Abstract
Domestication has resulted in immense phenotypic changes in animals despite their relatively short evolutionary history. The European rabbit is one of the most recently domesticated animals, but exhibits distinct morphological, physiological, and behavioral differences from their wild conspecifics. A previous study revealed that sequence variants with striking allele frequency differences between wild and domestic rabbits were enriched in conserved noncoding regions, in the vicinity of genes involved in nervous system development. This suggests that a large proportion of the genetic changes targeted by selection during domestication might affect gene regulation. Here, we generated RNA-sequencing data for four brain regions (amygdala, hypothalamus, hippocampus, and parietal/temporal cortex) sampled at birth and revealed hundreds of differentially expressed genes (DEGs) between wild and domestic rabbits. DEGs in amygdala were significantly enriched for genes associated with dopaminergic function and all 12 DEGs in this category showed higher expression in domestic rabbits. DEGs in hippocampus were enriched for genes associated with ciliary function, all 21 genes in this category showed lower expression in domestic rabbits. These results indicate an important role of dopamine signaling and ciliary function in the evolution of tameness during rabbit domestication. Our study shows that gene expression in specific pathways has been profoundly altered during domestication, but that the majority of genes showing differential expression in this study have not been the direct targets of selection.
Keywords: transcriptome, domestication, the European rabbit, evolution of tameness, dopamine, amygdala
Significance
A hallmark of domestic animals is a tame behavior. In a previous study, we reported that genetic differences between domestic and wild rabbits are particularly common in the vicinity of genes expressed during brain development. In the present study, we have compared gene expression between wild and domestic rabbits in four regions of the brain. We find hundreds of genes that show statistically significant differences in expression levels between the two types of rabbits. Our results indicate that changes in dopamine signaling and ciliary function contributed to evolution of tameness.
Introduction
Domestication, the evolutionary process during which organisms adapt to living closely with humans, is accompanied by pronounced morphological, physiological, and behavioral changes. It is striking that various phenotypic traits, such as decreased brain size, shorter muzzle, floppy ears, depigmented coat color, prolonged or even continuous breeding season, and increased tameness, are shared across different domestic animals. This common set of changes has been termed “the domestication syndrome.” Above all, tameness, enabled by reduced aggressiveness and increased tolerance toward humans, is the hallmark of domestication and one of the first phenotypic changes to evolve in domestic animals. However, little is known about the initial genetic changes that facilitated adaptation to captivity.
The European rabbit (Oryctolagus cuniculus) is among the most recently domesticated animals (∼1,400 years ago) (Clutton-Brock 1999), and the most closely related wild progenitors are still present in southern France (Carneiro et al. 2011, 2014). As described in “The Origin of Species,” where Charles Darwin stated that “no animal is more difficult to tame than the young of the wild rabbit; scarcely any animal is tamer than the young of the tame rabbit” (Darwin 1859), behavioral differences between wild and domestic rabbits are striking. These behavioral differences provide an opportunity to reveal the genetic substrates underlying rapid behavioral evolution during domestication.
Our previous comparison between the genomes of wild and domestic rabbits revealed several important results (Carneiro et al. 2014). First, the genetic basis of domestication must be highly polygenic, because we observed shifts in allele frequencies at many loci rather than complete fixation. Second, highly differentiated loci were enriched in conserved noncoding regions. Finally, genes involved in brain and neuronal development were enriched around region displaying strong genetic differentiation. These results support a scenario where shifts in allele frequency at many regulatory loci have affected gene expression regulating brain and neuronal development, presumably associated with tame behavior. A brain imaging study also revealed sharp differences in brain architecture between wild and domestic rabbits (Brusini et al. 2018). For example, domestic rabbits exhibited reduced amygdala volume and enlarged medial frontal cortex volume, a pattern consistent with the expected brain changes associated with a diminished fearful response in domestic rabbits. Taken together, these studies strongly suggest that changes in transcriptional regulation in the brain have a critical role for the behavioral differences between wild and domestic rabbits.
Gene expression studies comparing the brain of wild and domesticated animals are scarce and have mostly focused on the frontal cortex (Albert et al. 2012; Wang et al. 2018), whereas multiple other brain regions involved in emotional circuits, stress response or neurogenesis, which are likely relevant for domestication, have been poorly characterized. Here, we present a comparative transcriptomic study investigating several brain regions (amygdala, hypothalamus, hippocampus, and parietal/temporal cortex) in newborn wild and domestic rabbits. Differentially expressed genes (DEGs) in each brain region were examined and intersected with the molecular signatures of selection previously reported (Carneiro et al. 2014), probing possible evolutionary mechanisms associated with rabbit domestication.
Results
Differentially Expressed Genes between Wild and Domestic Rabbits
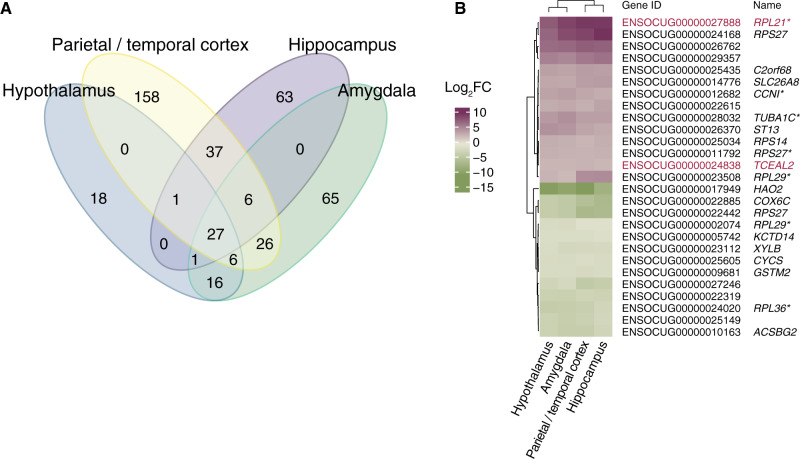
RNA-seq analysis was carried out for four brain regions (amygdala, hypothalamus, hippocampus, and parietal/temporal cortex) in newborn wild (n = 3) and domestic (n = 3) rabbits (supplementary table S1, Supplementary Material online). Comparing gene expression between wild and domestic rabbits resulted in the following number of DEGs: Amygdala (147 genes, 92 with higher expression in domestic individuals), hypothalamus (69 genes, 24 with higher expression in domestic individuals), hippocampus (135 genes, 21 with higher expression in domestic individuals), and parietal/temporal cortex (261 genes, 154 with higher expression in domestic individuals) (fig. 1a and supplementary table S2, fig. S1, and database S1, Supplementary Material online). Twenty-seven DEGs were shared among all four brain regions, including eight coding for ribosomal proteins (fig. 1b). Two of these, RPL21 and TCEAL2, showed signals of selection in our previous comparison of wild and domestic rabbits (Carneiro et al. 2014). Permutation tests confirmed the consistent differential expression across domestic brains and the significant enrichment of genes encoding ribosomal proteins (supplementary database S2, Supplementary Material online).
Fig. 1.
(A) Overlap among genes showing differential expression between wild versus domestic rabbits in four brain regions. (B) Twenty-seven differentially expressed genes were found to be shared among all brain regions. Genes with selection signal between domestic and wild rabbits are highlighted in red. Purple and green represent higher expression in wild and domestic rabbits, respectively. Genes with an asterisk denote manually curated genes with insufficient annotation of their names.
Gene Ontology Enrichment Analyses
To identify enriched terms among our list of DEGs, we used the Genomic Regions Enrichment of Annotations Tool (GREAT) (HyperBonfP<0.05 and HyperFdrQ<0.05) and the Database for Annotation, Visualization and Integrated Discovery (DAVID) (false discovery rate [FDR]<0.05) software with human orthologs as input. The results are shown in table 1 and supplementary table S3, Supplementary Material online, respectively, and detailed results of significantly enriched terms are summarized in supplementary database S3, Supplementary Material online. We found a significant overrepresentation of genes involved in dopaminergic neurotransmission in the amygdala as well as dynein complex and ciliary axoneme in hippocampus. No significant enrichment of functional categories was found for DEGs in hypothalamus, whereas only DAVID detected significant enrichment for the keywords “Glycoprotein” or “secreted” among DEGs in parietal and temporal cortex (supplementary table S3, Supplementary Material online). Given the consistent enrichment and insightful functions of DEGs in terms of rabbit domestication, we put particular focus on DEGs in amygdala and hippocampus hereafter.
Table 1.
List of All Functional Terms Significantly Overrepresented (FDR Q value < 0.05) in the Set of Differentially Expressed Genes (DEGs) Analyzed Using the GREAT Software
| Brain Region | Term | The Number of DEGs in the Category | Total Number of Expressed Genes in the Category | Fold Enrichment | FDR Q Value |
|---|---|---|---|---|---|
| Amygdala | GO biological process | ||||
| Locomotory behavior | 12 | 152 | 6.0 | 7.8×10−3 | |
| Startle response | 5 | 14 | 25.5 | 7.9×10−3 | |
| Behavior | 19 | 436 | 3.4 | 8.3×10−3 | |
| Neuromuscular process | 8 | 71 | 8.7 | 8.9×10−3 | |
| Response to amphetamine | 5 | 18 | 21.3 | 1.0×10−2 | |
| Hypothalamus | Not detected | ||||
| Hippocampus | GO biological process | ||||
| Cilium movement | 9 | 43 | 15.4 | 7.2×10−5 | |
| Axoneme assembly | 7 | 43 | 11.8 | 1.2×10−2 | |
| GO cellular component | |||||
| Ciliary plasm | 11 | 90 | 9.5 | 1.5×10−5 | |
| Axoneme | 11 | 89 | 9.5 | 2.9×10−5 | |
| Axoneme part | 6 | 28 | 16.5 | 8.3×10−4 | |
| Dynein complex | 6 | 42 | 10.7 | 8.1×10−3 | |
| Axonemal dynein complex | 4 | 14 | 22.9 | 8.1×10−3 | |
| GO molecular function | |||||
| Dynein intermediate chain binding | 5 | 23 | 16.5 | 2.5×10−2 | |
| Dynein light chain binding | 5 | 22 | 16.9 | 4.3×10−2 | |
| Parietal and temporal cortex | Not detected |
Protein–Protein Interaction Networks of the DEGs
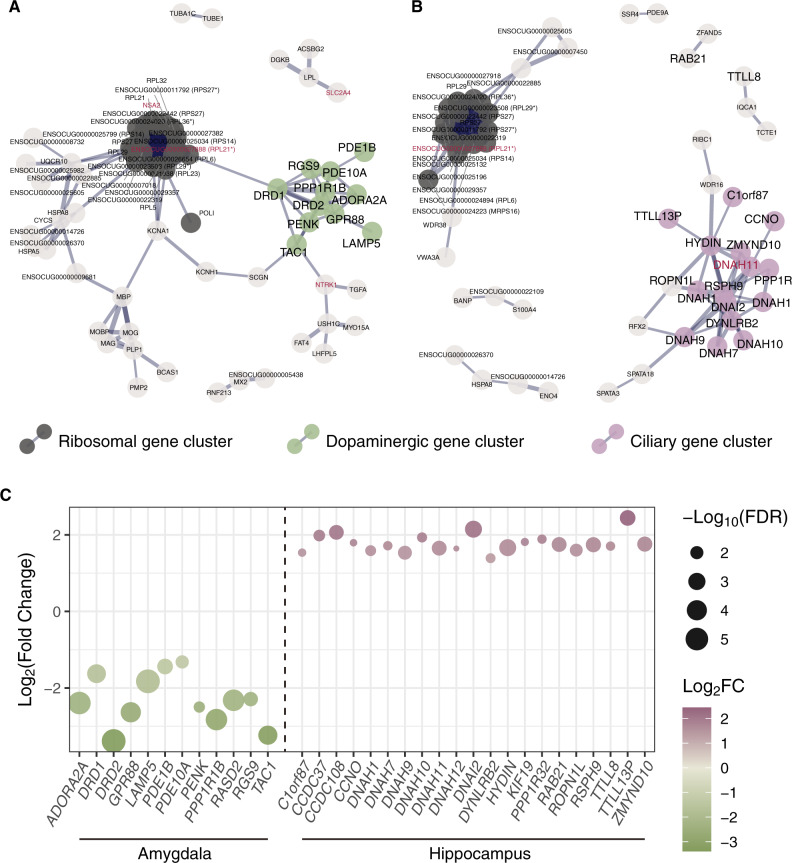
We next investigated protein–protein interaction (PPI) networks using our list of DEGs for amygdala and hippocampus. The subnetworks show that the major clusters consist of a large number of ribosomal genes in both brain regions, in addition to the dopaminergic and ciliary genes in amygdala and in hippocampus, respectively (fig. 2). Here, we defined the genes related to dopaminergic neurotransmission and ciliary function based on the clustering and the annotation in the enrichment analysis and examined more closely the expression profiles of individual genes. As a result, we found a strikingly consistent pattern among these DEGs: 12 out of 12 dopaminergic genes showed higher expression in amygdala from domestic rabbits, and 21 out of 21 DEGs associated with ciliary function showed low expression in the hippocampus of domestic rabbits (fig. 2c and supplementary fig. S2, Supplementary Material online). One of these genes associated with ciliary function, DNAH11, shows signatures of selection between wild and domestic rabbits (Carneiro et al. 2014).
Fig. 2.
Protein–protein interaction networks among differentially expressed genes detected in (A) amygdala and (B) hippocampus. Only genes connected in networks are shown, and ones belonging to clustered subnetworks estimated by Cytoscape are highlighted in different colors. Black represents ribosomal gene clusters in both figures. Clusters corresponding to the genes involved in dopaminergic neurotransmission in amygdala and dynein complex and cilium axoneme in hippocampus are highlighted in green and purple, respectively. Defined dopaminergic or ciliary genes based on the clustering and enrichment analysis (DAVID and GREAT) are shown with large symbols. Genes associated with signals of selection are highlighted in red. The thickness of the line indicates the strength of data support analyzed by STRING. Some of the genes without annotation by STRING are shown with the name of orthologs in parentheses (an asterisk denotes manual curation). (C) Changes in expression levels of all the dopaminergic genes showing differential expression in amygdala and all the ciliary genes showing differential expression in hippocampus. Positive values on the y-axis represent higher expression in wild rabbits, and high expression in domestic and wild rabbits are shown in green and purple, respectively.
Intersection of DEGs with Genetic Signatures of Selection
To explore a possible overlap between differential expression and the previously identified signals of selection between wild and domestic rabbits (Carneiro et al. 2014), we intersected DEGs with selective sweep regions and regions in the near vicinity (±100 kb) of SNPs with high delta allele frequencies (dAF>0.9); dAF is the previously reported allele frequency difference between wild and domestic rabbits (Carneiro et al. 2014). In the latter case, we defined putative genes under selection as those with three or more SNPs with high dAF.
However, we found that DEGs were not significantly associated with these candidate signals of selection (table 2 and supplementary table S3, Supplementary Material online). In addition, we did not observe any significant changes in the expression levels of genes in the near vicinity of SNPs in highly conserved noncoding regions with high dAF (supplementary fig. S3, Supplementary Material online). We further examined if the SNPs located within ±100 kb from transcription start sites (TSS) of DEGs exhibited a trend toward elevated dAFs, as expected if the differential expression is driven by cis-acting regulatory mutations. However, there was no such trend in our data (supplementary fig. S4, Supplementary Material online). Taken together, these results indicate that a large proportion of DEGs do not exhibit differential expression due to their association with cis-acting regulatory variants showing strong genetic differentiation between wild and domestic rabbits, but are more likely to reflect genetic changes in genes that directly or indirectly affect the expression of DEGs in our data set.
Table 2.
Number of DEGs Intersected with Selective Sweep Signals
| Brain Region | Sweep Regiona | Number of DEGs | Number of Non-DEGs | Odds Ratiob | P Valuec |
|---|---|---|---|---|---|
| Amygdala | Selected | 6 | 472 | ||
| Not selected | 141 | 12,819 | 1.16 | 0.65 | |
| Hypothalamus | Selected | 1 | 492 | ||
| Not selected | 68 | 13,081 | 0.39 | 0.52 | |
| Hippocampus | Selected | 3 | 459 | ||
| Not selected | 132 | 12,411 | 0.61 | 0.64 | |
| Parietal and temporal cortex | Selected | 11 | 447 | ||
| Not selected | 250 | 12,260 | 1.21 | 0.50 |
From Carneiro et al. (2014).
>1 means enrichment of selection signal in DEGs.
Fisher’s exact test.
Discussion
Our comparative transcriptomic analyses detected hundreds of DEGs possibly involved in the striking changes in behavior and brain morphology between wild and domestic rabbits (Brusini et al. 2018). Our previous genomic screen demonstrated that SNPs at noncoding, evolutionary conserved sites, located in the vicinity of genes expressed during the development of the brain and the neuronal system, played a key role during domestication (Carneiro et al. 2014). This strongly suggests that many of these SNPs most likely are associated with altered gene regulation during development. However, very few of the genes associated with signals of selection showed differential expression in the present study, and there was no significant enrichment in regions inferred to be under selection. Our results from this study highlight that it may be exceedingly challenging to reveal differential gene expression associated with a noncoding sequence variant because altered expression may occur in a small subset of cells and during a short period of development. This is well illustrated by three cis-acting regulatory mutations (Pea-comb, Rose-comb, and Duplex-comb) causing altered comb morphology in chicken (Wright et al. 2009; Imsland et al. 2012; Dorshorst et al. 2015). All three mutations are structural changes (intronic copy number expansion, inversion, and duplication, respectively) causing ectopic expression of transcription factor genes restricted to a few days of development. One possible interpretation is that the majority of DEGs detected in the present study are not caused by cis-acting regulatory mutations affecting the expression of DEGs directly, but likely indirectly through the action of upstream genes which may have happened during development before our sampling of four brain regions at birth.
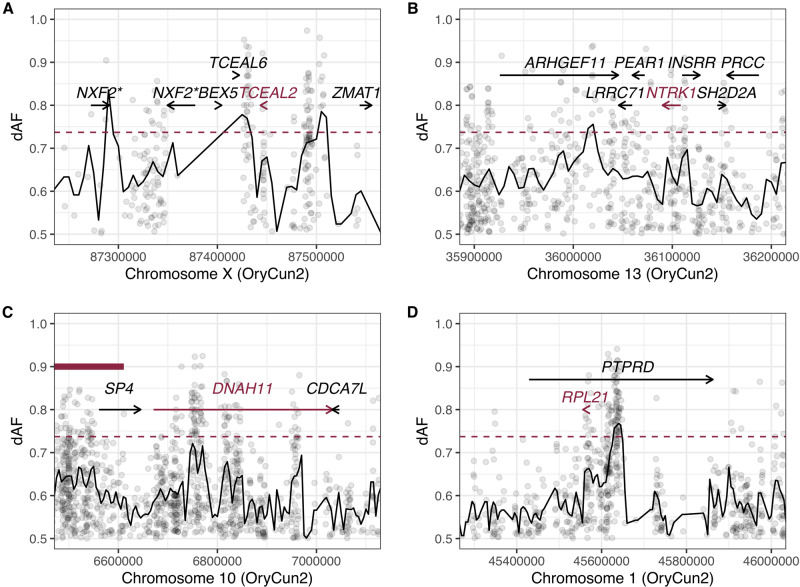
TCEAL2 was one of the few genes showing differential expression and associated with a genetic signature of selection during domestication. TCEAL2 was differentially expressed in all the four brain regions, and we detected many highly divergent SNPs between wild and domestic individuals near this gene (fig. 3a). Although its detailed function is unknown, it is a transcription factor mainly expressed in the cerebral cortex (The human protein atlas; https://www.proteinatlas.org/ENSG00000184905-TCEAL2/tissue; last accessed on August 12, 2020) and a recent study has found variants of this gene shared among patients suffering from major depressive disorder (Subaran et al. 2016). Deletion of the chromosomal region covering TCEAL2 also causes mental retardation and behavioral disorders in humans (Grillo et al. 2010). These studies suggest an important regulatory role for TCEAL2 in factors controlling emotional and/or cognitive function, and TCEAL2 is thus a candidate gene underlying dynamic transcriptomic changes in the rabbit brain during domestication.
Fig. 3.
Signatures of selection for genomic regions harboring (A) TCEAL2, (B) NTRK1, (C) DNAH11, and (D) RPL21. Black lines and points represent window-based (±5 kb) and per-site dAF values, respectively. Red dashed lines indicate top 1% of dAF calculated from its distribution across the genome. A red bold line in (C) represents a selective sweep reported in Carneiro et al. (2014). Both genes denoted with asterisk in (A) seem to be orthologs of human NXF2.
One of the significant characteristics of domestic rabbit brain is the reduced amygdala volume relative to wild conspecifics (Brusini et al. 2018). Since amygdala has been recognized as an evolutionarily primitive region that governs fear responses (LeDoux 2007; Choi and Kim 2010), the reduction in its volume is likely associated with the reduced fear response of domestic rabbits (Brusini et al. 2018). In the present study, we uncovered a significant enrichment of pathways related to dopaminergic neurotransmission, behavior, and startle response among the detected DEGs in amygdala, including two important paralogs of dopamine receptors (DRD1 and DRD2) that are major mediators of dopamine signaling in the central nervous system. The differential expression of dopaminergic genes may reflect the anatomical changes in the domestic rabbit brain mentioned above. Although a reduction of amygdala volume was observed, the proportion of dopaminergic neurons may have increased, leading to the consistent upregulation of dopaminergic genes. Given that dopamine signaling in amygdala is generally considered to regulate fear responses (Abraham et al. 2014; Bergman et al. 2014; Lee et al. 2017), our results strongly suggest dopamine signaling as a candidate pathway that contributed to the evolution of tameness in domestic rabbits. Strikingly, we observed that all the differentially expressed dopaminergic genes were upregulated in domestic individuals (fig. 2c and supplementary fig. S2a, Supplementary Material online). This is in fact consistent with previous studies showing the effects of dopaminergic regulation in amygdala on fear response, such as that enhanced dopaminergic signaling, as represented by DRD1, DRD2, and PPP1R1B, in medial prefrontal cortex and amygdala rescued deficient fear extinction of mice in a context-independent manner (Whittle et al. 2016). Furthermore, chronic social stress led to decreased expression of ADORA2A, DRD2, GPR88, RGS9, and PPP1R1B in amygdala associated with increased fear acquisition of mice (Azzinnari et al. 2014), indicating that upregulation of dopaminergic genes in amygdala, which likely promotes dopamine signaling, could be associated with the reduced fear responses of domestic rabbits.
Although signals of selection were not generally associated with these dopaminergic genes, we found that SLC17A8, which encodes vesicular glutamate transporter 3 and detected as a DEG in this study, was located in a selective sweep region in domestic rabbits (supplementary table S2, Supplementary Material online). Glutamate is an excitatory neurotransmitter working in the central nervous system affecting dopamine signaling (Sakae et al. 2015; Nishi et al. 2017). Glutamate receptors, such as GRM3 (glutamate metabotropic receptor 3) and GRIN2D (glutamate ionotropic receptor NMDA type subunit 2D) have been suggested to be under selection during the evolution of tameness in domesticated fox (Wang et al. 2018). Moreover, GRIK2 (glutamate receptor, ionotropic, kainate 2) was shown to be associated with a selective sweep in domestic rabbits (Carneiro et al. 2014). Compared with such receptors, SLC17A8 may be more closely linked to mechanisms regulating synaptic neurotransmitter release and can greatly contribute to the magnitude of signaling. NTRK1 (neurotrophic receptor tyrosine kinase 1) could also be of particular interest as it is located in the vicinity of dopaminergic gene clusters in the PPI network (fig. 2a) and has highly differentiated SNPs in adjacent genomic regions (fig. 3b). Neurotrophins are a family of neurotrophic factors known to play crucial roles during neuronal maintenance and plasticity (Huang and Reichardt 2001) and they have a significant impact on brain development.
Cilia and flagella are found in various types of cells and fulfill important functions in motility, sensory reception, and signaling (Wheatley et al. 1996; Satir and Christensen 2007). Genes encoding proteins in the dynein complex and ciliary axoneme were significantly enriched among DEGs detected in hippocampus, and they consistently exhibited lower expression in domestic rabbits than in wild rabbits (table 1 and supplementary table S2, Supplementary Material online; fig. 2c and supplementary fig. S2b, Supplementary Material online). These genes include those encoding axonemal proteins of the outer or inner dynein arm and its docking complex (DNAH1, DNAH7, DNAH9, DNAH11, DNAI2, DYNLRB2, and ZMYND10), the radial spoke heads (RSPH9), the central-pair apparatus (HYDIN), and the one regulating and promoting centriole amplification and maturation (CCNO), many of which seem to have important roles in motile cilia (Reiter and Leroux 2017). Motile cilia are microtubule-based structures protruding from the apical surface of sperm, epithelial cells in the airways, the oviduct, and the brain ventricles (Spassky 2013). In the brain ventricles, ciliary beating of ependymal cells generates cerebrospinal fluid flow, which not only transports nutrients and signaling molecules, and removes waste products (Del Bigio 2010) but also supports neural stem cell proliferation (Petrik et al. 2018) and neuronal migration (Sawamoto 2006). On the other hand, neurons and astrocytes have immotile primary cilia growing from their basal bodies, and primary cilia have been found to regulate hippocampal neurogenesis through sonic hedgehog signaling (Breunig et al. 2008). Our network analysis and intersection with selection signals detected DNAH11 as the most promising candidate which may have driven evolutionary changes in ciliary gene expression, given its central location in the ciliary gene network (fig. 2b) and the observed selection signal upstream and within the intronic region of the gene (fig. 3c). TTC21B, encoding retrograde intraflagellar transport protein IFT139, and one of the prime candidate genes under selection during domestication with a missense mutation at a residue that is universally conserved among mammals except in domestic rabbits (Carneiro et al. 2014), could also affect the expression levels of ciliary DEGs including its interacting paralog TTC21A (supplementary table S2, Supplementary Material online). Since TTC21B regulates sonic hedgehog activity and forebrain development in mice (Tran et al. 2008; Stottmann et al. 2009) and TTC21B mutations cause ciliopathies in humans (Davis et al. 2011), it is possible that the missense mutation in TTC21B has a causal relationship with the consistent downregulation of many ciliary genes in the hippocampus of domestic rabbits (fig. 2c and supplementary fig. S2b, Supplementary Material online). The downregulation of ciliary genes might in turn cause reduced neurogenesis in hippocampus, consistent with the reduced volume of hippocampus and the suggested reduced neural speed and/or compromised information processing in the brain of domestic rabbits (Brusini et al. 2018).
Lastly, genes encoding ribosomal proteins were significantly enriched in the DEGs across all brain regions (figs. 1 and 2;supplementary database S2, Supplementary Material online), although the enrichment was not detected in DAVID or GREAT analysis due to the lack of accurate annotation and orthologous information for those genes. One of the genes, RPL21, stood out as it was among the 27 DEGs across the four brain regions. This gene exhibited the most prominent fold change (fig. 1b) and showed a signature of selection in the adjacent genomic region (fig. 3d), though its specific function is unknown. Given the fundamental roles in protein synthesis and translation within cells, the differential expression of genes encoding ribosomal proteins could profoundly affect the function of a large number of genes. A series of recent studies have shown a link between changes in ribosomal gene expression across various areas of the brain and behavior associated with changes in brain neurotransmitter activities, caused by social defeat or fighting experience in mice (Smagin et al. 2016, 2018). Since the mechanism underlying the involvement of ribosomal genes in brain function is still poorly understood, unraveling the functional significance of the concerted expression changes of ribosomal genes in the domestic rabbit brain is of particular interest.
Overall, our study revealed differential expression between wild and domestic newborn rabbits in genes involved in hippocampal ciliary function and amygdaloid dopamine signaling. These genes were all expressed in a very concerted way, which could be derived from dynamic trans-regulatory changes rather than cis-regulatory changes in each gene, given that the majority of DEGs were not associated with signals of selection. Considering that our samples were obtained from newborns, it is noteworthy that the pathways related to neuronal development and/or flight response already exhibit significant differentiation in transcriptional regulation at this very early age of development. Given its limited sample size, it is possible that the present study failed to capture subtle differences in expression changes between wild and domestic brains. In future work, it would be worthwhile to investigate differential gene expression at a higher resolution using more samples and multiple time points during development and with a higher spatial resolution using spatial transcriptomics or single-cell sequencing. Furthermore, gene editing may be used to investigate the possible phenotypic effects caused by the most obvious candidate mutations, such as the missense mutation in TTC21B.
Materials and Methods
Sample Information, RNA Extraction, and Library Preparation
We sampled three wild (Oryctolagus cuniculus cuniculus) and three domestic newborn rabbits (New Zealand white). All wild rabbits were sampled from a single litter, whereas all domestic rabbits came from different litters. Rabbits were kept under standard conditions of housing with unrestricted access to food and water at the Research Center of Wild Lagomorphs (Córdoba, Spain). All experimental procedures complied with European Union (Directive No. 2010/63/UE) and the corresponding permits were issued by the Ethical Committee of the Consejo Superior de Investigaciones Cientificas (CSIC). Newborn rabbits were deeply sedated with a mixture of xylacin (Rompun, 8 mg/kg; Bayer) and ketamine (Imalgene 1000, 40 mg/kg; Merial) administered intramuscularly. Newborn decapitation was performed following induction of anesthesia. Four regions were dissected for expression analyses (amygdala, hypothalamus, hippocampus, and parietal/temporal cortex). The dissection of the different brain parts was guided by an atlas for the adult rat brain (Palkovits and Brownstein 1988), and all samples were dissected by the same person. Wild and domestic rabbits were sampled 1 week apart. The extracted brains were frozen using liquid nitrogen and kept at −80 °C until RNA was extracted. The total RNA was isolated with the AllPrep DNA/RNA/miRNA Universal kit (Qiagen). The entire tissue was homogenized using the TissueLyser LT (Qiagen) and the appropriate volume of lysate was used for RNA isolation. The RNA quality and concentration were measured by the RNA ScreenTape assay (TapeStation, Agilent Technologies).
Strand-specific mRNA sequencing libraries were generated using the SENSE RNA-Seq Library Prep kit (Lexogen). Briefly, 1 µg of total RNA was poly-A selected using magnetic beads. Illumina-compatible linker sequences were introduced to the mRNA by random hybridization. The amplified libraries were size-selected for an average insert size of ∼320 bp. The libraries were sequenced as 125-bp paired-end reads using an Illumina HiSeq instrument. The hippocampus and parietal and temporal cortex libraries were sequenced at Centre for Genomic Regulation, Spain, whereas the amygdala and hypothalamus libraries were sequenced at SciLifeLab-Uppsala, Sweden. Except for the dissection, samples from wild and domestic rabbits were processed in parallel in the two sequencing batches. Thus, for each tissue all samples (wild and domestic) were run on the same lane.
Data Processing and Differential Expression Analysis
We evaluated the quality of the raw reads using FastQC (Andrews 2010) to perform downstream filtering and trimming. We first trimmed RNA-seq reads using trimmomatic (version 0.3) (Bolger et al. 2014) using default parameters and keeping only pairs with at least 36-bp length. About 94–97% of the reads passed trimming and were used for alignment to the OryCun2 genome assembly. Rabbit is a highly polymorphic species, and here, we compare wild and domestic individuals. Therefore, in order to avoid mapping bias, we aligned the reads with the Genomic Short-read Nucleotide Alignment Program (GSNAP, version 2014-12-23) (Wu and Nacu 2010) using a catalog of previously identified SNPs (Carneiro et al. 2014). Approximately 80% of the reads were successfully aligned to the OryCun2 genome assembly (supplementary table S1, Supplementary Material online; Carneiro et al. 2014). As for mitochondrial genes, chrUn1133 was a misassembled sequence of mitochondrial genome which influenced the alignment of reads representing the mitochondrial genome. In order to overcome mapping biases, we realigned reads on only mitochondrial genome and performed differential expression analyses of mitochondrial genes together with nuclear genes following the pipeline described below.
Principal component analysis of the overall profiles divided the samples into two major clusters: One group consisting of samples representing amygdala and hypothalamus and other consisting of samples from hippocampus and parietal/temporal cortex (supplementary fig. S5, Supplementary Material online). We did not observe a clear distinction between the two brain regions within each major group. This subdivision into major groups is likely explained by batch effects, since the two groups of samples were sequenced at two different sequencing platforms (see above; supplementary table S1, Supplementary Material online). However, this technical artifact is not expected to affect our inferences of gene expression differences between wild and domestic rabbits since all comparisons were performed within and not between brain regions and therefore used samples sequenced at the same platform.
We assigned the reads to Ensembl76 annotation and generated fragment counts by featureCounts (v1.4.5-p1) (Liao et al. 2014) extracting fragment counts where reads with mapping quality >20, which were uniquely mapped reads in essence, and only pairs that are properly aligned on the same chromosome. We used fragment counts to perform differential expression analysis using edgeR (Robinson et al. 2010) with pairwise comparison (exactTest) for each region of brain. After normalizing all the count data for effective library size as a single matrix based on the trimmed mean of M values method, we kept genes with >1 count per million (CPM) in at least three samples in each comparison. We determined significant DEGs by three criteria: 1) FDR (corrected by the Benjamini and Hochberg method) ≤0.05, 2) |log2-fold changes|≥1, and 3) SD per mean of CPM (STD/MeanCPM) in a given group <1 and no overlaps in CPM between wild and domestic rabbits. To further confirm the consistency in the differential expression across regions of brain, we conducted permutation tests by shuffling the label (wild and domestic) for each animal.
DEGs were further evaluated by performing enrichment analysis for GO pathways, and MGI phenotypes by using the DAVID (version 6.8) (Dennis et al. 2003) and the GREAT (version 4.0.4) (McLean et al. 2010). One-to-one human orthologs and their coordinates corresponding to all the genes expressed in each region of rabbit brain were obtained from Ensembl76 and used as background list of genes in both analyses. Moreover, PPIs among DEGs were visualized through STRING (version 11.0; Szklarczyk et al. 2019) and clustered subnetworks were detected by the MCODE module (version 1.5.1) of Cytoscape (version 3.8.0; Shannon 2003). In this analysis, we used all the 141 (out of 147) DEGs in amygdala and all the 125 (out of 135) DEGs in hippocampus that are included in the STRING database.
Intersection with Genes Showing Signals of Selection in Domestic Rabbits
To investigate the overlap between DEGs and genes showing strong differentiation between wild and domestic rabbits, we used the data from our previous study (Carneiro et al. 2014). Briefly, the study involved resequencing of six pools of domestic rabbits and 14 pools of wild rabbits with the aim to detect genomic regions under selection during domestication. In the present study, we first intersected DEGs with ±100 kb of each selective sweep region reported in a previous study (Carneiro et al. 2014). We further utilized dAF data between wild and domestic rabbits (Carneiro et al. 2014) for SNPs with dAF>0.5 and conducted the same analysis using a threshold of three or more SNPs with dAF>0.9 in a gene region ±100 kb based on Ensembl76 annotation. For genes of particular interest, the calculation of dAF on a window basis (window size: 10 kb; step size: 5 kb; the average number of SNPs per window was 10.0) was conducted to reveal significant deviations from the genome-wide average. Subsequently, we evaluated the possible association between divergent SNPs in putative regulatory regions and differences in gene expression between wild and domestic individuals. This was done by comparing the absolute difference (log2-fold changes) in gene expression levels between genes with or without a highly divergent SNP (dAF≥0.8) located on conserved noncoding elements within 1 Mb from the TSS. This annotation was conducted based on 29 mammalian sequences and Ensembl annotations (Carneiro et al. 2014). Moreover, comparing the relative abundance (M values) of SNPs within ±100 kb from TSS of DEGs and non-DEGs across dAF bins, we examined if the SNPs located within ±100 kb from TSS of DEGs exhibited a trend toward high dAFs. M values were calculated as follows:
where is the observed number of SNPs in the given bini (ranges from 0.50–0.55 to 0.95–1.00) and categoryj, classified by whether they are located near DEGs or not for each comparison of four brain regions. is the expected number of SNPs in the given bini (from 0.50–0.55 to 0.95–1.00) and categoryj, and was calculated as below:
where is the total number of SNPs located within ±100 kb from TSS of any genes across genome (n = 963,548) and and are the total number of SNPs in the given bini or categoryj.
Supplementary Material
Acknowledgments
We thank Mats Fredrikson for valuable comments on the article. The National Genomics Infrastructure (NGI)/Uppsala Genome Center and UPPMAX provided service in massive parallel sequencing and computational infrastructure. Work performed at NGI/Uppsala Genome Center has been funded by RFI/VR and Science for Life Laboratory, Sweden. The study was supported by Knut and Alice Wallenberg Foundation (to L.A.), Vetenskapsrådet (to L.A.), and by the Fundação para a Ciência e Tecnologia (FCT) through POPH-QREN funds from the European Social Fund and Portuguese MCTES (FCT Investigator Grants to M.C.; IF/00283/2014/CP1256/CT0012).
Author Contributions
L.A., M.C., and F.H. conceived the study. M.C., J.A.B.-A., and R.V. developed the animal material. H.R. dissected all brain samples. S.Y. and C.F. performed experimental work. D.X.S. and N.R. were responsible for the bioinformatics analysis. C.J.R. contributed to bioinformatic analysis. D.X.S., N.R., and L.A. wrote the article with contributions from all other authors. All authors approved the article before submission.
These authors contributed equally to this work.
Data deposition: This project has been deposited at SRA under the accession number PRJNA603201. The codes used in the present study are available on https://github.com/nimarafati/Rabbit_Domestication_Brain_RNA_Seq.
Literature Cited
- Abraham AD, Neve KA, Lattal KM. 2014. Dopamine and extinction: a convergence of theory with fear and reward circuitry. Neurobiol Learn Mem. 108:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert FW, et al. 2012. A comparison of brain gene expression levels in domesticated and wild animals. PLoS Genet. 8(9):e1002962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed August 12, 2020.
- Azzinnari D, et al. 2014. Mouse social stress induces increased fear conditioning, helplessness and fatigue to physical challenge together with markers of altered immune and dopamine function. Neuropharmacology 85:328–341. [DOI] [PubMed] [Google Scholar]
- Bergman O, et al. 2014. Association between amygdala reactivity and a dopamine transporter gene polymorphism. Transl Psychiatry. 4(8):e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig JJ, et al. 2008. Primary cilia regulate hippocampal neurogenesis by mediating sonic hedgehog signaling. Proc Natl Acad Sci U S A. 105(35):13127–13132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusini I, et al. 2018. Changes in brain architecture are consistent with altered fear processing in domestic rabbits. Proc Natl Acad Sci U S A. 115(28):7380–7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M, et al. 2011. The genetic structure of domestic rabbits. Mol Biol Evol. 28(6):1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M, et al. 2014. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science 345(6200):1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J-S, Kim JJ. 2010. Amygdala regulates risk of predation in rats foraging in a dynamic fear environment. Proc Natl Acad Sci U S A. 107(50):21773–21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock J. 1999. The natural history of domesticated mammals. Cambridge: Cambridge University Press. [Google Scholar]
- Darwin C. 1859. On the origins of species by means of natural selection or the preservation of favoured races in the struggle for life. London: John Murray. [PMC free article] [PubMed] [Google Scholar]
- Davis EE, et al. 2011. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet. 43(3):189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bigio MR. 2010. Ependymal cells: biology and pathology. Acta Neuropathol. 119(1):55–73. [DOI] [PubMed] [Google Scholar]
- Dennis G, et al. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4(9):R60. [PubMed] [Google Scholar]
- Dorshorst B, et al. 2015. A genomic duplication is associated with ectopic eomesodermin expression in the embryonic chicken comb and two Duplex-comb phenotypes. PLoS Genet. 11(3):e1004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo L, et al. 2010. Familial 1.1 Mb deletion in chromosome Xq22.1 associated with mental retardation and behavioural disorders in female patients. Eur J Med Genet. 53(2):113–116. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. 2001. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 24(1):677–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imsland F, et al. 2012. The Rose-comb mutation in chickens constitutes a structural rearrangement causing both altered comb morphology and defective sperm motility. PLoS Genet. 8(6):e1002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux J. 2007. The amygdala. Curr Biol. 17:868–874. [DOI] [PubMed] [Google Scholar]
- Lee JH, Lee S, Kim J-H. 2017. Amygdala circuits for fear memory: a key role for dopamine regulation. Neuroscientist 23(5):542–553. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30(7):923–930. [DOI] [PubMed] [Google Scholar]
- McLean CY, et al. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 28(5):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi A, et al. 2017. Glutamate counteracts dopamine/PKA Signaling via dephosphorylation of DARPP-32 Ser-97 and alteration of its cytonuclear distribution. J Biol Chem. 292(4):1462–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palkovits M, Brownstein MJ. 1988. Maps and guide to microdissection of the rat brain. New York: Elsevier Science Publ. Co. [Google Scholar]
- Petrik D, et al. 2018. Epithelial sodium channel regulates adult neural stem cell proliferation in a flow-dependent manner. Cell Stem Cell 22(6):865–878. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Leroux MR. 2017. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 18(9):533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakae DY, et al. 2015. The absence of VGLUT3 predisposes to cocaine abuse by increasing dopamine and glutamate signaling in the nucleus accumbens. Mol Psychiatry. 20(11):1448–1459. [DOI] [PubMed] [Google Scholar]
- Satir P, Christensen ST. 2007. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 69(1):377–400. [DOI] [PubMed] [Google Scholar]
- Sawamoto K. 2006. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science 311(5761):629–632. [DOI] [PubMed] [Google Scholar]
- Shannon P. 2003. Cytoscape: a software environment for integrated models. Genome Res. 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagin DA, et al. 2016. Dysfunction in ribosomal gene expression in the hypothalamus and hippocampus following chronic social defeat stress in male mice as revealed by RNA-Seq. Neural Plast. 2016:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagin DA, et al. 2018. Heterogeneity of brain ribosomal genes expression following positive fighting experience in male mice as revealed by RNA-Seq. Mol Neurobiol. 55(1):390–401. [DOI] [PubMed] [Google Scholar]
- Spassky N. 2013. Motile cilia and brain function: ependymal motile cilia development, organization, function and their associated pathologies In: Tucker L, Caspary T, editors. Cilia and nervous system development and function. Dordrecht (The Netherlands): Springer; p. 193–207. [Google Scholar]
- Stottmann RW, Tran PV, Turbe-Doan A, Beier DR. 2009. Ttc21b is required to restrict sonic hedgehog activity in the developing mouse forebrain. Dev Biol. 335(1):166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subaran RL, Odgerel Z, Swaminathan R, Glatt CE, Weissman MM. 2016. Novel variants in ZNF34 and other brain-expressed transcription factors are shared among early-onset MDD relatives. Am J Med Genet. 171(3):333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, et al. 2019. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47(D1):D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PV, et al. 2008. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet. 40(4):403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. 2018. Genomic responses to selection for tame/aggressive behaviors in the silver fox (Vulpes vulpes). Proc Natl Acad Sci U S A. 115(41):10398–10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley DN, Wang AM, Strugnell GE. 1996. Expression of primary cilia in mammalian cells. Cell Biol Int. 20(1):73–81. [DOI] [PubMed] [Google Scholar]
- Whittle N, et al. 2016. Enhancing dopaminergic signaling and histone acetylation promotes long-term rescue of deficient fear extinction. Transl Psychiatry. 6(12):e974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D, et al. 2009. Copy number variation in intron 1 of SOX5 causes the Pea-comb phenotype in chickens. PLoS Genet. 5(6):e1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26(7):873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.