Abstract
Coronavirus disease 2019 (COVID-19), an acute respiratory infection, is largely associated with dysregulation and impairment of the immune system. This study investigated how the immune system changes were related to disease severity in COVID-19 patients. The frequencies of different immune cells and levels of pro- and anti-inflammatory cytokines in whole blood of participants were determined by flow cytometry and enzyme-linked immunosorbent assay, respectively. The values of other inflammatory agents were also studied. In the late recovery stage, unlike CD56high CD16+/− NK cells and monocytes, CD56low CD16+ NK cell numbers were increased (P < 0.0001–0.05). Th1, Th2, and Th17 cell percentages were significantly lower in patients than healthy control (P < 0.0001–0.05), while their frequencies were increased following disease recovery (P < 0.0001–0.05). The numbers of Tregs, activated CD4+ T cells, and exhausted CD8+ T cells were significantly decreased during a recovery (P < 0.0001–0.05). No significant change was observed in exhausted CD4+ T cell number during a recovery (P > 0.05). B cell showed an increased percentage in patients compared to healthy subjects (P < 0.0001–0.05), whereas its number was reduced following recovery (P < 0.0001–0.05). IL-1α, IL-1β, IL-6, TNF-α, and IL-10 levels were significantly decreased in the late recovery stage (P < 0.0001–0.05). However, TGF-β1 level was not significantly changed during the recovery (P > 0.05). Lymphocyte numbers in patients were significantly decreased (P < 0.001), unlike ESR value (P < 0.001). Lymphocyte number was negatively correlated to ESR value and Th2 number (P < 0.05), while its association with monocyte was significantly positive at the first day of recovery (P < 0.05). The immune system changes during the disease recovery to improve and regulate immune responses and thereby may associate with the reduction in disease severity.
Keywords: COVID-19, disease recovery, disease severity, immune system, immunodysregulation
Introduction
In December 2019, the world experienced the outbreak of 2019 novel coronavirus disease (COVID-19) caused by a novel coronavirus, named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2).1–4 This virus is a highly infectious and causes upper and lower respiratory tract pneumonia resulting in acute respiratory distress syndrome (ARDS).5–9 The fatality rate of COVID-19 is approximately 3.4%, but it may be increased in some subjects with health problems, for example, diabetes, immunodeficiency disorders, hypertension, end-stage renal failure, and cardiovascular disease. It is not the first time that coronavirus family causes a severe respiratory disease. Genetic analysis of SARS-CoV-2 genomes showed that there are two types of this virus, including L type (70%) and S type (30%) which are aggressive and ancestral, respectively.3,10
Although the pathogenesis of COVID-19 is not well understood yet, defects in function and/or regulation of the immune system such as the storm of inflammatory cytokines and lymphopenia can contribute to the intensity of pathogenic coronavirus infections.11–13 In despite of some reports pointing to impacts of immune responses in the pathogenesis of COVID-19,14 the accurate roles of immune cells in developing or inhibiting the disease are unknown. In viral infections, the innate immunity mainly produces some antiviral components such as defensins and interferon type I (IFN-I) which inhibits viral replication through degrading virus RNA and inducing adaptive immune responses.15 However, some reports have indicated that IFN-I plays a dual role in protective and destructive responses in coronavirus infections.16 SARS-CoV-2 probably induces delayed IFN-I responses that enhance virus invasion in early phases of the disease.17 It is reported that dysregulation and/or impairments of the innate and adaptive immune systems may participate in tissue damage and disease severity in patients with COVID-19 through reducing numbers and defecting functions of CD4+ T helper (Th) cells, CD8+ T cells, natural killer (NK) cells, and B cells.16,17 The increased percentage and enhanced function of neutrophil have been also proposed as other mechanisms involved in the pathogenesis of the disease.18,19 The increased productions of some inflammatory agents can contribute to ARDS in patients, including C-reactive protein (CRP), IL-1β, IL-1α, IL-6, and TNF-α.11,20
Extensive data from the literature have demonstrated that Th1 cells play critical role in the main antiviral mechanism(s) of adaptive immunity. Th1 cell-produced cytokines enhance the functions of cytotoxic T cells and productions of antibodies, especially IgG and IgM of B cells.17 Previous studies on similar viral infections to SARS-CoV2 have indicated that downregulation of Th1 and Th2 cell functions may be involved in infection severity and ARDS progression in patients with Middle East Respiratory Syndrome-coronavirus (MERS-CoV) infection.21 In addition, these studies have shown that the increased frequencies of Th1 and CD8+ T cells in patients with MERS-CoV infection were correlated to the good prognosis of the disease, whereas Th2 cytokines were associated with higher mortality.20,21 Others have revealed that Th1 and Th17 cells can stimulate and participate in inducing inflammatory reactions in patients with COVID-19.19 Another immune cell associated with pathogenesis of COVID-19 may be regulatory T cells (Tregs), the immune regulatory cells critical for inhibiting inflammation and immune response-associated tissue damage. Some studies have reported that COVID-19 patients had the reduced number of Tregs in peripheral blood.22
Given that clinical data have provided evidence to reveal that severe cases of COVID-19 are largely associated with overreactive immune responses resulting in a cytokine storm and dysregulation of the immune system,22 the aim of this study was whether changes of the innate (monocytes and different subsets of NK cells) and adaptive immune cells (activated CD4+ T cells, activated CD8+ T cells, exhausted CD4+ T cells, exhausted CD8+ T cells, Th1 cells, Th2 cells, Th17 cells, Tregs, B cells) during a recovery were correlated to the disease severity. In this regard, the plasma levels of pro- and anti-inflammatory cytokines including IL-1α, IL-1β, TNF-α, IL-6, TGF-β1 and IL-10 along with other inflammatory agents were also measured.
Materials and methods
Study subjects
This is an analytical observational (case-control) study performed on 57 patients with COVID-19, who were referred to a COVID-19 center, Isfahan, Iran from March 2020 to April 2020, and 40 healthy individuals without any the signs and symptoms of acute respiratory infections and other health problems affected the immune system. The diagnosis of COVID-19 was confirmed by the specialist according to clinical and laboratory criteria including: (1) clinical evaluation; (2) real time-polymerase chain reaction (RT-PCR) assay; (3) CT scan imaging. All patients had pulmonary involvement and were not on treatment with drugs influencing the immune system and antibodies production (i.e. steroids, sulfasalazine, phenytoin, and antimalarial drugs) prior to study initiation. The patients suffered from clinical signs and symptoms of COVID-19 at least 3–5 days before going to a COVID-19 center to start therapeutic approaches. The study was approved by the Ethics Committee of Isfahan University of Medical Sciences (protocol number: 52409) and performed in accordance with the declaration of Helsinki for medical research involving human subjects. The informed consent was obtained from the participants and legally authorized representatives of dead patients before the study initiation. According to the SD values observed in similar studies,23,24 sample sizes for two groups of study were calculated by the following statistical formula:
α (study accuracy) = 95%; β (study power) = 80%; d (effect size) = 80%; Zα = 1.96; Zβ = 0.89; S1 = 1.6; S2 = 1.4.
Sample collection and cell counting
At the first day (the early recovery stage) and 10 days of initiation of therapeutic methods (the late recovery stage), heparinized blood samples (5 ml) were obtained from patients. To determine the immune situation of patients, the blood sampling (5 ml) from healthy subjects was also performed. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by Ficoll-Paque centrifugation according to the manufacturer’s instructions (Lymphodex, Germany). The isolated cells were washed twice with phosphate buffered saline (PBS) at 300 × g for 10 min. The cells were then counted using a Burker Neubauer chamber (haemocytometer, 40443001, Hecht Assistent, Germany) and cell viability was measured by trypan blue dye exclusion.
The number of lymphocytes in peripheral blood of COVID-19 patients in the early and late stages of recovery and healthy subjects were assessed by an automated cell counter system UF-100® (Sysmex, Kobe, Japan) within 3 h after collecting blood samples.
Flow cytometry
To determine the percentages of activated T cells, exhausted T cells, Th1 cells, Th2 cells, Th17 cells, Tregs, B cells, NK cells, and monocytes in peripheral blood of COVID-19 patients (the first day and 10 days of initiation of therapeutic approaches) and healthy subjects, PBMCs were stained with different monoclonal antibodies or matched to isotype control IgG for 30 min at 4○C. Isotype-matched control antibodies were used as negative controls. Fixation and permebilization of the cells were performed for staining some intracellular molecules with different antibodies according to the manufacturer’s guideline (eBiosciences, USA). The stained cells were washed twice with PBS and centrifuged at 300 × g for 10 min at room temperature. The percentages of the stained cells were measured by a FACSCalibur system (Becton Dickinson, San Jose, CA). The cell markers used to determine the frequencies of the stained cells are indicated in Table 1. In this regard, the FlowJo software (v10.1, FlowJo, Ashland, OR, USA) was used to gate lymphocyte population using forward and side scatter to exclude debris or dead cells from the analysis of different cells. Afterwards, the lymphocyte population was gated to assess the frequencies of the CD4+ cells which were used to determine the percentages of Th1 cells (CD4+ T-bet+ IFN-γ+ cells), Th2 cells (CD4+ IL-4+ GATA3+ cells), Th17 cells (CD4+ IL-17α+ RORγt+ cells), Tregs (CD4+ CD127low FoxP3+ cells), and activated CD4+ T cells (CD4+ CD25+ CD69+ cells). The CD3+ cell population was also determined using the gating of lymphocyte population and was then used to measure the percentages of B cells (CD3− CD19+ CD22+ cells), exhausted CD4+ T cells (CD3+ CD4+ PD-1+ cells), exhausted CD8+ T cells (CD3+ CD8+ PD-1+ cells), CD56low CD16+ NK cells (CD3− CD56low CD16+ cells), and CD56high CD16+/− NK cells (CD3−CD56high CD16+/− cells). In this study, CD8+ CD25+ CD69+ cells and CD14+ CD16+ CD11b+ cells were respectively considered as the activated CD8+ T cells and monocytes. The monoclonal and their isotype control antibodies are shown in Table 1.
Table 1.
Antibodies used for determing the changes of the immune system of COVID-19 patients by flow cytometry.
| Fluorochrome/antibody | Isotype | Clone | Company (all from USA) |
|---|---|---|---|
| CD3-FITC antibody | Mouse IgG2a, κ | HIT3a | BioLegend |
| CD4-PE/CY5 antibody | Rat IgG2a, κ | H129.19 | BioLegend |
| CD8-PE/CY5 antibody | Mouse IgG1, κ | HIT8a | BioLegend |
| CD14-FITC antibody | Mouse IgG1, κ | 63D3 | BioLegend |
| CD16-PE antibody | Mouse IgG1, κ | 3G8 | BioLegend |
| CD56-PE/CY5 antibody | Mouse IgG1, κ | 5.1H11 | BioLegend |
| CD19- PE/CY5 antibody | Mouse IgG1, κ | HIB19 | BioLegend |
| CD22-PE antibody | Mouse IgG1, κ | HIB22 | BioLegend |
| CD25-FITC antibody | Mouse IgG1, κ | BC96 | BioLegend |
| CD69-PE antibody | Mouse IgG1, κ | FN50 | BioLegend |
| PD1-PE antibody | Mouse IgG2b, κ | A17188B | BioLegend |
| CD127-FITC antibody | Mouse IgG1, κ | A019D5 | BioLegend |
| Tbet-FITC antibody | Mouse IgG1, κ | 4B10 | BioLegend |
| IFNg-PE antibody | Hamster IgG | 2HUB-159 | BioLegend |
| GATA3-PE antibody | Mouse IgG2b, κ | 16E10A23 | BioLegend |
| IL4- PerCP/Cyanine5.5 antibody | Rat IgG1, κ | MP4-25D2 | BioLegend |
| IL17-PE antibody | Mouse IgG1, κ | BL168 | BioLegend |
| FOXO3-PE antibody | Mouse IgG1, κ | 206D | BioLegend |
| RORγt-PE antibody | – | – | BioLegend |
Investigations of cytokines and other inflammatory factors
To determine cytokine profiles in COVID-19 patients, the plasma samples were obtained from whole blood (5 ml) of participants and the levels of IL-1α, IL-1β, TNF-α, IL-6, TGF-β1, and IL-10 cytokines were measured using an Enzyme-linked immunoasorbent assay (ELISA) kit (Karmania Pars Gene, Iran) according the manufacturer’s instructions.
The levels of erythrocyte sediment rate (ESR) and C-reactive protein (CRP) of COVID-19 patients were measured using the erythrocyte sedimentation rate (ESR) analyzer (Parsian Teb, Iran) and Mindray BS-800 automated biochemistry analyzer (Shenzhen Mindray Bio-Medical Electronics, China), respectively.
Statistical analysis
Data were analyzed by GraphPad Prism 6 (GraphPad Software, USA) and are expressed as the mean standard error of the mean (SEM) and mean ± standard deviation (SD). The normal distribution of data was determined by Kolmogrov–Smirnov test. The groups with normal distribution were compared by One-way analysis of variance (ANOVA) and unpaired t-tests, while Mann–Whitney and Kruskal–Wallis tests were used to compare ones with non-normal distribution. P < 0.05 was considered statistically significant.
Results
Subject descriptions
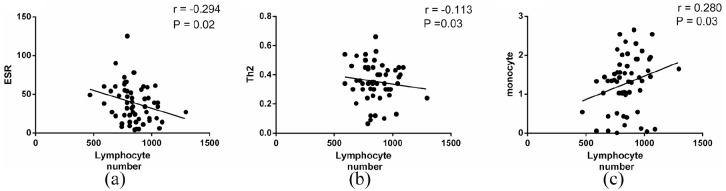
In this study, the mean ± SD of age of patients was 67.8 ± 15.18, while it was 66.01 ± 7.11 in healthy subjects. Of the 57 patients, 51 (89.48%) were discharged from hospital and 6 (10.52%) died during the study. Some patients had fatigue, mild shortness of breath, myalgia, loss of weight, smell, and taste in the late recovery stage. The mean ± SD of hemoglobin was significantly lower in patients than healthy groups (11.28 ± 2.21 g/dl vs 14.44 ± 0.89 g/dl, P < 0.001). The mean ± SD of lymphocyte number was 847.26 ± 142.44 cells/μl blood (11.45% from 1 × 104 counted WBCs) in the early stage of recovery, while it was 2203 ± 157.69 cells/μl blood (30.45% from 1 × 104 counted WBCs) in the late stage of recovery. The number of lymphocytes was negatively correlated to ESR value and Th2 number (Figure 1(a) and (b), P < 0.05, r = −0.2948, 95% confidence interval (CI) = −0.5216 to −0.0292; P < 0.05, r = −0.4398, 95% CI = −0.6928 to −0.0903, respectively); however, this association was positive for monocyte at the first day of recovery (Figure 1(c), P < 0.05, r = 0.2803, 95% CI = 0.0134–0.5100). Table 2 depicts the demographic and other characteristics of COVID-19 and healthy subjects.
Figure 1.
Correlations of lymphocyte numbers with the value of ESR and numbers of Th2 cells and monocytes in COVID-19 patients. (a and b) Results of Spearman test showed lymphocyte number was negatively associated with ESR value and Th2 cell percentage in COVID-19 patients (P < 0.05). (c) There was a positive correlation between lymphocyte and monocyte numbers in COVID-19 patients (P < 0.05).
Table 2.
The demographic, laboratory, and clinical characteristics of COVID-19 and healthy subjects.
| Patients (N = 57) | Control (N = 40) | P value | |
|---|---|---|---|
| Sex | Male: 31 (54.4%) | Male: 21 (52.5%) | 0.85 |
| Female: 26 (45.6%) | Female: 19 (47.5%) | ||
| Smoking | 17 (29.82%) | 12 (30%) | 0.58 |
| Lymphocyte count (cell/μl) | 847 ± 142.44 | 3072 ± 814.57 | <0.001 |
| CRP | Negative: 2 (3.5%) | Negative: 40 (100%) | |
| Positive: 55 (96.5%) | |||
| 1+: 12 (21.05%) | |||
| 2+: 19 (33.34%) | |||
| 3+: 17 (29.83%) | |||
| 4+: 7 (12.28%) | |||
| ESR(mm/h) | 38.96 ± 24.36 | 7.7 ± 4.42 | <0.001 |
| Background disease | Diabetes: 7 (25.92%) | – | |
| Kidney disease (ESRD & CKD): 8 (29.62%) | |||
| Lung disease (COPD, asthma): 6 (22.22%) | |||
| Hypertension: 18 (31.58%) | |||
| Hypothyroidism: 1 (3.7%) | |||
| Colon cancer: 1 (3.7%) | |||
| IHD: 5 (18.51%) | |||
| CVA: 1 (3.7%) | |||
| RA: 1 (3.7%) | |||
| Window period (day) | 8.59 ± 7.24 | – | |
| Treatment | Anti-viral drugs: 19 (33.33%) | – | |
| Anti-inflammatory drugs (prednisolone): 6 (10.52%) | |||
| Antibiotics: 32 (56.15%) |
RT-PCR: real time polymerase chain reaction; CRP: C-reactive protein; ESR: erythrocyte sedimentation rate; ESRD: end stage renal disease; CKD: chronic kidney disease; COPD: chronic obstructive pulmonary disease; IHD: ischemic heart disease; CVA: cerebrovascular accident; RA: rheumatoid arthritis.
Assessments of innate immune cells in patients with COVID-19 during a recovery stage
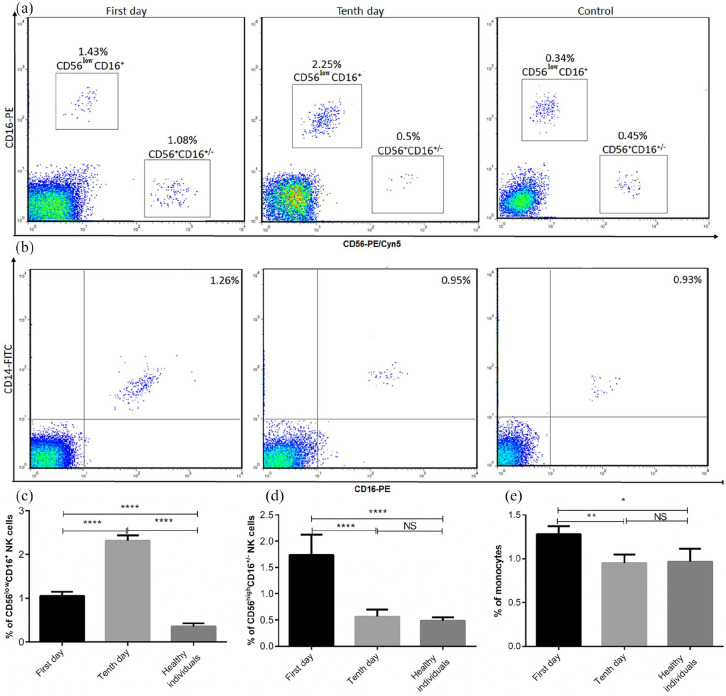
The number of CD56low CD16+ NK cells in patients was significantly increased compared to healthy subjects. Its frequency was significantly higher in the late recovery stage than early recovery stage (Figure 2(a) and (c), P < 0.0001). The frequency of CD56high CD16+/− NK cells in the early stage of recovery was significantly increased in comparison with the late stage of recovery and healthy individuals (Figure 2(a) and (d), P < 0.0001). Others results indicated that patients, at the first day of initiation of therapeutic methods, had a significant increase in the percentage of monocyte compared to those at 10 days of treatment and healthy subjects (Figure 2(b) and (e), P < 0.05).
Figure 2.
The frequencies of innate immune cells in COVID-19 and control subjects. The percentages of CD56low CD16+ NK cells, CD56high CD16+/− NK cells, and monocytes were studied by flow cytometry (a and b) and then analyzed (c–e). The results are representative of 57 independent experiments for COVID-19 patients at the first day of treatment, 51 independent experiments for COVID-19 patients in 10 days of treatment, and 40 independent experiments for healthy individuals. Each bar in (c–e) shows mean ± SEM.
*P < 0.05, **P < 0.01, ****P < 0.0001.
Investigations of adaptive immune cells in patients with COVID-19 during a recovery stage
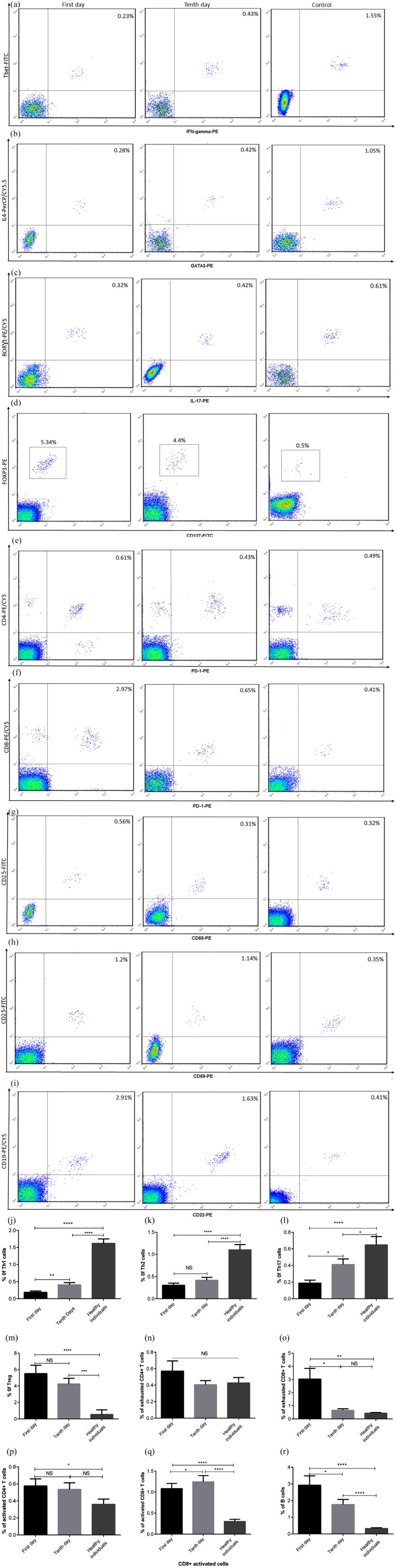
To determine the situations of humoral and cellular immunity in patients with COVID-19, the frequencies of Th1, Th2, Th17, Treg, activated CD4+T cells, activated CD8+ T cells, exhausted CD4+ T cells, exhausted CD8+ T cells, and B cells in COVID-19 patients were investigated after 1 and 10 days of initiation of therapeutic methods. Our data revealed that the percentages of Th1, Th2, and Th17 cells were significantly lower in patients than healthy control (Figure 3(a)–(c) and (j)–(l), P < 0.0001–0.05). However, the numbers of Tregs, exhausted CD4+ T cells, exhausted CD8+ T cells, activated CD4+ T cells, activated CD8+ T cells were increased in patients compared to healthy subjects, although the increased percentage of exhausted CD4+ T cells was not statistically significant (Figure 3(d)–(h) and (m)–(q), P < 0.0001–0.05). After 10-day recovery, the frequency of exhausted CD8+ T cells were significantly reduced, unlike activated CD8+ T cells (Figure 3(f), (h), (o), and (q), P < 0.05). No significant changes were observed in the frequencies of Tregs, exhausted CD4+ T cell, and activated CD4+ T cells between patients in the early stage and late stage of recovery (Figure 3(d), (e), (g), (m), (n), and (p)). B cells showed an increased percentage in patients compared to healthy subjects, while this increase was significantly reduced in the late stage of recovery (Figure 3(i) and (r), P < 0.0001–0.05).
Figure 3.
The percentages of adaptive immune cells in COVID-19 and healthy individuals. PBMCs were isolated from healthy subjects and COVID-19 patients and then stained with different monoclonal antibodies. The percentages of Th1, Th2, Th17, Tregs, exhausted CD4+ T cells, exhausted CD8+ T cells, activated CD4+ T cells, activated CD8+ T cells, and B cells were assessed using flow cytometry (a–i) and then analyzed (j–r). The depicted results are representative of 57 independent experiments for COVID-19 patients at the first day of treatment, 51 independent experiments for COVID-19 patients in 10 days of treatment, and 40 independent experiments for healthy groups. Data show mean ± SEM.
*P < 0.05, **P < 0.01, ****P < 0.0001.
The cytokine profiles of patients with COVID-19
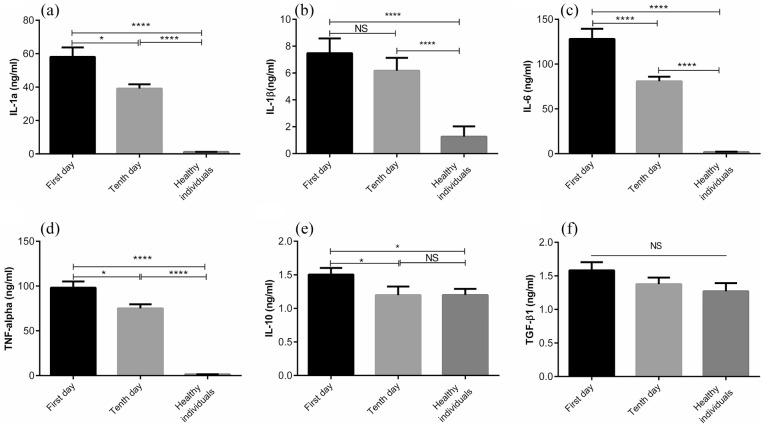
Having considered that severe COVID-19 is largely related to a cytokine storm, cytokine profiles of COVID patients were assessed during a recovery. As shown in Figure 4(a)–(d), statistically significant reduction in the levels of pro-inflammatory cytokines (IL-1α, IL-1β, IL-6, and TNF-α) in patients were observed during a recovery, with the exception of IL-1β level (P < 0.0001–0.05). The same trend was observed in IL-10 so that its level was significantly higher in the early recovery stage than the late recovery stage (Figure 4(e), P < 0.05). However, there was no significant difference in TGF-β1 level between patients and healthy individuals, although a reduction was observed during a recovery (Figure 4(f)).
Figure 4.
The plasma concentrations of pro-and anti-inflammatory cytokines in COVID-19 and control groups. The levels of IL-1α, IL-1β, IL-6, IL-10, TNF-α, and TGF-β1 were measured by ELISA (a–f). The depicted results are representative of 40 independent experiments for control group, 57 independent experiments for COVID-19 patients at the first day of treatment, and 51 independent experiments for COVID-19 patients in 10 days of treatment. All data show mean ± SEM.
*P < 0.05, **P < 0.01, ****P < 0.0001.
Discussion
COVID-19, as a pandemic disease, is responsible for considerable mortality and morbidity.25 Immune system functions have fundamental role in the pathogenesis and outcome of disease.26 Therefore, the current study focused on determining how immune system changes during a recovery were correlated to disease severity.
Having considered that innate immunity provides the early line of defense against viral infections, some innate immune cells were studied in the course of 10 days after initiation of treatment, which is almost a period that the disease is deteriorated and may result in death or recovery from COVID-19.27,28 Our data showed that CD56lowCD16+ NK cell number was significantly lower in the early stage of recovery than the late stage of recovery; however its frequency was noticeably increased compared to healthy subjects. In line with these findings, Zheng et al. reported that the total number of NK cells was dramatically decreased in patients in a severe stage of the disease in comparison with others in the mild stage of the disease and healthy subjects, which this reduction was accompanied by functional exhaustion of these cells.29 Furthermore, it was demonstrated that the reduced number and functional exhaustion of these cells were restored following disease recovery.29 According to the levels of CD16 and CD56 expression, NK cells are classified into two distinguished subsets including: CD56highCD16+/− and CD56lowCD16+ NK cells. The CD56lowCD16+ NK cells have high expression levels of killer inhibitory receptors, the maturation marker (CD57), and natural and antibody-dependent cellular cytotoxicity which is mediated by releasing high levels of perforin and enhanced killing.16,30,31 These findings suggest that the reduced number of CD56lowCD16+ NK cells may contribute to disease susceptibility in the early stages of disease. Other results of the current study revealed that the percentage of another subset of NK cells (CD56highCD16+/− NK cells) was significantly increased at the first day of recovery. CD56highCD16+/− NK cells are described by NKG2A, low level of perforin, and are primarily characterized by cytokine production.16,30,31 Therefore, it is likely that the changes in the frequencies of two subsets of NK cells during the disease recovery are protective mechanisms to eliminate the SARS-COV2 and thereby reducing inflammation occurred in the early stages of disease. In an attempt to discover the frequency of other cells of innate immunity, the number of monocytes was also assessed. We observed that COVID-19 patients had significantly higher percentage of monocytes in the early stage of recovery than those in the late stage of recovery and healthy subjects. This observation was in contrast with previous study showing severe cases of COVID-19 tend to have lower percentages of monocytes.24 This discrepancy may be attributed to disease stage which patients were evaluated.
In the next step, the adaptive immune system of COVID-19 subjects was studied after 1 and 10 days of initiation of therapeutic methods. The results indicated that patients had the reduced number of lymphocyte in comparison with healthy subjects. In line with this finding, Qin et al. declared that patients with COVID-19 had a reduction in T cell number accompanied by the severity of the disease. Moreover, Qin et al. indicated that suppressor and helper T cell percentages were lower in patients than normal group. The authors have also demonstrated that the frequency of naive Th cells increased in severe cases, unlike memory Th cells and Tregs which were more obviously decreased in severe cases.24 Other studies have revealed that the whole numbers of T cells, CD4+ T cells, and CD8+ T cells were significantly decreased in patients with COVID-19.29,32 Wen et al. by transcriptional analyzes of PBMCs of recovered patients with COVID-19 showed that patients had higher frequencies of B cells and T cells in the late recovery stage than those in the early recovery stage. Moreover, the authors have shown that CD4+ and CD8+ T cell numbers were notably decreased.17 In agreement with previous study, our data revealed that the frequencies of Th cells (Th1, Th2, and Th17 cells) in patients were significantly lower in the early recovery stage than the late recovery stage and healthy individuals. More interestingly, the percentages of exhausted CD4+ T cells and exhausted CD8+ T cells were higher in the early stage of recovery than the late stage of recovery. These findings were consistent with other reports indicating the number of CD8+ T cells was markedly decreased and its function was exhausted in COVID-19 patients.29 In contrast with the percentage of activated CD4+ T cell which was increased in the early stage of recovery, the activated CD8+ T cell had the reduced frequency; however its number was significantly increased in the late stage of recovery, unlike activated CD4+ T cell number. In disagreement with other reports showing increased frequency of B cells in the late stage of recovery,17 we observed that the percentage of this cell was decreased following recovery. It is thought that the reduced numbers of activated CD4+ T cells and B cells are related to different therapeutic approaches used to reduce inflammation and their impacts.
Regarding the impact of the cytokine storm in COVID-19 pathogenesis, the levels of some pro- and anti-inflammatory cytokines were investigated. The plasma levels of IL-1a, IL-1β, IL-6, TNF-α, and IL-10 were significantly higher in COVID-19 patients than the control group. However, no significant correlation was observed between the levels of these cytokines and other inflammatory factors such as ESR which was notably increased in the early recovery stage. Moreover, other data indicated that the levels of these cytokines were reduced during the disease recovery. In agreement with these findings, it is indicated that patients with MERS-CoV infection, which is the same as SARS-CoV2 in pathogenesis, had the increased expression level of IL-10.33 Similar to our results, Chen et al., in a study on 48 patients with COVID-19, revealed that IL-6 level was dramatically increased.34 Others have shown that the levels of IL-6, IL-10, and TNF-α were increased in COVID-19 patients.32 In despite of the changes in the levels of IL-1a, IL-1β, IL-6, TNF-α, and IL-10 during a 10-day recovery period, there was no significant difference in TGF-β1 level between the early recovery and late recovery stages, although a reduction was observed during a recovery. A limitation of the study was the lack of determination of immune system differences between alive and dead patients with COVID-19 during a recovery period. Therefore, this limitation needs to be addressed in future studies.
Conclusion
The results of this study provide evidence to show that COVID-19 patients, who need to hospitalization, had some changes in the immune system during the disease recovery to improve and regulate immune responses. These changes may play a fundamental role in reducing disease severity through regulating cytokine productions involved in the inflammation and functions of various immune cells. Nevertheless, larger and more multicenter studies are needed to validate these conclusions.
Acknowledgments
The authors thank all subjects who participated in this study.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was financially supported by Isfahan University of Medical Sciences.
Ethics approval: Ethical approval for this study was obtained from *the Ethics Committee of Isfahan University of Medical Sciences (protocol number: 52409)*.
Informed consent: Written informed consent was obtained from the participants and legally authorized representatives of dead patients before the study initiation. This requirement was performed according to protocols of the Ethics Committee of Isfahan University of Medical Sciences.
ORCID iD: Nahid Eskandari  https://orcid.org/0000-0003-1910-9359
https://orcid.org/0000-0003-1910-9359
References
- 1. Qiao J. (2020) What are the risks of COVID-19 infection in pregnant women? The Lancet 395(10226): 760–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shi S, Qin M, Shen B, et al. (2020) Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiology 5(7): 802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lillie PJ, Samson A, Li A, et al. (2020) Novel coronavirus disease (Covid-19): The first two patients in the UK with person to person transmission. Journal of Infection 80(5): 578–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Su S, Wong G, Shi W, et al. (2016) Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends in Microbiology 24(6): 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu X, Zhang L, Du H, et al. (2020) SARS-CoV-2 infection in children. New England Journal of Medicine 382(17): 1663–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heymann DL, Shindo N. (2020) COVID-19: What is next for public health? The Lancet 395(10224): 542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang C, Wang Y, Li X, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 395(10223): 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corman VM, Landt O, Kaiser M, et al. (2020) Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 25(3): 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carlos WG, Dela Cruz CS, Cao B, et al. (2020) Novel Wuhan (2019-nCoV) coronavirus. American Journal of Respiratory and Critical Care Medicine 201(4): P7–P8. [DOI] [PubMed] [Google Scholar]
- 10. Tang X, Wu C, Li X, et al. (2020) On the origin and continuing evolution of SARS-CoV-2. National Science Review 6(7): 1012–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li X, Geng M, Peng Y, et al. (2020) Molecular immune pathogenesis and diagnosis of COVID-19. Journal of Pharmaceutical Analysis 2(10): 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, et al. (2013) Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: A descriptive study. The Lancet Infectious Diseases 13(9): 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yin Y, Wunderink RG. (2018) MERS, SARS and other coronaviruses as causes of pneumonia. Respirology 23(2): 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prompetchara E, Ketloy C, Palaga T. (2020) Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pacific Journal of Allergy and Immunology 38(1): 1–9. [DOI] [PubMed] [Google Scholar]
- 15. Gineau L, Cognet C, Kara N, et al. (2012) Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. The Journal of Clinical Investigation 122(3): 821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cao X. (2020) COVID-19: Immunopathology and its implications for therapy. Nature Reviews Immunology 20(5): 269–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wen W, Su W, Tang H, et al. (2020) Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discovery 6(1): 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cao X. (2020) COVID-19: Immunopathology and its implications for therapy. Nature Reviews Immunology 20(5): 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prompetchara E, Ketloy C, Palaga T. (2020) Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pacific Journal of Allergy and Immunology 38(1): 1–9. [DOI] [PubMed] [Google Scholar]
- 20. Hu Q, Hao C, Tang S. (2020) From sepsis to acute respiratory distress syndrome (ARDS): Emerging preventive strategies based on molecular and genetic researches. Bioscience Reports 40(5): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alosaimi B, Hamed ME, Naeem A, et al. (2020) MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine 126: 154895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bonam SR, Kaveri SV, Sakuntabhai A, et al. (2020) Adjunct immunotherapies for the management of severely ill COVID-19 patients. Cell Reports Medicine 1(2): 100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng M, Gao Y, Wang G, et al. (2020) Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cellular & Molecular Immunology 17(5): 533–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qin C, Zhou L, Hu Z, et al. (2020) Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clinical Infectious Diseases 71(15): 762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rundle AG, Park Y, Herbstman JB, et al. (2020) COVID-19 related school closings and risk of weight gain among children. Obesity 28(6): 1008–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wen W, Su W, Tang H, et al. (2020) Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. medRxiv 6(1): 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baud D, Qi X, Nielsen-Saines K, et al. (2020) Real estimates of mortality following COVID-19 infection. The Lancet Infectious Diseases 20(7): 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang W, Zhao Y, Zhang F, et al. (2020) The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The experience of clinical immunologists from China. Clinical Immunology 108393: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zheng M, Gao Y, Wang G, et al. (2020) Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cellular & Molecular Immunology 17: 533–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mace EM, Hsu AP, Monaco-Shawver L, et al. (2013) Mutations in GATA2 cause human NK cell deficiency with specific loss of the CD56bright subset. Blood 121(14): 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stabile H, Fionda C, Gismondi A, et al. (2017) Role of distinct natural killer cell subsets in anticancer response. Frontiers in Immunology 8: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Diao B, Wang C, Tan Y, et al. (2020) Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Frontiers in Immunology 11: 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li G, Fan Y, Lai Y, et al. (2020) Coronavirus infections and immune responses. Journal of Medical Virology 92(4): 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen X, Zhao B, Qu Y, et al. (2020) Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clinical Infectious Diseases 10(1): 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]