| Istradefylline | 22 | –6.1 ± 0.07 | 29.039–32.061 | Cys145 | Cys145 (3.867 Å) |
| Cys145* (3.720 Å/3.936 Å) | |||||
| His41* (3.753 Å) | |||||
| Dyphylline | 89 | –6.0 ± 0.34 | 2.009–4.637 | Cys145 | His41 (3.619 Å/3.673 Å/3.713 Å). |
| Glu166 | Cys145 (6 side contacts, distance range: 3.091–4.027 Å) | ||||
| Glu166 (3.015 Å). | |||||
| Caffeine | 67 | –5.6 ± 0.30 | 0.000–0.000 | Glu166 | His41 (3.720 Å/3.807 Å). |
| Cys145* | Cys145 (3.629 Å/3.694 Å). | ||||
| Glu166 (3.504 Å/ 3.912 Å/2.988 Å). | |||||
| Bromotheophylline | 44 | –5.6 ± 0.26 | 0.000–0.000 | Glu166 | His41 (5 side contacts, distance range 2.733 Å to 3.565 Å). |
| Cys145 (3.708 Å/3.817 Å) | |||||
| Glu166 (3.385 Å/3.417/ 3.612 Å). | |||||
| Pentoxifylline | 67 | –5.4 ± 0.19 | 2.485–4.751 | Cys145 | His41 (5 side contacts, distance range 3.585–3.906 Å). |
| Glu166 (7 side contacts, distance range: 3.211–3.767 Å). | |||||
| Theophylline | 67 | –4.9 ± 0.17 | 1.526–3.155 | Glu166 | His41 (3.583 Å/3.668 Å/3.717 Å/3.764 Å). |
| Cys145 (3.566 Å/3.733 Å/ 3.746 Å). | |||||
| Glu166 (3.003 Å/3.951 Å). |
Binding percentage based on the number of conformations attached to the active sites of CLpro (total conformations was nine).
SD based on the others score energies of conformations.
Different representing mode of interactions.
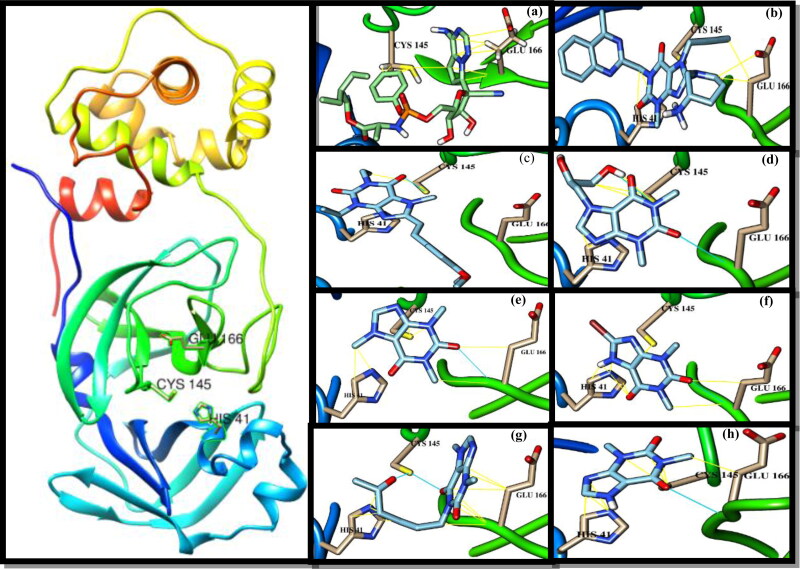
Remdesivir has been docked as a reference drug with 3CLpro. It shows van der Waal’s interactions with the 3CLpro with the lowest binding percentage (Table 1). Linagliptin among all tested drugs has shown the best binding affinity with relatively high binding percentage and the lowest RMSD value. Istradefylline shows the lowest binding percentage and the highest RMSD value with regardless to Remdesivir. The interactions of these drugs and other 3CPs with CLpro including van der Waal’s in yellow stripes and hydrogen bonds blue stripes are depicted in Figure 1. Linagliptin and istradefylline were approved in 2011 and 2019 to remediate type II diabetes and Parkinson’s disease, respectively (Deacon & Holst, 2010; Hauser et al., 2003). Dyphylline, caffeine and theophylline may be of particular importance as they are used to treat respiratory diseases such as asthma, cardiac dyspnea and bronchitis (Aranda, et al., 1977; Cooper, et al., 2004; El-said & Hashem, 1991; Furukawa, et al., 1983; Holbert et al., 1955; Jilani et al., 2019; Msimanga et al., 1997; Muir & McGuirk, 1987; Speer et al., 2017). The binding affinity of dyphylline was followed by caffeine, bromotheophylline, pentoxifylline and theophylline. These medications have shown a tendency to form a hydrogen bond with the active residues of 3CLpro (Table 1).
Figure 1.
Crystal structure of 3-chymotrypsin-like protease (3CLpro) of SARS-CoV-2 (PDB ID: 6Y2E) and ligands docked with 3CLpro and its contact sites with HIS41, CYS145 and GLU166. (a) Remdesivir; (b) linagliptin; (c) istradefylline; (d) dyphylline; (e) caffeine; (f) bromotheophylline; (g) pentoxifylline; (h) theophylline. Hydrocarbon skeleton cyan, nitrogen atoms are blue, oxygens red.
3.2. MD simulation
The RMSD, with reference to the initial structure, was computed along the trajectories as it can be seen in Figure 2. It could be observed that the ligands significantly affected the equilibration states of 3CLpro, as the majority of the tested systems reached their equilibrium after ∼160 ns. The average values of the RMSDs lie between 0.33 and 0.39 Å throughout the simulation (Table 2), elucidating the favorable behavior of 3CPs to form stable complexes with 3CLpro.
Figure 2.
The RMSD values of the simulated complexes of 3CLpro and ligands throughout the 200 ns of production runs.
Table 2.
Maximum and mean values of RMSD and RMSF for simulation trajectories throughout the 200 ns of production runs.
| CLpro complex type | RMSD |
RMSF |
||
|---|---|---|---|---|
| Max | Mean | Max | Mean | |
| Remdesivir | 0.51 | 0.39 | 0.53 | 0.15 |
| Linagliptin | 0.49 | 0.39 | 0.45 | 0.15 |
| Istradefylline | 0.55 | 0.39 | 0.84 | 0.17 |
| Dyphylline | 0.58 | 0.41 | 0.86 | 0.16 |
| Caffeine | 0.56 | 0.41 | 0.63 | 0.16 |
| Bromotheophylline | 0.52 | 0.38 | 0.57 | 0.16 |
| Pentoxifylline | 0.51 | 0.39 | 0.57 | 0.15 |
| Theophylline | 0.47 | 0.33 | 0.38 | 0.14 |
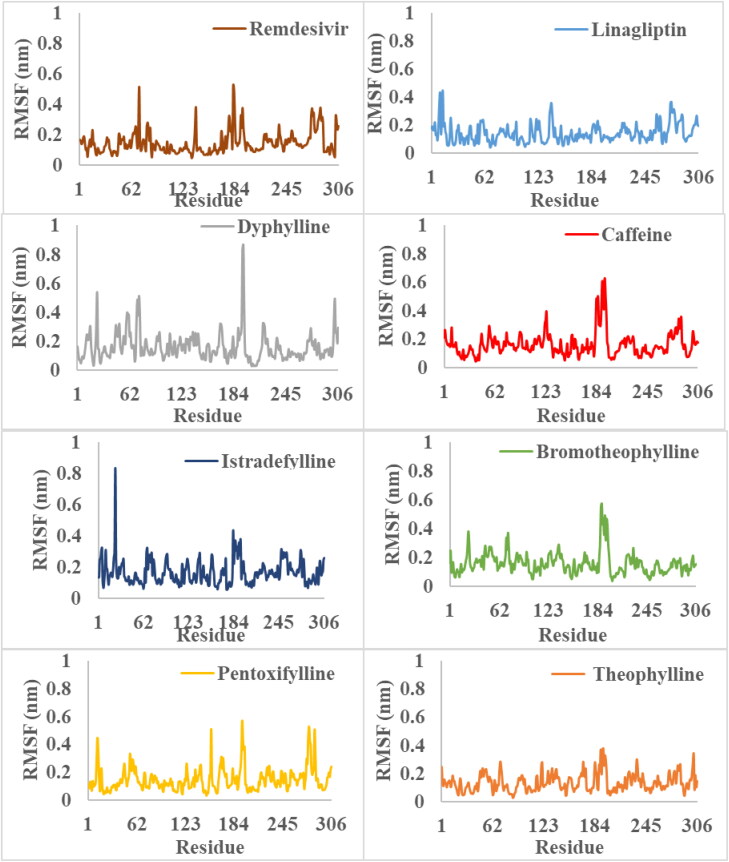
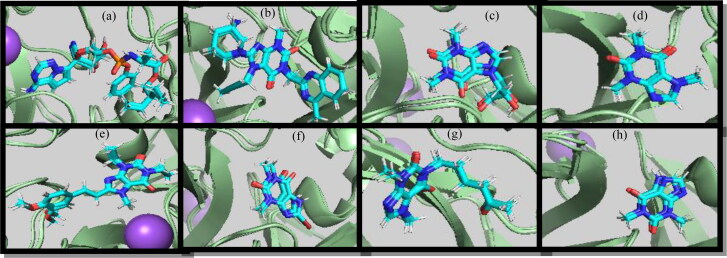
Furthermore, the RMSF was performed to analyze the fluctuations of the backbone residues of 3CLpro structure (Figure 3). The RMSF values of 3CLpro/3CP showed relatively similar fluctuations (Table 2). The fluctuations in the active site regions of HIS41, CYS145 and GLU166 were minor, indicating that the flexibility of these regions was lost by binding to the 3CP. Linagliptin and theophylline showed the lowest RMSF values, which indicates the greater stability of the complex formed. In general, the RMSF values elucidate the active site residues remained in stable conformation during the simulation (Figure 4), and therefore, the uses of 3CP as 3CLpro inhibitors are recommended.
Figure 3.
The RMSF values of the simulated complexes of 3CLpro and ligands throughout the 200 ns of production runs.
Figure 4.
(a) Remdesivir, (b) linagliptin, (c) dyphylline, (d) caffeine, (e) istradefylline, (f) bromotheophylline, (g) pentoxifylline, and (h) theophylline at 1 ns, 133 ns, and 200 ns throughout the production runs.
3.3. The binding free energies
The values of binding free energies of 3CPs as 3CLpro inhibitors are tabulated in Table 2. The study is specifying van der Waals interactions as the driving force for the inhibitors to bind to 3CLpro. The complex formation is favorable for electrostatic interactions. Whereas, the solvation energies show disfavorable complexation in both the case of MMPBSA and MMGBSA. The calculated MMGBSA binding free energies are better than the corresponding MMPBSA. The overall results suggest that the Linagliptin is the more potent inhibitor against 3CLpro followed by caffeine and dyphylline compared with other 3CPs (Table 3).
Table 3.
The MMPBSA and MMGBSA terms for the binding of 3CPs to 3CLPro of SARS-CoV-2.
| CLpro complex type | Evdw | Eelec | Esol (gb) | Esol (pb) | –TΔS | ΔGbind,(gb) (kcal/mol) | ΔGbind,(pb) (kcal/mol) |
|---|---|---|---|---|---|---|---|
| Remdesivir | –52.06 ± 0.01 | –46.89 ± 0.06 | 41.30 ± 0.04 | 50.63 ± 0.22 | 25.08 | –32.57 ± 0.03 | –23.25 ± 0.26 |
| Linagliptin | –66.39 ± 0.10 | 0.00 ± 0.0 | 10.05 ± 0.03 | 24.90 ± 0.10 | 24.51 | –31.83 ± 0.07 | –16.98 ± 0.18 |
| Istradefylline | –46.48 ± 0.08 | –13.78 ± 0.18 | 22.73 ± 0.03 | 35.55 ± 0.17 | 24.00 | –13.53 ± 0.09 | –0.71 ± 0.16 |
| Dyphylline | –28.96 ± 0.04 | –24.16 ± 0.09 | 22.68 ± 0.03 | 26.26 ± 0.09 | 22.49 | –7.95 ± 0.08 | –4.37 ± 0.12 |
| Caffeine | –30.38 ± 0.01 | –17.38 ± 0.01 | 17.11 ± 0.01 | 21.29 ± 0.08 | 21.73 | –8.91 ± 0.02 | –4.73 ± 0.07 |
| Bromotheophylline | –30.59 ± 0.04 | –12.44 ± 0.03 | 17.85 ± 0.01 | 23.69 ± 0.19 | 22.43 | –2.75 ± 0.01 | 3.09 ± 0.20 |
| Pentoxifylline | –30.95 ± 0.01 | –23.40 ± 0.02 | 28.45 ± 0.01 | 31.08 ± 0.09 | 22.98 | –2.92 ± 0.02 | –0.29 ± 0.11 |
| Theophylline | –22.59 ± 0.02 | –25.78 ± 0.03 | 25.59 ± 0.01 | 27.84 ± 0.15 | 21.51 | –1.27 ± 0.03 | 0.98 ± 0.14 |
Perhaps the most interesting of these drugs are caffeine and theophylline for their high hydrophilicity properties because such drugs cross the cell’s biological membrane easily as well as when secreted outside the body upon completion of its job (Ndikuryayo et al., 2019). Furthermore, caffeine and theophylline are naturally available in many plant species such as cocoa beans, kola nuts, tea leaves and coffee beans (Petimar et al., 2019; Risner, 2008). Consequently, it is easy to make it available to a large number of patients, regardless of their whereabouts or living standards, which is difficult or impossible-like for some other medications such as Remdesivir. However, the in vitro experiments can determine more precisely which of these drugs are the most therapeutic and act to stop this pandemic COVID-19.
Lastly, the dipyrimidinone ring in the 3CPs has a high tendency to form hydrogen bonds with the active residues of Glu166 and Cys145 of CLpro with lower RMSDs. The 3CPs are small molecules, devoid of chiral centers and produced in large commercial quantities. This study suggests the repurposing of these drugs with a special focus on linagliptin, and caffeine for COVID-19 treatment after a suitable validation and clinical trials. To our knowledge, this study demonstrates for the first time very inexpensive drugs as 3CLpro inhibitors.
Glossary
Abbreviations
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- 3CLpro
3-chymotrypsin-like protease
- 3CPs
caffeine and caffeine-containing pharmaceuticals
- MMPBSA
molecular mechanics-Poisson–Boltzmann surface area
- MMGBSA
molecular mechanics-generalized-Born surface area
- RMSD
root mean square deviation
- RMSF
root mean square fluctuation
Disclosure statement
The author declares that he has no conflict of interest.
Data availability statement
The data will be available upon request.
References
- Ahmad, S., Abbasi, H. W., Shahid, S., Gul, S., & Abbasi, S. W. (in press). Molecular docking, simulation and MM-PBSA studies of Nigella sativa compounds: A computational quest to identify potential natural antiviral for COVID-19 treatment. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1775129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R., & Hilgenfeld, R. (2003). Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science (New York, N.Y.), 300(5626), 1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- Aranda, J. V., Gorman, W., Bergsteinsson, H., & Gunn, T. (1977). Efficacy of caffeine in treatment of apnea in the low-birth-weight infant. The Journal of Pediatrics, 90(3), 467–472. 10.1016/S0022-3476(77)80718-X [DOI] [PubMed] [Google Scholar]
- Beck, B. R., Shin, B., Choi, Y., Park, S., & Kang, K. (2020). Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Computational and Structural Biotechnology Journal, 18, 784–790. 10.1016/j.csbj.2020.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, D. A., Darden, T. A., Cheatham, T. E., III, Simmerling, C. L., Wang, J., Duke, R. E., Luo, R., Merz, K. M., Pearlman, D. A., Crowley, M., & Walker, R. C. (2006). AMBER 9 (p. 45). University of California. [Google Scholar]
- Chang, D., Lin, M., Wei, L., Xie, L., Zhu, G., Cruz, C. S. D., & Sharma, L. (2020). Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan, China. JAMA, 323(11), 1092–1093. 10.1001/jama.2020.1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.-C., Tung, Y.-A., Lee, K.-H., Chen, T.-F., Hsiao, Y.-C., Chang, H.-C., Hsieh, T.-T., Su, C.-H., Wang, S.-S., & Yu, J.-Y. (2020). Potential therapeutic agents for COVID-19 based on the analysis of protease and RNA polymerase docking. 10.20944/preprints202002.0242.v2
- Cooper, A., Mikhail, A., Lethbridge, M. W., Kemeny, D. M., & Macdougall, I. C. (2004). Pentoxifylline improves hemoglobin levels in patients with erythropoietin-resistant anemia in renal failure. Journal of the American Society of Nephrology, 15(7), 1877–1882. 10.1097/01.ASN.0000131523.17045.56 [DOI] [PubMed] [Google Scholar]
- Da Silva, A. W. S., & Vranken, W. F. (2012). ACPYPE – AnteChamber PYthon Parser interfacE. BMC Research Notes, 5(1), 367. 10.1186/1756-0500-5-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, S., Sarmah, S., Lyndem, S., & Singha Roy, A. (in press). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon, C. F., & Holst, J. J. (2010). Linagliptin, a xanthine-based dipeptidyl peptidase-4 inhibitor with an unusual profile for the treatment of type 2 diabetes. Expert Opin Investig Drugs, 19(1), 133–140. 10.1517/13543780903463862 [DOI] [PubMed] [Google Scholar]
- de Leeuw, S. W., Perram, J. W., & Smith, E. R. (1980). Simulation of electrostatic systems in periodic boundary conditions. I. Lattice sums and dielectric constants. Proceedings of the Royal Society of London A: Mathematical and Physical Sciences, 373(1752), 27–56. 10.1098/rspa.1980.0135 [Google Scholar]
- Duan, Y., Zhu, H.-L., & Zhou, C. (2020). Advance of promising targets and agents against COVID-19 in China. Drug Discovery Today, 25(5), 810–812. 10.1016/j.drudis.2020.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., & Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics (Oxford, England), 27(16), 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Said, Y., & Hashem, F. (1991). In-vitro evaluation of sustained-release dyphylline tablets. Drug Development and Industrial Pharmacy, 17(2), 281–293. 10.3109/03639049109043825 [Google Scholar]
- Elzupir, A. O. (2020). Inhibition of SARS-CoV-2 main protease 3CLpro by means of α-ketoamide and pyridone-containing pharmaceuticals using in silico molecular docking. Journal of Molecular Structure, 1222, 128878. 10.1016/j.molstruc.2020.128878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani, M., Hasaniazad, M., Faezi, S., Guklani, H., Davoodian, P., Ahmadi, N., Einakian, M. A., Karmostaji, A., & Ahmadi, K. (in press). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [Google Scholar]
- Furukawa, C. T., Shapiro, G. G., Pierson, W. E., & Bierman, C. W. (1983). Dyphylline versus theophylline: A double-blind comparative evaluation. Journal of Clinical Pharmacology, 23(10), 414–418. 10.1002/j.1552-4604.1983.tb01784.x [DOI] [PubMed] [Google Scholar]
- Hauser, R. A., Hubble, J. P., Truong, D. D., & Group, I. U.-S.; Istradefylline US-001 Study Group (2003). Randomized trial of the adenosine A(2A) receptor antagonist istradefylline in advanced PD. Neurology, 61(3), 297–303. 10.1212/01.wnl.0000081227.84197.0b [DOI] [PubMed] [Google Scholar]
- Herten-Crabb, A., & Davies, S. E. (2020). Why WHO needs a feminist economic agenda. The Lancet, 395(10229), 1018–1020. 10.1016/S0140-6736(20)30110-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann, D. L., & Shindo, N. (2020). COVID-19: What is next for public health? The Lancet, 395(10224), 542–545. 10.1016/S0140-6736(20)30374-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbert, J., Grote, I., & Smith, H. (1955). Some new soluble salts of 8-bromotheophylline. Journal of the American Pharmaceutical Association, 44(6), 355–357. 10.1002/jps.3030440614 [DOI] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Jilani, T. N., Preuss, C. V., & Sharma, S. (2019). Theophylline. In StatPearls [Internet ]. StatPearls Publishing. [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [Google Scholar]
- Lai, C.-C., Shih, T.-P., Ko, W.-C., Tang, H.-J., & Hsueh, P.-R. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): The epidemic and the challenges. International Journal of Antimicrobial Agents, 55(3), 105924. 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, J. A., Martinez, C., Kasavajhala, K., Wickstrom, L., Hauser, K. E., & Simmerling, C. (2015). ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. Journal of Chemical Theory and Computation, 11(8), 3696–3713. 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, B. R. III, McGee, T. D. Jr., Swails, J. M., Homeyer, N., Gohlke, H., & Roitberg, A. E. (2012). MMPBSA.py: An efficient program for end-state free energy calculations. Journal of Chemical Theory and Computation, 8(9), 3314–3321. 10.1021/ct300418h [DOI] [PubMed] [Google Scholar]
- Msimanga, H. Z., Charles, M. J., & Martin, N. W. (1997). Simultaneous determination of aspirin, salicylamide, and caffeine in pain relievers by target factor analysis. Journal of Chemical Education, 74(9), 1114. 10.1021/ed074p1114 [Google Scholar]
- Muir, W. W. III, & McGuirk, S. (1987). Cardiovascular drugs: Their pharmacology and use in horses. The Veterinary Clinics of North America. Equine Practice, 3(1), 37–57. 10.1016/S0749-0739(17)30690-9 [DOI] [PubMed] [Google Scholar]
- Nair, K. P. (in press). The caffeine, methylxanthines, and behavior linkages. In Food and human responses. 10.1007/978-3-030-35437-4_10 [Google Scholar]
- Ndikuryayo, F., Kang, W.-M., Wu, F.-X., Yang, W.-C., & Yang, G.-F. (2019). Hydrophobicity-oriented drug design (HODD) of new human 4-hydroxyphenylpyruvate dioxygenase inhibitors. European Journal of Medicinal Chemistry, 166, 22–31. 10.1016/j.ejmech.2019.01.032 [DOI] [PubMed] [Google Scholar]
- Nelson, M. T., Humphrey, W., Gursoy, A., Dalke, A., Kalé, L. V., Skeel, R. D., & Schulten, K. (1996). NAMD: A parallel, object-oriented molecular dynamics program. The International Journal of Supercomputer Applications and High Performance Computing, 10(4), 251–268. 10.1177/109434209601000401 [Google Scholar]
- Numata, J., Wan, M., & Knapp, E. W. (2007). Conformational entropy of biomolecules: Beyond the quasi-harmonic approximation. Genome Informatics, 18, 192–205. 10.11234/gi1990.18.192 [PubMed] [Google Scholar]
- Petimar, J., O’Reilly, É., Adami, H. O., Brandt, P. A., Buring, J., English, D. R., Freedman, D. M., Giles, G. G., Håkansson, N., Kurth, T., Larsson, S. C., Robien, K., Schouten, L. J., Weiderpass, E., Wolk, A., & Smith-Warner, S. A. (2019). Coffee, tea, and caffeine intake and amyotrophic lateral sclerosis mortality in a pooled analysis of eight prospective cohort studies. European Journal of Neurology, 26(3), 468–475. 10.1111/ene.13840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera – A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Phillips, J. C., Braun, R., Wang, W., Gumbart, J., Tajkhorshid, E., Villa, E., Chipot, C., Skeel, R. D., Kale, L., & Schulten, K. (2005). Scalable molecular dynamics with NAMD. Journal of Computational Chemistry, 26(16), 1781–1802. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risner, C. H. (2008). Simultaneous determination of theobromine,(+)-catechin, caffeine, and (−)-epicatechin in standard reference material baking chocolate 2384, cocoa, cocoa beans, and cocoa butter. Journal of Chromatographic Science, 46(10), 892–899. 10.1093/chromsci/46.10.892 [DOI] [PubMed] [Google Scholar]
- Roe, D. R., & Cheatham, T. E. III. (2013). PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. Journal of Chemical Theory and Computation, 9(7), 3084–3095. 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Rothan, H. A., & Byrareddy, S. N. (2020). The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. Journal of Autoimmunity, 109, 102433. 10.1016/j.jaut.2020.102433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino-Silva, R., Jardim, A. C. G., & Siqueira, W. L. (2020). Coronavirus COVID-19 impacts to dentistry and potential salivary diagnosis. Clinical Oral Investigations, 24(4), 1619–1621. 10.1007/s00784-020-03248-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereen, M. A., Khan, S., Kazmi, A., Bashir, N., & Siddique, R. (2020). COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research, 24, 91–98. 10.1016/j.jare.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sk, M. F., Roy, R., Jonniya, N. A., Poddar, S., & Kar, P. (in press). Elucidating biophysical basis of binding of inhibitors to SARS-CoV-2 main protease by using molecular dynamics simulations and free energy calculations. Journal of Biomolecular Structure and Dynamic. 10.1080/07391102.2020.1768149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer, E. M., Dowling, D. J., Ozog, L. S., Xu, J., Yang, J., Kennady, G., & Levy, O. (2017). Pentoxifylline inhibits TLR- and inflammasome-mediated in vitro inflammatory cytokine production in human blood with greater efficacy and potency in newborns. Pediatric Research, 81(5), 806–816. 10.1038/pr.2017.6 [DOI] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ul Qamar, M. T., Alqahtani, S. M., Alamri, M. A., & Chen, L.-L. (2020). Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. Journal of Pharmaceutical Analysis, 10(4), 313–319. 10.1016/j.jpha.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- US-Food & drug Administration. (2020). Fact sheet for health care providers: Emergency use authorization (EUA) of Remdesivir (GS-5734™). US-Food & Drug Administration. [Google Scholar]
- Velavan, T. P., & Meyer, C. G. (2020). The COVID-19 epidemic. Tropical Medicine & International Health, 25(3), 278–280. 10.1111/tmi.13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahedi, H. M., Ahmad, S., & Abbasi, S. W. (in press). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Wang, J. (2020). Fast identification of possible drug treatment of Coronavirus disease-19 (COVID-19) through computational drug repurposing study. Journal of Chemical Information and Modeling, 60(6), 3277–3286. 10.1021/acs.jcim.0c00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., & Case, D. A. (2004). Development and testing of a general amber force field. Journal of Computational Chemistry, 25(9), 1157–1174. 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2020). Coronavirus disease 2019 (COVID-19): Situation report (p. 67). World Health Organization. [Google Scholar]
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica. B, 10(5), 766–788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (in press). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W., China Novel Coronavirus Investigating and Research Team . (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data will be available upon request.