The polyproline-rich domain of Tau, the core constituent of neurofibrillary tangles, can undergo liquid–liquid phase separation in living cells. Tau PRD condensation drives clustering of Tau on microtubules under the control of phosphorylation and can form a co-condensate with EB1, a regulator of plus-end microtubule dynamic instability.
Abstract
Tau protein in vitro can undergo liquid–liquid phase separation (LLPS); however, observations of this phase transition in living cells are limited. To investigate protein state transitions in living cells, we attached Cry2 to Tau and studied the contribution of each domain that drives the Tau cluster in living cells. Surprisingly, the proline-rich domain (PRD), not the microtubule binding domain (MTBD), drives LLPS and does so under the control of its phosphorylation state. Readily observable, PRD-derived cytoplasmic condensates underwent fusion and fluorescence recovery after photobleaching consistent with the PRD LLPS in vitro. Simulations demonstrated that the charge properties of the PRD predicted phase separation. Tau PRD formed heterotypic condensates with EB1, a regulator of plus-end microtubule dynamic instability. The specific domain properties of the MTBD and PRD serve distinct but mutually complementary roles that use LLPS in a cellular context to implement emergent functionalities that scale their relationship from binding α-beta tubulin heterodimers to the larger proportions of microtubules.
Introduction
Many proteins have domain structures in which a contiguous amino acid sequence can operationally serve a discrete function but, in the context of the entire protein, become regulated within a larger sequence space. The domain structure of Tau is well recognized, but the functions of these domains, and particularly the internal control they exert over Tau function, are poorly understood. Tau protein undergoes a liquid-to-solid phase transition in numerous neurodegenerative conditions, most prominently Alzheimer’s disease, frontotemporal dementia, and chronic traumatic encephalopathy, as well as more rare conditions such as Pick’s disease, progressive supranuclear palsy, corticobasal degeneration, Neiman–Pick type 3, and subacute sclerosing encephalitis, among others (Wang and Mandelkow, 2016). In the past few years, liquid–liquid phase-separated states, called condensates, in cells have become associated with several proteins that accumulate in neurodegeneration, including FUS (Maharana et al., 2018; Patel et al., 2015), TDP-43 (Gasset-Rosa et al., 2019; Mann et al., 2019), HNRNPA1 (Gui et al., 2019; Lin et al., 2015), and DDX (Nott et al., 2015), as well as Tau (Zhang et al., 2017). An important feature of condensates is the very high concentration of protein within a small volume compared with the solute phase, while the protein constituents of these compartments are hydrated and exchange freely. The thermodynamics of liquid–liquid phase separation (LLPS) of biopolymers from solution to a condensed state has been studied extensively using polymer physics models and tools (Brangwynne et al., 2015; Lin et al., 2019), but the correlates of these events in living cells are mostly unknown. LLPS in vitro can be well described as simple or complex coacervates; however, condensates in living cells involve multiple diverse components whose binding affinities and concentration are dynamically regulated by an “in vivo phase diagram” with a staggering complexity of phases and intrinsic variables that control the transitions among them (Riback et al., 2020). While in vitro studies can prove the sufficiency of molecules to undergo LLPS, they strip the system of its inherent complexity. A complementary technique for grasping the function of phase transitions and how they are regulated in vivo is to make acute changes in the valency of a core scaffolding protein by fusing it to a photo-sensitive oligomerization domain, thereby gaining the ability to abruptly alter the location in phase space within the cytoplasm.
One way to achieve this light-mediated change in protein valency is to use the optogenetic protein Cry2, the photolyase homology region of a cryptochrome from Arabidopsis thaliana, that undergoes light-sensitive self-association upon blue light exposure (Taslimi et al., 2014). Cry2WT-mCherry (Cry2 wild type fused to mCherry) by itself will not undergo LLPS in heterologous cells, but when fused to the intrinsically disordered region (IDR) of FUS, HNRNPA1, or DDX4, these proteins can drive droplet formation (Shin et al., 2017). LLPS of these proteins likely involves additional cellular factors that lower the free energy for LLPS that the blue light–activated Cry2 mimics. Tau is an intrinsically disordered protein, binds RNA with multivalent interactions and, like the IDR of many RNA-binding proteins (Elbaum-Garfinkle et al., 2015; Lin et al., 2015; Molliex et al., 2015; Patel et al., 2015), can undergo phase separation in vitro (Zhang et al., 2017). Here we demonstrate the potential of Tau for LLPS in living cells and identify the Tau proline-rich domain (PRD) as the regulator of condensate formation.
Results
Tau condensation facilitates microtubule (MT) binding
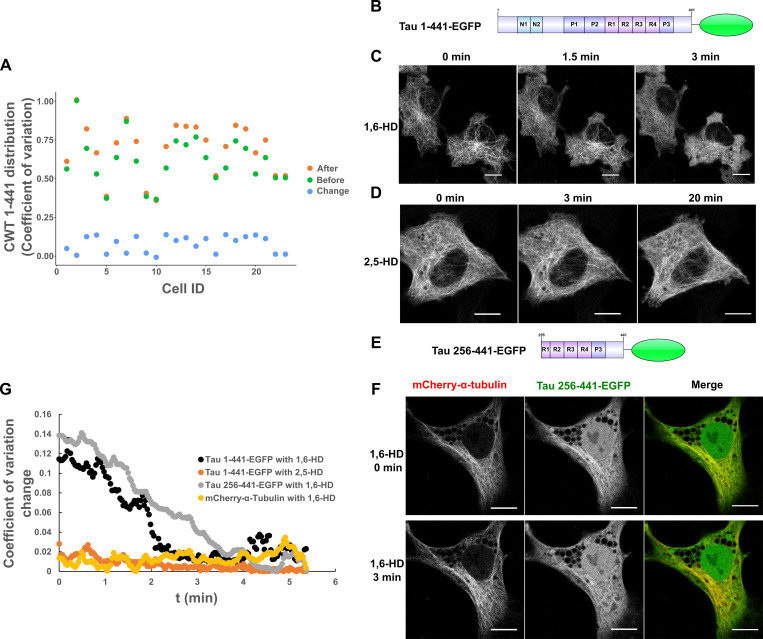
We cloned a full-length Tau 2N4R construct fused to Cry2WT-mCherry (CWT 1–441) and, in the absence of blue-light activation, we observed the expected MT pattern and some diffuse signal in the cytoplasm. With Cry2 activation by blue light, the signal from the diffuse pool was markedly reduced, and the MT signal increased (Fig. 1, A–C). The increased Tau binding to MT was quantified by computing the pixel coefficient of variation (CV) across the cell, with the lower value indicating a more uniform distribution and a higher value indicating discrete localization (Fig. S1 A). This result suggested that Tau condensate enhanced its binding to MTs. To support this observation, we expressed Tau 1–441-EGFP (Fig. S1 B) in HeLa cells and treated cells with 1,6-hexanediol (1,6-HD) which interferes with weak hydrophobic protein–protein or protein–RNA interactions that can promote the formation of dynamic, liquid-like assemblies (Kroschwald et al., 2017). 3 min after 1,6-HD treatment, the EGFP signal appeared diffuse in the cytoplasm (Fig. S1 C). The control, 2,5-HD, had no effect even after 20 min (Fig. S1 D). MTs were unaffected by these treatments as indicated by 1,6-HD treatment of cells labeled with mCherry-α-tubulin (Fig. S1, E and F). Similar results were observed with Tau 256–441-EGFP (Fig. S1, E–G).
Figure 1.
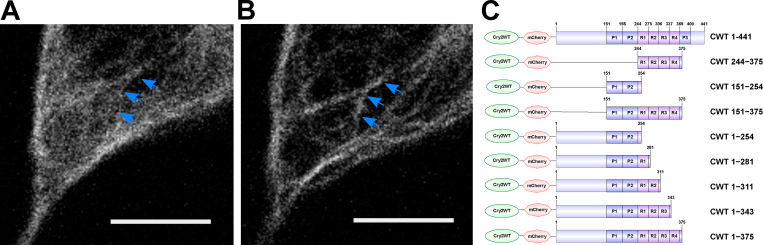
Tau condensation mediates binding to MTs. (A and B) Representative Z-stack images of Cry2WT-mCherry-Tau 2N4R 1–441 (CWT 1–441) overexpressed in neuroblastoma SH SY5Y cell before (A) and 50 s after activation (B). Signal from the diffuse pool was reduced and the MT signal (blue arrows) increased. Scale bar, 5 µm. (C) Schematic diagram of Cry2WT-mCherry-Tau constructs studied, the Cry2WT fused to mCherry, and various Tau fragments.
Figure S1.
Quantification of Cry2WT-mCherry-Tau (CWT 1–441) distribution in SH SY5Y before and after blue-light activation and Tau dissociates from MTs after 1,6-HD treatment in HeLa cells. (A) Cry2WT-mCherry-Tau (CWT 1–441) distribution in SH SY5Y before activation (green dots), after activation (orange dots), and change by activation (blue dots) were quantified using pixel CV (y axis) for each cell (x axis). The CV of the Tau signal after blue-light activation is significantly higher than before, n = 23, P = 7.0 × 10−7, paired t test. (B–D) Full-length Tau 1–441-EGFP is sensitive to 1,6-HD but not to 2,5-HD. (B) Protein schematic diagram of Tau 1–441-EGFP. (C) Tau 1–441-EGFP expressed in HeLa cells was treated with 1,6-HD, which resulted in the loss of a MT-binding pattern to a diffuse pattern. Selected images at 0 min, 1.5 min, and 3 min are presented. (D) Tau 1–441-EGFP was treated with 2,5-HD in HeLa cell; selected images at 0 min, 3 min, and extended time to 20 min are presented. Tau binds to MT even after extended treatment of 2,5-HD. (E and F) Tau truncations containing MTBD and the extreme C terminus remain sensitive to 1,6-HD treatment, and MT was not affected. (E) Protein schematic diagram of Tau 256–441-EGFP. (F) Tau 256–441-EGFP (green) and mCherry-α-tubulin (red) were overexpressed in HeLa cells, treated with 1,6-HD. Montage of selected images at 0 min and 3 min treatment are presented. From left to right, mCherry-α-tubulin, Tau 256–441-EGFP, and the merge. While truncated Tau became diffuse after 3 min, mCherry-α-tubulin still maintained the MT pattern. Note that there is a nuclear population of EGFP signal for Tau 256–441-EGFP overexpression, which is commonly seen for EGFP fused with proteins of low molecular weight. (G) Quantification of Tau distribution upon 1,6-HD or 2,5-HD treatment based upon CV. Distribution of Tau 1–441-EGFP treated with 1,6-HD (black), Tau 1–441-EGFP treated with 2,5-HD (orange), Tau 256–441-EGFP treated with 1,6-HD (gray), and mCherry-a-tubulin treated with 1,6-HD (yellow) was evaluated and plotted over the time. ID, identification; t, time.
The Tau PRD mediates Tau condensation in cells
With the finding that Tau phase separation as a condensate can enhance its binding to MTs, we sought to determine which domains of Tau drive condensation-assisted MT binding and began by analyzing them individually. The Tau domains consist of the N-terminal domain, the PRD subdivided as P1 and P2 with ∼25% prolines, the MT-binding domain (MTBD) with four repeat regions, and the C-terminal domain from amino acid 369 to 441, which includes a short P3 region flanking the MTBD (Fig. 1 C; Gustke et al., 1994; Mukrasch et al., 2005). Although the MTBD can undergo LLPS in vitro (Ambadipudi et al., 2017; Zhang et al., 2017), when this domain was fused to Cry2WT-mCherry (designated CWT 244–375) and transfected into SH SY5Y cells, blue light did not induce the cytoplasmic condensates characteristic of LLPS. In contrast, the PRD domain fused to Cry2WT-mCherry (designated CWT 151–254) expressed in SH SY5Y underwent condensation within 5 s of blue-light activation (Fig. 2, A and B). Cry2WT-mCherry alone expressed in SH SY5Y cells at a comparable concentration did not cluster following blue-light activation (Fig. 2 B; Lee et al., 2014; Taslimi et al., 2014). The extent of light-activated condensation of PRD was quantified by computing the pixel CV, which evaluates the overall distribution across the cell (Fig. 2 B).
Figure 2.
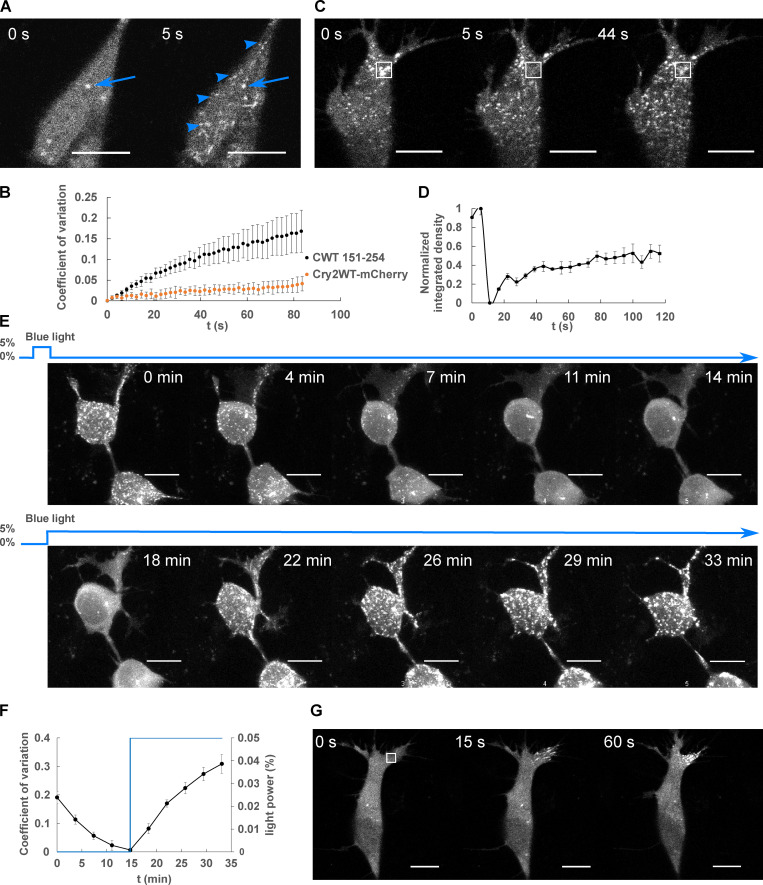
LLPS of Cry2WT-mCherry-Tau 151–254 (CWT 151–254). (A) CWT 151–254 overexpressed in SH SY5Y cells activated by shallow blue light (488 nm, 5% power) rapidly induced condensates (blue arrowheads). Representative fluorescent images at 0 and 5 s after activation are shown. Occasional preactivation foci are sometimes present and do not participate in light-activated phase separation (blue arrow). (B) Temporal evolution of phase separation of CWT 151–254 (black points) and Cry2WT-mCherry (orange points) was monitored on a single confocal plane, quantified by CV across the cell and plotted over time. n = 7 for CWT 151–254 and n = 5 for Cry2WT-mCherry; error bar in SE. (C and D) FRAP of CWT 151–254. (C) Representative time-lapse images of FRAP indicated by the 3 × 3-µm bleach site is marked by the white square. See Video 1. (D) Integrated intensity density for the bleached areas was monitored over time during FRAP. 55% recovery was achieved after bleaching. n = 3. (E and F) CWT 151–254 phase separation is reversible. (E) Time-lapse images of maximum intensity Z-projection of CWT 151–254 show phase separation is reversible after the withdrawal of blue light (0–14 min) and reassembled when subject to blue light reactivation (14–33 min). (F) Quantification of CWT 151–254 from disassembly to reassembly as light power changes. CWT 151–254 distribution quantified by CV (black) and light activation indicated by the blue light power (blue) were plotted over time. n = 8; error bar in SE. (G) Time-lapse images of local shallow light activation of CWT 151–254 demonstrate localized progression toward phase separation. A rectangular area of 3 × 3-µm (white square) was stimulated with blue light. See also Video 2 for local activation and the field activation followed. Scale bar, 10 µm. t, time.
To probe the bulk exchange dynamics of light-induced CWT 151–254 condensates, we performed FRAP by bleaching the mCherry signal (Fig. 2, C and D; and Video 1). CWT 151–254 can exhibit up to 55 ± 15% (n = 3) fluorescence recovery. Droplets formed by blue-light activation of CWT 151–254 disassembled within a few minutes in the absence of blue light, then reassembled when exposed again to blue light (Fig. 2, E and F), indicating that droplet formation of this construct is initially reversible. Local activation of CWT 151–254 by directing the light to a 3 × 3-µM square demonstrated that phase separation can be spatially localized (Fig. 2 G and Video 2 for local field activation). As expected for light-activated phase separation, Tau condensation likely involves weak multivalent interactions that can be strengthened and aided in their detection by Cry2 (Bracha et al., 2019). Taken together, these data show that the PRD can undergo light-induced condensation independent of the MTBD.
Video 1.
FRAP of Cry2WT-mCherry-Tau 151–254 (CWT 151–254) in SH SY5Y cell. This video shows the selected area (white rectangle) of CWT 151–254 overexpressed in SH SY5Y cell was bleached by UV light and followed by fluorescence recovery.
Video 2.
Local and whole-field activation of Cry2WT-mCherry-Tau 151–254 (CWT 151–254) in SH SY5Y cell. CWT 151–254 overexpressed in SH SY5Y cells first formed local phase separation after being activated at the selected area (white rectangle) by blue light. Cell-wide phase separation was then observed when the whole viewing field was subjected to blue light.
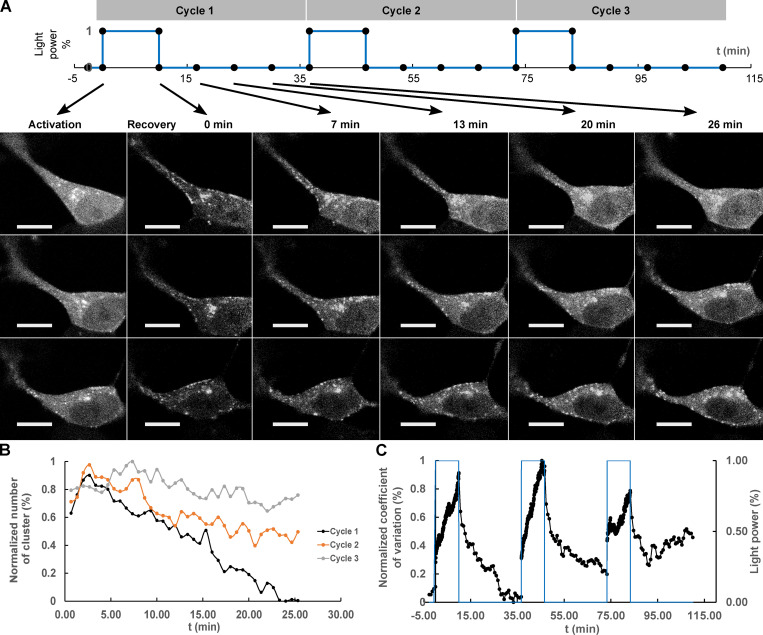
We sought to determine the reversibility of Tau PRD condensates over longer time periods by cycling through multiple illumination periods. A sequence of blue light exposure followed by a recovery phase with three activation cycles was performed (Fig. 3 A; Shin et al., 2017). After the first cycle, the condensates were mostly dissolved, but residual condensates increased progressively after each cycle. The result was quantified by counting the condensates and normalizing the result (see Materials and methods). The percentage of condensates that remained was 0%, 42%, and 73% after the first, second, and third cycle, respectively (Fig. 3 B). The kinetics of cluster assembly and disassembly were also analyzed by pixel CV (Fig. 3 C). As expected, the values increased during assembly and decreased during disassembly. Through the recovery, the rate of the disassembly gradually slowed and eventually reached a plateau. Upon each cycle, the recovery plateaued progressively earlier and further from the initial state. These findings resemble an “aging” process in which the liquid state transitions to a nonexchangeable gel or a solid phase (Shin et al., 2017).
Figure 3.
Repeated deep blue-light activation reduced recovery of Cry2WT-mCherry-Tau 151–254 (CWT 151–254) biocondensates. (A) Top: A sequence of three cycles of deep blue-light activation, 488 nm, 100% power for 10 min, each followed by 26 min of recovery time applied to SH SY5Y cells, in which CWT 151–254 was overexpressed. Bottom: Time-lapse of single Z-stack images shows CWT 151–254 biocondensate assembly upon light activation and disassembly when light was removed following the sequence on the top. For each cycle, the image at initial activation and recovery at 0, 7, 13, 20, and 26 min are shown. (B) The number of the clusters post-activation was counted and plotted. (C) Redistribution of CWT 151–254 through the time course is quantified by CV and change in light power over the time course. Scale bar, 10 µm. t, time.
The PRD is the most heavily phosphorylated region in Tau, and its phosphorylation is controlled by numerous kinases in diverse signaling pathways (Martin et al., 2013). Phosphorylation in this region will modulate the charge distribution and affect the weak electrostatic interactions that mediate condensation. Placing phosphomimetic mutations within the PRD completely abolished blue light–induced phase separation of CWT (Table 1). Mono-, di-, or triple-phosphomimetic mutations at T231E or S235E, T231E/S235E or T212E/S214E/T217E, or S202D/T205D/S208D all abolished light-induced phase separation. In contrast, LLPS of CWT 151–254 with alanine mutations at the same phosphorylation sites was not abolished. Upon light activation, CWT 151–254 S202A/T205A/S208A and CWT 151–254 S202A/T205A/S208A/T212A/S214A still phase separated.
Table 1. Phosphomimetics in PRD as the regulation of condensationa.
| Construct | LLPS |
|---|---|
| CWT 151-254 | Yes |
| CWT 151-254 T231E | No |
| CWT 151-254 S235E | No |
| CWT 151-254 T231E/S235E | No |
| CWT 151-254 T212E/S214E/T217E | No |
| CWT 151-254 S202D/T205D/S208D | No |
| CWT 151-254 S202A/T205A/S208A | Yes |
| CWT 151-254 S202A/T205A/S208A/T212A/S214A | Yes |
All experiments were performed at the same expression levels and at constant light levels.
PDR condensates promote Tau association with MTs
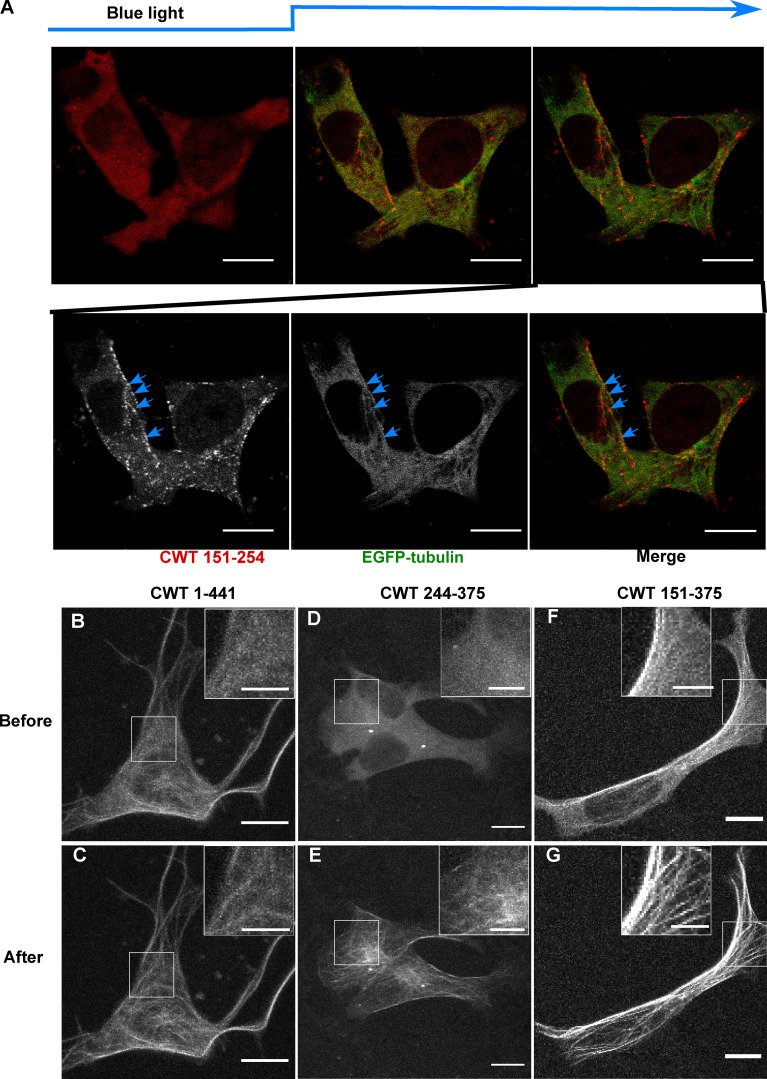
Having characterized the isolated domain behavior of the PRD, we next sought to determine its relationship to the MT and its behavior in the context of the neighboring MTBD. Light-induced PRD condensates (CWT 151–254) were frequently observed to align along MTs (Fig. 2 A and Fig. 4 A). The MTBD on its own (CWT 244–375) and transfected into SH SY5Y cells did not bind MTs nearly as well as full-length Tau (Fig. 4, B–E). In contrast, CWT 151–375 (Fig. 4, F and G), which contains both the PRD and the MTBD, bound to MTs similarly to full-length Tau (CWT 1-441) before activation and increased its binding further after light activation, also similar to full-length Tau (Fig. 4, B and C).
Figure 4.
Cry2WT-mCherry-Tau fragments associate with MT. (A) Association of PRD condensates with MTs. EGFP-α-tubulin (green) and CWT 151–254 (red) were coexpressed in neuroblastoma SH SY5Y cells. Time-lapse images show CWT 151–254 forms LLPS upon blue-light activation starting from the second time frame. Montage of two fluorescent channels in the third time frame demonstrates CWT 151–254 foci are attached to EGFP-α-tubulin on MT, like beads on a thread (blue arrow). (B–G) Cry2WT-mCherry fused to MTBD without and with PRD were overexpressed in SH SY5Y cells, and signal distribution was studied before and after blue light. Representative maximum intensity Z-projection images before and after blue light of each construct are shown, CWT 1–441 before (B) and after (C), CWT 244–375 before (D) and after (E), CWT 151–375 before (F) and after (G). Insets (scale bar, 5 µm) within the white rectangle show more details of the diffuse pool and the MT-binding population in each construct. Scale bar unless specifically mentioned, 10 µm.
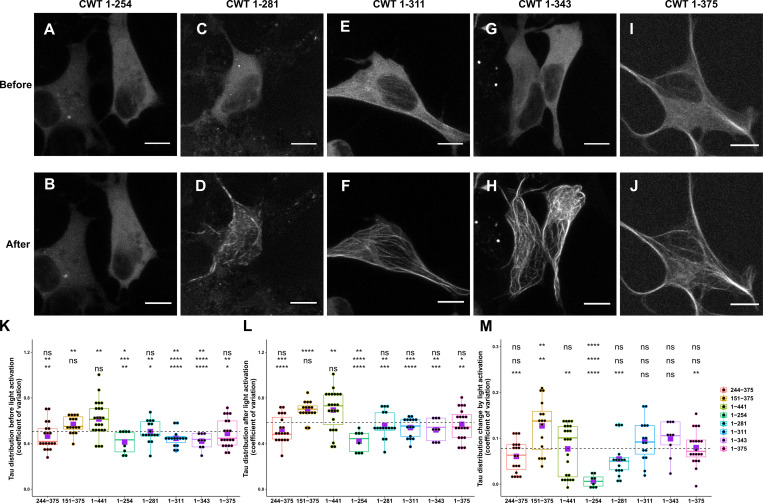
Inclusion of the N-terminal domain with the PRD (CWT 1–254) had an inhibitory effect over Tau condensation. Unlike the PRD domain alone, this construct showed only minimal association with MTs both before and after light activation (Fig. S2, A and B). This result suggested that the N-terminal 1–150 region negatively regulates PRD phase separation, likely owing to its many negatively charged residues, and is consistent with the negative regulation of the N-terminal region on PRD binding to soluble tubulin and slowing polymerization (McKibben and Rhoades, 2019) and previous work showing that removal of the N-terminal region increased the affinity of Tau for MTs (Gustke et al., 1994).
Figure S2.
Comparison of Cry2WT-mCherry-Tau distribution before and after light activation for various Tau constructs. (A–J) Cry2WT-mCherry fused various Tau constructs was overexpressed in SH SY5Y cells. Signal distribution was studied before and after blue light. Representative maximum intensity Z-projection images before and after blue light of each construct are shown, CWT 1–254 before (A) and after (B), CWT 1–281 before (C) and after (D), CWT 1–311 before (E) and after (F), CWT 1–343 before (G) and after (H), and CWT 1–375 before (I) and after (J). Scale bar, 10 µm. (K–M) Quantifications of Cry2WT-mCherry-Tau distribution before activation (K), after activation (L), and change by activation (M) are shown in boxplot. Constructs from left to right along the x axis are CWT 244–375, CWT 151–375, CWT 1–441, CWT 1–254, CWT 1–281, CWT 1–311, CWT 1–343, and CWT 1–375. Distribution of Cry2WT-mCherry-Tau is evaluated by pixel CV over the cell shown along the y axis. n = 19, 16, 23, 8, 16, 15, 9, and 20 for constructs from left to right along the x axis. Mean for each group is indicated by a purple square, also listed in Table S1. Base-mean for all the constructs as a whole is indicated as a horizontal dotted line. Statistical significance by pairwise t test of each independent group is indicated at the top of each plot, from top to bottom, against all the constructs as a whole, against CWT 1–441, and against CWT 151–375, correspondingly. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
As additional MTBDs were sequentially added to the CWT 1–254 construct, MT binding increased, and the diffuse labeling decreased (Fig. 1 C; Fig. S2, C–M; Table S1; and Materials and methods). A construct from the N terminus that included all of the MT-binding repeats (CWT 1–375) further increased association with MTs after blue light, but was still not maximized until we used a full-length construct that included the P3 (376–400) domain (Fig. S2, I and J). The P3 domain flanks the MTBD and also undergoes proline-directed phosphorylation (Mukrasch et al., 2005). These data demonstrate that condensation of the PRD alone can drive association with MTs and that association is inhibited by the N-terminal domain but enhanced by the MTBD as well as the C-terminal domain.
The Tau PRD phase separates with end-binding protein (EB) 1
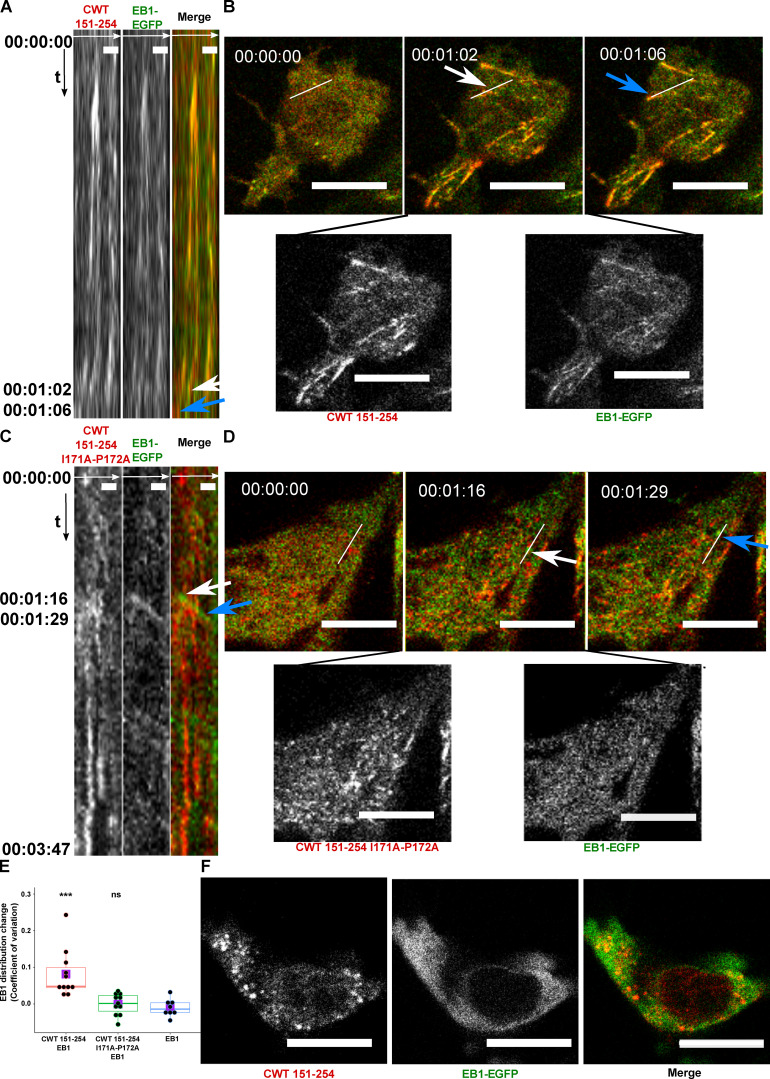
Plus-end–tracking proteins (+TIPs) form a complex interaction network at MT plus ends, where they control MT dynamics and MT anchorage to subcellular targets such as vesicles and kinetochores (Galjart, 2010). EB1 is a major integrator of the +TIP complex that appears as “comets” at MT ends (Akhmanova and Steinmetz, 2010). Tau forms a complex with EB1 through an interaction between the MT-binding repeats and the P2 region of the PRD interacting with the acidic C-terminal tail of EB1 (Ramirez-Rios et al., 2016). The observation that CWT 151–254 moved along MTs as a comet-shaped structure during the initial 30 s (Fig. 2 G), before it formed a condensate, prompted us to study the interaction of CWT 151–254 with EB1. We coexpressed CWT 151–254 and EB1-EGFP in SH SY5Y cells activated by blue light. Kymograph tracking the fluorescent “comets” (Mimori-Kiyosue et al., 2000) of EB1-EGFP showed spatiotemporal colocalization with CWT 151–254 after blue-light activation in which light activation first induced comet formation that rapidly extended along MTs (Fig. 5, A and B; and Video 3).
Figure 5.
Cry2WT-mCherry-Tau 151–254 (CWT 151–254) tracks with EB1 after blue-light activation. (A and B) CWT 151–254 overexpressed with EB1-EGFP in SH SY5Y cells and activated by shallow blue light (488 nm, 5% power) induced colocalization. (A) Kymograph of CWT 151–254 (red) and EB1-EGFP (green) is generated from time-lapse imaging for 66 s at 2-s imaging intervals, with the x axis the selected line crossing the image and the y axis the time of recording. Fluorescence imaging of two channels shows the formed CWT 151–254 (red) condensate colocalized with moving EB1-EGFP (green). Scale bar, 2 µm. (B) Representative time-lapse images at the times of 00:00:00, 00:01:02, and 00:01:06 are shown in the top panel, where the line used to generate the kymograph is shown in white. The kymograph in A moves from left to right. Selected moving foci on the line are indicated with white or blue arrows on both the kymograph and the corresponding time-lapse images. Two fluorescent channels of the middle image at 00:01:02, CWT 151–254 (red, left), EB1-EGFP (green, right), are presented at the bottom panel. See also Video 3. (C and D) CWT 151–254 with I171A and P172A double mutant and EB1-EGFP overexpressed in SH SY5Y cells activated by shallow blue light (488 nm, 5% power). The induced double mutant CWT 151–254 (red) condensate does not colocalize with EB1-EGFP. (C) Kymograph of CWT 151–254 with I171A and P172A double mutant (red) and EB1-EGFP (green) generated from a time-lapse imaging for 3 min and 47 s at 2-s imaging intervals, with the x axis being the selected line crossing the image and the y axis the time of recording. Fluorescence imaging of two channels shows the formed CWT 151–254 with I171A and P172A double mutant (red) condensate does not colocalize with moving EB1-EGFP (green). Scale bar, 2 µm. (D) Representative time-lapse images at the times of 00:00:00, 00:01:16, and 00:01:29 are shown in the top panel, where the line used to generate the kymograph is shown in white. Selected moving foci on the line are indicated with white or blue arrows on both the kymograph and the corresponding time-lapse images. Two fluorescent channels the middle image at 00:01:16, CWT 151–254 I171A and P172A (red, left), EB1-EGFP (green, right), are presented in the bottom panel. (E) Quantification of EB1-EGFP distribution after light activation. EB1 was expressed in SH SY5Y cells with CWT 151–254, or with CWT 151–254 I171A/P172A double mutant, or with no co-overexpression. Distribution of EB1 is evaluated by the change in the CV after 60 s of light activation shown along the y axis. n = 11, 11, and 8 for the groups from left to right along the x axis. Mean for each group is indicated by a purple square. Statistical significance by pairwise t test of each independent group against EB1 without CWT 151–254 is indicated at the top of the plot. (F) CWT 151–254 (red) and EB1-EGFP (green) overexpressed in SH SY5Y cell, treated with 20 µM nocodazole for 2 h, activated by blue light. EB1-EGFP appeared diffuse in the cytoplasm and not colocalized with the CWT 151–254 light-induced spherical condensates. Representative montage image is shown. See also Video 4. Montage of two fluorescent channels is presented from the left, the mCherry channel, the GFP channel separate and the merged. Scale bar, 10 µm unless specifically mentioned. ns, not significant. ***, P < 0.001.
Video 3.
Cry2WT-mCherry-Tau 151–254 (CWT 151–254, red) and EB1-EGFP (green) overexpressed in SH SY5Y cell activated by shallow blue light (488 nm, 5% power). The induced CWT 151–254 (red) condensate colocalizes with moving EB1-EGFP.
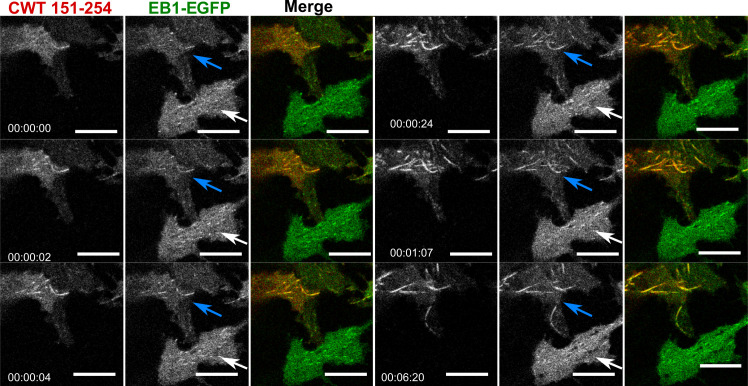
We could not employ FRAP for these condensates because they moved too rapidly, but EB1 comets dissolved following 1,6 hexanediol treatment. Colocalized CWT 151–254/EB1 was also observed along the MT lattice (Fig. S3) similar to EB1 when overexpressed alone (Skube et al., 2010), suggesting activated CWT 151–254 helped to concentrate EB1 on MT. Quantification of EB1 as diffuse signal and bundles (the remainder being puncta) shows that EB1-GFP distribution is significantly different in the presence of CWT 151–254. Over time, bundles were enhanced upon light activation of CWT 151–254, while the diffuse signal did not change in the absence or presence of CWT 151–254 (Table S2). This result suggests that CWT 151–254 enhances clustering on MT without varying the ratio of EB1 on or off MTs. The Tau PRD contains one S/TxIP (169TRIP172), the consensus motif for EB1 binding (Honnappa et al., 2009). A double mutation in this consensus-binding motif of Tau (I171A and P172A) abolished colocalization with EB1-EGFP (Fig. 5, C and D). Quantification of EB1-EGFP distribution following light activation confirmed the presence of this complex (Fig. 5 E).
Figure S3.
Cry2WT-mCherry-Tau 151–254 (CWT 151–254)/EB1-EGFP bound along the MT lattice of MT bundles upon light activation. Time-lapse of EB1-EGFP (green) when overexpressed with Cry2WT-mCherry-Tau 151–254 (CWT 151–254, red) in SH SY5Y cells activated by shallow blue light (488 nm, 5% power). Note that besides binding MT +TIPs and moving as comets, upon light activation, some CWT 151–254/EB1-EGFP extended the binding along the MT lattice and MT bundles (following the blue arrows in the time-lapse) while the cell at the bottom, which also has expression of EB1-EGFP but no CWT 151–254, does not show shift of EB1 localization from MT +TIPS, remaining as a comet (white arrows). Montage of two fluorescent channels is presented in the order from left, the mCherry channel, the GFP channel separate, and merged. Scale bar, 10 µm.
The EB1-EGFP/CWT 151–254 interaction was disrupted by the same phosphorylation sites that disrupted CWT 151–254 alone. Multiple mono-, di-, or triple phosphomimetic mutations, T231E or S235E, or T231E/S235E or T212E/S214E/T217E, or S202D/T205D/S208D, all completely abolished CWT 151–CWT 151254 comets (Table 1), whereas alanine mutations at the same phosphorylation sites did not abolish comets.
The association of the CWT 151–254/EB1 complex with MTs prompted us to examine the colocalization of the complex following treatment with nocodazole (Fig. 5 F and Video 4). Under these conditions, the EB1 signal became diffuse, and CWT 151–254 formed spherical condensates no longer colocalized with EB1. Thus, CWT 151–254/EB1 complexes were dependent upon the presence of polymerized MTs.
Video 4.
Cry2WT-mCherry-Tau 151–254 (CWT 151–254, red) and EB1-EGFP (green) overexpressed in SH SY5Y cells, treated with 20 µM nocodazole for 2 h. Activation by blue light induces spherical condensates of CWT 151–254, while EB1-EGFP remains diffused in the cytoplasm. No colocalization of CWT 151–254 and EB1-EGFP is observed.
In vitro Tau PRD phase separation
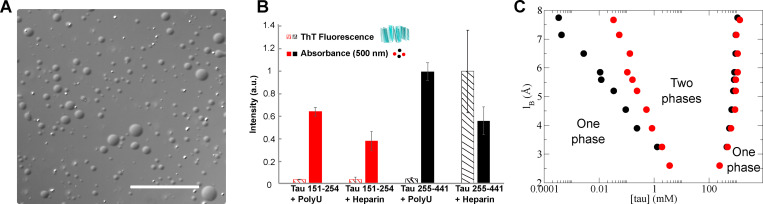
The distinct properties of the PRD and the MTBD when fused to Cry2 prompted further investigation of these constructs in vitro. This comparison was especially important because we previously showed that an MTBD construct, Tau 255–441, can readily undergo phase separation when using RNA or heparin as a cofactor (Lin et al., 2020; Zhang et al., 2017) This construct included the sequence observed in the insoluble Tau fibril by cryoelectron microscopy (Fitzpatrick et al., 2017). Tau 151–254 protein with a 6xHIS tag at the N terminus was purified. This protein was mixed with the cofactor in a 1:1 charge ratio. The occurrence of phase separation was evaluated by measuring turbidity via absorbance at 500 nm, and fibril formation measured by fluorescence of thioflavin T (ThT), which binds to β-sheets of stacked Tau proteins. Directly after addition of polyuridylic acid (PolyU) to Tau 151–254, droplets formed (Fig. 6 A) that were capable of fusing and deforming as they wet the glass surface. Tau 255–441 mixed with the heparin cofactor not only formed droplets that fused and deformed as they wet the glass surface, but also induced significant ThT fluorescence (Lin et al., 2020). In contrast to Tau 255–441, Tau 151–254 did not show any ThT fluorescence (Fig. 6 B). Transmission electron microscopy imaging confirmed that no fibrils were formed by Tau 151–254. Although both Tau 151–254 and Tau 255–441 readily formed droplets in vitro with the addition of the negatively charged cofactors, PolyU or heparin (Fig. 6 A), condensation was insufficient for the PRD to make fibrils. This distinction may be related to energetics involved in the difference between Tau 255–441 and Tau 151–254 to form fibrils.
Figure 6.
Similar to Tau 255–441, Tau 151–254 can form droplets in vitro. (A) Representative bright-field images of Tau 151–254/polyU droplets. Scale bar is 50 µm. (B) Direct comparison of Tau 151–254 (red) and Tau 255–441(black) shows phase separation occurs for both protein segments with addition of PolyU or heparin. In contrast to the Tau 255–441 heparin complex, Tau 151–254 does not show the capacity to form fibrils with either PolyU or heparin. Turbidity was evaluated at 500 nm (filled bar); fibrilization was evaluated by ThT fluorescence (striped bar). Protein concentration was 50 µM, PolyU concentration (where applicable) was 125 µg/ml, and heparin concentration was 12.5 µM. All data were normalized by the largest measured value or absorbance or fluorescence. n = 3; error bar in SD. (C) Predicted Tau 151–254/RNA binodal phase coexistence points modeled using FTS. Calculations performed for Tau 151–254 (red dots) are compared with our previously reported (Lin et al., 2019) phase diagram for Tau 255–441 (black dots), showing that both sequences have a stable two-phase region under similar electrostatic environments and solvent conditions. The conditions for stable binodal phase coexistence depend on the strength of the electrostatic interaction, characterized in our model by the lB as the input parameter. The region defined by the coexistence points is the region at which the system forms a thermodynamically stable droplet phase in coexistence with a solution phase. Areas corresponding to one-phase or two-phase are shown in the diagram.
To test if the charge patterning of the PRD (Tau 151–254) leads to a similar charge-driven phase separation by the complex coacervation mechanism (Zhang et al., 2017) as compared with the MTBD (Tau 255–441), we computed a phase diagram for the LLPS of a polymer physics model of Tau 151–254 using a numerical method called field theoretical simulation (FTS; Lin et al., 2019). The details of the FTS calculations can be found in the Method of FTS section. In our computational model, the protein is described as a bead-spring polymer with a charge sequence determined by the primary amino acid sequence of Tau at pH 7.4 in the presence of RNA (Fig. 6 C).
The phase diagram depends on the strength of the electrostatic interaction, characterized in our model by the Bjerrum length (lB) as the input parameter which controls the strength of the Coulombic interaction according to the pair potential energy where σiand σj are the charges on residue i and j at pH 7.4, and r is the distance between the two residues. All simulations were performed in implicit solvent with statistical segments interacting through a weak excluded volume potential. Binodal points are plotted as a function of Tau concentration versus the lB that characterizes the extent of electrostatic screening.
At low lB, the electrostatic strength in the model is not sufficient for phase separation, but at lB > 2.5 Å, our models of both Tau 151–254 and Tau 255–441 have a region where phase separation occurs (Fig. 6 C). The left points show the lower concentration limit for a stable droplet in solution, and the right points show the upper concentration window. We found that Tau 255–441 and Tau 151–254 have a similar propensity to form droplets, as manifest by a similar phase diagram (Fig. 6 C). As a negative control, both the N-terminal region of Tau (Tau 1–150) and the C-terminal region of Tau (376–441) have been modeled and found not to show any stable two-phase window at these values of lB. Thus, the biophysical properties of the Tau PRD and MTBD are similar in terms of their ability to phase-separate; however, their differences are revealed in the biological context of a living cell.
Discussion
The discovery that Tau is capable of LLPS in vitro (Ambadipudi et al., 2017; Hernández-Vega et al., 2017; Siahaan et al., 2019; Tan et al., 2019; Wegmann et al., 2018; Zhang et al., 2017) raised the question of whether similar phase states of Tau are present in living cells. Although both the full-length Tau and MTBD are capable of phase separation in vitro (Fichou et al., 2018; Lin et al., 2019), when expressed in cells, these constructs did not form LLPS droplets under any of a variety of conditions. We amplified the weak multivalent interactions of Tau in cells by fusing it to a light-activatable self-associating Cry2WT motif while ascertaining that Cry2WT alone did not phase-separate under the same conditions. The fusion of “sticky” IDRs to the photolyase homology region of A. thaliana Cry2 (Bugaj et al., 2013) has been used to demonstrate condensation of aggregation domains in RNA-binding proteins (Shin et al., 2017). For example, the IDR of FUS fused to Cry2WT as well as HNRNPA1 or the N-terminal IDR of DDX4 (Shin et al., 2017); an IDR–Cry2 fusion protein can confer tunable light dependence to its multivalent interactions (Shin et al., 2017). In our hands, this approach identified the unique function of the Tau PRD (151–254) in driving condensation under a variety of experimental conditions. Focused blue light at specific subcellular locations can locally induce CWT 151–254 phase separation (Fig. 2 G), which undergoes FRAP (Fig. 2, C and D). The multivalent interactions of these condensates were controlled by the Tau phosphorylation state (Table 1); therefore, Tau condensates could be tuned by the complex phosphorylation patterns of Tau. The majority of the >40 known Tau phosphorylation sites are located in the PRD (Gong et al., 2005) and are mediated by kinases that participate in multiple signaling pathways.
Our in vitro and in silica data support that the PRD can form droplets with the addition of negatively charged cofactors, similar to the MTBD. Interestingly, the experimental and theoretical biophysics show that the LLPS propensities of MTBD and PRD are indistinguishable; therefore, their difference lies in their biological properties. Among these properties is complex multiple phosphorylation of the PRD, which results in vastly more charge variations to work in concert with the MTBD.
Considerable interest has mounted in the detailed relationship of Tau to MTs. Cryoelectron microscopic images of the Tau MTBDs demonstrate an interaction with the MT surface on which single repeats bind across an αβ-tubulin dimer (Kellogg et al., 2018). In a surprising finding, McKibben and Rhoades (2019) showed that the isolated PRD bound tubulin tightly compared with the MTBD, exhibited a higher affinity for taxol-stabilized MTs, and was the only isolated domain capable of significantly polymerizing tubulin. These authors found that under saturable stoichiometric binding conditions, one Tau bound to two tubulin dimers. Therefore, based solely on the observations in both studies, the number of Tau molecules that bind to MTs is in the range of 400–800 copies within a 1-μm segment of MT given a 13-protofilament structure with ∼1,600 αβ-tubulin heterodimers (Löwe et al., 2001). However, it is widely agreed that numerous factors will alter the number of bound Tau molecules including Tau splicing in the repeat region, the state of Tau phosphorylation and acetylation, the specific locus of binding along the MT lattice, and the tubulin concentration, among others (Lippens et al., 2016). Despite the difficulty of determining precise numbers of Tau molecules at the scale of polymerized MTs in cells, certainly hundreds or thousands of Tau molecules are concentrated within a small volume for MT polymerization and stability. Phase separation provides high local concentrations of Tau exceeding that of the solute phase, in which molecular collisions would make precisely oriented Tau–MT binding improbable. Rather, Tau proteins can collectively implement their function at the larger scale of MTs by securing the assembly of multiple αβ-tubulin dimers nearly instantaneously.
McKibben and Rhoades (2019) suggest that the PRD serves as the core tubulin-binding domain, binding to two tubulin dimers as a critical step toward initiating polymerization, and multiple weak tubulin-binding sites in the MTBD increase the local concentration of tubulin to accelerate MT growth. Thus, the differential properties of these two domains ultimately lead to the assembly and stability of a MT. Unfortunately, the PRD was not visualized in the cryoelectron microscopy of the Tau–MT interaction (Kellogg et al., 2018). Tan et al. (2019) used bacterially expressed, GFP-tagged full-length human Tau to bind to taxol-stabilized MTs in vitro. In this study, Tau molecules initially bound diffusely along the entire MT lattice and progressed to equilibrium as condensates. Interestingly, Tau condensates had increased average dwell time along the MT lattice (Tan et al., 2019), thereby taking advantage of condensate/solute phase equilibrium properties that will sustain a high local concentration of Tau in proximity to the MT. Phase-separated droplets can also regulate their interfacial tension and thereby wet the solid MT surface.
Tan et al. (2019) also show that Tau condensation only formed on GDP MT lattices, suggesting that Tau condensation was gated by the nucleotide state of the MT lattice and therefore does not condense in the region of the GTP cap. The inhibition of Tau condensation at the GTP cap may function in conjunction with the co-condensation of Tau CWT 151–254 and EB1 that we observed. EB proteins control the protofilament number and the length of taper at MT tips and mechanically stiffen the MT (Lopez and Valentine, 2014; Vitre et al., 2008; Zhang et al., 2015) as well as track the ends of growing MTs (Bieling et al., 2007, 2008; Komarova et al., 2009) by recognizing a nucleotide-dependent conformation that is transiently formed during MT assembly of two adjoining tubulin subunits (Roth et al., 2018). Exchange between CWT 151–254/EB1 complexes and Tau condensates on the MT lattice may coordinate the nucleotide state of the MT with its polymerization capacity within the region of the EB comet.
In route to achieving an authentic in vivo understanding of Tau LLPS, overexpression studies can serve as a heuristic model, particularly in the analysis of protein domains. Overexpression studies can reveal specific domain functions that are obscured in more in vivo settings due to competition with other domains in the same protein, as is the case here regarding a requirement to set a balance between the contrasting roles of the PRD and the N-terminal domain. However, observations of LLPS through protein overexpression in live cells do not reflect in vivo Tau concentrations (Alberti et al., 2019). Conclusions from such methods will require novel approaches to more closely approximate the in vivo setting. In a living cell, proteins retain many of their fundamental biophysical properties that become highly regulated and specified to achieve precision functions in the context of numerous dynamic conditions. The PRD has a prominent role in regulating Tau LLPS in living cells, a function that we detected by combining in vitro and in vivo studies along with the optogenetic probe Cry2. Tau condensates mediated by the PRD can create finely tuned molecular locales to regulate cellular architectures under the control of MTs.
Materials and methods
Reagents
Nocodazole and PolyU (RNA; mol wt 800–1,000 kD) were purchased from Sigma-Aldrich, 1,6-HD and 2,5-HD from Sigma-Aldrich, and heparin from Galen Laboratory Supplies.
Cell culture
SH-SY5Y neuroblastoma cells (American Type Culture Collection, CRL-2266) in DMEM/F12/GlutaMAX medium (Gibco) with 10% FBS or HeLa cells in DMEM medium with 10% FBS were cultured as monolayers at 2 × 104 per well on eight-well µ-Slides (ibidi) at 37°C with 5% CO2 in a humidified incubator. The population of neuroblast-like cells may decrease with increased passage number (Kovalevich and Langford, 2013); therefore, the passage number was kept <30, and differentiation induced by nutritional deprivation was carefully avoided.
Plasmid and recombinant protein
Plasmids for mammalian expression
Plasmid Cry2olig-mCherry-Tau 1–441 was prepared by inserting a DNA fragment encoding the full-length Tau into the linearized Cry2olig-mCherry (Addgene, 60032) backbone at the C terminus of mCherry using the Gibson assembly Cloning kit (New England BioLab Int.). Cry2WT-mCherry-Tau 1–441 is then produced by reversing the E490G mutation of Cry2olig. Cry2WT-mCherry-truncated Tau plasmids including Cry2WT-mCherry-Tau 244–375, 151–254, 1–375, 1–343, 1–311, 1–281, 1–254, and 151–375 were prepared from Cry2WT-mCherry-Tau 1–441 using the Q5 Site-Directed Mutagenesis Kit (New England BioLab Int.). Plasmids Cry2WT-mCherry-Tau 151–254 psuedo-phosphorylated mutants or alanine substitutes were prepared from Cry2WT-mCherry-Tau 151–254 using the same mutagenesis kit. Unless otherwise mentioned, all the transformations were done in a high-efficiency competent cell (New England BioLab Int.), and all the resulting plasmids were partially sequenced to confirm the coding sequence. Tau 1–441-EGFP and Tau 256–441-EGFP were gifts from G. Schmitt-Ulms (University of Toronto, Toronto, Canada). The plasmids encoding EGFP-α-tubulin (Addgene, 12298), pmCherry-α-tubulin (Addgene, 21043), and EB1-EGFP (Addgene, 39299) were purchased.
Human Tau protein (2N4R) with 36-nt annealing arms
5′-TCCGGACTCAGATCTCGAGCTCAAGCTTCGAATTCAATGGCTGAGCCCCGCCAGGAGTTCGAAGTGATGGAAGATCACGCTGGGACGTACGGGTTGGGGGACAGGAAAGATCAGGGGGGCTACACCATGCACCAAGACCAAGAGGGTGACACGGACGCTGGCCTGAAAGAATCTCCCCTGCAGACCCCCACTGAGGACGGATCTGAGGAACCGGGCTCTGAAACCTCTGATGCTAAGAGCACTCCAACAGCGGAAGATGTGACAGCACCCTTAGTGGATGAGGGAGCTCCCGGCAAGCAGGCTGCCGCGCAGCCCCACACGGAGATCCCAGAAGGAACCACAGCTGAAGAAGCAGGCATTGGAGACACCCCCAGCCTGGAAGACGAAGCTGCTGGTCACGTGACCCAAGCTCGCATGGTCAGTAAAAGCAAAGACGGGACTGGAAGCGATGACAAAAAAGCCAAGGGGGCTGATGGTAAAACGAAGATCGCCACACCGCGGGGAGCAGCCCCTCCAGGCCAGAAGGGCCAGGCCAACGCCACCAGGATTCCAGCAAAAACCCCGCCCGCTCCAAAGACACCACCCAGCTCTGGTGAACCTCCAAAATCAGGGGATCGCAGCGGCTACAGCAGCCCCGGCTCCCCAGGCACTCCCGGCAGCCGCTCCCGCACCCCGTCCCTTCCAACCCCACCCACCCGGGAGCCCAAGAAGGTGGCAGTGGTCCGTACTCCACCCAAGTCGCCGTCTTCCGCCAAGAGCCGCCTGCAGACAGCCCCCGTGCCCATGCCAGACCTGAAGAATGTCAAGTCCAAGATCGGCTCCACTGAGAACCTGAAGCACCAGCCGGGAGGCGGGAAGGTGCAGATAATTAATAAGAAGCTGGATCTTAGCAACGTCCAGTCCAAGTGTGGCTCAAAGGATAATATCAAACACGTCCCGGGAGGCGGCAGTGTGCAAATAGTCTACAAACCAGTTGACCTGAGCAAGGTGACCTCCAAGTGTGGCTCATTAGGCAACATCCATCATAAACCAGGAGGTGGCCAGGTGGAAGTAAAATCTGAGAAGCTTGACTTCAAGGACAGAGTCCAGTCGAAGATTGGGTCCCTGGACAATATCACCCACGTCCCTGGCGGAGGAAATAAAAAGATTGAAACCCACAAGCTGACCTTCCGCGAGAACGCCAAAGCCAAGACAGACCACGGGGCGGAGATCGTGTACAAGTCGCCAGTGGTGTCTGGGGACACGTCTCCACGGCATCTCAGCAATGTCTCCTCCACCGGCAGCATCGACATGGTAGACTCGCCCCAGCTCGCCACGCTAGCTGACGAGGTGTCTGCCTCCCTGGCCAAGCAGGGTTTGTGAGGATCCACCGGATCTAGATAACTGATCATAATC-3′. The primers are detailed in Table 2.
Table 2. Primers used for generating the listed plasmids.
| Plasmids | Forward primer | Reverse primer |
|---|---|---|
| Cry2 G490E | 5′-GAAGCACAGATCATGATCGGAG-3′ | 5′-ACGGGTTCTTGAAATAGCTTTAGC-3′ |
| Cry2WT-mCherry-Tau 1-375 | 5′-TGAGGATCCACCGGATCTAGATAAC-3′ | 5′-CTTGTGGGTTTCAATCTTTTTATTTCCTCCG-3′ |
| Cry2WT-mCherry-Tau 1-343 | 5′-TGAGGATCCACCGGATCTAGATAAC-3′ | 5′-CTTCTCAGATTTTACTTCCACCTGG-3′ |
| Cry2WT-mCherry-Tau 1-311 | 5′-TGAGGATCCACCGGATCTAGATAAC-3′ | 5′-TTTGTAGACTATTTGCACACTGCC-3′ |
| Cry2WT-mCherry-Tau 1-281 | 5′-TGAGGATCCACCGGATCTAGATAAC-3′ | 5′-CTTCTTATTAATTATCTGCACCTTCCCG-3′ |
| Cry2WT-mCherry-Tau 1-254 | 5′-TGAGGATCCACCGGATCTAGATAAC-3′ | 5′-CTTCAGGTCTGGCATGGG-3′ |
| Cry2WT-mCherry-Tau 244-375 | 5′-CAGACAGCCCCCGTGCCC-3′ | 5′-CATTGAATTCGAAGCTTGAGCTCG-3′ |
| Cry2WT-mCherry-Tau 151-375 | 5′-ATCGCCACACCGCG-3′ | 5′-CATTGAATTCGAAGCTTGAGCTCG-3′ |
| Cry2WT-mCherry-Tau 151-254 | 5′-TGAGGATCCACCGGATCTAGATAAC-3′ | 5′-CTTCAGGTCTGGCATGGG-3′ |
| Cry2WT-mCherry-Tau 151-254 T231E | 5′-CCGTCTTCCGCCAAGAGCC-3′ | 5′-CGACTTGGGTGGCTCACGGACCACTGCCACCTTC-3′ |
| Cry2WT-mCherry-Tau 151-254 S235E | 5′-CCGTCTTCCGCCAAGAGCC-3′ | 5′-CTCCTTGGGTGGAGTACGGACCACTGCCACCTTC-3′ |
| Cry2WT-mCherry-Tau 151-254 T231E/S235E | 5′-CCGTCTTCCGCCAAGAGCC-3′ | 5′-CTCCTTGGGTGGCTCACGGACCACTGCCACCTTC-3′ |
| Cry2WT-mCherry-Tau 151-254 T212E/S214E/T217E | 5′-CCCACCCGGGAGCCCAAGAAG-3′ | 5′-TGGTTCTGGAAGTTCCGGTTCGCGGGAGCGGCTGCCG-3′ |
| Cry2WT-mCherry-Tau 151-254 S202D/T205D/S208D | 5′-GACCCCGGCGACCGCTCCCGCACCCCG-3′ | 5′-GCCTGGATCGCCGGGGCTGCTGCTGTAGCC-3′ |
| Cry2WT-mCherry-Tau 151-254 S202A/T205A/S208A | 5′-GCTCCCGGCGCCCGCTCCCGCACCCCG-3′ | 5′-GCCTGGTGCGCCGGGGCTGCTGTAGCC-3′ |
| Cry2WT-mCherry-Tau 151-254 S202A/T205A/S208A/T212A/S214A | 5′-CTTCCAACCCCACCCACCCG-3′ | 5′-TGCCGGTGCGCGGGAGCGTGCGCCGGGTGCGCCTGGTGCGCCGGGGCTGCTGTAGCC-3′ |
| Cry2WT-mCherry-Tau 151-254 I171A/P172A | 5′-GCAAAAACCCCGCCCGC-3′ | 5′-TGCTGCCCTGGTGGCGTTGGCCT-3′ |
Plasmids for bacterial expression and recombinant proteins
cDNA of Tau 151–254 was inserted into a pMCSG17 plasmid using HiFi assembly (New England BioLab Int., E2621S). The resulting vector encoded a protein consisting of a hexahistidine tag followed by a tobacco etch virus enzymatic cleavage site attached to the Tau 151–254 sequence. The plasmid was transformed into DH5a and BL21DE3 cells.
Tau 255–441 was purified as previously described (Zhang et al., 2017). A similar procedure was used for purification of Tau 151–254; however, the 65°C heat-shock stage of purification was not used. Following nickel-nitrilotriacetic acid (Ni-NTA) affinity purification, size-exclusion chromatography was used. The elution fraction from the Ni-NTA was collected and concentrated, then loaded onto a Biorad ENrich 70 SEC column preequilibrated with 20 mM Hepes and 40 mM NaCl buffer. Fractions were collected and pooled based on UV absorption at 280 nm. The species identity was confirmed with SDS-PAGE and matrix-assisted laser desorption/ionization–time-of-flight mass spectroscopy. The concentration was determined with a bicinchoninic acid assay, or from the absorption at 280 nm.
Live cell imaging
Cells were plated on no. 1.5H 170-µm glass-bottom eight-well µ-Slides (ibidi) and grown typically overnight in normal growth medium to reach ∼50% confluency. Live cell imaging was performed using a 60× glycerol immersion objective on a Leica Sp8 laser scanning confocal microscope equipped with a temperature stage at 37°C and supplied with 5% CO2. Cells were imaged typically using two laser wavelengths (488 nm for Cry2 activation and GFP channel imaging/555 nm for mCherry) and sequentially scanned by line. For activation, unless specifically mentioned, three 488-nm light powers of 5%, 30%, and 100% were used for shallow, intermediate, and deep activation conditions, respectively. Cells were monitored over time in either xyt or xyzt mode. For local activation of CWT 151–254, a square of 3 × 3 µm was stimulated with blue light, and the dynamics of the region were quantified.
For FRAP, cells were first activated by 488 nm with an appropriate blue light intensity. Immediately after termination of the activation phase, light-induced clusters were bleached by UV light with 100% power on a 3 × 3–µm square of for a period of time, depending on the signal intensity of the site, and their fluorescence recovery was monitored while maintaining identical activation conditions for the clustering. It is practically helpful to bleach the site to the point that it is still noticeable, so the signal recovery can be precisely followed up.
Live cell imaging analysis
Phase separation quantified by single-cell CV of labeled proteins
Phase separation results in nonuniform distributions of phase-separated proteins in cells. When labeled proteins are diffusely distributed, their CV (i.e., the SD in pixel intensity from all measured points in the cell over the mean signal across the entire cells) is an excellent measure of the nonuniformity. The images were analyzed in Fiji-ImageJ.
FRAP
Integrated intensity density of the area for single Z-stacks was monitored, corrected for photobleaching with a reference area of the same size, and normalized with prebleaching intensity.
Reversibility of clustering
Z-projection of the whole cell, applying the SD method embedded within ImageJ, was monitored over time. The CV of the region of interest (ROI) was plotted over the time course. For repeated activation and recovery experiments, images of single Z-stacks were monitored and analyzed by ImageJ by both counting the particles and analyzing the CV of the pixel distribution in each image. To count the particles, raw images were smoothed through Gaussian blur. Then the Threshold was set based on the preactivation intensity. The clusters formed after activation in each frame were counted through the ImageJ function of Analyze Particles. The number of clusters in each image was then normalized according to the maximum and minimum number of clusters in all three cycles.
Analysis of Tau condensation on MT binding
Cells were typically imaged by use of two laser wavelengths (488 nm for Cry2 activation/555 nm for mCherry imaging). To quantify Cry2WT-mCherry Tau relocation, Z-stack imaging was performed systemically in five cycles. For the first cycle, the imaging was done with only the 555-nm laser and the following four cycles with both 488-nm and 555-nm lasers in a sequential “between lines” manner. Each cycle took 60 ± 5 s, and 10% power of the 488-nm laser was used for activation. The images in the first cycle and the third cycle were selected for image analysis, corresponding to Cry2WT Tau before and 60 s after blue-light activation. Images were generated from cells plated and transfected on different days.
The relocation of CWT from the diffused state to the MT-binding state in a cell was monitored by CV. Custom-written macros were used to automate the image analysis and quantification process. Briefly, a raw 3D image was first Z-projected into a 2D image, applying the SD method embedded in ImageJ. Each individual cell was outlined as the ROI with particle-analyzing tools after the Z-projection 2D image was subjected to background subtraction and Gaussian filter. The CV within the ROI was then measured and reported for Cry2WT-mCherry Tau distribution before and after blue-light activation in each cell.
We applied the second image analysis method to visualize Tau distribution change in a cell. Using the built-in plugin “image calculator” with the operation of subtraction, the difference of the Z-projection 2D image before and after activation was calculated. With proper thresholding, a binary 2D image was generated depicting the area where the CV was increased after activation. The detected distribution shift in each cell was examined manually to validate the CV values.
To quantify the EB1 Punctate versus MT-Bound versus Diffuse, we did image analysis with an additional preprocessing segmentation step to partition a digital image into multiple segments. To define the diffuse and nondiffuse (puncta plus bundles) population of EB1, each cell was outlined and auto-thresholded into a 2D binary image using the ImageJ embedded Yen method. Next, we applied a particle analysis method to separate bundles from puncta according to segment size and shape.
In vitro turbidimetry and brightfield microscopy
Turbidity of samples at room temperature were measured by OD at a 500 nm wavelength (OD500) using a BioTek synergy2 plate reader. The amount of coacervates in a sample was approximated to be proportional to its maximum OD500. Brightfield microscopy images of condensates were taken with a Leica BX51 or IX51 compound microscope.
In vitro Tau PRD and PolyU condensate formation
Unless otherwise stated, condensates of Tau PRD with PolyU RNA or heparin were formed with 50 µM Tau PRD and 125 µg/ml PolyU, and heparin concentration was 12.5 µM. This corresponds to a positive:negative charge ratio of 1:1. Ambient temperature and 20 mM Hepes, 40 mM NaCl, pH 7, buffer were used.
Computational modeling of the MTBD and PRD charge pattern
The strength of the Coulombic interaction was parameterized according to the potential energy where lB controls the strength of the Coulombic interaction (for reference, lB = 0.71 nm for water at 300 K). All simulations were performed in implicit solvent with statistical segments interacting through a weak excluded volume potential. Binodal points were plotted as a function of Tau concentration versus the lB that characterizes the extent of electrostatic screening. Simulations are performed by numerically propagating a complex Langevin (CL) equation of motion for the complex-valued fields. See the Method of FTS section for detailed methods and parameterization. All simulations were performed on NVIDIA Tesla M2070 or K80 graphics processing units using the PolyFTS code maintained by the Fredrickson group at the University of California, Santa Barbara, and licensed through the Complex Fluid Design Consortium.
Method of FTS (Fig. 6 C)
Recently we developed a coarse-grained polyampholyte model for studying charge-sequence–dependent phase behavior of Tau in the presence of RNA (Lin et al., 2019). A numerical method from the polymer physics community known as FTS was introduced to overcome the limitations of traditional molecular dynamics simulation. The method begins by transforming the canonical partition function of the coarse-grained model into a complex-valued statistical field theory by an exact Hubbard–Stratonovich transformation.
The fluctuating chemical and electrostatic fields are sampled using CL simulation. The fields were sampled on a spatial grid of 343 sites. An exponential time difference algorithm was used to numerically propagate the CL equations of motion with a timestep of 0.02. All simulations were performed in a cubic box of length 5.55 nm using periodic boundary conditions.
In this method, each amino acid is represented by a single interaction site of size b. All microscopic site densities are smeared over a finite volume by convolution with a Gaussian profile. The segment size b determines the length scale of the system. To parameterize our model, we set b = 0.4 nm, which approximates the distance between carbons in the protein backbone. Charged residues i and j interact via the electrostatic potential where σi is the charge on residue i at pH 7.4. The parameter lB is defined as with e the unit of electronic charge, the vacuum permittivity, and the relative dielectric strength of the solvent. To parameterize the model, we set for water. In our model, lB is an input parameter, which characterizes the strength of the electrostatic interaction energy. For reference, at 300 K, for water, lB = 0.71 nm. By running multiple simulations each at different values of lB, we can determine the phase coexistence points as a function of lB and compare how changes in the primary amino acid sequence widens or narrows the window for coacervate through charge-sequence effects. Besides Coulomb interactions, the polymer segments in implicit water interact via a short-ranged excluded volume repulsion potential of strength v. To compare among Tau constructs, we include a negatively charged polyelectrolyte species with the same degree of polymerization in order to model co-coacervating species such as single-stranded RNA. For Tau 255–441 and Tau 151–254 regions, the volume fraction was chosen to maintain a 1:1 charge ratio between polyampholyte and polyanion species. For the N terminus Tau 1–150 and C terminus Tau 376–441 sequences, which are net negatively charged, total charge neutrality was maintained by adding positively charged sodium ions to balance the net negative charge from the polymer species.
To compare the droplet stability of the PRD (151–254) with the N terminus (1–150) region, we performed FTS simulations in the Gibbs ensemble (Riggleman and Fredrickson, 2010). In these simulations, a dense (coacervate) phase and a dilute (solution) phase are simultaneously simulated in separate cells. Together these simulation cells form a canonical ensemble for which both the total mass and total volume are conserved. Chemical and mechanical equilibrium is achieved by performing mass and volume swaps between the two simulations cells while imposing charge neutrality constraints. The concentration and relative box volumes are updated periodically until convergence is achieved. The results of a Gibbs ensemble simulation performed with and which corresponds roughly to T = 293 K in our model, shows that the PRD has a dilute phase and concentrated (droplet) phase that reach a stable coexistence, while the simulation cells for the N terminus region converge to a single phase of homogenous bulk density, corresponding to the case in which the droplet vanishes.
Quantification and statistical analysis
Statistical parameters are indicated in the legends of each figure, including the definitions of error bars (standard error [SE] or SD) and the number experimental replication, denoted by n. Data distribution was assumed to be normal, but this was not formally tested. For light activation–induced change of Tau distribution with each independent construct, a pairwise t test was performed. For EB1 distribution, two-way multivariate ANOVA was performed. Statistics was performed with R-Software (3.6.1).
Contact for reagent and resource sharing
Further information and requests for reagents may be directed to K.S. Kosik (Kenneth.Kosik@ucsb.edu).
Data and code availability
Source data for figures in the paper are available upon request.
Online supplemental material
Fig. S1 shows the quantification of Cry2WT-mCherry-Tau (CWT 1–441) distribution in SH SY5Y before and after blue-light activation, and Tau dissociates from MTs after 1,6-HD treatment in HeLa cells. Fig. S2 shows the comparison of Cry2WT-mCherry-Tau distribution before and after light activation for various Tau constructs. Fig. S3 shows Cry2WT-mCherry-Tau 151–254 (CWT 151–254)/EB1-EGFP bound along the MT lattice of MT bundles upon light activation. Table S1 summarizes the mean and SE for each Cry2WT-mCherry-Tau construct, in cases of before activation, after activation, and change by activation. Table S2 summarizes the mean and SD for EB1-EGFP distribution in the absence or presence of CWT 151–254 as diffuse or bundles. Video 1 shows FRAP of Cry2WT-mCherry-Tau 151–254 (CWT 151–254) in SH SY5Y cells. Video 2 shows local and whole-field activation of Cry2WT-mCherry-Tau 151–254 (CWT 151–254) in SH SY5Y cells. Video 3 shows Cry2WT-mCherry-Tau 151–254 (CWT 151–254, red) and EB1-EGFP (green) overexpressed in SH SY5Y cells activated by shallow blue light (488 nm, 5% power). Video 4 shows Cry2WT-mCherry-Tau 151–254 (CWT 151–254, red) and EB1-EGFP (green) overexpressed in SH SY5Y cells, treated with 20 µM of nocodazole for 2 h.
Supplementary Material
summarizes the mean and SE for each Cry2WT-mCherry-Tau construct, in cases of before activation, after activation, and change by activation.
summarizes the mean and SD for EB1-EGFP distribution in the absence or presence of CWT 151–254 as diffuse or bundles.
Acknowledgments
We acknowledge Dr. Carol Vandenberg and Dr. Israel Hernandez at the Neuroscience Research Institute, University of California, Santa Barbara, for valuable discussions.
This study was funded by U54 NS (100717; K.S. Kosik), the Tau Consortium (K.S. Kosik and S. Han), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (K.S. Kosik and, M.Z. Wilson), the Larry L. Hillblom Foundation (K.S. Kosik), the Edward N. and Della L. Thome Memorial Foundation (K.S. Kosik), and the National Institutes of Health (R01AG056058; K.S. Kosik, S. Han, J. Shea, and J. McCarty). G.H. Fredrickson and J. Shea acknowledge the Materials Research Science and Engineering Centers Program of the National Science Foundation (DMR 1720256), and J. Shea acknowledges the National Science Foundation (MCB-1716956). The computational part of this research used resources of the Extreme Science and Engineering Discovery Environment (supported by National Science Foundation Project TG-MCA05S027 and the Center for Scientific Computing from the California NanoSystems Institute, University of California, Santa Barbara, available through the Materials Research Laboratory; the National Science Foundation Materials Research Science and Engineering Centers (DMR-1720256) and the National Science Foundation (CNS-1725797). We acknowledge the use of the Neuroscience Research Institute, Molecular, Cell and Developmental Biology Microscopy Facility, and the Resonant Scanning Confocal Microscope supported by the National Science Foundation Major Research Instrumentation Program (DBI-1625770).
The authors declare no competing financial interests.
Author contributions: K.S. Kosik and X. Zhang conceived the project. X. Zhang performed the cellular experiments. M. Vigers performed the in vitro experiments. J. McCarty performed in silico simulation. J.N. Rauch and M.Z. Wilson advised with methodologies. X. Zhang, M. Vigers, J. McCarty, S. Han, and K.S. Kosik wrote the manuscript. All authors edited and reviewed the manuscript. K.S. Kosik supervised the cellular work. S. Han supervised the in vitro work. J-E. Shea and G.H. Fredrickson supervised the in silico simulation.
References
- Akhmanova A., and Steinmetz M.O.. 2010. Microtubule +TIPs at a glance. J. Cell Sci. 123:3415–3419. 10.1242/jcs.062414 [DOI] [PubMed] [Google Scholar]
- Alberti S., Gladfelter A., and Mittag T.. 2019. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell. 176:419–434. 10.1016/j.cell.2018.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambadipudi S., Biernat J., Riedel D., Mandelkow E., and Zweckstetter M.. 2017. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun. 8:275 10.1038/s41467-017-00480-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E.L., Sandblad L., Dogterom M., Brunner D., and Surrey T.. 2007. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 450:1100–1105. 10.1038/nature06386 [DOI] [PubMed] [Google Scholar]
- Bieling P., Kandels-Lewis S., Telley I.A., van Dijk J., Janke C., and Surrey T.. 2008. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 183:1223–1233. 10.1083/jcb.200809190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha D., Walls M.T., and Brangwynne C.P.. 2019. Probing and engineering liquid-phase organelles. Nat. Biotechnol. 37:1435–1445. 10.1038/s41587-019-0341-6 [DOI] [PubMed] [Google Scholar]
- Brangwynne C.P., Tompa P., and Pappu R.V.. 2015. Polymer physics of intracellular phase transitions. Nat. Phys. 11:899–904. 10.1038/nphys3532 [DOI] [Google Scholar]
- Bugaj L.J., Choksi A.T., Mesuda C.K., Kane R.S., and Schaffer D.V.. 2013. Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods. 10:249–252. 10.1038/nmeth.2360 [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C.C.-H., Eckmann C.R., Myong S., and Brangwynne C.P.. 2015. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA. 112:7189–7194. 10.1073/pnas.1504822112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichou Y., Lin Y., Rauch J.N., Vigers M., Zeng Z., Srivastava M., Keller T.J., Freed J.H., Kosik K.S., and Han S.. 2018. Cofactors are essential constituents of stable and seeding-active tau fibrils. Proc. Natl. Acad. Sci. USA. 115:13234–13239. 10.1073/pnas.1810058115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick A.W.P., Falcon B., He S., Murzin A.G., Murshudov G., Garringer H.J., Crowther R.A., Ghetti B., Goedert M., and Scheres S.H.W.. 2017. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature. 547:185–190. 10.1038/nature23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galjart N. 2010. Plus-end-tracking proteins and their interactions at microtubule ends. Curr. Biol. 20:R528–R537. 10.1016/j.cub.2010.05.022 [DOI] [PubMed] [Google Scholar]
- Gasset-Rosa F., Lu S., Yu H., Chen C., Melamed Z., Guo L., Shorter J., Da Cruz S., and Cleveland D.W.. 2019. Cytoplasmic TDP-43 De-mixing Independent of Stress Granules Drives Inhibition of Nuclear Import, Loss of Nuclear TDP-43, and Cell Death. Neuron. 102:339–357.e7. 10.1016/j.neuron.2019.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C.X., Liu F., Grundke-Iqbal I., and Iqbal K.. 2005. Post-translational modifications of tau protein in Alzheimer’s disease. J. Neural Transm. (Vienna). 112:813–838. 10.1007/s00702-004-0221-0 [DOI] [PubMed] [Google Scholar]
- Gui X., Luo F., Li Y., Zhou H., Qin Z., Liu Z., Gu J., Xie M., Zhao K., Dai B., et al. 2019. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat. Commun. 10:2006 10.1038/s41467-019-09902-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustke N., Trinczek B., Biernat J., Mandelkow E.M., and Mandelkow E.. 1994. Domains of tau protein and interactions with microtubules. Biochemistry. 33:9511–9522. 10.1021/bi00198a017 [DOI] [PubMed] [Google Scholar]
- Hernández-Vega A., Braun M., Scharrel L., Jahnel M., Wegmann S., Hyman B.T., Alberti S., Diez S., and Hyman A.A.. 2017. Local Nucleation of Microtubule Bundles through Tubulin Concentration into a Condensed Tau Phase. Cell Rep. 20:2304–2312. 10.1016/j.celrep.2017.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S.M., Weisbrich A., Damberger F.F., Bhavesh N.S., Jawhari H., Grigoriev I., van Rijssel F.J.A., Buey R.M., Lawera A., et al. 2009. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 138:366–376. 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Kellogg E.H., Hejab N.M.A., Poepsel S., Downing K.H., DiMaio F., and Nogales E.. 2018. Near-atomic model of microtubule-tau interactions. Science. 360:1242–1246. 10.1126/science.aat1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y., De Groot C.O., Grigoriev I., Gouveia S.M., Munteanu E.L., Schober J.M., Honnappa S., Buey R.M., Hoogenraad C.C., Dogterom M., et al. 2009. Mammalian end binding proteins control persistent microtubule growth. J. Cell Biol. 184:691–706. 10.1083/jcb.200807179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalevich J., and Langford D.. 2013. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 1078:9–21. 10.1007/978-1-62703-640-5_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S., Maharana S., and Simon A.. 2017. Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. Matters (Zur.). 10.19185/matters.201702000010 [DOI] [Google Scholar]
- Lee S., Park H., Kyung T., Kim N.Y., Kim S., Kim J., and Heo W.D.. 2014. Reversible protein inactivation by optogenetic trapping in cells. Nat. Methods. 11:633–636. 10.1038/nmeth.2940 [DOI] [PubMed] [Google Scholar]
- Lin Y., Protter D.S., Rosen M.K., and Parker R.. 2015. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell. 60:208–219. 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., McCarty J., Rauch J.N., Delaney K.T., Kosik K.S., Fredrickson G.H., Shea J.-E., and Han S.. 2019. Narrow equilibrium window for complex coacervation of tau and RNA under cellular conditions. eLife. 8 e42571 10.7554/eLife.42571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Fichou Y., Zeng Z., Hu N.Y., and Han S.. 2020. Electrostatically Driven Complex Coacervation and Amyloid Aggregation of Tau Are Independent Processes with Overlapping Conditions. ACS Chem. Neurosci. 11:615–627. 10.1021/acschemneuro.9b00627 [DOI] [PubMed] [Google Scholar]
- Lippens G., Landrieu I., Smet C., Huvent I., Gandhi N.S., Gigant B., Despres C., Qi H., and Lopez J.. 2016. NMR Meets Tau: Insights into Its Function and Pathology. Biomolecules. 6:28 10.3390/biom6020028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez B.J., and Valentine M.T.. 2014. Mechanical effects of EB1 on microtubules depend on GTP hydrolysis state and presence of paclitaxel. Cytoskeleton (Hoboken). 71:530–541. 10.1002/cm.21190 [DOI] [PubMed] [Google Scholar]
- Löwe J., Li H., Downing K.H., and Nogales E.. 2001. Refined Structure of Alpha Beta-Tubulin at 3.5 Å Resolution In J. Mol. Biol. Vol. 313 pp. 1045–1057. [DOI] [PubMed] [Google Scholar]
- Maharana S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillén-Boixet J., Franzmann T.M., et al. 2018. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 360:918–921. 10.1126/science.aar7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann J.R., Gleixner A.M., Mauna J.C., Gomes E., DeChellis-Marks M.R., Needham P.G., Copley K.E., Hurtle B., Portz B., Pyles N.J., et al. 2019. RNA Binding Antagonizes Neurotoxic Phase Transitions of TDP-43. Neuron. 102:321–338.e8. 10.1016/j.neuron.2019.01.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L., Latypova X., Wilson C.M., Magnaudeix A., Perrin M.-L., Yardin C., and Terro F.. 2013. Tau protein kinases: involvement in Alzheimer’s disease. Ageing Res. Rev. 12:289–309. 10.1016/j.arr.2012.06.003 [DOI] [PubMed] [Google Scholar]
- McKibben K.M., and Rhoades E.. 2019. Independent tubulin binding and polymerization by the proline-rich region of Tau is regulated by Tau’s N-terminal domain. J. Biol. Chem. 294:19381–19394. 10.1074/jbc.RA119.010172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Shiina N., and Tsukita S.. 2000. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10:865–868. 10.1016/S0960-9822(00)00600-X [DOI] [PubMed] [Google Scholar]
- Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., and Taylor J.P.. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 163:123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukrasch M.D., Biernat J., von Bergen M., Griesinger C., Mandelkow E., and Zweckstetter M.. 2005. Sites of tau important for aggregation populate beta-structure and bind to microtubules and polyanions. J. Biol. Chem. 280:24978–24986. 10.1074/jbc.M501565200 [DOI] [PubMed] [Google Scholar]
- Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D., et al. 2015. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 57:936–947. 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M., et al. 2015. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 162:1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Ramirez-Rios S., Denarier E., Prezel E., Vinit A., Stoppin-Mellet V., Devred F., Barbier P., Peyrot V., Sayas C.L., Avila J., et al. 2016. Tau antagonizes end-binding protein tracking at microtubule ends through a phosphorylation-dependent mechanism. Mol. Biol. Cell. 27:2924–2934. 10.1091/mbc.E16-01-0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback J.A., Zhu L., Ferrolino M.C., Tolbert M., Mitrea D.M., Sanders D.W., Wei M.-T., Kriwacki R.W., and Brangwynne C.P.. 2020. Composition-dependent thermodynamics of intracellular phase separation. Nature. 581:209–214. 10.1038/s41586-020-2256-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggleman R.A., and Fredrickson G.H.. 2010. Field-theoretic simulations in the Gibbs ensemble. J. Chem. Phys. 132 024104 10.1063/1.3292004 [DOI] [PubMed] [Google Scholar]
- Roth D., Fitton B.P., Chmel N.P., Wasiluk N., and Straube A.. 2018. Spatial positioning of EB family proteins at microtubule tips involves distinct nucleotide-dependent binding properties. J. Cell Sci. 132 jcs219550 10.1242/jcs.219550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Berry J., Pannucci N., Haataja M.P., Toettcher J.E., and Brangwynne C.P.. 2017. Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell. 168:159–171.e14. 10.1016/j.cell.2016.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siahaan V., Krattenmacher J., Hyman A.A., Diez S., Hernández-Vega A., Lansky Z., and Braun M.. 2019. Kinetically distinct phases of tau on microtubules regulate kinesin motors and severing enzymes. Nat. Cell Biol. 21:1086–1092. 10.1038/s41556-019-0374-6 [DOI] [PubMed] [Google Scholar]
- Skube S.B., Chaverri J.M., and Goodson H.V.. 2010. Effect of GFP tags on the localization of EB1 and EB1 fragments in vivo. Cytoskeleton (Hoboken). 67:1–12. 10.1002/cm.20409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R., Lam A.J., Tan T., Han J., Nowakowski D.W., Vershinin M., Simó S., Ori-McKenney K.M., and McKenney R.J.. 2019. Microtubules gate tau condensation to spatially regulate microtubule functions. Nat. Cell Biol. 21:1078–1085. 10.1038/s41556-019-0375-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taslimi A., Vrana J.D., Chen D., Borinskaya S., Mayer B.J., Kennedy M.J., and Tucker C.L.. 2014. An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 5:4925 10.1038/ncomms5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B., Coquelle F.M., Heichette C., Garnier C., Chrétien D., and Arnal I.. 2008. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 10:415–421. 10.1038/ncb1703 [DOI] [PubMed] [Google Scholar]
- Wang Y., and Mandelkow E.. 2016. Tau in physiology and pathology. Nat. Rev. Neurosci. 17:5–21. 10.1038/nrn.2015.1 [DOI] [PubMed] [Google Scholar]
- Wegmann S., Eftekharzadeh B., Tepper K., Zoltowska K.M., Bennett R.E., Dujardin S., Laskowski P.R., MacKenzie D., Kamath T., Commins C., et al. 2018. Tau Protein Liquid–liquid Phase Separation Can Initiate Tau Aggregation In EMBO J. Vol. 37 p. e98049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Alushin G.M., Brown A., and Nogales E.. 2015. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell. 162:849–859. 10.1016/j.cell.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lin Y., Eschmann N.A., Zhou H., Rauch J.N., Hernandez I., Guzman E., Kosik K.S., and Han S.. 2017. RNA stores tau reversibly in complex coacervates. PLoS Biol. 15 e2002183 10.1371/journal.pbio.2002183 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
summarizes the mean and SE for each Cry2WT-mCherry-Tau construct, in cases of before activation, after activation, and change by activation.
summarizes the mean and SD for EB1-EGFP distribution in the absence or presence of CWT 151–254 as diffuse or bundles.
Data Availability Statement
Source data for figures in the paper are available upon request.