Gliech and Holland discuss the guiding design principles of biological clocks across a variety of model systems.
Abstract
Biological timekeeping enables the coordination and execution of complex cellular processes such as developmental programs, day/night organismal changes, intercellular signaling, and proliferative safeguards. While these systems are often considered separately owing to a wide variety of mechanisms, time frames, and outputs, all clocks are built by calibrating or delaying the rate of biochemical reactions and processes. In this review, we explore the common themes and core design principles of cellular clocks, giving special consideration to the challenges associated with building timers from biochemical components. We also outline how evolution has coopted time to increase the reliability of a diverse range of biological systems.
Introduction
To consider cellular processes outside the context of time is to lose touch with the physical universe in which cells reside. Indeed, temporal accounting has been tallied for every biochemical reaction in every cellular process by every cell. Rather than simply allowing all biological programs to proceed at their fastest possible pace, cells harness time to guide, sense, and modulate biological outcomes. This deliberate temporal usage has increased the fidelity and scope of cellular processes by enabling the sequential execution of events (Pourquie, 2001; Delgado and Torres, 2016; Raff, 2007), timed responses (Renner and Schmitz, 2009; Heinzel et al., 2017; Thornquist et al., 2020), selectivity to input signals (Gerardin et al., 2019; Qian et al., 2019), delay sensing (Lambrus and Holland, 2017; Hellmuth and Stemmann, 2020), and organismal synchronization with the environment (Diernfellner and Brunner, 2020; Shalit-Kaneh et al., 2018; Buhr and Takahashi, 2013).
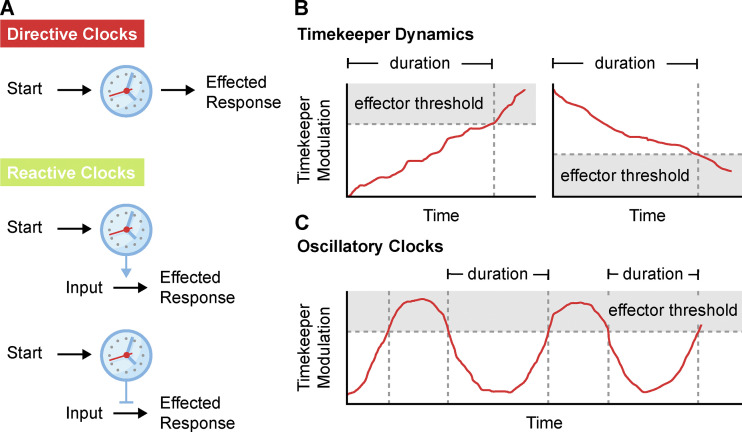
Biological clocks (see Definitions) can be broadly classified into directive and reactive clocks (Fig. 1 A and Definitions). Directive clocks set temporal constraints on a process and serve to direct predetermined outcomes. Imagine for example the fuse on a firework. Once lit, the fuse delays the inevitable launch and explosion so that a reveler has time to escape unharmed. In a biological context, the negative feedback loops in the ERK1/2 MAPK signaling network act as a directive clock. Following receptor activation, a signal down-regulation program is set in motion through inhibitory phosphorylation, receptor internalization, transcription of negative regulators, and phosphatase activity to turn off the signal (Lake et al., 2016). The time it takes to enact this program has been sculpted by evolution to allow for short or oscillatory bursts of signaling activity. In addition to negative feedback regulation in kinase signaling (Lake et al., 2016; Renner and Schmitz, 2009), directive clocks are also found in developmental programs (Raff, 2007), circadian biology (Bell-Pedersen et al., 2005), and other instances in which biological processes require a strict time frame.
Figure 1.
The fundamentals of cellular clocks. (A) Schematic of directive and reactive clocks. Directive clocks introduce a time delay between a trigger and its effected response. Reactive clocks create a time constraint that either promotes (top) or inhibits (bottom) an effected response from a secondary input. (B) Predictable timekeeper modulation generates effected cellular responses only after crossing a critical threshold. Clock duration is the time taken from the start of the timekeeper modulation to the triggering of the response. (C) Oscillatory clocks experience repeated cycles of timekeeper behavior. The downstream activity in oscillatory clocks is also gated by a critical effector threshold.
Reactive clocks are comparatively rare in biology and allow cells to make decisions based on the timing of inputs. Unlike the directive timers, the outcomes of these biological processes are not predetermined and rely on both temporal and external cues (Fig. 1 A). Imagine a contestant participating in a trivia game show. If the contestant answers the question correctly before time runs out, they win a prize. However, the wrong answer within the time frame, or the right answer outside of the set time frame, fails to produce the same reward. In this case, both the answer to the question and the time at which it is provided are accounted for. The mitotic surveillance pathway is an example of a reactive clock that acts to arrest cells only after experiencing abnormally long mitosis (Uetake and Sluder, 2010; Lambrus and Holland, 2017). In this case, cell fate is determined by both the time on the clock and the cue of mitotic exit. Since extending mitosis increases the frequency of mitotic errors, this timing mechanism protects cell populations from the detrimental consequences arising as a result of erroneous cell divisions.
In this review, we discuss similarities and differences in the designs of timers (see Definitions) across a broad range of biological processes. We also consider the common strategies, hurdles, and modifications that have been developed to tailor cellular clocks to the needs of the system in which they operate.
The fundamentals of cellular clocks
Timekeepers
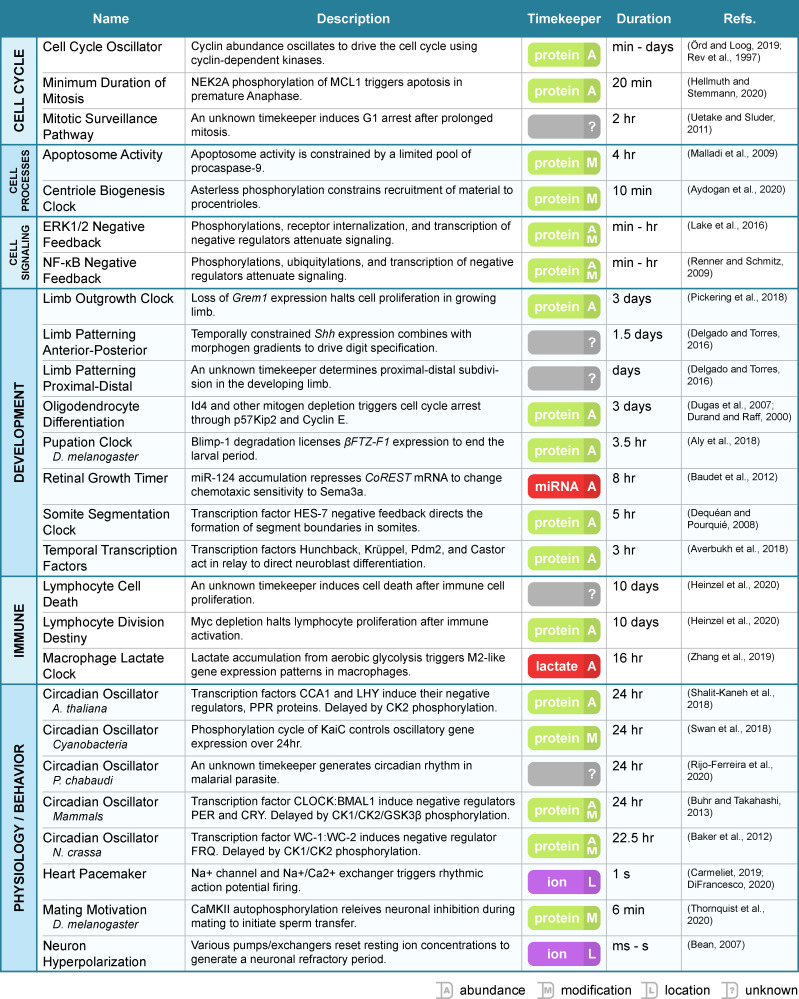
Biological clocks use a timekeeper to measure duration (see Definitions). Like sand in an hourglass, the timekeeper serves as a physical manifestation of elapsed time. The dynamics of this element set the overall clock pace and length (Fig. 1 B). In principle, a timekeeper can be anything with the ability to predictably change over time. Ions (Kohajda et al., 2020), metabolites (Zhang et al., 2019), microRNAs (Baudet et al., 2011), and promoter elements (Heinzel et al., 2017) have all been proposed as timekeeper elements, but biology primarily assigns timekeeping duties to proteins (Aly et al., 2018; Baker et al., 2012; Shalit-Kaneh et al., 2018; Buhr and Takahashi, 2013; Pickering et al., 2018; Fig. 2). In this case, the state of protein posttranslational modification, abundance, conformation, or complexing modulates over time to serve as a temporal marker.
Figure 2.
Summary of biological clocks. A compilation of the clock examples from this review organized by function. Clock name, a brief description, timekeeper type, timer duration, and key references are listed for each entry.
Consider, for example, the clock that constrains Drosophila melanogaster’s mating behavior (Thornquist et al., 2020). In this clock, mating motivation is directly linked to the protein state of Ca2+/calmodulin-dependent protein kinase II (CaMKII) in the specialized Crz neurons. At the onset of mating, CaMKII rapidly activates and prevents Crz neurons from firing. Over the next 6 min, a gradual increase in CAMKII inhibitory autophosphorylation serves to suppress its own kinase activity. Once CAMKII activity is sufficiently depleted, neuronal inhibition is released, and sperm transfer takes place. Here, the timekeeper is CaMKII and the measured dynamic is kinase activity, a proxy for the state of protein posttranslational modification.
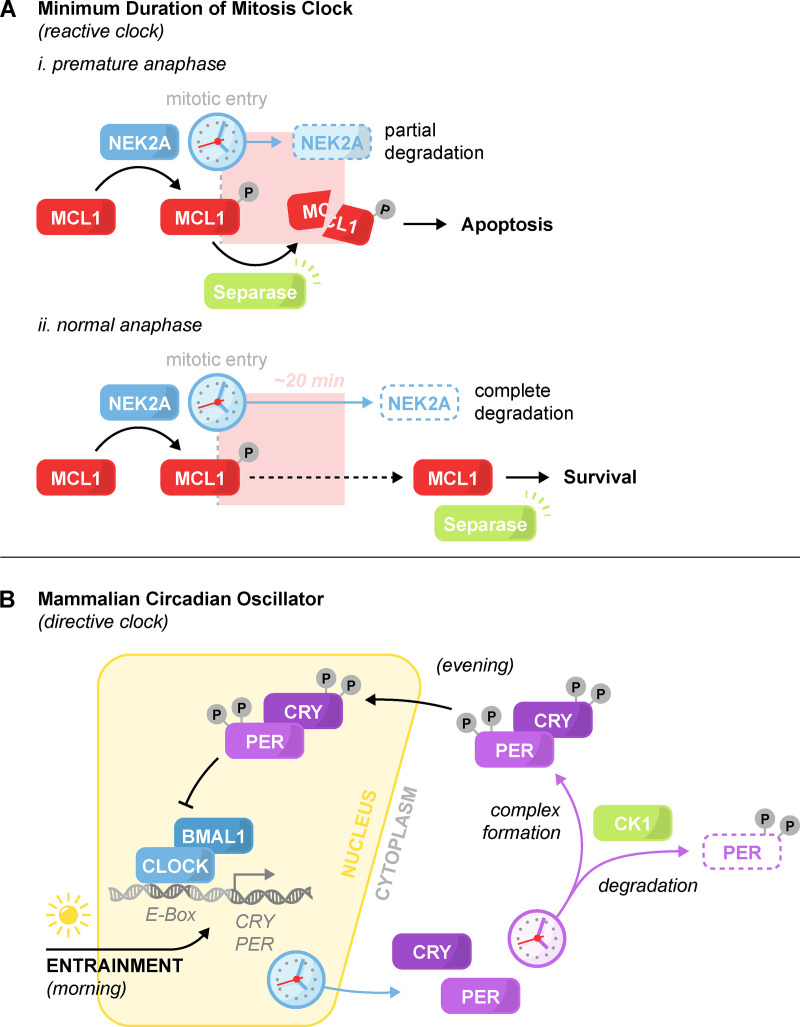
A second example comes from the clock that enforces a minimum duration in mammalian mitoses. This clock functions by generating a period at the onset of mitosis during which the activity of the protease Separase is toxic to cells (Hellmuth and Stemmann, 2020). Separase is required to initiate chromosome separation at anaphase, and cells that reach anaphase prematurely induce apoptosis through the Separase-dependent cleavage of the antiapoptotic protein MCL1 (induced myeloid leukemia cell differentiation 1). The time-sensitive toxicity of Separase activity is controlled by ongoing phosphorylation of MCL1 by NEK2A (NimA-related protein kinase 2). Mitotic entry triggers the gradual depletion of NEK2A and subsequent shift of MCL1 to a dephosphorylated state that is resistant to Separase cleavage (Fig. 3 A). The duration of this timer is therefore tied to the abundance and degradation rate of the timekeeper NEK2A.
Figure 3.
Minimum duration of mitosis clock and the mammalian circadian oscillator. The architecture of a reactive and directive clock. (A) Minimum duration of mitosis clock. The timekeeper NEK2A degrades at the onset of mitosis. Phosphorylation of the antiapoptotic protein MCL1 by NEK2A sensitizes MCL1 to Separase cleavage. (i) In the event of premature anaphase onset, Separase activation triggers apoptosis. (ii) In normal mitosis, degradation of NEK2A relieves MCL1 phosphorylation and the sensitivity to Separase activation, thereby enabling mitotic progression. (B) Simplified schematic of the core mammalian circadian oscillator. At dawn, the transcription factor CLOCK:BMAL1 initiates the transcription and accumulation of its negative regulators PER and CRY. In the evening, nuclear translocation of PER and CRY lead to CLOCK:BMAL1 repression and PER and CRY depletion. Depletion of PER and CRY de-represses CLOCK:BMAL1 and restarts the cycle. Dozens of gradual phosphorylations by CK1 promote the degradation, complexing, and nuclear translocation of PER and CRY. Light-induced transcription of PER and CRY is the primary mechanism for clock entrainment. In addition to what is shown, other feedback loops and kinases also help maintain the clock’s pace.
The end point of cellular clocks is determined by timekeeper modulation breaching an effector threshold (Fig. 1 B). To use the above examples, this would be the depletion of CaMKII activity until it can no longer inhibit neuronal firing or the degradation of NEK2A until it can no longer maintain MCL1 phosphorylation. In the case of directive clocks such as in Drosophila mating, the depletion of the timekeeper serves to commit cells to a specific response. With reactive clocks such as the minimum duration of mitosis, the time taken to deplete the timekeeper creates a window that licenses a stimulus to generate a response.
In special cases, clocks are characterized by repeated cycles of activity. These so-called oscillators (see Definitions) rely on a cyclical design with a built-in reset of the timekeeper (Fig. 1 C). Circadian oscillators (Bell-Pedersen et al., 2005) and Cyclin/CDK cell cycle oscillators (Örd and Loog, 2019) are perhaps the best known examples of this style of clock, but oscillators are also found in other contexts in which biological processes are regularly repeated or require periodicity. Take, for example, the formation of a new procentriole during centriole duplication in Drosophila embryos (Aydogan et al., 2020). The amount of material integrated into the new procentriole is controlled by oscillations in the recruitment of the master regulator Polo-like kinase 4 (PLK4) by its receptor Asterless. Asterless keeps time in this system by limiting the residency of PLK4 at the parent centriole through the accumulation of inhibitory phosphorylations. Repeated cycles of phosphatase activation during mitosis dephosphorylate Asterless to reset the clock and coordinate centriole duplication with the cell cycle oscillator.
Effectors
Timekeeper dynamics must be effected into a cellular response. Broadly speaking, timekeepers are either directly wired to the system they control, such as CaMKII controlling neuronal firing in Drosophila mating, or will feed into other existing cellular programs through the use of transcription factors. Transcription factors can also be used directly as the timekeeper, combining both schemes into a minimal clock architecture. In mammalian development, for example, segmentation of the presomitic mesoderm (eventual vertebrae, ribs, and skeletal muscle) is controlled by the oscillatory abundance of the HES7 (Hes family bHLH transcription factor 7) transcription factor (Kageyama et al., 2007; Bessho et al., 2001). Delayed inhibitory feedback of HES7 on its own promoter leads to repetitive periodic transcription of HES7 mRNA and waves of protein abundance. However, HES7 also binds and represses the promoter of the gene LFNG, which itself is an inhibitor of Notch signaling. In this way, HES7 abundance indirectly dictates the period of Notch pathway activity to control segmentation.
In the case of reactive clocks, effectors must be sensitive to both timing and a secondary cue to trigger a cellular response. Both clocks that regulate the minimum and maximum duration of mammalian mitoses make use of this principle. To reiterate briefly, the minimum duration of mitosis clock activates apoptosis when cells experience a premature onset of anaphase. MCL1, the specialized effector of cell death, integrates both a timed cue (NEK2A levels) with a secondary input (anaphase-induced Separase activation). Cell death occurs only if both NEK2A levels are high and Separase is activated (Fig. 3 A). The second timer, the mitotic surveillance pathway, induces cell cycle arrest if cells have taken too long to complete mitosis (Uetake and Sluder, 2010; Lambrus et al., 2016; Meitinger et al., 2016; Fong et al., 2016). Cells must exit mitosis within a certain time period to avoid activation of this pathway. Although the underlying molecular underpinnings are unclear, the activation of p53-mediated cell cycle arrest must be sensitive to a timed cue modulated by a mitotic timekeeper and a second input that depends on mitotic exit.
Time intervals of biological clocks
The most important consideration for biological clocks is maintaining agreement between the duration of the process being measured and the timekeeper dynamics. In other words, the process that drives a timer must be optimized to match the clock length (the time of day, for example, cannot be reliably pinpointed with an hourglass or a calendar). This constraint plays a major role in dictating the composition and architecture of the timing mechanism. At their shortest durations, biological clocks are limited by the maximum optimized rate of the underlying timekeeper reaction or process. At their longest, extrinsic noise and issues with robustness can overpower a timekeeper’s ability to behave predictably. Below, we discuss three examples of fast (seconds), medium (hours), and slow (days) timekeeping mechanisms that are well matched to the time frame of the biological process that they control.
A small cluster of cells called the sinoatrial node sets the rate of the beating heart. This process is tightly regulated by a pacemaker cellular clock. A human heart needs to beat on average once per second, and the fast pace of ion flow across membranes provides a well-suited timekeeping mechanism. The clock’s pace is set by specialized Na+ ion channels and a Na+/Ca2+ cation exchanger that work together to generate cyclical depolarizations of the neuron (Kohajda et al., 2020; DiFrancesco, 2020; Carmeliet, 2019; Bean, 2007).
As one of many layers of regulation in apoptosis, the function of the Apoptotic protease-activating factor 1/Caspase-9 apoptosome holoenzyme is temporally constrained. To execute cell death, Caspase-9 must associate with the apoptosome to process the effector Caspase-3. However, the binding affinity of Caspase-9 to the apoptosome is extremely weak. As a result, the apoptosome relies on the high-affinity binding of the precursor procaspase-9 and its processing in situ. The newly activated Caspase-9 leads to a short burst of apoptosome activity before it rapidly dissociates from the complex. For sustained activity, the holoenzyme requires many cycles of procaspase-9 binding, processing, and dissociation. Once the reserve of procaspase-9 in the cell is depleted, the holoenzyme shuts off. Complete processing of cellular procaspase-9 through the apoptosome takes several hours, providing a maximal time window for the cell to execute programmed cell death (Malladi et al., 2009; Li et al., 2017).
T and B lymphocytes experience a proliferative burst as part of the immune response to foreign invasion. This growth is constrained temporally by division destiny, a cellular clock that limits lymphocyte proliferation and prevents unbridled immune activation. Over the course of 10 d, the pro-proliferative protein Myc gradually depletes within lymphocytes until its level is no longer sufficient to promote cell division. While the exact mechanism is unclear, epigenetic changes at the MYC promoter have been proposed as the timekeeper in this system. The gradual loss of MYC transcriptional activity provides a defined time window for the expansion of immune cell populations (Heinzel et al., 2017).
A thorough understanding of the time ranges accessible to the variety of timekeeper architectures can help develop better models for unknown timer mechanisms. Consider the mammalian circadian oscillator, which was originally thought to be timed purely from the cyclical negative feedback of the heterodimeric transcription factor CLOCK:BMAL1 (Clock circadian regulator: Brain and muscle ARNT-like 1) and its targets Period (PER) and Cryptochrome (CRY; Buhr and Takahashi, 2013). In a simplified form, CLOCK:BMAL1 promotes the transcription of the PER and CRY proteins (Fig. 3 B). These target proteins then serve to negatively regulate and direct the degradation of CLOCK:BMAL1. Subsequently, PER/CRY transcription is attenuated, and CLOCK:BMAL1 is de-repressed to restart the ∼24-h cycle.
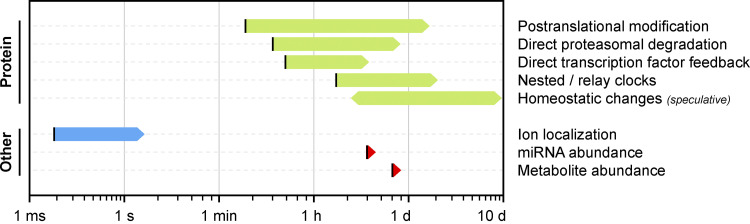
Based on other known clocks, direct transcriptional feedback loops such as these usually operate within the time frame of a few hours (Averbukh et al., 2018; Kageyama et al., 2007; Matsuda et al., 2020; Renner and Schmitz, 2009; Lake et al., 2016; Figs. 2 and 4). While extremely weak transcription factor binding could, in principle, extend the measured timer duration to a full 24-h cycle, such an architecture is likely to suffer from issues with intrinsic and extrinsic noise (Balázsi et al., 2011; Singh and Soltani, 2013). Therefore, one might predict that an additional time delay exists to slow the accumulation of the inhibitory PER/CRY complex. Indeed, it was later found that PER and CRY complex formation is dependent on dozens of phosphorylation events over the course of many hours by the extremely inefficient kinases Casein kinase 1 (CK1), CK2, and Glycogen synthase kinase 3β (Ode and Ueda, 2018). In addition, phosphorylation promotes PER degradation, which further serves to reduce the rate of accumulation of the PER/CRY complex. The mammalian circadian clock is therefore set by two nested timers: an abundance-based transcriptional timer containing within it a second phosphorylation-based delay timer (Fig. 3 B).
Figure 4.
Time frames of biological clocks. Approximate time frames for which timekeepers are used in biological clocks. Lower bounds are illustrated as black lines to represent the minimum clock duration that is set by the fastest rate of the biological process. Upper limits are illustrated as arrows to represent the ambiguous upper bound set by timekeeper robustness and clock reliability.
Methods for improving clock robustness
Cellular clocks are challenged by the internal and external noise of the systems in which they reside (Balázsi et al., 2011). For instance, the pace of a degradation-based timer may fluctuate based on the initial abundance of the timekeeper protein, the temperature-sensitive catalysis of polyubiquitylation, or competition for the proteasome by other substrates. Biological clocks have evolved to mitigate this variability and improve timekeeping robustness.
The degree of timing variability that can be tolerated will vary depending on the biological context. As such, not all clocks require immutable underpinnings. To draw from a previous example, it may not be critical for cells experiencing delayed divisions to undergo a cell cycle arrest after exactly 2 h in mitosis. Single-cell data show that the decision to arrest or continue proliferating varies between 1.5 and 2.5 h (Uetake and Sluder, 2010; Lambrus et al., 2016; Meitinger et al., 2016). By contrast, a circadian clock that fluctuates between 18 and 30 h, a similar percentage deviation, would be ineffective.
While the nature of the confounding variables is unique to different clock architectures, the strategies used to steady timekeeping are generalizable. In this section, we provide an overview of molecular mechanisms that ensure robustness in cellular clocks.
Timer design in response to robustness
It is reasonable to assume that the type of timekeeping mechanism (e.g., protein degradation vs. phosphorylation) and the layout of those mechanisms (e.g., single vs. multiple feedback loops) can affect the ability of a clock to reliably measure time and effect a cellular response. Robustness therefore can apply important evolutionary pressure when determining the overall architecture of timers.
Computational modeling has proven to be a valuable tool to evaluate components that improve the reliability of clocks. For example, in an exhaustive in silico screen of three-node enzymatic networks used for kinetic filtering of noise in cell signaling, it was found that the most effective and robust architectures were frequently used in biological systems (Gerardin et al., 2019). In other words, biological timers generally converge toward the most effective and robust layouts. In another example, different species of cyanobacteria rely on two closely related circadian clocks. The first is a free-running oscillator that can sustain periodicity for several days without external cues, while the loss of a single protein in the second has converted this oscillator to an hourglass-style clock that must be reset daily by environmental cues. Computational simulations of these two clocks revealed that their architectures make them particularly resistant to either intrinsic or extrinsic noise, respectively. The additional need for robustness in either category presumably played a role in determining which design was ultimately evolved by each species (Pittayakanchit et al., 2018).
Safety in numbers: Intercellular synchronization
A major contributor to noise in biological clocks is intrinsic variability at the single-cell level (Balázsi et al., 2011). Many cells running the same clock can mask this noise by using the averaging effect of large numbers. Interconnecting cellular oscillators therefore represent an appealing strategy for steadying clock pace. Whether setting the beating heart (Kohajda et al., 2020), circadian oscillations in the mammalian brain (Buhr and Takahashi, 2013; Yamaguchi et al., 2003), or the development of somites (Dequéant and Pourquié, 2008; Oates, 2020), overall clock pace is set by groups of synchronized cellular oscillators. Coordination between cells is achieved primarily through gap junction channels in the first two cases and intercellular Notch signaling in the third. In this way, intrinsic noise within individual cells is simply averaged away as groups of cells converge to a single harmonious oscillation.
Entrainment
Circadian oscillators have the additional luxury and challenge of mimicking a preexisting environmental clock, namely the alternation of day and night. Without external cues, biological mimicry of a 24-h cycle with exact precision is all but impossible. Entrainment is the process of using one set of oscillations, such as the fluctuations of light and temperature, to set the pace of another oscillator such as the biological circadian clock (Golombek and Rosenstein, 2010). Circadian oscillators then benefit organisms through their ability to physiologically anticipate cyclic changes in the environment (Bell-Pedersen et al., 2005). Biological oscillators can also entrain each other to create an oscillator hierarchy. This is the case with the circadian oscillators in peripheral organs, which take their cue from the master circadian oscillator in the brain (Brown et al., 2019), and the PLK4-driven centriole biogenesis oscillator, which is entrained by the cell-cycle oscillator (Aydogan et al., 2020).
Direct compensation of clocks
When biological clocks reach the reliable limits of their timekeeping architecture, they resort to the inclusion of specialized targeted modifications to increase robustness. These compensatory mechanics serve to directly counterbalance specific environmental influences to maintain a more consistent clock pace. Unlike the other robustness measures that have been discussed, direct compensation acts to mitigate the effects of a single noisy environmental variable rather than reduce the influence of biological noise as a whole. This focused approach is therefore unique to each clock system. Circadian oscillators offer several examples of how a diverse set of direct compensations steady the pace of biological clocks in response to temperature fluctuations.
As previously mentioned, the pace of the mammalian circadian oscillator is set in large part by kinase activity. Under uncompensated conditions, high temperatures increase phosphorylation and protein degradation rates and threaten to shorten the clock’s period. While humans are homeothermic, the body still experiences minute temperature fluctuations throughout the day which could affect the pace of the circadian oscillator (Kräuchi, 2002). Additionally, states of torpor and hibernation in mammals such as rodents can lower body temperature tens of degrees Celsius (Körtner and Geiser, 2000). To counterbalance this effect, PER2, a key transcription factor in the circadian feedback loop, uses temperature to switch between slow and fast degradation pathways. By using two kinases that are differentially sensitive to temperature, phosphorylation of PER2 at either its β-TrCP (β-Transducin repeat–containing protein) or FASP (Familial advanced sleep phase) domain provides a phosphoswitch that commits the protein to faster or slower degradation rates, respectively. Temperature changes shift the population of PER2 using each kinetic pathway so that the bulk degradation of the protein maintains 24-h clock periodicity (Narasimamurthy and Virshup, 2017).
The Arabidopsis circadian oscillator uses a similar counterbalancing principle, although at a different step in the transcription factor feedback cycle. Unlike homeothermic mammals, this plant requires stable timekeeping over a much wider temperature range. As with many sequence-specific protein–DNA interactions, the central transcription factor of the circadian oscillator Circadian clock associated 1 (CCA1) increases its affinity to transcriptional targets with elevated temperatures (Liu et al., 2008). At the same time, increased temperature causes the antagonistic kinase CK1 to increase its phosphorylation of CCA1 and inhibit DNA binding. Consequently, the binding of CCA1 to downstream promoters is relatively uniform over a broad range of temperatures (Portolés and Más, 2010).
The circadian rhythm of Cyanobacteria is generated by the cyclical phosphorylation and dephosphorylation of the kinase KaiC over 24 h (Swan et al., 2018; Nakajima et al., 2005). Unlike previous examples, no transcription is required for timing. Instead, the pace of this clock is set by the rate of ATP hydrolysis in KaiC hexameric complexes, which would normally be hastened at warmer temperatures. This effect is counterbalanced, however, by temperature-sensitive inhibitory KaiC autophosphorylation (Murakami et al., 2008). By negatively regulating its own activity through autoinhibition, this complex produces a stable ATP hydrolysis rate and phosphorylation kinetics to steady timer pace over a broad temperature range.
In each of these cases, compensation occurs at steps in the timing mechanism that are both critical for pace and particularly temperature labile. Taking note of the elements in biological clocks that are directly compensated can provide insight into which determinants play important roles in setting a timer’s pace.
Focusing on what is important: Timekeeper/effector integration
Some clocks circumvent the need for extreme precision in timing by directly using the timekeeper as the effector. For instance, developmental programs often prioritize the robust sequential execution of events rather than the specific time frame in which they occur. In one example, embryonic Drosophila neuroblasts rely on the sequential, timed decay of temporal transcription factors in neural progenitor cells to direct the proper patterning and differentiation of neurons (Averbukh et al., 2018). The transcription of genes required for differentiation is directly induced by a relay of activating and repressive transcription factor timekeepers. Developmental biology is replete with transcription factor–based clocks, presumably because of evolutionary pressure favoring the reliability of sequential gene expression patterns over exact timing (Delgado and Torres, 2016; Kageyama et al., 2007; Pickering et al., 2018; Aly et al., 2018; Roselló-Díez et al., 2014).
Timer uses in biology
Cellular clocks are found throughout biology and operate over a vast range of timescales to control activities ranging from cell-autonomous processes such as mitotic timing to broad organismal changes such as developmental patterning. The pervasiveness of timer usage by cells serves as a testament to how clocks help biological systems become more efficient, diverse, and robust. Below, we explore how cellular clocks offer unique solutions to a variety of biological problems.
Clocks function as noise filters and information decoders
Biological noise presents a significant challenge for intercellular signaling networks. The errant binding of a lone growth factor molecule to a receptor, for example, must be prevented from triggering downstream signal transduction and amplification. To address this issue, cells make use of a principle called kinetic filtering, which places a time delay on pathway activation to ensure that the incoming signal is sustained and robust before enacting a downstream response (Gerardin et al., 2019). These types of clocks erase signals from rapid and sporadic receptor activity, enabling cells to make better use of signaling networks for intentional communication.
Time encoding of intercellular communication can also be used to elicit differential responses. For instance, acute, oscillatory, or sustained activation of the nuclear factor-κB pathway sets in motion alternative transcriptional programs enabled by two clocks (Lane et al., 2017). In one instance, transcription initiation in a subset of target genes is gated by a slow chromatin regulatory step that requires sustained pathway activation. In the other, stable mRNA transcripts from target genes gradually accumulate over time to generate high levels of translation only after a sustained response (Sen et al., 2020; Purvis and Lahav, 2013; Fig. 5 A).
Figure 5.
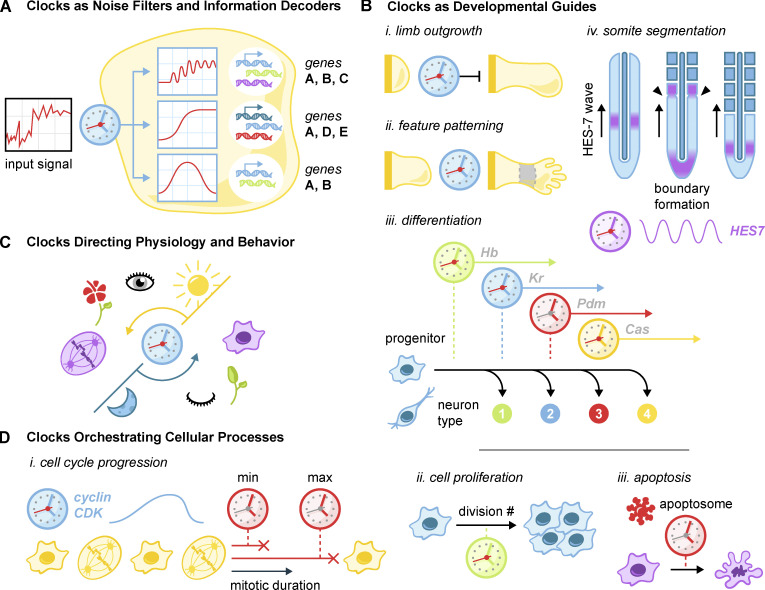
Uses of biological clocks. (A) Clocks are used as noise filters and information decoders in intercellular signaling networks. Kinetic filtering and timed negative feedback programs convert noisy input signals into acute, sustained, or oscillatory pathway activation. These dynamic responses elicit different gene expression patterns. (B) Varied clock usage in development. (i) Clocks determine the outgrowth size of developing limbs. (ii) Clocks act in concert with morphogen gradients to specify digits as well as particular developmental zones. (iii) Temporal sequential expression and decay of transcription factors in neural progenitor cells lead to the specification of multiple sets of differentiated neurons. (iv) Oscillations of HES genes traveling along the developing paraxial mesoderm intersect with a wavefront to generate segmented somites. (C) Circadian oscillations direct a wide variety of behaviors such as sleep/wake, cell division, and flower blooming. (D) Clocks manage many basic cellular processes. (i) The cell cycle is driven by the cyclin/CDK oscillator: a minimum- and maximum-duration clock set the appropriate duration of mitosis. (ii) Clocks limit programmed proliferative bursts such as that which occurs in lymphocytes following infection. (iii) The procaspase-9 clock limits the duration of apoptosome activity in programmed cell death.
Clocks serve as developmental guides
During development, organisms orchestrate the size and architecture of their features through the encoded behavior of individual cells. This raises the challenge of translating the intricacies of the developing body into single-cell decisions to proliferate and differentiate. Consider that the cells at the end of a developing arm must know to stop dividing despite having no immediate way to ascertain arm length. Cells in the developing mammalian embryo must robustly generate segments that will become the spine, ribs, and skeletal muscle, without access to a master blueprint. Progenitor cells in the brain must produce the correct number of multiple specific cell types with no access to an ongoing cellular census. In these circumstances, time becomes a metric to approximate what single cells cannot otherwise sense. Outgrowth time is used as a proxy for limb size to constrain total growth (Pickering et al., 2018; Sheeba et al., 2014). In development, temporally controlled transcription factor oscillations intersect with a morphogen gradient to inform cells of their location and direct the creation of segment boundaries (Dequéant and Pourquié, 2008; Kageyama et al., 2007; Hubaud and Pourquié, 2014). Finally, proliferation time approximates the number of divisions to force oligodendrocyte precursors to differentiate and generate an appropriate population size (Durand and Raff, 2000; Dugas et al., 2007).
In each instance, a cellular clock controls these programs by measuring the elapsed developmental time (Fig. 5 B). This solves the problem of informational downscaling by using temporal cues from within individual cells to achieve complex organization and patterning on an organismal scale.
Clocks direct physiology and behavior
Circadian oscillators serve as a link between many organisms and the day/night cycle of the planet. In humans, these oscillators alone account for changes in sleep schedule (Jagannath et al., 2017), metabolism (Zhu et al., 2017), body temperature (Panda, 2016), and hormone levels (Morris et al., 2012), among others. Disruption of circadian rhythms has been linked to a suite of pathologies, including decreased longevity, metabolic syndromes, immune dysfunction, cardiovascular disorders, and cancer (Evans and Davidson, 2013; Fig. 5 C). This wide array of circadian dysfunctions serves to highlight how heavily humans rely on the clock that synchronizes our diurnal species with cycles of day and night.
Across the tree of life, there are many examples of organisms tying their behavior to day/night cycles. The malarial parasite, for instance, makes use of an intrinsic circadian oscillator to coordinate its asexual cell cycle behavior with its mammalian host during infection (Rijo-ferreira et al., 2020). The fungus Neurospora crassa uses circadian timing to coordinate cycles of fungal growth (Baker et al., 2012), while cyanobacteria use it to control bioluminescence (Murakami et al., 2008). Many varieties of plants use the circadian cycle to coordinate flower opening and closing (Samach and Coupland, 2000). Plants also use circadian oscillators to measure the length of the day and relate it to seasonal changes that are used to direct flowering and fruiting behaviors (Samach and Coupland, 2000; Singh et al., 2020). This wide variety of circadian applications demonstrates the central role of these oscillators in providing a robust basis for diverse behaviors.
Although rare, the timing of noncircadian behaviors in multicellular organisms can also be achieved using biological clocks. One example is the previously discussed clock that dictates motivation during Drosophila mating (Thornquist et al., 2020). The rarity of this phenomenon suggests that clocks are generally ill suited for the direction of complex behaviors. Unlike the comparatively simple choice for single cells to proliferate, differentiate, or arrest, behavioral decisions in response to an unpredictable environment are better suited to neuronal networks.
Clocks manage and safeguard cellular processes
Clocks serve a specialized role in reining in the detrimental effects of runaway biological programs. This is achieved by setting strict temporal limits and using time as an indicator of processes gone awry. For instance, the body is shielded from lymphocyte hyperproliferation by two clocks: one that sets the lymphocyte proliferative potential and one that ensures eventual death (Heinzel et al., 2017). Macrophage response to infection is metered by a lactate-driven clock to switch from anaerobic to aerobic processes (Zhang et al., 2019). The apoptosome clock prevents cells from erroneously committing to programmed cell death (Malladi et al., 2009), while the accumulation and degradation of cyclins set temporal constraints on much of the eukaryotic cell cycle (Örd and Loog, 2019; Morgan, 1997). Finally, two clocks regulate the minimum and maximum allowable durations in mitosis to maintain genome stability (Lambrus and Holland, 2017; Hellmuth and Stemmann, 2020; Fig. 5 D).
These safeguards use one of two general mechanisms to regulate cells. In one instance, ongoing processes are strictly temporally constrained, as is the case with lymphocyte proliferation, the cell cycle oscillator, or the macrophage response. In the other instance, cells circumvent the need for direct sensing of errors by using temporal cues as a proxy. In this way, a single response pathway can serve to mitigate a wide variety of problems that all lead to a similar timing defect. For instance, the mitotic surveillance pathway can respond to extended mitoses caused by failed mitotic spindle formation or weak kinetochore–microtubule attachments alike.
Conclusion
Continued discoveries suggest that our census of biological clocks is far from complete. Additionally, while this review primarily focuses on systems for which we understand the molecular underpinnings, many clocks have been identified for which the driving mechanism remains to be determined. In several developmental clocks, for example, timekeeper proteins accumulate and deplete over the course of days to weeks. How such gradual and robust changes are possible remains unclear. Similarly, a more granular understanding of biological clocks may reveal potential therapeutic targets. For instance, bright light therapy in which the sun is simulated to better entrain the circadian clock to day/night cycles is widely used for treating mood disorders, despite providing only modestly beneficial outcomes (Nussbaumer-Streit et al., 2019). Modulating the circadian clock directly at a molecular level would likely prove more effective and is an area of ongoing research (Huang et al., 2020).
Biological clocks are as diverse in architecture as they are in function. This design plasticity indicates that timing mechanisms can be arrived at by many different means. However, while clock designs are often distinct, all timers follow the same generic blueprint: a dynamic timekeeper gradually modulates until it triggers a directive or reactive cellular response. Issues of pathway integration, time frame, and robustness all play a role in shaping the timer architecture to best suit the needs of the system in which they reside. The ubiquity of clocks underscores the broad influence timing has on the proper function of single cells and whole organisms. This suggests that the evolution of molecular timers represents a robust strategy for the directing and sensing of biological processes.
Definitions
Biological clock: A complete biological system for measuring time and effecting a cellular response. Timer: The underlying components of a biological clock that measure time. Oscillator: A cyclical biological clock characterized by repeated cycles of activity. Timekeeper: The physical entity at the core of a timer whose state reports on elapsed time. Directive clock: A clock in which the cellular outcome is predetermined. Reactive clock: A clock in which the cellular outcome is dependent on both temporal and external cues.
Acknowledgments
This work was supported by National Institutes of Health grants R01GM114119 and R01GM133897, an American Cancer Society Scholar Grant (RSG-16-156-01-CCG), and an American Cancer Society Mission Boost Grant (MBG-19-173-01-MBG).
The authors declare no competing financial interests.
References
- Aly H., Akagi K., and Ueda H.. 2018. Proteasome activity determines pupation timing through the degradation speed of timer molecule Blimp-1. Dev. Growth Differ. 60:502–508. 10.1111/dgd.12569 [DOI] [PubMed] [Google Scholar]
- Averbukh I., Lai S.L., Doe C.Q., and Barkai N.. 2018. A repressor-decay timer for robust temporal patterning in embryonic Drosophila neuroblast lineages. eLife. 7 e38631 10.7554/eLife.38631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydogan M.G., Steinacker T.L., Mofatteh M., Wilmott Z.M., Zhou F.Y., Gartenmann L., Wainman A., Saurya S., Novak Z.A., Wong S.S., et al. 2020. An autonomous oscillator times and executes centriole biogenesis. Cell. 181:1566–1581.e27. 10.1016/j.cell.2020.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C.L., Loros J.J., and Dunlap J.C.. 2012. The circadian clock of Neurospora crassa. FEMS Microbiol. Rev. 36:95–110. 10.1111/j.1574-6976.2011.00288.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázsi G., van Oudenaarden A., and Collins J.J.. 2011. Cellular decision making and biological noise: from microbes to mammals. Cell. 144:910–925. 10.1016/j.cell.2011.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudet M.L., Zivraj K.H., Abreu-Goodger C., Muldal A., Armisen J., Blenkiron C., Goldstein L.D., Miska E.A., and Holt C.E.. 2011. miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat. Neurosci. 15:29–38. 10.1038/nn.2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean B.P. 2007. The action potential in mammalian central neurons. Nat. Rev. Neurosci. 8:451–465. 10.1038/nrn2148 [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., and Zoran M.J.. 2005. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6:544–556. 10.1038/nrg1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y., Sakata R., Komatsu S., Shiota K., Yamada S., and Kageyama R.. 2001. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 15:2642–2647. 10.1101/gad.930601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.J., Pendergast J.S., and Yamazaki S.. 2019. Peripheral circadian oscillators. Yale J. Biol. Med. 92:327–335. 10.1016/b978-0-12-396971-2.00004-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr E.D., and Takahashi J.S.. 2013. Molecular components of the Mammalian circadian clock. Handb. Exp. Pharmacol. 217:3–27. 10.1007/978-3-642-25950-0_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet E. 2019. Pacemaking in cardiac tissue. From IK2 to a coupled-clock system. Physiol. Rep. 7 e13862 10.14814/phy2.13862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado I., and Torres M.. 2016. Gradients, waves and timers, an overview of limb patterning models. Semin. Cell Dev. Biol. 49:109–115. 10.1016/j.semcdb.2015.12.016 [DOI] [PubMed] [Google Scholar]
- Dequéant M.L., and Pourquié O.. 2008. Segmental patterning of the vertebrate embryonic axis. Nat. Rev. Genet. 9:370–382. 10.1038/nrg2320 [DOI] [PubMed] [Google Scholar]
- Diernfellner A.C.R., and Brunner M.. 2020. Phosphorylation Timers in the Neurospora crassa Circadian Clock. J. Mol. Biol. 432:3449–3465. 10.1016/j.jmb.2020.04.004 [DOI] [PubMed] [Google Scholar]
- DiFrancesco D. 2020. A Brief History of Pacemaking. Front. Physiol. 10:1599 10.3389/fphys.2019.01599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas J.C., Ibrahim A., and Barres B.A.. 2007. A crucial role for p57(Kip2) in the intracellular timer that controls oligodendrocyte differentiation. J. Neurosci. 27:6185–6196. 10.1523/JNEUROSCI.0628-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B., and Raff M.. 2000. A cell-intrinsic timer that operates during oligodendrocyte development. BioEssays. 22:64–71. [DOI] [PubMed] [Google Scholar]
- Evans J.A., and Davidson A.J.. 2013. Health Consequences of Circadian Disruption in Humans and Animal Models In Prog. Mol. Biol. Transl. Sci. Vol. 119 pp. 283–323. [DOI] [PubMed] [Google Scholar]
- Fong C.S., Mazo G., Das T., Goodman J., Kim M., O’Rourke B.P., Izquierdo D., and Tsou M.F.B.. 2016. 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. eLife. 5 e16270 10.7554/eLife.16270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerardin J., Reddy N.R., and Lim W.A.. 2019. The Design Principles of Biochemical Timers: Circuits that Discriminate between Transient and Sustained Stimulation. Cell Syst. 9:297–308.e2. 10.1016/j.cels.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golombek D.A., and Rosenstein R.E.. 2010. Physiology of circadian entrainment. Physiol. Rev. 90:1063–1102. 10.1152/physrev.00009.2009 [DOI] [PubMed] [Google Scholar]
- Heinzel S., Binh Giang T., Kan A., Marchingo J.M., Lye B.K., Corcoran L.M., and Hodgkin P.D.. 2017. A Myc-dependent division timer complements a cell-death timer to regulate T cell and B cell responses. Nat. Immunol. 18:96–103. 10.1038/ni.3598 [DOI] [PubMed] [Google Scholar]
- Hellmuth S., and Stemmann O.. 2020. Separase-triggered apoptosis enforces minimal length of mitosis. Nature. 580:542–547. 10.1038/s41586-020-2187-y [DOI] [PubMed] [Google Scholar]
- Huang S., Jiao X., Lu D., Pei X., Qi D., and Li Z.. 2020. Recent advances in modulators of circadian rhythms: an update and perspective. J. Enzyme Inhib. Med. Chem. 35:1267–1286. 10.1080/14756366.2020.1772249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubaud A., and Pourquié O.. 2014. Signalling dynamics in vertebrate segmentation. Nat. Rev. Mol. Cell Biol. 15:709–721. 10.1038/nrm3891 [DOI] [PubMed] [Google Scholar]
- Jagannath A., Taylor L., Wakaf Z., Vasudevan S.R., and Foster R.G.. 2017. The genetics of circadian rhythms, sleep and health. Hum. Mol. Genet. 26(R2):R128–R138. 10.1093/hmg/ddx240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., and Kobayashi T.. 2007. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 134:1243–1251. 10.1242/dev.000786 [DOI] [PubMed] [Google Scholar]
- Kohajda Z., Loewe A., Tóth N., Varró A., and Nagy N.. 2020. The Cardiac Pacemaker Story-Fundamental Role of the Na+/Ca2+ Exchanger in Spontaneous Automaticity. Front. Pharmacol. 11:516 10.3389/fphar.2020.00516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körtner G., and Geiser F.. 2000. The temporal organization of daily torpor and hibernation: circadian and circannual rhythms. Chronobiol. Int. 17:103–128. 10.1081/CBI-100101036 [DOI] [PubMed] [Google Scholar]
- Kräuchi K. 2002. How is the circadian rhythm of core body temperature regulated? Clin. Auton. Res. 12:147–149. 10.1007/s10286-002-0043-9 [DOI] [PubMed] [Google Scholar]
- Lake D., Corrêa S.A.L., and Müller J.. 2016. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell. Mol. Life Sci. 73:4397–4413. 10.1007/s00018-016-2297-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus B.G., and Holland A.J.. 2017. A New Mode of Mitotic Surveillance. Trends Cell Biol. 27:314–321. 10.1016/j.tcb.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus B.G., Daggubati V., Uetake Y., Scott P.M., Clutario K.M., Sluder G., and Holland A.J.. 2016. A USP28-53BP1-p53-p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J. Cell Biol. 214:143–153. 10.1083/jcb.201604054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane K., Van Valen D., DeFelice M.M., Macklin D.N., Kudo T., Jaimovich A., Carr A., Meyer T., Pe’er D., Boutet S.C., et al. 2017. Measuring Signaling and RNA-Seq in the Same Cell Links Gene Expression to Dynamic Patterns of NF-κB Activation. Cell Syst. 4:458–469.e5. 10.1016/j.cels.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhou M., Hu Q., Bai X.C., Huang W., Scheres S.H.W., and Shi Y.. 2017. Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme. Proc. Natl. Acad. Sci. USA. 114:1542–1547. 10.1073/pnas.1620626114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.C., Richard A.J., Datta K., and LiCata V.J.. 2008. Prevalence of temperature-dependent heat capacity changes in protein-DNA interactions. Biophys. J. 94:3258–3265. 10.1529/biophysj.107.117697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malladi S., Challa-Malladi M., Fearnhead H.O., and Bratton S.B.. 2009. The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 28:1916–1925. 10.1038/emboj.2009.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M., Yamanaka Y., Uemura M., Osawa M., Saito M.K., Nagahashi A., Nishio M., Guo L., Ikegawa S., Sakurai S., et al. 2020. Recapitulating the human segmentation clock with pluripotent stem cells. Nature. 580:124–129. 10.1038/s41586-020-2144-9 [DOI] [PubMed] [Google Scholar]
- Meitinger F., Anzola J.V., Kaulich M., Richardson A., Stender J.D., Benner C., Glass C.K., Dowdy S.F., Desai A., Shiau A.K., et al. 2016. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J. Cell Biol. 214:155–166. 10.1083/jcb.201604081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D.O. 1997. Cyclin-Dependent Kinases: Engines, Clocks, and Microprocessors In Annu. Rev. Cell Dev. Biol. Vol. 13 pp. 261–291. [DOI] [PubMed] [Google Scholar]
- Morris C.J., Aeschbach D., and Scheer F.A.J.L.. 2012. Circadian system, sleep and endocrinology. Mol. Cell. Endocrinol. 349:91–104. 10.1016/j.mce.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami R., Miyake A., Iwase R., Hayashi F., Uzumaki T., and Ishiura M.. 2008. ATPase activity and its temperature compensation of the cyanobacterial clock protein KaiC. Genes Cells. 13:387–395. 10.1111/j.1365-2443.2008.01174.x [DOI] [PubMed] [Google Scholar]
- Nakajima M., Imai K., Ito H., Nishiwaki T., Murayama Y., Iwasaki H., Oyama T., and Kondo T.. 2005. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 308:414–415. 10.1126/science.1108451 [DOI] [PubMed] [Google Scholar]
- Narasimamurthy R., and Virshup D.M.. 2017. Molecular mechanisms regulating temperature compensation of the circadian clock. Front. Neurol. 8:161 10.3389/fneur.2017.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaumer-Streit B., Forneris C.A., Morgan L.C., Van Noord M.G., Gaynes B.N., Greenblatt A., Wipplinger J., Lux L.J., Winkler D., and Gartlehner G.. 2019. Light therapy for preventing seasonal affective disorder. Cochrane Database Syst. Rev. 3 CD011269 10.1002/14651858.CD011269.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates A.C. 2020. Waiting on the Fringe: cell autonomy and signaling delays in segmentation clocks. Curr. Opin. Genet. Dev. 63:61–70. 10.1016/j.gde.2020.04.008 [DOI] [PubMed] [Google Scholar]
- Ode K.L., and Ueda H.R.. 2018. Design principles of phosphorylation-dependent timekeeping in eukaryotic circadian clocks. Cold Spring Harb. Perspect. Biol. 10 a028357 10.1101/cshperspect.a028357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Örd M., and Loog M.. 2019. How the cell cycle clock ticks. Mol. Biol. Cell. 30:169–172. 10.1091/mbc.E18-05-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S. 2016. Circadian physiology of metabolism. Science. 354:1008–1015. 10.1126/science.aah4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering J., Rich C.A., Stainton H., Aceituno C., Chinnaiya K., Saiz-Lopez P., Ros M.A., and Towers M.. 2018. An intrinsic cell cycle timer terminates limb bud outgrowth. eLife. 7 e37429 10.7554/eLife.37429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittayakanchit W., Lu Z., Chew J., Rust M.J., and Murugan A.. 2018. Biophysical clocks face a trade-off between internal and external noise resistance. eLife. 7 e37624 10.7554/eLife.37624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portolés S., and Más P.. 2010. The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 6 e1001201 10.1371/journal.pgen.1001201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourquie O. 2001. The vertebrate segmentation clock. J. Anat. 199:169–175. 10.1046/j.1469-7580.2001.19910169.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis J.E., and Lahav G.. 2013. Encoding and decoding cellular information through signaling dynamics. Cell. 152:945–956. 10.1016/j.cell.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Gelens L., and Bollen M.. 2019. Coordination of Timers and Sensors in Cell Signaling. BioEssays. 41 e1800217 10.1002/bies.201800217 [DOI] [PubMed] [Google Scholar]
- Raff M. 2007. Intracellular developmental timers. Cold Spring Harb. Symp. Quant. Biol. 72:431–435. 10.1101/sqb.2007.72.007 [DOI] [PubMed] [Google Scholar]
- Renner F., and Schmitz M.L.. 2009. Autoregulatory feedback loops terminating the NF-kappaB response. Trends Biochem. Sci. 34:128–135. 10.1016/j.tibs.2008.12.003 [DOI] [PubMed] [Google Scholar]
- Rijo-ferreira F., Acosta-rodriguez V.A., Abel J.H., and Kornblum I.. 2020. The malaria parasite has an intrinsic clock. Science. 368:746–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselló-Díez A., Arques C.G., Delgado I., Giovinazzo G., and Torres M.. 2014. Diffusible signals and epigenetic timing cooperate in late proximo-distal limb patterning. Development. 141:1534–1543. 10.1242/dev.106831 [DOI] [PubMed] [Google Scholar]
- Samach A., and Coupland G.. 2000. Time measurement and the control of flowering in plants. BioEssays. 22:38–47. [DOI] [PubMed] [Google Scholar]
- Sen S., Cheng Z., Sheu K.M., Chen Y.H., and Hoffmann A.. 2020. Gene Regulatory Strategies that Decode the Duration of NFκB Dynamics Contribute to LPS- versus TNF-Specific Gene Expression. Cell Syst. 10:169–182.e5. 10.1016/j.cels.2019.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalit-Kaneh A., Kumimoto R.W., Filkov V., and Harmer S.L.. 2018. Multiple feedback loops of the Arabidopsis circadian clock provide rhythmic robustness across environmental conditions. Proc. Natl. Acad. Sci. USA. 115:7147–7152. 10.1073/pnas.1805524115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba C.J., Andrade R.P., and Palmeirim I.. 2014. Limb patterning: from signaling gradients to molecular oscillations. J. Mol. Biol. 426:780–784. 10.1016/j.jmb.2013.11.022 [DOI] [PubMed] [Google Scholar]
- Singh A., and Soltani M.. 2013. Quantifying intrinsic and extrinsic variability in stochastic gene expression models. PLoS One. 8 e84301 10.1371/journal.pone.0084301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.K., Bhalerao R.P., and Eriksson M.E.. 2020. Growing in time: Exploring the molecular mechanisms of tree growth. Tree Physiol. tpaa065 10.1093/treephys/tpaa065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan J.A., Golden S.S., LiWang A., and Partch C.L.. 2018. Structure, function, and mechanism of the core circadian clock in cyanobacteria. J. Biol. Chem. 293:5026–5034. 10.1074/jbc.TM117.001433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornquist S.C., Langer K., Zhang S.X., Rogulja D., and Crickmore M.A.. 2020. CaMKII Measures the Passage of Time to Coordinate Behavior and Motivational State. Neuron. 105:334–345.e9. 10.1016/j.neuron.2019.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., and Sluder G.. 2010. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr. Biol. 20:1666–1671. 10.1016/j.cub.2010.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Isejima H., Matsuo T., Okura R., Yagita K., Kobayashi M., and Okamura H.. 2003. Synchronization of Cellular Clocks in the Suprachiasmatic Nucleus. Science. 302:1408–1412. 10.1126/science.1089287 [DOI] [PubMed] [Google Scholar]
- Zhang D., Tang Z., Huang H., Zhou G., Cui C., Weng Y., Liu W., Kim S., Lee S., Perez-Neut M., et al. 2019. Metabolic regulation of gene expression by histone lactylation. Nature. 574:575–580. 10.1038/s41586-019-1678-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Zhang Q., Pan Y., Mace E.M., York B., Antoulas A.C., Dacso C.C., and O’Malley B.W.. 2017. A Cell-Autonomous Mammalian 12 hr Clock Coordinates Metabolic and Stress Rhythms. Cell Metab. 25:1305–1319.e9. 10.1016/j.cmet.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]