Abstract

Natural or synthetic polycations are used as biocides or as drug/gene carriers. Understanding the interactions between these macromolecules and cell membranes at the molecular level is therefore of great importance for the design of effective polymer biocides or biocompatible polycation-based delivery systems. Until now, details of the processes at the interface between polycations and biological systems have not been fully recognized. In this study, we consider the effect of strong polycations with quaternary ammonium groups on the properties of anionic lipid membranes that we use as a model system for protein-free cell membranes. For this purpose, we employed experimental measurements and atomic-scale molecular dynamics (MD) simulations. MD simulations reveal that the polycations are strongly hydrated in the aqueous phase and do not lose the water shell after adsorption at the bilayer surface. As a result of strong hydration, the polymer chains reside at the phospholipid headgroup and do not penetrate to the acyl chain region. The polycation adsorption involves the formation of anionic lipid-rich domains, and the density of anionic lipids in these domains depends on the length of the polycation chain. We observed the accumulation of anionic lipids only in the leaflet interacting with the polymer, which leads to the formation of compositionally asymmetric domains. Asymmetric adsorption of the polycation on only one leaflet of the anionic membrane strongly affects the membrane properties in the polycation–membrane contact areas: (i) anionic lipid accumulates in the region near the adsorbed polymer, (ii) acyl chain ordering and lipid packing are reduced, which results in a decrease in the thickness of the bilayer, and (iii) polycation–anionic membrane interactions are strongly influenced by the presence and concentration of salt. Our results provide an atomic-scale description of the interactions of polycations with anionic lipid bilayers and are fully supported by the experimental data. The outcomes are important for understanding the correlation of the structure of polycations with their activity on biomembranes.
Introduction
Synthetic polycations have been proposed for many biomedical applications, such as biocides1 and carriers for the delivery of nucleic acids and proteins.2 For example, polyethylenimines (PEIs) (weak polyelectrolytes possessing amino groups) are characterized by their excellent ability to complex and transfect genes.3 Furthermore, polymers containing quaternary ammonium groups (strong polyelectrolytes) can be used as effective biocides against various microorganisms.4,5 Unfortunately, all polycations exhibit in vitro cytotoxicity, which is a major limitation in their clinical applications.6 The cytotoxicity of polycations is most often associated with their effects on cellular membranes. Polycations have been shown to damage plasma membranes and induce necrosis in human cells.6,7 Therefore, research into the effects of polycations on such membranes is important to the design of effective polymer biocides or biocompatible polycation-based delivery systems.
Interactions between polycations and anionic lipid membranes have been extensively investigated using experimental methods8,9 as well as computer simulations.10,11 In experiments, liposomes and supported lipid bilayers with lipid compositions mimicking those of biomembranes are commonly used as model membranes. It was shown that polycations can associate with, or penetrate into, a negatively charged lipid bilayer.8,12 However, the polycation–anionic liposome interactions depend on many factors, such as the type of polycation (weak or strong polyelectrolyte), the total charge of the macromolecule, the charge density on the lipid membrane (anionic lipid fraction), and salt concentration. For weak polycations, the results indicate the remarkable dependence of the stability of polycation-decorated anionic liposomes on the pH, temperature, and initial size of the liposomes.13 Zhang et al. showed that PEIs are able to induce lipid translocation across bilayers at physiological temperature.14 Our recent studies have shown that weak polycations, such as PEIs, interact with negative membranes mainly by forming hydrogen bonds with the lipid headgroups.10 These interactions induce substantial reorganization of the bilayer near the polymer due to the reorientation of lipid molecules. In contrast, strong polycations containing quaternary ammonium groups associate with anionic membranes mainly due to electrostatic interactions. The origin of this predominantly electrostatic binding is the entropic gain of translational freedom of the counterions released upon association of the polymer at the membrane.8 This gain in the free energy balances the loss of freedom of multicharged assemblies, which have a limited conformational space even in their unbound state, namely, the lipids in the membrane and the units of the polycation chains. Anionic lipids are concentrated in the vicinity of the adsorbed polycation chains by lateral diffusion, and in some cases, they flip-flop. Consequently, the negatively charged lipids form a domain around the polycation. Recently, Yaroslavov et al. showed that the adsorption of a strong polycation induces the grouping of all anionic lipids, initially evenly distributed in the membrane, in the outer membrane leaflet.15 Polycation adsorbed on the surface of charged vesicles can be completely removed from the membrane by increasing the salt concentration or by adding soluble polyanion. Therefore, the dissociation of the electrostatic liposome–polycation complexes is controlled by the salt concentration in the surrounding solution. However, many aspects of the strong polycation–anionic membrane interaction are still not recognized, especially the effect of the cationic polymer on the molecular organization of lipids in the bilayer.
Herein, we used atomistic molecular dynamics (MD) simulations and experimental methods to perform an extensive investigation of interactions between strong polycations with different chain lengths and anionic bilayers. Two strong polycations, poly([3-(methacryloylamino)propyl]trimethylammonium chloride) (PMAPTAC) with different molecular weights, were synthesized (Figure S1). The model lipid membrane consisted of zwitterionic 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and anionic 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS). Phosphatidylserines, which bear a negative charge, are important components of cell membranes and play a key role in cell cycle signaling, particularly in relation to apoptosis. In this study, we particularly focused on changes in the electrostatic potential at the membrane surface and lipid organization in the POPC/POPS membrane induced by the polycation adsorption. We determined the hydration and conformation of PMAPTAC chains in the aqueous phase and adsorbed on the POPC/POPS bilayer and lipid packing in polymer–membrane contact areas. The results emphasize that although PMAPTACs do not penetrate inside the membrane, their adsorption causes significant changes in its organization. In addition, we show how the presence of salt influences the strength of polycation–membrane interactions. Altogether, our results provide an atomic-scale image of the polycation adsorption on the anionic membrane and insights into how the anionic membrane is perturbed by polycations.
Experimental Section
Materials
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, ≥99.0%), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (sodium salt) (POPS, ≥99.0%), calcein, and chloroform were obtained from Sigma–Aldrich and used as received. 3-(Methacrylamido)propyl trimethylammonium chloride (MAPTAC, 95%) and 4,4′-azobis (4-cyanopentanoic acid) (V-501, 98%) from Wako Pure Chemical were used as received. 4-Cyanopentanoic acid dithiobenzoate (CPD) was synthesized according to the previously described method.16 Methanol was distilled after drying for 1 day on molecular sieves. Millipore-quality water was used in all experiments.
Synthesis of PMAPTAC16 and PMAPTAC147
PMAPTACy (y is the degree of polymerization, DP) was prepared via RAFT polymerization as shown in Figure S1. As an example, PMAPTAC147 was obtained as follows. MAPTAC (5.00 g, 22.7 mmol), V-501 (21.2 mg, 0.0757 mmol), and CPD (42.3 mg, 0.151 mmol) were dissolved in a mixture of MeOH (2.3 mL) and water (20.5 mL). The solution was degassed by purging with Ar gas for 30 min. Polymerization was carried out at 70 °C for 18 h. The reaction mixture was dialyzed against pure water for 1 day. PMAPTAC147 was recovered by freeze-drying (4.54 g, 90.9%). Mn(NMR) and DP estimated from 1H NMR (Figure S2) and Mw/Mn estimated from GPC (Figure S3) for the polymers are summarized in Table S1.
Preparation of POPC/POPS Liposomes
Large unilamellar vesicles (LUVs) were prepared from a mixture of POPC and POPS (20 wt %) by extrusion as described previously.17 Briefly, POPC was weighed into a glass flask and dissolved in chloroform. A chloroform solution of POPS was added to a 4:1 lipid ratio. The solvent was evaporated under a gentle stream of nitrogen to form a dry lipid film. PBS was added (usually the lipid concentration was 2.5 mg/mL), and the sample was vortex mixed for 5 min. The resulting multilamellar vesicle dispersion was subjected to five freeze–thaw cycles from liquid-nitrogen temperature to 60 °C and then extruded six times through a 100 nm membrane filter using a gas-pressure extruder. To prepare calcein-loaded liposomes, a solution of calcein in PBS (0.06 mol/dm3) was added to the lipid film. The calcein-loaded liposomes were separated from the nonencapsulated calcein by size-exclusion chromatography as previously described.18
Calcein-Release Studies
The experiments were performed in triplicate as previously described.19 Fluorescence spectra of the released calcein were recorded at 25 °C using a PerkinElmer LSD 50B spectrofluorometer. The amount of calcein released after time t, RF(t), was calculated according to the equation
| 1 |
where I0, It, and Imax are fluorescence intensities measured for the calcein-loaded liposomes before the polymer addition, at time t after the polymer introduction, and after the Triton X-100 addition (corresponding to complete calcein release), respectively.
Dynamic Light Scattering (DLS) Experiments
DLS and ζ-potential measurements were performed as described previously using a Malvern Nano ZS light-scattering apparatus (Malvern Instrument Ltd.).20 The time-dependent autocorrelation function of the photocurrent was acquired every 10 s with 15 acquisitions for each run at 25 °C. The z-averaged hydrodynamic mean diameter (dz), polydispersity index (PDI), and distribution profile of each sample were calculated using the software provided by the manufacturer.
MD Simulations
System Preparation
Tables 1 and S2 summarize the simulated systems. Two types of bilayers were prepared: (i) one containing 288 POPC molecules (bilayer POPC) and (ii) one containing 228 POPC and 60 POPS (20 mol %) molecules (bilayer POPC/POPS). Both membranes were hydrated with ∼30 000 water molecules and equilibrated for 50 ns. Oligomers consisting of 20 and 40 units (PMAPTAC20 and PMAPTAC40) were used as PMAPTAC models (shown in Figure 1). In the case of PMAPTAC20, five systems containing the POPC/POPS bilayer and N = 1, 2, 3, 4, and 6 oligomer molecules were prepared. For PMAPTAC40, one system with the POPC/POPS bilayer and one oligomer molecule was prepared. Oligomer molecules were manually placed close to the upper leaflet of the bilayer, which corresponds to the experimental setup in which a polycation is added to liposomes and only one lipid leaflet is accessible for the polymer. The systems were neutralized by introducing chloride counterions and subjected to energy minimization, followed by equilibration for 50 ns. The equilibrium simulations revealed that maximally two PMAPTAC20 molecules were able to adsorb on the upper leaflet in all of the systems studied (Figures S4–S6). Therefore, only systems with N = 1 and 2 PMAPTAC20 molecule(s) were considered for production simulations.
Table 1. Summary of the Main Simulated Systemsa.
| system | PMAPTAC molecules (N) | POPC/POPS molecules | water molecules | K+/Cl– ions | dunit/lipidb | simulation time (ns) |
|---|---|---|---|---|---|---|
| POPC/POPS | 0 | 230/58 | 15 802 | 58/0 | 0 | 3 × 1000 |
| M20-1 | 1 × PMAPTAC20 | 230/58 | 26 961 | 38/0 | 0.07 | 3 × 1000 |
| M20-2 | 2 × PMAPTAC20 | 230/58 | 30 139 | 18/0 | 0.14 | 3 × 1000 |
| M40-1 | 1 × PMAPTAC40 | 230/58 | 39 733 | 18/0 | 0.14 | 3 × 1000 |
| M40-1-22 | 1 × PMAPTAC40 | 230/58 | 39 433 | 168/150 | 0.14 | 3 × 1000 |
| [KCl] = 0.22 mol/dm3 | ||||||
| M40-1-04 | 1 × PMAPTAC40 | 230/58 | 39 673 | 48/30 | 0.14 | 3 × 500 |
| [KCl] = 0.04 mol/dm3 | ||||||
| M20-1-04 | 1 × PMAPTAC20 | 230/58 | 26 901 | 58/20 | 0.07 | 3 × 500 |
| [KCl] = 0.04 mol/dm3 | ||||||
| M20-2-04 | 2 × PMAPTAC20 | 230/58 | 30 079 | 40/22 | 0.14 | 3 × 500 |
| [KCl] = 0.04 mol/dm3 |
The number of PMAPTAC (N), POPC, POPS, and water molecules and ions in each system, the number of repetitions for equilibrium simulations, and the corresponding simulation times.
dunit/lipid is the number of PMAPTAC repeating units per lipid molecule in the system.
Figure 1.
(A) Single PMAPTAC oligomer consisting of 20 3-(methacrylamido)propyl trimethylammonium units. Nitrogen and oxygen atoms are shown in blue and red, respectively. (B) Snapshot of the simulation box containing 20 units of oligomer and Cl– anions (green spheres) in water (cyan) taken after 50 ns MD simulations. (C) Distributions of the end-to-end distance of the 20- (yellow) and 40-unit (orange) oligomers calculated from the simulations (4 × 50 ns for each system). The average value is indicated by vertical lines along with illustrative conformations. (D) gN–Cl(r) and gN–OW(r) radial distribution functions (RDFs) for the atomic pairs of ammonium nitrogens (N) and Cl– anions and ammonium nitrogens and water oxygens (OW), respectively. The gN–OW curves were multiplied by a factor of 3 for better visualization. The functions were averaged over four simulations, and shades represent standard errors calculated from independent runs. (E, F) Time evolution of the interaction energy between the PMAPTAC oligomer and water or Cl– anions, for 20- and 40-unit oligomers, respectively.
MD simulations were performed to analyze the influence of different polymer coverage (the number of PMAPTAC repeating units per lipid molecule in the system, dunit/lipid) and the oligomer length on the properties of the bilayer. To this end, three independent 1- or 0.5-μs-long production simulations were performed for each system. For reference, the POPC/POPS bilayer was simulated for 1 μs (system POPC/POPS) in three copies. The two-dimensional (2D) area per lipid (APL), membrane thickness, order parameter (|SCD|) for selected carbon atoms from the lipid tails, and the POPS density were calculated using the g_lomepro software.21 The method of calculating 2D density maps was implemented in g_lomepro for this work. The |SCD| values, averaged over all lipids, were calculated for the palmitoyl (sn-1) chain of POPC and POPS using GROMACS tools. To elucidate the depth of penetration of polycation molecules into the membrane, mass density profiles for ammonium nitrogens (N) of the polymer and for POPC double bonds (C=C) and phosphate groups (P8) were calculated using the GROMACS tools. The electrostatic potential across the box and the water ordering along the Z axis were calculated using the GROMACS tools, while an in-house software tool was developed to calculate the water ordering around the oligomer chains. Instantaneous interaction energies were extracted from the edr files after reanalyzing the trajectories.
Force Field Parameters and Simulations Details
All molecules were described using the OPLS-AA force field.22 POPC and POPS topologies were parametrized using the improved parameters for lipids.23 PMAPTAC oligomers were parametrized using the available OPLS-AA parameters; the quaternary ammonium group was described with the same parameters as used for the headgroup of phosphatidylcholine. The appropriate OPLS parameters were used for K+ and Cl– ions.24 Water was represented by the TIP3P model.25
All simulations were performed with the GROMACS 2018 simulation software.26−28 The SETTLE algorithm29 was used to constrain water bonds and angles. All other hydrogen-containing bonds were constrained using LINCS.30 All simulations were performed at 323.15 K with the Nosé–Hoover31,32 thermostat. The pressure was controlled at 1 bar using a semi-isotropic Parrinello–Rahman barostat33 (τ = 1 ps). Dispersive interactions and short-range repulsion were described by the Lennard-Jones potential with a cutoff of 1.4 nm. Electrostatic interactions were calculated using the particle-mesh Ewald (PME) method34 with a 1.4 nm real-space cutoff and a 0.12 nm Fourier grid. The dispersion correction for energy and pressure was applied.
Results
MD Simulations
Hydration and Conformation of PMAPTAC Chains in Water
We first considered the behavior of PMAPTAC in an aqueous medium. As an example, the final configuration of the PMAPTAC20 oligomer simulated in the water box is shown in Figure 1B. Figure 1C shows that both oligomers adopt bent conformations. To analyze the associations of counterions and water molecules with the PMAPTAC chains, we calculated the radial distribution function (RDF) for the ammonium nitrogen (N)–Cl– anion (gN–Cl) and ammonium nitrogen (N)–water oxygen (OW) (gN–OW) pairs (Figure 1D). All RDF functions were normalized by the density calculated over the whole simulation box, which allowed us to obtain a number of counterions and water molecules in solvation shells. In addition, information on the formation and spatial distribution of primary, secondary, or more interaction shells around the polycation chains can be extracted from these functions.
In the case of the N–OW pairs, the shape and the maxima locations of the RDF curves are very similar for both oligomers. The maximum at 0.5 nm shows that there is a well-defined first hydration shell around the N(CH3)3+ groups consisting of ca. 6 water molecules. As quaternary ammonium groups are not able to form hydrogen bonds, water molecules interact with them through electrostatic interactions between a positively charged N atom (partial charge +0.34e in our simulations) and negatively charged O atoms in water (partial charge −0.83e).
The gN–Cl radial distribution functions show a small maximum at about 0.4 nm, which corresponds to Cl– anions tightly bound to the polymer chain. The integration of these peaks resulted in an average number of counterions per polymer unit equal to approximately 0.04. This result indicates that a slight fraction of counterions is located directly at the polymeric ammonium groups. Nevertheless, most chloride anions are located less than 2–2.5 nm from the polymer chains, forming an ionic cloud around PMAPTAC molecules. The interaction energies between PMAPTAC and both water and chloride anions (Figure 1E,F) confirm that the polycations interact with them via strong Coulombic attraction.
Interaction of PMAPTAC with the POPC/POPS Bilayer
We performed MD simulations of polycation bilayer systems to investigate the behavior of polycations on the anionic membrane and determine the influence of polymer chain length on interactions with such a membrane. Figure 2 shows snapshots for systems: M20–1 and M40–1 (both with a single PMAPTAC oligomer) and M20–2 (with two PMAPTAC oligomers). In all of these systems, the oligomers readily adsorb onto the bilayer surface within less than 10 ns and remain on the surface until the end of the simulations (Figures 2 and S4). On the contrary, in systems containing more oligomers (M20-3, M20-4, and M20-6), we observed the adsorption of only two oligomers on the upper leaflet while the remaining molecules were repelled from this lipid layer, and due to periodic boundary conditions, they were able to migrate to and interact with the lower leaflet or remain not adsorbed (Figures S4–S6). Since only one lipid leaflet was exposed to polycations in the experiments, we focused on the simulations of systems M20-1, M40-1, and M20-2 for further analysis.
Figure 2.
Snapshots of systems M20-1 (A) and M40-1 (G) (with a single PMAPTAC oligomer) and M20-2 (D) (with two PMAPTAC oligomers) taken at the end (t = 1000 ns) of MD simulations. The PMAPTAC oligomers are shown as yellow/orange sticks with the nitrogen and oxygen atoms in blue and red, respectively. POPC and POPS are shown as gray and pink sticks, respectively, with their headgroups shown as spheres. Water is shown as a cyan surface. Distributions of the end-to-end distance of the 20- and 40-unit oligomers deposed on the POPC/POPS membrane are shown in panels B, E, and H. The average values are indicated by vertical lines. The radial distribution functions (RDFs) for the atomic pairs of PMAPTAC ammonium nitrogens and serine oxygens, POPS and POPC phosphate phosphors and water oxygens, and POPC ammonium nitrogens and water oxygens are shown in panels C, F, and I, respectively. The functions were averaged over the last 500 ns and three copies. Shaded areas represent standard deviations calculated from three independent runs.
The end-to-end distances of the oligomer chains adsorbed on the anionic membrane are on average smaller compared to the values for PMAPTAC in water, indicating that the polycations adopt more compact conformations on the membrane than in water (Figure 2B,E,H). To further analyze the polycation–membrane interactions, we calculated the RDF functions for atomic pairs: ammonium nitrogens of oligomer and serine oxygens (N–OS), phosphorus atoms of POPS (N–PS) and POPC (N–PC), and water oxygens (N–OW) for the M20-1, M40-1, and M20-2 systems (Figure 2). We also calculated the RDF function between the POPC ammonium atoms and water oxygens (NPC–OW) to compare the hydration of the polycations with that of the membrane. All of the N–OW RDF functions are similar to those in water, and the hydration number of the polymer N(CH3)3+ groups is similar to that in water (about six molecules). This shows that the polymer does not dehydrate when it settles on the membrane. On the other hand, our calculations show that the hydration of the POPC N(CH3)3+ groups is greater (about 11 water molecules at a distance of 0.5 nm from the nitrogen atom) than for the same groups in the polymer, indicating greater hydration of the membrane. PMAPTAC interacts strongly with the negatively charged O atoms of the serine group and does not penetrate deeper into the lipid membrane. Interactions with POPC are much weaker, suggesting the preferential interactions of the polymer with POPS lipids and their possible accumulation around the polymer molecule. (See also Figure S7 for interaction energies showing preferential interactions of PMAPTAC with POPS lipids.)
To further quantify the hydration of membrane-bound PMAPTAC oligomers, we calculated the orientation of the water molecules with respect to the membrane normal (Z axis, Figure S8), using the average cosine of the angle formed between the water dipole and the membrane normal. We observed a preferential ordering of water molecules at the membrane–water interfaces (for −3 nm < Z coordinate < – 1.5 and 1.5 nm < Z coordinate < 3 nm); however, it seems that such ordering is not affected by the presence of the polycation, which occupies only one interface (Z coordinate = ∼3 nm). Surprisingly, we noticed additional water ordering in all systems containing oligomers in the bulk water (that is, for −8 nm < Z coordinate < −4 and 4 nm < Z coordinate < 8 nm), while this effect did not occur in simulations of the pure POPC/POPS bilayer. (The average cosine of the angle between the water dipole and the membrane normal for bulk water is 0.) We suspected that this water ordering effect was a consequence of the very low concentration of mobile ions (only the counterions needed to neutralize the system were used) in a periodic box setup (Discussion section). To test this hypothesis, we performed additional simulations of polymer-containing systems at [KCl] = 0.04 mol/dm3 (Table 1, systems M20-1-04, M20-2-04, and M40-1-04) and of the M40-1 system at [KCl] = 0.22 mol/dm3 (system M4001-22). Indeed, using the lower KCl concentration was enough to remove the water ordering effect in bulk water (Figure S8B).
We also calculated the radial orientation of the water molecules around the oligomers in systems M20-1, M20-2, and M40-1 (Figure S9) by considering each unit in each polycation chain independently and computing the dipolar alignment order parameters35 as a function of the distance from the center of mass of the corresponding PMAPTAC unit. After averaging the calculated order parameters for each system, we observed a preferential orientation of water molecules around the PMAPTAC oligomer within 5 nm from the polymer chain, which is unaffected by the presence of KCl. The individual water molecules bound to the oligomers showed a quick exchange with the bulk since 99.98% of the residence times of water molecules around the oligomers were less than 50 ps.
Effect of Adsorbed PMAPTAC on the POPC/POPS Bilayer
We first consider the changes in lipid distribution induced by the oligomers adhered to the POPC/POPS membrane. Figure 3 depicts 2D profiles of the density of POPS molecules. In the case of the M20-1 and M40-1 systems, containing only one oligomer molecule, the density increases largely in polymer–membrane contact areas, reaching a value of 0.91 molecule nm–2 (for the POPC/POPS system, the average density of POPS is 0.29 molecule nm–2), which corresponds to the POPS/POPC ratio of 4:1. This result clearly demonstrates that the polycation adsorption process leads to the accumulation of the anionic lipids in the vicinity of the polymer chains (formation of POPS-rich domains). For the M20-2 system, with two oligomer molecules, the increase in the POPS density in the contact areas is lower than in the M20-1 system and is below 0.75 POPS molecule nm–2. This indicates that two PMAPTAC20 oligomers are able to accumulate fewer anionic lipids since they compete for available POPS molecules and also repel each other. Furthermore, a comparison of systems M20-2 and M40-1 that have the same dunit/lipid shows that PMAPTAC40 is able to attract POPS molecules more effectively than two PMAPTAC20 oligomers. In addition, as shown in Figure 3B,C, the shorter oligomer adopts more spherical conformations on the POPC/POPS membrane while PMAPTAC40 has a more elongated shape; therefore, the polymer–membrane contact areas are smaller in the M20-2 system (see also Figure 2B,E,H). As a result, the PMAPTAC20 can adsorb onto the surface of the bilayer, but the polymer–bilayer interactions are weaker compared to the longer polycation. Importantly, it should be noted that in the lower leaflet no changes in the POPS density were observed in any of the simulated systems, so the local accumulation of POPS lipids is limited to the leaflet that is in direct contact with PMAPTAC.
Figure 3.

Two-dimensional density profiles of POPS molecules in systems M20-1 (A), M20-2 (B), and M40-1 (C). The polymers are shown as yellow and orange licorice, while the average position of the grid corresponding to the lipid phosphorus atoms is shown by color-coded spheres. The value of 0.29 nm–1 on the color bar represents the average POPS density of the POPC/POPS bilayer without the polymer.
The adhesion of polycations to one anionic membrane leaflet can lead to a difference in electrostatic potential between leaflets. Therefore, the electrostatic potentials along the bilayer normal (Z axis) were calculated for M20-1, M20-2, and M40-1 systems. Figure 4 shows that the POPC/POPS membrane has a positive potential with respect to the aqueous phase and its electrostatic potential profile is characterized by three distinct maxima. Similar profile shapes for other anionic membranes have been reported previously.36,37 The adsorption of PMAPTAC oligomers on the membrane surface increases the positive potential in the upper leaflet. A local gradient of the electrostatic potential appears between the leaflets, which creates the potential difference between the two leaflets, referred to as interleaflet voltage.19 However, as M20-1, M20-2, and M40-1 systems contain very few mobile counterions needed to neutralize the overall charge of the system, we suspect that the calculated potential in a periodic box might not be representative of the actual potential, which can then lead to unexpected effects. (See water ordering above and the Discussion section.) Of special interest is that the addition of 40 mM KCl removes any effects on the potential caused by the polymer (Figure S15).
Figure 4.

Electrostatic potential profiles for the POPC/POPS bilayer and in the presence of polymer molecules (systems M20-1, M20-2, and M40-1). The shaded area indicates the standard deviation. Polymer molecules are adsorbed on the upper leaflet of the studied bilayers.
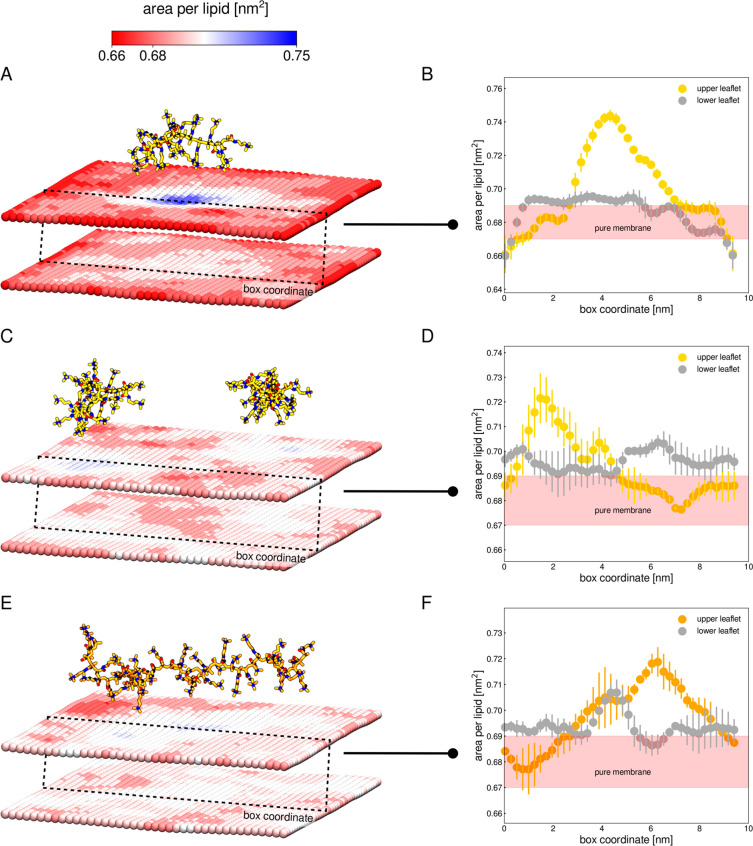
We next calculated 2D distributions of the area per lipid (APL) and bilayer thickness to assess the impact of the polycation adsorption on the membrane properties. Figure S10 shows that the pure POPC/POPS bilayer has almost uniform thickness, and fluctuations in the area occupied by the lipid molecule are small. The average values of APL and thickness are 0.68 ± 0.01 nm2 and 3.89 ± 0.02 nm, respectively, for the POPC/POPS system. The adsorbed PMAPTAC significantly changes the molecular organization of the POPC/POPS bilayer (Figures 5 and 6). In the areas where the polymer adheres to the surface, the bilayer becomes much thinner (membrane thickness decreases to ca. 3.6 nm) and more loosely packed (APL increases to ca. 0.75 nm2 in the upper leaflet in the M20-1 system, Figure 5A,C,E). We were interested to determine if this local increase in APL in the upper leaflet is also visible in the lower leaflet, which in turn could suggest interleaflet coupling. To this end, the APL values along one arbitrarily chosen box coordinate for the upper and lower leaflets were plotted (Figure 5B,D,E). The APL increase in the lower leaflet is much smaller and more uniform than in the upper one. Only in the M40-1 system, in which the oligomer interacts most strongly with the membrane, is a somewhat higher correlation in APL changes between leaflets observed. These results suggest that the adsorption of the polycation on the membrane may cause interleaflet coupling; however, the magnitude of such coupling is probably dependent on the strength of the polycation–membrane interaction.
Figure 5.
Two-dimensional profiles of the area per lipid (A, C, and E) and a comparison of the area per lipid in the upper and lower leaflets along a selected box coordinate (B, D, and E). Systems M20-1 (A and B), M20-2 (C and D), and M40-1 (E and F) are shown. The middle value in the color bar shows the average value of the area per lipid in the POPC/POPS system. The polymers are shown in yellow and orange licorice. The average position of grid elements corresponding to each lipid center of mass is shown by color-coded spheres.
Figure 6.

Two-dimensional profiles of the membrane thickness for systems M20-1 (A), M20-2 (B), and M40-1 (C). The middle value in the color bar shows the average value of the area per lipid in the POPC/POPS system. The polymers are shown in yellow and orange licorice. The average position of grid elements corresponding to the lipid phosphate atoms is shown as color-coded spheres.
To assess the conformation of the lipid acyl chains in the polymer-treated membranes, we calculated the deuterium order parameters, |SCD|, for the palmitoyl chains of all of the POPC and POPS molecules. Figure S11 shows that PMAPTAC adsorption causes a slight decrease in the ordering averaged over the whole membrane. More information is provided by 2D profiles of |SCD| for the third and ninth carbon atoms of the POPS sn-1 chains (Figure 7). The influence of PMAPTAC on the arrangement of hydrocarbon chains depends on the length of the polymer chain and dunit/lipid and thus on the strength of the polycation–membrane interactions. In systems with one oligomer molecule (M20-1 and M40-1), the adsorbed polycation largely reduces the value of the order parameter in the contact areas in the upper leaflet. Also, in the lower leaflet, the membrane ordering decreases to some extent, which suggests a slight coupling between the two leaflets. However, in the system with two PMAPTAC20 molecules, this influence is somewhat reduced, which is consistent with the lower effect of two PMAPTAC20 oligomers on APL and the membrane thickness. It should be emphasized that PMAPTAC mainly reduces the ordering of the upper segment of the lipid chain (segment 3) (i.e., the most ordered part of the membrane). The impact of the polycation on |SCD| in deeper regions of the bilayer is much smaller.
Figure 7.
Two-dimensional profiles of order parameter |SCD| for carbon atoms 3 (left) and 9 (right) from the sn-1 chain of POPS lipids. Systems M20-1 (A and B), M20-2 (C and D), and M40-1 (E and F) are shown. The middle values in the color bars show average order parameters for a given carbon atom in the POPC/POPS system. The polymers are shown in yellow and orange licorice. The average position of grid elements representing carbon atoms 3 (left) and 9 (right) is shown as color-coded spheres.
Effect of Salt Concentration on the Interaction of PMAPTAC with the POPC/POPS Bilayer
The effect of salt concentration on the polycation–anionic membrane interaction is well understood in the literature.15 The presence of salt weakens the electrostatic interactions between the polycation and anionic liposomes, and the adsorbed polycation can be completely removed from the surface of charged vesicles by increasing the ionic strength.15 Therefore, the dissociation of the electrostatic liposome–polycation complexes is controlled by the salt concentration in the surrounding solution. To investigate the salt effect in our systems, we simulated all three polycation-containing systems at [KCl] = 0.04 mol/dm3 (systems M20-1-04, M20-2-04, and M40-1-04; see Table 1) as well as system POPC/POPS at [KCl] = 0.22 and 0.65 mol/dm3 (systems POPC/POPS22 and POPC/POPS65, Table S2). Additionally, we simulated a system containing the longer oligomer at [KCl] = 0.22 mol/dm3 (system M40-1-22, Table 1). The results of our simulations are generally in good agreement with experimental observations. For systems containing only the POPC/POPS membrane, slight salt-concentration-dependent condensation of the membrane (decrease in APL and increase in thickness, Figure S10) was observed.
In polymer-containing systems, at lower salt concentrations (0.04 mol/dm3), PMAPTAC molecules remain bound to the membrane (Figure S12). Accordingly, the oligomers affect the membrane properties (Figures S13 and S14) in a manner similar to that observed in systems without salt (M20-1, M20-2, and M40-1); however, the magnitude of such changes is reduced in the presence of salt. The POPS accumulation, APL, and membrane thickness are 60–80% of those seen in salt-free systems. Consequently, the interleaflet coupling is hardly detectable in these systems. In addition, the effect of the adsorbed polymer removed any effect on the interleaflet voltage possibly brought about by the polymer in all systems containing 0.04 mol/dm3 salt (Figure S15), suggesting the important role of mobile ions in modulating the membrane’s electrostatic properties.
At a KCl concentration of 0.22 mol/dm3 (Figure S16), the PMAPTAC40 oligomer interacts with the membrane even more weakly and is able to repeatedly attach to and detach from the bilayer surface. Even when bound to the membrane, the oligomer remains, on average, further away from the lipid headgroups (Figure S16 B) compared to the spacing in system M40-1, indicating a screening effect of the polycation by chloride anions that compete with negatively charged POPS lipids for electrostatic interactions with PMAPTAC (Figure 8). Consequently, the cationic polymer has almost no effect on membrane properties such as the APL, thickness, and local density of the POPS molecules as well as the electrostatic potential (Figure S16C–F).
Figure 8.

Average energy of electrostatic interactions between PMAPTAC40 and POPS lipids in all three systems containing PMAPTAC40 with different salt contents (Table 1). The values presented are the averages over three simulations, and standard errors are shown as error bars.
DLS and Zeta Potential Measurements
It is known that the introduction of linear polycations into the dispersion of anionic liposomes can induce the reversible aggregation of vesicles, and stable isolated polycation-covered liposomes are formed after the addition of sufficient amounts of the polymers.38,39 To check the effect of our polymers on the aggregation behavior of the POPC/POPS liposomes, we used DLS and zeta potential measurements (Table 2). A series of samples containing a constant lipid concentration (c = 1 mg/mL) and various mass fractions of the polycation were prepared. The size of the bare POPC/POPS liposomes (4:1 ratio) was monodisperse (around 100 nm), and their surface potential was negative due to the presence of negatively charged lipids. The addition of PMAPTACs caused an increase in the zeta potential of the liposomes, which indicates that both PMAPTAC147 and PMAPTAC16 adsorbed on the surface of POPC/POPS vesicles (Table 2). However, the polymer chain length affected the observed increase in the ζ potential and liposome aggregation. In the case of PMAPTAC147, after introducing a small amount of the polycation, drastic increases in the size and PDI were observed, indicating polycation-induced liposome aggregation.38 Increasing the polycation concentration resulted in gradual reductions of dz and PDI and thus the dissociation of aggregates and the formation of isolated polycation-coated vesicles, as previously shown using direct microscopic observations.38 The ζ value increased with increasing PMAPTAC147 content and reached a constant value of ca. 43 mV at a mass fraction higher than 9%. On the contrary, the POPC/POPS liposomes treated with PMAPTAC16 remained aggregated even at high polymer concentrations, as indicated by the dz values. In addition, the zeta potential became positive at a similar polymer content, but the ζ value was 4 times lower than that of the longer polycation.
Table 2. Values of the Mean Hydrodynamic Diameter (dz), Polydispersity Index (PDI), and Zeta Potential (ζ) of the POPC/POPS (4:1 Weight Ratio) Liposomes Dispersed in a PBS Solutiona and Treated with PMAPTACsb.
| system | polycation concentration (μg/mL) | polycation content (wt %) | dz (nm) (n = 5) | PDI (n = 5) | ζ (mV) (n = 8) |
|---|---|---|---|---|---|
| anionic LUVs | 0 | 0 | 106.2 ± 0.2 | 0.07 ± 0.02 | –52.1 ± 2.3 |
| anionic LUVs | 6 | 1.2 | 173.9 ± 3.8 | 0.38 ± 0.01 | –43.4 ± 2.2 |
| with PMAPTAC147 | 12 | 2.4 | 672.4 ± 182.0 | 1.0 | –41.9 ± 2.6 |
| 18 | 3.6 | >103 | 1.0 | –22.2 ± 1.6 | |
| 24 | 4.8 | >104 | 1.0 | 17.8 ± 1.5 | |
| 30 | 6.0 | 265.7 ± 28.4 | 0.37 ± 0.02 | 34.1 ± 1.2 | |
| 42 | 8.4 | 205.1 ± 2.5 | 0.27 ± 0.04 | 40.5 ± 2.5 | |
| 48 | 9.6 | 189.2 ± 1.5 | 0.25 ± 0.03 | 43.4 ± 2.3 | |
| 54 | 10.8 | 183.9 ± 2.5 | 0.26 ± 0.04 | 42.8 ± 2.5 | |
| 60 | 12.0 | 176.5 ± 4.2 | 0.24 ± 0.03 | 43.0 ± 2.4 | |
| 66 | 13.2 | 156.2 ± 1.5 | 0.18 ± 0.02 | 43.0 ± 2.7 | |
| anionic LUVs | 10 | 2 | 324.0 ± 22.9 | 0.56 ± 0.14 | –34.8 ± 2.2 |
| with PMAPTAC16 | 25 | 5 | >104 | 1.0 | 4.5 ± 1.0 |
| 50 | 10 | >104 | 1.0 | 20.0 ± 0.6 | |
| 75 | 15 | >103 | 1.0 | 23.7 ± 1.7 | |
| 100 | 20 | >103 | 1.0 | 26.5 ± 2.3 | |
| 250 | 50 | >103 | 1.0 | 29.3 ± 1.9 | |
| 500 | 100 | 872.6 ± 700.4 | 0.84 ± 0.24 | 31.7 ± 1.7 | |
| 750 | 150 | 421.9 ± 67.9 | 0.80 ± 0.24 | 31.4 ± 1.6 |
Total lipid concentration of c = 1 mg/mL.
Values are the mean ± standard deviation.
Calcein-Release Studies
The ability of polymers to porate lipid membranes can be quantified by monitoring the leakage of a fluorescent dye (e.g., calcein) from liposomes.18,40 Liposomes, containing calcein at a concentration causing its self-quenching, are treated with the polymer, and then the fluorescence of calcein is measured. If the polymer increases the permeability of the membrane for hydrophilic compounds (e.g., by poration), then calcein may leak from the liposomes and fluoresce due to dilution in the bulk aqueous phase. To study the membrane activity of PMAPTACs, the calcein-loaded POPC/POPS vesicles were incubated with the polycations at concentrations of 30 and 13.2 wt % with respect to the lipid contents for PMAPTAC16 and PMAPTAC147, respectively. The released calcein fraction (RF) was calculated according to eq 1 (Figure 9). The presence of polycations at the concentrations tested did not increase the fluorescence intensity of calcein over time. Therefore, it can be assumed that PMAPTACs are not able to form pores in lipid membranes at such concentrations, which is consistent with the lack of penetration of the bilayer by the polymer molecules observed in the MD simulations.
Figure 9.

Time course of calcein release (RF) from the POPC/POPS liposomes treated with PMAPTAC16 (30 wt %) and PMAPTAC147 (13.2 wt %).
Discussion
We examined the interaction between the linear polycations (PMAPTACs) having quaternary ammonium groups and the anionic lipid membrane. It is well known that polycations strongly adhere to biomembranes, which are most typically negatively charged, forming soft coronae tenuously adsorbed or anchored on the membrane.8,15 However, the molecular landscape of such interactions is not fully understood. Therefore, we applied a combination of atomistic-scale MD simulations and experimental techniques to explore several aspects of the association of strong polycations to anionic lipid membranes. To this end, we first examined the behavior of the polycation in the aqueous phase. Next, we characterized the molecular mechanism of the interaction of PMAPTAC with the POPC/POPS membrane. In addition, we focused on perturbations in membrane properties induced by PMAPTAC adsorption. Finally, we examined the effects of different salt concentrations on the interactions of PMAPTAC with the POPC/POPS membrane.
PMAPTAC Is Highly Hydrated in Aqueous Solution
The degree of hydration of a polycation is one of the most important factors affecting its interaction with lipid membranes. Surprisingly, the hydration of polycations with quaternary ammonium groups has not yet been studied. To examine the behavior of PMAPTAC in aqueous media, we simulated both oligomers in the box of water (Figure 1). The results indicate that PMAPTACs are highly hydrated in aqueous solution, with six water molecules in the first hydration shell. Chloride anions are in close proximity to the polymer chain, but few of them are in direct contact with the positively charged PMAPTAC groups. Our results are consistent with MD simulations of the hydration of choline groups in phospholipids. The simulations of water around fully hydrated phospholipids showed a mobile clathrate-like hydration shell of approximately five to six water molecules around the positively charged N+(CH3)3 choline moiety.41
Despite strong hydration and repulsion between the charged units along the polymer chain, the PMAPTAC chains have some flexibility and are not fully elongated, as indicated by the end-to-end distance (Figure 1). The charged groups of PMAPTAC are linked to the polymer chain by a long spacer and are located away from the polymer backbone. In addition, repulsive interactions between the N+(CH3)3 groups are partially screened by the chloride counterions. As a result, PMAPTAC can adopt a bent conformation with the charged groups located asymmetrically around the polymer backbone. For comparison, Ivanov et al. simulated a short (20 unit) derivative of polymethacrylate with pendant NH3+ groups in aqueous solution for 7.5–9.5 ns.42 It was found that this oligomer adopted extended helical shapes, which was explained by the electrostatic repulsion of the charged side chains; however, the end-to-end distance was not analyzed. We believe that strong polycations are relatively flexible due to the screening effect of the counterions, but this issue should be further addressed in a long simulation of a much longer polymer chain. It is also worth noting here that the dimensions of solvated polymers without a well-defined structure exhibit a strong force field dependence.43
PMAPTACs Strongly Adhere to the POPC/POPS Membrane via Electrostatic Interactions
Most polycations adhere to biological membranes via (1) hydrophobic interactions between the hydrophobic moiety in amphiphilic polycations and lipid bilayers, (2) hydrogen bonding between polymer donor groups and the phosphate group in phosphorylcholine heads, and (3) electrostatic attraction between the positive charges of polycations and the negative surface charges of cells.2,10 PMAPTAC has no purely hydrophobic moieties in its structure; therefore it may interact with biomembranes mainly through electrostatic attraction. In addition, PMAPTAC units contain amide bonds that can serve as a proton donor in forming a hydrogen bond (H bond). Our previous computational modeling of PEIs interacting with POPC membranes showed that PEIs were capable of forming H bonds with polar groups of zwitterionic lipids.10 To determine the ability of PMAPTAC to adhere to the zwitterionic membrane, we tested systems containing a bilayer made out of only POPC lipids and PMAPTAC oligomers (Figure S18). The simulations show that the PMAPTAC molecules do not interact appreciably with the POPC membrane in any of the systems studied. To estimate the interaction between PMAPTAC and the pure POPC membrane, we calculated the energies of electrostatic interaction between the polymer and lipid molecules (Figure S18). Positive values of these energies indicate that the interactions are repulsive. The zwitterionic headgroups of phosphatidylcholines, although neutral, are oriented in bilayers so that their negative phosphates lie deeper than their positively charged choline groups that are exposed to the aqueous phase.
Our MD simulations and zeta potential measurements clearly show that the PMAPTAC molecules readily associate with the POPC/POPS membrane. In contrast to the POPC membrane, the energy of electrostatic interactions between the polycation and both POPC and POPS lipids in systems containing the POPC/POPS membrane is negative (attractive), as shown in Figure S7. The polycation approaching the POPC/POPS membrane triggers a local increase in the concentration of the anionic lipids that results in the formation of negatively charged domains in the bilayer, around the polymer. Lateral lipid diffusivity allows for a rapid change in the local composition. It has been reported that polycations can interact selectively with the membrane in the fluid phase (lipid diffusion is possible), whereas below the fluid–gel transition temperature no association took place.44 Thus, the formation of highly charged patches effectively strengthens the binding of polyelectrolytes to the membrane. The phase separation of anionic lipids, when anionic membranes are mixed with a polycation, has been previously demonstrated experimentally using calorimetric45,46 and 1H NMR measurements.47 An important result from our simulations is that the polycation chains remain strongly hydrated when settled down on the POPC/POPS bilayer. Therefore, PMAPTAC is preferentially located in the polar region, close to the headgroups, and does not penetrate into the center of the bilayer (Figure S4), in contrast to hydrophobically modified polycations.18,19,48
PMAPTAC adsorption is strongly dependent on the length of the polymer chain. We experimentally observed that PMAPTAC16 interacts much less effectively with the POPC/POPS membrane compared to PMAPTAC147. This is consistent with previous observations. Franzin et al.47 investigated the interaction among three polylysines (weak polycations) with 5, 30, and 100 lysine residues per chain and POPC/POPS (70:30 mol/mol) membranes. They observed that only the two longer polymers added to the vesicles caused the formation of polylysine-bound domains enriched in POPS, in coexistence with polylysine-free domains depleted in POPS. At physiological salt concentrations, only the longest polylysine was able to laterally segregate POPS from mixtures with POPC and form POPS-enriched domains. Also, in the case of a strong polycation, polymers with higher degrees of polymerization were shown to have a greater ability to induce the lateral segregation of anionic lipids in mixed membranes.49 The authors also showed that strong polycations completely bind to the liposomes up to a certain concentration, and at higher concentrations, the appearance of free (unbound) polycations is observed. The results of our MD simulations are consistent with experimental findings and can provide deeper insight into the polycation–membrane interactions. The adsorption of PMAPTAC40 induces a significant accumulation of POPS lipids in the contact areas (even up to 80 mol %, although the POPS density in these domains is not uniform; see Figure 3C). In the case of system M20-2 having the same dunit/lipid as system M40-1, we observed much lower POPS densities in the anionic domains. Therefore, the two PMAPTAC20 oligomers interact more weakly with the membrane, and the additional molecules are not able to adsorb on the upper leaflet (Figures S4–S6). This observation is consistent with the view that longer, polyvalent polymers interact more strongly with surfaces than shorter molecules, most likely for entropic reasons.50 In addition, polycations adsorb to the surface of anionic liposomes only to a limited value of dunit/lipid, which is independent of the length of the polymer chain. The mass density profiles of K+ cations (Figure S17) show that the adsorption of the polycation on the upper leaflet of the membrane causes the release of the monocations from the bilayer surface. At a polymer coverage dunit/lipid greater than 0.14, the PMAPTAC oligomers completely replace the potassium cations.
Adsorbed PMAPTAC Has a Significant Impact on the Membrane Properties
As a result of strong Coulombic associations, PMAPTAC creates POPS-rich domains whose features differ significantly from the rest of the membrane. Importantly, our simulations show that the approaching PMAPTAC chain induces the formation of the anionic domains only in the upper leaflet. We did not observe a change in the POPS density in the lower monolayer in any of the simulated systems. The adsorption of PMAPTAC has a considerable effect on the ordering of the acyl chains and lipid packing in the polymer–membrane contact areas. As indicated by the significant increase in APL (Figure 5), the presence of the polycation causes the lipid molecules to move away from each other, which is accompanied by a decrease in the order of the upper segments of the acyl chains (close to the carbonyl groups). As a consequence, the membrane thickness in the contact areas decreases considerably. Furthermore, a comparison of the M20-2 and M40-1 systems that have the same dunit/lipid reveals that PMAPTAC40 adopts more elongated conformations at the membrane surface than the shorter oligomers. Therefore, its polymer–bilayer contact areas are much larger compared to the two PMAPTAC20 oligomers. As a consequence, the impact of the longer-chain polycations on the membrane properties is substantially greater than that exerted by the shorter cationic polymers.
The adsorbed polymer induces the described changes mainly in the upper leaflet of the membrane, which is in direct contact with the polymer. We also observed slight changes in the lipid organization of the lower leaflet. For example, APL increases and the order parameter decreases for lipids located in the areas of the lower leaflet adjacent to the POPS-rich domains in the upper leaflet. This is a further contribution to the observed reduction in the thickness of the whole bilayer. It is known that the formation of domains in one leaflet has some impact on the properties of the opposite leaflet.51 This phenomenon is called interleaflet coupling or transbilayer coupling. Several possible mechanisms of the coupling between two leaflets have been proposed, in which acyl-chain interdigitation into opposing leaflets and surface tension in the midplane seem possible in our systems.51,52 To assess the extent of interdigitation between both lipid monolayers, we calculated the mass density profiles of the terminal carbon atoms in the hydrocarbon tails. Figure S19 show that the acyl chains in the polycation-treated bilayer do not protrude into the opposite layer more deeply than those in the pure membrane. Therefore, we believe that the interleaflet coupling observed in our systems results from the minimalization of so-called interleaflet tension (the surface tension at the midplane interface between the two monolayers of a membrane domain).53 This tension is predicted to exist between opposing leaflets if the packing density of the hydrocarbon chains changes across the midplane interface. To avoid this apparent energy cost, the lipids in the lower leaflet alter their organization so that the packing density of their tails is similar to that in the polycation-decorated upper leaflet (similar on both sides of the interface). It is worth noting that in bilayers simulated with periodic boundary conditions, changes in APL in one leaflet will cause changes in APL in the other leaflet (the total area of both leaflets is related to each other). However, without interleaflet coupling, uniform changes in the APL distribution in the lower leaflet should be expected.
We did not observe any rupture of the POPC/POPS bilayer during the simulations. The reason for this is likely that PMAPTAC interacts only with the membrane surface and does not penetrate it. As the mass density profiles show (Figure S17), the polycation main chains are located in the aqueous phase near the membrane, and a large part of the positively charged groups are directed to the bulk solution. Our findings are consistent with the calcein-release measurements, showing that PMAPTAC did not increase the membrane permeability for hydrophilic compounds, thus the presence of PMAPTAC does not induce pores in the anionic membranes.
Increasing Ionic Strength Significantly Reduces the PMAPTAC–Anionic Membrane Interaction
Simulating membrane systems with a minimal salt concentration and a large, charged molecule adsorbed on the bilayer poses a technically intricate challenge. The described situation resembles the simulation of an asymmetric lipid membrane in a periodic environment. Such asymmetric membranes present an electric dipole, which in turn generates a transmembrane potential as demonstrated in a double-bilayer setup in MD simulations.54 On the contrary, in the case of a single asymmetric membrane in a simulation box, the potential drop across the box is constrained to 0 V due to the imposed periodic boundary conditions. This constraint is easily satisfied by the redistribution of ions in solution. However, for simulations with a low salt concentration, an additional contribution is observed from the ordering of water dipole moments along the normal to the membrane surface (Figure S8). Even a slight increase in the ion concentration (0.04 mol/dm3 KCl) was enough to completely remove this water-ordering effect (Figure S8). Therefore, we recommend the use of salt (preferably KCl, due to potential strong interactions of Na+ with lipid membranes) for all simulations of membranes, especially asymmetric ones, to avoid the membrane potential and/or water-ordering related effects. A detailed description and quantification of this artifact will be provided in a separate work.
MD simulations of systems containing KCl at a concentration of 0.04 mol/dm3 allowed us to observe the effect of salt on polycation–membrane interactions. As expected, even a low salt concentration is able to strongly modulate such interactions (Figure 8). The oligomers still adsorb to the membrane, but the changes in membrane organization, packing, and electrostatic properties are decreased compared to systems without the addition of salt. The interleaflet potential is reduced to 0 mV in the presence of salt. At an even higher salt concentration (0.22 mol/dm3 KCl), PMAPTAC40 does not appreciably interact with the membrane (Figure 8). These observations show that the interactions of polycations with anionic membranes can be modulated by the manipulation of ionic strength.
Force Field Inaccuracies
MD simulations of multicomponent, highly charged systems are often difficult due to inherent inaccuracies in the force field parametrization. In the present work, we used the OPLS-AA force field for lipids, which is commonly used to describe lipid membranes.55 However, the inaccuracies in the lipid headgroup structure, often resulting in inaccurate membrane properties such as APL, exist in any force field.56 The binding of alkali cations to PC and PS lipid headgroups is particularly problematic, which is not precisely described in any of the existing force fields.56,57 Partially because of these issues, we initially decided to perform our simulations at a minimal possible ion concentration (counterions only), and later added only a moderate amount of salt (0.04 mol/dm3 KCl). In our simulations of the pure POPC/POPS bilayer, at different salt concentrations, we observed only a moderate condensation effect (Figure S10), even at a high salt concentration (>0.5 mol/dm3). With the PMAPTAC molecule, which possesses quaternary ammonium groups, the issue is less severe than for the alkali ions because quaternary ammonium ions have a lower charge density.58 New developments in force field parametrization open up possibilities for further investigations of ion–membrane interactions.56,57
Unfortunately, we were unable to find an experimental APL value for the POPC/POPS bilayer. For the POPC bilayer, the experimental APL determined by solid-state NMR is 0.705 ± 0.042 nm2 at 48 °C,59 which is consistent with our simulations (0.671 ± 0.001 nm2 at 323 K), suggesting that the force field that is used captures the main structural properties of the membranes studied.
Conclusions
In this work, we investigated how strong polycations with quaternary ammonium groups (PMAPTAC) behave at anionic lipid membranes and affect their properties. We found that PMAPTAC binds to the POPC/POPS bilayer due to electrostatic interactions and adopts more compact conformations; however, the polymer–membrane interactions depend strongly on the length of the polycation. The polycation adsorption is strongly associated with the formation of the POPS-rich domains and the longer polycations are able to form domains with the higher density of anionic lipids. The anionic lipid accumulation occurs only in the upper leaflet, leading to the formation of compositionally asymmetric domains. Our experimental results strongly support this finding, showing that the polycations can adsorb on the surface of the POPC/POPS liposomes. In addition, even though the polycations do not penetrate the membrane, they cause a significant change in its properties, such as the local area per lipid, membrane thickness, and acyl chain ordering, especially in the membrane–polymer contact regions. The interactions of the polycation with POPC/POPS membranes are modulated by the presence of salt in a concentration-dependent manner. Our MD simulations and fluorescence experiments indicate that the polycations with quaternary ammonium groups are unable to open pores in the POPC/POPS liposomes. Therefore, we believe that the mechanism of cytotoxicity of such polycations is more complicated than simple pore opening, which warrants further study.
Acknowledgments
The project was financed by the National Science Centre, Poland (grant no. 2016/21/B/ST5/00250). W.K. was supported by the German Research Foundation DFG through FOR 2518 “DynIon”, Project P5.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.langmuir.0c01062.
Figures S1–S19 and Tables S1 and S2 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Chen A.; Peng H.; Blakey I.; Whittaker A. K. Biocidal Polymers: A Mechanistic Overview. Polym. Rev. 2017, 57, 276–310. 10.1080/15583724.2016.1223131. [DOI] [Google Scholar]
- Kim K.; Chen W. C.W.; Heo Y.; Wang Y. Polycations and their biomedical applications. Prog. Polym. Sci. 2016, 60, 18–50. 10.1016/j.progpolymsci.2016.05.004. [DOI] [Google Scholar]
- Boussif O.; Lezoulach F.; Zanta M.; Mergny M.; Scherman D.; Demeneix B.; Behr J. P. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 7297–7301. 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Zolotarskaya O. Y.; Nair S. S.; Ehrhardt C. J.; Ohman D. E.; Wynne K. J.; Yadavalli V. K. Real-Time Observation of Antimicrobial Polycation Effects on Escherichia coli: Adapting the Carpet Model for Membrane Disruption to Quaternary Copolyoxetanes. Langmuir 2016, 32, 2975–2984. 10.1021/acs.langmuir.5b04247. [DOI] [PubMed] [Google Scholar]
- Wytrwal M.; Koczurkiewicz P.; Wojcik K.; Michalik M.; Kozik B.; Zylewski M.; Nowakowska M.; Kepczynski M. Synthesis of strong polycations with improved biological properties. J. Biomed. Mater. Res., Part A 2014, 102A, 721–731. 10.1002/jbm.a.34744. [DOI] [PubMed] [Google Scholar]
- Fischer D.; Li Y.; Ahlemeyer B.; Krieglstein J.; Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials 2003, 24, 1121–1131. 10.1016/S0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- Moghimi S. M.; Symonds P.; Murray J. C.; Hunter A. C.; Debska G.; Szewczyk A. A two-stage poly(ethylenimine)-mediated cyto-toxicity: implications for gene transfer/therapy. Mol. Ther. 2005, 11, 990–995. 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Tribet C.; Vial F. Flexible macromolecules attached to lipid bilayers: impact on fluidity, curvature, permeability and stability of the membranes. Soft Matter 2008, 4, 68–81. 10.1039/B708431P. [DOI] [PubMed] [Google Scholar]
- McGeachy A. C.; Dalchand N.; Caudill E. R.; Li T.; Dogangun M.; Olenick L. L.; Chang H.; Pedersen J. A.; Geiger F. M. Interfacial electrostatics of poly(vinylamine hydrochloride), poly(diallyldimethylammonium chloride), poly-L-lysine, and poly-L-arginine interacting with lipid bilayers. Phys. Chem. Chem. Phys. 2018, 20, 10846–10856. 10.1039/C7CP07353D. [DOI] [PubMed] [Google Scholar]
- Kwolek U.; Jamróz D.; Janiczek M.; Nowakowska M.; Wydro P.; Kepczynski M. Interactions of Polyethylenimines with Zwitterionic and Anionic Lipid Membranes. Langmuir 2016, 32, 5004–5018. 10.1021/acs.langmuir.6b00490. [DOI] [PubMed] [Google Scholar]
- Kostritskii A. Y.; Kondinskaia D. A.; Nesterenko A. M.; Gurtovenko A. A. Adsorption of Synthetic Cationic Polymers on Model Phospholipid Membranes: Insight from Atomic-Scale Molecular Dynamics Simulations. Langmuir 2016, 32, 10402–10414. 10.1021/acs.langmuir.6b02593. [DOI] [PubMed] [Google Scholar]
- Ding L.; Chi E. Y.; Schanze K. S.; Lopez G. P.; Whitten D. G. Insight into the Mechanism of Antimicrobial Conjugated Polyelectrolytes: Lipid Headgroup Charge and Membrane Fluidity Effects. Langmuir 2010, 26, 5544–5550. 10.1021/la9038045. [DOI] [PubMed] [Google Scholar]
- Sabín J.; Vazquez-Vazquez C.; Prieto G.; Bordi F.; Sarmiento F. Double Charge Inversion in Polyethylenimine-Decorated Liposomes. Langmuir 2012, 28, 10534–10542. 10.1021/la3019259. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Wu F.-G.; Hu P.; Chen Z. Interaction of Polyethylenimine with Model Cell Membranes Studied by Linear and Nonlinear Spectroscopic Techniques. J. Phys. Chem. C 2014, 118, 12195–12205. 10.1021/jp502383u. [DOI] [Google Scholar]
- Yaroslavov A. A.; Sybachin A. V.; Efimova A. A. Stabilization of electrostatic polymer-colloid complexes. Colloids Surf., A 2018, 558, 1–7. 10.1016/j.colsurfa.2018.08.042. [DOI] [Google Scholar]
- Mitsukami Y.; Donovan M. S.; Lowe A. B.; McCormick C. L. Water-Soluble Polymers. 81. Direct Synthesis of Hydrophilic Styrenic-Based Homopolymers and Block Copolymers in Aqueous Solution via RAFT. Macromolecules 2001, 34, 2248–2256. 10.1021/ma0018087. [DOI] [Google Scholar]
- Lewandowska J.; Kȩpczyński M.; Bednar J.; Rzad E.; Moravcikova V.; Jachimska B.; Nowakowska M. Silicone-stabilized liposomes. Colloid Polym. Sci. 2010, 288, 37–45. 10.1007/s00396-009-2124-y. [DOI] [Google Scholar]
- Kepczynski M.; Jamróz D.; Wytrwal M.; Bednar J.; Rzad E.; Nowakowska M. Interactions of a Hydrophobically Modified Polycation with Zwitterionic Lipid Membranes. Langmuir 2012, 28, 676–688. 10.1021/la203748q. [DOI] [PubMed] [Google Scholar]
- Awasthi N.; Kopec W.; Wilkosz N.; Jamróz D.; Hub J. S.; Zatorska M.; Petka R.; Nowakowska M.; Kepczynski M. Molecular Mechanism of Polycation-Induced Pore Formation in Biomembranes. ACS Biomater. Sci. Eng. 2019, 5, 780–794. 10.1021/acsbiomaterials.8b01495. [DOI] [PubMed] [Google Scholar]
- Kepczynski M.; Nawalany K.; Jachimska B.; Romek M.; Nowakowska M. Pegylated tetraarylporphyrin entrapped in liposomal membranes. A possible novel drug-carrier system for photodynamic therapy. Colloids Surf., B 2006, 49, 22–30. 10.1016/j.colsurfb.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Gapsys V.; de Groot B. L.; Briones R. Computational analysis of local membrane properties. J. Comput.-Aided Mol. Des. 2013, 27, 845–858. 10.1007/s10822-013-9684-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Maxwell D. S.; Tirado-Rives J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. 10.1021/ja9621760. [DOI] [Google Scholar]
- Róg T.; Orłowski A.; Llorente A.; Skotland T.; Sylvänne T.; Kauhanen D.; Vattulainen I. Data including GROMACS input files for atomistic molecular dynamics simulations of mixed, asymmetric bilayers including molecular topologies, equilibrated structures, and force field for lipids compatible with OPLS-AA parameters. Data in Brief 2016, 7, 1171–1174. 10.1016/j.dib.2016.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aaqvist J. Comment on “Transferability of Ion Models. J. Phys. Chem. 1994, 98, 8253–8255. 10.1021/j100084a049. [DOI] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Van Der Spoel D.; Lindahl E.; Hess B.; Groenhof G.; Mark A. E.; Berendsen H. J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Pronk S.; Pall S.; Schulz R.; Larsson P.; Bjelkmar P.; Apostolov R.; Shirts M. R.; Smith J. C.; Kasson P. M.; van der Spoel D.; Hess B.; Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–54. 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Miyamoto S.; Kollman P. A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. 10.1002/jcc.540130805. [DOI] [Google Scholar]
- Hess B.; Bekker H.; Berendsen H. J. C.; Fraaije J. G. E. M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. . [DOI] [Google Scholar]
- Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. 10.1063/1.447334. [DOI] [Google Scholar]
- Hoover W. G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A: At., Mol., Opt. Phys. 1985, 31, 1695–1697. 10.1103/PhysRevA.31.1695. [DOI] [PubMed] [Google Scholar]
- Parrinello M.; Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. 10.1063/1.328693. [DOI] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Persson F.; Söderhjelm P.; Halle B. The spatial range of protein hydration. J. Chem. Phys. 2018, 148, 215104. 10.1063/1.5031005. [DOI] [PubMed] [Google Scholar]
- Enkavi G.; Mikkolainen H.; Güngör B.; Ikonen E.; Vattulainen I. Concerted regulation of npc2 binding to endosomal/lysosomal membranes by bis(monoacylglycero)phosphate and sphingomyelin. PLoS Comput. Biol. 2017, 13, e1005831 10.1371/journal.pcbi.1005831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobyov I.; Allen T. W. On the role of anionic lipids in charged protein interactions with membranes. Biochim. Biophys. Acta, Biomembr. 2011, 1808, 1673–1683. 10.1016/j.bbamem.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Wytrwal M.; Bednar J.; Nowakowska M.; Wydro P.; Kepczynski M. Interactions of serum with polyelectrolyte-stabilized liposomes: Cryo-TEM studies. Colloids Surf., B 2014, 120, 152–159. 10.1016/j.colsurfb.2014.02.040. [DOI] [PubMed] [Google Scholar]
- Quemeneur F.; Rinaudo M.; Maret G.; Pepin-Donat B. Decoration of lipid vesicles by polyelectrolytes: mechanism and structure. Soft Matter 2010, 6, 4471–4481. 10.1039/c0sm00154f. [DOI] [Google Scholar]
- Eren T.; Som A.; Rennie J. R.; Nelson C. F.; Urgina Y.; Nusslein K.; Coughlin E. B.; Tew G. N. Antibacterial and Hemolytic Activities of Quaternary Pyridinium Functionalized Polynorbornenes. Macromol. Chem. Phys. 2008, 209, 516–524. 10.1002/macp.200700418. [DOI] [Google Scholar]
- Lopez C. F.; Nielsen S. O.; Klein M. L.; Moore P. B. Hydrogen Bonding Structure and Dynamics of Water at the Dimyristoylphosphatidylcholine Lipid Bilayer Surface from a Molecular Dynamics Simulation. J. Phys. Chem. B 2004, 108, 6603–6610. 10.1021/jp037618q. [DOI] [Google Scholar]
- Ivanov I.; Vemparala S.; Pophristic V.; Kuroda K.; DeGrado W. F.; McCammon J. A.; Klein M. L. Characterization of Nonbiological Antimicrobial Polymers in Aqueous Solution and at Water-Lipid Interfaces from All-Atom Molecular Dynamics. J. Am. Chem. Soc. 2006, 128, 1778–1779. 10.1021/ja0564665. [DOI] [PubMed] [Google Scholar]
- Rauscher S.; Gapsys V.; Gajda M. J.; Zweckstetter M.; de Groot B. L.; Grubmüller H. Structural Ensembles of Intrinsically Disordered Proteins Depend Strongly on Force Field: A Comparison to Experiment. J. Chem. Theory Comput. 2015, 11, 5513–5524. 10.1021/acs.jctc.5b00736. [DOI] [PubMed] [Google Scholar]
- Mecke A.; Lee D. K.; Ramamoorthy A.; Orr B. G.; Banaszak Holl M. M. Synthetic and Natural Polycationic Polymer Nanoparticles Interact Selectively with Fluid-Phase Domains of DMPC Lipid Bilayers. Langmuir 2005, 21, 8588–8590. 10.1021/la051800w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaroslavov A. A.; Efimova A. A.; Lobyshev V. I.; Kabanov V. A. Reversibility of structural rearrangements in the negative vesicular membrane upon electrostatic adsorption/desorption of the polycation. Biochim. Biophys. Acta, Biomembr. 2002, 1560, 14–24. 10.1016/S0005-2736(01)00453-9. [DOI] [PubMed] [Google Scholar]
- Yaroslavov A. A.; Rakhnyanskaya A. A.; Yaroslavova E. G.; Efimova A. A.; Menger F. M. Polyelectrolyte-coated liposomes: Stabilization of the interfacial complexes. Adv. Colloid Interface Sci. 2008, 142, 43–52. 10.1016/j.cis.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Franzin C. M.; Macdonald P. M. Polylysine-Induced 2H NMR-Observable Domains in Phosphatidylserine/Phosphatidylcholine Lipid Bilayers. Biophys. J. 2001, 81, 3346–3362. 10.1016/S0006-3495(01)75968-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkosz N.; Jamróz D.; Kopeć W.; Nakai K.; Yusa S.; Wytrwal-Sarna M.; Bednar J.; Nowakowska M.; Kepczynski M. Effect of Polycation Structure on Interaction with Lipid Membranes. J. Phys. Chem. B 2017, 121, 7318–7326. 10.1021/acs.jpcb.7b05248. [DOI] [PubMed] [Google Scholar]
- Ivashkov O. V.; Sybachin A. V.; Efimova A. A.; Pergushov D. V.; Orlov V. N.; Schmalz H.; Yaroslavov A. A. The Influence of the Chain Length of Polycations on their Complexation with Anionic Liposomes. ChemPhysChem 2015, 16, 2849–2853. 10.1002/cphc.201500474. [DOI] [PubMed] [Google Scholar]
- Mammen M.; Choi S.-K.; Whitesides G. M. Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angew. Chem., Int. Ed. 1998, 37, 2754–2794. . [DOI] [PubMed] [Google Scholar]
- Nickels J. D.; Smith J. C.; Cheng X. Lateral organization, bilayer asymmetry, and inter-leaflet coupling of biological membranes. Chem. Phys. Lipids 2015, 192, 87–99. 10.1016/j.chemphyslip.2015.07.012. [DOI] [PubMed] [Google Scholar]
- Róg T.; Orłowski A.; Llorente A.; Skotland T.; Sylvänne T.; Kauhanen D.; Ekroos K.; Sandvig K.; Vattulainen I. Interdigitation of long-chain sphingomyelin induces coupling of membrane leaflets in a cholesterol dependent manner. Biochim. Biophys. Acta, Biomembr. 2016, 1858, 281–288. 10.1016/j.bbamem.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Perlmutter J. D.; Sachs J. N. Interleaflet Interaction and Asymmetry in Phase Separated Lipid Bilayers: Molecular Dynamics Simulations. J. Am. Chem. Soc. 2011, 133, 6563–6577. 10.1021/ja106626r. [DOI] [PubMed] [Google Scholar]
- Gurtovenko A. A.; Vattulainen I. Membrane Potential and Electrostatics of Phospholipid Bilayers with Asymmetric Transmembrane Distribution of Anionic Lipids. J. Phys. Chem. B 2008, 112, 4629–4634. 10.1021/jp8001993. [DOI] [PubMed] [Google Scholar]
- Róg T.; Orłowski A.; Llorente A.; Skotland T.; Sylvänne T.; Kauhanen D.; Ekroos K.; Sandvig K.; Vattulainen I. Data including GROMACS input files for atomistic molecular dynamics simulations of mixed, asymmetric bilayers including molecular topologies, equilibrated structures, and force field for lipids compatible with OPLS-AA parameters. Data in Brief 2016, 7, 1171–1174. 10.1016/j.dib.2016.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antila H.; Buslaev P.; Favela-Rosales F.; Ferreira T. M.; Gushchin I.; Javanainen M.; Kav B.; Madsen J. J.; Melcr J.; Miettinen M. S.; Määttä J.; Nencini R.; Ollila O. H. S.; Piggot T. J. Headgroup Structure and Cation Binding in Phosphatidylserine Lipid Bilayers. J. Phys. Chem. B 2019, 123, 9066–9079. 10.1021/acs.jpcb.9b06091. [DOI] [PubMed] [Google Scholar]
- Catte A.; Girych M.; Javanainen M.; Loison C.; Melcr J.; Miettinen M. S.; Monticelli L.; Maatta J.; Oganesyan V. S.; Ollila O. H. S.; Tynkkynen J.; Vilove S. Molecular electrometer and binding of cations to phospholipid bilayers. Phys. Chem. Chem. Phys. 2016, 18, 32560–32569. 10.1039/C6CP04883H. [DOI] [PubMed] [Google Scholar]
- Kashefolgheta S.; Verde A. V. Developing force fields when experimental data is sparse: AMBER/GAFF-compatible parameters for inorganic and alkyl oxoanions. Phys. Chem. Chem. Phys. 2017, 19, 20593–20607. 10.1039/C7CP02557B. [DOI] [PubMed] [Google Scholar]
- Leftin A.; Molugu T. R.; Job C.; Beyer K.; Brown M. F. Area per Lipid and Cholesterol Interactions in Membranes from Separated Local-Field 13C NMR Spectroscopy. Biophys. J. 2014, 107, 2274–2286. 10.1016/j.bpj.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.