Brandt et al. establish mechanisms that target Polo-like kinase during meiotic prophase in C. elegans. CDK-1 phosphorylates a synaptonemal complex component, SYP-1, to generate docking sites for PLK-2, whose association is prevented until crossover formation to ensure homologue pairing, synapsis, and chromosome remodeling.
Abstract
Polo-like kinases (PLKs) play widely conserved roles in orchestrating meiotic chromosome dynamics. However, how PLKs are targeted to distinct subcellular localizations during meiotic progression remains poorly understood. Here, we demonstrate that the cyclin-dependent kinase CDK-1 primes the recruitment of PLK-2 to the synaptonemal complex (SC) through phosphorylation of SYP-1 in C. elegans. SYP-1 phosphorylation by CDK-1 occurs just before meiotic onset. However, PLK-2 docking to the SC is prevented by the nucleoplasmic HAL-2/3 complex until crossover designation, which constrains PLK-2 to special chromosomal regions known as pairing centers to ensure proper homologue pairing and synapsis. PLK-2 is targeted to crossover sites primed by CDK-1 and spreads along the SC by reinforcing SYP-1 phosphorylation on one side of each crossover only when threshold levels of crossovers are generated. Thus, the integration of chromosome-autonomous signaling and a nucleus-wide crossover-counting mechanism partitions holocentric chromosomes relative to the crossover site, which ultimately defines the pattern of chromosome segregation during meiosis I.
Introduction
The production of haploid gametes during meiosis is achieved by a single round of DNA replication followed by two consecutive nuclear divisions. Unique to meiosis is its protracted prophase I, in which chromosomes pair and synapse with their homologues through the formation of a zipper-like protein structure called the synaptonemal complex (SC; Bhalla and Dernburg, 2008; Page and Hawley, 2004). Concomitantly, meiotic recombination initiates with the generation of programmed DNA double-strand breaks (DSBs; de Massy, 2013). A subset of these breaks is repaired to form crossovers, which, together with sister chromatid cohesion, physically link each pair of homologues. During meiosis I, sister kinetochores are attached by microtubules from the same spindle pole (monoorientation), and the cohesion at centromeres is protected from cleavage (Petronczki et al., 2003). These rearrangements of chromosomes then enable the separation of homologues in meiosis I and later sister chromatids in meiosis II.
Polo-like kinases (PLKs) play widely conserved but diverse roles in regulating the complex chromosome behavior during meiosis. The budding yeast Cdc5 is central to crossover recombination and SC disassembly in late pachytene (Argunhan et al., 2017; Clyne et al., 2003; Sourirajan and Lichten, 2008). It also controls the monoorientation of sister kinetochores (Galander et al., 2019) and stepwise loss of cohesin during meiosis (Attner et al., 2013; Brar et al., 2006; Katis et al., 2010). In Caenorhabditis elegans, a meiosis-specific PLK-2 is localized to special chromosomal regions known as pairing centers and promotes homologue pairing and synapsis (Harper et al., 2011; Labella et al., 2011). PLK-2 subsequently relocates to the SC in pachytene and controls DSB formation, SC disassembly, and meiotic chromosome segregation (Harper et al., 2011; Nadarajan et al., 2017; Sato-Carlton et al., 2018). Similarly, mammalian PLK-1 localizes along the SC and functions in SC disassembly (Jordan et al., 2012).
The multifaceted functions of PLKs raise the question of how PLKs are dynamically targeted to distinct structures during meiotic progression. PLKs are recruited to specific subcellular locations using their Polo-box domain (PBD), which binds a phosphorylated protein through a short peptide motif (S-[pS/pT]-P/X; Elia et al., 2003). Thus, generation of these phosphosites by a priming kinase provides an initial layer of PLK regulation. In C. elegans, recruitment of PLK-2 to pairing centers is primed by CHK-2, a meiosis-specific Ser/Thr kinase, which phosphorylates a family of zinc-finger proteins (HIM-8, ZIM-1, ZIM-2, and ZIM-3; Kim et al., 2015) that bind specific DNA sequences enriched at pairing centers (Phillips and Dernburg, 2006; Phillips et al., 2005). PLK-2 localization to pairing centers is also promoted by the nucleoplasmic protein complex HAL-2/3, although the mechanism of its action remains unclear (Roelens et al., 2019; Zhang et al., 2012). Recent work has demonstrated that recruitment of PLK-2 to the SC occurs through binding a PBD-binding motif in the C-terminal domain of an SC component, SYP-1 (Nadarajan et al., 2017; Pattabiraman et al., 2017; Sato-Carlton et al., 2018; Fig. 1, A and B). However, the identity of the upstream kinase responsible for SYP-1 phosphorylation is unknown.
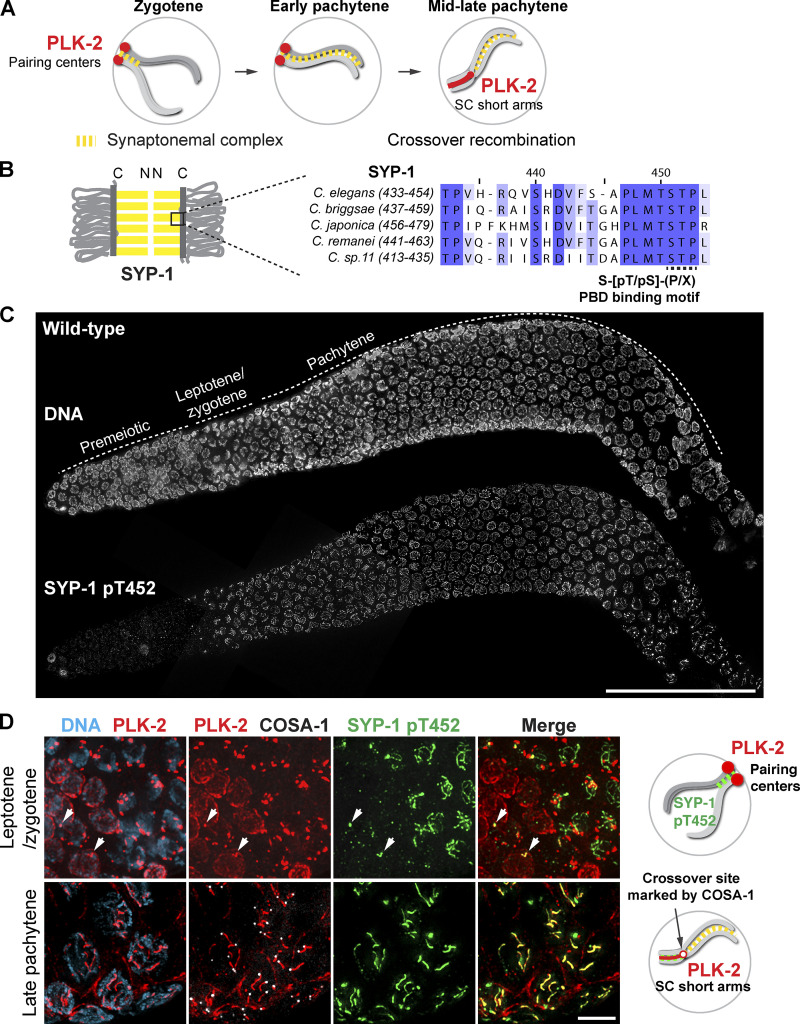
Figure 1.
PLK-2 is preferentially targeted to pairing centers in early meiotic prophase despite the presence of SYP-1 T452 phosphorylation in the C. elegans germline. (A) A schematic illustrating the dynamic localization of PLK-2 during meiotic prophase in C. elegans. (B) A schematic and sequence alignment showing the conserved PBD-binding motif within the C-terminal region of SYP-1. C, C-terminus. N, N-terminus. (C) Composite projection images of a whole gonad dissected from wild-type hermaphrodite and stained for DNA and SYP-1 pT452. Scale bar, 50 µm. (D) Immunofluorescence images of leptotene/zygotene nuclei (upper panels) and late pachytene nuclei (lower panels) showing DNA (blue), PLK-2 (red), COSA-1 (white), and SYP-1 pT452 (green). Arrows indicate the localization of PLK-2 to polycomplexes in early meiotic prophase. Scale bar, 5 µm. Diagrams illustrating the results are shown on the right.
In the holocentric C. elegans, a single crossing-over event partitions the chromosome into the short and long arms and distributes key chromosome-associated proteins asymmetrically relative to the crossover site, eventually defining where cohesion will be released during meiosis I (Altendorfer et al., 2020; Martinez-Perez et al., 2008; Nabeshima et al., 2005). While two axis components, HTP-3 and HIM-3, remain associated with all arms of chromosomes (Zetka et al., 1999; Goodyer et al., 2008; Severson et al., 2009), the other axis-associated proteins, HTP-1/2 and LAB-1, become restricted to the long arm of a bivalent (de Carvalho et al., 2008; Martinez-Perez et al., 2008). The SC components in C. elegans (SYP-1, -2, -3, -4, -5, and -6; Colaiácovo et al., 2003; Hurlock et al., 2020; MacQueen et al., 2002; Smolikov et al., 2007; Smolikov et al., 2009; Zhang et al., 2020) and two meiotic RING finger proteins, ZHP-1/2 (Zhang et al., 2018), are reciprocally restricted to the short arm. The short arm ultimately recruits the Aurora B kinase, AIR-2, in maturing oocytes in a manner dependent on Haspin kinase and the cyclin-dependent kinase CDK-1 (Ferrandiz et al., 2018), which plays an essential role in the releasing of sister chromatid cohesion during meiosis I (Kaitna et al., 2002; Rogers et al., 2002). Recent evidence has shown that the N-terminus of HTP-1/2 is required for localization of LAB-1 to the long arm (Ferrandiz et al., 2018), which in turn recruits protein phosphatase 1 (PP1) to antagonize AIR-2 recruitment (de Carvalho et al., 2008; Tzur et al., 2012). Thus, chromosome remodeling and two-step cohesion loss during meiosis are regulated by the interplay and spatial distribution of kinases and phosphatases in C. elegans.
A key unsolved question is what the initial trigger is that signals the presence of crossovers to alter chromosome architecture. One of the earliest events that differentiate the long versus short arm is the enrichment of PLK-2 on the SC short arm, which mediates axis remodeling and asymmetric SC disassembly (Harper et al., 2011; Pattabiraman et al., 2017; Sato-Carlton et al., 2018). PLK-2 can also be recruited to the crossover site (Woglar and Villeneuve, 2018), thus making it an ideal candidate to signal the presence of crossovers. However, the mechanism by which PLK-2 is targeted to the crossover site and the SC during meiotic prophase remains poorly understood.
Here, we demonstrate that CDK-1 directs the recruitment of PLK-2 to the SC through phosphorylation of SYP-1. We also identify mechanisms that prevent precocious association of PLK-2 with the SC in early meiotic prophase to ensure proper axis assembly, homologue pairing, and synapsis. Interestingly, the spatial and temporal control of asymmetric PLK-2 localization is provided by the enrichment of SYP-1 phosphorylation on the short arm, which requires PLK-2 kinase activity itself and threshold levels of crossover designation within the nucleus. Thus, the integration of chromosome-autonomous signaling and a nucleus-wide crossover-counting mechanism drives the spatial partitioning of holocentric chromosomes relative to the crossover site, which ultimately defines the pattern of chromosome segregation during meiosis I.
Results
PLK-2 docking sites are generated at the SC in early meiotic prophase
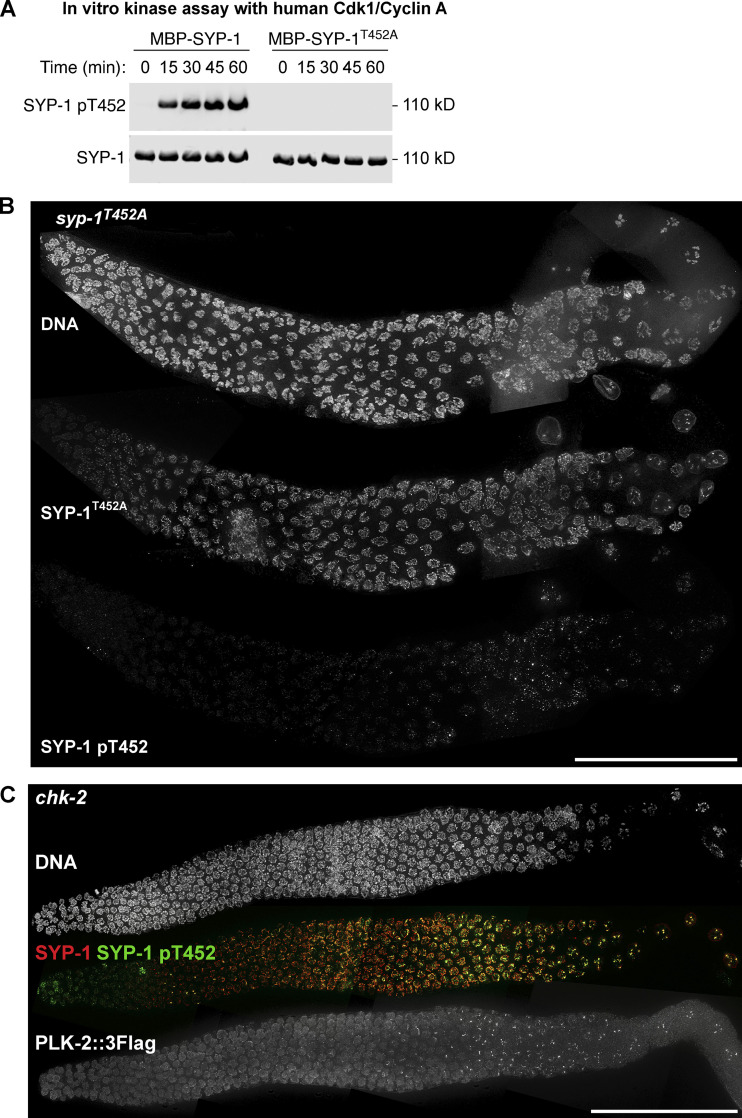
To identify the signaling cascade controlling the recruitment of PLK-2 to the SC, we generated a polyclonal antibody against the phosphopeptide surrounding SYP-1 T452 (Fig. 1 B). The affinity-purified SYP-1 pT452 antibody recognized recombinant SYP-1 only in the presence of human CDK-1/cyclin A, and recognition of phosphorylated SYP-1 was abolished by the T452A mutation (Fig. S1 A). Thus, SYP-1 T452 is readily phosphorylated by a CDK in vitro, and our antibody is specific to phosphorylated SYP-1 T452. Consistent with a previous report (Sato-Carlton et al., 2018), phosphorylation of SYP-1 at T452 was robustly detected along the SC just before meiotic entry and was later restricted to one side of each crossover, marked by a cyclin-like protein, COSA-1, which mirrors the localization of PLK-2 to the short arm in late pachytene (Fig. 1, C and D). The phospho-signal was abolished in animals homozygous for the syp-1T452A mutation (Fig. S1 B), further validating the specificity of our antibody.
Figure S1.
Phospho-specificity of the SYP-1 pT452 antibody used in this study, and the analysis of SYP-1 T452 phosphorylation and PLK-2 localization in chk-2 mutants. (A) In vitro kinase assays using human CDK-1/cyclin A to phosphorylate MBP–SYP-1 with or without the T452A mutation. Western blot using antibodies against SYP-1 and phosphorylated SYP-1 at T452 is shown. (B) Composite immunofluorescence images of a whole gonad dissected from syp-1T452A mutants showing DNA, SYP-1, and SYP-1 pT452 staining. Scale bar, 50 µm. (C) Composite immunofluorescence images of a whole CHK-2–depleted gonad showing DNA (white), SYP-1 (red), SYP-1 pT452 (green), and PLK-2::3Flag staining. Scale bar, 50 µm.
Prior to the onset of meiosis, PLK-2 was enriched at the nuclear envelope (Fig. 1 D). PLK-2 is then primarily recruited to pairing centers upon meiotic entry through PBD-binding motifs within the pairing center proteins HIM-8, ZIM-1, ZIM-2, and ZIM-3 (Harper et al., 2011; Kim et al., 2015; Labella et al., 2011). PLK-2 was occasionally detected with SC aggregates, known as polycomplexes, that were positive for SYP-1 pT452 staining in early prophase (Fig. 1 D, arrows in top panels). However, it was only after crossover designation in late pachytene that PLK-2 became localized along the SC (Fig. 1 D), despite the presence of its binding sites on SYP-1 at earlier stages.
Constrained PLK-2 activity at the pairing centers is essential for proper axis assembly, homologue pairing, and synapsis
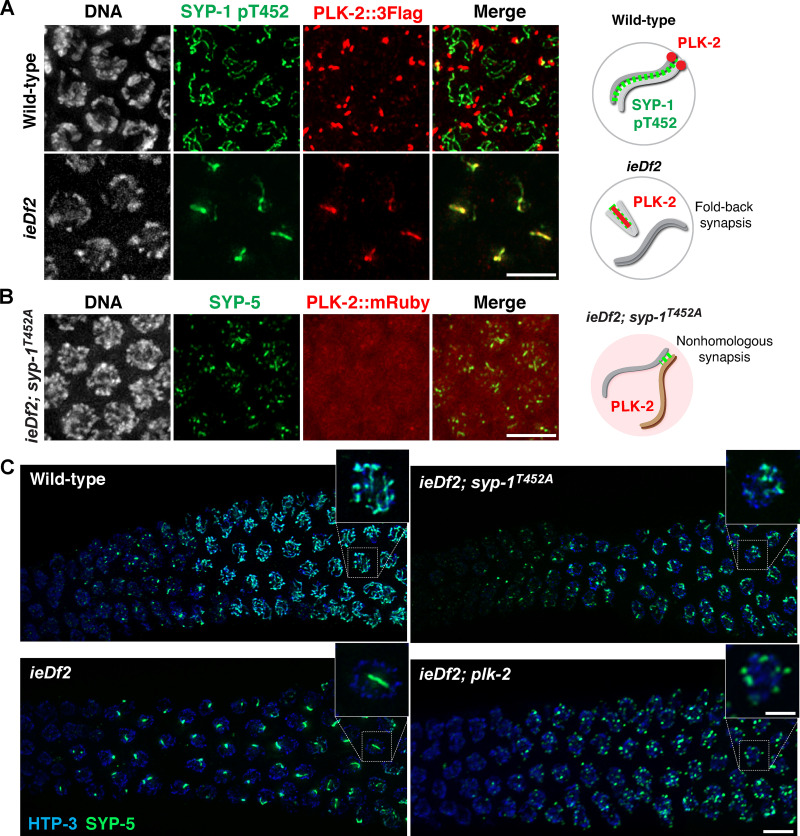
We hypothesized that pairing centers might prevent the recruitment of PLK-2 to the SC. To test this, we examined the localization of PLK-2 in the absence of pairing centers using a strain (ieDf2) that lacks all four pairing center proteins (HIM-8/ZIMs; Harper et al., 2011). In agreement with a recent report (Roelens et al., 2019), PLK-2 was immediately localized along phosphorylated SYP-1 stretches in ieDf2 animals (Fig. 2 A), which has previously been shown to represent intrachromosomal fold-back synapsis within unpaired chromosomes (Harper et al., 2011). Thus, the SC can recruit PLK-2 in the absence of pairing centers. When PLK-2 docking sites were deleted on both pairing centers and the SC by introducing the SYP-1 T452A mutation in the ieDf2 background, PLK-2 was largely detected in the nucleoplasm (Fig. 2 B). Therefore, pairing centers and the SC are the major PLK-2 docking sites available at the onset of meiosis; however, PLK-2 is preferentially targeted to pairing centers in early prophase.
Figure 2.
Constrained PLK-2 activity at the pairing centers is essential for proper synapsis. (A) Immunofluorescence images of early pachytene nuclei from wild-type and ieDf2 mutants stained for DNA (white), SYP-1 pT452 (green), and PLK-2::3Flag (red). Scale bar, 5 µm. (B) Immunofluorescence images of mid-pachytene nuclei from ieDf2; syp-1T452A mutants stained for DNA (white), SYP-1 (green), and PLK-2::mRuby (red). Scale bar, 5 µm. Diagrams illustrating the results are shown on the right. (C) Composite projection images of gonad sections from indicated genotypes, spanning from meiotic entry to mid-pachytene, showing HTP-3 (blue) and SYP-5 (green). Scale bar, 5 µm. Insets show zoomed-in view of a representative nucleus from the boxed regions. Scale bar, 2 µm.
Interestingly, we noticed that many short SC stretches appear in ieDf2; syp-1T452A animals (Fig. 2, B and C), suggesting that the fold-back synapsis in ieDf2 animals might be due to precocious association of PLK-2 with the SC. Indeed, SYP proteins existed as multiple small aggregates and exhibited a delay in forming linear stretches in ieDf2; plk-2 double mutants (Fig. 2 C), supporting the conclusion that uncontrolled action of PLK-2 on the SC causes premature loading of SYP proteins between nonhomologous chromosomes.
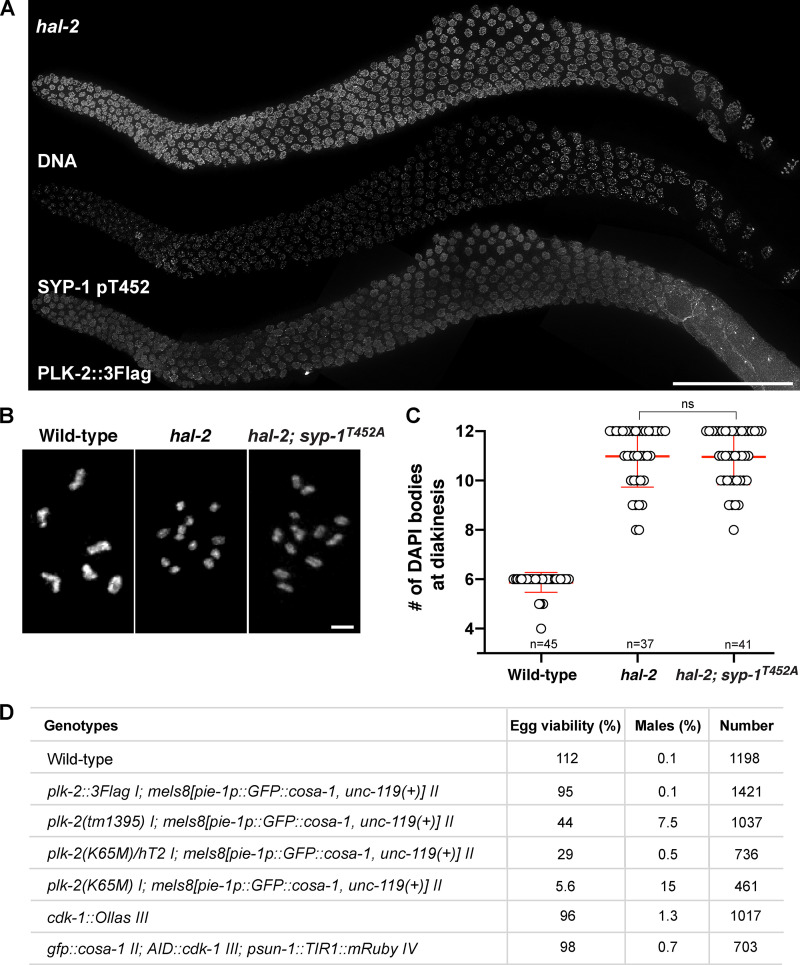
Recruitment of PLK-2 to pairing centers is primed by CHK-2–dependent phosphorylation of the pairing center proteins (Kim et al., 2015). In CHK-2–depleted animals, phosphorylation of SYP-1 T452 was diminished in early meiotic prophase, and PLK-2 was found in the nucleoplasm (Fig. S1 C). Nevertheless, robust phospho-signals for SYP-1 T452 were detected on SC stretches later in the germline, which ultimately recruited PLK-2 (Fig. S1 C). Targeting of PLK-2 to pairing centers also depends on the nucleoplasmic HAL-2/3 complex, which functions to promote full activation of CHK-2 and to antagonize the localization and activity of PLK-2 in the nucleoplasm (Roelens et al., 2019; Zhang et al., 2012). In hal-2 mutants, PLK-2 remained largely nucleoplasmic as previously reported (Roelens et al., 2019), and SYP-1 T452 phosphorylation was detected along the SC (Fig. S2 A), which has been shown to assemble onto unpaired chromosome axes (Zhang et al., 2012). Thus, PLK-2 docking sites on the SC are generated independently of CHK-2 and HAL-2/3.
Figure S2.
The SYP-1 T452A mutation does not rescue the failure to form crossovers in hal-2 mutants. (A) Composite immunofluorescence images of a whole gonad from hal-2 mutants showing DNA, SYP-1 pT452, and PLK-2::3Flag staining. Scale bar, 50 µm. (B) Oocyte nuclei at diakinesis from indicated genotypes were stained with DAPI. Scale bar, 3 µm. (C) Graph showing the number of DAPI-staining bodies in oocytes at diakinesis (mean ± SD). Numbers of nuclei scored are indicated (n = 45 for wild type; n = 37 for hal-2; n = 41 for hal-2; syp-1T452A). ns, not significant (P > 0.05) by two-tailed Mann-Whitney test. (D) Table showing the percent viable and male progeny from C. elegans hermaphrodites of indicated genotypes.
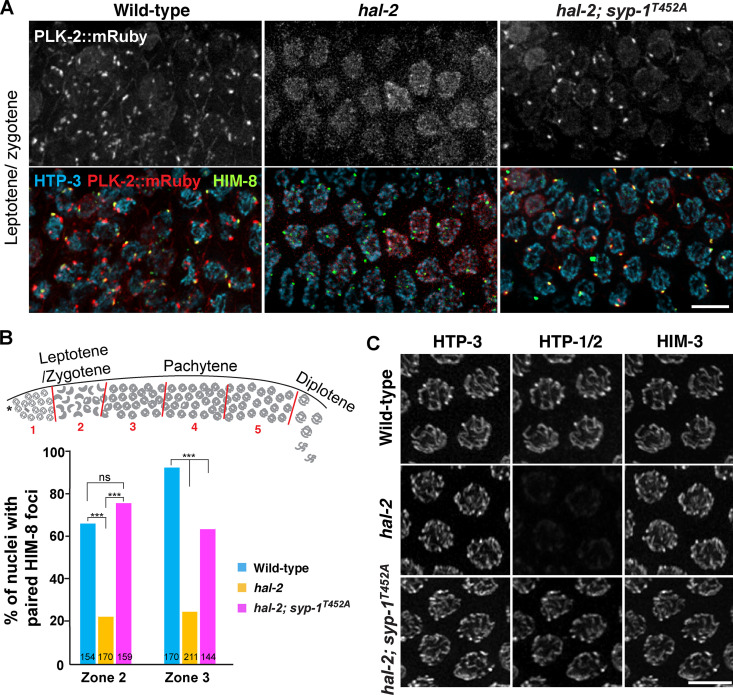
Previous evidence has shown that defects in nuclear reorganization and homologue pairing in hal-2 mutants are rescued by removal of SYP proteins (Zhang et al., 2012). We reasoned that this might be due to the loss of PLK-2 binding to the SC such that PLK-2 is then free to localize to pairing centers, restoring the nuclear reorganization required for homologue pairing. To test this hypothesis, we combined the null allele of hal-2 with the T452A mutation in SYP-1 and examined the localization of PLK-2. Strikingly, bright PLK-2 foci were detected at the X chromosome pairing center in hal-2; syp-1T452A mutants (Fig. 3 A), which largely rescued the pairing of X chromosomes (Fig. 3, A and B). In hal-2 mutants, loading of HTP-1/2 onto the chromosome axis is impaired, which is also restored by removal of SYP proteins (Zhang et al., 2012). We observed a similar rescue in chromosomal localization of HTP-1/2 in hal-2; syp-1T452A mutants throughout the germline (Fig. 3 C), indicating that a key function of the HAL-2/3 complex is to prevent precocious interactions between the SC and PLK-2 in early prophase. However, PLK-2 was rarely detected at the autosomal pairing centers in hal-2; syp-1T452A mutants, and the SYP-1 T452A mutation only partially restored the crescent-shaped nuclear morphology characteristic to the leptotene/zygotene stage (Fig. 3 A). We attribute this to the requirement of HAL-2/3 in full activation of CHK-2 (Roelens et al., 2019). Phosphorylation of HIM-8 by CHK-2 occurs more robustly than for the ZIM proteins at the autosomal pairing centers (Kim et al., 2015). Thus, the X chromosome pairing centers are more likely to recruit PLK-2 when CHK-2 activity is reduced in hal-2; syp-1T452A mutants. The SYP-1 T452A mutation also did not rescue the failure to form crossovers in hal-2 mutants (Fig. S2, B and C), consistent with the requirement of CHK-2 in generation of meiotic DSBs (MacQueen and Villeneuve, 2001). Taken together, our results demonstrate that constrained PLK-2 activity at the pairing centers is essential for proper axis assembly, homologue pairing, and synapsis.
Figure 3.
The SYP-1 T452A mutation partially rescues targeting of PLK-2 to pairing centers in hal-2 mutants. (A) Immunofluorescence images of leptotene/zygotene nuclei from wild-type, hal-2, and hal-2; syp-1T452A mutants showing PLK-2::mRuby (white or red), HTP-3 (blue), and HIM-8 (green). Scale bar, 5 µm. (B) Graph showing the percentage of nuclei with paired HIM-8 foci from the indicated genotypes. To score pairing of the X chromosome pairing center, the germline was divided into five zones as shown in the diagram. Zone 2 contains leptotene/zygotene nuclei (n = 154 for wild type; n = 170 for hal-2; n = 159 for hal-2; syp-1T452A), and zone 3 contains nuclei in early pachytene (n = 170 for wild type; n = 211 for hal-2; n = 144 for hal-2; syp-1T452A). ***, P < 0.0001; ns, not significant (P > 0.05) by χ2 analysis. (C) Immunofluorescence images of pachytene nuclei from wild-type, hal-2, and hal-2; syp-1T452A mutants showing the staining for HTP-3, HTP-1/2, and HIM-3. Scale bar, 5 µm.
Asymmetric enrichment of SYP-1 phosphorylation requires the kinase activity of PLK-2 and threshold levels of crossover designation
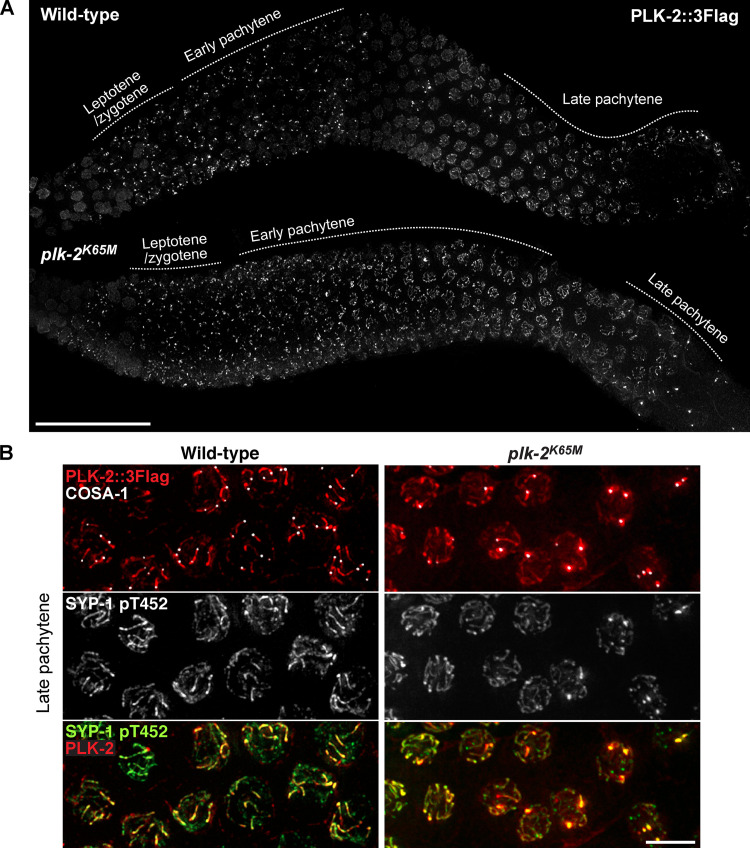
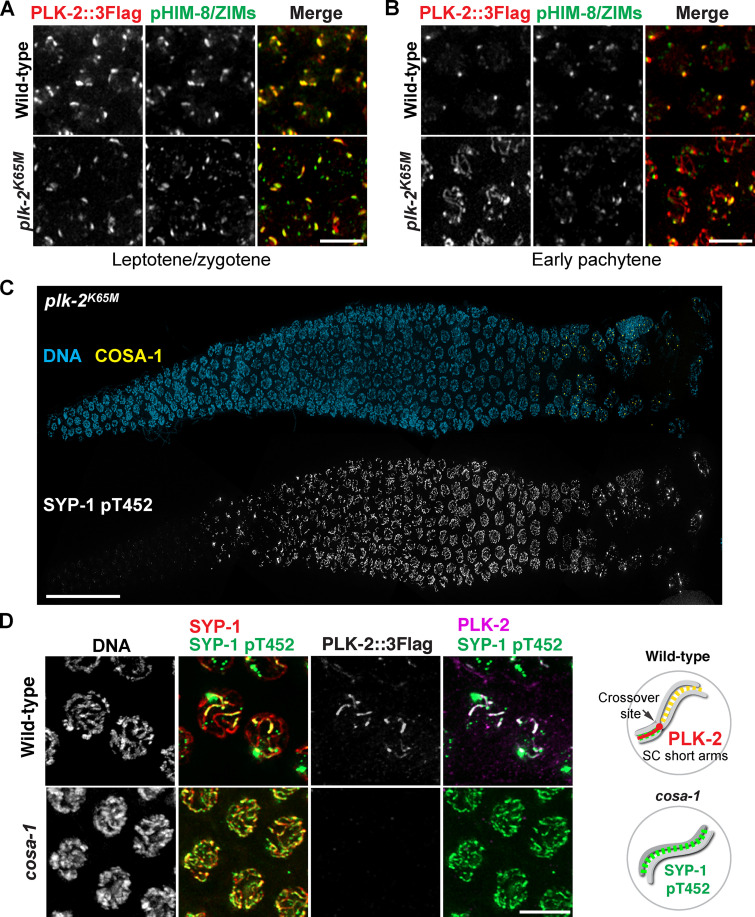
Phosphorylation of SYP-1 T452 is robustly detected upon meiotic entry in the absence of PLK-2 (Sato-Carlton et al., 2018), indicating that PLK-2 is not the kinase responsible for the initial SYP-1 T452 phosphorylation. However, the phospho-signal for SYP-1 was significantly weakened on chromosome arms after crossover designation in plk-2–null animals, suggesting that PLK-2 is necessary for enforcing its own binding to the SC (Sato-Carlton et al., 2018). To test whether PLK-2 kinase activity is required, we generated a worm strain expressing a kinase-dead form of PLK-2 (K65M; Link et al., 2018). The K65M mutation was semi-dominant, as only ∼30% of embryos from the heterozygous animal were viable (Fig. S2 D), suggesting that the presence of PLK-2K65M may interfere with the ability of PLK-1 to compensate for PLK-2 functions (Harper et al., 2011; Labella et al., 2011). Animals homozygous for the plk-2K65M mutation exhibited more aggravated defects, further reducing the embryo viability to ∼6%. The kinase-dead PLK-2 localized normally to the nuclear envelope in the premeiotic region and to pairing centers in early prophase (Fig. 4 A). However, the kinase-dead PLK-2 precociously spread to SC stretches in early pachytene (Fig. S3, A and B), hinting that PLK-2 kinase activity is required for sustaining its association with the pairing centers. Consistent with the results from plk-2–null animals, the initial phosphorylation of SYP-1 T452 was robustly detected in plk-2 K65M mutants (Fig. S3 C). However, this phospho-signal was reduced from chromosome arms after crossover designation, and both SYP-1 phosphorylation and PLK-2 were concentrated at crossover sites in plk-2K65M animals (Fig. 4, A and B; and Fig. S3 C). Thus, we conclude that PLK-2 kinase activity is required to enforce asymmetric phosphorylation of SYP-1 T452.
Figure 4.
The kinase activity of PLK-2 is required for enrichment of SYP-1 T452 phosphorylation along chromosomes that have designated crossovers. (A) Composite immunofluorescence images of whole gonads dissected from wild-type and plk-2K65M hermaphrodites and stained for PLK-2::3Flag. Scale bar, 50 µm. (B) Immunofluorescence images of late pachytene nuclei from wild-type and plk-2K65M mutants showing PLK-2::3Flag (red), COSA-1 (white), and SYP-1 pT452 (green) staining. Scale bar, 5 µm.
Figure S3.
Enrichment of SYP-1 phosphorylation on the short arm requires PLK-2 kinase activity and crossover formation. (A and B) Zoomed-in images of nuclei from leptotene/zygotene (A) and early pachytene (B) showing PLK-2::3Flag (red) and phospho-HIM-8/ZIMs (green) staining. Scale bars, 5 µm. (C) Composite immunofluorescence images of a whole gonad dissected from the plk-2K65M (kinase dead) homozygote animal showing DNA (blue), COSA-1 (yellow), and SYP-1 pT452 staining (white). Scale bar, 50 µm. (D) Late-pachytene nuclei from wild-type and cosa-1(tm3298) mutants were stained for DNA, SYP-1 (red), SYP-1 pT452 (green), and PLK-2 (magenta). Scale bar, 5 µm. Diagrams illustrating the results are shown on the right.
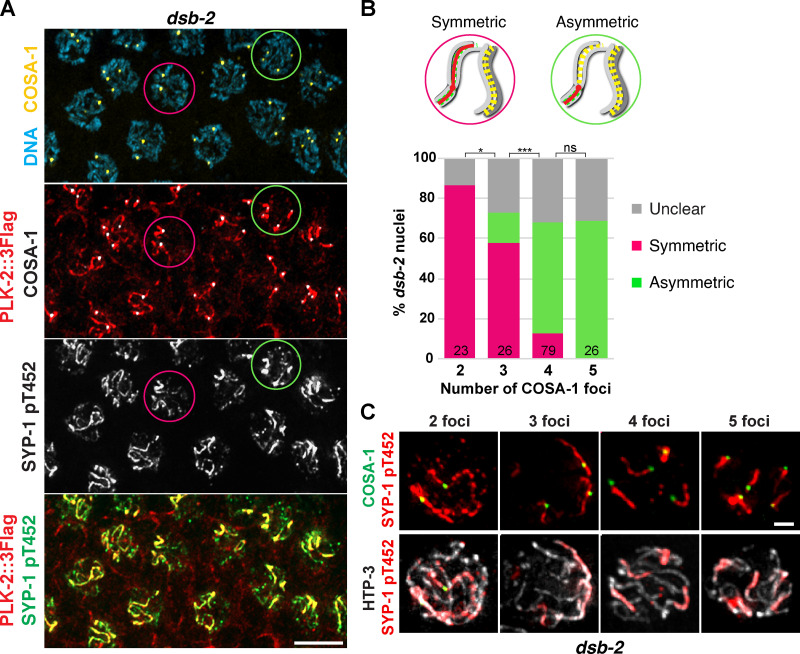
Enrichment of PLK-2 to the SC short arm depends on crossover formation, and this is important for chromosome remodeling required for homologue separation during meiosis I (Nadarajan et al., 2017; Pattabiraman et al., 2017; Sato-Carlton et al., 2018). Thus, we investigated how SYP-1 T452 phosphorylation is regulated by crossover designation. In crossover-defective cosa-1 mutants (Yokoo et al., 2012), SYP-1 T452 phosphorylation persisted along the SC, and yet PLK-2 was not recruited (Fig. S3 D). We also examined SYP-1 phosphorylation in dsb-2 mutants, in which formation of meiotic DSBs is substantially reduced, leading to crossover designation only on a few chromosomes (Rosu et al., 2013). Consistent with PLK-2 localization in dsb-2 mutants (Pattabiraman et al., 2017), phosphorylation of SYP-1 T452 was detected along chromosomes harboring COSA-1, while it was lost from chromosomes that had failed to designate crossovers (Fig. 5 A). Interestingly, we observed that the asymmetric appearance of PLK-2 and SYP-1 phosphorylation correlates with the number of crossover-designated sites within each nucleus. While most nuclei with two to three COSA-1 foci exhibited symmetric distribution of SYP-1 phosphorylation along the chromosome length (87% in nuclei with two COSA-1; 58% in nuclei with three COSA-1), the majority of nuclei with four to five COSA-1 foci showed asymmetric SYP-1 phosphorylation on one side of each crossover (56% in nuclei with four COSA-1; 69% in nuclei with five COSA-1; Fig. 5, B and C). Thus, signals from crossover-designated sites are transmitted in cis to enrich for SYP-1 phosphorylation, and asymmetric remodeling of chromosomes requires a threshold level of crossover designation within the nucleus.
Figure 5.
Threshold levels of crossovers are required for asymmetric enrichment of SYP-1 T452 phosphorylation. (A) Immunofluorescence images of late pachytene nuclei from wild-type and dsb-2 mutants showing DNA (blue), COSA-1 (yellow), PLK-2 (red), and SYP-1 pT452 (green) staining. Circles represent examples of nuclei showing either symmetric (magenta) or asymmetric (green) SYP-1 pT452/PLK-2 localization relative to COSA-1. Scale bar, 5 µm. (B) Diagrams illustrating nuclei with the symmetric versus asymmetric SYP-1 pT452 staining and a graph showing the distribution of SYP-1 pT452 staining in dsb-2 nuclei with two COSA-1 foci (n = 23), three COSA-1 foci (n = 26), four COSA-1 foci (n = 79), and five COSA-1 foci (n = 26). *, P < 0.05; ***, P < 0.0001; ns, not significant (P > 0.05) by Fisher’s exact test. (C) Late pachytene nuclei from dsb-2 mutants were stained for SYP-1 pT452 (red), COSA-1 (green), and HTP-3 (white) and categorized by the number of COSA-1 foci. Scale bar, 1 µm.
CDK-1 is responsible for SYP-1 phosphorylation at T452
We next sought to identify the upstream kinase responsible for the phosphorylation of SYP-1 T452. Docking sites for PLKs often overlap with consensus phosphorylation motifs for CDKs (Archambault and Glover, 2009), and SYP-1 T452 is indeed an excellent substrate for recombinant human CDK-1 in vitro (Fig. S1 A). Thus, we hypothesized that a CDK might phosphorylate SYP-1 to prime the recruitment of PLK-2 to the SC.
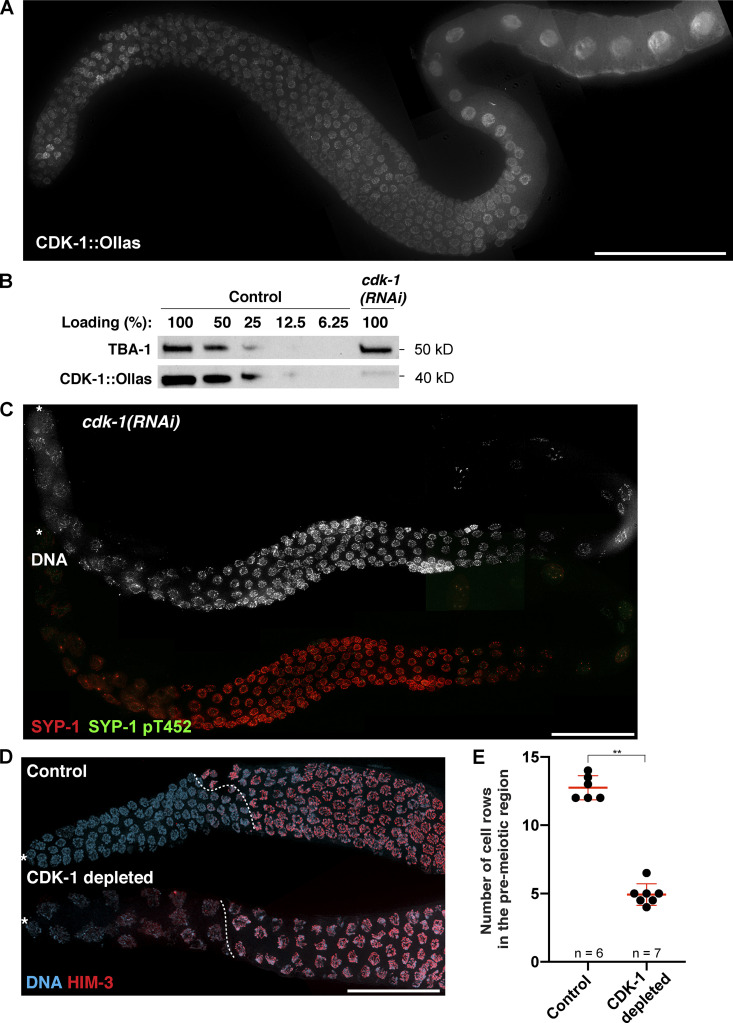
Recent evidence has shown that CDK-1 controls the recruitment of AIR-2 to the short arm (Ferrandiz et al., 2018), raising the possibility that CDK-1 might play a role in chromosome remodeling upon crossover designation. Thus, we tagged the endogenous CDK-1 with a small epitope Ollas at its C-terminus and examined its expression by immunofluorescence. Consistent with its potential functions in the germline, CDK-1 was robustly expressed in nuclei at all stages of meiotic prophase (Fig. S4 A). We first tested its significance by RNAi. The knockdown efficiency was determined to be >85% by comparing the lysates of RNAi-treated worms to serially diluted control samples in a Western blot (Fig. S4 B). Animals treated with cdk-1 RNAi displayed fewer enlarged nuclei in the premeiotic region, a phenotype characteristic of endoduplication after failed mitotic entry (Edgar and Orr-Weaver, 2001), and defects in chromosome condensation in diplotene (Fig. S4 C). Importantly, knockdown of cdk-1 completely abolished SYP-1 T452 phosphorylation (Fig. S4 C), demonstrating that CDK-1 is required for the phosphorylation of SYP-1 T452.
Figure S4.
Knockdown of CDK-1 by RNAi abolishes the phosphorylation of SYP-1 at T452 and abrogates PLK-2 targeting to the SC. (A) Composite immunofluorescence image of a full-length gonad dissected from a worm strain expressing CDK-1::Ollas. Scale bar, 50 µm. (B) Western blots showing the knockdown of CDK-1 by RNAi. The level of CDK-1::Ollas in cdk-1(RNAi) worms was compared with serially diluted worm lysates from control animals. Tubulin (TBA-1) was used as a loading control. Molecular weights for both proteins are indicated on the right. (C) Composite immunofluorescence images of a full-length gonad from a cdk-1 RNAi–treated worm showing DNA (white), SYP-1 (red), and SYP-1 pT452 staining (green). Asterisks indicate the distal tip of the germline. Scale bar, 50 µm. (D) Immunofluorescence images of distal germlines from control and CDK-1–depleted animals showing DNA (blue) and HIM-3 (red) staining. Asterisks indicate the distal tip of the germline, and dotted lines indicate the meiotic entry. Scale bar, 50 µm. (E) Graph showing the number of cell rows in the premeiotic region in control versus CDK-1–depleted germline. Mean ± SD is shown. Numbers of gonads scored are indicated on the bottom (n = 6 for control; n = 7 for CDK-1 depleted). **, P < 0.01 by two-tailed Mann-Whitney test.
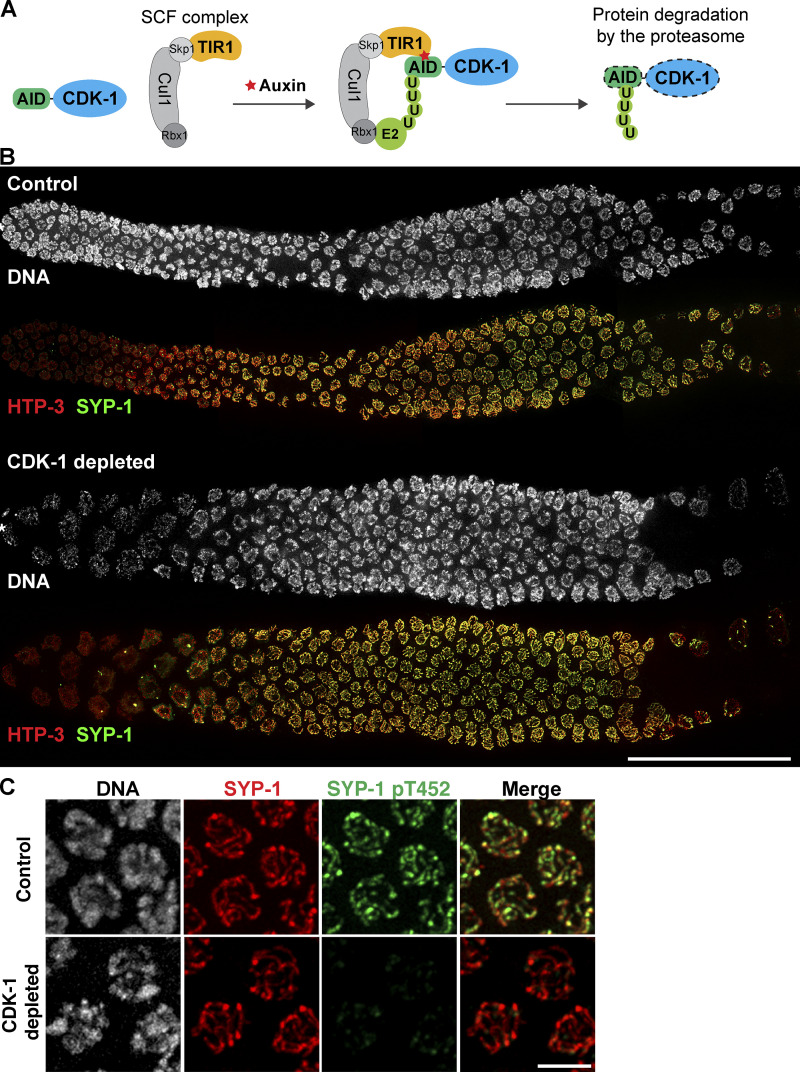
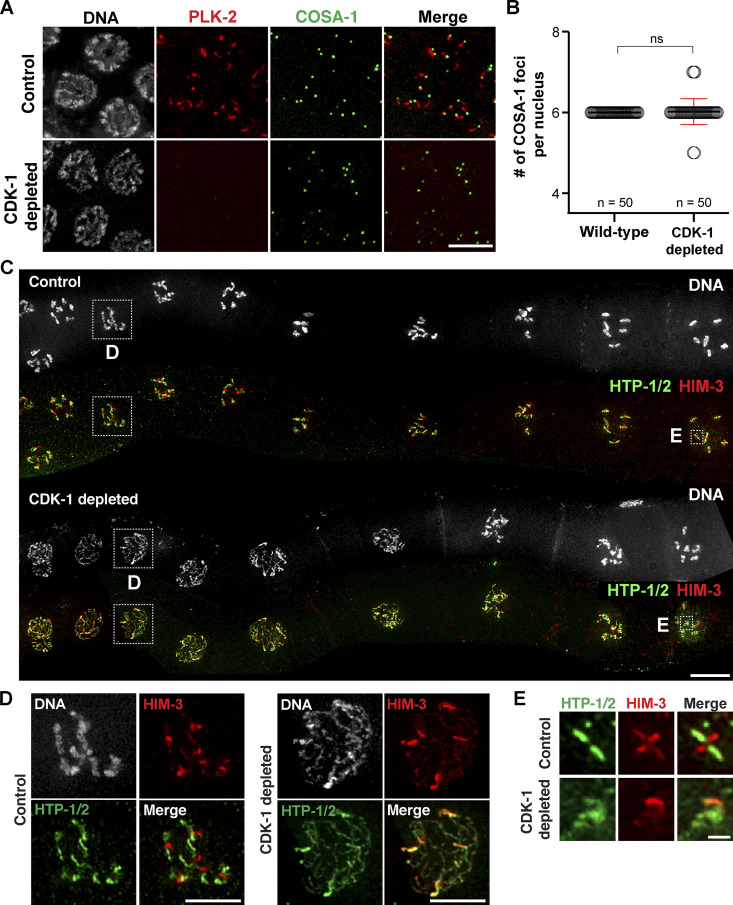
We also employed the auxin-inducible degradation (AID) system (Zhang et al., 2015) to confirm the requirement for CDK-1 in SYP-1 phosphorylation. To this end, we inserted the AID tag at the N-terminus of endogenous CDK-1 in a genetic background that expresses the plant F-box protein TIR1 from the germline-specific sun-1 promoter (Fig. 6 A). Self-progeny of this worm strain was largely viable (98% egg viability; Fig. S2 D), indicating that the AID tag does not interfere with CDK-1 functions. Consistent with the RNAi results, fewer enlarged nuclei were observed in the premeiotic region of CDK-1–depleted germlines (Fig. S4, D and E). Depletion of CDK-1 did not alter the recruitment of PLK-2 to pairing centers (Fig. S5, A and B), nor did it affect synapsis and crossover formation (Fig. 6 B), as germlines lacking CDK-1 displayed the requisite six COSA-1 foci on average in late pachytene and six DAPI-staining bodies at diakinesis (Fig. 7, A–C). However, phosphorylation of SYP-1 T452 was completely eliminated in CDK-1–depleted animals (Fig. 6 C), corroborating the conclusion that CDK-1 is responsible for phosphorylating SYP-1 at T452.
Figure 6.
CDK-1 is required for chromosome remodeling after crossover formation. (A) A schematic illustrating the strategy to deplete CDK-1 in the adult germline using the AID system. U, ubiquitination. (B) Whole gonads dissected from control and CDK-1–depleted animals were stained for DNA (white), HTP-3 (red), and SYP-1 (green). Composite immunofluorescence images are shown. Scale bar, 50 µm. (C) Immunofluorescence of mid-pachytene nuclei from control and CDK-1–depleted animals showing DNA (white), SYP-1 (red), and SYP-1 pT452 (green). Scale bar, 5 µm.
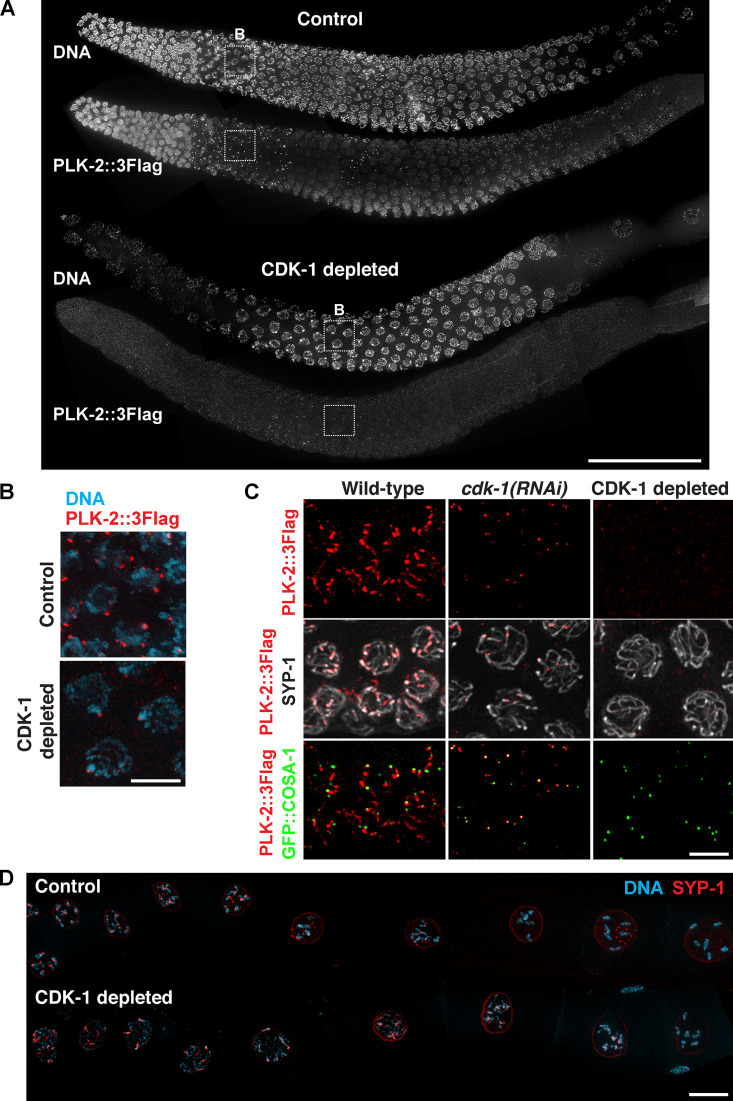
Figure S5.
Depletion of CDK-1 in the germline by AID system leads to failures in targeting PLK-2 to distinct subnuclear structures. (A) Full-length gonads dissected from control versus CDK-1–depleted animals were stained for DNA and PLK-2::3Flag. Scale bar, 50 µm. (B) Zoomed-in images of the boxed regions in A showing DNA (blue) and PLK-2 (red) staining from control and CDK-1–depleted germlines during leptotene/zygotene. Scale bar, 5 µm. (C) Immunofluorescence of late-pachytene nuclei from wild-type, cdk-1(RNAi), and CDK-1–depleted worms showing SYP-1, PLK-2 (red), and COSA-1 (green). Scale bar, 5 µm. (D) Composite immunofluorescence images of diakinesis nuclei from control versus CDK-1–depleted germline showing DNA (blue) and SYP-1 (red). Scale bar, 5 µm.
Figure 7.
CDK-1 is required for chromosome remodeling after crossover formation. (A) Immunofluorescence images of late-pachytene nuclei from control and CDK-1–depleted animals showing DNA (white), PLK-2 (red), and COSA-1 (green) staining. Scale bar, 5 µm. (B) Graph showing the quantification of COSA-1 number per nucleus in wild-type (n = 50) and CDK-1–depleted animals (n = 50). Mean ± SD is shown. ns, not significant (P > 0.05) by two-tailed Mann-Whitney test. (C) Composite immunofluorescence images of diakinesis nuclei from control and CDK-1–depleted animals showing DNA (white), HTP-1/2 (green), and HIM-3 (red) staining. Scale bar, 10 µm. (D) Zoomed-in images of a diplotene nucleus from control and CDK-1–depleted germlines as indicated in C. Scale bars, 5 µm. (E) Zoomed-in images of an individual chromosome in diakinesis from control and CDK-1–depleted germlines as indicated in C. Scale bar, 1 µm.
CDK-1 is required for targeting PLK-2 to the SC short arm and for chromosome remodeling
As a consequence of the failure to phosphorylate SYP-1 T452 in CDK-1–depleted germlines, PLK-2 was unable to localize to the SC even after crossover designation (Fig. 7 A and Fig. S5, A and C). In addition, PLK-2 was no longer observed at the nuclear periphery in the premeiotic region of these animals (Fig. S5 A), indicating that CDK-1 is also responsible for targeting PLK-2 to the nuclear envelope, similar to the PLK-1 regulation in mitotic prophase (Linder et al., 2017; Martino et al., 2017). Interestingly, recruitment of PLK-2 to crossover sites was also abolished when CDK-1 was depleted by auxin (Fig. 7 A), while this was not observed in cdk-1 RNAi-treated animals (Fig. S5 C). We attribute this to a stronger CDK-1 depletion by the AID system and conclude that CDK-1 primes the recruitment of PLK-2 not only to the SC, but also to the crossover site.
Recruitment of PLK-2 to the SC short arm is essential for SC disassembly and axis remodeling in late prophase (Harper et al., 2011; Sato-Carlton et al., 2018). In the wild type, HTP-1/2 was removed from the short arm in diplotene (Martinez-Perez et al., 2008), whereas HIM-3 and HTP-3 remained on all chromosome arms (Goodyer et al., 2008; Zetka et al., 1999). In CDK-1–depleted animals, however, HTP-1/2 persisted on the short arm (Fig. 7, C and D), and SC disassembly was delayed until diakinesis (Fig. S5 D). Thus, CDK-1 is required for targeting PLK-2 to the SC during meiotic prophase, which drives the asymmetric remodeling of holocentric chromosomes. While chromosomes were continually compacted in the wild type to eventually yield the cruciform-shaped bivalent structure, chromosomes in CDK-1–depleted oocytes appeared diffuse and lacked distinct bivalent structure in diakinesis (Fig. 7, C and E). This is distinct from the phenotypes observed in syp-1T452A animals and likely reflects the additional requirement of CDK-1 in chromosome condensation (Bazile et al., 2010).
Discussion
The dynamic localization of PLK-1 during mitosis is largely controlled by the interaction between the PBD within PLK-1 to a short peptide motif that has been phosphorylated by CDK-1 (Archambault and Glover, 2009). Here, we demonstrate that this general paradigm also applies to PLK-2 targeting during meiosis in C. elegans. Previous work using RNAi had shown a requirement for CDK-1 in germ cell mitosis; however, it did not reveal its functions during meiotic prophase (Boxem et al., 1999). We have now established that CDK-1 primes the recruitment of PLK-2 to the nuclear envelope in the premeiotic region as well as to crossover sites and the SC in pachytene, thereby driving the chromosome remodeling essential for proper chromosome segregation during meiosis I (Fig. 8).
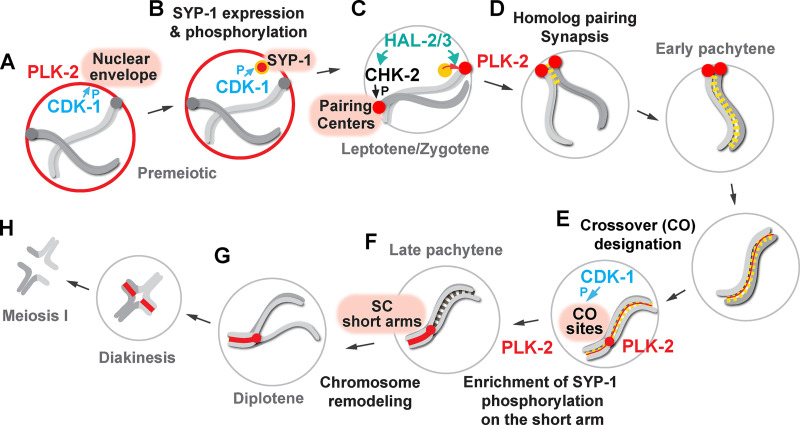
Figure 8.
Model for targeting PLK-2 to distinct subnuclear structures during meiotic prophase in C. elegans. (A) In the premeiotic region of the germline, CDK-1 primes the localization of PLK-2 to the nuclear envelope. P, phosphorylation. (B) CDK-1 phosphorylates newly expressed SYP-1 at T452 just before the meiotic onset, and a pool of PLK-2 can localize to SC polycomplexes. (C) Upon meiotic entry, CHK-2 becomes active and phosphorylates the pairing center proteins (Kim et al., 2015), which serve as the preferred docking sites for PLK-2. The nucleoplasmic HAL-2/3 complex ensures the PLK-2 localization to pairing centers by promoting CHK-2 activity and by preventing premature association of PLK-2 to SYP proteins. (D) Constrained PLK-2 activity at the pairing centers is essential for proper axis assembly, homologue pairing, and synapsis. (E) CDK-1 is also responsible for targeting PLK-2 to the crossover-designated sites. (F) SYP-1 phosphorylation and PLK-2 are enriched on the SC short arm relative to the crossover site, and this requires PLK-2 kinase activity and a threshold level of crossover within the nucleus. (G) PLK-2 drives the asymmetric SC disassembly. (H) The short arm becomes the site of cohesion loss during meiosis I.
Our evidence shows that two major PLK-2 docking sites are generated in early meiotic prophase: (1) the SC, which is primed by CDK-1–dependent phosphorylation of SYP-1, and (2) the pairing centers, which are primed by CHK-2–dependent phosphorylation of HIM-8/ZIMs (Kim et al., 2015). Interestingly, PLK-2 is preferentially targeted to the pairing centers, where it acts locally to drive homologue pairing and synapsis (Harper et al., 2011; Labella et al., 2011). Uncontrolled action of PLK-2 on the SC is deleterious, as exemplified in ieDf2 and hal-2 mutants, leading to erroneous loading of SYP proteins between unpaired chromosome axes (Harper et al., 2011; Roelens et al., 2019; Zhang et al., 2012). Remarkably, mutating the PBD-binding motif on SYP-1 rescues PLK-2 targeting to the pairing centers in hal-2 mutants, which restores HTP-1/2 loading, homologue pairing, and synapsis (Fig. 3). Therefore, combined with its requirement for full activation of CHK-2 (Roelens et al., 2019), the HAL-2/3 complex ensures the recruitment of PLK-2 to the pairing centers by preventing premature association of PLK-2 with SYP-1 (Fig. 8). The HAL-2/3 complex is largely nucleoplasmic but has been shown to colocalize with SYP aggregates when axis assembly is disrupted (Roelens et al., 2019; Zhang et al., 2012). Thus, we speculate that the HAL-2/3 complex acts through the nucleoplasmic pool of SYP proteins to regulate their association with PLK-2.
CDK-1–mediated phosphorylation of SYP-1 occurs shortly before meiotic entry and persists along the entire length of chromosomes unless crossover designation occurs. Thus, the spatiotemporal control to recruit PLK-2 to the SC is not provided by SYP-1 phosphorylation by CDK-1. Rather, the signal must emanate from crossover-designated sites to recruit PLK-2, which is shown here to also require CDK-1 (Fig. 8). It remains to be determined when and which proteins are phosphorylated by CDK-1 to generate docking sites for PLK-2 at the crossover site. We have demonstrated that the kinase-dead mutant of PLK-2 accumulates at crossover sites but fails to localize along the SC due to the loss of SYP-1 phosphorylation from chromosome arms (Fig. 4). This is consistent with the model in which PLK-2 is initially recruited to the crossover site and then spreads along the SC in a manner that is dependent on its own kinase activity (Fig. 8). One interesting possibility raised by these results is that PLK-2 might phosphorylate SYP-1 and reinforce its own docking sites after crossover formation, similar to the self-priming of PLK-1 recruitment to the central spindle in late mitosis (Neef et al., 2007). However, SYP-1 T452 does not conform to the consensus motif for PLK phosphorylation, and PLK-2 was unable to phosphorylate SYP-1 T452 in our in vitro kinase assays (data not shown). Therefore, we think it more likely that PLK-2 activity is required to preserve SYP-1 phosphorylation through an indirect mechanism affecting the turnover of SYP proteins and/or by sustaining the activity of CDK-1 through phosphorylating its upstream regulators, such as CDC25 phosphatases or WEE1 kinases (Gheghiani et al., 2017; Watanabe et al., 2004).
Our evidence corroborates the previous findings that crossover-designated sites transmit a signal to recruit PLK-2 and stabilize the SCin cis. This change in the dynamic state of the SC has been shown to depend on PLK-2 (Machovina et al., 2016; Nadarajan et al., 2017; Pattabiraman et al., 2017), which retains SYP-1 phosphorylation on chromosomes harboring crossovers. We have now shown that the symmetry breaking of SYP-1 phosphorylation and PLK-2 recruitment requires a threshold level of crossover designation within the nucleus. In nuclei with sub-threshold crossovers, SYP-1 phosphorylation is retained along the length of chromosomes that have designated crossovers, while it is lost from chromosomes without crossovers. On the other hand, SYP-1 phosphorylation and PLK-2 are enriched on the SC short arm when sufficient numbers of crossovers (more than four) are generated (Fig. 5 B). We speculate that crossover-designated recombination intermediates nucleate and compete for limiting factors (e.g., PLK-2) to stabilize the SC in a chromosome-autonomous manner. Sub-threshold crossovers would recruit high levels of such SC-stabilizing factors, which then spread along the entire length of chromosomes by reinforcing SYP-1 phosphorylation. As previously suggested (Pattabiraman et al., 2017), this may also deplete the soluble pools of PLK-2 or SYP proteins below the critical concentration required to maintain the SC on chromosomes without crossovers, leading to their desynapsis (Machovina et al., 2016). As crossover designation arises above the threshold, the concentration of SC-stabilizing factors recruited to each chromosome will be decreased. Then the self-reinforcing mechanism for PLK-2 targeting and SYP-1 phosphorylation might yield a faster equilibrium and their accumulation on the SC short arm.
An alternative explanation for the asymmetric enrichment of SYP-1 phosphorylation is the presence of an opposing phosphatase that antagonizes PLK-2 and dephosphorylates SYP-1 on the long arm. PP1 is recruited to the long arm by HTP-1/2 and LAB-1 to antagonize the recruitment of AIR-2 in maturing oocytes (de Carvalho et al., 2008; Ferrandiz et al., 2018). Thus, we explored the possibility that PP1 might be responsible for dephosphorylating SYP-1 from the long arm upon crossover designation. However, both SYP-1 phosphorylation and PLK-2 localization exhibited the asymmetric appearance relative to the crossover-designated site in lab-1(tm1791) or PP1GSP-1/2-depleted animals (data not shown), ruling out the contribution of PP1 to symmetry breaking in pachytene. Taken together, our evidence suggests that differentiation of the short versus long arms is controlled by intrinsic properties of PLK-2, which is targeted to chromosomes harboring crossovers and whose effects depend on the number of crossover-designated sites within the nucleus. Thus, the integration of nucleus-wide and chromosome-autonomous signaling partitions the holocentric chromosome relative to the crossover site, which ultimately defines the pattern of chromosome segregation during meiosis I (Fig. 8).
Materials and methods
C. elegans strains and CRISPR-mediated genome editing
All strains were maintained at 20°C following standard protocols (Brenner, 1974). N2 Bristol was used as the wild-type strain. The following mutations and balancers were used: ieDf2 IV, hal-2(tm4960) III, dsb-2(me96) II, cosa-1(tm3298) III, plk-2(tm1395) I, lab-1(tm1791), hT2 [bli-4(e937) let-?(q782)] qls48] (I,III), qC1[dpy-19(e1259) glp-1(a339)] III, and mIs11[myo-2p::GFP + pes-10p::GFP + F22B7.9::GFP] IV.
The strain expressing the kinase-dead PLK-2 (K65M) was generated by CRISPR-mediated genome editing as previously described (Dokshin et al., 2018). Young adult YKM20 (plk-2(kim24[plk-2::3flag]) I; mels8[pie-1p::GFP::cosa-1, unc-119(+)] II) hermaphrodites were injected with 0.25 µg/µl of Cas9 complexed with 10 µM trans-activating CRISPR RNA (tracrRNA)/CRISPR RNA (crRNA) oligos (Integrated DNA Technologies [IDT]), pRF4::rol-6(su1006) (40 ng/µl; WormBase WBCnstr00004720; Mello et al., 1991), and a single-strand DNA (ssDNA) oligo (200 ng/µl; IDT) with 35 bp homology arms on both sides as a repair template (Table S2). F1 Roller animals were lysed and screened for insertion by PCR using a K65M-specific primer. The plk-2K65M allele was verified by sequencing and maintained as a heterozygote balanced by hT2.
Strains expressing CDK-1::Ollas and AID::CDK-1 were generated by CRISPR-mediated genome editing (Paix et al., 2015). N2 or CA1199 (unc-119(ed3) III; ieSi38[sun-1p::TIR1::mRuby::sun-1 3′UTR + Cbr-unc-119(+)] IV) were injected with 16 µM Cas9 protein complexed with 16 µM tracrRNA/crRNA oligos (IDT), pCFJ104 (5 ng/µl; Addgene #19328), pCFJ90 (2.5 ng/µl; Addgene #19327), and an ssDNA oligo (100 ng/µl; IDT) as a repair template. The Ollas tag was inserted at the 3′ end of the endogenous cdk-1 coding sequence with a Gly–Ser linker (GGATCG). The AID tag and a Gly–Gly–Ser–Gly linker (GGAGGCTCAGGA) were inserted at the 5′ end of the endogenous cdk-1 in two chunks using ssDNA oligos. F1 progeny were lysed and screened for successful insertion by PCR, and the correct insertion was validated by sequencing.
For testing the contribution of PP1, we generated a worm strain expressing GSP-1::AID and/or GSP-2::AID by CRISPR. YKM490 (plk-2(kim23[plk-2::3FLAG]) I; meIs8 [pie-1p::GFP::cosa-1 + unc-119(+)] II; ieSi38 [sun-1p::TIR1::mRuby::sun-1 3′UTR + Cbr-unc-119(+)] IV) was injected with 16 µM Cas9 protein complexed with 16 µM tracrRNA/crRNA oligos (IDT), pCFJ104 (5 ng/µl; Addgene #19328), pCFJ90 (2.5 ng/µl; Addgene #19327), and two gBlock fragments (50 ng/µl; IDT) for gsp-1::AID and gsp-2::AID as a repair template. F1 progeny were lysed and screened for successful insertion by PCR, and the correct insertion was validated by sequencing.
Antibody production
A synthetic phosphopeptide of SYP-1 flanking T452 (SAPLMTSpTPLTAATRC; Biomatik) was coupled to the maleimide-activated keyhole limpet hemocyanin (KLH, Thermo Scientific) and injected into rabbits (Pocono Rabbit Farm & Laboratory). Polyclonal SYP-1 pT452 antibodies were affinity purified by passing the immune serum through SulfoLink Coupling Resins (Thermo Scientific) coupled to nonphospho-peptide (SAPLMTSTPLTAATRC) and subsequently binding to phosphopeptide-coupled resins. Specificities of the antibodies were tested by dot blots using phospho- and nonphospho-peptides, by in vitro kinase assays using recombinant MBP–SYP-1 and CDK-1/cyclin A, and by staining dissected gonads from animals expressing SYP-1T452A.
Protein expression and purification
The full-length open reading frame of SYP-1 was amplified from a C. elegans cDNA library and cloned into the pMAL vector (New England BioLabs) to express maltose-binding protein (MBP)–tagged SYP-1 fused to a 6His tag. To make MBP–SYP-1T452A–6His, the Thr at residue 452 was mutated to Ala (ACA → GCA) by Q5 mutagenesis (New England BioLabs). Protein expression was induced at 15°C for ∼16 h with 50 µM IPTG in Rosetta (DE3) pLysS. Bacterial pellets were resuspended in lysis buffer (PBS, 500 mM NaCl, 20 mM imidazole, and 1 mM DTT) and lysed by three freeze/thaw cycles and sonication after lysozyme treatment (0.25 mg/ml) on ice for 30 min. After centrifugation at 15,000 rpm (JA-17) for 30 min, the supernatant was incubated with nickel-nitrilotriacetic acid resins (Qiagen) for 1 h at 4°C. The protein was further purified by HiTrap SP HP (GE Healthcare) using a 100 mM to 1 M NaCl gradient elution in PBS with 1 mM DTT. The peak fraction for MBP–SYP-1 was supplemented with glycerol (20% final) and snap frozen in liquid nitrogen.
In vitro kinase assays
In vitro kinase assays were performed at room temperature in 20 mM Hepes, pH 7.4, 25 mM KCl, 1 mM MgCl2, and 1 mM DTT in the presence of 0.2 mM MgATP. 2 µM wild-type and T452A mutant MBP–SYP-1–6His were incubated with 20 U of recombinant human CDK-1/cyclin A (Sigma SRP5008), and the reactions were stopped at the indicated time points with sample buffer and analyzed by Western blot.
Egg count
L4 hermaphrodites were picked onto individual nematode growth medium (NGM) plates and transferred to new plates every 12 h for a total of 4–5 d. Eggs and hatched L1s were counted immediately after transfer, and surviving progeny and males on each plate were counted when F1 reached adulthood.
RNAi
cdk-1 RNAi was performed by feeding YKM154 (cdk-1::Ollas III) animals with the Escherichia coli strain HT115 (DE3), either carrying an Ahringer RNAi library clone of cdk-1 (Kamath et al., 2003) or the empty L4440 vector. Concentrated bacterial cultures were plated onto RNAi plates (NGM, 25 µg/ml carbenicillin, and 1 mM IPTG), left for 1 h at room temperature to dry, and incubated overnight at 37°C to induce expression. L4 hermaphrodites were picked onto RNAi plates and left for 48 h before dissection for immunofluorescence.
Auxin-mediated degradation of CDK-1 and GSP-1/2 (PP1 orthologues)
Auxin-mediated degradation of CDK-1 and GSP-1/2 from the C. elegans germline was performed as previously described (Zhang et al., 2015). Briefly, auxin plates were prepared by diluting a 400 mM auxin solution (indole-3-acetic acid in ethanol) into the NGM, cooled after autoclaving, to a final concentration of 1 mM. Plates were allowed to dry at room temperature and were stored at 4°C for up to 1 mo. Plates were spread with the E. coli strain OP50-1 1 d before use and incubated overnight at 37°C. Young adult aid::cdk-1 animals with and without the Psun-1::TIR1::mRuby transgene were picked onto auxin plates and left for 18 h at 20°C before immunofluorescence. For PP1GSP-1/2 depletion, worm strains expressing GSP-1::AID and/or GSP-2::AID were treated with 1 mM auxin for 24 h at 20°C before immunofluorescence.
Immunofluorescence
Hermaphrodite germlines were dissected from 24 h post-L4 adults in egg buffer (25 mM Hepes, pH 7.4, 118 mM NaCl, 48 mM KCl, 2 mM EDTA, 5 mM EGTA, 0.1% Tween-20, and 15 mM NaN3) and fixed in 1% formaldehyde before freezing in liquid nitrogen. Dissected germlines were further fixed in methanol at −20°C for 1 min and rehydrated with PBS with 0.1% Tween-20. Samples were then blocked with blocking reagent (Roche 11096176001) for 1 h and incubated with primary antibodies overnight at 4°C. Primary antibodies were used at the following dilutions in blocking buffer (Roche 11096176001) in PBS with 0.1% Tween-20: FLAG (mouse, 1:500; Sigma M2), Ollas (rat, 1:500; Thermo Fisher MA5-16125), SYP-1 (goat, 1:500; MacQueen et al., 2005), SYP-1 pT452 (rabbit, 1:500), SYP-5 (rabbit, 1:1,000; Hurlock et al., 2020), HTP-3 (guinea pig, 1:500; MacQueen et al., 2005), HIM-3 (chicken, 1:500; Hurlock et al., 2020), GFP Booster (1:200; Chromotek gb2AF488), HTP-1/2 (rabbit, 1:500; Martinez-Perez et al., 2008), HIM-8 (rat, 1:500; Phillips et al., 2005), and phospho-HIM-8/ZIMs (rabbit, 1:1,000; Kim et al., 2015). The following secondary antibodies were purchased from Invitrogen or Jackson ImmunoResearch and used at 1:200 dilution: donkey anti-rabbit Alexa Fluor 555, donkey anti-rabbit Alexa Fluor 647, donkey anti-mouse Alexa Fluor 488, donkey anti-mouse Alexa Fluor 555, donkey anti-guinea pig fluorescein, donkey anti-goat Alexa Fluor 647, and donkey anti-rat Alexa Fluor 594.
Fixed slides were imaged at room temperature on a DeltaVision Elite system (GE Healthcare) with an Olympus 100 × 1.4 NA oil-immersion objective and a scientific complementary metal-oxide semiconductor camera (PCO). 3D image stacks were collected at 0.2-µm intervals, processed by iterative deconvolution (enhanced ratio, 20 cycles), and projected by the Volume Viewer tool using SoftWoRx suite (GE Healthcare). Composite images were assembled and colored in Adobe Photoshop.
Statistical analysis
Statistical analysis was performed using Graphpad Prism 8. For the analysis HIM-8 pairing (Fig. 3 B), a χ2 test was used to determine the differences between wild-type and mutant animals within individual gonad zones. For the analysis of SYP-1 pT452 asymmetry (Fig. 5 B), a two-tailed Fisher’s exact test was used to compare the proportion of nuclei exhibiting either symmetric or asymmetric SYP-1 pT452 staining. For the analysis of the number of COSA-1 foci (Fig. 7 B), number of DAPI bodies (Fig. S2 C), and number of cell rows in the premeiotic region (Fig. S4 E), P values were calculated using a two-tailed Mann-Whitney U test, which is a nonparametric statistical analysis. For all tests performed, the number of data points (n) is included at the base of the graph.
Online supplemental material
Fig. S1 shows validation of the SYP-1 pT452 antibody and SYP-1 pT452 staining in a CHK-2–depleted germline. Fig. S2 shows images and quantification showing the number of DAPI-staining bodies in diakinesis oocytes from the wild-type, hal-2, and hal-2; syp-1T452A mutants, and a table showing egg viability and percent males from strains generated in this study. Fig. S3 shows PLK-2 and SYP-1 pT452 staining in plk-2K65M and cosa-1 mutants. Fig. S4 shows the level of CDK-1 knockdown and the abolishment of SYP-1 pT452 staining upon cdk-1 RNAi treatment. Fig. S5 shows the impact of CDK-1 depletion on PLK-2 localization and SC disassembly. Table S1 lists the alleles generated in this study. Table S2 lists the crRNAs, repair templates, and genotyping primers for mutant alleles generated in this study. Table S3 lists the worm strains used in this study.
Supplementary Material
lists the alleles generated in this study.
lists the crRNAs, repair templates, and genotyping primers for mutant alleles generated in this study.
lists the worm strains used in this study.
Acknowledgments
We thank A.F. Dernburg (University of California, Berkeley, Berkeley, CA), O. Rog (University of Utah, Salt Lake City, UT), and A. Villeneuve (Stanford University, Palo Alto, CA) for antibodies and strains and A. Alessi and J. Kim (Johns Hopkins University, Baltimore, MD) for the C. elegans RNAi library. We also thank J. Kim and M. Hurlock for critical reading of the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Program (P40OD010440).
This work was supported by funding from the National Institutes of Health to Y. Kim (R35GM124895).
The authors declare no competing financial interests.
Author contributions: J.N. Brandt and Y. Kim conceived and designed the study. J.N. Brandt performed most experiments. K.A. Hussey and Y. Kim generated and characterized the SYP-1 pT452 antibody. J.N. Brandt and Y. Kim wrote and revised the manuscript.
References
- Altendorfer E., Láscarez-Lagunas L.I., Nadarajan S., Mathieson I., and Colaiácovo M.P.. 2020. Crossover Position Drives Chromosome Remodeling for Accurate Meiotic Chromosome Segregation. Curr. Biol. 30:1329–1338.e7: E7. 10.1016/j.cub.2020.01.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault V., and Glover D.M.. 2009. Polo-like kinases: conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 10:265–275. 10.1038/nrm2653 [DOI] [PubMed] [Google Scholar]
- Argunhan B., Leung W.-K., Afshar N., Terentyev Y., Subramanian V.V., Murayama Y., Hochwagen A., Iwasaki H., Tsubouchi T., and Tsubouchi H.. 2017. Fundamental cell cycle kinases collaborate to ensure timely destruction of the synaptonemal complex during meiosis. EMBO J. 36:2488–2509. 10.15252/embj.201695895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attner M.A., Miller M.P., Ee L., Elkin S.K., and Amon A.. 2013. Polo kinase Cdc5 is a central regulator of meiosis I. Proc. Natl. Acad. Sci. USA. 110:14278–14283. 10.1073/pnas.1311845110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazile F., St-Pierre J., and D’Amours D.. 2010. Three-step model for condensin activation during mitotic chromosome condensation. Cell Cycle. 9:3263–3275. 10.4161/cc.9.16.12620 [DOI] [PubMed] [Google Scholar]
- Bhalla N., and Dernburg A.F.. 2008. Prelude to a Division. Annu. Rev. Cell Dev. Biol. 24:397–424. 10.1146/annurev.cellbio.23.090506.123245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxem M., Srinivasan D.G., and van den Heuvel S.. 1999. The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development. 126:2227–2239. [DOI] [PubMed] [Google Scholar]
- Brar G.A., Kiburz B.M., Zhang Y., Kim J.-E., White F., and Amon A.. 2006. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 441:532–536. 10.1038/nature04794 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne R.K., Katis V.L., Jessop L., Benjamin K.R., Herskowitz I., Lichten M., and Nasmyth K.. 2003. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 5:480–485. 10.1038/ncb977 [DOI] [PubMed] [Google Scholar]
- Colaiácovo M.P., MacQueen A.J., Martinez-Perez E., McDonald K., Adamo A., La Volpe A., and Villeneuve A.M.. 2003. Synaptonemal Complex Assembly in C. elegans Is Dispensable for Loading Strand-Exchange Proteins but Critical for Proper Completion of Recombination. Dev. Cell. 5:463–474. 10.1016/S1534-5807(03)00232-6 [DOI] [PubMed] [Google Scholar]
- de Carvalho C.E., Zaaijer S., Smolikov S., Gu Y., Schumacher J.M., and Colaiacovo M.P.. 2008. LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev. 22:2869–2885. 10.1101/gad.1691208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B. 2013. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 47:563–599. 10.1146/annurev-genet-110711-155423 [DOI] [PubMed] [Google Scholar]
- Dokshin G.A., Ghanta K.S., Piscopo K.M., and Mello C.C.. 2018. Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics. 210:781–787. 10.1534/genetics.118.301532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., and Orr-Weaver T.L.. 2001. Endoreplication cell cycles: more for less. Cell. 105:297–306. 10.1016/S0092-8674(01)00334-8 [DOI] [PubMed] [Google Scholar]
- Elia A.E., Cantley L.C., and Yaffe M.B.. 2003. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 299:1228–1231. 10.1126/science.1079079 [DOI] [PubMed] [Google Scholar]
- Ferrandiz N., Barroso C., Telecan O., Shao N., Kim H.-M., Testori S., Faull P., Cutillas P., Snijders A.P., and Colaiácovo M.P.. 2018. Spatiotemporal regulation of Aurora B recruitment ensures release of cohesion during C. elegans oocyte meiosis. Nat. Commun. 9:834 10.1038/s41467-018-03229-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galander S., Barton R.E., Borek W.E., Spanos C., Kelly D.A., Robertson D., Rappsilber J., and Marston A.L.. 2019. Reductional Meiosis I Chromosome Segregation Is Established by Coordination of Key Meiotic Kinases. Dev. Cell. 49:526–541.e5. 10.1016/j.devcel.2019.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheghiani L., Loew D., Lombard B., Mansfeld J., and Gavet O.. 2017. PLK1 activation in late G2 sets up commitment to mitosis. Cell Rep. 19:2060–2073. 10.1016/j.celrep.2017.05.031 [DOI] [PubMed] [Google Scholar]
- Goodyer W., Kaitna S., Couteau F., Ward J.D., Boulton S.J., and Zetka M.. 2008. HTP-3 Links DSB Formation with Homolog Pairing and Crossing Over during C. elegans Meiosis. Dev. Cell. 14:263–274. 10.1016/j.devcel.2007.11.016 [DOI] [PubMed] [Google Scholar]
- Harper N.C., Rillo R., Jover-Gil S., Assaf Z., Bhalla N., and Dernburg A.F.. 2011. Pairing Centers Recruit a Polo-like Kinase to Orchestrate Meiotic Chromosome Dynamics in C. elegans. Dev. Cell. 21:934–947. 10.1016/j.devcel.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlock M.E., Čavka I., Kursel L.E., Haversat J., Wooten M., Nizami Z., Turniansky R., Hoess P., Ries J., Gall J.G., et al. . 2020. Identification of novel synaptonemal complex components in C. elegans. J. Cell Biol. 219 e201910043 10.1083/jcb.201910043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P.W., Karppinen J., and Handel M.A.. 2012. Polo-like kinase is required for synaptonemal complex disassembly and phosphorylation in mouse spermatocytes. J. Cell Sci. 125:5061–5072. 10.1242/jcs.105015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitna S., Pasierbek P., Jantsch M., Loidl J., and Glotzer M.. 2002. The Aurora B Kinase AIR-2 Regulates Kinetochores during Mitosis and Is Required for Separation of Homologous Chromosomes during Meiosis. Curr. Biol. 12:798–812. 10.1016/S0960-9822(02)00820-5 [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. . 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421:231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Katis V.L., Lipp J.J., Imre R., Bogdanova A., Okaz E., Habermann B., Mechtler K., Nasmyth K., and Zachariae W.. 2010. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev. Cell. 18:397–409. 10.1016/j.devcel.2010.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kostow N., and Dernburg A.F.. 2015. The Chromosome Axis Mediates Feedback Control of CHK-2 to Ensure Crossover Formation in C. elegans. Dev. Cell. 35:247–261. 10.1016/j.devcel.2015.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella S., Woglar A., Jantsch V., and Zetka M.. 2011. Polo Kinases Establish Links between Meiotic Chromosomes and Cytoskeletal Forces Essential for Homolog Pairing. Dev. Cell. 21:948–958. 10.1016/j.devcel.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Linder M.I., Köhler M., Boersema P., Weberruss M., Wandke C., Marino J., Ashiono C., Picotti P., Antonin W., and Kutay U.. 2017. Mitotic disassembly of nuclear pore complexes involves CDK1-and PLK1-mediated phosphorylation of key interconnecting nucleoporins. Dev. Cell. 43:141–156.e7. 10.1016/j.devcel.2017.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link J., Paouneskou D., Velkova M., Daryabeigi A., Laos T., Labella S., Barroso C., Piñol S.P., Montoya A., and Kramer H.. 2018. Transient and partial nuclear lamina disruption promotes chromosome movement in early meiotic prophase. Dev. Cell. 45:212–225.e7. 10.1016/j.devcel.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machovina T.S., Mainpal R., Daryabeigi A., McGovern O., Paouneskou D., Labella S., Zetka M., Jantsch V., and Yanowitz J.L.. 2016. A surveillance system ensures crossover formation in C. elegans. Curr. Biol. 26:2873–2884. 10.1016/j.cub.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., and Villeneuve A.M.. 2001. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 15:1674–1687. 10.1101/gad.902601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Colaiácovo M.P., McDonald K., and Villeneuve A.M.. 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16:2428–2442. 10.1101/gad.1011602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Phillips C.M., Bhalla N., Weiser P., Villeneuve A.M., and Dernburg A.F.. 2005. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 123:1037–1050. 10.1016/j.cell.2005.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Schvarzstein M., Barroso C., Lightfoot J., Dernburg A.F., and Villeneuve A.M.. 2008. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 22:2886–2901. 10.1101/gad.1694108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino L., Morchoisne-Bolhy S., Cheerambathur D.K., Van Hove L., Dumont J., Joly N., Desai A., Doye V., and Pintard L.. 2017. Channel nucleoporins recruit PLK-1 to nuclear pore complexes to direct nuclear envelope breakdown in C. elegans. Dev. Cell. 43:157–171.e7. 10.1016/j.devcel.2017.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C.C., Kramer J.M., Stinchcomb D., and Ambros V.. 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10:3959–3970. 10.1002/j.1460-2075.1991.tb04966.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A.M., and Colaiácovo M.P.. 2005. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J. Cell Biol. 168:683–689. 10.1083/jcb.200410144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S., Lambert T.J., Altendorfer E., Gao J., Blower M.D., Waters J.C., and Colaiácovo M.P.. 2017. Polo-like kinase-dependent phosphorylation of the synaptonemal complex protein SYP-4 regulates double-strand break formation through a negative feedback loop. eLife. 6 e23437 10.7554/eLife.23437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R., Gruneberg U., Kopajtich R., Li X., Nigg E.A., Sillje H., and Barr F.A.. 2007. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 9:436–444. 10.1038/ncb1557 [DOI] [PubMed] [Google Scholar]
- Page S.L., and Hawley R.S.. 2004. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20:525–558. 10.1146/annurev.cellbio.19.111301.155141 [DOI] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D., and Seydoux G.. 2015. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics. 201:47–54. 10.1534/genetics.115.179382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman D., Roelens B., Woglar A., and Villeneuve A.M.. 2017. Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS Genet. 13 e1006670 10.1371/journal.pgen.1006670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Siomos M.F., and Nasmyth K.. 2003. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 112:423–440. 10.1016/S0092-8674(03)00083-7 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., and Dernburg A.F.. 2006. A Family of Zinc-Finger Proteins Is Required for Chromosome-Specific Pairing and Synapsis during Meiosis in C. elegans. Dev. Cell. 11:817–829. 10.1016/j.devcel.2006.09.020 [DOI] [PubMed] [Google Scholar]
- Phillips C.M., Wong C., Bhalla N., Carlton P.M., Weiser P., Meneely P.M., and Dernburg A.F.. 2005. HIM-8 Binds to the X Chromosome Pairing Center and Mediates Chromosome-Specific Meiotic Synapsis. Cell. 123:1051–1063. 10.1016/j.cell.2005.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelens B., Barroso C., Montoya A., Cutillas P., Zhang W., Woglar A., Girard C., Martinez-Perez E., and Villeneuve A.M.. 2019. Spatial Regulation of Polo-Like Kinase Activity During Caenorhabditis elegans Meiosis by the Nucleoplasmic HAL-2/HAL-3 Complex. Genetics. 213:79–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E., Bishop J.D., Waddle J.A., Schumacher J.M., and Lin R.. 2002. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 157:219–229. 10.1083/jcb.200110045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S., Zawadzki K.A., Stamper E.L., Libuda D.E., Reese A.L., Dernburg A.F., and Villeneuve A.M.. 2013. The C. elegans DSB-2 Protein Reveals a Regulatory Network that Controls Competence for Meiotic DSB Formation and Promotes Crossover Assurance. PLoS Genet. 9 e1003674 10.1371/journal.pgen.1003674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato-Carlton A., Nakamura-Tabuchi C., Chartrand S.K., Uchino T., and Carlton P.M.. 2018. Phosphorylation of the synaptonemal complex protein SYP-1 promotes meiotic chromosome segregation. J. Cell Biol. 217:555–570. 10.1083/jcb.201707161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A.F., Ling L., van Zuylen V., and Meyer B.J.. 2009. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 23:1763–1778. 10.1101/gad.1808809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Eizinger A., Schild-Prufert K., Hurlburt A., McDonald K., Engebrecht J., Villeneuve A.M., and Colaiácovo M.P.. 2007. SYP-3 Restricts Synaptonemal Complex Assembly to Bridge Paired Chromosome Axes During Meiosis in Caenorhabditis elegans. Genetics. 176:2015–2025. 10.1534/genetics.107.072413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Schild-Prufert K., and Colaiácovo M.P.. 2009. A Yeast Two-Hybrid Screen for SYP-3 Interactors Identifies SYP-4, a Component Required for Synaptonemal Complex Assembly and Chiasma Formation in Caenorhabditis elegans Meiosis. PLoS Genet. 5 e1000669 10.1371/journal.pgen.1000669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourirajan A., and Lichten M.. 2008. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 22:2627–2632. 10.1101/gad.1711408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur Y.B., De Carvalho C.E., Nadarajan S., Van Bostelen I., Gu Y., Chu D.S., Cheeseman I.M., and Colaiácovo M.P.. 2012. LAB-1 targets PP1 and restricts Aurora B kinase upon entrance into meiosis to promote sister chromatid cohesion. PLoS Biol. 10 e1001378 10.1371/journal.pbio.1001378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Arai H., Nishihara Y., Taniguchi M., Watanabe N., Hunter T., and Osada H.. 2004. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFβ-TrCP. Proc. Natl. Acad. Sci. USA. 101:4419–4424. 10.1073/pnas.0307700101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woglar A., and Villeneuve A.M.. 2018. Dynamic Architecture of DNA Repair Complexes and the Synaptonemal Complex at Sites of Meiotic Recombination. Cell. 173:1678–1691.e16. 10.1016/j.cell.2018.03.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R., Zawadzki K.A., Nabeshima K., Drake M., Arur S., and Villeneuve A.M.. 2012. COSA-1 Reveals Robust Homeostasis and Separable Licensing and Reinforcement Steps Governing Meiotic Crossovers. Cell. 149:75–87. 10.1016/j.cell.2012.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka M.C., Kawasaki I., Strome S., and Muller F.. 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13:2258–2270. 10.1101/gad.13.17.2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Miley N., Zastrow M.S., MacQueen A.J., Sato A., Nabeshima K., Martinez-Perez E., Mlynarczyk-Evans S., Carlton P.M., and Villeneuve A.M.. 2012. HAL-2 promotes homologous pairing during Caenorhabditis elegans meiosis by antagonizing inhibitory effects of synaptonemal complex precursors. PLoS Genet. 8 e1002880 10.1371/journal.pgen.1002880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ward J.D., Cheng Z., and Dernburg A.F.. 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 142:4374–4384. 10.1242/dev.129635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Koehler S., Rillo-Bohn R., and Dernburg A.F.. 2018. A compartmentalized signaling network mediates crossover control in meiosis. eLife. 7 e30789 10.7554/eLife.30789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Xie S., Wang R., Guo S., Zhao Q., Nie H., Liu Y., Zhang F., Chen M., Liu L., et al. . 2020. Multivalent weak interactions between assembly units drive synaptonemal complex formation. J. Cell Biol. 219 e201910086 10.1083/jcb.201910086 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the alleles generated in this study.
lists the crRNAs, repair templates, and genotyping primers for mutant alleles generated in this study.
lists the worm strains used in this study.