Abstract
Theories predict that directional selection during adaptation to a novel habitat results in elevated meiotic recombination rate. Yet the lack of population-level recombination rate data leaves this hypothesis untested in natural populations. Here, we examine the population-level recombination rate variation in two incipient ecological species, the microcrustacean Daphnia pulex (an ephemeral-pond species) and Daphnia pulicaria (a permanent-lake species). The divergence of D. pulicaria from D. pulex involved habitat shifts from pond to lake habitats as well as strong local adaptation due to directional selection. Using a novel single-sperm genotyping approach, we estimated the male-specific recombination rate of two linkage groups in multiple populations of each species in common garden experiments and identified a significantly elevated recombination rate in D. pulicaria. Most importantly, population genetic analyses show that the divergence in recombination rate between these two species is most likely due to divergent selection in distinct ecological habitats rather than neutral evolution.
Keywords: single-sperm genotyping, genetic maps, habitat transition, Daphnia pulex, Daphnia pulicaria, Qst, Pst
Significance
Whether directional selection during adaptation to a novel habitat results in elevated meiotic recombination remains largely untested in natural populations. This work examines the population-level recombination rate in two closely related microcructacean species Daphnia pulex and Daphnia pulicaria using single-sperm genotyping approach. Recombination rate data from two linkage groups show elevated recombination rates in D. pulicaria whose divergence from D. pulex is accompanied by a habitat shift. Importantly, population genetic analysis suggests that this divergence of recombination is likely adaptive rather than neutral.
Introduction
Meiotic recombination is a hallmark of meiosis as it occurs in the majority of sexually reproducing eukaryotes (Cavalier-Smith 2002; Otto and Lenormand 2002). Although it remains contested as to why recombination originated in the last common ancestor of eukaryotes (Kondrashov 1988; Cavalier-Smith 2002), recombination plays an essential role in repairing the actively induced double-strand DNA breaks in the prophase I of meiosis (Pâques and Haber 1999). The presence of at least one recombination event (i.e., crossover event) per chromosome arm between homologous chromosomes ensures the correct segregation of chromosomes into daughter cells, preventing chromosome nondisjunction and aneuploidy (Hassold and Hunt 2001).
Besides its well-known role in creating new haplotypes and in facilitating adaptation (Rice 2002), meiotic recombination is an important evolutionary force shaping the eukaryotic genomic architectures. Recombination rate is a determinant of the distribution of genetic diversity in the genomes (Begun and Aquadro 1992; Charlesworth et al. 1993; Lercher and Hurst 2002; Booker et al. 2017). Recombination reduces selection interference between linked sites (Hill and Robertson 1966; Felsenstein and Yokoyama 1976; Barton 1995b; Cutter and Payseur 2013) and slows down the accumulation of deleterious mutations and of transposable elements (Lynch et al. 1993; Rizzon et al. 2002; Dolgin and Charlesworth 2008; Kent et al. 2017). Moreover, recombination and associated biased gene conversion can influence codon usage bias (Comeron et al. 1999; Pouyet et al. 2017) and base composition (Duret and Arndt 2008; Mugal et al. 2015).
Meiotic recombination rate varies greatly at multiple biological levels, e.g., within genome, between individuals and populations, and between species (Smukowski and Noor 2011; Ortiz-Barrientos et al. 2016; Dapper and Payseur 2017; Ritz et al. 2017). Understanding the genetic basis and evolutionary forces underlying such variation is a major challenge to biologists. Striking progress has been made in mapping the genetic factors responsible for within-genome variation and for between-individual variation. For example, the zinc finger domain protein PRDM9 is a major determinant of recombination hotspots in the genomes of human and mice (Baudat et al. 2010; Myers et al. 2010; Grey et al. 2011; Brick et al. 2012). In addition, promoters and transcription start sites have been identified to be associated with elevated recombination rate in dogs (Auton et al. 2013), the yeast Saccharomyces cerevisiae (Pan et al. 2011), birds (Singhal et al. 2015), and Arabidopsis (Choi et al. 2013). On the individual level, several meiosis-related genes (e.g., Rnf212, Cplx1, Rec8, Prdm9) have been identified to be responsible for variation of recombination rates in mammalian species including humans, cattle, and Soay sheep (Kong et al. 2008; Chowdhury et al. 2009; Sandor et al. 2012; Johnston et al. 2016; Halldorsson et al. 2019). However, it should be noted that these loci explain only a small portion (∼3–11%) of the phenotypic variance between individuals (Kong et al. 2014; Johnston et al. 2016).
In contrast, the genetic factors governing the interspecific variation of recombination rate remain understudied (Dapper and Payseur 2017), although many studies have compared recombination rate differences between closely related species at different genomic scales (Smukowski and Noor 2011). We note that this research area has drawn increasing amount of attention, with a dicistronic gene mei-217/mei-218 recently identified to be responsible for recombination rate difference between Drosophila melanogaster and Drosophila mauritiana (Brand et al. 2018) and TEX11 and other genes involved in synaptonemal complex suggested as candidates driving the evolution of recombination rate in mammals (Dapper and Payseur 2019). On the other hand, another equally understudied question is whether natural selection plays a role in shaping the between-species divergence. Despite numerous evolutionary theories have examined how natural selection can modulate the evolution and divergence of recombination rates between species, the lack of in-depth population-level data (see below) leaves these theories untested in natural systems, severely limiting our understanding of the evolutionary forces driving recombination rate divergence.
In populations undergoing divergence and incipient speciation, recombination rates could be driven to increase if the breakdown of overrepresented association of alleles, i.e., linkage disequilibrium, is beneficial. Generally speaking, three different situations can lead to the buildup of linkage disequilibrium and determine how recombination rates respond to natural selection. In the presence of linkage disequilibrium caused by weak negative epistasis, selection favors increased recombination in a large population in stable environment (Otto and Lenormand 2002). Genetic drift can also lead to the accumulation of linkage between beneficial alleles and deleterious alleles in finite populations, and the increase of recombination rate would be favored by selection to bring together beneficial alleles (Otto 2009). Furthermore, temporal fluctuations in the environment favor different combinations of alleles, which could lead to increased recombination rate in environments with rapid and consistent temporal variation (Charlesworth 1976; Barton 1995a; Otto and Michalakis 1998), whereas in the absence of fluctuations, recombination is selected against.
Despite the diverse views on the relative importance of these evolutionary forces in shaping the evolution of recombination rate, it is consistently predicted that transition to a novel environment would lead to an increase of recombination rate due to directional selection (Butlin 2005). Empirical work on indirect selection of physiology-related traits in Drosophila supports this view (Korol and Iliadi 1994; Aggarwal et al. 2015). However, for domesticated animals that underwent strong directional selection, there seems to be no increase of recombination rate (Munoz-Fuentes et al. 2015), contradicting previous views of elevated recombination in domesticated plants (Ross-Ibarra 2004) and animals (Burt and Bell 1987; Poissant et al. 2010).
Notably, few studies have directly addressed whether habitat shift in natural populations results in elevation of recombination rate. A key challenge is that, for model organisms where recombination is heavily investigated, for example, human, mice, Drosophila, and yeast, little is known about the ecological changes involved in speciation. Thus, the interspecific difference between Drosophila species, for example, ∼2-fold difference between Drosophila melanogaster and Drosophila mauritiana (Brand et al. 2018), and the difference between yeast species, for example, 40% lower recombination rate in Saccharomyces paradoxus than in S. cerevisiae (Liu et al. 2019), are unfortunately decoupled from the consideration of ecology.
Another challenge in understanding the relationship between ecological shifts, directional selection, and recombination rate evolution is that multipopulation data on recombination rate is largely lacking (but see Saleem et al. 2001 and Samuk et al. 2020). Recombination rate is laborious to measure, which usually involves producing and genotyping hundreds of recombinant progenies with a large number of genetic markers to generate only a single genetic map. Such practice is difficult to scale up to population-level studies. Thus, current estimates of recombination rates for most species are derived from the average recombination rates in the two lineages used for crossing-based map construction. Often the number of genetic maps for a single species remains below a handful except for some heavily studied model organisms and economically important crops and animals, yielding low statistical power for rigorously investigating the driving forces of interspecific differentiation of recombination rate in a population genetic framework.
If a genetic map is constructed using computational methods based on linkage disequilibrium through population sequencing, we can obtain estimates of population recombination rates (4×Ne×c), which is an average of two sexes over large span of evolutionary time (McVean et al. 2004). The fact that population recombination rate is scaled by effective population size (Ne) makes it difficult to directly estimate recombination rate and can confound comparisons between diverging populations that may have distinct demographic histories (Rogers 2014; Dapper and Payseur 2018). Promising advances to incorporate demographic changes into this approach have emerged in recent years (Kamm et al. 2016; Spence and Song 2019). However, studies using this method to compare multiple populations remain rare beyond the classic models, mostly with a focus on intragenomic variation (Auton et al. 2012). Therefore, we argue that the lack of understanding on the population- and individual-level variation of recombination rates ought to be addressed if we are to dissect the genetic basis of recombination rate variation.
The emergence of novel genomic sequencing techniques such as whole-genome sequencing of single-sperm cells (Xu et al. 2015) provides an efficient solution to estimating population-level recombination variation (albeit it only measures male-specific recombination rate). Taking advantage of this approach to investigate how ecological shifts and directional selection impact recombination rate, this study examines the male-specific recombination rate in two ecologically distinct, incipient microcrustacean species Daphnia pulex and D. pulicaria.
A well-known characteristic of the Daphnia system resides in its cyclically parthenogenetic reproduction. Under favorable environmental conditions, female Daphnia produces directly developing embryos (i.e., live birth of neonates released from brood pouch) via apomictic parthenogenesis, generating genetically identical, diploid daughters. However, stressful conditions, e.g., food shortage (Deng 1996) and decrease in temperature, trigger Daphnia females to switch to sexual reproduction and also to parthenogenetically produce males via environmental sex determination (Olmstead and Leblanc 2002). The parthenogenetic production of males allows us to amass a large number of males and sperm cells of the same genotype to examine recombination rate.
As members of the D. pulex species complex, D. pulex and D. pulicaria are estimated to have started diverging from 800,000 to 2,000,000 years ago (Colbourne and Hebert 1996; Omilian and Lynch 2009; Cristescu et al. 2012). These two species are morphologically nearly indistinguishable (Brandlova et al. 1972) but occupy distinct, overlapping freshwater habitats in North America, with D. pulex mostly living in ephemeral fishless ponds and D. pulicaria inhabiting stratified permanent lakes. Importantly, population genetic data suggest that the divergence of D. pulicaria from D. pulex most likely involved a habitat transition event from pond to lake systems (Cristescu et al. 2012).
As stratified permanent lakes and ephemeral ponds pose distinct selection regimes (e.g., distinct predators, environmental factors), these two species have most likely undergone strong local adaptation and divergent selection in their distinct habitats, resulting in clear physiological and behavioral differences. For example, compared with D. pulicaria, D. pulex grows faster to a larger size, reproduces at an earlier age. In addition, D. pulicaria exhibits diel vertical migration in lakes, whereas D. pulex displays no such behavior. Interestingly, the frequency of sexual reproduction is also different between the two. Daphnia pulex goes through sexual reproduction producing resting eggs before ponds dry up in early summer every year, whereas D. pulicaria can persist in lakes largely without sex for a few years (Dudycha and Tessier 1999; Cáceres and Tessier 2004; Dudycha 2004). Notably, prezygotic isolation has developed between these two species (Deng 1997), with D. pulex switching to sexual reproduction at long-day hours (16 h/day) and D. pulicaria switching to sexual at short-day hours (10 h/day). Despite these differences, D. pulex and D. pulicaria can still generate fertile cyclically parthenogenetic F1 offspring in laboratory crossing experiments, indicating the absence of complete reproductive isolation (Heier and Dudycha 2009).
In this pilot study, we examine whether neutral evolution (i.e., genetic drift) is sufficient to explain the divergence of meiotic recombination rate between these two species, with the alternative hypothesis being that directional selection involved in ecological shifts better explains the between-species divergence. As our pilot experiment, we estimated recombination rate for a 1.5-Mb region on linkage group 8 and a 0.5-Mb region on linkage group 9 in three geographically isolated populations of each species. Most interestingly, our results yield strong support for significantly higher recombination rate in D. pulicaria than in D. pulex, and the between-species divergence in recombination rate cannot be accounted for by genetic drift and is most likely due to directional selection.
Results
Recombination Rate Estimates
We performed microsatellite genotyping on whole-genome amplified single-sperm cells to estimate recombination rates for a 1.5-Mb region on linkage group 8 and a 0.5-Mb region on linkage group 9 in three populations of D. pulex and D. pulicaria each. Our recombination rate estimates show that D. pulicaria tends to recombine at a higher rate than D. pulex (figs. 1 and 2). For the region on linkage group 8 alone, although the mean recombination rate is higher in D. pulicaria, no statistically significant difference (t-test P = 0.10) is found between the mean of D. pulex (16.9 cM, SD = 8.4) and that of D. pulicaria (28.4 cM, SD = 4.5). However, for the region on linkage group 9, the average recombination rate of D. pulicaria (mean = 24.5, SD = 1.4) is higher (t-test P = 0.046) than that of D. pulex (mean = 13.0, SD = 6.8). When we compared the recombination rates of both linkage groups in these two species, D. pulicaria has an overall significantly higher recombination rate than D. pulex (t-test P = 0.006).
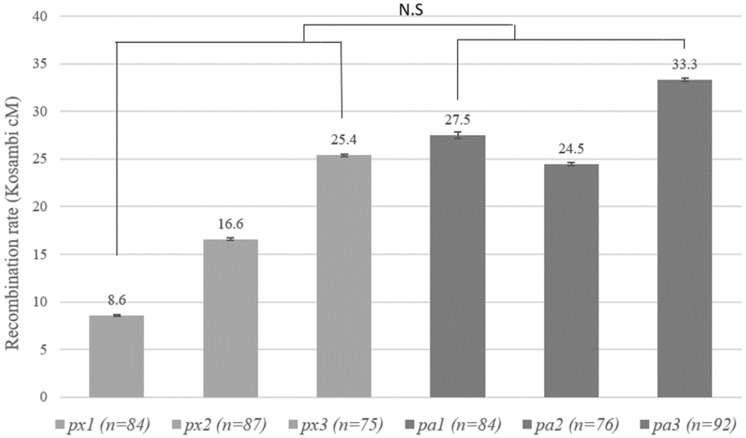
Fig. 1.
Estimated recombination rates for a 1.5-Mb region on linkage group 8 for three isolates of Daphnia pulex (px) and Daphnia pulicaria (pa) each (n represents the number of genotyped sperm). Each gray bar represents the recombination estimate from a specific Daphnia isolate with error bar representing SE. The average recombination rate between these two species is not significantly different (NS, not significant).
Remarkably, our recombination rate estimates show that the within-species recombination rate variation is markedly higher in D. pulex than in D. pulicaria for regions on both linkage groups. In D. pulex, recombination rates of the three sampled populations range from 8.6 to 25.4 Kosambi cM for the 1.5-Mb region on linkage group 8 with a nearly 3-fold difference (fig. 1). On the other hand, within-species variation in D. pulicaria for the interval in linkage group 8 is much lower, ranging from 24.5 to 33.3 Kosambi cM among the three examined populations (fig. 1).
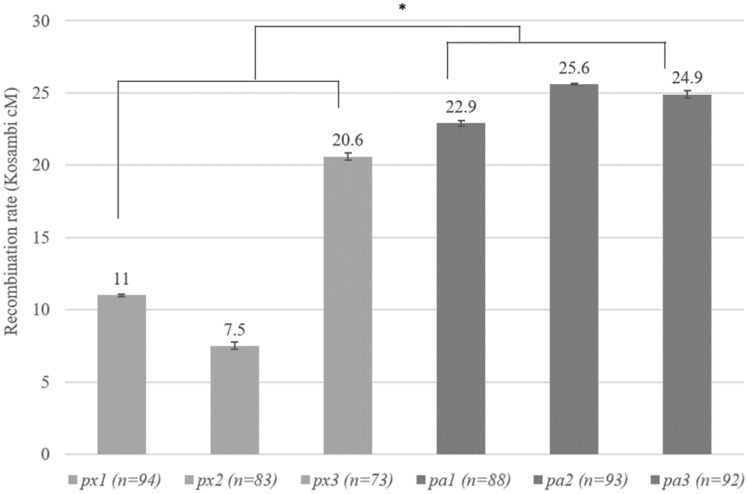
A resembling pattern of distinct within-species variation is also observed for linkage group 9. For D. pulex, the map distance of the 0.5-Mb region on linkage group 9 range between 7.5 cM and 20.6 Kosambi cM, showing a nearly 3-fold difference among populations (fig. 2). However, for D. pulicaria, the recombination rates of the same interval in linkage group 9 from the examined populations show little variation between 22.9 and 25.6 Kosambi cM (fig. 2).
Fig. 2.
Estimated recombination rates for a 0.5-Mb region on linkage group 9 for three isolates of Daphnia pulex (px) and Daphnia pulicaria (pa) each (n represents the number of genotyped sperm). Each gray bar represents the recombination estimate from a specific Daphnia isolate with error bar representing SE. The average recombination rate between these two species is significantly different (*P < 0.05).
P st–Fst Comparison
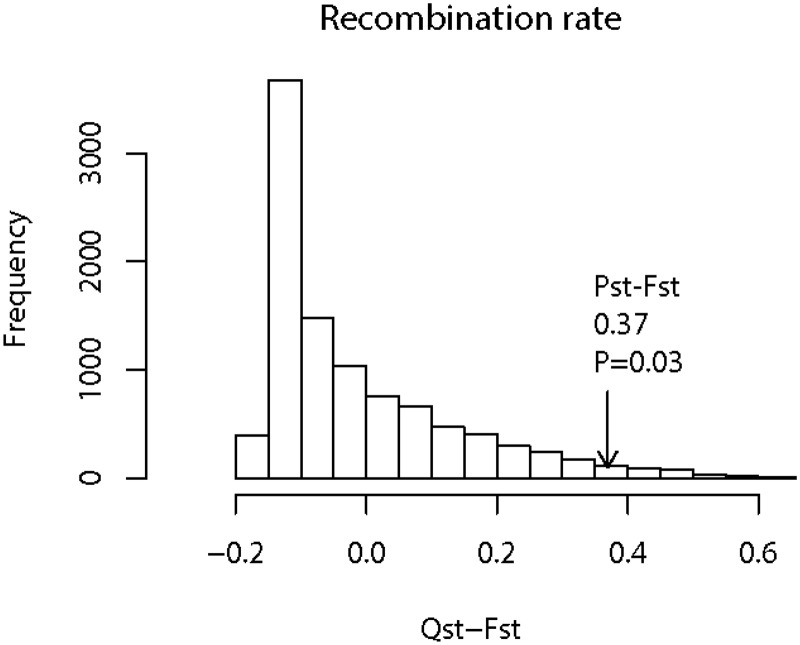
An important approach for determining whether the divergence of phenotypic traits is neutral is to compare Qst of phenotypic traits and Fst of neutral molecular markers. As Fst for molecular markers, Qst is a metric measuring the population differentiation for phenotypic traits (Prout and Barker 1993; Spitze 1993). In theory, the Qst of neutral traits on average should be equal to the mean Fst of neutral molecular markers (Rogers and Harpending 1992; Whitlock and Mccauley 1999; Whitlock 2008). We calculated Pst (Leinonen et al. 2006), a surrogate of Qst, based on the recombination rates of both linkage groups 8 and 9 (see Discussion for the implications of using Pst). The mean Pst for recombination rate is 0.52 based on our ANOVA analyses of 1,000 bootstrap replicates. We also estimated that the average genome-wide Fst of 4-fold degenerate sites between D. pulex and D. pulicaria is 0.15 (0.12 and 0.20 for the interval on linkage groups 8 and 9, respectively). Based on the distribution of Fst of genome-wide 4-fold degenerate sites, we simulated the Qst of a neutrally evolving trait to estimate the distribution of the test statistic Qst–Fst. Interestingly, our Pst–Fst value is significantly higher than the Qst–Fst values of the simulated neutrally evolving trait (fig. 3, P = 0.03), leading us to reject the neutral hypothesis and to conclude that recombination rate divergence between these two species is adaptive.
Fig. 3.
The simulated distribution of Qst–Fst for a neutral trait and the observed Pst–Fst (indicated by an arrow).
Discussion
A New Model System for Studying Recombination Rate Divergence
Meiotic recombination is one of the most laborious genetic parameters to estimate, with most species having no more than a handful of genetic maps each. Due to the lack of data on population-level recombination rate variation, many theories on the evolution of recombination remain untested in natural populations. In the past two decades, only a handful of studies surveyed the recombination rate variation within and between populations from different environments. Prior work on the fungus Sordaria fimicola revealed heritable genetic variation in recombination rate between strains inhabiting harsh and mild habitat, with higher recombination rates found in the harsh habitat than in the mild environment (Saleem et al. 2001). Using novel single-sperm genotyping approach, the current study supports the hypothesis that directional selection coupled with habitat shift leads to elevated recombination rate in the model system of microcrustacean Daphnia. One could argue that the permanent lake habitat seems more stable and less harsh than ephemeral pond habitats and lower recombination rate would evolve in the lake species D. pulicaria. We suggest that stability of lake environment may not necessarily mean a benign environment for Daphnia. Other factors such as predator abundance may also determine the harshness of environment to Daphnia. Moreover, the higher recombination rate in D. pulicaria may result from the fact that D. pulicaria has lower frequency of sexual reproduction than D. pulex so that higher recombination rate evolves to produce genetically diverse offspring. Interestingly, based on Pst–Fst comparison analysis, we find strong evidence that the divergence of recombination rates between these two species is adaptive and unlikely to be explained by genetic drift. With overall significantly higher recombination rates observed in D. pulicaria than in D. pulex, we argue that the directional selection that led to the local adaptation of D. pulicaria to permanent lake habitats (e.g., physiology, life history, see Introduction) most likely shaped the recombination rate divergence, providing support to the theory that directional selection leads to elevated recombination rate. However, with current data, we cannot exclude the possibly that selection reduces recombination rate in D. pulex. We also suggest that the different recombination rates of the surveyed regions in these two species are unlikely due to translocation events (e.g., in one species the surveyed region moved from the tip of chromosome to a more central location). This is because the interspecific F1s, F2s, and backcrosses have normal fertility (Heier and Dudycha 2009), suggesting no major chromosomal structure alterations affecting recombination.
It should be noted that different recombination rate between these two species is unlikely to be due to phenotypic plasticity as the habitat transition event most likely occurred ∼1–2 Ma, and the examined Daphnia isolates have been acclimatized to lab conditions and were measured for recombination rates in a common garden experiment. On the other hand, as the current study is based on a relatively small set of populations and on two linkage groups, it remains to be seen whether the observed pattern holds true for the genome-wide recombination rate variation for a larger set of D. pulex and D. pulicaria populations.
Although prior work identified some empirical support for the theory that directional selection leads to elevated recombination rate, these work has largely been restricted to comparing domesticated animals, plants, and fungi (Saleem et al. 2001; Ross-Ibarra 2004; Munoz-Fuentes et al. 2015) with their wild progenitors and to examining laboratory populations (Korol and Iliadi 1994; Aggarwal et al. 2015). Although studies investigating intra- and inter-specific recombination rate divergence are not uncommon (reviewed in Smukowski and Noor [2011]), this kind of studies are usually deficient in an ecological understanding of the speciation process or lack the population-level sampling required for inferring the driving forces of recombination rate differentiation. However, a recent study examining two ecologically different populations of Drosophila pseudoobscura found that the divergence of genome-wide recombination rate is due to natural selection (Samuk et al. 2020). Our study is valuable in providing solid evidence in support of this hypothesis from the perspective of incipient species pairs undergoing ecological speciation. Notably, as Daphnia is different from the other examined species in that their sexual reproduction is triggered by environmental conditions (e.g., they only reproduce under certain environmental conditions and the frequency of sex is less frequent in D. pulicaria than in D. pulex), it is likely that recombination rate in Daphnia is subject to stronger selection to produce genetically diverse offspring than other species that engages in regular sexual reproduction. The lower frequency of sexual reproduction in D. pulicaria may also contribute to its higher recombination rate than D. pulex. We therefore argue that the well-understood ecology (distinct ephemeral pond vs. permanent habitats) and evolutionary history (speciation associated with transition from pond to lake habitats) of D. pulex and D. pulicaria set up an excellent framework for future in-depth investigation of the evolutionary and genetic basis of divergence in recombination rates.
Single-Sperm Sequencing for Studying Recombination Rate Variation in Emerging Systems
A major hurdle in studying recombination rate variation is the laborious process of generating genetic maps. This study greatly benefited from the novel whole-genome sequencing technique developed for single-sperm cells (Xu et al. 2015; Xu and Young 2017). Single-sperm sequencing emerged in 1980s as a methodology for estimating localized recombination rates (Li et al. 1988; Cui et al. 1989). Nonetheless, empowered by whole-genome sequencing technologies, this technique has recently been applied to human and mouse to examine whole-genome recombination patterns (Lu et al. 2012; Wang et al. 2012; Hinch et al. 2019).
We note that as collecting a large number of sperm/pollen cells is feasible in many species, our experimental procedure for single-sperm sequencing/genotyping can be applied to other emerging model systems. Even for species with large genome sizes, we have seen multiple studies using single-sperm sequencing to examine the regulation of recombination events in mouse (Hinch et al. 2019) and human (Bell et al. 2020). Although our protocol uses flow cytometry to isolate single cells and relies on whole-genome amplification of single cells, these are currently common laboratory procedures. We hope that an increasing number of researchers will take advantage of this approach to examine the divergence of recombination rates in a diverse set of emerging model systems with interesting ecological attributes. Nonetheless, the sperm sequencing approach does not allow to evaluate sexual dimorphism and fine-scale variation in recombination rate.
P st–Fst Comparison
We used Pst–Fst comparison to determine whether the divergence of recombination rate between D. pulex and D. pulicaria is adaptive. This test is based on the observation that for neutral phenotypic traits that are controlled by purely additive genes the mean Qst (we used Pst as a surrogate for Qst) is equal to the mean Fst of neutral genetic loci (Lande 1992; Whitlock 1999). Although Qst for a quantitative trait is calculated using additive genetic variance obtained through breeding experiments, Pst is based on total phenotypic variance, which could be inflated due to environmental factors, thus complicating the interpretation of Pst–Fst comparison. Although the observation Qst=Fst for a neutral trait is based on several assumptions and this study likely violated some of them, we argue that the strong evidence pointing to the adaptive nature of the observed divergence in recombination rate is unlikely compromised (see below).
An important assumption of Qst=Fst for neutral phenotypic traits is that the loci from which Fst is derived should be neutral. Although there have been concerns about whether Qst=Fst when the Fst is based on markers such as microsatellites that have high mutation rate (Hendry 2002), the use of SNPs in our study alleviates this concern. Furthermore, despite that in other species 4-fold degenerate sites have been shown to experience purifying selection, population genomic analyses of D. pulex show that these sites evolve in a nearly neutral fashion (Lynch et al. 2017). Therefore, our use of genome-wide 4-fold degenerate sites (n = 94,711) should provide a meaningful estimate of the mean Fst of neutral sites.
Our analysis differs from the standard Qst–Fst analysis in the use of Pst as a substitute of Qst. However, this study differs significantly from studies directly collecting phenotypic data from the field because the recombination rates were estimated in a common garden experiment. As recombination rate is known to be of great phenotypic plasticity due to biotic and abiotic factors such as age and temperature (Hunter, Robinson, et al. 2016; Lloyd et al. 2018), we estimated recombination rates from 2-week-old males that were maintained under controlled temperature and photoperiod. Therefore, the obtained Pst value is unlikely to be inflated by environmental effects.
Because Qst is defined based on additive variance of traits, one may wonder whether the Pst–Fst test in this study is biased toward rejecting the neutral hypothesis. Based on previous work that examines how dominance and epistatic effects may affect this test, we argue that our results are unlikely to be biased. Although the genetic basis of recombination rate variation (e.g., relative contribution of additive variance, dominance effects, and epistasis) is poorly understood, we consider the potential impact of epistasis and dominance effects in turns. It is true that our Pst estimates could be affected by dominance and epistasis. However, it has been shown that epistasis tends to produce Qst values less than neutral Fst (Whitlock 1999). Similarly, dominance makes Qst equal or less than neutral Fst under the assumption of an island model (Goudet and Büchi 2006; Goudet and Martin 2007). Even though dominance under limited demographic circumstances can make Qst of neutral traits exceed neutral Fst, this is unlikely for traits affected by multiple loci (Goudet and Martin 2007) such as recombination rates. Taking all these into consideration, we argue that our use of Pst in this study makes our test likely conservative.
Due to limited resources, this study only examined the recombination variation of two genomic regions in males. Despite the promising evidence that the positive selection is responsible for the divergence in recombination rate between D. pulex and D. pulicaria, it remains unclear whether this is true for the genome-wide variation in males and whether this is true for female-specific recombination rate. Sex-specific difference in recombination rates often shows that females recombine more frequently than males (see Sardell and Kirkpatrick 2020). For example, human females on an average recombine 1.6 times as much as males (Kong et al. 2010), and sticklebacks show a similar pattern (Sardell et al. 2018). It will be interesting to examine the female recombination rate divergence between D. pulex and D. pulicaria and determine whether male and female recombination rate evolution is shaped by the same evolutionary forces.
Within-Species Recombination Rate Divergence
With much of the focus of this study probing whether between-species divergence in recombination rate is adaptive, it is necessary to provide some explanation about the contrasting pattern of within-species divergence in these two species. As mentioned in Results, the intraspecific recombination rate of D. pulex varies by nearly 3-fold for both linkage groups 8 and 9, whereas the intraspecific variation within D. pulicaria is much lower with a ∼1.3-fold difference on linkage group 8 and little variation on linkage group 9 (figs. 1 and 2). The within-species divergence in D. pulex is larger than all the currently available within-species divergence (reviewed in Ritz et al. 2017), such as ∼1.6-fold variation in both sexes of human (Coop et al. 2008), 1.9-fold in mice (Dumont et al. 2009), 1.1- to 2-fold in Drosophila (Brooks and Marks 1986; Hunter, Huang, et al. 2016), 1.3-fold in Arabidopsis (Sanchez-Moran et al. 2002), whereas the within-species divergence in D. pulicaria is in line with these available estimates.
One plausible explanation for this drastic difference between these two species is that selection pressure for maintaining recombination rates among different D. pulicaria populations is much more uniform than among D. pulex populations. To better understand this, we can use results of previous work on how spatially heterogeneous selection pressure influences the evolution recombination rate (Lenormand and Otto 2000). Regardless of the forms of epistasis, linkage disequilibrium, and the amount of linkage between recombination rate modifier and the selected loci, when environmental selection pressures vary between populations with frequent migration it is predicted that more variation in recombination rate is expected in populations inhabiting highly spatially variable environments (Lenormand and Otto 2000). Although it is often said that the typical habitat of D. pulex is ephemeral pond habitats, we have to acknowledge that ecological conditions of each pond population probably differ substantially in terms of pond sizes, depths, hydrological conditions, habitat heterogeneity, predators, and other biotic and abiotic factors. On the other hand, the ecology of the different stratified permanent lake habitats of D. pulicaria may differ to a lesser extent. We therefore hypothesize that the greater variability of recombination within D. pulex is likely due to the greater amount of heterogeneity among the pond habitats. This hypothesis is certainly worth future investigation by examining a large number of populations of each species, which can provide insight into how spatially heterogeneous selection shapes the evolution of recombination rates.
Materials and Methods
Daphnia Culture and Sperm Extraction
Males were collected for three isolates of D. pulex and D. pulicaria each (table 1). Each isolate represents a distinct population and clonally produced males (i.e., genetically identical excluding rare mutations) of each isolate were collected. To avoid maternal effect on recombination rate, females of each isolates were maintained in the same conditions for two generations. To induce the clonal production of males, mature females of the third generation with early sign of carrying broods were collected and cultured at 20 °C in artificial lake water (Kilham et al. 1998) containing 400 nM methyl farnesoate, a juvenile hormone that determines the sex of Daphnia offspring (Olmstead and Leblanc 2002). They were fed ad libitum with a suspension of Scenedesmus obliquus, and the offspring were screened for males. A total of 15–18 males were collected from each clone (same genotype) and were maintained in the lab for 2 weeks before sperm collection.
Table 1.
Summary of the Daphnia Isolates Used for the Recombination Rate Estimates, with Sampling Locations and NCBI SRA Accession Numbers for Whole-Genome Sequencing Raw Reads
| Species | Isolates | SRA (NCBI) | Lab Code | Location |
|---|---|---|---|---|
| Daphnia pulex | px1 | SRX4386564 | SW4 | Illinois |
| px2 | SRX4386576 | LPB17 | Long point, Ontario, Canada | |
| px3 | SRX4386574 | Tex21 | 42°12, −83°12, Textile Road, Michigan | |
| Daphnia pulicaria | pa1 | SRS1024794 | Little Curtis | 45°43, −122°44, Curtis Lake, Oregon |
| pa2 | SRS1024791 | RLSD26 | 44°57, −96°49, Round Lake, South Dakota | |
| pa3 | SRS1024797 | AroMoose | 44°50, −69°16, Sebasticook Lake, Maine |
For analyzing recombination rate of each Daphnia isolate, we collected sperm from all the identified males because they have identical genotype. To extract sperm, each male immersed in a drop of double-distilled water (ddH2O) was gently pressed with a cover-slip on a microscope slide. The ddH2O surrounding each individual was collected using Sigmacote-washed capillary needles and mouth pipettes into a 1.5-ml microcentrifuge tubes containing 50 μl of 1× PBS solution (Xu et al. 2015). To facilitate the sorting of single sperm cells by flow cytometry, we stained sperm cells using 8 μl of Hoechst 33342 (100 μg/ml) (Sigma–Aldrich) and incubated the sample in the dark at room temperature for 2 h.
Single-Sperm Cell Sorting
A BD FACS Aria-II cell sorter was used to isolate single sperm cells into individual wells of 96-well PCR plates containing cell lysis buffer. The specific settings of the FACS Aria II instrument were 488 nm 100 mW laser for light scatter detection and 355 nm 20 mW for Hoechst detection. A nozzle of 70 mm was used at 45 psi, and FSC-PMT was used for optimal small particle discrimination.
Each well of the PCR plate contained 5 μl of lysis buffer consisting of Tris (30 mM), EDTA (2 μM), potassium chloride (20 μM), Triton (0.2%), DTT (40 mM), and protease/Proteinase K (2.5 μg/μl). Cell lysis was performed in a thermal cycler at 50 °C for 3 h, 75 °C for 20 min, and 80 °C for 5 min.
Whole-Genome Amplification
To obtain enough DNA from each sperm for genotyping, the lysed single sperm cell was used for MALBAC (multiple annealing and looping-based amplification) whole-genome amplification (Zong et al. 2012). MALBAC consists of a preamplification stage and a standard PCR amplification. The preamplification is initiated with random primers, each having a common 27-nucleotide sequence (5ʹ-GTGAGTGATGGTTGAGGTAGTGTGGAG-3ʹ) and eight variable nucleotides that can evenly hybridize to the templates.
Preamplification Stage
A solution of 3.0 μl ThermoPol buffer (New England Biolabs), 1 μl dNTPs (10 mM), 0.75 μl each of two primers NT and NG (10 μM), and 19.5 μl H2O was added to each sperm sample. The samples were incubated at 95 °C for 5 min and quenched immediately on ice. About 0.5 μl of Bst large fragment polymerase (New England Biolabs) was added to each sample and the following thermal amplification regime is performed: 10 °C for 45 s, 15 °C for 45 s, 20 °C for 45 s, 30 °C for 45 s, 40 °C for 45 s, 50 °C for 45 s, 65 °C for 2 min, 95 °C for 20 s, followed by quenching on ice. Subsequently, five cycles of preamplification cycles were performed, consisting of 10 °C for 45 s, 15 °C for 45 s, 20 °C for 45 s, 30 °C for 45 s, 40 °C for 45 s, 50 °C for 45 s, 65 °C for 2 min, 95 °C for 20 s, and 58 °C for 40 s, followed by quenching on ice. About 0.5 μl Bst large fragment polymerase was added to each sample before carrying out the next cycle.
Standard PCR Amplification Stage
A standard PCR amplification was performed on the amplicons from the preamplification stage using the 27mer as primer (5ʹ-GTGAGTGATGGTTGAGGTAGTGTGGAG-3ʹ) to generate the 1–2 μg DNA required for downstream genotyping. Each reaction consisted of the product from the preamplification, 3 μl ThermoPol Buffer (New England Biolabs), 1 μl dNTPs (10 mM), 23.5 ml H2O, 1.5 μl 27mer (10 μM), and 1 μl DeepVentR exo-polymerase (New England Biolabs). The PCR thermal regime consisted of 22 rounds of 94 °C for 20 s, 59 °C for 20 s, 65 °C for 1 min, 72 °C for 2 min, which was followed by 72 °C for 5 min.
Recombination Rate Estimation
To examine recombination rate variation in D. pulex and D. pulicaria, we focused on two regions that are at the tip of the linkage groups and have ∼20 cM genetic distance on linkage groups 8 and 9 from the microsatellite-based genetic map by Cristescu et al. (2006). For linkage group 8, located between the microsatellite markers d077 and d068, the interval is 1.5 Mb, whereas on linkage group 9, the region spans ∼0.5 Mb lying between the microsatellite markers d171 and d118.
For detecting recombination events, two heterozygous markers are required. However, the four mapped microsatellite markers (i.e., d077, d068, d171, d118) are not heterozygous in all the Daphnia isolates. In cases where any of these markers are homozygous in any isolate, new heterozygous microsatellites were identified within a 50-kb window centered at the mapped marker and were used for estimating recombination (supplementary table 1, Supplementary Material online). The web-based platform WebSat (Martins et al. 2009) was used for identifying microsatellite markers and primer designs.
Our microsatellite genotyping followed the strategy outlined by Schuelke (2000). Briefly, a M13 tail is added to the 5′-end of the forward primer, and a M13 sequenced labeled with one of the NED, PET, FAM, and VIC fluorescent dye was used in the PCR. The thermal cycling program for microsatellite amplification consisted of 3 min at 95 °C, ten cycles of 35 s at 95 °C, 35 s at 56 °C (the temperature increased by 1 °C for each cycle) and 45 s at 72 °C, 30 cycles of 35 s at 95 °C, 35 s at 48 °C, 45 s at 72 °C, and a final 10 min at 72 °C. Fragment analysis was performed on an ABI 3130 Genetic Analyzer (Life Technologies) using 20× diluted PCR product. The four different M13 dyes allowed the pooled genotyping of different markers labeled with different dyes. The genotypes were called using the software GeneMapper 4.0 (Life Technologies).
To estimate the recombination rate for the two intervals of interests, 2-locus genotypes were examined for the pool of genotyped sperm for each Daphnia isolate. The number of sperm genotyped for each Daphnia isolate ranged from 73 to 94. The two most abundant genotypes were identified as the parental genotypes, whereas the two rare genotypes were derived from recombination events. For example, the two locus genotypes for d077 (alleles: 227 and 232 bp) and d068 (alleles: 337 and 343 bp) are 227/337 (ten sperm cells), 232/343 (ten sperm cells), 232/337 (40 sperm cells), and 227/343 (40 sperm cells). Then, the genotypes 227/337 and 232/343 are recognized as recombinant genotypes with a recombinant frequency of 0.2. The frequency of recombinants is converted to Kosambi cM map distance. The SE of recombination was calculated as , where the p represents proportion of recombinant sperm cells and n represents the number of sampled sperm cells.
P st–Fst Comparison
To investigate whether the divergence of recombination rate between these two species is adaptive, we performed Pst–Fst comparison analysis. As the divergence of quantitative traits can be shaped by mutation, selection, and genetic drift, various methods have been developed for deciphering whether the divergence of phenotypic traits is neutral (i.e., can be adequately explained by drift alone) or adaptive. An important approach among these is the comparison of Qst and Fst values.
Analogous to the famous Fst for measuring population differentiation using molecular markers (reviewed in Holsinger and Weir [2009]), Qst (Prout and Barker 1993; Spitze 1993) is established as a measure of the genetic differentiation among populations for phenotypic traits. For a neutral quantitative trait with additive genetic basis, its Qst value on an average should be equivalent to the mean Fst of neutral loci (Rogers and Harpending 1992; Whitlock and Mccauley 1999; Whitlock 2008), providing an important means for distinguishing between neutral and adaptive divergence. Therefore, if the Qst of a trait is significantly higher than the mean Fst of neutral loci, it would indicate divergent selection on this trait. On the contrary, if Qst of a trait is significantly smaller than the mean Fst of neutral loci, it would indicate stabilizing selection on the trait in the presence of drift. Moreover, identical values of Qst and Fst would indicate no evidence for selection acting in a spatially heterogeneous manner.
As specific breeding experimental designs in a common garden environment are required for estimating additive variance that is required for calculating Qst, many studies on wild populations used another metric Pst that is a surrogate to Qst (Leinonen et al. 2006). Pst is a metric measuring total phenotypic variance (rather than additive variance) among populations, which could be confounded by environmental effects for phenotype data directly collected from the field. Although our recombination rates were measured in a controlled environment and in same-aged males, our experiments did not allow us to estimate the additive variance. Thus, we decided to use Pst as a surrogate for Qst in this analysis.
To estimate Pst of recombination rate, we used recombination rate data for both chromosomes 8 and 9 to quantify within- and between-species variances using ANOVA in R. This strategy gave us a larger sample size and more statistical power than examining single-linkage groups alone. The between-species variance was calculated using the equation Var(s)=(MSs−MSe)/n, where MSs and MSe represent the mean squares of between- and within-species, respectively, and n represents the number of data points for each species (n = 6). The within-species variance Var(e) is equal to MSe, which is the mean squares of within-species. The Pst value is calculated using the equation Var(s)/[Var(s)+2Var(e)]. A total of 1,000 bootstrap replicates were generated and analyzed using ANOVA to estimate the distribution and mean value of Pst.
To determine whether the divergence of recombination rates between D. pulex and D. pulicaria is adaptive, we followed the approach of Whitlock and Guillaume (2009) to examine the difference between Qst and Fst with Qst–Fst as the test statistic. This approach rests upon the notion that the mean Qst value of neutral traits is expected to be the same as the mean Fst of neutral makers under certain assumptions (Whitlock and Guillaume 2009). The Fst between D. pulex and D. pulicaria was estimated using genome-wide 4-fold degenerate sites (n = 94,711) extracted from the whole-genome sequences of these isolates from Tucker et al. (2013).
To simulate the distribution of the Qst of a neutral trait, we calculated the expected between-species variance Var(s) using the formula Var(s)=2Fst.bootstrap×Var(e)/(1−Fst.bootstrap), where Fst.bootstrap is the mean value of a bootstrap sample of 4-fold degenerate sites and Var(e) is the observed within-species variance. Then we simulated the between- and within-species variance, Var(s).hat and Var(e).hat, respectively. Var(e).hat was calculated as multiplied by a random number drawn from a chi-square distribution with the degree of freedom at within-species level (i.e., DFwithin), whereas Var(s).hat was simulated as multiplied by a random number drawn from a chi-square distribution with the degree of freedom at between-species level (i.e., DFbetween). Furthermore, the simulated Qst was calculated as Var(s).hat/[Vars(s).hat+2Var(e).hat]. The simulation was repeated for 10,000 times to obtain a distribution of the test metric Qst–Fst. Lastly, we determined whether the observed Pst–Fst differs significantly from the neutral expectations by identifying the quantile of simulated distribution that had higher values than the observation, which gave us the P value of the test. This procedure was perform using a R script slightly modified from Lind et al. (2011).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Marelize Snyman, Trung Huynh, Hongjun Wang, and Thinh Pham for their help and constructive comments on this article. This work is partly supported by start-up funds from University of Texas at Arlington and is partly supported by National Institutes of Health (Grant No. R35GM133730 to S.X.).
Literature Cited
- Aggarwal DD, et al. 2015. Experimental evolution of recombination and crossover interference in Drosophila caused by directional selection for stress-related traits. BMC Biol. 13(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, et al. 2012. A fine-scale chimpanzee genetic map from population sequencing. Science 336(6078):193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, et al. 2013. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 9(12):e1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. 1995. a. A general model for the evolution of recombination. Genet Res. 65(2):123–144. [DOI] [PubMed] [Google Scholar]
- Barton N. 1995. b. Linkage and the limits to natural selection. Genetics 140:821–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, et al. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Nord Stud Alcohol Drugs. 327(5967):836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356(6369):519–244. [DOI] [PubMed] [Google Scholar]
- Bell AD, et al. 2020. Insights into variation in meiosis from 31,228 human sperm genomes. Nature 583(7815):259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker TR, Ness RW, Keightley PD. 2017. The recombination landscape in wild house mice inferred using population genomic data. Genetics 207(1):297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand CL, Cattani MV, Kingan SB, Landeen EL, Presgraves DC. 2018. Molecular evolution at a meiosis gene mediates species differences in the rate and patterning of recombination. Curr Biol. 28(8):1289–1295.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandlova J, Brandl Z, Fernando CH. 1972. The Cladocera of Ontario with remarks on some species and distribution. Can J Zool. 50(11):1373–1403. [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485(7400):642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks LD, Marks RW. 1986. The organization of genetic variation for recombination in Drosophila melanogaster. Genetics 114(2):525–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A, Bell G. 1987. Mammalian chiasma frequencies as a test of two theories of recombination. Nature 326(6115):803–805. [DOI] [PubMed] [Google Scholar]
- Butlin RK. 2005. Recombination and speciation. Mol Ecol. 14(9):2621–2635. [DOI] [PubMed] [Google Scholar]
- Cáceres CE, Tessier AJ. 2004. Incidence of diapause varies among populations of Daphnia pulicaria. Oecologia 141(3):425–431. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2002. Origins of the machinery of recombination and sex. Heredity (Edinb). 88(2):125–141. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. 1976. Recombination modification in a fluctuating environment. Adv Appl Probab. 8(1):2–4. [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134(4):1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, et al. 2013. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet. 45(11):1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Bois PRJ, Feingold E, Sherman SL, Cheung VG. 2009. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 5(9):e1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, Hebert PDN. 1996. The systematics of North American Daphnia (Crustacea: Anomopoda): a molecular phylogenetic approach. Philos Trans R Soc Lond B Biol Sci. 351(1337):349–360. [DOI] [PubMed] [Google Scholar]
- Comeron JM, Kreitman M, Aguadé M. 1999. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics 151(1):239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. 2008. High resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among Humans. Science 319(5868):1395–1398. [DOI] [PubMed] [Google Scholar]
- Cristescu ME, Colbourne JK, Radivojac J, Lynch M. 2006. A microsatellite-based genetic linkage map of the waterflea, Daphnia pulex: On the prospect of crustacean genomics. Genomics 88(4):415–430. [DOI] [PubMed] [Google Scholar]
- Cristescu M, Constantin A, Bock DG, Cáceres CE, Crease TJ. 2012. Speciation with gene flow and the genetics of habitat transitions. Mol Ecol. 21(6):1411–1422. [DOI] [PubMed] [Google Scholar]
- Cui XF, et al. 1989. Single-sperm typing: determination of genetic distance between the Gγ-globin and parathyroid hormone loci by using the polymerase chain reaction and allele-specific oligomers. Proc Natl Acad Sci U S A. 86(23):9389–9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Payseur BA. 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat Rev Genet. 14(4):262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapper AL, Payseur BA. 2017. Connecting theory and data to understand recombination rate evolution. Philos Trans R Soc B. 372(1736):20160469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapper AL, Payseur BA. 2018. Effects of demographic history on the detection of recombination hotspots from linkage disequilibrium. Mol Biol Evol. 35(2):335–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapper AL, Payseur BA. 2019. Molecular evolution of the meiotic recombination pathway in mammals. Evolution 73(12):2368–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H-W. 1996. Environmental and genetic control of sexual reproduction in Daphnia. Heredity (Edinb). 76(5):449–458. [Google Scholar]
- Deng H-W. 1997. Photoperiodic response of sexual reproduction in the Daphnia pulex group is reversed in two distinct habitats. Limnol Oceanogr. 42:609–611. [Google Scholar]
- Dolgin ES, Charlesworth B. 2008. The effects of recombination rate on the distribution and abundance of transposable elements. Genetics 178(4):2169–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudycha JL. 2004. Mortality dynamics of Daphnia in contrasting habitats and their role in ecological divergence. Freshwater Biol. 49(5):505–514. [Google Scholar]
- Dudycha JL, Tessier AJ. 1999. Natural genetic variation of life span, reproduction, and juvenile growth in Daphnia. Evolution 53(6):1744. [DOI] [PubMed] [Google Scholar]
- Dumont BL, Broman KW, Payseur BA. 2009. Variation in genomic recombination rates among heterogeneous stock mice. Genetics 182(4):1345–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Arndt PF. 2008. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 4(5):e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J, Yokoyama S. 1976. The evolutionary advantage of recombination. II. Individual selection for recombination. Genetics 83(4):845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J, Büchi L. 2006. The effects of dominance, regular inbreeding and sampling design on Qst, an estimator of population differentiation for quantitative traits. Genetics 172(2):1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J, Martin G. 2007. Under neutrality, Qst≤Fst when there is dominance in an island model. Genetics 176(2):1371–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey C, et al. 2011. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 9(10):e1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halldorsson BV, et al. 2019. Human genetics: characterizing mutagenic effects of recombination through a sequence-level genetic map. Science 363(6425):eaau1043. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P. 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2(4):280–291. [DOI] [PubMed] [Google Scholar]
- Heier CR, Dudycha JL. 2009. Ecological speciation in a cyclic parthenogen: sexual capability of experimental hybrids between Daphnia pulex and Daphnia pulicaria. Limnol Oceanogr. 54(2):492–502. [Google Scholar]
- Hendry AP. 2002. QST > = ≠ ST? Trends Ecol Evol. 17(11):502. [Google Scholar]
- Hill WG, Robertson A. 1966. The effect of linkage on limits to artificial selection. Genet Res. 8(3):269–294. [PubMed] [Google Scholar]
- Hinch AG, et al. 2019. Factors influencing meiotic recombination revealed by whole-genome sequencing of single sperm. Science 363(6433):eaau8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger KE, Weir BS. 2009. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat Rev Genet. 10(9):639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CM, Huang W, Mackay TFC, Singh ND. 2016. The genetic architecture of natural variation in recombination rate in Drosophila melanogaster. PLoS Genet. 12(4):e1005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CM, Robinson MC, Aylor DL, Singh ND. 2016. Genetic background, maternal age, and interaction effects mediate rates of crossing over in Drosophila melanogaster females. G3 (Bethesda) 6:1409–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SE, Bérénos C, Slate J, Pemberton JM. 2016. Conserved genetic architecture underlying individual recombination rate variation in a wild population of Soay sheep (Ovis aries). Genetics 203(1):583–598. [DOI] [PMC free article] [PubMed]
- Kamm JA, Spence JP, Chan J, Song YS. 2016. Two-locus likelihoods under variable population size and fine-scale recombination rate estimation. Genetics 203(3):1381–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent TV, Uzunović J, Wright SI. 2017. Coevolution between transposable elements and recombination. Philos Trans R Soc B Biol Sci. 372: 20160458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilham SS, Kreeger DA, Lynn SG, Goulden CE, Herrera L. 1998. COMBO: a defined freshwater culture medium for algae and zooplankton. Hydrobiologia 377(1/3):147–159. [Google Scholar]
- Kondrashov AS. 1988. Deleterious mutations and the evolution of sex. Nature 336(6198):435–440. [DOI] [PubMed] [Google Scholar]
- Kong A, et al. 2008. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319(5868):1398–1401. [DOI] [PubMed] [Google Scholar]
- Kong A, et al. 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467(7319):1099–1103. [DOI] [PubMed] [Google Scholar]
- Kong A, et al. 2014. Common and low-frequency variants associated with genome-wide recombination rate. Nat Genet. 46(1):11–16. [DOI] [PubMed] [Google Scholar]
- Korol AB, Iliadi KG. 1994. Increased recombination frequencies resulting from directional selection for geotaxis in Drosophila. Heredity (Edinb). 72(1):64–68. [DOI] [PubMed] [Google Scholar]
- Lande R. 1992. Neutral theory of quantitative genetic variance in an island model with local extinction and colonization. Evolution 46(2):381–389. [DOI] [PubMed] [Google Scholar]
- Leinonen T, Cano JM, Mäkinen H, Merilä J. 2006. Contrasting patterns of body shape and neutral genetic divergence in marine and lake populations of threespine sticklebacks. J Evol Biol. 19(6):1803–1812. [DOI] [PubMed] [Google Scholar]
- Lenormand T, Otto SP. 2000. The evolution of recombination in a heterogeneous environment. Genetics 156:423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Hurst LD. 2002. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18(7):337–340. [DOI] [PubMed] [Google Scholar]
- Li H, et al. 1988. Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature 335(6189):414–417. [DOI] [PubMed] [Google Scholar]
- Lind MI, Ingvarsson PK, Johansson H, Hall D, Johansson F. 2011. Gene flow and selection on phenotypic plasticity in an island system of rana temporaria. Evolution 65(3):684–697. [DOI] [PubMed] [Google Scholar]
- Liu H, Maclean CJ, Zhang J. 2019. Evolution of the yeast recombination landscape. Mol Biol Evol. 36(2):412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A, Morgan C, Franklin FCH, Bomblies K. 2018. Plasticity of meiotic recombination rates in response to temperature in Arabidopsis. Genetics 208(4):1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, et al. 2012. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science 338(6114):1627–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Butcher D, Bürger R, Gabriel W. 1993. The mutational meltdown in asexual populations. J Hered. 84(5):339–344. [DOI] [PubMed] [Google Scholar]
- Lynch M, et al. 2017. Population genomics of Daphnia pulex. Genetics 206(1):315–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins W, Lucasa DCS, Neves KFD, Bertioli DJ. 2009. WebSat – a web software for microsatellite marker development. Bioinformation 3(6):282–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GAT et al. 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304(5670):581–584. [DOI] [PubMed] [Google Scholar]
- Mugal CF, Weber CC, Ellegren H. 2015. GC-biased gene conversion links the recombination landscape and demography to genomic base composition: GC-biased gene conversion drives genomic base composition across a wide range of species. BioEssays 37(12):1317–1326. [DOI] [PubMed] [Google Scholar]
- Munoz-Fuentes V, et al. 2015. Strong artificial selection in domestic mammals did not result in an increased recombination rate. Mol Biol Evol. 32:510–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, et al. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327(5967):876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmstead AW, Leblanc GA. 2002. Juvenoid hormone methyl farnesoate is a sex determinant in the crustacean Daphnia magna. J Exp Zool. 293(7):736–739. [DOI] [PubMed] [Google Scholar]
- Omilian AR, Lynch M. 2009. Patterns of intraspecific DNA variation in the Daphnia nuclear genome. Genetics 182(1):325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Engelstädter J, Rieseberg LH. 2016. Recombination rate evolution and the origin of species. Trends Ecol Evol. 31(3):226–236. [DOI] [PubMed] [Google Scholar]
- Otto SP. 2009. The evolutionary enigma of sex. Am Nat. 174:S1–S14.. [DOI] [PubMed] [Google Scholar]
- Otto SP, Lenormand T. 2002. Resolving the paradox of sex and recombination. Nat Rev Genet. 3(4):252–261. [DOI] [PubMed] [Google Scholar]
- Otto SP, Michalakis Y. 1998. The evolution of recombination in changing environments. Trends Ecol Evol 13(4):145–151. [DOI] [PubMed] [Google Scholar]
- Pan J, et al. 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144(5):719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F, Haber JE. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 63(2):349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poissant J, et al. 2010. Genetic linkage map of a wild genome: genomic structure, recombination and sexual dimorphism in bighorn sheep. BMC Genomics 11(1):524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyet F, Mouchiroud D, Duret L, Sémon M. 2017. Recombination, meiotic expression and human codon usage. Elife 6:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prout T, Barker JSF. 1993. F statistics in Drosophila buzzatii: selection, population size and inbreeding. Genetics 134:369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. 2002. Experimental tests of the adaptive significance of sexual recombination. Nat Rev Genet. 3(4):241–251. [DOI] [PubMed] [Google Scholar]
- Ritz KR, Noor MAF, Singh ND. 2017. Variation in recombination rate: adaptive or not? Trends Genet. 33(5):364–374. [DOI] [PubMed] [Google Scholar]
- Rizzon C, Marais G, Gouy M, Biemont C. 2002. Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 12(3):400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AR. 2014. How population growth affects linkage disequilibrium. Genetics 197(4):1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AR, Harpending H. 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 9:552–569. [DOI] [PubMed] [Google Scholar]
- Ross-Ibarra J. 2004. The evolution of recombination under domestication: a test of two hypotheses. Am Nat. 163(1):105–112. [DOI] [PubMed] [Google Scholar]
- Saleem M, Lamb BC, Nevo E. 2001. Inherited differences in crossing over and gene conversion frequencies between wild strains of Sordaria fimicola from ‘Evolution Canyon’. Genetics 159(4):1573–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuk K, Manzano-Winkler B, Ritz KR, Noor MAF. 2020. Natural selection shapes variation in genome-wide recombination rate in Drosophila pseudoobscura. Curr Biol. 30(8):1517–1528.e6. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E, Armstrong SJ, Santos JL, Franklin FCH, Jones GH. 2002. Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162(3):1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor C, et al. 2012. Genetic variants in REC8, RNF212, and PRDM9 influence male recombination in cattle. PLoS Genet. 8(7):e1002854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardell JM, et al. 2018. Sex differences in recombination in sticklebacks. G3 (Bethesda) 8:1971–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardell JM, Kirkpatrick M. 2020. Sex differences in the recombination landscape. Am Nat. 195(2):361–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuelke M. 2000. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol. 18(2):233–234. [DOI] [PubMed] [Google Scholar]
- Singhal S, et al. 2015. Stable recombination hotspots in birds. Science 350(6263):928–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smukowski CS, Noor MAF. 2011. Recombination rate variation in closely related species. Heredity (Edinb). 107(6):496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JP, Song YS. 2019. Inference and analysis of population-specific fine-scale recombination maps across 26 diverse human populations. Sci Adv. 5(10):eaaw9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitze K. 1993. Population structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics 135(2):367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AE, Ackerman MS, Eads BD, Xu S, Lynch M. 2013. Population-genomic insights into the evolutionary origin and fate of obligately asexual Daphnia pulex. Proc Natl Acad Sci U S A. 110(39):15740–15745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fan HC, Behr B, Quake SR. 2012. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150(2):402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC. 1999. Neutral additive genetic variance in a metapopulation. Genet Res. 74(3):215–221. [DOI] [PubMed] [Google Scholar]
- Whitlock MC. 2008. Evolutionary inference from QST. Mol Ecol. 17(8):1885–1896. [DOI] [PubMed] [Google Scholar]
- Whitlock MC, Guillaume F. 2009. Testing for spatially divergent selection: comparing QST to FST. Genetics 183(3):1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC, Mccauley DE. 1999. Indirect measures of gene flow and migration: fst≠1/(4Nm+1). Heredity (Edinb). 82(2):117–125. [DOI] [PubMed] [Google Scholar]
- Xu S, et al. 2015. A male-specific genetic map of the microcrustacean Daphnia pulex based on single-sperm whole-genome sequencing. Genetics 201(1):31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Young K. 2017. Whole-genome haplotyping of single sperm of Daphnia pulex (Crustacea, Anomopoda). In: Tiemann-Boege I, Betancourt A, editors. Haplotyping: Methods and Protocols. New York (NY): Springer New York. p. 147–157. [DOI] [PubMed]
- Zong C, Lu S, Chapman AR, Xie X. 2012. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338(6114):1622–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.