Abstract
The ribosome’s common core, comprised of ribosomal RNA (rRNA) and universal ribosomal proteins, connects all life back to a common ancestor and serves as a window to relationships among organisms. The rRNA of the common core is similar to rRNA of extant bacteria. In eukaryotes, the rRNA of the common core is decorated by expansion segments (ESs) that vastly increase its size. Supersized ESs have not been observed previously in Archaea, and the origin of eukaryotic ESs remains enigmatic. We discovered that the large ribosomal subunit (LSU) rRNA of two Asgard phyla, Lokiarchaeota and Heimdallarchaeota, considered to be the closest modern archaeal cell lineages to Eukarya, bridge the gap in size between prokaryotic and eukaryotic LSU rRNAs. The elongated LSU rRNAs in Lokiarchaeota and Heimdallarchaeota stem from two supersized ESs, called ES9 and ES39. We applied chemical footprinting experiments to study the structure of Lokiarchaeota ES39. Furthermore, we used covariation and sequence analysis to study the evolution of Asgard ES39s and ES9s. By defining the common eukaryotic ES39 signature fold, we found that Asgard ES39s have more and longer helices than eukaryotic ES39s. Although Asgard ES39s have sequences and structures distinct from eukaryotic ES39s, we found overall conservation of a three-way junction across the Asgard species that matches eukaryotic ES39 topology, a result consistent with the accretion model of ribosomal evolution.
Keywords: Asgard, ribosomal RNA, archaea, expansion segments
Significance
Eukaryotes possess longer and more complex ribosomal RNAs (rRNAs) than Bacteria or Archaea, including eukaryotic-specific rRNA expansion segments (ESs). The origin and evolution of ESs has long remained a mystery. We show that two recently discovered Asgard archaeal phyla, Lokiarchaeota and Heimdallarchaeota, contain large subunit rRNA with ESs that are “supersized” (>100 nt) compared with all other prokaryotes studied to date. Asgard ESs are structurally distinct from ESs of eukaryotes, but share a common three-way junction out of which the ES grew. Our findings raise the possibility that supersized ESs existed on the ribosomal surface before the last eukaryotic common ancestor, opening the question of whether ribosomal complexity is more deeply rooted than previously thought.
Introduction
The ribosome connects all life on Earth back to the Last Universal Common Ancestor (LUCA) (Woese and Fox 1977). The small ribosomal subunit (SSU) decodes mRNA and the large ribosomal subunit (LSU) links amino acids together to produce coded protein. Both subunits are made of ribosomal RNA (rRNA) and ribosomal protein (rProtein). All cytoplasmic ribosomes contain a structurally conserved common core, comprised 2,800 nt and 28 rProteins, and including the peptidyl transferase center (PTC) in the LSU and the decoding center (DCC) in the SSU (Melnikov et al. 2012; Bernier et al. 2018). The rRNA of the common core is a reasonable structural approximation of the rRNA in LUCA and is similar to rRNA of extant bacteria (Melnikov et al. 2012; Petrov et al. 2014b; Bernier et al. 2018).
In Eukarya, the rRNA of the common core is elaborated by expansion segments (ESs, fig. 1) (Veldman et al. 1981; Clark et al. 1984; Hassouna et al. 1984; Gonzalez et al. 1985; Michot and Bachellerie 1987; Bachellerie and Michot 1989; Gutell 1992; Lapeyre et al. 1993; Gerbi 1996; Schnare et al. 1996). ESs emerge from a small number of conserved sites on the common core and are excluded from regions of essential ribosomal function such as the DCC, the PTC, and the subunit interface (Ben-Shem et al. 2010; Anger et al. 2013). Some archaea exhibit short ESs (µ-ESs) protruding from the same regions as eukaryotic ESs (Armache et al. 2013). Halococcus morrhuae, a halophilic archaeon, contains a large rRNA insertion in an LSU region that lacks ESs in eukaryotes (Tirumalai et al. 2020). ESs are larger and more numerous on the LSU than on the SSU; across phylogeny, size variation of SSU rRNA is around 10% of that of LSU rRNA (Gutell 1992; Gerbi 1996; Bernier et al. 2018).
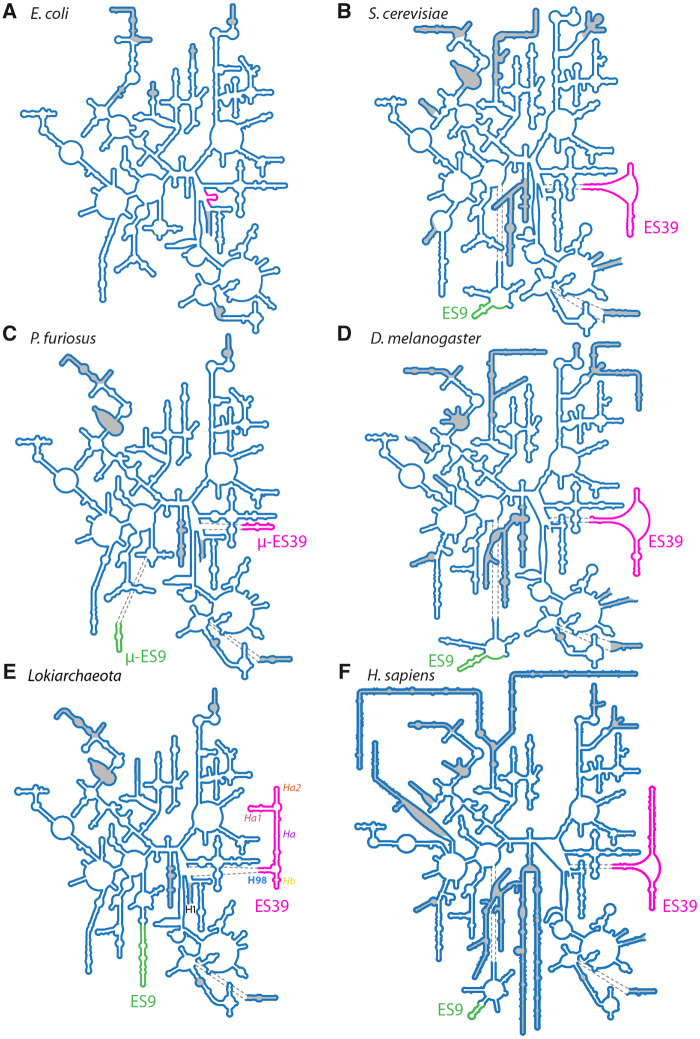
Fig. 1.
Secondary structures of the LSU rRNA from Bacteria, Archaea, and Eukarya. (A) E. coli (Bacteria), (B) S. cerevisiae (fungus, Eukarya); (C) P. furiosus (Archaea); (D) D. melanogaster (insect, Eukarya); (E) Lokiarchaeota B5 (Archaea); (F) H. sapiens (primate, Eukarya). Secondary structures in panels A–D and F are taken from Petrov et al. (2014a). Secondary structure in panel E is from this study. Universal rRNA common core is shown in blue lines (not shaded). ES9 is green. ES39 is magenta. Helices in panel E are abbreviated as Helix 1 (H1), helix 98 (H98), helix a (Ha), helix a1 (Ha1), helix a2 (Ha2), and helix b (Hb). ESs and helices not present in the common core are shaded in gray. Sizes of secondary structures are to scale. The numbering scheme of Noller et al. (1981) and Leffers et al. (1987) was used to label the helices and ESs.
ESs mutate quickly, at rates much greater than those of common core rRNA (Gonzalez et al. 1985, 1988; Ajuh et al. 1991; Gerbi 1996). For example, ES7 of Drosophila melanogaster (fruit fly) is AU-rich and lacks sequence homology with the GC-rich ES7 of Aedes albopictus (Asian tiger mosquito). The high rate of mutation is indicated by the sequence differences in species of the order Diptera that diverged only 200–250 Ma (Aris-Brosou and Yang 2002; Kumar et al. 2017). Although sequence conservation of ESs is low, secondary and tertiary structures are conserved and are excellent sources of information on homology and evolution. For this reason, early studies used rRNA secondary structures to establish phylogenetic relationships (Woese, Kandler, et al. 1990).
The recent discovery and characterization of the Asgard archaeal superphylum suggests that the last Asgard and eukaryotic common ancestor (LAsECA) contained key components of eukaryotic cellular systems (Spang et al. 2015, 2019; Klinger et al. 2016; Eme et al. 2017; Zaremba-Niedzwiedzka et al. 2017; Narrowe et al. 2018). Eukaryotic signature proteins (ESPs) found in Asgard archaea are involved in cytoskeleton, trafficking, ubiquitination, and translation (Spang et al. 2015; Zaremba-Niedzwiedzka et al. 2017; Akıl and Robinson 2018; Melnikov et al. 2020; Stairs and Ettema 2020). Asgard archaea also contain several homologs of eukaryotic rProteins (Hartman and Fedorov 2002; Spang et al. 2015; Zaremba-Niedzwiedzka et al. 2017). Metazoan rRNAs contain supersized ESs of hundreds of nucleotides (nt); hereafter, we define “supersized” ESs as those with >100 nt. Prior to this study, it was not known if Asgard rRNAs could contain features such as supersized ESs. Supersized ESs have not been observed previously in Bacteria or Archaea and were considered unique to eukaryotes (Ware et al. 1983; Clark et al. 1984; Hassouna et al. 1984; Gerbi 1996; Melnikov et al. 2012).
Here, we use computational and experimental approaches to characterize structure and evolution of Asgard rRNAs. We find that rRNAs of Lokiarchaeota and Heimdallarchaeota, both from the Asgard phyla, are elaborated on the LSU by supersized ESs. In size and complexity, Lokiarchaeota and Heimdallarchaeota LSU ESs exceed those of diverse protist rRNAs and rival those of metazoan rRNAs. No ESs were found in SSU rRNA of Lokiarchaeota. Based on our observations, we explore possible evolutionary pathways of ES growth in Asgard archaea and Eukarya.
Results
Comparative Analysis Reveals Broad Patterns of Complete LSU rRNA Size Relationships
Previously, we developed the SEREB MSA (Sparse and Efficient Representation of Extant Biology, Multiple Sequence Alignment) for comparative analysis of sequences from the translation system (Bernier et al. 2018). The SEREB MSA is a sparse and unbiased alignment containing representations of all major phyla and is designed specifically to understand phenomena that are universal to all of biology. This MSA was manually curated and extensively cross-validated and is useful as a seed to study a variety of evolutionary phenomena. Recently, we augmented the SEREB MSA to include additional metazoan sequences, allowing us to characterize ESs and their evolution in metazoans (Mestre-Fos et al. 2019a, 2019b). Here, we augmented the SEREB MSA to include 21 sequences from the Asgard superphylum and 12 sequences from deeply branching eukaryotes (supplementary data sets S1 and S2, Supplementary Material online).
The SEREB MSA indicates that LSU rRNA size relationships are governed by the general pattern: Bacteria [2,725–2,960 nt, n = 61 {n is number of species}] < non-Asgard archaea (2,886–3,094 nt, n = 43) < Eukarya (3,300–5,200 nt, n = 41; supplementary fig. S1, Supplementary Material online). This pattern is broken by eukaryotic parasites (3,047–4,086 nt, n = 2, which are anomalously small) and Asgard archaea (3,038–3,300 nt, n = 12, which are anomalously large). The SEREB MSA confirms previous observations that archaeal rRNAs frequently contain µ-ESs at positions of attachment of eukaryotic ESs. For example, in Archaea, µ-ES39 is connected at the site of attachment of the large ES39 in eukaryotes (Armache et al. 2013).
Lokiarchaeota Are Intermediate between Eukarya and Archaea in LSU rRNA Size
The Asgard augmentation of the SEREB MSA revealed unexpectedly large Lokiarchaeota LSU rRNAs. Lokiarchaeota LSU rRNAs range from 3,100 to 3,300 nt (n = 7). Lokiarchaeota LSU rRNAs are close to or within the observed size range of eukaryotic LSU rRNAs (supplementary fig. S1, Supplementary Material online). Other Asgard LSU rRNA sizes are in the upper archaeal range (from 3,038 to 3,142 nt, n = 6). Supersized archaeal ESs, which rival eukaryotic ESs, are observed in Lokiarchaeota and Heimdallarchaeota spp. These ESs connect to the universal common core rRNA at the sites of attachment of eukaryotic ES9 and ES39 and archaeal µ-ES9 and µ-ES39 (fig. 1). Here, we explored the Asgard augmentation of the SEREB MSA to investigate the structure, distribution, and evolution of rRNA expansions of Asgard archaea.
ES9 and ES39 in Some Asgard Archaea Are Larger Than µ-ESs of Other Archaea and ESs of Various Protists
The MSA shows that ES39 in Lokiarchaeota ranges in size from 95 to 200 nt and in Heimdallarchaeota ranges from 115 to 132 nt. Other Asgard species have shorter ES39s: Thorarchaeota, Odinarchaeota, and Freyarchaeota ES39 sequences are between 50 and 60 nt. These Asgard ES39 sizes are well within the range of eukaryotic ES39, which is around 80 nt in a variety of protists, 138 nt in Saccharomyces cerevisiae, 178 nt in D. melanogaster, and 231 nt in Homo sapiens (fig. 2 and supplementary data S3, Supplementary Material online). For Candidatus Lokiarchaeota archaeon 1244-F3-H4-B5 (Lokiarchaeota B5), the primary focus of our work here, ES39 is 191 nt (figs. 2 and 3). The first cultured Lokiarchaeota (Prometheoarchaeum syntropicum) and one other Lokiarchaeota sequence have 95 nt ES39s (fig. 2 and supplementary data S3, Supplementary Material online), slightly below our definition of a supersized ES, but still more than twice as long as the largest archaeal µ-ES39, Pyrococcus furiosus (45 nt, fig. 1C and supplementary data set S3, Supplementary Material online). All Asgard have larger ES39 sequences than other archaea. Archaea from the Euryarchaeota phylum, except for the classes Methanococci and Thermococci, lack ES39 entirely (less than 5 nt) and µ-ES39 of species from other non-Asgard archaea varies between 0 and 45 nt (supplementary data set S3, Supplementary Material online).
Fig. 2.
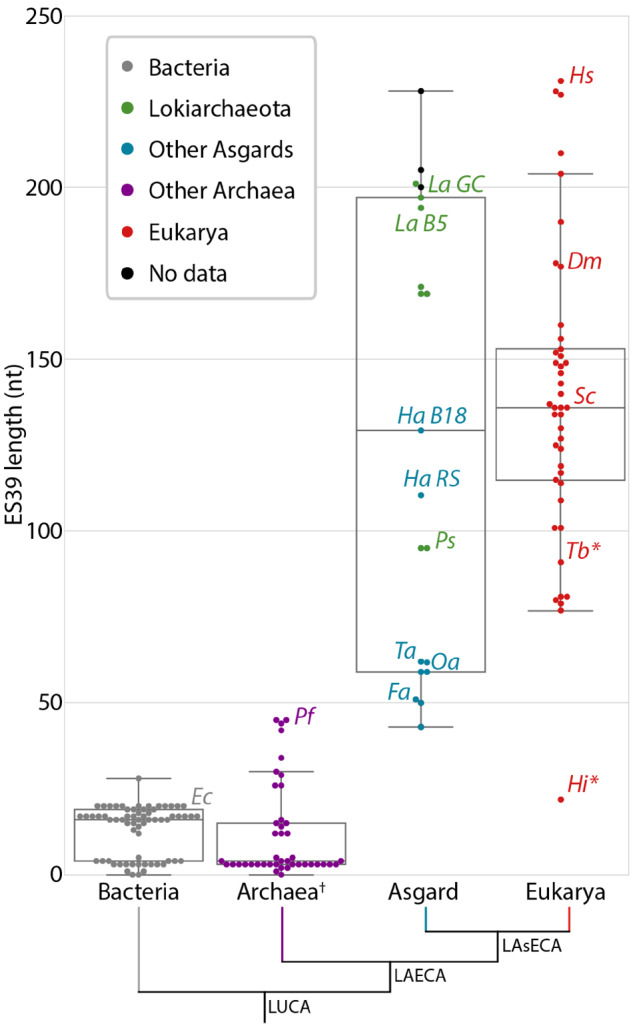
Distribution of ES39 lengths for species within each of the three domains of life, with incorporating Asgard archaea. The number of nucleotides was calculated between alignment positions 4891 and 5123 (H. sapiens numbering) of the LSU alignment for each species (supplementary data set S1, Supplementary Material online). The box shows the quartiles of the data set. Whiskers extend to show the rest of the distribution, except for points that are determined to be outliers using a function of the interquartile range. Bacteria sequences are gray, Lokiarchaeota sequences are green, other Asgard sequences are blue, other archaeal sequences are purple, eukaryotic sequences are red, and sequences from metatranscriptomic contigs (supplementary data set S2, Supplementary Material online) for which there is no species determination are black. Ec, Escherichia coli; Dm, Drosophila melanogaster; Fa, Freyarchaeota; Ha B18, Heimdallarchaeota B18G1; Ha RS, Heimdallarchaeota RS678; Hi, Hexamita inflata; Hs, Homo sapiens; La B5, Lokiarchaeota B5; La GC, Lokiarchaeota GC14_75; Oa, Odinarchaeota LCB4; Pf, Pyrococcus furiosus; Ps, Prometheoarchaeum syntropicum; Sc, Saccharomyces cerevisiae; Ta, Thorarchaeota; Tb, Trypanosoma brucei. *Parasitic representatives of the deeply branching eukaryotic group Excavata. †Excluding the Asgard superphylum.
Among Asgard archaea, some Lokiarchaeota spp. exhibit large ES9s. ES9 ranges from 29 (P. syntropicum) to 103 nt ( Lokiarchaeota B5), compared with 29 nt in S. cerevisiae, 44 nt in D. melanogaster and H. sapiens, and 111 nt in Guillardia theta (supplementary fig. S2, Supplementary Material online). In other non-Asgard archaea, ES9 varies between 20 and 30 nt. ES9 and ES39 are the primary contributors to the large size of Lokiarchaeota LSU rRNAs compared with the LSU rRNAs of other archaea. Outside of the Asgard superphylum, archaea lack supersized ESs.
Supersized ESs of Lokiarchaeota Are Transcribed In Situ
To assess whether Lokiarchaeota ESs are transcribed, we assembled metatranscriptomic reads from sediment from the Gulf of Mexico, which is known to contain Lokiarchaeota sequences (Yergeau et al. 2015). We did not find metatranscriptomes that contain sequences from Heimdallarchaeota. Multiple transcripts from Lokiarchaeota-like LSU ribosomes contain ES9 and ES39 sequences, confirming that Lokiarchaeota ESs are indeed transcribed in situ (fig. 4D andsupplementary data set S3, Supplementary Material online).
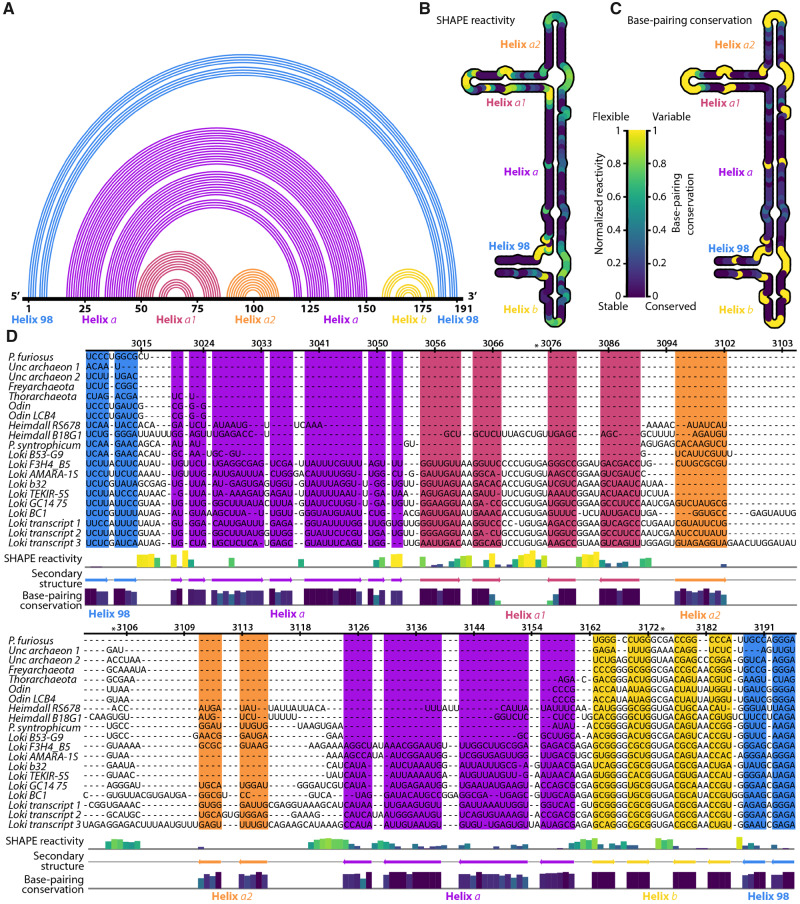
Fig. 4.
Secondary structure of Lokiarchaeota B5 ES39 from experiment and computation. (A) 1D topology map of base pairs. The primary sequence of ES39 is on the horizontal. Arcs indicate base pairs. Each helix is a distinct color. (B) SHAPE reactivity for ES39 mapped onto the secondary structure. Darker color indicates less flexible (paired) rRNA. (C) Base pairing conservation within the Asgard superphylum mapped on the secondary structure. Darker color indicates covarying (paired) rRNA across Asgard. Unpaired rRNA, for which no covariation data can be calculated, is gold. (D) SHAPE reactivity and base-pairing conservation mapped onto the ES39 MSA of Asgard sequences. Lokiarchaeota B5 numbering was used. The secondary structure is indicated with colored arrows below the alignment and as colored background. SHAPE reactivity is indicated with a bar graph above the secondary structure annotation, colors of the bars are consistent with panel B. Base-pairing conservation is indicated with a bar graph below the secondary structure annotation; colors of the bars are consistent with panel C. Panel D was generated with Jalview (Waterhouse et al. 2009). Helices are labeled with colored text in each panel; blue, helix 98; violet, helix a; pink, helix a1; orange, helix a2; yellow, helix b. Full sequence names and sequencing project identifiers are available in supplementary data set S2, Supplementary Material online. Both SHAPE reactivity and covariation are normalized. *Positions of conserved GNRA tetraloops.
Asgard LSU rRNA Contain the Common Core
We determined the extent of structural similarity between LSU rRNAs of Lokiarchaeota and those of eukaryotes. We combined computation and experiment to model the secondary structure of Lokiarchaeota B5 LSU rRNA (fig. 1E andsupplementary fig. S3, Supplementary Material online). Secondary and 3D structures of several eukaryotic and archaeal ribosomes provided a basis for modeling by homology. Like all other LSU rRNAs, Lokiarchaeota LSU rRNA contains the common core, which is trivial to model because the positions of backbone atoms of the common core are highly conserved in all cytosolic ribosomes.
ES39 Has a Well-Defined Structural Core in Eukaryotes
Before comparing eukaryotic and archaeal ES39s, we characterized the conserved core of eukaryotic ES39, which we call the ES39 signature fold. We compared experimental 3D structures of rRNAs in a wide range of eukaryotic ribosomes (Ben-Shem et al. 2010; Klinge et al. 2011; Khatter et al. 2015; Li et al. 2017). The ES39 signature fold, which is common to all eukaryotic ES39s, consists of helix 98 (H98; 20–30 nt), helix b (Hb; 40–50 nt), and their linkage by three unpaired 15 nt long segments of rRNA (supplementary fig. S4, Supplementary Material online). These unpaired segments are tightly associated with the ribosomal core. In all eukaryotes, two of the unpaired segments interact with eukaryotic-specific α-helical extensions on rProteins uL13 and eL14 (supplementary fig. S6, Supplementary Material online). The third unpaired segment interacts with ES7 and rProtein aL33 (supplementary figs. S4, S7, and S8, Supplementary Material online) (Khatter et al. 2015). Hb is terminated by a conserved GNRA tetraloop (supplementary figs. S4 and S5, Supplementary Material online). The ES39 signature fold is conserved in structure but not in sequence.
The ES39 Signature Fold Can Be Decorated by an Additional Helix
Many eukaryotes possess a third helix (Ha) that projects from the ES39 signature fold (supplementary fig. S4, Supplementary Material online). Ha is variable in length, it is shortest in unicellular eukaryotes, such as Tetrahymena thermophila (no helix), Toxoplasma gondii (10 nt), and S. cerevisiae (18 nt). Metazoan eukaryotes have the longest Ha, such as D. melanogaster (20 nt) and representatives of Chordata (106 nt for H. sapiens; supplementary fig. S4, Supplementary Material online).
Initial Lokiarchaeota B5 ES39 Secondary Structure Models Were Predicted by Two Methods
A preliminary secondary structural model of ES39 of Lokiarchaeota B5 was generated using mfold (Zuker 2003) (fig. 3). The program mfold predicts a minimum free energy secondary structures using experimental nearest-neighbor parameters. We selected the mfold model with lowest free energy for further investigation. A preliminary secondary structural model assumed Lokiarchaeota ES39 was structurally homologous with ES39 of H. sapiens. The mfold model was confirmed to be correct, and the H. sapiens homology model was determined to be incorrect by covariation analysis and Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) reactivity data as described below.
Fig. 3.
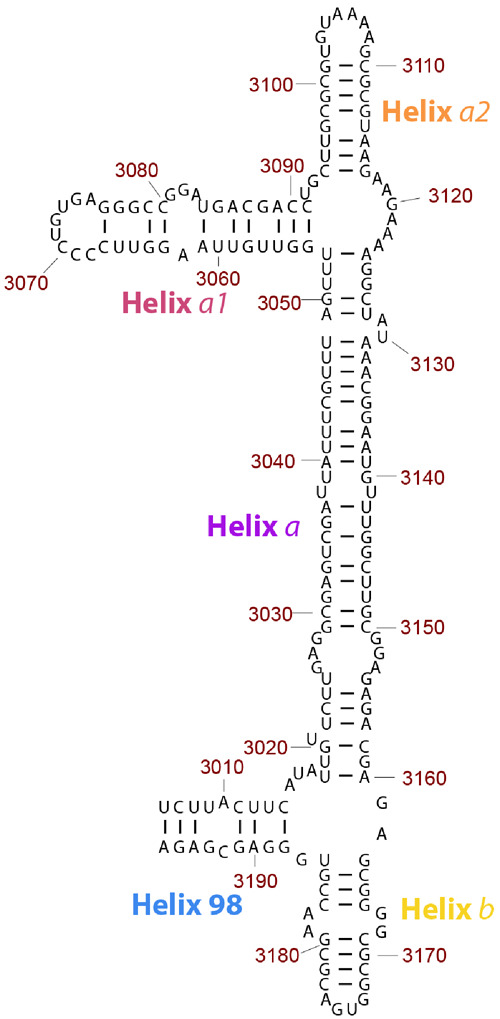
Secondary structure model of ES39 from Lokiarchaeota B5. ES39 spans nucleotide positions 3,006–3,196 of the Lokiarchaeota B5 LSU rRNA. Canonical base-pairing positions are indicated with black lines. Helices are annotated with colored labels: blue—Helix 98, purple—Helix a, pink—Helix a1, orange—Helix a2, gold—Helix b. This figure was generated with RiboVision (Bernier et al. 2014).
Covariation Supports the Mfold Model for the Secondary Structure of ES39 of Lokiarchaeota B5
Covariation, or coupled changes in paired nucleotides across phylogeny, can help reveal RNA secondary structure (Levitt 1969; Ninio et al. 1969; Woese et al. 1980; Noller et al. 1981; Gutell et al. 1993, 1994). Base-pairing relationships can be detected through covariation. We compared covariation predictions of each secondary model with observed covariation in the MSAs. The secondary structure predicted by mfold is consistent with observed covariation (fig. 4) whereas secondary structure predicted by H. sapiens homology model is not. The observation of covarying nucleotides supports the model determined by mfold.
Chemical Footprinting Confirms the Mfold Model for Secondary Structure of Lokiarchaeota B5 ES39
We further tested the secondary structural model of ES39 from Lokiarchaeota B5 using SHAPE. This experimental method provides data on RNA flexibility at nucleotide resolution (Merino et al. 2005; Wilkinson et al. 2006). SHAPE reactivity is generally high for unpaired nucleotides, which are more flexible, and low for paired nucleotides, which are less flexible. SHAPE has been widely used to probe the structure of rRNA (Leshin et al. 2011; Lavender et al. 2015; Gomez Ramos et al. 2017; Lenz et al. 2017) and other RNAs (Wilkinson et al. 2005; Gilbert et al. 2008; Stoddard et al. 2008; Watts et al. 2009; Novikova et al. 2012; Spitale et al. 2013; Huang et al. 2014). The SHAPE results from Lokiarchaeota B5 ES39 rRNA (supplementary fig. S9, Supplementary Material online) are in agreement with the secondary structure based on mfold, which, as shown above, is consistent with observed covariation. Reactivity is low for paired nucleotides in the mfold model and is high in loops and bulges (fig. 4B). The accuracy of the SHAPE data is supported by the observation of relatively high reactivity at the vast majority of unpaired nucleotides and low reactivity for most paired nucleotides of the mfold model. The Lokiarchaeota SHAPE data are not consistent with models that force Lokiarchaeota ES39 to conform to the H. sapiens secondary structure.
Asgard ES39 Deviates from the Eukaryotic ES39 Signature Fold
In eukaryotic ES39, the junction of helices H98, Ha, and Hb contains three unpaired segments, each 15 nt. In Lokiarchaeota B5, ES39 lacks unpaired segments greater than 8 nt and contains fewer unpaired nucleotides (fig. 3). Lokiarchaeota B5 ES39 is composed of four short helices, each up to 38 nt (H98; Helix a1: Ha1; Helix a2: Ha2; and Hb), and one long helix (Ha: 72 nt). H98 and Hb connect in a three-way junction with Ha at the base of ES39. Ha1 and Ha2 split Ha at the top of ES39 in a three-way junction (fig. 3).
We modeled and visualized secondary structures of ES39 sequences from additional Asgard species (fig. 5 and supplementary fig. S10, Supplementary Material online). None of these secondary structures exhibited unpaired regions longer than 10 nt. ES39 of all Asgard archaea contain a three-way junction that connects H98, Ha, and Hb, as observed in ES39 of Lokiarchaeota B5 (fig. 3). Such three-way junctions can indicate locations at which accretions in the ribosomal structure took place (Petrov et al. 2014b). Furthermore, ES39 in Heimdallarchaeota B18G1 has an additional branching of Ha into Ha1 and Ha2, mirroring the morphology of ES39 in Lokiarchaeota B5 (fig. 5 and supplementary fig.S10, Supplementary Material online). Despite the common branching morphology, the length of the individual helices varies substantially between different species (figs. 4D and 5).
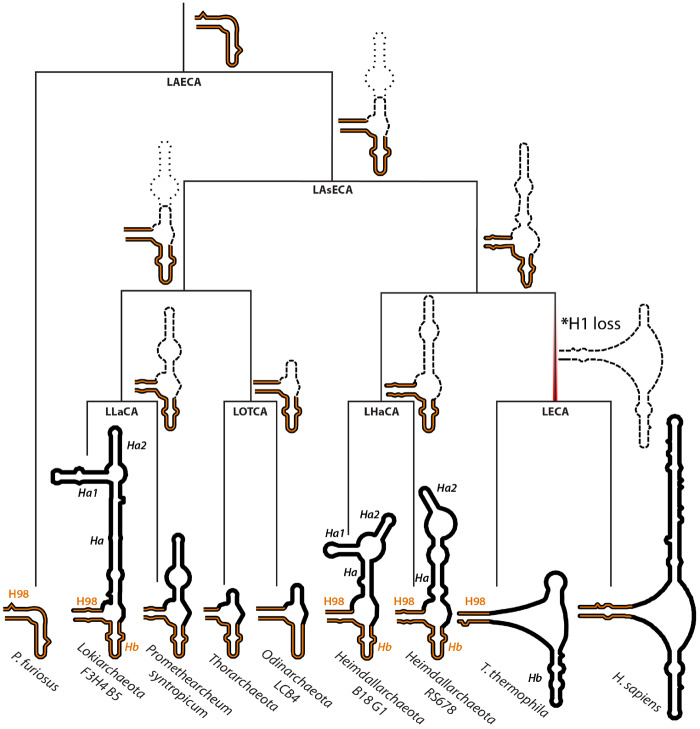
Fig. 5.
ES39 secondary structures mapped on Asgard phylogeny. Secondary structures of P. furiosus (Archaea), Lokiarchaeota B5 (Asgard archaea), P. syntropicum (Asgard archaea), Thorarchaeota (Asgard archaea), Odinarchaeota LCB4 (Asgard archaea), Heimdallarchaeota B18 G1 and RS678 (Asgard archaea), T. thermophila (Eukarya), and H. sapiens (Eukarya). Helix abbreviations are the same as in figure 1E. Helices 98 and b are highlighted in orange. The phylogenetic tree topology is from Williams et al. (2020). Ancestral clades on the phylogenetic tree are labeled as LAECA, Last Archaeal and Eukaryotic Common Ancestor; LAsECA, Last Asgardian and Eukaryotic Common Ancestor; LECA, Last Eukaryotic Common Ancestor; LHaCA, Last Heimdallarchaeota Common Ancestor; LLaCA, Last Lokiarchaeal Common Ancestor; LOTCA, Last Odin- and Thor-archaeota Common Ancestor. Likely ancestral structures are indicated next to ancestral clades. The minimal common structure from related extant species is shown with bold dashes. The uncertainty in ancestral sizes of helix a are shown with light dashes. *Occurrence of strand dissociation in ES39 at LECA due to loss of helix 1 is indicated with a red gradient.
Asgard ES39 Is Located within an Archaeal Structural Environment in the Ribosome
ES39 in Eukarya protrudes from helices 94 and 99 of the ribosomal common core (fig. 6). In three dimensions, ES39 is close to ES7 and rProteins uL13, eL14, and aL33 (supplementary figs. S4, S6, and S7, Supplementary Material online). These elements in all Asgard species are more similar to Archaea than to Eukarya. In addition, ribosomes of both Lokiarchaeota and Heimdallarchaeota, like those of all Archaea, contain helix 1 (H1), which is in direct contact with H98 at the base of ES39, whereas eukaryotes lack H1. Combined with the large size of Lokiarchaeota and Heimdallarchaeota ES39, these characteristics predict that ribosomes from these two Asgard groups have a unique structure in this region.
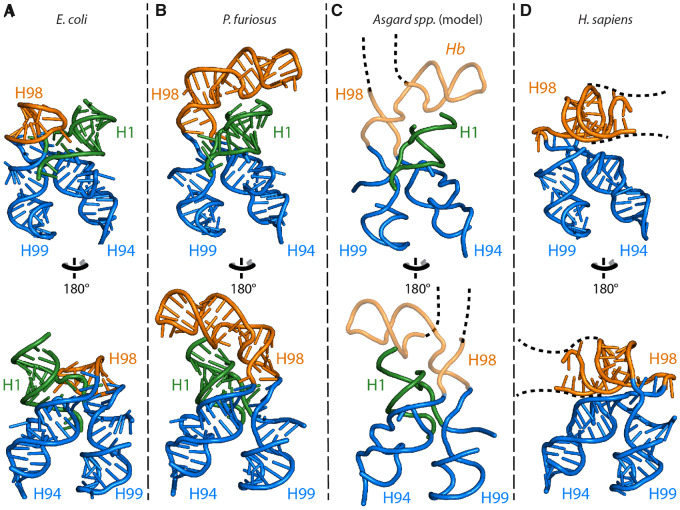
Fig. 6.
3D structures of ES39 and its neighborhood from representative species of the major life domains and Asgard archaea. (A) E. coli (Bacteria), (B) P. furiosus (Archaea), (C) Asgard archaea, and (D) H. sapiens (Eukarya). Helices are abbreviated as helix 1 (H1), helix 94 (H94), helix 98 (H98), helix 99 (H99), and helix b (Hb). Helix 98 is orange, helix 1 is green, helices 94 and 99 are blue. The P. furiosus structure is used as model for the Asgard structures. Likely structure of Asgard helices 98 and b are shown with backbone trace. The likely position and direction of the Asgard ES39 continuation is indicated with a black dashed line. The direction of eukaryotic ES39 continuation is indicated with a black dashed line.
The Pathway of ES39 Evolution Appears to Be Unique
In general, ESs have increased in size over evolution via accretion. Growth by insertion of one rRNA helix into another leaves a structural mark. These growth processes are indicated by the joining of an rRNA helical trunk to an rRNA helical branch at a highly localized three- or four-way junction (an insertion fingerprint) (Petrov et al. 2014b, 2015). Basal structure is preserved when new rRNA is acquired. For instance, ES7 shows continuous growth over phylogeny, expanding from LUCA to Archaea to diverse protist groups to metazoans to mammals (Armache et al. 2013; Petrov et al. 2014b; Bernier et al. 2018). The accretion model predicts that H98, at the base of ES39, would superimpose in Bacteria, Archaea, and Eukarya, but in fact H98 does not overlap in superimposed 3D structures (fig. 6). The archaeon P. furiosus has a slightly extended and bent H98 compared with the bacterium Escherichia coli (fig. 6). This spatial divergence is likely due to the difference in how E. coli H98 and P. furiosus H98 interact with H1 of the LSU. Escherichia coli H98 interacts within the H1 minor groove through an A-minor interaction, whereas in P. furiosus H98 is positioned on top of H1 (fig. 6). H1 is absent in eukaryotes (fig. 1), allowing H98 to occupy the position of H1 (fig. 6) and cause a strand dissociation in the eukaryotic ES39 structure.
Structurally, Lokiarchaeota and Heimdallarchaeota ES39 May Extend in a Different Direction Than Eukaryotic ES39
Lokiarchaeota and Heimdallarchaeota spp. have larger ES39 than other archaea (fig. 2) and possess H1, unlike Eukarya (fig. 6). We predict that Lokiarchaeota and Heimdallarchaeota ES39 have an archaeal-like interaction with H1 through H98 and Hb (fig. 6). In Asgard species, the highly variable Ha (figs. 4D and 5) likely grows out from the three-way junction between H98 and Hb, perpendicular to the eukaryotic Ha (fig. 6). Although Ha of eukaryotic ES39 is pointed in the direction of the sarcin–ricin loop, the large Ha of Lokiarchaeota and Heimdallarchaeota is likely pointed towards the central protuberance or the exit tunnel. The difference in directionality between Asgard and eukaryotic ES39 arises from the loss of H1 in Eukarya, where H98 and Hb partially occupy the vacated space.
Weak Sequence Homology between ES39 Sequences
We searched for ES39 homology between Asgards and deeply branching protist groups. MSAs between Asgard (n = 16) and eukaryotic (n = 15) ES39 sequences were compared with eukaryotic-only and Asgard-only MSAs. The percent identities in the eukaryotic-only ES39 MSA varied from 6% to 90% (supplementary fig. S11, Supplementary Material online). As expected, more closely related eukaryotes had greater identities (40–90% within Chordata; supplementary fig. S11, Supplementary Material online). Common core rRNA, with up to 80% identity even between distantly related species, was much more conserved than ES rRNA, (supplementary fig. S12, Supplementary Material online). Within the Lokiarchaeota, ES39 sequences shared 40–60% identity (supplementary fig. S13, Supplementary Material online). The extent of similarity between Asgard and eukaryotic ES39 sequences (10–30% identities; supplementary fig. S14, Supplementary Material online) was similar to the extent of similarity between distantly related eukaryotes (supplementary fig. S11, Supplementary Material online).
Conservation of a GNRA-Tetraloop Capping ES39 Hb between Eukarya and Asgard
The MSA of Hb from ES39 reveals a GNRA tetraloop, which is a conserved motif that caps Hb in both eukaryotes and Asgard archaea. Three sequences of deeply branching eukaryotic representatives exhibit sequence similarity to Hb sequences from Asgard archaea (supplementary fig. S5, Supplementary Material online).
Discussion
The recent discovery of the archaeal Asgard superphylum, which contain multitudes of ESPs, has redefined our understanding of eukaryotic evolution (Spang et al. 2015; Zaremba-Niedzwiedzka et al. 2017). Incorporation of Asgard species into phylogenies has changed our perspective on the relationship of Archaea and Eukarya in the tree of life (Hug et al. 2016; Fournier and Poole 2018; Doolittle 2020; Williams et al. 2020). Here, we incorporate rRNA secondary structure and 3D interactions into comparative analyses. Our work extends structure-based methods of comparative analysis to Lokiarchaeota and Heimdallarchaeota rRNAs, and mechanistic models for the evolution of common rRNA features in Eukarya and some Asgard archaea. In the following sections, we explore possible evolutionary pathways for this rRNA region.
Lokiarchaeota and Heimdallarchaeota rRNAs Adhere to Most Rules of Ribosomal Evolution
The ribosome is a window to relationships among organisms (Woese and Fox 1977; Hillis and Dixon 1991; Olsen and Woese 1993; Fournier and Gogarten 2010; Hug et al. 2016) and was a driver of ancient evolutionary processes (Agmon et al. 2005; Smith et al. 2008; Bokov and Steinberg 2009; Fox 2010; Petrov et al. 2015; Melnikov et al. 2018; Venkataram et al. 2020). Previous work has revealed rules of ribosomal variation over phylogeny (Hassouna et al. 1984; Gerbi 1996; Melnikov et al. 2012; Bernier et al. 2018), as well as mechanisms of ribosomal change over evolutionary history (Petrov et al. 2014b, 2015; Kovacs et al. 2017; Melnikov et al. 2018). Building off those studies, we assessed the extent to which Lokiarchaeota and Heimdallarchaeota ribosomes follow or deviate from previously established rules of ribosomal sequence and structure variation (Bowman et al. 2020). We found that Lokiarchaeota and Heimdallarchaeota ribosomes:
contain the universal common core of rRNA and rProteins (this work) (Spang et al. 2015; Bernier et al. 2018),
confine rRNA variability of structure and size to ESs/µ-ESs (this work),
restrict ESs to universally conserved sites on the common core (Ware et al. 1983; Clark et al. 1984; Hassouna et al. 1984; Michot and Bachellerie 1987; Bachellerie and Michot 1989; Lapeyre et al. 1993; Gerbi 1996),
avoid ES attachment from the ribosomal interior or near functional centers (Ben-Shem et al. 2010; Anger et al. 2013), and
concentrate variability in structure and size on LSU rRNA, not SSU rRNA (Gerbi 1996; Bernier et al. 2018).
The ribosomes of Lokiarchaeota, but not of Heimdallarchaeota, violate the established pattern of increasing rRNA length in the order: Bacteria < Archaea ≪ Eukarya (Melnikov et al. 2012; Petrov et al. 2014b; Bernier et al. 2018). Lokiarchaeota LSU rRNAs are much longer than predicted for archaea; in fact, Lokiarchaeota rRNA eclipses the length of rRNA in many deeply branching eukaryotes. Notably, the ES9 size of the only yet cultured Lokiarchaeota, P. syntropicum (Imachi et al. 2020) is an outlier to other Lokiarchaeota metagenome-derived sequences. Prometheoarchaeum syntropicum ES9 has a similar size to non-Asgard archaea (supplementary fig. S2, Supplementary Material online). ES39 in all studied Lokiarchaeota are larger than in any other archaea known to date. Some Lokiarchaeota exhibit ES39 larger than ES39 of most eukaryotes.
Two Scenarios for the Evolution of ES39
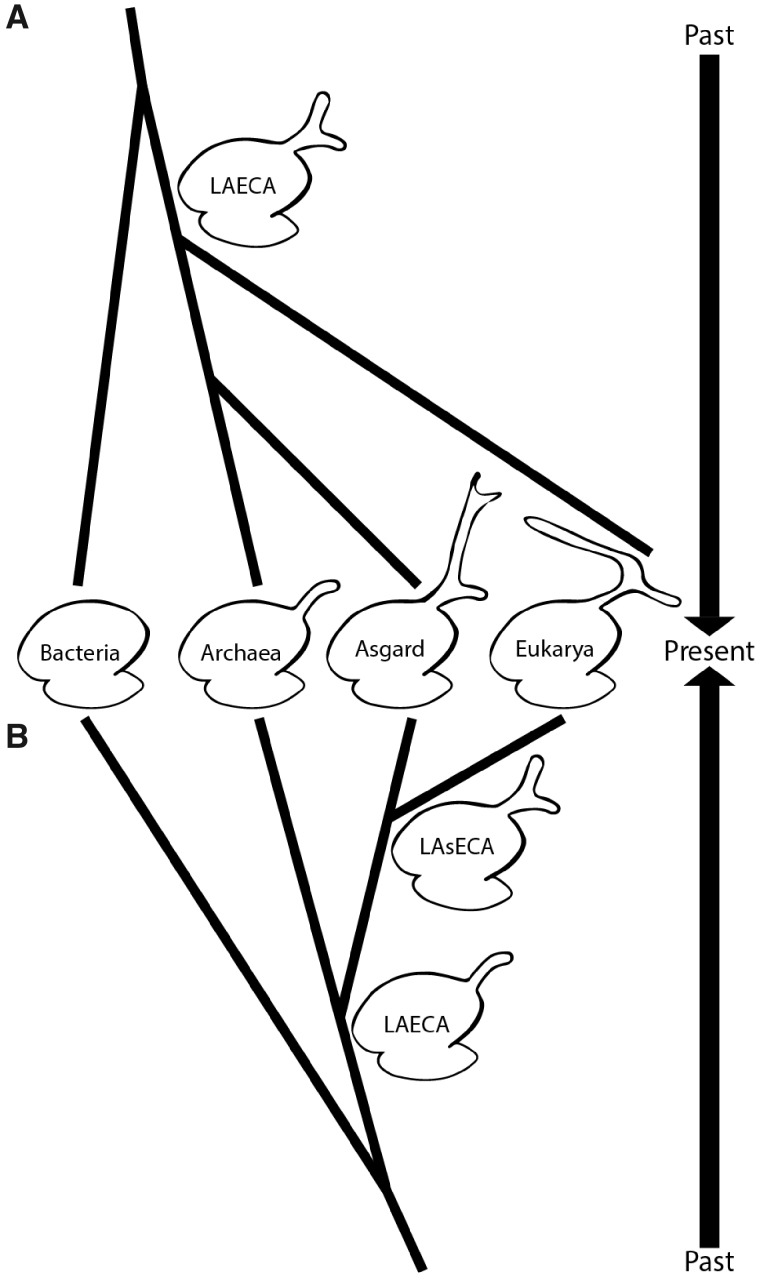
Two scenarios could explain the similarities of ES39 in Eukarya and Asgard archaea. One scenario is based on a three-domain tree of life and predicts parallel evolution of ES39 (fig. 7A), whereas the other assumes close common ancestry of Asgard archaea and Eukarya (fig. 7B). Our results do not exclude either one completely; therefore, we will discuss both.
Fig. 7.
Two scenarios of ES39 evolution illustrated by the tree of life topology. (A) Parallel independent growth of the ES39 region between Asgard archaea and Eukarya with a three-helical ES39 in LAECA. (B) Shared ancestry of three-helical ES39 between Asgard and Eukarya. Ribosomes of extant species are shown in the middle and past phylogenetic relationships extend up and down following the two scenarios. ES sizes are exaggerated for illustrative purposes.
Parallel Evolution of ES39 between Asgard Archaea and Eukarya
The difference in directionality of Ha in Asgard and Eukarya combined with the high evolutionary rates in ESs, and the lack of structural or sequence similarity, implies parallel evolution of ES39 with differential functions. Previous research has shown that archaeal ribosomes can contain µ-ESs (Armache et al. 2013; Petrov et al. 2014b) and large ES in a novel LSU region (Tirumalai et al. 2020). It is possible that Last Archaeal and Eukaryotic Common Ancestor (LAECA) had a larger ES39 (fig. 7A), which shrank in most archaea and was independently developed in Asgard archaea and eukaryotes. This theory is supported by the large structural diversity of ES39 in Asgard archaea. Loss of archaeal ES also matches previously documented reductions of archaeal rProteins (Lecompte et al. 2002) and ribosome biogenesis factors (Ebersberger et al. 2014). The parallel growth of LSU ESs in Asgard and Eukarya could also be the result of similar selection pressures. An explanation of similar ES sizes among Lokiarchaeota, Heimdallarchaeota, and higher eukaryotes (e.g. Chordata) may be slow growth rates and small population sizes, which are associated with characteristics that would otherwise be eliminated by purifying selection (Lynch 2007; Rajon and Masel 2011). Both Lokiarchaeota and Chordata have low growth rates and small populations (Imachi et al. 2020), which might lead to larger size of ES rRNA.
Possible Common Ancestry of ES39 in Asgard and Eukarya
Supersized ESs were thought to be unique to eukaryotic ribosomes. We now know that to be incorrect: two Asgard phyla exhibit supersized ESs. ES sequences are highly variable, even among closely related species (supplementary fig. S11, Supplementary Material online), making ESs impractical for sequence-based determination of ancient ancestry. Secondary and 3D structures are more conserved than sequences, allowing a more accurate detection of distant ancestry between species (Woese, Kandler, et al. 1990). By integrating the accretion model of ribosomal evolution (Petrov et al. 2014b, 2015) and structure-informed phylogeny, we discuss the ancestral relationship of Eukarya and Archaea.
The Accretion Model Suggests ES39 in LAsECA Contained Three Helices
An important structural difference between Asgard and other archaea is the three-helical topology of ES39. All Asgard have three helices in ES39, whereas other Archaea either lack ES39 altogether or contain a short-bent helix consisting of H98 and Hb (figs. 2, 5, and 6). The accretion model predicts that ES39 of the LAECA contained H98 and Hb (fig. 5) because they comprise the minimal form of ES39 shared between Asgard and other archaea (figs. 4 and 5). The insertion of Ha between H98 and Hb at LAsECA is consistent with the minimal ES39 (fig. 5). Subsequently, the three-helical topology would be inherited by all Asgard species, consistent with variable Ha sizes (figs. 4 and 5). Similarly, eukaryotes have highly conserved H98 and Hb whereas Ha is variable (supplementary figs. S4 and S5, Supplementary Material online). Eukarya differ from Asgard archaea in the unpaired rRNA segments that connect H98, Hb, and Ha (supplementary fig. S4, Supplementary Material online). The absence of insertion fingerprints precludes a detailed model for the relative ages of eukaryotic H98, Ha, and Hb. It is reasonable to assume that LECA had a three-helical ES39 inherited from one of its Asgardian sister clades. This ancient three-helical ES39 underwent restructuring and strand dissociation upon the loss of H1 (figs. 5 and 6). Subsequently, the unpaired structure of ES39 was inherited by all eukaryotes (fig. 5).
The specific roles of µ-ESs and ESs over phylogeny are unknown but are likely complex, polymorphic, and pleotropic. The observation of µ-ESs in Archaea, ESs in Eukarya, and supersized ESs in Asgard species can be explained by either a two- or three-domain tree of life. In a two-domain model, the ES39 size and topology can be explained by a gradual growth and accretion of ribosomal complexity, consistent with a close relationship between Asgard archaea and Eukarya seen in Williams et al. (2020). In a three-domain model, a large ES39 in LAECA could have been parallelly inherited in Asgard archaea and Eukarya without them necessarily being related. The rest of the archaeal domain experienced shrinkage of this ES region.
Conclusions
Lokiarchaeota and Heimdallarchaeota ribosomes contain supersized ES39s with structures that are distinct from eukaryotic ES39s. Lokiarchaeota ES9s are larger than eukaryotic ES9s. To date, Lokiarchaeota and Heimdallarchaeota are the only prokaryotic phyla with supersized ESs, bringing the size of their LSUs close to those of Eukarya. Lokiarchaeota and Heimdallarchaeota ES39 likely grow outward from the ribosomal surface in a different direction than eukaryotic ES39s. Our findingsopen the possibility that supersized ESs decorated some ribosomal surfaces before LECA, suggesting that ribosomal complexity may be more deeply rooted than previously thought.
Materials and Methods
Genome Sequencing, Assembly, and Binning
Sample Collection
Sediments were cored from the deep seafloor at ODP site 1244 (44°35.1784′N; 125°7.1902′W; 895 m water depth) on the eastern flank of Hydrate Ridge ∼3 km northeast of the southern summit on ODP Leg 204 in 2002 (Trehu et al. 2003) and stored at −80 °C at the IODP Gulf Coast Repository.
DNA Extraction
DNA was extracted from sediment from core F3-H4 (18.1 meters below the seafloor) using a MO-BIO PowerSoil total RNA Isolation Kit with the DNA Elution Accessory Kit, following the manufacturer protocol without beads. Approximately 2 g of sediments were used for the extraction from six extractions (12 g total) and DNA pellets from the two replicates from each depth were pooled together. DNA concentrations were measured using a Qubit 2.0 fluorometer with dsDNA High Sensitivity reagents (Invitrogen, Grand Island, NY). DNA yield was 7.5 ng per gram of sediment.
Multiple Displacement Amplification, Library Preparation, and Sequencing
Genomic DNA was amplified using a REPLI-g Single Cell Kit (Qiagen, Germantown, MD) using UV-treated sterile plasticware and reverse transcription-PCR grade water (Ambion, Grand Island, NY). Quantitative PCR showed that the negative control began amplifying after 5 h of incubation at 30 °C, and therefore, the 30 °C incubation step was shortened to 5 h using a Bio-Rad C1000 Touch thermal cycler (Bio-Rad, Hercules, CA). DNA concentrations were measured by Qubit. Two micrograms of MDA-amplified DNA were used to generate genome libraries using a TruSeq DNA PCR-Free Kit following the manufacturer’s protocol (Illumina, San Diego, CA). The resulting libraries were sequenced using a Rapid-Run on an Illumina HiSeq 2500 to obtain 100 bp paired-end reads. Metagenomic sequences were deposited into NCBI as accession numbers SAMN07256342–07256348 (BioProject PRJNA390944).
Metagenome Assembly, Binning, and Annotation
Demultiplexed Illumina reads were mapped to known adapters using Bowtie2 in local mode to remove any reads with adapter contamination. Demultiplexed Illumina read pairs were quality trimmed with Trim Galore (Martin 2011) using a base Phred33 score threshold of Q25 and a minimum length cutoff of 80 bp. Paired-end reads were then assembled into contigs using SPAdes assembler (Bankevich et al. 2012) with −meta option for assembling metagenomes, iterating over a range of k-mer values (21, 27, 33, 37, 43, 47, 51, 55, 61, 65, 71, 75, 81, 85, 91, 95). Assemblies were assessed with reports generated with QUAST (Gurevich et al. 2013). Features on contigs were predicted through the Prokka pipeline with RNAmmer for rRNA, Aragorn for tRNA, Infernal and Rfam for other noncoding RNA and Prodigal for protein coding genes (Laslett and Canback 2004; Lagesen et al. 2007; Hyatt et al. 2010; Nawrocki and Eddy 2013; Seemann 2014; Kalvari et al. 2018). Each genomic bin was searched manually for 23S rRNA sequences. The Lokiarchaeota F3-H4-B5 bin (estimated 2.8% completeness and 0% contamination) was found to contain a 3,300 nt 23S rRNA sequence. The Lokiarchaeota F3-H4-B5 bin was deposited into NCBI as BioSample SAMN13223206 and GenBank genome accession number WNEK00000000.
Environmental 23S rRNA Transcript Assembly and Bioinformatic Analysis
Species Selection for Analysis
To efficiently sample the tree of life, we used species from the SEREB database (Bernier et al. 2018) as a starting set. These species were selected to sparsely sample all major phyla of the tree of life where allowed by available LSU rRNA sequences. We further added LSU sequences from species of recently discovered phyla in the archaeal and eukaryotic domains (supplementary data set S2, Supplementary Material online). These additional species were again selected as representatives from major phyla. To gain a higher level of evolutionary resolution in the Asgard superphylum, we added complete LSU sequences from several available Asgard species and constructed assemblies from available meta-transcriptomes. All species used in our analysis with their respective phylogenetic groups are listed in supplementary data set S3, Supplementary Material online.
Assembly
Publicly available environmental meta-transcriptomic reads were downloaded from NCBI BioProject PRJNA288120 (Yergeau et al. 2015). Quality evaluation of the reads was performed with FastQC (Andrews 2012) and trimming was done with TrimGalore (Martin 2011). Assembly of SRR5992925 was done using the SPADES (Bankevich et al. 2012) assembler with −meta and −rna options, to evaluate which performs better. Basic statistic measures such as Nx, contig/transcript coverage and length were compared (supplementary data sets S4 and S5, Supplementary Material online) yielding better results for the rnaSPAdes assembler. All subsequent meta-transcriptomic data sets were assembled with rnaSPAdes.
Identifying rRNA Sequences
BLAST databases were constructed (Altschul et al. 1990) from the resulting contig files and they were queried for ribosomal regions characteristic of the Asgardian clade (ES39/ES9 sequences from GC14_75). In addition, the program quast (Gurevich et al. 2013) with −rna-finding option was used.
SEREB MSA Augmentation
High scoring transcripts, as well as genomic sequences from added species, were included in the SEREB MSA (Bernier et al. 2018) using the program mafft (Katoh and Standley 2013) with the −add option. Known intronic regions (Cannone et al. 2002) were removed from new sequences. The highly variable region of ES39 was manually aligned using structural predictions from mfold (Zuker 2003).
Defining ES Regions within Asgard Archaea
To compare eukaryotic, archaeal, and Asgard ES regions we defined the basal helices of ES growth for ES9 and ES39. We calculated ES sizes by including all nucleotides within the start and end of the basal helices. In eukaryotes ES9 is a region of a cumulative expansion among helices 29, 30, and 31 [see domain D3 in Hassouna et al. {1984}]. For ES9 we used helix 30 because the growth in Lokiarchaeota was contained within that helix. For ES39 we used H98, consistent with previous observations of the variability within that region (Gerbi 1996).
Sequence Homology between Asgard and Eukaryotic ES39 Sequences
To uncover any sequence conservation within the ES39 region of the LSU between Asgard archaea and eukaryotes, we used the augmented SEREB MSA. Scripts used for generation of data figures and tables, as well as MSAs of the ES39 region are available at https://github.com/petaripenev/Asgard_LSU_project (last accessed August 21, 2020). The ES39 sequences for all eukaryotes and Asgard archaea were extracted from the global LSU alignment. Chordate eukaryotic sequences were removed as they were too GC rich, parasitic eukaryotic sequences were also removed. Multiple different automated alignment methods were used (Do et al. 2005; Sievers et al. 2011; Katoh and Standley 2013). Following the automated methods, we applied manual alignment guided by secondary structure predictions. Pairwise percent identities for the alignments where calculated with Ident and Sim from the Sequence Manipulation Suite (Stothard 2000).
LSU Size Comparison
The LSU size comparison was based on the transcribed gene for the LSU, which comprised a single uninterrupted rRNA sequence for bacteria and archaea (fig. 1A, C, and E), and comprised multiple concatenated rRNA sequences for the fragmented eukaryotic rRNA gene (fig. 1B, D, and F). The 5S rRNA, which is essentially constant, is excluded from the size calculation. The comparison takes into account whether the rRNAs are from endosymbionts and pathogens, which tend to contain reduced genomes, metabolisms, and translation systems (Peyretaillade et al. 1998; Moran 2002; McCutcheon and Moran 2012).
Secondary Structure Models
To model the secondary structure of Candidatus Lokiarchaeota archaeon 1244-F3-H4-B5 LSU rRNA, we used the secondary structure of P. furiosus (Petrov et al. 2014a) and a MSA of archaeal LSU rRNAs broadly sampled over the phylogeny (supplementary data set S1, Supplementary Material online). Locations of ESs were unambiguously identified from the MSA. Due to the low percent identity (<50%) (Bernhart and Hofacker 2009) we applied ab initio modeling for ES regions. GNRA tetraloops are highly conserved structural motifs in rRNA structure (Woese, Winker, et al. 1990; Heus and Pardi 1991; Nissen et al. 2001). Conserved GNRA tetraloop sequences from the Asgard sequences were used to determine the limits of variable helices in ES regions. The secondary structures of the ESs were predicted by mfold (Zuker 2003).
Covariation
To verify the secondary structures of the highly variable ES regions base-pairing conservation was calculated with the program Jalview (Waterhouse et al. 2009). Gaps from the MSA were ignored in the calculation to produce comparable results about available regions. The base-pairing model of secondary structures of ES9 (supplementary fig. S15, Supplementary Material online) and ES39 (fig. 4C and D) was generated in the Jalview annotation format and used for the base-pairing conservation calculation.
Defining the Eukaryotic ES39 Signature Fold
To identify the structurally invariant part of ES39 in eukaryotes, we used superimposition based on the common core within domain VI of the ribosomal structures from 4 eukaryotes (T. thermophila, T. gondii, S. cerevisiae, H. sapiens; supplementary fig. S4, Supplementary Material online). Initially the D. melanogaster ribosomal structure (PDB ID: 4V6W) (Anger et al. 2013) was used in identifying the core. However, as it has additional loops elongating the unpaired regions, we excluded it from our analysis. Drosophila melanogaster is known to have AU-enriched ESs; therefore, it is not surprising that it has perturbations in its ES39.
Structural Analysis of ES39 Region in Bacteria, Archaea, and Eukarya
To identify the likely direction of Asgard ES39 we compared the ES39 region among Bacteria, Archaea, and Eukarya from available ribosomal structures. The structure used for Bacteria was from E. coli (PDB ID: 4V9D) (Dunkle et al. 2011), for Archaea was from P. furiosus (PDB ID: 4V6U) (Armache et al. 2013), and for Eukarya was from H. sapiens (PDB ID: 4V88) (Ben-Shem et al. 2010). The model for ES39 Hb for Asgard ES39 is based on ES39 Hb from the P. furiosus ribosomal structure.
ES39 rRNA SHAPE Analysis
Synthesis of Lokiarchaeota ES39 rRNA
pUC57 constructs containing T7 promoter and the gene encoding Lokiarchaeota ES39 rRNA was linearized using HindIII restriction enzyme. Lokiarchaeota ES39 rRNA was synthesized by in vitro transcription using HiScribe T7 High Yield RNA Synthesis Kit; New England Biolabs. RNA was then precipitated in ethanol/ammonium acetate and purified by G25 size exclusion chromatography (illustraTMNAPTM-10, GE Healthcare). RNA purity was assayed by denaturing gel electrophoresis.
SHAPE Reaction
Selective SHAPE ( Wilkinson et al. 2006) was performed to chemically probe local nucleotide flexibility in ES39 rRNA. In vitro-transcribed ES39 rRNA was added to folding buffer [180 mM NaOAc, 50 mM Na-HEPES {pH 8.0} and 1 mM 1,2-diaminocyclohexanetetraacetic acid {DCTA}] to obtain 400 nM RNA in total volume of 80 μl. RNA was annealed by cooling from 75 to 25 °C at 1 °C/min. RNA modification reaction was performed with final concentration of 100 mM benzoyl cyanide (Sigma) prepared in dimethyl sulfoxide (DMSO). Nonmodified RNA samples were incubated with DMSO. Reactions were carried out for 2 min at room temperature. Modified RNAs and a control sample were purified by precipitation in ethanol and ammonium acetate at 20 °C for 2 h. RNA samples were centrifuged at 4 °C for 10 min. The RNA pellets were washed with 100 μl of 80% ethanol for two times and dried out using a SpeedVac vacuum concentrator. TE buffer [22 μl of 1 mM EDTA and 10 mM Tris-Cl {pH 8.0}] was added to each sample to resuspend the pellet.
Reverse Transcription
Reverse transcription was conducted on 20 μl of modified RNAs and unmodified RNA sample as a control, in presence of 8 pmol 5′[6-FAM] labeled primer (5′-GAACCGGACCGAAGCCCG-3′) 2 mM DTT, 625 μM of each deoxynucleotidetriphosphate (dNTP), and 5 μl of reverse transcription (RT) 5X first-strand buffer [250 mM Tris-HCl {pH 8.3}, 375 mM KCl, 15 mM MgCl2]. To anneal the primer, samples were heated at 95 °C for 30 s, held at 65 °C for 3 min, and then 4 °C for 10 min. RT mixtures were incubated at 52 °C for 2 min before addition of 1 μl (200 U) of Superscript III Reverse transcriptase (Invitrogen) and reactions were incubated at 55 °C for 2 h. later, RT terminated by heating samples at 70 °C for 15 min. Chain termination sequencing reaction was performed on 10 pmol unmodified RNA prepared in TE buffer, 8 pmol 5′[6-FAM] labeled primer, with a ratio of 1:1 dideoxynucleotide (ddNTP) to dNTP. A sequencing reaction was performed with the same condition without ddNTPs.
Capillary Electrophoresis of RT Reaction Products and Data Analysis
Capillary electrophoresis of RT reactions was performed as described previously (Hsiao et al. 2013). For each reaction 0.6 μl DNA size standard (Geneflo 625), 17.4 μl Hi-Di Formamide (Applied Biosystems), and 2 μl of RT reaction mixture were loaded in a 96-well plate. Samples were heated at 95 °C for 5 min before electrophoresis and the RT products were resolved using applied biosystems. SHAPE data were processed using a Matlab scripts as described previously (Athavale et al. 2012). SHAPE profile was mapped onto ES39 rRNA secondary structure with the RiboVision program (Bernier et al. 2014).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Cecilia Kretz and Piyush Ranjan for sample preparation and genome binning. We thank Brett Baker for providing an unpublished Heimdallarchaeota sequence. We thank Jessica Bowman, Santi Mestre-Fos, Aaron Engelhart, Anthony Poole, and Betul Kacar for helpful conversations. This research was supported by National Aeronautics and Space Administration grants NNX16AJ29G, the Center for the Origin of Life grant 80NSSC18K1139, and a Center for Dark Energy Biosphere Investigations (C-DEBI) Small Research Grant. This is C-DEBI contribution 536. This research was supported in part through research cyberinfrastructure resources and services provided by the Partnership for an Advanced Computing Environment (PACE) at the Georgia Institute of Technology, Atlanta, GA, USA.
Author Contributions
A.S.P., L.D.W., and J.B.G. conceptualized the research. P.I.P. curated the data and performed computational analysis. A.S.P., L.D.W., and J.B.G. acquired the funding and administered the project. S.F.-A. performed the SHAPE experiments. J.J.C. and V.J.P. performed analyses. A.S.P. prepared 2D diagrams for the LSU rRNA of Lokiarchaeota and performed the analysis of introns. A.S.P., L.D.W., R.R.G., and J.B.G. supervised the research. R.R.G validated the research. P.I.P. and V.J.P. prepared the figures. P.I.P., J.B.G., and L.D.W. wrote the manuscript with input from all authors.
Data deposition: The project has been deposited at GenBank under the accession WNEK01000002.1; the Lokiarchaeota F3-H4-B5 23S rRNA gene is the reverse complement of nucleotide positions 251-3579.
Literature Cited
- Agmon I, Bashan A, Zarivach R, Yonath A. 2005. Symmetry at the active site of the ribosome: structural and functional implications. Biol Chem. 386(9):833–844. [DOI] [PubMed] [Google Scholar]
- Ajuh PM, Heeney PA, Maden BE. 1991. Xenopus borealis and Xenopus laevis 28S ribosomal DNA and the complete 40S ribosomal precursor RNA coding units of both species. Proc Royal Soc B 245(1312):65–71. [DOI] [PubMed] [Google Scholar]
- Akıl C, Robinson RC. 2018. Genomes of Asgard archaea encode profilins that regulate actin. Nature 562(7727):439–443. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Andrews S. 2012. FastQC: a quality control tool for high throughput sequence data. Available from: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed August 21, 2020.
- Anger AM, et al. 2013. Structures of the human and Drosophila 80S ribosome. Nature 497(7447):80–85. [DOI] [PubMed] [Google Scholar]
- Aris-Brosou S, Yang Z. 2002. Effects of models of rate evolution on estimation of divergence dates with special reference to the metazoan 18S ribosomal RNA phylogeny. Syst Biol. 51(5):703–714. [DOI] [PubMed] [Google Scholar]
- Armache J-P, et al. 2013. Promiscuous behaviour of archaeal ribosomal proteins: implications for eukaryotic ribosome evolution. Nucleic Acids Res. 41(2):1284–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athavale SS, et al. 2012. Domain III of the T. thermophilus 23S rRNA folds independently to a near-native state. RNA 18(4):752–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellerie JP, Michot B. 1989. Evolution of large subunit rRNA structure. The 3′ terminal domain contains elements of secondary structure specific to major phylogenetic groups. Biochimie 71(6):701–709. [DOI] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M. 2010. Crystal structure of the eukaryotic ribosome. Science 330(6008):1203–1209. [DOI] [PubMed] [Google Scholar]
- Bernhart SH, Hofacker IL. 2009. From consensus structure prediction to RNA gene finding. Brief Funct Genomics 8(6):461–471. [DOI] [PubMed] [Google Scholar]
- Bernier CR, et al. 2014. RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss. 169:195–207. [DOI] [PubMed] [Google Scholar]
- Bernier CR, Petrov AS, Kovacs NA, Penev PI, Williams LD. 2018. Translation: the universal structural core of life. Mol Biol Evol. 35(8):2065–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokov K, Steinberg SV. 2009. A hierarchical model for evolution of 23S ribosomal RNA. Nature 457(7232):977–980. [DOI] [PubMed] [Google Scholar]
- Bowman JC, Petrov AS, Frenkel-Pinter M, Penev PI, Williams LD. 2020. Root of the tree: the significance, evolution, and origins of the ribosome. Chem Rev. 120(11):4848–4878. [DOI] [PubMed] [Google Scholar]
- Cannone JJ, et al. 2002. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3(2):1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark CG, Tague BW, Ware VC, Gerbi SA. 1984. Xenopus laevis 28S ribosomal RNA: a secondary structure model and its evolutionary and functional implications. Nucleic Acids Res. 12(15):6197–6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. 2005. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 15(2):330–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. 2020. Evolution: two domains of life or three? Curr Biol. 30(4):R177–179. [DOI] [PubMed] [Google Scholar]
- Dunkle JA, et al. 2011. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332(6032):981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersberger I, et al. 2014. The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res. 42(3):1509–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG. 2017. Archaea and the origin of eukaryotes. Nat Rev Microbiol. 15(12):711–723. [DOI] [PubMed] [Google Scholar]
- Fournier GP, Gogarten JP. 2010. Rooting the ribosomal tree of life. Mol Biol Evol. 27(8):1792–1801. [DOI] [PubMed] [Google Scholar]
- Fournier GP, Poole AM. 2018. A briefly argued case that Asgard archaea are part of the eukaryote tree. Front Microbiol. 9:1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox GE. 2010. Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol. 2(9):a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi S. 1996. Expansion segments: regions of variable size that interrupt the universal core secondary structure of ribosomal RNA In: Zimmermann RA, Dahlberg AE, editors. Ribosomal RNA: structure, evolution, processing and function in protein synthesis. Boca Raton (FL: ): Telford–CRC Press; p. 87. [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT. 2008. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 15(2):177–182. [DOI] [PubMed] [Google Scholar]
- Gomez Ramos LM, et al. 2017. Eukaryotic ribosomal expansion segments as antimicrobial targets. Biochemistry 56(40):5288–5299. [DOI] [PubMed] [Google Scholar]
- Gonzalez IL, et al. 1985. Variation among human 28S ribosomal RNA genes. Proc Natl Acad Sci U S A. 82(22):7666–7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez IL, Sylvester JE, Schmickel RD. 1988. Human 28S ribosomal RNA sequence heterogeneity. Nucleic Acids Res. 16(21):10213–10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29(8):1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR. 1992. Evolutionary characteristics of 16S and 23S rRNA structure In: Hartman M.atsuno K, editors. The Origin and Evolution of the Cell. New Jersey: World Scientific Publishing Co. p. 243–309. [Google Scholar]
- Gutell RR, Gray MW, Schnare MN. 1993. A compilation of large subunit (23S and 23S-like) ribosomal RNA structures: 1993. Nucleic Acids Res. 21(13):3055–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Larsen N, Woese CR. 1994. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 58(1):10–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman H, Fedorov A. 2002. The origin of the eukaryotic cell: a genomic investigation. Proc Natl Acad Sci U S A. 99(3):1420–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassouna N, Mithot B, Bachellerie J-P. 1984. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 12(8):3563–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus HA, Pardi A. 1991. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253(5016):191–194. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Dixon MT. 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol. 66(4):411–453. [DOI] [PubMed] [Google Scholar]
- Hsiao C, et al. 2013. Molecular paleontology: a biochemical model of the ancestral ribosome. Nucleic Acids Res. 41(5):3373–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, et al. 2014. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat Chem Biol. 10(8):686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, et al. 2016. A new view of the tree of life. Nat Microbiol. 1(5):16048. [DOI] [PubMed] [Google Scholar]
- Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imachi H, et al. 2020. Isolation of an archaeon at the prokaryote–eukaryote interface. Nature 577(7791):519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, et al. 2018. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46(D1):D335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. 2015. Structure of the human 80S ribosome. Nature 520(7549):640–645. [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. 2011. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with Initiation actor 6. Science 334(6058):941–948. [DOI] [PubMed] [Google Scholar]
- Klinger CM, Spang A, Dacks JB, Ettema TJG. 2016. Tracing the archaeal origins of eukaryotic membrane-trafficking system building blocks. Mol Biol Evol. 33(6):1528–1541. [DOI] [PubMed] [Google Scholar]
- Kovacs NA, Petrov AS, Lanier KA, Williams LD. 2017. Frozen in time: the history of proteins. Mol Biol Evol. 34(5):1252–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, TimeTrees, and divergence times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Lagesen K, et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35(9):3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B, Michot B, Feliu J, Bachellerie J-P. 1993. Nucleotide sequence of the Schizosaccharomyces pombe 25S ribosomal RNA and its phylogenetic implications. Nucleic Acids Res. 21(14):3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavender CA, et al. 2015. Model-free RNA sequence and structure alignment informed by SHAPE probing reveals a conserved alternate secondary structure for 16S rRNA. PLoS Comput Biol. 11(5):e1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecompte O, Ripp R, Thierry JC, Moras D, Poch O. 2002. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 30(24):5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers H, Kjems J, Østergaard L, Larsen N, Garrett RA. 1987. Evolutionary relationships amongst archaebacteria: a comparative study of 23 S ribosomal RNAs of a sulphur-dependent extreme thermophile, an extreme halophile and a thermophilic methanogen. J Mol Biol. 195(1):43–61. [DOI] [PubMed] [Google Scholar]
- Lenz TK, Norris AM, Hud NV, Williams LD. 2017. Protein-free ribosomal RNA folds to a near-native state in the presence of Mg2+. RSC Adv. 7(86):54674–54681. [Google Scholar]
- Leshin JA, Heselpoth R, Belew AT, Dinman J. 2011. High throughput structural analysis of yeast ribosomes using hSHAPE. RNA Biol. 8(3):478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. 1969. Detailed molecular model for transfer ribonucleic acid. Nature 224(5221):759–763. [DOI] [PubMed] [Google Scholar]
- Li Z, et al. 2017. Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii. Cell Res. 27(10):1275–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007. The origins of genome architecture. Sunderland (MA: ): Sinauer Associates, Inc. [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12. [Google Scholar]
- McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10(1):13–26. [DOI] [PubMed] [Google Scholar]
- Melnikov S, et al. 2012. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 19(6):560–567. [DOI] [PubMed] [Google Scholar]
- Melnikov S, et al. 2020. Archaeal ribosomal proteins possess nuclear localization signal-type motifs: implications for the origin of the cell nucleus. Mol Biol Evol. 37(1):124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Manakongtreecheep K, Söll D. 2018. Revising the structural diversity of ribosomal proteins across the three domains of life. Mol Biol Evol. 35(7):1588–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc. 127(12):4223–4231. [DOI] [PubMed] [Google Scholar]
- Mestre-Fos S, et al. 2019. a. Profusion of G-quadruplexes on both subunits of metazoan ribosomes. PLoS One 14(12):e0226177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre-Fos S, et al. 2019. b. G-quadruplexes in human ribosomal RNA. J Mol Biol. 431(10):1940–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B, Bachellerie JP. 1987. Comparisons of large subunit rRNAs reveal some eukaryote-specific elements of secondary structure. Biochimie 69(1):11–23. [DOI] [PubMed] [Google Scholar]
- Moran NA. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108(5):583–586. [DOI] [PubMed] [Google Scholar]
- Narrowe AB, et al. 2018. Complex evolutionary history of translation Elongation Factor 2 and diphthamide biosynthesis in Archaea and Parabasalids. Genome Biol Evol. 10(9):2380–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29(22):2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J, Favre A, Yaniv M. 1969. Molecular model for transfer RNA. Nature 223(5213):1333–1335. [DOI] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. 2001. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci U S A. 98(9):4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF, et al. 1981. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 9(22):6167–6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova IV, Hennelly SP, Sanbonmatsu KY. 2012. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 40(11):5034–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen GJ, Woese CR. 1993. Ribosomal RNA: a key to phylogeny. FASEB J. 7(1):113–123. [DOI] [PubMed] [Google Scholar]
- Petrov AS, et al. 2014. a. Secondary structures of rRNAs from all three domains of life. PLoS One 9(2):e88222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, et al. 2014. b. Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci U S A. 111(28):10251–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, et al. 2015. History of the ribosome and the origin of translation. Proc Natl Acad Sci U S A. 112(50):15396–15401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyretaillade E, et al. 1998. Microsporidian Encephalitozoon cuniculi, a unicellular eukaryote with an unusual chromosomal dispersion of ribosomal genes and a LSU rRNA reduced to the universal core. Nucleic Acids Res. 26(15):3513–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajon E, Masel J. 2011. Evolution of molecular error rates and the consequences for evolvability. Proc Natl Acad Sci U S A. 108(3):1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnare MN, Damberger SH, Gray MW, Gutell RR. 1996. Comprehensive comparison of structural characteristics in eukaryotic cytoplasmic large subunit (23 S-like) ribosomal RNA. J Mol Biol. 256(4):701–719. [DOI] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069. [DOI] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Lee JC, Gutell RR, Hartman H. 2008. The origin and evolution of the ribosome. Biol Direct 3(1):16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, et al. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521(7551):173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, et al. 2019. Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of Asgard archaeal metabolism. Nat Microbiol. 4(7):1138–1148. [DOI] [PubMed] [Google Scholar]
- Spitale RC, et al. 2013. RNA SHAPE analysis in living cells. Nat Chem Biol. 9(1):18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stairs CW, Ettema TJG. 2020. The archaeal roots of the eukaryotic dynamic actin cytoskeleton. Curr Biol. 30(10):R521–526. [DOI] [PubMed] [Google Scholar]
- Stoddard CD, Gilbert SD, Batey RT. 2008. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA 14(4):675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P. 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 28(6):1102–1104. [DOI] [PubMed] [Google Scholar]
- Tirumalai MR, Kaelber JT, Park DR, Tran Q, Fox GE. 2020. Cryo-electron microscopy visualization of a large insertion in the 5S ribosomal RNA of the extremely halophilic archaeon Halococcus morrhuae. FEBS OpenBio. DOI: 10.1002/2211-5463.12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trehu AM, et al. 2003. ODP Leg 204; gas hydrate distribution and dynamics beneath southern Hydrate Ridge. JOIDES J. 29:5–8. [Google Scholar]
- Veldman GM, et al. 1981. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 9(24):6935–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataram S, Monasky R, Sikaroodi SH, Kryazhimskiy S, Kacar B. 2020. Evolutionary stalling and a limit on the power of natural selection to improve a cellular module. Proc Natl Acad Sci U S A. 117(31):18582–18590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware VC, et al. 1983. Sequence analysis of 28S ribosomal DNA from the amphibian Xenopus laevis. Nucleic Acids Res. 11(22):7795–7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JM, et al. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460(7256):711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. 2005. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNAAsp transcripts. J Am Chem Soc. 127(13):4659–4667. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc. 1(3):1610–1616. [DOI] [PubMed] [Google Scholar]
- Williams TA, Cox CJ, Foster PG, Szöllősi GJ, Embley TM. 2020. Phylogenomics provides robust support for a two-domains tree of life. Nat Ecol Evol. 4(1):138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, et al. 1980. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 8(10):2275–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 74(11):5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eukarya. Proc Natl Acad Sci U S A. 87(12):4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Winker S, Gutell RR. 1990. Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc Natl Acad Sci U S A. 87(21):8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau E, et al. 2015. Microbial community composition, functions and activities in the Gulf of Mexico, one year after the Deepwater Horizon accident. Appl Environ Microbiol. 81(17):5855–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba-Niedzwiedzka K, et al. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541(7637):353–358. [DOI] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31(13):3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.