Abstract
A novel coronavirus (SARS-CoV2) has caused a major outbreak in humans around the globe, and it became a severe threat to human healthcare than all other infectious diseases. Researchers were urged to discover and test various approaches to control and prevent such a deadly disease. Considering the emergency and necessity, we screened reported antiviral compounds present in the traditional Indian medicinal plants for the inhibition of SARS-CoV2 main protease. In this study, we used molecular docking to screen 41 reported antiviral compounds that exist in Indian medicinal plants and shown amentoflavone from the plant Torreyanucifera with a higher docking score. Furthermore, we performed a 40 ns atomic molecular dynamics simulation and free binding energy calculations to explore the stability of the top five protein–ligand complexes. Through the article, we insist that the amentoflavone, hypericin and Torvoside H from the traditional Indian medicinal plants may be used as a potential inhibitor of SARS-CoV2 main protease and further biochemical experiments could shed light on understanding the mechanism of inhibition by these plant-derived antiviral compounds.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV2, main protease, medicinal plants, molecular docking and dynamics, drug design
Introduction
Infectious diseases caused by pathogens like parasites, fungi, bacteria or virus are one among the leading causes of mortality around the world. The recent outbreak of coronavirus disease (COVID-19) has shown the devastative effect on the humanity globally (World Health Organization, 2020). Currently, according to WHO the total COVID-19 confirmed cases have reached approximately 26.9 million and more than 880 thousand infected people died globally. Even though some countries are panning out this pandemic, several are still struggling to control the virus spreading. The first novel coronavirus (SARS-CoV2) was identified and reported in Wuhan city, China, at the end of December 2019 and on 30 January 2020 WHO declared a public health emergency of international concern (PHEIC) regarding the COVID-19 outbreak (Ji et al., 2020; Lee & Hsueh, 2020; Malik et al., 2020; Xu et al., 2020).
Human coronaviruses are predominantly concomitant with upper respiratory tract illnesses ranging from mild to moderate including common cold. Most of the people may be infected with one or more of these viruses at some point in their lifetime (Killerby et al., 2018). The SARS-CoV and MERS-CoV are the two major causes of severe pneumonia in humans (Song et al., 2019). The COVID-19 is known to show symptoms slowly over an incubation period of around 2 weeks. The virus replicates in the upper and lower respiratory tract, forming lesions (Chan, Yuan, et al., 2020). The general symptoms observed in the infected individuals are fever, cough, dyspnoea and lesion in the lungs (Huang et al., 2020). In the advanced stage, the symptoms of this virus show pneumonia which progresses to severe pneumonia and acute respiratory distress syndrome (ARDS) which results in the need for life-support to sustain the patient's life (Lai et al., 2020).
It has been reported that the SARS-CoV and SARS-CoV2 have similar kind of receptors, especially the receptor-binding domain (RBD) and the receptor-binding motif (RBM) in the viral genome (Tai et al., 2020; Yin & Wunderink, 2018; T. Zhang et al., 2020). During the SARS infection, the RBM of the S protein gets directly attached to the Angiotension-Converting Enzyme 2 (ACE2) in the human or the host cells (Nikhat & Fazil, 2020). The ACE2 protein is expressed in various organs of the human body mainly in the lungs, kidney and intestine, the prime targets of the coronavirus (Zhao et al., 2020). SARS-CoV-2 shares homology with the SARS-CoV (70% of genes), but the rate of transmission and infectivity of the SARS-CoV2 has been remarkable; this accelerated spreading rate may be due to a gain of function mutation, making this novel virus different from the SARS-CoV virus.
Several therapeutic strategies have been tested throughout the world to mitigate the infection, however, at least for now it is limited to prevention of the infection and further complications such as multi-organ failure through supportive therapies. Few preliminary studies have explored the therapeutic potential of opinavir/ritonavir: a protease inhibitor commonly used in HIV treatment and others have reported the use of remdesivir: a nucleoside analogue, nucleoside reverse transcriptase inhibitor (NRTI) such as lamivudine and tenofovir disoproxilfumarate, and neuraminidase inhibitors (NAIs) such as peramivir, zanamivir and oseltamivirfor treating COVID-19. Recently, FDA approved remdesivir to be used as an emergency drug for treating acute COVID-19 cases (Blanco et al., 2020; Chang et al., 2020; Gordon et al., 2020; Liao et al., 2020). SARS-CoV2 is genetically similar to SARS-CoV sharing around 70% of genes. Chymotrypsin-like protease (3CLpro)/main protease (Mpro) is an essential component for the replication of coronavirus and it is a potential target for designing anti-viral drugs against SARS-CoV2 (Anand et al., 2003).
Traditional Indian medicine use plants, minerals and animal products for curing human diseases. Traditional knowledge regarding the plant sources and their usage are essential to use them accurately and for the right condition (Tabuti et al., 2003). About 25,000 plant-based formulations have been used in folk remedies in Indian medicine (Adhikari & Paul, 2018). Owing to their use in traditional medicine, many plant molecules have been studied and subsequently modulated into drugs for various diseases (Bharadwaj et al., 2019; Dwivedi et al., 2020; Fabricant & Farnsworth, 2001; Li-Weber, 2009).
Plant-derived compounds such as phytochemicals possess immense therapeutic potential with a wide range of bioactivities including anti-microbial, anti-fungal, anti-viral, anti-inflammatory, and anti-cancer properties. The SARS-CoV2 main protease is considered as a promising drug target in this study, since it is dissimilar to human proteases and its crucial role in the viral replication cycle (Ullrich & Nitsche, 2020). With the knowledge gained on the structure and inhibitors of the main protease from previous epidemical coronaviruses, we strongly believe the main protease as one of the most attractive viral targets for antiviral drug discovery against SARS-CoV2. Hence, in the present study, we investigated potential anti-viral candidates found in Indian medicinal plants that may inhibit SARS-CoV2 main protease using molecular docking and molecular dynamics simulation approaches.
Material and methods
Protein preparation
The three-dimensional structure of SARS-CoV2 main protease was considered as a major therapeutic target for COVID-19 (Jin et al., 2020; Muralidharan et al., 2020; H. Zhang et al., 2020) and obtained from the Protein Data Bank (PDB-6Y2G) (Berman et al., 2000; L. Zhang et al., 2020). The protein chain is made up of 306 amino acid residues and experimentally solved by the X-ray diffraction method at 2.20Å resolution. Protein structure retrieved was minimized by adding missing residues and hydrogen bonds by Autodock tools (Morris, Ruth, et al., 2009). Active site identification remains a challenging task because of the presence of numerous cavities or pockets in a protein. In this case, the native cocrystallized ligand is used as a control for molecular docking experiments. To make a comparative study of the binding mode of main protease–ligand complexes, initially we performed the docking studies with the known binder ((∼{tert}-butyl ∼{N}-[1-[(2 ∼ {S})-3-cyclopropyl-1-oxidanylidene-1-[[(2 ∼ {S},3 ∼ {R})-3-oxidanyl-4-oxidanylidene-1-[(3 ∼ {S})-2-oxidanylidenepyrrolidin-3-yl] − 4- [(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate) (co-crystallized) with docking score −7.60 using autodock.
Ligand preparation
Plant-based antiviral compounds were manually identified from the literature carefully and retrieved their structure from PubChem database (Kim et al., 2019). Subsequently, all the compounds were converted to three-dimensional coordinates by using open babel molecular converter program and saved in PDB format (O’Boyle et al., 2011). Particularly, the native ligand and heteroatoms were removed from the receptor before docking, the compounds were optimized by adjusting rotamers preferences and energy minimization. The list of 41 antiviral compounds from Indian medicinal plants is shown in Table 1.
Table 1.
Predicted binding energy of plants derived antiviral compounds with SARS-CoV2 main protease.
| S. No | Compound name | Plant name | Family | Binding energy (kcal/mol) |
|---|---|---|---|---|
| 1 | Amentoflavone | Torreyanucifera | Taxaceae | −10.0 |
| 2 | Lectin | Momordiacharantia | Cucurbitaceae | −8.5 |
| 3 | glycyrrhizicacid | Glycyrrhizaglabra | Fabaceae | −8.5 |
| 4 | Hypericin | Hypericumperforatum | Hypericaceae | −8.4 |
| 5 | Torvoside H | Solanum torvum | Solanaceae | −8.4 |
| 6 | Galloylglucose 1 | Terminalia chebula | Combretaceae | −8.3 |
| 7 | Ocotillone | Ailanthus allisima | Simaroubaceae | −8.0 |
| 8 | Saikosaponin B2 | Bupleurum sp. | Apiaceae | −8.0 |
| 9 | Berberine | Berberisaristata | Berberidaceae | −7.9 |
| 10 | Camelliatannin H | Camellia japonica | Theaceae | −7.7 |
| 11 | Bryophyllin A | Kalanchoepinnata | Crassulaceae | −7.6 |
| 12 | lupeol | Strobilanthuscusia | Acanthaceae | −7.6 |
| 13 | Quercetin | Polygonumviscosum | Polygonaceae | −7.6 |
| 14 | Silymarin | Silybummarianum | Asteraceae | −7.6 |
| 15 | Myricetin | Cochlospermum religiosum | Bixaceae | −7.5 |
| 16 | Torvanol A | Solanum torvum | Solanaceae | −7.5 |
| 17 | Mimusopicacid | Mimusopselengli | Sapotaceae | −7.4 |
| 18 | Scopadulcic_acid | Scopariadulcis | Scrophulariaceae | −7.4 |
| 19 | Scutellarein | Oroxylum indicum | Bignoniaceae | −7.4 |
| 20 | Daphnoretin | Wickstroemiaindica | Thymelaceae | −7.3 |
| 21 | Apigenin | Ranunculus scleratus | Ranunculaceae | −7.1 |
| 22 | Iridoid | Barleriaprionitis | Acanthaceae | −7.1 |
| 23 | catechin | Ephedra sinica | Ephedraceae | −7.1 |
| 24 | lycorine | Hymenocallis littoralis | Amarylidaceae | −7.0 |
| 25 | Periglaucine | Pericampylusglaucus | Menispermaceae | −6.9 |
| 26 | andrographolide | Andrographispaniculata | Acanthaceae | −6.9 |
| 27 | Cepharadione B | Piper betel | Piperaceae | −6.8 |
| 28 | Ovatodiolide | Anisomelesindica | Lamiaceae | −6.8 |
| 29 | Paederoside | Paedariascandens | Rubiaceae | −6.8 |
| 30 | Catalpol | Picrorhizakurroa | Scrophulariaceae | −6.8 |
| 31 | Coclaurine | Nelumbonucifera | Nymphaceae | −6.6 |
| 32 | Curcumin | Curcuma longa | Zingiberaceae | −6.5 |
| 33 | Nuciferine | Nelumbonucifera | Nymphaceae | −6.4 |
| 34 | Rhuscholide A | Rhussinensis | Anacardiaceae | −6.3 |
| 35 | Odorinol | Aglaia roxburghiana | Meliaceae | −6.3 |
| 36 | Parthenolide | Taracetium vulgare | Asteraceae | −6.0 |
| 37 | Furomollugin | Rubiacardifolia | Rubiaceae | −5.9 |
| 38 | Illicinone-A | Illiciumverum | Illiaceae | −5.0 |
| 39 | Naphthoquinone | Rubiacardifolia | Rubiaceae | −4.9 |
| 40 | Piperitenone | Lippiajavanica | Verbenaceae | −4.3 |
| 41 | Trypsin | Milletia pinnata | Fabaceae | −3.0 |
Among the small molecules, amentoflavone found to show stand as the topmost based on the energy value.
Molecular docking of antiviral compounds from Indian medicinal plants
Molecular docking was carried out to predict the affinity between the plants derived antiviral compounds against the main protease of SARS-CoV2 using Auto Dock 4.2 program (Morris, Goodsell, et al., 2009). Polar hydrogen atoms were individually added and merged to the protein structure. Kollman charges and solvation parameters were determined by default. Gasteiger charges were added to the minimized ligand structures, and all bonds were made rotatable and flexible by allowing the detection of root torsion. Grid maps with grid spacing of 0.375 Å in the x, y and z-dimensions of 64 × 66 × 66 points were set to cover the entire protein. The Lamarckian Genetic Algorithm (LGA) was used to search for the lowest binding energy by implementing local minimization of the genetic algorithm, to enable modification of the gene population. LGA parameters were set as follows: 100 search (docking) runs; population size of 150; 25,000,000 of energy evaluations; 27,000 numbers of generations; a mutation rate of 0.02 and crossover rate of 0.8. Docking calculation was performed in the Auto Dock 4.2 software.
Molecular dynamics simulation of top-scoring main protease–amentoflavone complex
GROMACS 4.6 software was used to perform atomic molecular dynamics simulations with an all atom AMBER99SB force field (Case et al., 2005) for the top five protein–ligand complexes sorted based on docking scores. ACPYPE, which depends on Antechamber was used to generate ligand structural topology (Sousa Da Silva & Vranken, 2012). The protein–ligand complex system was solvated in a cubic box (size determined by keeping the closest protein atom to the box boundary at 1 nm) of TIP3P (transferable intermolecular potential with 3 points) water (Jorgensen et al., 1983) using minimization of the steepest descent algorithm. The net charge of the system was neutralized by adding counter ions. Charge interactions (electrostatic) were computed by the Particle Mesh Ewald method (Darden et al., 1993). After refining the complex by energy minimization, a 1-ns isothermal-isovolumetric ensemble simulation was carried out to equilibrate the water box with a force constant of 1000 kJ/(mol·Å) in x, y and z dimensions. A 1-ns NpT (isobaric-isothermic) ensemble simulation was used to equilibrate the water box in 1 atm pressure with a force constant of 1000 kJ/(mol·Å) in each dimension. Subsequently, another 40 ns NpT ensemble MD simulation was performed for production simulation with a fixed temperature of 308 K at 1 atm pressure. Anisotropic diagonal position scaling on time step interval of 0.002 ps was employed to maintain a constant pressure during MD simulations. Additionally, the Berendsen algorithm and Lennard-Jones cut-off value were fixed at 0.2 constant and 9 Å, respectively. Hydrogen bond formation was analyzed with Gromacs program modules. Analysis and plotting of results were carried out by using VMD and other standard inbuilt tools in Gromacs software (Humphrey et al., 1996). The trajectory files are saved appropriately for further computations.
Computation of binding free energy by MM-PBSA
The free binding energy of protein–ligand complex structures was computed from Molecular dynamics trajectories by the Molecular Mechanics/Poisson-Boltzmann Surface Area (MMPBSA) approach (Wang et al., 2018). The MM-PBSA free energy for all the top five protein–ligand complexes was predicted. The last 20 ns trajectory (20 frames from each nanosecond) of the 40 ns normal NPT MD simulation was adopted to calculate the binding free energy by the MM/PBSA method in the g_mmpsba tool (Kumari et al., 2014). The contribution of each residue toward the binding free energy was calculated and analyzed in detail.
Results and discussion
Molecular docking plays a vibrant role in deciphering lead compounds identification. The ligand-binding sites identified from the crystal structure were selected as flexible residues for molecular docking studies. The docking of 41 plants based antiviral compounds against the main protease was carried out using Auto Dock 4.2. Protein and small molecules were given input by loading as macromolecule and ligands. In order to continue docking, hetero atoms were removed from the main protease structure and Kollman united atom charges, solvation parameters and polar hydrogens were added. Gasteiger charges and non-polar hydrogens were assigned to ligands. The grid was set by choosing residues in the identified pockets over main protease. Grid values set were −10.764722, 12.509171 and 68.968991 for X, Y and Z axis, respectively. The Lamarckian genetic algorithm was preferred to examine the conformers in protein and maximum conformers were chosen as default poses for every compound during docking. Conformations (poses) of the protein–ligand complex were observed from lowest to highest binding free energy (ΔG) (Table 1). From the result of docking, conformational similarity, intermolecular energy and RMSD were observed. The output was clustered based on the root-mean-square deviation (RMSD) tolerance of 2.0 Å.
The binding energy of 41 plant-derived antiviral compounds was shown in Table 1. The compound amentoflavone from Torreyanucifera plant is shown to have the highest binding energy (−10.0 kcal/mol). Amentoflavone is a widely studied, naturally occurring polyphenolic biflavonoid found in many plants with wide bioactivities including anti-viral, anti-inflammatory, anti-cancer, anti-diabetic, anti-oxidant and anti-microbial properties (Chan, Yip, et al., 2020; Li et al., 2019). Selaginellabryopteris and Calophylluminophyllum are two other notable plants containing amentoflavone used in Indian traditional medicines. The top five compounds including amentoflavone such as lectin (Momordiacharantia), glycyrrhizicacid (Glycyrrhizaglabra), hypericin (Hypericumperforatum) and torvoside H (Solanum torvum) respectively are considered for further analysis.
The interactions like hydrogen, hydrophobic and other non-bonded terms between the top five compounds sorted by docking scores and main protease are visualized using Discovery Studio Visualizer software and was depicted in Figure 1. The interactions between protein and ligand are dominated by van der Waals hydrophobic interactions, which is indicated in light green color balls in the figure. The dark green color balls in the figure indicate conventional hydrogen bond contributions of amino acid residues. The red color ball in the figure indicates unfavorable hydrogen bonds. From the figure, it is observed that amentoflavone form the highest number of amino acid residue interactions (19 interactions) with the main protease than other four top compounds. The compounds lectin and hypericin form 16 amino acid residue interactions with main protease whereas other two compounds (glycyrrhizic acid and Torvoside H) form 18 and 17 amino acid residue interactions respectively. Interestingly, His41 contributes cation–pi interaction and Cys44 contributes disulphide bridge formation to form strong and stable interactions.
Figure 1.
Schematic 3D and 2D interaction diagram of top five compounds with SARS-CoV2 main protease. Dark green balls indicate van der Waals interactions whereas light green color indicates hydrogen bonds. Yellow and red color indicates cation-pi and unfavorable hydrogen bonds between protein and ligand [Amentoflavone (A), Lectin (B), glycyrrhizicacid (C), Hypricin (D) and Torvoside H (E)].
The drug-likeness prediction of 41 Indian medicinal plants derived antiviral compounds were performed by using the program available at http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp and tabulated in Supplementary Table 1. For the top five compounds, an important Lipinski filter high lipophilicity (expressed as LogP less than 5) is satisfied. The molecular mass and hydrogen bond donors/acceptors of glycyrrhizicacid and Torvoside H are high compared to the other three compounds. Overall, the drug-likeness prediction results indicate the amentoflavone and hypericin as a likely drug.
Atomic molecular dynamics simulations of the main protease–ligand complexes
Molecular dynamics simulations were carried out on the top five main protease–ligand complexes (Amentoflavone, Lectin, glycyrrhizicacid, Hypricin and Torvoside H) obtained from the molecular docking program. The dynamic stability of the protein–ligand complexes was estimated by observing RMSD changes during the simulation time as plotted in Figure 2. The fluctuation of protein complex structures was measured by balancing overall conformations during 40 ns simulations. The root-mean-square deviation (RMSD) of the main protease–amentoflavone complex along a 40-ns molecular dynamics simulation is highly stable which is evident from the average RMSD (<0.5 nm). The RMSD of the other ligands is relatively dynamic compared to the protein RMSD (between 0.5 and 1 nm), which is evident from the figure. In contrast, the RMSD of the main protease–amentoflavone complex with a fitting reference indicates that the ligand itself undergoes a small degree of conformation change during binding. The backbone RMSD of the protein is acceptable for all the top five complexes and provides insights into the order of conformational changes.
Figure 2.
The root mean square deviation of protein backbone and ligand (Amentoflavone (A), Lectin (B), glycyrrhizicacid (C), Hypricin (D) and Torvoside H (E)) for 40 ns simulation time is presented.
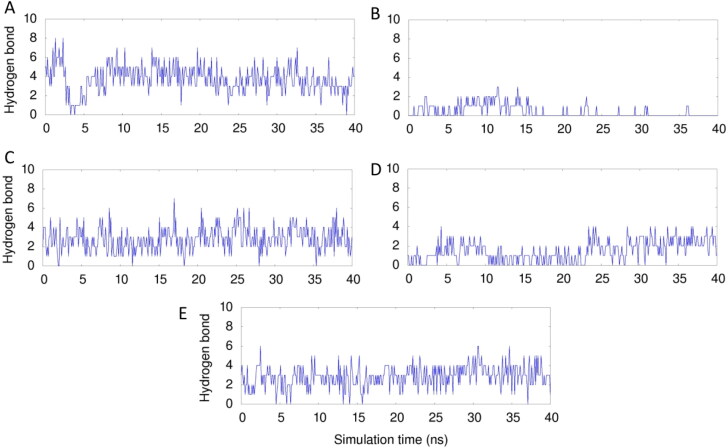
Protein–ligand interactions are typically stabilized by hydrogen, hydrophobic and electrostatic interactions. The formation of hydrogen bonds between protein and ligand for the 40 ns simulation time is shown in Figure 3. An average of 6 and 4 hydrogen bonds are formed by amentoflavone and hypericin respectively with the main protease during the simulation which is high compared to the other three complexes. Interestingly, lectin forms very less number of hydrogen bonds with the main protease. The hydrogen bond number increased in the later stage of simulation indicating that enhanced hydrogen bonding is possible to form a stable protein–ligand complex in the case of amentoflavone and hypericin complexes.
Figure 3.
The number of hydrogen bonds formed between main protease-top five ligands (Amentoflavone (A), Lectin (B), glycyrrhizicacid (C), Hypericin (D) and Torvoside H (E)) for 40 ns simulation time is presented.
Binding free energy of protein–ligand complexes
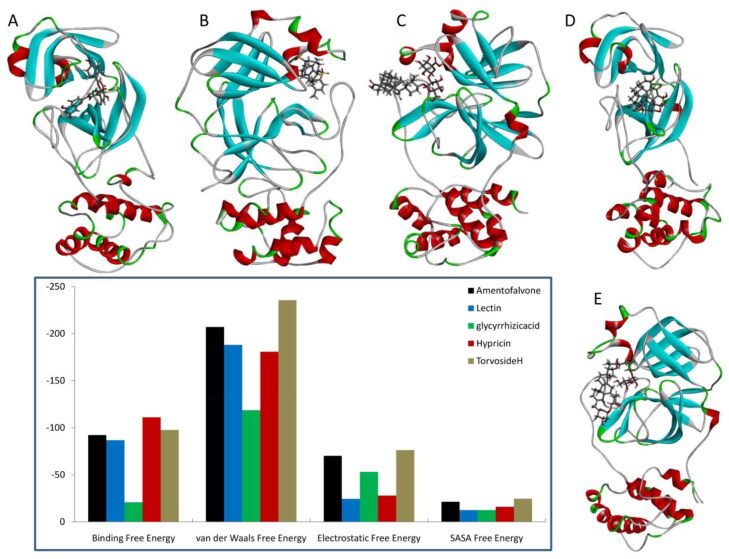
The computation of the binding free energy of protein and ligand complex by molecular mechanics—Poisson-Boltzmann surface area continuum salvation (MM-PBSA) is the most commonly used methods to support the results of molecular docking. The physical property of protein and ligand complexes were calculated using the MM_PBSA approach. The binding energies were predicted through the MD trajectories. The snapshot of the last frame during free energy calculations was extracted and presented in Figure 4. The free binding energy of protein with hypericin (−111.172 kJ/mol) and amentoflavone (−92.314 kJ/mol) are higher than the other three protein–ligand complexes which is presented in Figure 4. Interestingly, the main protease–glycyrrhizicacid complex has very low free binding energy (−20.903 kJ/mol). Other stabilizing physical free energies such as van der Waals, electrostatic and Solvent Accessible Surface Area (SASA) were also presented in Figure 4. The main protease–Torvoside H complex shows higher van der Waals (−235.71 kJ/mol), electrostatic (−76.324 kJ/mol) and SASA free energies (−24.658 kJ/mol). Through our observations, amentaflavone, hypericin and Torvoside H complexed with SARS-CoV2 main protease show better free energies predicted as evidenced from Figure 4. The importance and antiviral activity of the above three compounds is reviewed in the literature (Dhawan, 2012). The analysis of per residue-free energy of main protease suggests the importance of amino acid residues in ligand binding which is presented in Supplementary Figure 1. From the figure, it is observed that the hydrophobic residues (Leu141, Thr45 and Thr190), charged residues (Asn142 and Glu166) and Salt bridges (Cys44) forms stabilizing interactions between protein and ligands.
Figure 4.
The snapshots of last frame of top five complexes (Amentoflavone (A), Lectin (B), glycyrrhizicacid (C), Hypericin (D) and Torvoside H (E)) were presented. Free binding energy (kJ/mol) calculated using MMGBSA method for above top five protein–ligand complexes after molecular docking simulation.
Conclusion
In conclusion, we screened 41 plant-derived compounds from Indian medicinal plants that may inhibit SARS-CoV2 main protease using the molecular docking approach. In our molecular docking study, we found amentoflavone, lectin, glycyrrhizicacid, hypericin and torvoside H showed high binding energy. However, among them, amentoflavone from Torreyanucifera plant showed highest binding energy of −10.0 kcal/mol with the SARS-COV2 main protease and further In vitro and In Vivo studies are required to understand the underlying molecular mechanisms and signaling pathways involved in SARS-COV2 main protease inhibition. In the literature, there are reports based on insilico studies that the amentoflavone can act as a potential inhibitor of SARS-CoV2 (Mishra et al., 2020). Furthermore, we performed molecular dynamics simulations and free energy calculations on the top five compounds to validate our findings from the molecular docking procedure. A careful analysis of results suggests that the compounds such as amentaflavone, hypericin, and Torvoside H respectively complexed with SARS-CoV2 main protease show better binding with stabilizing interactions. Our results provide detailed protein–ligand interactions of five Indian medicinal plants derived antiviral compounds by a 40 ns atomic molecular dynamics simulations. We believe our research would provide valuable information in developing new, novel and natural anti-viral drugs for COVID-19.
Acknowledgements
HS and KKV would like to acknowledge the DST FIST facility for this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Adhikari, P. P., & Paul, S. B. (2018). History of Indian traditional medicine: A medical inheritance. Asian Journal of Pharmaceutical and Clinical Research, 11(1), 421. 10.22159/ajpcr.2018.v11i1.21893 [DOI] [Google Scholar]
- Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R., & Hilgenfeld, R. (2003). Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science (New York, N.Y.), 300(5626), 1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., & Bourne, P. E. (2000). The protein data bank. Nucleic Acids Research, 28(1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj, S., Lee, K. E., Dwivedi, V. D., Yadava, U., Panwar, A., Lucas, S. J., Pandey, A., & Kang, S. G. (2019). Discovery of Ganoderma lucidum triterpenoids as potential inhibitors against Dengue virus NS2B-NS3 protease. Scientific Reports, 9(1). 10.1038/s41598-019-55723-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco, J. L., Ambrosioni, J., Garcia, F., Martínez, E., Soriano, A., Mallolas, J., & Miro, J. M. (2020). COVID-19 in patients with HIV: Clinical case series. The Lancet HIV, 7(5), e314–e316. 10.1016/S2352-3018(20)30111-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, D. A., Cheatham, T. E., Darden, T., Gohlke, H., Luo, R., Merz, K. M., Onufriev, A., Simmerling, C., Wang, B., & Woods, R. J. (2005). The Amber biomolecular simulation programs. Journal of Computational Chemistry, 26(16), 1668–1688. 10.1002/jcc.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F.-W., Yuan, S., Kok, K.-H., To, K. K.-W., Chu, H., Yang, J., Xing, F., Liu, J., Yip, C. C.-Y., Poon, R. W.-S., Tsoi, H.-W., Lo, S. K.-F., Chan, K.-H., Poon, V. K.-M., Chan, W.-M., Ip, J. D., Cai, J.-P., Cheng, V. C.-C., Chen, H., Hui, C. K.-M., & Yuen, K.-Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. The Lancet, 395(10223), 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F. W., Yip, C. C. Y., To, K. K. W., Tang, T. H. C., Wong, S. C. Y., Leung, K. H., Fung, A. Y. F., Ng, A. C. K., Zou, Z., Tsoi, H. W., Choi, G. K. Y., Tam, A. R., Cheng, V. C. C., Chan, K. H., Tsang, O. T. Y., & Yuen, K. Y. (2020). Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. Journal of Clinical Microbiology, 58(5), e00310-20. 10.1128/JCM.00310-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y., Tung, Y., Lee, K., Chen, T., Hsiao, Y., Chang, C., Hsieh, T., Su, C., Wang, S., Yu, J., Lin, Y., Lin, Y., Tu, Y. E., & Tung, C. (2020). Potential therapeutic agents for COVID-19 based on the analysis of protease and RNA polymerase docking. Preprint. 10.20944/preprints202002.0242.v1 [DOI] [Google Scholar]
- Darden, T., York, D., & Pedersen, L. (1993). Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. The Journal of Chemical Physics, 98(12), 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- Dhawan, B. N. (2012). Anti-viral activity of Indian plants. Proceedings of the National Academy of Sciences India Section B – Biological Sciences, 82(1), 209–224. 10.1007/s40011-011-0016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi, V. D., Bharadwaj, S., Afroz, S., Khan, N., Ansari, M. A., Yadava, U., Tripathi, R. C., Tripathi, I. P., Mishra, S. K., & Kang, S. G. (2020). Anti-dengue infectivity evaluation of bioflavonoid from Azadirachta indica by dengue virus serine protease inhibition. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1734485 [DOI] [PubMed] [Google Scholar]
- Fabricant, D. S., & Farnsworth, N. R. (2001). The value of plants used in traditional medicine for drug discovery. Environmental Health Perspectives, 109 (Suppl 1), 69–75. 10.1289/ehp.01109s169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, C. J., Tchesnokov, E. P., Feng, J. Y., Porter, D. P., & Gotte, M. (2020). The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. The Journal of Biological Chemistry, 295(15), 4773–4779. 10.1074/jbc.AC120.013056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., Fan, C., Li, M., Nie, H. L., Wang, F. B., Wang, H., Wang, R., Xia, J., Zheng, X., Zuo, X., & Huang, J. (2020). COVID-19: A call for physical scientists and engineers. ACS Nano, 14(4), 3747–3754. 10.1021/acsnano.0c02618 [DOI] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Ji, W., Wang, W., Zhao, X., Zai, J., & Li, X. (2020). Cross-species transmission of the newly identified coronavirus 2019-nCoV. Journal of Medical Virology, 92(4), 433–440. 10.1002/jmv.25682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Zhao, Y., Sun, Y., Zhang, B., Wang, H., Wu, Y., Zhu, Y., Zhu, C., Hu, T., Du, X., Duan, Y., Yu, J., Yang, X., Yang, X., Liu, X., Guddat, L. W., Xiao, G., Zhang, L., Yang, H., & Rao, Z. (2020). Structural basis for the inhibition of COVID-19 virus main protease by carmofur, an antineoplastic drug. Nature Structural & Molecular Biology, 27, 529–532. https//doi.org/10.1038/s41594-020-0440-6 [DOI] [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Killerby, M. E., Biggs, H. M., Haynes, A., Dahl, R. M., Mustaquim, D., Gerber, S. I., & Watson, J. T. (2018). Human coronavirus circulation in the United States 2014–2017. Journal of Clinical Virology: The Official Publication of the Pan American Society for Clinical Virology, 101, 52–56. 10.1016/j.jcv.2018.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., Li, Q., Shoemaker, B. A., Thiessen, P. A., Yu, B., Zaslavsky, L., Zhang, J., & Bolton, E. E. (2019). PubChem 2019 update: Improved access to chemical data. Nucleic Acids Research, 47(D1), D1102–D1109. 10.1093/nar/gky1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari, R., Kumar, R., Consortium, O. S. D. D., & Lynn, A, Open Source Drug Discovery Consortium (2014). g _ mmpbsa – A GROMACS tool for MM-PBSA and its optimization for high-throughput binding energy calculations. Journal of Chemical Information and Modeling, 54(7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Lai, C. C., Shih, T. P., Ko, W. C., Tang, H. J., & Hsueh, P. R. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. International Journal of Antimicrobial Agents, 55(3), 105924. 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, P. I., & Hsueh, P. R. (2020). Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. Journal of Microbiology, Immunology and Infection, 53(3), 365–367. 10.1016/j.jmii.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Weber, M. (2009). New therapeutic aspects of flavones: The anticancer properties of Scutellaria and its main active constituents Wogonin, Baicalein and Baicalin. Cancer Treatment Reviews, 35(1), 57–68. 10.1016/j.ctrv.2008.09.005 [DOI] [PubMed] [Google Scholar]
- Li, F., Song, X., Su, G., Wang, Y., Wang, Z., Jia, J., Qing, S., Huang, L., Wang, Y., Zheng, K., & Wang, Y. (2019). Amentoflavone inhibits HSV-1 and ACV-resistant strain infection by suppressing viral early infection. Viruses, 11(5), 466. 10.3390/v11050466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, J., Way, G., & Madahar, V. (2020). Target virus or target ourselves for COVID-19 drugs discovery?-Lessons learned from anti-influenzas virus therapies. World Journal of Virology, 9(3), 19–26. 10.1016/j.medidd.2020.100037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, Y. S., Sircar, S., Bhat, S., Sharun, K., Dhama, K., Dadar, M., Tiwari, R., & Chaicumpa, W. (2020). Emerging novel coronavirus (2019-nCoV)—Current scenario, evolutionary perspective based on genome analysis and recent developments. The Veterinary Quarterly, 40(1), 68–76. 10.1080/01652176.2020.1727993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, A., Pathak, Y., Tripathi, V. (2020). Natural compounds as potential inhibitors of novel coronavirus (COVID-19) main protease: An in silico study. Preprint. 10.21203/rs.3.rs-22839/v1. [DOI]
- Morris, G. M., Ruth, H., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). Software news and updates AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Nikhat, S., & Fazil, M. (2020). Overview of Covid-19; its prevention and management in the light of Unani medicine. The Science of the Total Environment, 728, 138859. 10.1016/j.scitotenv.2020.138859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel: An open chemical toolbox. Journal of Cheminformatics, 3. 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Z., Xu, Y., Bao, L., Zhang, L., Yu, P., Qu, Y., Zhu, H., Zhao, W., Han, Y., & Qin, C. (2019). From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses, 11(1), 59. 10.3390/v11010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa Da Silva, A. W., & Vranken, W. F. (2012). ACPYPE – AnteChamber PYthon Parser interfacE. BMC Research Notes, 5(1), 367. 10.1186/1756-0500-5-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuti, J. R. S., Lye, K. A., & Dhillion, S. S. (2003). Traditional herbal drugs of Bulamogi, Uganda: Plants, use and administration. Journal of Ethnopharmacology, 88(1), 19–44. 10.1016/S0378-8741(03)00161-2 [DOI] [PubMed] [Google Scholar]
- Tai, W., He, L., Zhang, X., Pu, J., Voronin, D., Jiang, S., Zhou, Y., & Du, L. (2020). Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cellular and Molecular Immunology, 17(6), 613–620. 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich, S., & Nitsche, C. (2020). The SARS-CoV-2 main protease as drug target. Bioorganic & Medicinal Chemistry Letters, 30(17), 127377. 10.1016/j.bmcl.2020.127377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Greene, D., Xiao, L., Qi, R., & Luo, R. (2018). Recent developments and applications of the MMPBSA method. Frontiers in Molecular Biosciences, 4, 87. 10.3389/fmolb.2017.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2020). Coronavirus (COVID-19) events as they happen. Who.
- Xu, X., Chen, P., Wang, J., Feng, J., Zhou, H., Li, X., Zhong, W., & Hao, P. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences, 63(3), 457–460. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., & Wunderink, R. G. (2018). MERS, SARS and other coronaviruses as causes of pneumonia. Respirology (Carlton, Vic.), 23(2), 130–137. 10.1111/resp.13196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Saravanan, K. M., Yang, Y., & Hossain, T. (2020). Deep learning based drug screening for novel coronavirus 2019-nCov. Interdiscip Sci, Jun 1, 1–9. 10.1007/s12539-020-00376-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T., Wu, Q., & Zhang, Z. (2020). Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Current Biology, 30(7), 1346–1351.e2. 10.1016/j.cub.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Yuan, Q., Wang, H., Liu, W., Liao, X., Su, Y., Wang, X., Yuan, J., Li, T., Li, J., Qian, S., Hong, C., Wang, F., Liu, Y., Wang, Z., He, Q., He, B., Zhang, T., Ge, S., … Zhang, Z. (2020). Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clinical Infectious Diseases. 10.1093/cid/ciaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]