Abstract
COVID-19 and its causative organism SARS-CoV2 that emerged from Wuhan city, China have paralyzed the world. With no clinically approved drugs, the global health system is struggling to find an effective treatment measure. At this crucial juncture, screening of plant-derived compounds may be an effective strategy to combat COVID-19. The present study investigated the binding affinity of phytocompounds with 3-Chymotrypsin-like (3CLpro) and Papain-like proteases (PLpro) of SARS-CoV2 using in-silico techniques. A total of 32 anti-protease phytocompounds were investigated for the binding affinity to the proteins. Docking was performed in Autodock Vina. Pharmacophore descriptors of best ligands were studied using LigandScout. Molecular dynamics (MD) simulation of apo-protein and ligand-bound complexes was carried out in YASARA software. The druglikeness properties of phytocompounds were studied using ADMETlab. Out of 32 phytochemicals, amentoflavone and gallocatechin gallate showed the best binding affinity to 3CLpro (–9.4 kcal/mol) and PLpro (–8.8 kcal/mol). Phytochemicals such as savinin, theaflavin-3,3-digallate, and kazinol-A also showed strong affinity. MD simulation revealed ligand-induced conformational changes in the protein with decreased surface area and higher stability. The RMSD/F of proteins and ligands showed stability of the protein suggesting the effective binding of the ligand in both the proteins. Both amentoflavone and gallocatechin gallate possess promising druglikeness property. The present study thus suggests that Amentoflavone and Gallocatechin gallate may be potential inhibitors of 3CLpro and PLpro proteins and effective drug candidates for SARS-CoV2. However, the findings of in silico study need to be supported by in vivo studies to establish the exact mode of action.
Communicated by Ramaswamy H. Sarma
Keywords: Phytocompounds, 3CLpro, PLpro, SARS-CoV2, docking
1. Introduction
COVID-19 pandemic is a rapidly spreading dreadful disease caused by a novel coronavirus SARS-CoV2 with more than 24.02 million positive cases across the world till date (2:44 p.m. CEST, 27 August 2020, WHO). According to the latest WHO update, the global death rate of COVID-19 now stands at 3.42% (https://covid19.who.int/). SARS-CoV2 is a positive-sense single-stranded RNA virus belonging to the family Coronaviridae that originated from the Wuhan city of China in 2019 (Tang et al., 2020). It is considered to be the seventh human coronavirus (HCoV) under the same family after 229E, NL63, OC43, HKU1, SARS-CoV, and MERS-CoV (Benvenuto et al., 2020). Like SARS and MERS coronaviruses, SARS-CoV2 is a highly pathogenic and contagious virus that causes severe respiratory complications and even death (Cui et al., 2019). Sequence analysis reveals that all the human coronaviruses have a common origin (from bats?) except HKU1 and OC43 which are believed to be originated from rodents (Forni et al., 2017). The attack and pathogenicity of SARS-CoV2 progress with four transmission stages—asymptomatic, moderate, extreme, and clinical which starts at the lower respiratory tract followed by invasion of pulmonary epithelial cells and hijacking the entire host cell machinery. The most common symptoms of COVID-19 include cough, fever, malaise, gastrointestinal symptoms, loss of smell, sore throat, heart failure, and acute kidney injury (Gandhi et al., 2020).
Currently, there is no clinically approved drug/vaccine for the treatment of COVID-19 and SARS-CoV2. However, several clinical trials are in progress to find out possible therapies for COVID-19. Recently, hydroxychloroquine, remdesivir, and ivermectin have been positioned as a possible treatment for COVID-19 (Caly et al., 2020; Wang et al., 2020). Several researchers are looking at the repurposing of existing antiviral drugs to test the efficacy on SARS-CoV2 (Elfiky, 2020; Kandeel & Al-Nazawi, 2020). Meanwhile, it is also crucial to understand the pathogenic mechanism of SARS-CoV2 to develop effective drugs or vaccines against the virus. Based on the virus pathogenesis and clinical manifestations, several studies have put forwarded potential therapeutic drug targets of SARS-CoV2 (Omolo et al., 2020). Among the several drug targets, coronavirus 3CLpro and PLpro are considered to be crucial targets of COVID-19 management (Zhou et al., 2020). 3CLpro and PLpro are encoded by ORF-1 of virus genome which plays a major role in the processing of inactive polyproteins (pp1a and pp1ab) into active non-structural proteins (nsps). These nsps finally helps in the replication, generation, as well as infection of the virus (Zhou et al., 2019). Inhibition of these proteases can be crucial for further replication and infection of SARS-CoV2 (Wu et al., 2020). Recent studies revealed that 3CLpro and PLpro share 96% and 83% sequence identity (at protein level) between SARS-CoV and –CoV2 (Morse et al., 2020). However, no such distinct structural differences were found in the active-site pockets of the enzymes which therefore suggest that 3CLpro and PLpro could be a potential drug target in SARS-CoV2. Therefore, drugs or compounds that are used as protease inhibitors in SARS-CoV may also be investigated for its effectiveness against SARS-CoV2.
Plants have always been a rich source of bioactive compounds having tremendous medicinal values. Unlike synthetic drugs that are costly, and possess undesirable side-effects, phytocompounds are less expensive, readily available, and contain less side-effects. Several medicinal plants and isolated compounds have been established as anti-virus and protease inhibitors by many researchers (Liu et al., 2020; Park et al., 2013). Ryu et al. (2010) reported the SARS-CoV protease inhibitor property of apigeni and luteolin isolated from Tripterygium regelii. Similarly, major bioactive compound rhamnazin 3-O-rutinoside isolated from Sarcocornia fruticosa strongly inhibited the protease activity of Hepatitis-C virus (Hawas et al., 2019). Several phytochemicals of known and unknown biological functions have been investigated by many authors in a computer-based approach to find out lead compounds that can effectively inhibit therapeutic targets in SARS-CoV2 including 3CLpro and PLpro (Chojnacka et al., 2020; Vardhan & Sahoo, 2020). The present study investigated the binding affinity of anti-protease phytochemicals with 3-chymotrypsin- and papain-like proteases of SARS-CoV2 using molecular docking and molecular dynamics simulation approach. Furthermore, we also investigated the in silico druglikeness and ADMET properties of the phytochemicals.
2. Materials and methods
2.1. Selection of phytochemicals and ligand preparation
A total of 32 phytocompounds that are reported to have protease inhibitor property are collected from different pieces of literature. The name of the chemicals and the publication details are presented in Table 1. The sdf files of each chemical were downloaded from the Pubchem database (http://pubchem.ncbi.nlm.nih.gov/). The files were converted into pdb file using OpenBabel software (O'Boyle et al., 2011). Finally, chemicals were converted into pdbqt file using Autodock tool (Trott & Olson, 2010).
Table 1.
Anti-protease phytocompounds from different plants and target molecule.
| Name of the compound | Name of the plants | Virus | Target | References |
|---|---|---|---|---|
| Beta-sitosterol Sinigrin |
Isatis indigotica Fortune ex Lindl | SARS-CoV | Inhibits Mpro | (Lin et al., 2005) |
| Xanthoangelol E Xanthoangelol |
Angelica keiskei (Miq.) Koidz. | SARS-CoV | Inhibits Mpro and PLpro | (Park et al., 2016) |
| Savinin Betulinic acid |
Chamaecyparis obtusa var. formosana Hayata | SARS-CoV | Inhibits 3CLpro | (Wen et al., 2007) |
| Pristimerin Iguesterin |
Tripterygium regelii Sprague & Takeda | SARS-CoV | Inhibits 3CLpro | (Ryu et al., 2010) |
| Apigenin Biflavone Amentoflavone Luteolin |
Torreya nucifera (L.) Siebold & Zucc. | SARS-CoV | Inhibits 3CLpro | (Ryu et al., 2010) |
| Curcumin | Curcuma longa L. | SARS-CoV | Inhibits 3CLpro | (Wen et al., 2007) |
| Dihydrotanshinone-I Cryptotanshinone |
Salvia miltiorrhiza Bunge | SARS-CoV | Inhibits 3CL- and PLpro | (Park et al., 2012) |
| Broussochalcone A 4-Hydroxy-isolonchocarpin Papyriflavonol-A Kazinol-A |
Broussonetia papyrifera (L.) L'Her. ex Vent. | SARS-CoV | Inhibits 3CLpro | (Park et al., 2017) |
| Aurantiamide acetate | Artemisia annua Pall. | SARS-CoV | Inhibits 3CLpro | (Okebe et al., 2014) |
| Epigallocatechin Theaflavin-3,3′-digallate Gallocatechin gallate |
Camellia sinensis (L.) Kuntze | SARS-CoV | Inhibits 3CLpro | (Chen et al., 2005) |
| Baicalein | Scutellaria baicalensis Georgi | SARS-CoV2 | Inhibits 3CLpro | (Liu et al., 2020) |
| Hirsutinone | Alnus japonica (Thunb.) Steud. | SARS-CoV | Inhibits PLpro | (Park et al., 2012) |
| Ellagic acid | Lagerstroemia speciosa (L.) Pers. | HIV-1 | Inhibit proteases | (Nutan et al., 2013) |
| Isobavachalcone 4′-O-methylbavachalcone Psoralidin |
Psoralea corylifolia L. | SARS-COV | Inhibit PLpro | (Kim et al., 2014) |
| Cinnamic amide | Tribulus terrestris L. | SARS-COV | Inhibits PLpro | (Song et al., 2014) |
| 6,8-Diprenylgenistein | Erythrina senegalensis DC. | HIV-1 | Inhibits proteases | (Lee et al., 2009) |
| Rhamnazin 3-O-rutinoside | Sarcocornia fruticosa (L.) Moq. | Hepatitis C virus | inhibits proteases | (Hawas et al., 2019) |
2.2. Collection and preparation of proteins
Three-dimensional structures of the 3-Chymotrypsin-like protease (3CLpro) and Papain-like protease (PLpro) of SARS-CoV2 were downloaded from RCSB-PDB database (https://www.rcsb.org/). The pdb IDs of the crystal structures of proteases were 6M2N for 3CLpro and 7JN2 for PLpro. Both the protein structures were processed by removing the water and other hetatms. Next, polar hydrogens and Kollman charges were added to the pdb files, and finally converted into pdbqt file format using AutoDock Tools (Trott & Olson, 2010).
2.3. Molecular docking
After the compounds and enzymes were prepared docking was carried out using AutoDock Vina (Trott & Olson, 2010). The amino acid residues interacting with the co-crystallized ligands of both the 6M2N and 7JN2 were taken as the active site residues and accordingly docking grid parameters were set. The active pocket amino acids residues in 6M2N were His41, Cys44, Met49, Leu141, Asn142, Gly143, Ser144, Cys145, His164, Met165, Glu166, Asp187, and Gln189 while the active pocket of 7JN2 contains amino acid residues such as Leu162, Gly163, Asp164, Glu167, Pro247, Pro248, Tyr264, Tyr268, Gln269, Tyr273, and Thr301. The grid parameters were set as x, y, z size- and centre-coordinates: 40, 50, 72 and –32.939, –65.379, and 39.480 for 3CLpro and for PLpro the parameters were 48, 54, 42, and 51.130, 31.061, and 1.325 for x, y, z size- and centre-coordinate, respectively. The docking algorithm was carried out by keeping the default exhaustiveness at 8. After docking, the best poses scoring the lowest binding energy (kcal/mol) for both 3CLpro and PLpro were selected and visualize in Discovery Studio.
2.4. Molecular dynamics simulation
To validate the docking study and evaluate the change in protein conformation, molecular dynamics simulation was implemented. The YASARA dynamics commercial package (Krieger et al., 2003) was used to simulate the complex where AMBER14 force field was applied (Dickson et al., 2014). The water molecules were added and with 0.9% NaCl salt at 310 K temperature (Krieger et al., 2006). The periodic boundary condition was maintained where the Particle Mesh Ewald method was applied to calculate long range electrostatic interaction. A cubic simulation cell was created to simulate the protein which was 20Ǻ bigger than the protein. The Berendsen thermostat was used to control the temperature of the simulation system. The protein complex and apo structure was initially cleaned and minimized by steepest gradient approaches (5000 cycles). The normal simulation time was maintained with a time step of 1.25 fs (Krieger & Vriend, 2015). The simulation trajectory was saved for every 100ps to analyze the result. The simulation result was incorporated with the default script of YASARA. Finally, root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), solvent accessible surface area (SASA), and Hydrogen bonds (H-bonds) were analyzed (Islam et al., 2019; Khan et al., 2020; Mahmud et al., 2020a, 2020b). To have more correct result from molecular dynamics simulation, each complex were run thrice (n=3) and average result was used for analysis. To calculate the binding free energy, the Molecular Mechanics Poisson-Boltzmann Surface Area method was applied. The edited YASARA script was employed for this calculation. The following equation had been used for the calculation of MM-PBSA (Gilson & Honig, 1988).
Here, ΔGMM is the sum of van der Waals and electrostatic interaction, ΔGPB and ΔGSA is the polar and non-polar solvation energies, TΔS is the entropic contribution (Razzaghi-Asl et al., 2018). The 1000 trajectory files were considered for MM-PBSA calculation.
2.5. Pharmacophore modeling
Pharmacophore features of the two best binding compounds were determined by LigandScout software which demonstrated Structural Activity Relationship (Muchtaridi et al., 2017) and also suggested compound’s descriptors necessary for optimal molecular interactions with the biological targets. The optimized chemical structures (pdb) of all the compounds were loaded into Ligandscout working space and key pharmacophore features were determined including H-bond donor, H-bond acceptor, hydrophobic, aromatic, halogen bond donor, positively and negatively ionizable groups.
2.6. Analysis of druglikeness and ADMET properties
Top ten best binding phytochemicals with the 3CLpro and PLpro were selected and druglikeness properties were studied using SwissADME (Daina et al., 2017) and PubChem database. The druglikeness properties of selected drugs were evaluated based on Lipinski’s rule (Lipinski, 2004). Similarly, the absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of drugs were predicted using ADMETlab (Dong et al., 2018).
2.7. Statistical analysis
Statistical calculation were carried out in Microsoft excel and OriginPro softwares. Values are expressed in mean ± standard deviation.
3. Results
3.1. Phytocompounds selection
The search for drugs and vaccines for COVID-19 pandemic is the most urgent and challenging task for global researchers at this moment. In this drive of drug discovery, we have investigated 32 phytochemicals from 17 medicinal plants that are reported to have anti-protease property. Table 1 showed the name of the plants, isolated compounds, and the target proteins of the phytocompounds reported from different plants. All the selected phytocompounds are reported to possess in vitro protease inhibition properties. The compounds were found to have inhibitory activity against 3CLpro and PLpro enzymes of many viruses such as SARS-CoV, HIV, and Hepatitis C virus.
3.2. Molecular docking study
Three-dimensional structures of 3CLpro and PLpro have been retrieved from PDB database. Structurally, 3CLpro and PLpro contains 306 and 312 amino acid residues, respectively. The total numbers of atoms present in the protein structures were 4682 and 4898, respectively. The secondary structure of 3CLpro has 17.26% alpha helix and 25.01% beta strands and the protein secondary structure elements (SSE) were found to be 42.27%. Similarly, PLpro has 21.9% and 28.23% alpha helix and beta strand, respectively forming the total SSE 50.13%. The active site of the 3CLpro protein is found to be located at the cleft region between domain I and II region. Similarly, the active site of PLpro is found to be located at the groove region between the Thumb domain and Finger domain of the protein.
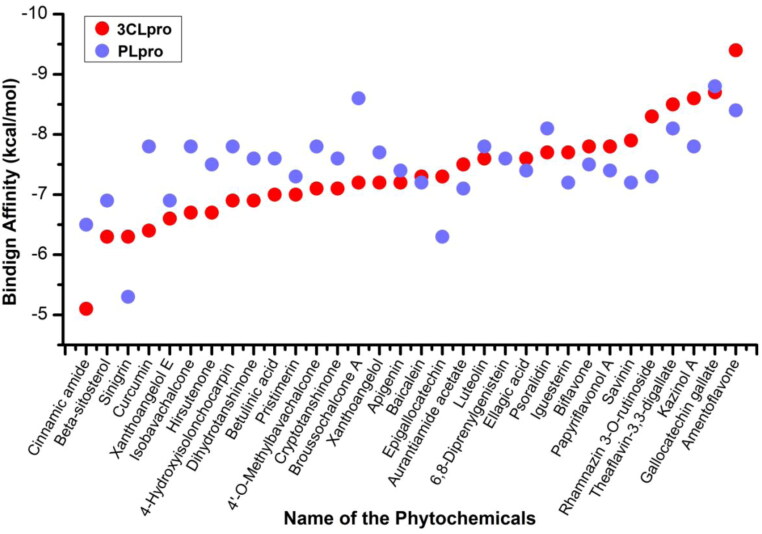
All the 32 phytocompounds were analyzed for the binding affinity with two key proteases of SARS-CoV2. Figure 1 showed the binding energies (kcal/mol) of all the 32 compounds with 3CLpro and PLpro proteins of the SARS-CoV2. In silico docking studies found that the binding affinities of anti-protease phytocompounds range from –5.1 to –9.4 kcal/mol in 3CLpro and –5.5 to –8.8 kcal/mol in PLpro proteins, respectively. The average binding energies were found to be –7.34 kcal/mol and –7.48 kcal/mol for 3CLpro and PLpro proteins, respectively. Half of the compounds were found to have –7.3 kcal/mol or higher binding affinity against 3CLpro while 18 compounds out of 32 were found to have stronger binding affinity than the average energy. Amentoflavone showed the strongest affinity to 3CLpro while cinnamic amide showed weakest binding energy. Similarly, gallocatechin gallate and sinigrin showed the strongest and weakest binding affinities to PLpro protein.
Figure 1.
Binding energies of anti-protease phytocompounds with 3-chymotrypsin-like and papain-like proteases of SARS-CoV2.
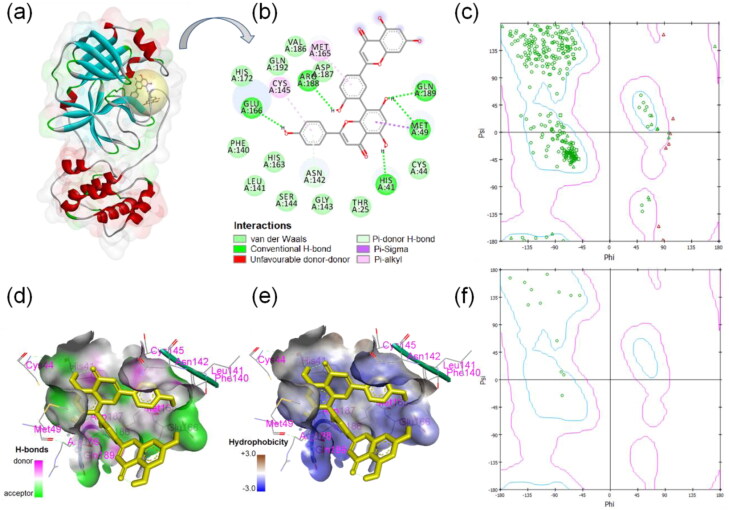
The two-dimensional display of binding interactions, ligand-protein interacting residues, hydrophobicity as well as Ramachandran plots are presented in Figure 2. The three-dimensional surface view structure with ligand binding sphere view shows that ligand amentoflavone binding site is in between the domain-I and -II region of the 3CLpro protein (Figure 2(a)). Thirteen amino acid residues of 3CLpro such as His41, Cys44, Met49, Phe140, Leu141, Asn142, Cys145, Met165, Glu166, Val186, Asp187, Arg188, and Gln189 were found to make interactions with the ligand. Out of 13 active site amino acid residues, 10 residues were found to make interaction with the ligand. Five H-bonds were formed between ligand and His41, Met49, Glu166, Arg188, and Gln189 amino acids of 3CLpro (Figure 2(b)). Figure 2(c) showed the amino acid distribution in the Ramachandran plot of complete 3Clpro protein while Figure 2(f) compares distribution of the amino acid residues that are interacting to the ligand only. Most of the amino acid residues (seven) that are interacting with the ligand amentoflavone were from coil-structure of 3CLpro while two amino acids each from helix, turn, and β-sheet structure that made the interaction with the ligand. Out of 13 ligand-interacting residues, six are strongly hydrophilic and remaining seven are hydrophobic in nature.
Figure 2.
Binding interactions of SARS-CoV2 3-chymotrypsin-like protease and amentoflavone. (a) Ligand binding sphere and surface view of protein, (b) two-dimensional display of ligand-3CLpro interactions, (c) Ramachandran plot of 3CLpro, (d) H-bond property of binding pocket, (e) hydrophobicity profile of binding pocket, and (f) Ramachandran plot of ligand-interacting amino acid residues.
Figure 3 showed the two-dimensional display of binding interactions, ligand-protein interactions, hydrophobicity, and Ramachandran plots of the ligand-interacting residues. The three-dimensional surface view structure with ligand binding sphere view showed that ligand gallocatechin gallate binding site lies between thumb domain and palm domain region of PLpro protein (Figure 3(a)). Fourteen amino acid residues of PLpro Asp164, Arg166, Glu167, Met208, Pro247, Pro248, Tyr264, Gly266, Asn267, Tyr268, Gln269, Cys270, Tyr273, and Thr301 were found to make interactions with the ligand. Out of 11 active site amino acid residues, nine residues were found to make interactions with the ligand. Five amino acids formed conventional H-bond with the ligand while other residues formed van der Waals interaction (11 residues), pi-bonds, and other non-covalent interactions (Figure 3(b)). Ramachandran plots displayed the amino acid distribution and conformation of amino acid residues in the plot (Figure 3(c,f)). Nine amino acids residues of PLpro are found to be distributed outside the allowed region of the plot. Out of 14 amino acid residues interacting with the ligand, 6 belongs to β-sheet structure, 4 to coil structure, and 2 each from turn and helix structure of the protein. Amino acid Gln269 formed unfavorable bond with the ligand and is place outside the allowed region in the Ramachandran plot (Figure 3(b,f)). The H-bond donor or acceptor residues as well as hydrophobicity property of the ligand-interacting amino acid residues are presented in Figure 3(d,e). The ligand surrounding amino acids are found to be entirely consist of hydrophilic residues except two amino acids Met209 and Cys270 that are hydrophobic in nature (Figure 4).
Figure 3.
Binding interactions of SARS-CoV2 Papain-like protease and gallocatechin gallate. (a) Ligand binding sphere and surface view of PLpro, (b) two-dimensional display of ligand-PLpro interactions, (c) Ramachandran plot of PLpro, (d) H-bond property of binding pocket, (e) hydrophobicity profile of binding pocket, and (f) Ramachandran plot of ligand-interacting amino acid residues.
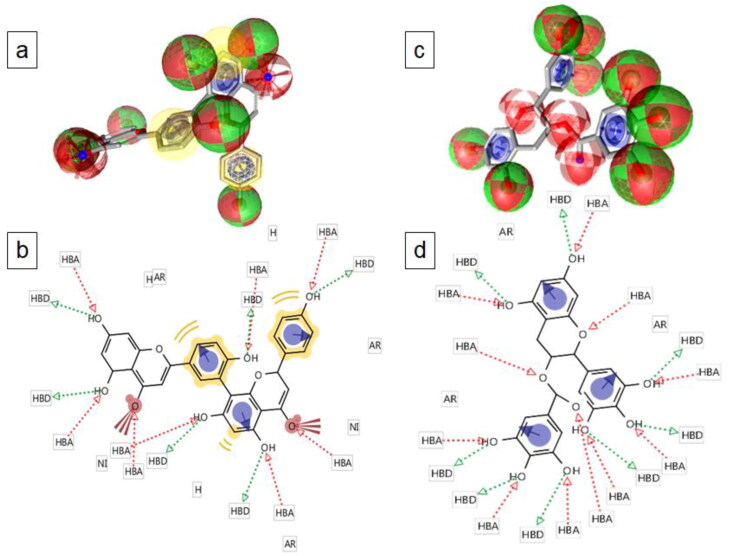
Figure 4.
Two-dimensional and three-dimensional display of pharmacophore feature of amentoflavone (a and b) and gallocatechin gallate (c and d). Green arrow, H-bond donor (HBD); red arrow, H-bond acceptor (HBA); yellow colour, hydrophobic group (H); blue, aromatic ring (AR); brown color with three spikes, non-ionising group (NI) of the compounds.
3.3. Molecular dynamics simulations
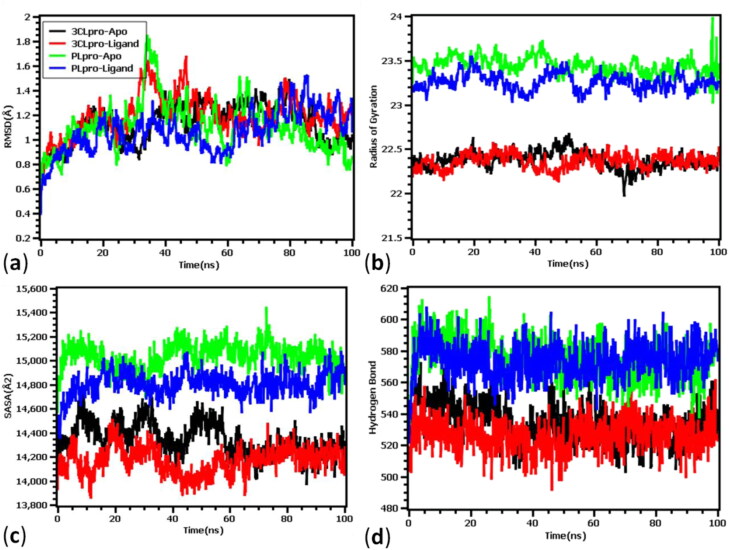
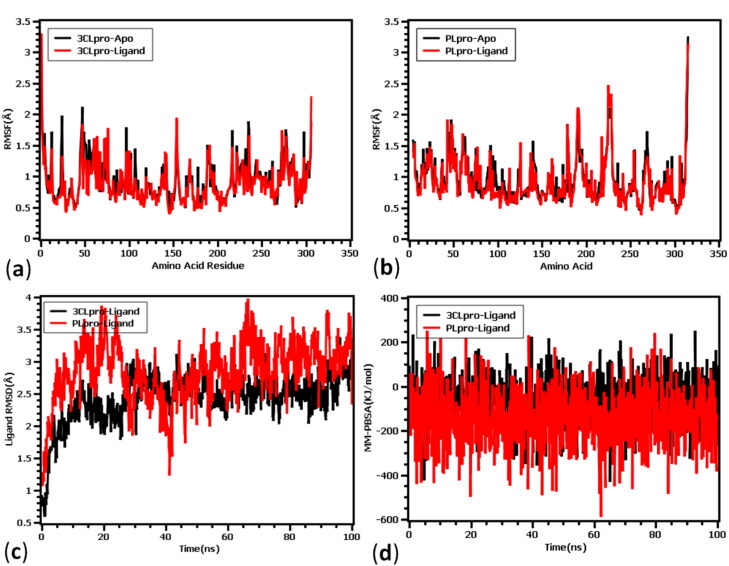
The RMSD of 3CLpro apo-protein (ligand-free) and ligand-bound complexes, Rg fluctuations, SARA values, and hydrogen bond profile during the period of simulation (100 ns) are presented in Figure 5. Figure 5(a) showed that the apo-protein and ligand bound 3CLpro and PLpro exhibit initial rise till 5 ns and thereafter showed stable nature. But after 40 ns all four complexes showed unstable due to the fluctuations in RMSD profile. Interestingly, the RMSD descriptors of the complexes never exceeded 2.5Ǻ value, which denotes the structural integrity of the proteins. The average RMSD descriptors of 3CLpro apo-protein and ligand-complex were found to be 1.13 ± 0.9 Å and 1.191 ± 0.2 Å, respectively. Similarly, the RMSD of apo and ligand-bound PLpro were found to be 1.11 ± 0.01 Å and 1.06 ± 0.02 Å, respectively. The MD simulation result from the 3CLpro and PLpro complex suggested that upon ligand binding no significant deviations or conformational changes were taken place in the protein structure. The radius of gyration of the protein complex denotes the degree of compactness and rigidness of the protein. Greater Rg value indicates higher flexibility and conformation of the protein whereas lower Rg value denotes more rigidity. Figure 5(b) showed that the 3CLpro protein has higher stability compared to PLpro. It has been observed that both the proteases have almost similar stability in apo-form as well as ligand bound-complex form. The average Rg values of 3CLpro were found to be 22.37 ± 0.90 Å and 22.36 ± 0.90 Å for apo and ligand bound complex. Similarly, Rg values were found to be 23.44 ± 0.30 Å and 23.25 ± 1.30 Å for apo- and ligand-bound PLpro protein, respectively. Figure 5(c) showed that the solvent-accessible surface areas (SARA) of 3CLpro and PLpro both in apo form and ligand-bound complex form. In both the proteases, ligand-bound complexes were found to have smaller SASA value compared to apo-form of the proteins. The average SASA values decreased from 14345.04 ± 0.20Ǻ2 in apo-form to 14168.25 ± 0.90Ǻ2 in ligand-bound complex of 3CLpro. However, after 60 ns both the apo and ligand-bound 3CLpro complex showed almost similar surface area. Like Rg, PLpro was found to have slightly bigger solvent accessible surface area than 3CLpro. The SASA values were found to be 15033.04±.02Ǻ2 and 14804.37 ± 0.9Ǻ2 for apo and ligand-bound structures of PLpro, respectively. Moreover, to understand the hydrogen bond and their contributions in gaining stability of the four systems, quantitative assessment of hydrogen bond was done. From Figure 5(d) it can be observed that, all four complexes did not deviate and similar number of hydrogen bond were found for apo-protein and ligand-bound complexes. The average numbers of hydrogen bonds were found to be 534.05 ± 0.1, 526.04 ± 0.9, 575.14 ± 0.30, and 573.77 ± 1.10 for 3CLpro apo, 3CLpro-Ligand, PLpro apo, and PLpro-ligand complexes, respectively. Upon binding of ligands to the proteins there were slight changes in the number of H-bonds.
Figure 5.
Molecular dynamics simulation of phytocompounds with 3CLpro and PLpro proteins of SARS-CoV2. (a) RMSD of apo- and ligand-bound 3CLpro and PLpro proteins, (b) Rg values, (c) fluctuations in the solvent accessibility surface area, and (d) nature of H-bonding during the period of simulation.
The fluctuations of amino acids of both the protease enzymes in apo- and ligand-bound forms, fluctuations of ligand-atoms, and binding energies during the period of simulations (100 ns) were presented in Figure 6. The root mean square fluctuation (RMSF) is useful for characterizing local changes along the protein chain. The amino acid residues Ser1 (gamma-turn), Glu55 (helix-strand), Ile59 (helix-strand), Gln244 (helix-strand), and Gln306 (gamma-turn) from SARS-Cov-2 3CLpro exhibited more fluctuations. While in case of PLpro protein, higher RMSF peaks were observed in His47 (beta-turn), Asn48 (beta-turn), Val66 (helix-strand), Leu113 (helix-strand), Lys190 (beta-turn), Lys218 (helix-strand), Lys228 (beta-turn), Gln229 (helix-strand), Cys270 (beta-turn), and Lys315 (helix-strand) amino acid residues. Overall, less flexible nature of residues stabilized both the protein-ligand complexes. Figure 6(c) showed the RMSDs both the ligands of 3CLpro and PLpro proteases. Ligand RMSD indicates how stable the ligand is with respect to the protein and its binding pocket. It was observed that there was a slight increase in fluctuation of both the ligands till 10 ns and reached almost stable state without much fluctuation. After 20 ns simulation time, RMSD profile reaches its stability and no instability was observed for 3CLpro complex. However, PLpro complex followed the similar trend at the initial phase but they stabilized after 40 ns simulation time. The comparative analysis between these two complexes suggested that 3CLpro complex has less ligand flexibility than the PLpro ligand. The binding free energy of the two docked complexes was calculated and the average binding energy was found to be –69.77 ± 1.10 kJ/mol and –142.56 ± 0.90 kJ/mol for 3CLpro and PLpro complexes, respectively. The 3CLpro complex had more favourable binding with the ligand and higher binding energy represents more tight binding. From Figure 6(d) it was observed that PLpro complex had similar MM-PBSA profile till 40 ns and some aberration were observed from 40 to 60 ns simulation time. However, during rest of the simulation period the complex, binding energy remained in similar pattern. On the other hand, 3CLpro complex had also some deviation in binding energy in 50 to 60 ns period and except that all simulation trajectory confirms its stability in MM-PBSA descriptor.
Figure 6.
(a) RMSD of amino acids of apo and ligand-bound 3CLpro, (b) RMSD of amino acids of apo and ligand-bound PLpro protein, (c) RMSD of ligand atoms and, (d) binding energies of protein-ligand complexes during the period of simulation (100 ns).
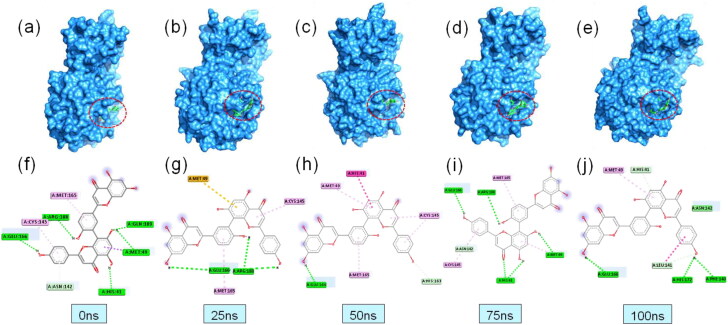
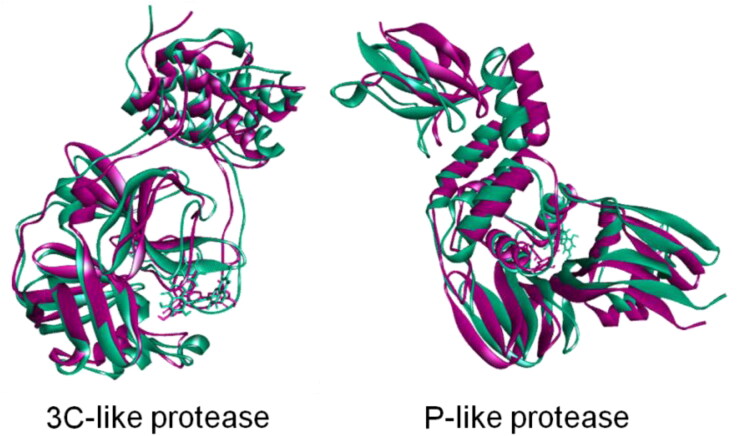
To understand the change and binding stability in the ligand-docked complexes, we also studied the protein-ligand interaction profile of the two complexes. Table 2 and Figures 7 and 8 showed the binding properties of protein-ligand complexes and the amino acid residues involved at different time intervals. Figures 7(a–e) and 8(a–e) showed the surface view of the topological changes of the protein-ligand complexes at 0, 25, 50, 75, and 100 ns. The study showed that binding of ligand amentoflavone to the 3CLpro protein is stabilized by 5, 2, 2, 5, and 5 hydrogen bonds at 0, 25, 50, 75, and 100 ns during simulation. The amino acid residues involved in the interactions, nature of bonding, and bond length were shown in Table 2 and Figures 7(f-j) and 8(f–j). Amino acid residue Glu166 was found to be a crucial residue forming H-bond with the ligand throughout the simulation period. Most of the residues were found to be repeated at different time intervals forming non-covalent interactions. After 100 ns of simulation, the amino acid residues involved in H-bond were found to be Glu166, Phe140, His172, Leu141 and His41 in 3CLpro protein. However, one amide-pi-stacked bond was observed at Asn142 and one pi-alkyl bond at Met49 position for 3CLpro complex. Interestingly, non-covalent interaction at the active groove of 3CLpro (His41, Leu141, Asn142, Met49, and Glu166) was observed after 100 ns simulation which correlates with possible inhibition of 3CLpro by the ligand molecules. However, at 100 ns simulation time, PLpro complex had two hydrogen bond at Glu167, Asp164, one electrostatic bond at Arg166, one Pi-pi-stacked bond at Tyr268, two pi-alkyl bond at Pro247 and Pro248 residue with gallocatechin gallate ligand. This PLpro complex also had non-bonded interaction at the active sites (Asp164, Pro248, and Tyr268) after dynamics simulation which is required for the inhibition of this protein. Moreover, complex from 25, 50, and 75 simulation trajectories also followed the similar trend binding pattern. Figure 9 showed the superimposition between pre-MD and post-MD structures of both the enzymes which showed structural changes with RMSD values 1.799 Å and 1.78 Å for PLpro and 3CLpro protein complexes, respectively. The RMSD profile from superimposition also confirmed the inflexibility of the docked complexes.
Table 2.
Different bonding interactions in 3CLpro and PLpro-ligand complex after 0, 25, 50, 75, and 100 ns of molecular dynamics simulation.
| Complex | Residue | Interaction type | Distance(Å) |
|---|---|---|---|
| 3CLpro-Ligand (0 ns) | Glu166 | Hydrogen bond | 2.37 |
| Arg188 | Hydrogen bond | 2.76 | |
| Gln189 | Hydrogen bond | 3.03 | |
| Met49 | Hydrogen bond | 3.77 | |
| His41 | Hydrogen bond | 2.37 | |
| Cys145 | Pi-Alkyl | 5.26 | |
| Met165 | Pi-Donor | 4.64 | |
| Asn142 | Pi-Sigma | 3.42 | |
| 3CLpro-Ligand (25 ns) | Arg188 | Hydrogen bond | 2.28 |
| Glu166 | Hydrogen bond | 1.94 | |
| Met49 | Pi-Sulfur | 5.56 | |
| Met165 | Pi-Alkyl | 4.95 | |
| Cys145 | Pi-Alkyl | 4.65 | |
| 3CLpro-Ligand (50 ns) | Glu166 | Hydrogen bond | 1.58 |
| His163 | Hydrogen bond | 2.73 | |
| His41 | Pi-Pi-T shape | 4.68 | |
| Met165 | Pi-Alkyl | 4.67 | |
| Cys145 | Pi-Alkyl | 5.3 | |
| Met49 | Pi-Alkyl | 5.29 | |
| 3CLpro-Ligand (75 ns) | His41 | Hydrogen bond | 2.72 |
| Arg188 | Hydrogen bond | 2.75 | |
| Met49 | Hydrogen bond | 2.98 | |
| Glu166 | Hydrogen bond | 2.37 | |
| Asn142 | Hydrogen bond | 2.51 | |
| Met165 | Pi-Alkyl | 4.63 | |
| Cys145 | Pi-Alkyl | 5.26 | |
| 3CLpro-Ligand (100 ns) | Glu166 | Hydrogen bond | 1.86 |
| Phe140 | Hydrogen bond | 3.01 | |
| His172 | Hydrogen bond | 1.86 | |
| Leu141 | Hydrogen bond | 2.73 | |
| His41 | Hydrogen bond | 2.75 | |
| Asn142 | Amide-Pi stacked | 3.83 | |
| Met49 | Pi-Alkyl | 5.44 | |
| PLpro-Ligand (0 ns) | Asp164 | Hydrogen bond | 1.99 |
| Arg166 | Hydrogen bond | 3.19 | |
| Glu167 | Hydrogen bond | 2.95 | |
| Tyr264 | Hydrogen bond | 2.29 | |
| Tyr273 | Hydrogen bond | 2.2 | |
| Pro247 | Pi-Alkyl | 5.33 | |
| Pro248 | Pi-Alkyl | 4.26 | |
| PLpro-Ligand (25 ns) | Arg166 | Hydrogen bond | 2.34 |
| Glu167 | Hydrogen bond | 2.95 | |
| Tyr264 | Hydrogen bond | 2.29 | |
| Asp164 | Hydrogen bond | 1.98 | |
| Tyr273 | Hydrogen bond | 2.19 | |
| Asn267 | Hydrogen bond | 2.67 | |
| Tyr268 | Hydrogen bond | 2.81 | |
| Pro247 | Alkyl | 4.26 | |
| Pro248 | Pi-Alkyl | 4.96 | |
| PLpro-Ligand (50 ns) | Arg166 | Hydrogen bond | 2.23 |
| Gln269 | Hydrogen bond | 2.5 | |
| Asp164 | Hydrogen bond | 1.58 | |
| Tyr273 | Hydrogen bond | 2.44 | |
| Asn267 | Hydrogen bond | 2.66 | |
| Tyr264 | Hydrogen bond | 4.35 | |
| Tyr268 | Pi-Pi Stacked | 5.93 | |
| Pro248 | Alkyl | 5.05 | |
| PLpro-Ligand (75 ns) | Glu167 | Hydrogen bond | 1.79 |
| Asp164 | Hydrogen bond | 1.94 | |
| Arg166 | Hydrogen bond | 3.98 | |
| Tyr268 | Pi-Pi Stacked | 4.6 | |
| Pro247 | Pi-Alkyl | 5.31 | |
| Pro248 | Pi-Alkyl | 5.06 | |
| PLpro-Ligand (100 ns) | Glu167 | Hydrogen bond | 1.79 |
| Asp164 | Hydrogen bond | 1.66 | |
| Arg166 | Electrostatic | 3.98 | |
| Tyr268 | Pi-Pi Stacked | 2.68 | |
| Pro247 | Pi-Alkyl | 5.31 | |
| Pro248 | Pi-Alky | 5.06 |
Figure 7.
Binding interactions between 3CLpro protein and amentoflavone complex at different time intervals of MD simulation (colour representations are same as Figure 2).
Figure 8.
Binding interactions between PLpro protein and gallocatechin gallate complex at different time intervals of MD simulation (colour representations are same as Figure 2).
Figure 9.
Superimpositions of pre-(pink) and post-MD simulations (green) of three-dimensional structures of 3CL-protease and PL-protease of SARS-CoV2 during.
3.4. Druglikeness and ADMET properties of phytocompounds
ADMET profile of a compound is an important property that predicts the druglikeness of a compound. Table 3 showed the various druglikeness properties of phytocompounds. According to Lipinski’s rule, a compound may be regarded to have druglikeness property if the molecular weight of a compound is ≤500 Da, LogP value ≤5, number of H-bond donor ≤5, and H-bond acceptor is ≤10. Table 2 showed that most of the compounds possess druglikeness property except few compounds such as rhamnazin-3-O-rutinoside and theaflavin-3,3-digallate which violated the Lipinski’s rule. The top binding ligands, amentoflavone and gallocatechin gallate showed violation in two parameters. Amentoflavone violated the rule in MW and HBD, while gallocatechin gallate violated in HBD and HBA. Topological polar surface areas of most of the compounds were below the 100Å2. Smaller the TPSA value higher the permeability of the molecule through the cell membrane, and therefore better distribution.
Table 3.
Druglikeness properties of phytocompounds.
| Phytochemicals | Pubchem CID | Molecular formula | MW (g/mol) | LogP < 5 | HBD < 5 | HBA < 10 | TPSA (Ų) |
|---|---|---|---|---|---|---|---|
| 4′-O-methylbavachalcone | 42607530 | C22H24O4 | 352.4 | 5.8 | 1 | 4 | 55.8 |
| 4-hydroxyisolonchocarpin | 5321800 | C20H18O4 | 322.4 | 4.5 | 2 | 4 | 66.8 |
| 6,8-Diprenylgenistein | 480783 | C25H26O5 | 406.5 | 6.5 | 3 | 5 | 87 |
| Amentoflavone | 5281600 | C30H18O10 | 538.5 | 5 | 6 | 10 | 174 |
| Apigeni | 5280443 | C15H10O5 | 270.24 | 1.7 | 3 | 5 | 87 |
| Aurantiamide acetate | 124319 | C27H28N2O4 | 444.5 | 4.4 | 2 | 4 | 84.5 |
| Baicalein | 5281605 | C15H10O5 | 270.24 | 1.7 | 3 | 5 | 87 |
| β-Sitosterol | 222284 | C29H50O | 414.7 | 9.3 | 1 | 1 | 20.2 |
| Sinigrin | 23682211 | C10H16KNO9S2 | 397.5 | 4 | 11 | 7 | 203 |
| Betulinic acid | 64971 | C30H48O3 | 456.7 | 8.2 | 2 | 3 | 57.5 |
| Biflavone | 9980790 | C30H18O4 | 442.5 | 6.1 | 0 | 4 | 52.6 |
| Broussochalcone A | 6438825 | C20H20O5 | 340.4 | 4.7 | 4 | 5 | 98 |
| Cinnamic amide | 5273472 | C9H9NO | 147.17 | 1.4 | 1 | 1 | 43.1 |
| Cryptotanshinone | 160254 | C19H20O | 296.4 | 3.8 | 0 | 3 | 43.4 |
| Curcumin | 969516 | C21H20O6 | 368.4 | 3.2 | 2 | 6 | 93.1 |
| Dihydrotanshinone | 5316743 | C18H14O3 | 278.3 | 3.2 | 0 | 3 | 43.4 |
| Ellagic acid | 5281855 | C14H6O8 | 302.19 | 1.10 | 4 | 8 | 141.34 |
| Epigallocatechin | 72277 | C15H14O7 | 306.27 | 0 | 6 | 7 | 130.61 |
| Gallocatechin gallate | 5276890 | C22H18O11 | 458.37 | 1.17 | 8 | 11 | 197.37 |
| Hirsutenone | 637394 | C19H20O5 | 328.36 | 3.09 | 4 | 5 | 97.99 |
| Iguesterin | 46881919 | C28H36O2 | 404.58 | 6.09 | 1 | 2 | 37.30 |
| Isobavachalcone | 5281255 | C20H20O4 | 324.37 | 5.10 | 3 | 4 | 77.76 |
| Kazinol-A | 442414 | C25H30O4 | 394.50 | 6.60 | 3 | 4 | 69.92 |
| Luteolin | 5280445 | C15H10O6 | 286.24 | 2.53 | 4 | 6 | 111.13 |
| Papyriflavonol A | 10343070 | C25H26O7 | 438.47 | 6.02 | 5 | 7 | 131.36 |
| Pristimerin | 159516 | C30H40O4 | 464.64 | 6.27 | 1 | 4 | 63.60 |
| Psoralidin | 5281806 | C20H16O5 | 336.34 | 4.69 | 2 | 5 | 83.81 |
| Rhamnazin 3-O-rutinoside | 5464499 | C29H34O16 | 638.57 | 0.32 | 8 | 16 | 247.43 |
| Savinin | 5281867 | C20H16O6 | 352.34 | 3.62 | 0 | 6 | 63.22 |
| Theaflavin-3,3-digallate | 135403795 | C43H32O20 | 868.70 | 4.71 | 13 | 20 | 351.12 |
| Xanthoangelol | 643007 | C21H22O6 | 370.40 | 4.43 | 3 | 6 | 96.22 |
| Xanthoangelol-E | 10022050 | C25H28O4 | 392.49 | 6.96 | 3 | 4 | 77.76 |
MW, molecular weight; HBD, hydrogen bond donor; HBA, hydrogen bond acceptor; TSPA, topological polar surface area.
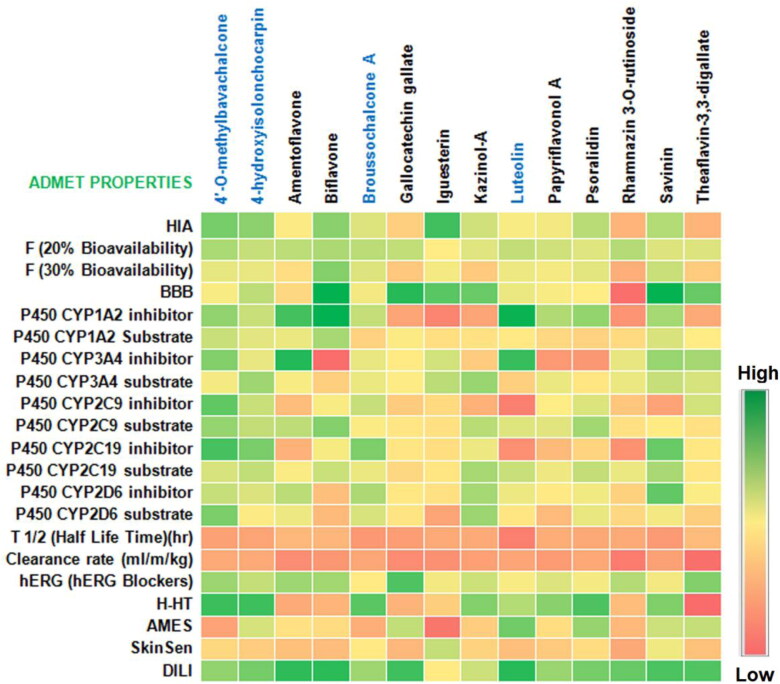
The heat map of the ADMET properties of the top ten best binding phytocompounds with 3CLpro and PLpro are presented in the Figure 10. It is observed from the study that almost all the phytocompounds have considerable ADMET properties. The absorption and permeability of all the top ten ligands showed moderate to high vales except few compounds such as rhamnazin-3-O-rutinoside and theaflavin-3-3-digallate which are predicted to have to permeability and absorption though human intestine. Amentoflavone showed low BBB property suggesting easy permeability and distribution to the brain cells while gallocatechin gallate showed high BBB value. Cytochrome-P450 is an important enzyme family necessary for detoxification and metabolism of drugs and toxic substances. Inhibition of this enzyme complex allows the drug molecules to reach the target site. 3CLpro top binding compound amentoflavone showed strong inhibitory property to Cyto-P450 enzyme family along with other phytocompounds. Gallocatechin gallate, on the other hand showed moderate cyto-P450 inhibitory property. The 3CLpro and PLpro best binding ligands were predicted to have low toxicity and carcinogenic property. Similarly, other phytocompounds are also predicted to have moderate to low toxicity activity.
Figure 10.
ADMET properties of the best-binding phytocompounds with 3CL protease and PL protease of SARS-CoV2. HIA, human intestinal absorption; BBB, blood-brain barrier; H-HT, human hepatotoxicity; AMES, Ames mutagenicity, SkinSen, skin sensitization; DILI, drug induced liver injury; blue colored chemicals; PL, protease docked compounds.
4. Discussion
The pandemic and severity of COVID-19 and its causative organism SARS-CoV2 have tremendously affected the health and wealth of the global community. Millions of lives have been lost so far and tens of millions are still undergoing treatment. With no clinically approved drugs, the current situation requires speedy investigation in the line of drug design to develop an effective drug to combat COVID-19. The molecular docking and molecular dynamics simulation are the most common and frequently used techniques to assess the interaction between ligand-protein complexes at the atomic level. By using these techniques novel inhibitors against disease-causing biological targets can be designed (Gogoi et al., 2019; Swargiary et al., 2020). In the present study, we have investigated the 3CLpro and PLpro inhibitory property of 32 phytocompounds reported to have anti-protease activity by several researchers. The study found that amentoflavone and gallocatechin gallate isolated from Torreya nucifera and Pichia pastoris possess strong binding affinity to 3CLpro and PLpro proteins, respectively.
Amentoflavone is a biflavonoid constituent of a number of plants including Torreya nucifera, Chamaecyparis obtusa, Ginkgo biloba, and many others (Pan et al., 2005; Ryu et al., 2010). The plant compound is known to possess several biological activities such as neuroprotector (Shin et al., 2006), anti-inflammatory activity (Banerjee et al., 2002), anticancer (Lee et al., 2013), and many others. Amentoflavone have also been reported by several researchers as inhibitor of viral protease enzyme (Sun et al., 1995; Jayadevappa et al., 2020). Furthermore, rhamnazin-3-O-rutinoside, theaflavin-3,3-digallate, and kazinol-A also showed good binding affinity to the target proteins. Several recent studies have reported many phytochemicals that are predicted to have considerable binding affinity to proteases and other target protein of SARS-CoV2 (Qamar et al., 2020; Pan et al., 2005). We have found that the highest binding energy of 3CLpro and PLpro was –9.4 kcal/mol and –8.8 kcal/mol with the top ligands. Similarly, most of the research findings showed that the binding energies of protein and ligand averages about –7.0 to –8.0 kcal/mol (Shin et al., 2006). In a similar study, in silico analysis by Strodel et al. (2020) revealed amentoflavone as one of the top SARS-CoV2 3Clpro (Mpro) binding ligand (–9.28 kcal/mol). The strong binding property of amentoflavone has been attributed to the presence of extended ring structures combined with functional group. It was also reported that one of the phenolic ring make H-bond with Ser46 residue of protein and 11 van der Waals interactions. However, in the present study we have found that all the five H-bonds (Glu166, Arg188, Gln189, Met49, and His41) were formed by the phenolic rings of the 3CLpro, and Ser46 did not form any contact with the protein. Similarly, Mishra et al. (2020) also revealed that amentoflavone made strongest interaction with the SARS-CoV2 main protease (–9.13 kcal/mol). In a 10 ns MD simulation they have reported that the peak RMSF was observed in amino acid residue-106, 181, 46 and 151 which is different from our findings. We observed that the amino acid residues Ser1, Glu55, Ile59, Gln244, and Gln306 from SARS-CoV2 3CLpro exhibited more fluctuations. Gallocatechin gallate is a derivative of catechin, a natural phenol having several medicinal values. In the present study gallocatechin gallate have been found to bind strongly with PLpro. Nguyen et al. (2012) also reported the inhibition property of epigallocatechin gallate and gallocatechin gallate against 3CLpro protein of SARS-CoV. Recent in silico study by Ghosh et al. (2020) revealed that gallocatechin gallate exhibited strong binding affinity to 3CLpro (–9.0 kcal/mol). In another study, a similar compound, epigallocatechin gallate showed strong binding interaction with PLpro (–13.91 kcal/mol). Amino acid residues His89, Lys92, Trp93, Trp106, Asp108, Cys155, Asn156, and Lys157 were found to involve in H-bonding with the ligand (Laskar & Choudhury, 2020). In the present study, gallocatechin gallate is found to make five H-bonds with PLpro pre-MD simulation.
It is also observed that the binding affinity of compounds depends upon the pharmacophore descriptors of the molecule. MD simulation studies have revealed that two best ligand molecules based on binding affinity showed more stability than the ligand-free protein structure. The higher RMSD values of apo structures revealed that, the 3CLpro and PLpro of SARS-CoV-2 have higher rigidity after the binding of the ligand suggesting the stability and effective binding of the ligands. Also, radius of gyration and solvent accessible surface area suggest the similar trend of the docked complex where two hit compounds exhibit less mobile nature and structural compactness. The tight packaging of the docked complex was illustrated and also no expansion of protein volume was maintained for both docked structure. Insilico druglikeness and ADMET properties are important parameters for the screening of the possible drug candidate. By using in silico tools it is now possible to predict the druglikeness of a compound based on its structure, physical, and chemical properties (Banerjee et al., 2002). Lipinski’s rule of five predicts whether a compound is orally absorbed or not and it depends on four main characteristics of a compound, molecular weight, lipophilicity, H-bond donor, and acceptor. According to the rule, a molecule will not be orally active if it violates two or more of the four rules (Lipinski, 2004). The present study found that the best binding ligands possess the considerable property of druglikeness. A good drug candidate must also possess other properties such as high absorption by gastrointestinal tract, easy distribution, metabolism, excretion or elimination, and less toxicity called ADMET profile (Lee et al., 2013). Similarly, the present study showed that amentoflavone and gallocatechin gallate possesses good ADMET properties suggesting the possibility of potential drug candidates against SARS-CoV2. The high absorbability by GI, high permeability through cell membrane, resistant to on route metabolism, and less cytotoxicity properties suggest therapeutic potentiality of the phytocompounds. Other phytocompounds such as iguesterin, psoralidin, rhamnazin 3-o-rutinoside, theaflavin-3,3-digallate, and kazinol-A also showed promising ADMET properties with less side effects.
5. Conclusion
Plants have been the source of healthcare needs since ancient times. In this crucial juncture of COVID-19 pandemic, a speedy approval of effective drug and medicine is indeed the need of the hour. Plants, being a rich source of phytocompounds may be an effective strategy to combat SARS-CoV2. The present study revealed the binding affinities of phytocompounds to the active sites of two crucial proteins 3-chymotrypsin- and papain-like proteases of SARS-CoV2. Phytocompounds amentoflavone and gallocatechin gallate not only bound strongly with the target proteins but also stabilized the three-dimensional conformations of the protein structures after binding. Furthermore, insilico druglikeness and ADMET profiling of the compounds showed promising therapeutic potential. The present study therefore may be taken as the basis for further studies and to finally establish the efficacy of the phytocompounds.
Acknowledgements
Authors thank Head, Department of Zoology, Bodoland University, India for providing necessary facility for carrying out this work. Authors also acknowledge the Head, Department of Genetic Engineering and Biotechnology, University of Rajshahi, Bangladesh, for providing necessary facilities.
Funding Statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure statement
Authors declare no conflict of interest.
References
- Banerjee, T., Valacchi, G., Ziboh, V. A., & van der Vliet, A. (2002). Inhibition of TNFα-induced cyclooxygenase-2 expression by amentoflavone through suppression of NF-κB activation in A549 cells. Molecular and Cellular Biochemistry, 238(1–2), 105–110. 10.1023/A:1019963222510 [DOI] [PubMed] [Google Scholar]
- Benvenuto, D., Giovanetti, M., Ciccozzi, A., Spoto, S., Angeletti, S., & Ciccozzi, M. (2020). The 2019-new Coronavirus epidemic: Evidence for virus evolution. Journal of Medical Virology, 92(4), 455–459. 10.1002/jmv.25688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., & Wagstaff, K. M. (2020). The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Research, 178 (2020), 104787. 10.1016/j.antiviral.2020.104787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.-N., Lin, C. P. C., Huang, K.-K., Chen, W.-C., Hsieh, H.-P., Liang, P.-H., & Hsu, J. T.-A. (2005). Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3,3'-digallate (TF3). Evidence-Based Complementary and Alternative Medicine: ECAM, 2(2), 209–215. 10.1093/ecam/neh081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chojnacka, K., Witek-Krowiak, A., Skrzypczak, D., Mikula, K., & Młynarz, P. (2020). Phytochemicals containing biologically active polyphenols as an effective agent against Covid-19-inducing coronavirus. Journal of Functional Foods, 73, 104146. 10.1016/j.jff.2020.104146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, J., Li, F., & Shi, Z. (2019). Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology, 17(3), 181–192. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717. 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, C. J., Madej, B. D., Skjevik, A. A., Betz, R. M., Teigen, K., Gould, I. R., & Walker, R. C. (2014). Lipid14: The amber lipid force field. Journal of Chemical Theory and Computation, 10(2), 865–879. 10.1021/ct4010307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMasi, J. A., Hansen, R. G., & Grabowski, H. G. (2003). The price of innovation: New estimates of drug development costs. Journal of Health Economics, 22(2), 151–185. 10.1016/S0167-6296(02)00126-1 [DOI] [PubMed] [Google Scholar]
- Dong, J., Wang, N.-N., Yao, Z.-J., Zhang, L., Cheng, Y., Ouyang, D., Lu, A.-P., & Cao, D.-S. (2018). ADMETlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. Journal of Cheminformatics, 10(1), 29. doi: 10.1155/2012/281078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sciences, 248, 117477. 10.1016/j.lfs.2020.117477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni, D., Cagliani, R., Clerici, M., & Sironi, M. (2017). Molecular evolution of human coronavirus genomes. Trends in Microbiology, 25(1), 35–48. 10.1016/j.tim.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi, R. T., Lynch, J. B., & Rio, C. (2020). Mild or moderate covid-19. New England Journal of Medicine. 10.1056/NEJMcp2009249. [DOI] [PubMed] [Google Scholar]
- Ghosh, R., Chakraborty, A., Biswas, A., & Chowdhuri, S. (2020). Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors – an in silico docking and molecular dynamics simulation study. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1779818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson, M. K., & Honig, B. (1988). Calculation of the total electrostatic energy of a macromolecular system: Solvation energies, binding energies, and conformational analysis. Proteins, 4(1), 7–18. 10.1002/prot.340040104 [DOI] [PubMed] [Google Scholar]
- Gogoi, A., Dutta, D., Verma, A. K., Nath, H., Frontera, A., Guha, A. K., & Bhattacharyya, M. K. (2019). Energetically favorable anti-electrostatic hydrogen bonded cationic clusters in Ni (II) 3,5-dimethylpyrazole complexes: Anticancer evaluation and theoretical studies. Polyhedron, 168, 113–126. 10.1016/j.poly.2019.04.043 [DOI] [Google Scholar]
- Guan, L., Yang, H., Cai, Y., Sun, L., Di, P., Li, W., Liu, G., & Tang, Y. (2019). ADMET-score - A comprehensive scoring function for evaluation of chemical drug-likeness. MedChemComm, 10(1), 148–157. 10.1039/c8md00472b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi, G. A., Ogunro, O. B., Adegunloye, A. P., Ogunyemi, O. M., & Afolabi, S. O. (2020). Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CLpro): An in silico screening of alkaloids and terpenoids from African medicinal plants. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1764868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawas, U. W., El-Kassem, L. T., Shaher, F., & Al-Farawati, R. (2019). In vitro inhibition of Hepatitis C virus protease and antioxidant by flavonoid glycosides from the Saudi costal plant Sarcocornia fruticosa. Natural Product Research, 33(23), 3364–3371. 10.1080/14786419.2018.1477153 [DOI] [PubMed] [Google Scholar]
- Islam, M. J., Parves, M. R., Mahmud, S., Tithi, F. A., & Reza, M. A. (2019). Assessment of structurally and functionally high-risk nsSNPs impacts on human bone morphogenetic protein receptor type IA (BMPR1A) by computational approach. Computational Biology and Chemistry, 80(2019), 31–45. 10.1016/j.compbiolchem.2019.03.00. [DOI] [PubMed] [Google Scholar]
- Jayadevappa, M. K., Karkera, P. R., Siddappa, R. Y., Telkar, S., & Karunakara, P. (2020). Investigation of plant flavonoids as potential dengue protease inhibitors. Journal of Herbmed Pharmacology, 9(4), 366–373. 10.34172/jhp.2020.46 [DOI] [Google Scholar]
- Kandeel, M., & Al-Nazawi, M. (2020). Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sciences, 251, 117627. 10.1016/j.lfs.2020.117627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M. A., Mahmud, S., Alam, A. S. M. R. U., Rahman, M. E., Ahmed, F., & Rahmatullah, M. (2020). Comparative molecular investigation of the potential inhibitors against SARS-CoV-2 main protease: A molecular docking study. Journal of Biomolecular Structure and Dynamics, 1-7. 10.1080/07391102.2020.1796813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. W., Seo, K. H., Curtis-Long, M. J., Oh, K. Y., Oh, J.-W., Cho, J. K., Lee, K. H., & Park, K. H. (2014). Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. Journal of Enzyme Inhibition and Medicinal Chemistry, 29(1), 59–63. 10.3109/14756366.2012.753591 [DOI] [PubMed] [Google Scholar]
- Krieger E., Elmar, G. V., & Spronk, C. (2003). YASARA – Yet another scientific artificial reality application. www.yasara.org
- Krieger, E., Nielsen, J. E., Spronk, C. A. E. M., & Vriend, G. (2006). Fast empirical pKa prediction by Ewald summation. Journal of Molecular Graphics & Modelling, 25(4), 481–486. 10.1016/j.jmgm.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Krieger, E., & Vriend, G. (2015). New ways to boost molecular dynamics simulations. Journal of Computational Chemistry, 36(13), 996–1007. 10.1002/jcc.23899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskar, M. A., & Choudhury, M. D. (2020). Search for therapeutics against COVID 19 targeting SARS-CoV-2 papain-like protease: An in silico study. ResearchSquare (Preprint). 10.21203/rs.3.rs-33294/v1. [DOI] [Google Scholar]
- Lee, J., Oh, W. K., Ahn, J. S., Kim, Y. H., Mbafor, J. T., Wandji, J., & Fomum, Z. T. (2009). Prenylisoflavonoids from Erythrina senegalensis as novel HIV-1 protease inhibitors. Planta Medica, 75(3), 268–270. 10.1055/s-0028-1088395 [DOI] [PubMed] [Google Scholar]
- Lee, J. S., Sul, J. Y., Park, J. B., Lee, M. S., Cha, E. Y., Song, I. S., Kim, J. R., & Chang, E. S. (2013). Fatty acid synthase inhibition by amentoflavone suppresses HER2/neu (erbB2) oncogene in SKBR3 human breast cancer cells. Phytotherapy Research: PTR, 27(5), 713–720. 0 10.1002/ptr.4778 [DOI] [PubMed] [Google Scholar]
- Lin, C.-W., Tsai, F.-J., Tsai, C.-H., Lai, C.-C., Wan, L., Ho, T.-Y., Hsieh, C.-C., & Chao, P.-D L. (2005). Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Research, 68(1), 36–42. 10.1016/j.antiviral.2005.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski, C. A. (2004). Lead- and drug-like compounds: The rule-of-five revolution. Drug Discovery Today Technologies, 1(4), 337–341. http:// 10.1016/j.ddtec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Liu, H., Ye, F., Sun, Q., Liang, H., Li, C., & Lu, R. (2020). Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in-vitro. BioRxiv. 10.1101/2020.04.10.035824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud, S., Parves, M. R., Riza, Y. M., Sujon, K. M., Ray, S., Tithi, F. A., Zaoti, Z. F., Alam, S., & Absar N. (2020. a). Exploring the potent inhibitors and binding modes of phospholipase A2 through in silico investigation. Journal of Biomolecular Structure and Dynamics, 38(14), 4221–4231. 10.1080/07391102.2019.1680440. [DOI] [PubMed] [Google Scholar]
- Mahmud, S., Uddin, M. A. R., Zaman, M., Sujon, K. M., Rahman, M. E., & Shehab, M. N. (2020. b). Molecular docking and dynamics study of natural compound for potential inhibition of main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1-9. 10.1080/07391102.2020.1796808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, A., Pathak, Y., Choudhir, G., Kumar, A., Mishra, S. K., & Tripathi, V. (2020). Natural compounds as potential inhibitors of novel coronavirus (COVID-19) main protease: An in silico study. ResearchSquare (Preprint). 10.21203/rs.3.rs-22839/v2. [DOI] [Google Scholar]
- Morse, J. S., Lalonde, T., Xu, S., & Liu, W. R. (2020). Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem: A European Journal of Chemical Biology, 21(5), 730–738. 10.1002/cbic.202000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchtaridi, M., Syahidah, H. N., Subarnas, A., Yusuf, M., Bryant, S. D., & Langer, T. (2017). Molecular Docking and 3d-pharmacophore modeling to study the interactions of chalcone derivatives with estrogen receptor alpha. Pharmaceuticals, 10(4), 81. 10.3390/ph10040081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T. T. H., Woo, H.-J., Kang, H.-K., Nguyen, V. D., Kim, Y.-M., Kim, D.-W., Ahn, S.-A., Xia, Y., & Kim, D. (2012). Flavonoid-mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnology Letters, 34(5), 831–838. 10.1007/s10529-011-0845-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutan, M. M., Goel, T., Das, T., Malik, S., & Suri, S. (2013). Ellagic acid & gallic acid from Lagerstroemia speciosa L. inhibit HIV-1 infection through inhibition of HIV-1 protease & reverse transcriptase activity. Indian Journal of Medical Research, 137, 540–548. [PMC free article] [PubMed] [Google Scholar]
- O'Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel: An open chemical toolbox. Journal of Cheminformatics, 1-14. 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okebe, J., Bojang, K., & D'Alessandro, U. (2014). Use of artemisinin and its derivatives for the treatment of malaria in children. The Pediatric Infectious Disease Journal, 33(5), 522–524. 10.1097/INF.0000000000000306 [DOI] [PubMed] [Google Scholar]
- Omolo, C. A., Soni, N., Fasiku, V. O., Mackraj, I., & Govender, T. (2020). Update on therapeutic approaches and emerging therapies for SARS-CoV-2 virus. European Journal of Pharmacology, 883, 173348. 10.1016/j.ejphar.2020.173348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., Tan, N., Zeng, G., Zhang, Y., & Jia, R. (2005). Amentoflavone and its derivatives as novel natural inhibitors of human Cathepsin B. Bioorganic & Medicinal Chemistry, 13(20), 5819–5825. 10.1016/j.bmc.2005.05.071 [DOI] [PubMed] [Google Scholar]
- Pandey, P., Rane, J. S., Chatterjee, A., Kumar, A., Khan, R., Prakash, A., & Ray, S. (2020). Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: An in silico study for drug development. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1796811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.-Y., Jeong, H. J., Kim, J. H., Kim, Y. M., Park, S.-J., Kim, D., Park, K. H., Lee, W. S., & Ryu, Y. B. (2012). Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biological & Pharmaceutical Bulletin, 35(11), 2036–2042. 10.1248/bpb.b12-00623 [DOI] [PubMed] [Google Scholar]
- Park, J.-Y., Kim, J. H., Kwon, J. M., Kwon, H.-J., Jeong, H. J., Kim, Y. M., Kim, D., Lee, W. S., & Ryu, Y. B. (2013). Dieckol, a SARS-CoV 3CL(pro) inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorganic & Medicinal Chemistry, 21(13), 3730–3737. 10.1016/j.bmc.2013.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.-Y., Ko, J.-A., Kim, D. W., Kim, Y. M., Kwon, H.-J., & Jeong, H. J. (2016). Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. Journal of Biomolecular Structure and Dynamics, 31. 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- Park, J.-Y., Yuk, H. J., Ryu, H. W., Lim, S. H., Kim, K. S., Park, K. H., Ryu, Y. B., & Lee, W. S. (2017). Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. Journal of Enzyme Inhibition and Medicinal Chemistry, 32(1), 504–512. 10.1080/14756366.2016.1265519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar, M. T., Alqahtani, S. M., Alamri, M. A., & Chen, L. L. (2020). Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. Journal of Pharmaceutical Analysis. 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzaghi-Asl, N., Mirzayi, S., Mahnam, K., & Sepehri, S. (2018). Identification of COX-2 inhibitors via structure-based virtual screening and molecular dynamics simulation. Journal of Molecular Graphics & Modelling, 83, 138–152. 10.1016/j.jmgm.2018.05.010 [DOI] [PubMed] [Google Scholar]
- Ryu, Y. B., Jeong, H. J., Kim, J. H., Kim, Y. M., Park, J.-Y., Kim, D., Nguyen, T. T. H., Park, S.-J., Chang, J. S., Park, K. H., Rho, M.-C., & Lee, W. S. (2010). Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorganic & Medicinal Chemistry, 18(22), 7940–7947. 10.1016/j.bmc.2010.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, Y. B., Park, S.-J., Kim, Y. M., Lee, J.-Y., Seo, W. D., Chang, J. S., Park, K. H., Rho, M.-C., & Lee, W. S. (2010). SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii. Bioorganic & Medicinal Chemistry Letters, 20(6), 1873–1876. 10.1016/j.bmcl.2010.01.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D. H., Bae, Y. C., Kim‐Han, J. S., Lee, J. H., Choi, I. Y., Ho, K., Sik Kang, S. S., Kim, W., & Han, B. Y. (2006). Polyphenol amentoflavone affords neuroprotection against neonatal hypoxic-ischemic brain damage via multiple mechanisms . Journal of Neurochemistry, 96(2), 561–572. 10.1111/j.1471-4159.2005.03582.x [DOI] [PubMed] [Google Scholar]
- Song, Y. H., Kim, D. W., Curtis-Long, M. J., Yuk, H. J., Wang, Y., Zhuang, N., Lee, K. H., Jeon, K. S., & Park, K. H. (2014). Papain-like protease (plpro) inhibitory effects of cinnamic amides from Tribulus terrestris fruits. Biological & Pharmaceutical Bulletin, 37(6), 1021–1028. 10.1248/bpb.b14-00026 [DOI] [PubMed] [Google Scholar]
- Strodel, B., Olubiyi, O., Olagunju, M., Keutmann, M., & Loschwitz, J. (2020). High throughput virtual screening to discover inhibitors of the main protease of the coronavirus SARS-CoV-2. Preprints. doi: 10.20944/preprints202004.0161.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, C. M., Yu, S. L., Ou, J. C., & Syu, W. J. (1995). Test of the biflavones from Selaginella moellendorffii on the in vitro inhibition of HIV-1 protease. Journal of Traditional Chinese Medicine, 6(3), 223–230. [Google Scholar]
- Swargiary, A., Verma, A. K., Singh, S., Roy, M. K., & Daimari, M. (2020). Antioxidant and antiproliferative activity of selected medicinal plants of lower Assam, India: An in vitro and in silico study. Anti-Cancer Agent in Medical Chemistry. 10.2174/1871520620666200719000449 [DOI] [PubMed] [Google Scholar]
- Tang, X., Wu, C., Li, X., Song, Y., Yao, X., Wu, X., Duan, Y., Zhang, H., Wang, Y., Qian, Z., Cui, J., & Lu, J. (2020). On the origin and continuing evolution of SARS-CoV-2. National Science Review, 7(6), 1012–1023. 10.1093/nsr/nwaa036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. http:// 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhan, S., & Sahoo, S. K. (2020). In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Computers in Biology and Medicine, 124, 103936. 10.1016/j.compbiomed.2020.103936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., Shi, Z., Hu, Z., Zhong, W., & Xiao, G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30(3), 269–271. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, C.-C., Kuo, Y.-H., Jan, J.-T., Liang, P.-H., Wang, S.-Y., Liu, H.-G., Lee, C.-K., Chang, S.-T., Kuo, C.-J., Lee, S.-S., Hou, C.-C., Hsiao, P.-W., Chien, S.-C., Shyur, L.-F., & Yang, N.-S. (2007). Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. Journal of Medicinal Chemistry, 50(17), 4087–4095. 10.1021/jm070295s [DOI] [PubMed] [Google Scholar]
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10(5), 766–788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H., Fang, Y., Xu, T., Ni, W., Shen, A., & Meng, X. (2020). Potential therapeutic targets and promising drugs for combating SARS-CoV-2. British Journal of Pharmacology, 177(14), 3147–3161. 10.1111/bph.15092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Fang, L., Yang, Z., Xu, S., Lv, M., Sun, Z., Chen, J., Wang, D., Gao, J., & Xiao, S. (2019). Identification of novel proteolytically inactive mutations in coronavirus 3C-like protease using a combined approach. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 33(12), 14575–14587. 10.1096/fj.201901624RR [DOI] [PubMed] [Google Scholar]